Abstract
Dendrites are the primary sites on neurons for receiving and integrating inputs from their presynaptic partners. Defects in dendrite development perturb the formation of neural circuitry and impair information processing in the brain. Extracellular cues are important for shaping the dendritic morphogenesis, but the underlying molecular mechanisms are not well understood. In this study, we examined the role of ARMS (ankyrin repeat-rich membrane spanning protein), also known as Kidins220 (kinase D-interacting substrate of 220 kDa), previously identified as a downstream target of neurotrophin and ephrin receptors, in dendrite development. We report here that knockdown of ARMS/Kidins220 by in utero electroporation impairs dendritic branching in mouse cerebral cortex, and silencing of ARMS/Kidins220 in primary rat hippocampal neurons results in a significant decrease in the length, number, and complexity of the dendritic arbors. Overexpression of cell surface receptor tyrosine kinases, including TrkB and EphB2, in ARMS/Kidins220-deficient neurons can partially rescue the defective dendritic phenotype. More importantly, we show that PI3K (phosphoinositide-3-kinase)- and Akt-mediated signaling pathway is crucial for ARMS/Kidins220-dependent dendrite development. Furthermore, loss of ARMS/Kidins220 significantly reduced the clustering of EphB2 receptor signaling complex in neurons. Our results collectively suggest that ARMS/Kidins220 is a key player in organizing the signaling complex to transduce the extracellular stimuli to cellular responses during dendrite development.
Introduction
Precise establishment of neural circuits requires proper navigation of axons to their targets and elaboration of dendrites in target neurons to integrate multiple synaptic inputs. Dendrites serve as the primary sites on neurons for receiving information from their presynaptic partners, and the complexity of dendritic branching controls functional connectivity between neurons (Jan and Jan, 2003, 2010). Thus, defects in dendrite branching result in alteration of neural circuits and impairment in neural functions. Indeed, defective dendritic arborization has been reported to be associated with various neurodegenerative diseases (e.g., Alzheimer's disease) and psychiatric disorders (e.g., schizophrenia) (Broadbelt et al., 2002; Baloyannis, 2009; Emoto, 2011).
Dendrite development is guided by an intrinsic differentiation program that is modified by extrinsic cues (Miller and Kaplan, 2003). For example, soluble factors, including brain-derived neurotrophic factor (BDNF) and other neurotrophins, as well as transmembrane ligands like ephrinB, regulate dendritic morphogenesis through their cell surface receptors in coordination with the signaling pathways triggered by neuronal activity (McAllister et al., 1995; Hoogenraad et al., 2005; Konur and Ghosh, 2005). These cell surface receptors regulate the growth or maintenance of dendrites through modulation of Ras–MEK–MAPK and PI3K (phosphoinositide-3-kinase)–Akt–mTOR (mammalian target of rapamycin)-regulated protein translation pathways, CREB-dependent gene transcription, and small Rho GTPase-mediated cytoskeletal reorganization (Dijkhuizen and Ghosh, 2005; Govek et al., 2005; Jaworski et al., 2005; Cheung et al., 2007; Finsterwald et al., 2010; Kwon et al., 2011). Nonetheless, the molecular mechanisms that relay the signals from the cell surface receptors to intracellular effectors during dendrite development remain unclear.
Previously identified as a downstream target of neurotrophin and ephrin receptors, ankyrin repeat-rich membrane spanning protein (ARMS), also known as Kidins220 (kinase D-interacting substrate of 220 kDa), is a multidomain transmembrane protein that not only transduces signaling mediated by the receptor tyrosine kinase (RTK), but also regulates the stability of cytoskeletal network through a number of cytoskeleton-associated proteins, including Septin5, MAPs (microtubule-associated proteins), and stathmin family members (Kong et al., 2001; Arévalo et al., 2004, 2006; Chang et al., 2004; Luo et al., 2005; Higuero et al., 2010). ARMS/Kidins220 plays an important role in the development of the nervous system, such as promoting neurite outgrowth, maintaining dendritic spine stability, and modulating synaptic transmission (Arévalo et al., 2004, 2010; López-Menéndez et al., 2009; Wu et al., 2009; Sutachan et al., 2010; Wu et al., 2010). In particular, reduced expression of ARMS/Kidins220 leads to a reduced dendritic branching in the barrel somatosensory cortex and dentate gyrus (Wu et al., 2009).
In this study, we report that loss of ARMS/Kidins220 in hippocampal neurons significantly reduces the complexity of dendritic trees. Similar to its role in cultured neurons, ARMS/Kidins220 is critical for sculpting the dendritic arborization of layer II–IV pyramidal neurons in mouse cerebral cortex. The activation of PI3K signaling pathway restores the normal dendritic phenotype in ARMS/Kidins220-depleted neurons, suggesting that the RTK–PI3K signaling pathway plays a critical role in ARMS/Kidins220-dependent dendritic morphogenesis. Together, our study reveals a novel mechanism by which ARMS/Kidins220 scaffolds the RTK signaling complex to shape the dendritic trees.
Materials and Methods
Animals.
All animals, including rats and mice, were bred in the Animal and Plant Care Facility of The Hong Kong University of Science and Technology and were handled in accordance with the Animals (Control of Experiments) Ordinance of Hong Kong.
Antibodies and constructs.
Antibodies against EphB2 and phosphorylated Ser522 of CRMP-2 were kindly provided by Professor Elena Pasquale at Sanford-Burnham Medical Research Institute (La Jolla, CA) and Professor Yoshio Goshima at Yokohama City University (Yokohama, Japan), respectively. GFP (polyclonal), GFP (monoclonal), MAP2, Cux1, and 4G10 antibodies were from MBL, Life Technologies, Sigma-Aldrich, Santa Cruz Biotechnology, and Millipore, respectively. ARMS/Kidins220 knock-down construct was generated by subcloning of the 64 bp fragment containing the shRNA target sequence into the pSUPER vector. The shRNA target sequence for rat ARMS/Kidins220 is 5′-GTCAATTGCTCCGACAAGT-3′.
Cell culture, transfection, and Western blot analysis.
HEK 293T cells were maintained in DMEM with 10% fetal bovine serum and transfected with Lipofectamine and PLUS reagents according to the protocols from the manufacturer (Life Technologies). Primary hippocampal neurons were prepared from embryonic day 18 (E18) rat or transgenic mice and maintained in Neurobasal medium with 2% B27 supplement. Primary neurons were transfected using calcium phosphate method as previously described (Fu et al., 2007). Knockdown of ARMS/Kidins220 expression by in utero electroporation was performed in E14 ICR mice and the neuronal morphology was examined at different postnatal stages (Ip et al., 2012). ARMS/kidins220 transgenic mouse brains at E17 were collected for Western blot analysis. Except for the pregnant female mice used in the studies of in utero electroporation, all the experiments were performed with animals of either sex.
Confocal imaging and quantification.
Images were captured by Olympus Fluoview300 or Fluoview1000 confocal microscopes with a 40× or 60× oil lens. The number and length of dendrites were quantified using ImageJ (National Institutes of Health) with NeuronJ plugin (Meijering et al., 2004). Sholl analysis, which measures the number of intersections of neuronal dendrites crossing a series of concentric circles from the cell body, was performed using ImageJ embedded with Sholl Analysis plugin (A. Ghosh, University of California San Diego, La Jolla, CA). Twenty to 40 neurons from three independent experiments were measured.
Results
Knockdown of ARMS/Kidins220 leads to impairment in dendrite development at postnatal stages
Although ARMS/Kidins220 homozygous knock-out mice are embryonic lethal, defective phenotype of dendrites could be observed in the heterozygous mice, suggesting that ARMS/Kidins220 is involved in the refinement and stabilization of dendritic arbor (Wu et al., 2009). Interestingly, we observed aberrant upregulation of Ser522-phosphorylated microtubule regulator CRMP-2 (collapsin response mediator protein-2) in ARMS+/− brains (Fig. 1A). Since the phosphorylation status of CRMP-2 is critical for the proper dendrite organization (Yamashita et al., 2011), our finding suggests that ARMS/Kidins220 may regulate dendrite development through modulation of microtubule dynamics (Fig. 1A). To further elucidate the role of ARMS/Kidins220 in dendrite development in vivo, we silenced the expression of ARMS/Kidins220 in mouse neocortex at E14 by in utero electroporation. Knockdown efficiency of the shRNA against ARMS/Kidins220 (ARMS shRNA) was confirmed in primary cortical neurons (Fig. 1B). Whereas essentially all of the neurons (∼97% of GFP+ cells) were able to migrate to the upper cortical plate (layers II–IV) of the control mouse brains at postnatal day 4 (P4), knockdown of ARMS/Kidins220 led to a disruption of radial migration (with ∼42% of the ARMS/Kidins220-deficient neurons trapped in layers V–VI and white matter; Fig. 1C,D), suggesting that ARMS/Kidins220 is involved in neuronal migration. The migration defect was also observed at different developmental stages (i.e., P1 and P20; data not shown). The neurons in layers II–IV of the control brains extended an obvious apical dendrite from the soma with multiple short dendrites at P4, and the dendritic complexity increased upon development, characterized by pyramidal-like morphology with a long apical dendrite and multiple basal dendrites at P20 (Fig. 1E,F). Intriguingly, for those ARMS/Kidins220-deficient neurons that were able to reach the layer II-IV, they displayed significantly reduced dendritic complexity. The ARMS/Kidins220-deficient neurons had a poorly branched dendritic arbor, with a simplification of basal dendrites, while the apical dendrites were not significantly affected (Fig. 1E,F). Both the number of dendrites and total dendrite length were decreased in the ARMS/Kidins220 knock-down neurons (Fig. 1G,H). This result strongly suggests that ARMS/Kidins220 is important for dendrite development in vivo.
Figure 1.
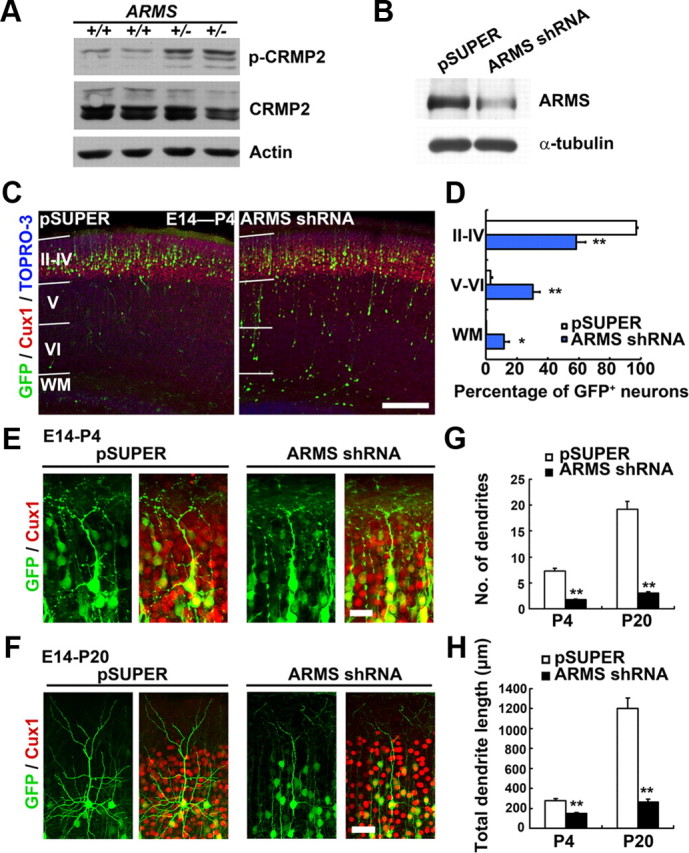
In utero knockdown of ARMS/Kidins220 leads to dendritic defect in postnatal stages. A, ARMS+/− mouse brains exhibits deregulation of CRMP-2 phosphorylation. B, ARMS shRNA suppressed the expression of ARMS/Kidins220 in primary cortical neurons. C–H, ARMS/Kidins220 in the mouse cerebral cortex was knocked down by in utero electroporation at E14. C, Knockdown of ARMS/Kidins220 resulted in impaired neuronal migration at P4. Scale bar, 200 μm. D, Quantification of the layer distribution of control and ARMS/Kidins220 knock-down neurons at P4 (GFP+ cells). WM, White matter. *p < 0.01, **p < 0.001, Student's t test. E, F, High-magnification images showing the morphology of ARMS/Kidins220 knock-down neurons in neocortex at P4 and P20. Scale bars: E, 20 μm; F, 50 μm. G, H, Quantitative analysis of the number and total length of dendrites in ARMS/Kidins220 knock-down cortex at P4 and P20. **p < 0.001, Student's t test.
ARMS/Kidins220 is required for dendrite arborization in hippocampal neurons
To investigate how ARMS/Kidins220 shapes neuronal dendrites, we used cultured hippocampal neurons, a well established model system for investigating neuronal differentiation (Dotti et al., 1988). The expression of ARMS/Kidins220 was silenced in hippocampal neurons by expressing the ARMS shRNA at 3 days in vitro (3 DIV) and 7 DIV, and the neuronal morphology was examined at 7 and 14 DIV, respectively (Fig. 2A,B). Sholl analysis revealed that knockdown of ARMS/Kidins220 significantly reduces the complexity of the dendritic trees (Fig. 2C,D). Unlike the control neurons, which have the peak number of intersections at 50–70 μm from the cell body at 14 DIV, the maximal number of intersections in ARMS/Kidins220 knock-down neurons shifted to 20–30 μm from the cell body, indicating a critical role of ARMS/Kidins220 in shaping the dendritic arbor (Fig. 2D). Both the number of dendrites and their average length also reduced significantly in these ARMS/Kidins22-knock-down neurons (Fig. 2E,F). The reduction of dendrite number in ARMS/Kidins220-deficient neurons could be observed since early developmental stages (e.g., at 7 DIV); however, the decrease in dendrite length was not obvious until the neurons become mature (Fig. 2E,F). The effect of ARMS/Kidins220 deficiency on reducing dendritic tree complexity was further confirmed by staining the ARMS/Kidins220-depleted neurons with the dendritic marker protein MAP2 (Fig. 2G,H). Furthermore, we cultured hippocampal neurons from the wild-type (WT) and ARMS+/− mice, and examined the dendrite morphology of individual neurons at 14 DIV by staining with MAP2. Compared with the wild-type control, ARMS+/− neurons exhibited much shorter (72.13 ± 3.76 μm in ARMS+/− vs 84.36 ± 2.60 μm in wild-type; p < 0.01, Student's t test) and fewer dendrites (10.59 ± 0.61 in ARMS+/− vs 12.52 ± 0.66 in wild-type; p < 0.05, Student's t test) (Fig. 2I,J). These results collectively suggest that ARMS/Kidins220 functions as a critical regulator in the development of neuronal dendrites.
Figure 2.
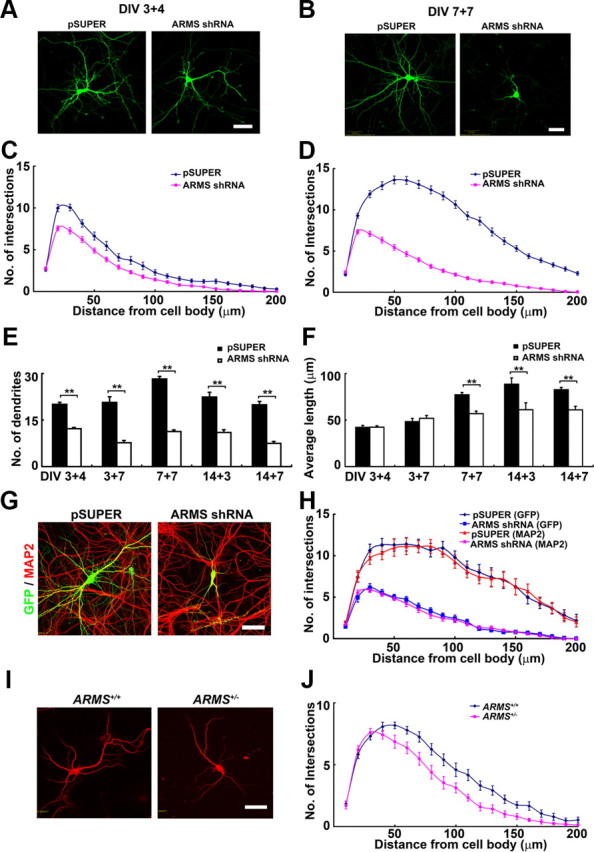
ARMS/Kidins220 is required for dendrite development in hippocampal neurons. A, Hippocampal neurons were transfected with pSUPER vector and ARMS shRNA at 3 DIV, and neuronal morphology was examined at 7 DIV (DIV 3 + 4). Scale bar, 50 μm. B, ARMS/Kidins220 was knocked down at 7 DIV, and the morphology of dendritic trees was investigated at 14 DIV (DIV 7 + 7). Scale bar, 50 μm. C, D, Sholl analysis of neurons transfected with pSUPER vector and ARMS shRNA shown in A and B, respectively. E, F, Measurement of dendrite number and length of ARMS/Kidins220 knock-down neurons at different stages. **p < 0.005, Student's t test. G, Hippocampal neurons transfected with pSUPER vector and ARMS shRNA (DIV 7 + 7) were stained with the dendritic marker MAP2. Scale bar, 50 μm. H, Sholl analysis of transfected neurons in G. I, Hippocampal neurons from wild-type and ARMS+/− mice cultured at low density were stained with MAP2 antibody. The dendrite morphology of individual neurons was examined. Scale bar, 50 μm. J, Sholl analysis of neurons in I.
PI3K/Akt pathway is required for ARMS/Kidins220-dependent dendrite development
It has been shown that PI3K and its downstream effector Akt transduce dendrite-promoting signals from the cell surface receptors to the protein translation machinery and cytoskeleton (Jaworski et al., 2005). Thus, we investigate whether PI3K and Akt are required for the ARMS/Kidins220-dependent dendrite development. While expression of the wild-type p110 catalytic subunit of PI3K did not significantly affect the dendritic trees in ARMS/Kidins220-deficient neurons, expression of the constitutively active form of PI3K (PI3K CA) increased the number of dendrites significantly (Fig. 3A–C). To further elucidate the role of PI3K in ARMS/Kidins220-regulated dendrite development, we expressed the PI3K CA in ARMS+/− neurons and found that activation of PI3K signaling fully restores the dendritic arbors in the ARMS/Kidins220-deficient neurons (Fig. 3D,E). Similarly, ARMS/Kidins220-deficient neurons overexpressing constitutively active Akt exhibited a significant improvement in dendritic complexity (Fig. 3F–H). In contrast, stimulating the MAPK pathway by overexpressing constitutively active MEK1 was unable to restore the dendritic trees in the ARMS/Kidins220 knock-down neurons (data not shown). Therefore, our results support that ARMS/Kidins220 regulates dendrite development through PI3K/Akt-mediated signaling pathway.
Figure 3.
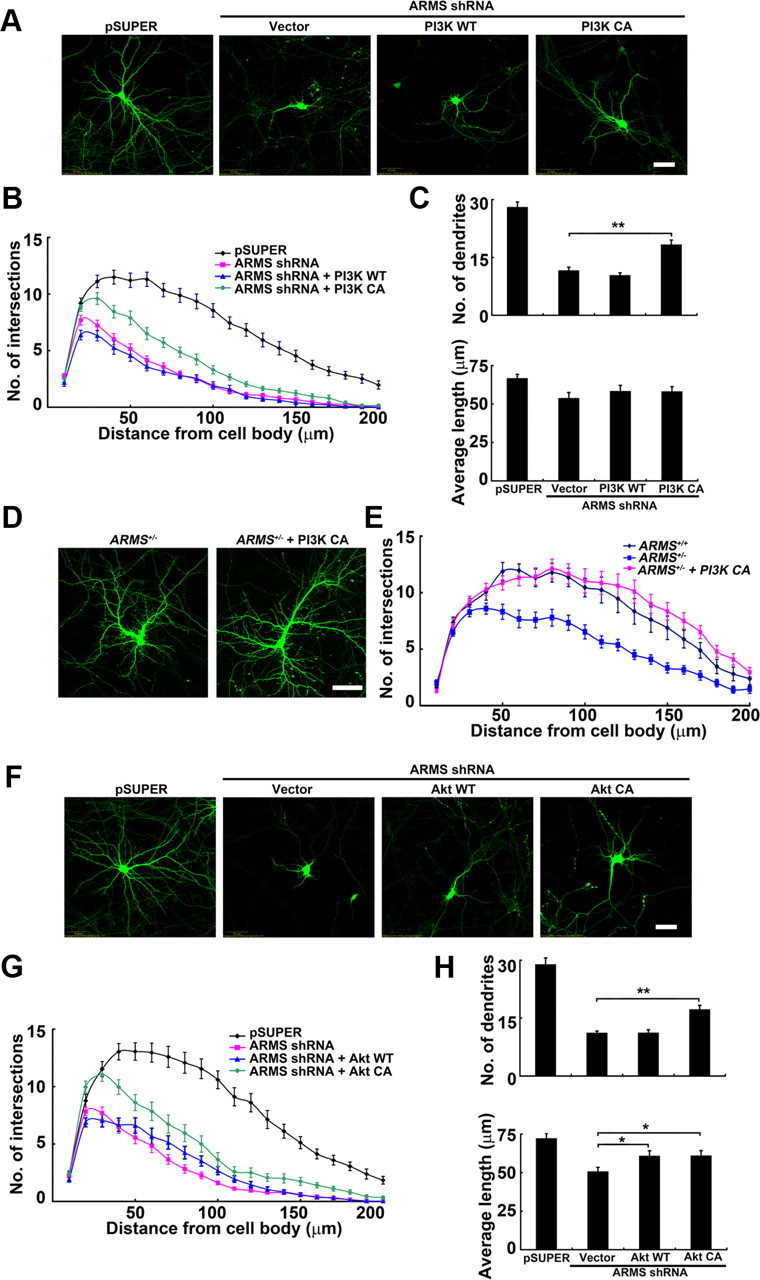
Activation of PI3K and Akt rescues the defective dendritic phenotype in ARMS/Kidins220 knock-down neurons. A, WT or CA form of PI3K was cotransfected with ARMS shRNA at 7 DIV. At 14 DIV, overexpression of PI3K CA increased number of dendrites in these knock-down neurons. Scale bar, 50 μm. B, C, Measurement of the dendritic complexity of neurons in A. **p < 0.005, one-way ANOVA, with Student–Newman–Keuls test. D, Overexpression of PI3K CA fully rescued the dendritic defect in ARMS+/− neurons. Scale bar, 50 μm. E, Sholl analysis. F, WT or constitutively active CA Akt partially rescued the aberrant dendritic morphology in ARMS/Kidins220 knock-down neurons. Scale bar, 50 μm. G, H, Quantitative analysis of neurons shown in F. Sholl analysis (G), number, and average length of dendrites (H). *p < 0.05, **p < 0.005, one-way ANOVA, with Student–Newman–Keuls test.
ARMS/Kidins220 regulates dendrite development through neurotrophin and ephrin receptors
ARMS/Kidins220 is tyrosine phosphorylated by the cell surface receptors TrkB and EphB (Kong et al., 2001). Considering the importance of these two receptors in activating PI3K/Akt signaling and promoting dendrite development (Delcroix et al., 2003; Hoogenraad et al., 2005; Jaworski et al., 2005; Pasquale, 2005), we investigated whether ARMS/Kidins220 regulates dendrite development through modulating the signaling pathways mediated by these cell surface receptors. Interestingly, we found that overexpression of TrkB, the receptor for BDNF, increased the number and length of dendrites in ARMS/Kidins220 knock-down neurons (Fig. 4A–C), although the rescue was not complete. However, overexpressing BDNF, which has been shown to promote dendritic growth, did not obviously increase the dendritic complexity in ARMS/Kidins220-deficient neurons, suggesting that the dendritic defect does not result from the lack of BDNF (Fig. 4A–C). We further confirmed that the protein level of BDNF is not altered in ARMS/Kidins220 knock-down neurons (125.3 ± 7.5% of control; p < 0.005, Student's t test), whereas the BDNF-induced clustering of TrkB was significantly reduced in these knock-down neurons (number: 61.9 ± 11.8% of control, p < 0.05; area: 74.2 ± 7.2% of control, p < 0.05; intensity: 70.0 ± 8.1% of control, p < 0.05; Student's t test).
Figure 4.
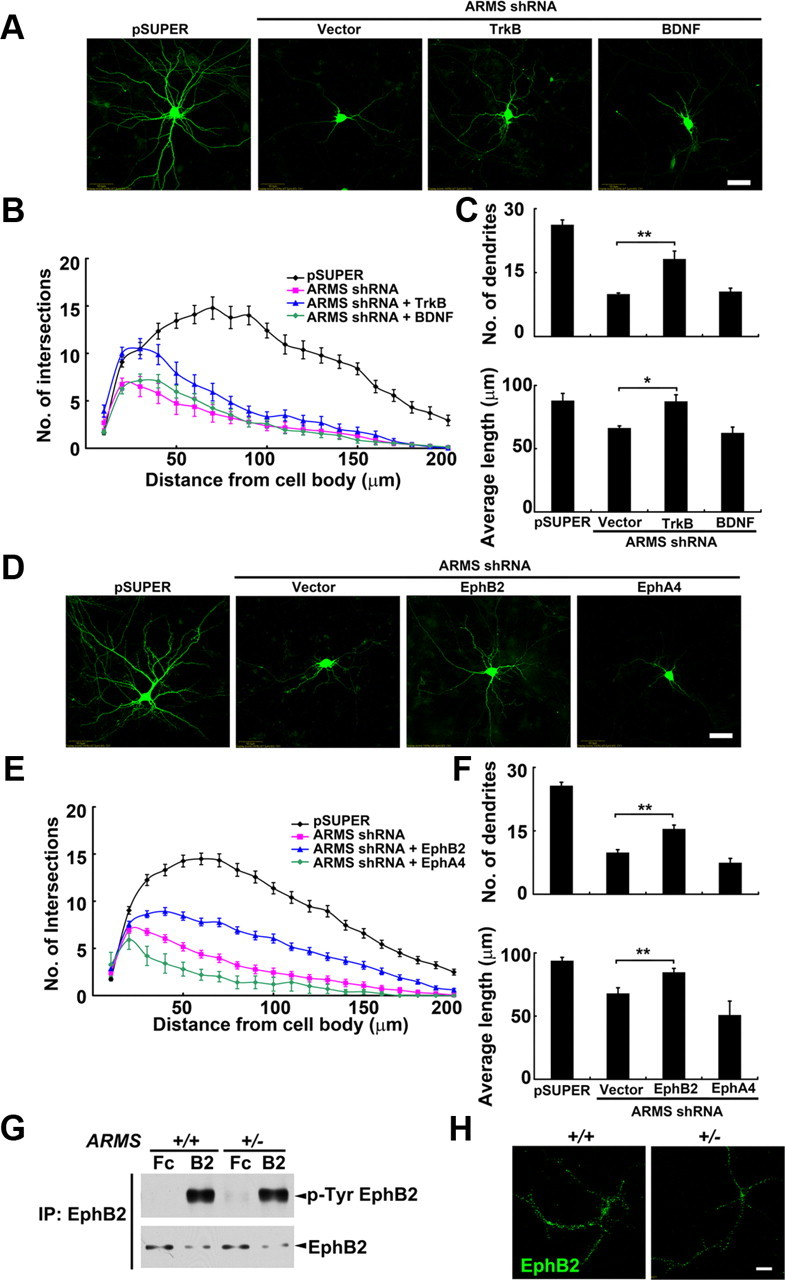
Activation of neurotrophin and ephrin signaling restores dendrite development in ARMS/Kidins220-deficient neurons. A, Representative images from cultured hippocampal neurons transfected with pSUPER vector, ARMS shRNA, ARMS shRNA + TrkB, and ARMS shRNA + BDNF. Scale bar, 50 μm. B, C, Quantitative analysis of neurons in A. *p < 0.05, **p < 0.005, one-way ANOVA, with Student–Newman–Keuls test. D, Expression of EphB2, but not EphA4, restored the defective dendrite development in ARMS/Kidins220 knock-down neurons. Scale bar, 50 μm. E, F, Quantitative analysis of neurons in D. **p < 0.005, one-way ANOVA, with Student–Newman–Keuls test. G, Treatment of ephrin-B2 (B2) induced the phosphorylation of EphB2 in cultured ARMS+/+ and ARMS+/− neurons. Fc fragment (Fc) was used as a control. H, Ephrin-B2-induced clustering of EphB2 was decreased in ARMS+/− neurons. Scale bar, 20 μm.
On the other hand, when EphB2, the receptor for ephrin ligands, was overexpressed in ARMS/Kidins220-deficient neurons, the reduction in the dendrite number and length in the deficient neurons was partially rescued, characterized by an increase in the complexity of dendritic trees (Fig. 4D–F). In contrast, EphA4, another member of the Eph receptor family that also physically interacts with ARMS/Kidins220, was not able to exhibit a similar rescue effect (Fig. 4D–F), indicating the specificity of EphB2 in ARMS/Kidins220-dependent dendrite development. To further understand the mechanism by which ARMS/Kidins220 regulates EphB2 signaling, we examined whether ARMS/Kidins220 regulates the phosphorylation and clustering of EphB2 receptor, two critical aspects for activation of the EphB2 signaling (Egea et al., 2005). We found that the ligand-induced phosphorylation of EphB2 was comparable in neurons prepared from brains of ARMS+/− mice and wild-type controls (Fig. 4G). However, a prominent reduction in the number (82.4 ± 5.2% of control, p < 0.005), size (79.7 ± 4.0% of control, p < 0.005), and intensity (78.5 ± 5.2% of control, p < 0.005; Student's t test) of ligand-induced EphB2 clusters was observed in ARMS/Kidins220-deficient neurons (Fig. 4H) (data not shown). These findings suggest that ARMS/Kidins220 is crucial for organizing the EphB2-containing signaling complex in regulating dendritic growth and stabilization.
Discussion
A central question in neuroscience is how neuronal networks are formed. It is therefore important to elucidate the mechanisms that underlie the precise regulation of neuronal structures. We report here that ARMS/Kidins220 is a key regulator of dendrite development, the primary structure for the establishment of neuronal circuit. More importantly, we provide evidence that the RTK signaling pathways mediated by neurotrophin and ephrin receptors are involved in ARMS/Kidins220-dependent dendrite development.
Changes of neuronal structures are achieved through the regulation of cytoskeleton. A plethora of molecules have been shown to regulate the stability of actin and microtubule networks under the control of cell surface neurotrophin and ephrin receptors during dendrite morphogenesis, such as PI3K, MAPK, Rac1, and Pak1 (Delcroix et al., 2003; Miller and Kaplan, 2003; Pasquale, 2005). Although PI3K and MAPK are both required for dendrite development, the activation of PI3K alone, but not MAPK, is sufficient to promote dendrite formation (Dijkhuizen and Ghosh, 2005). The failure of MAPK signaling to rescue the dendritic branching in ARMS/Kidins220-deficient neurons is consistent with the notion that loss of ARMS/Kidins220 impairs the PI3K-mediated dendrite formation. PI3K signaling enhances the growth of dendrites either through the regulation of cytoskeleton stability or protein synthesis. CRMP-2 is a downstream effector of PI3K and Akt that facilitates the microtubule assembly and stabilization, whose activity is tightly regulated in dendrite development (Fukata et al., 2002; Lin et al., 2011). The prominent reduction of CRMP-2 activity (because of enhanced phosphorylation at Ser522) in ARMS+/− brain suggests that PI3K/CRMP-2-dependent assembly and stabilization of microtubule cytoskeleton is involved in ARMS/Kidins220-dependent dendrite development. Whether the Akt-mTOR-dependent protein synthesis is involved in the process awaits further investigation.
Dendritic growth is promoted by the activation of cell surface receptors such as Trk and Eph receptors, both of which employ ARMS/Kidins220 to convey the extracellular stimuli to the intracellular effectors (Kong et al., 2001). We previously reported that the postsynaptic clustering of EphA4 as well as its downstream signaling cascades are cooperatively regulated by ARMS/Kidins220 and its interacting proteins (Luo et al., 2005). In this study, we found that clustering of other RTK members TrkB and EphB2 was reduced due to the loss of ARMS/Kidins220. As the oligomerization of RTKs upon ligand stimulation is vital for their signal transduction, overexpression of these cell surface receptors, which enhances their clustering, partially restores the defective signaling and dendrite development in ARMS/Kidins220-deficient neurons. In addition to the role in organizing receptor clustering, ARMS/Kidins220 is likely to act as a master scaffold for orchestrating signaling complexes of RTKs during the critical period of dendrite development. Glutamate receptor interacting protein 1 (GRIP1) and kinesin-1 are two interacting proteins that regulate the trafficking of EphB2 to facilitate dendrite formation (Hoogenraad et al., 2005). Interestingly, ARMS/Kidins220 interacts with kinesin-1 and undergoes a kinesin-1-dependent trafficking to the tips of growing neurites during neuronal differentiation (Bracale et al., 2007). In addition, GRIP1 possesses multiple PDZ domains that potentially bind to the class I PDZ-binding motif of ARMS/Kidins220 (Luo et al., 2005). It will be of interest to investigate further whether ARMS/Kidins220 forms a large complex with GRIP1 and kinesin-1 during the trafficking of EphB2 and whether the complex is critical for the presentation of EphB2 in appropriate subcellular compartments for dendrite development.
What is the functional significance of ARMS/Kidins220-regulated dendrite development? Since dendrites act as the scaffold for dendritic spines, which contain the postsynaptic apparatus of neuronal synapses, it is likely that ARMS/Kidins220 plays a key role in the regulation of neuronal communication. Indeed, we found that acute knockdown of ARMS/Kidins220 not only impaired dendrite development, but also resulted in the loss of dendritic spines (data not shown). Consistent with this notion, dendritic spines in ARMS+/− mice exhibit reduced stability in vivo, suggesting that ARMS/Kidins220 is essential for dendritic spine morphogenesis (Wu et al., 2009). The molecular mechanisms underlying the ARMS/Kidins220-regulated spine and synapse development await further investigation.
Footnotes
This study was supported in part by the Research Grants Council of Hong Kong (Grant HKUST661007), the Areas of Excellence Scheme of the University Grants Committee (Grant AoE/B-15/01), and Hong Kong Jockey Club. N.Y.I. was the recipient of the Croucher Foundation Senior Research Fellowship. We thank Kwok-Wang Hung, Yang Shen, Cara Kwong, Busma Butt, and Hayley Tsang for their excellent technical assistance. We also thank Dr. Kwok-On Lai for his critical comments on the manuscript, and members of the Ip laboratory for helpful discussions.
The authors declare no financial conflicts of interest.
References
- Arévalo JC, Yano H, Teng KK, Chao MV. A unique pathway for sustained neurotrophin signaling through an ankyrin-rich membrane-spanning protein. EMBO J. 2004;23:2358–2368. doi: 10.1038/sj.emboj.7600253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arévalo JC, Pereira DB, Yano H, Teng KK, Chao MV. Identification of a switch in neurotrophin signaling by selective tyrosine phosphorylation. J Biol Chem. 2006;281:1001–1007. doi: 10.1074/jbc.M504163200. [DOI] [PubMed] [Google Scholar]
- Arévalo JC, Wu SH, Takahashi T, Zhang H, Yu T, Yano H, Milner TA, Tessarollo L, Ninan I, Arancio O, Chao MV. The ARMS/Kidins220 scaffold protein modulates synaptic transmission. Mol Cell Neurosci. 2010;45:92–100. doi: 10.1016/j.mcn.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloyannis SJ. Dendritic pathology in Alzheimer's disease. J Neurol Sci. 2009;283:153–157. doi: 10.1016/j.jns.2009.02.370. [DOI] [PubMed] [Google Scholar]
- Bracale A, Cesca F, Neubrand VE, Newsome TP, Way M, Schiavo G. Kidins220/ARMS is transported by a kinesin-1-based mechanism likely to be involved in neuronal differentiation. Mol Biol Cell. 2007;18:142–152. doi: 10.1091/mbc.E06-05-0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbelt K, Byne W, Jones LB. Evidence for a decrease in basilar dendrites of pyramidal cells in schizophrenic medial prefrontal cortex. Schizophr Res. 2002;58:75–81. doi: 10.1016/s0920-9964(02)00201-3. [DOI] [PubMed] [Google Scholar]
- Chang MS, Arevalo JC, Chao MV. Ternary complex with Trk, p75, and an ankyrin-rich membrane spanning protein. J Neurosci Res. 2004;78:186–192. doi: 10.1002/jnr.20262. [DOI] [PubMed] [Google Scholar]
- Cheung ZH, Chin WH, Chen Y, Ng YP, Ip NY. Cdk5 is involved in BDNF-stimulated dendritic growth in hippocampal neurons. PLoS Biol. 2007;5:e63. doi: 10.1371/journal.pbio.0050063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcroix JD, Valletta JS, Wu C, Hunt SJ, Kowal AS, Mobley WC. NGF signaling in sensory neurons: evidence that early endosomes carry NGF retrograde signals. Neuron. 2003;39:69–84. doi: 10.1016/s0896-6273(03)00397-0. [DOI] [PubMed] [Google Scholar]
- Dijkhuizen PA, Ghosh A. BDNF regulates primary dendrite formation in cortical neurons via the PI3-kinase and MAP kinase signaling pathways. J Neurobiol. 2005;62:278–288. doi: 10.1002/neu.20100. [DOI] [PubMed] [Google Scholar]
- Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea J, Nissen UV, Dufour A, Sahin M, Greer P, Kullander K, Mrsic-Flogel TD, Greenberg ME, Kiehn O, Vanderhaeghen P, Klein R. Regulation of EphA 4 kinase activity is required for a subset of axon guidance decisions suggesting a key role for receptor clustering in Eph function. Neuron. 2005;47:515–528. doi: 10.1016/j.neuron.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Emoto K. Dendrite remodeling in development and disease. Dev Growth Differ. 2011;53:277–286. doi: 10.1111/j.1440-169X.2010.01242.x. [DOI] [PubMed] [Google Scholar]
- Finsterwald C, Fiumelli H, Cardinaux JR, Martin JL. Regulation of dendritic development by BDNF requires activation of CRTC1 by glutamate. J Biol Chem. 2010;285:28587–28595. doi: 10.1074/jbc.M110.125740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu WY, Chen Y, Sahin M, Zhao XS, Shi L, Bikoff JB, Lai KO, Yung WH, Fu AK, Greenberg ME, Ip NY. Cdk5 regulates EphA4-mediated dendritic spine retraction through an ephexin1-dependent mechanism. Nat Neurosci. 2007;10:67–76. doi: 10.1038/nn1811. [DOI] [PubMed] [Google Scholar]
- Fukata Y, Itoh TJ, Kimura T, Ménager C, Nishimura T, Shiromizu T, Watanabe H, Inagaki N, Iwamatsu A, Hotani H, Kaibuchi K. CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat Cell Biol. 2002;4:583–591. doi: 10.1038/ncb825. [DOI] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- Higuero AM, Sánchez-Ruiloba L, Doglio LE, Portillo F, Abad-Rodríguez J, Dotti CG, Iglesias T. Kidins220/ARMS modulates the activity of microtubule-regulating proteins and controls neuronal polarity and development. J Biol Chem. 2010;285:1343–1357. doi: 10.1074/jbc.M109.024703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad CC, Milstein AD, Ethell IM, Henkemeyer M, Sheng M. GRIP1 controls dendrite morphogenesis by regulating EphB receptor trafficking. Nat Neurosci. 2005;8:906–915. doi: 10.1038/nn1487. [DOI] [PubMed] [Google Scholar]
- Ip JP, Shi L, Chen Y, Itoh Y, Fu WY, Betz A, Yung WH, Gotoh Y, Fu AK, Ip NY. alpha2-chimaerin controls neuronal migration and functioning of the cerebral cortex through CRMP-2. Nat Neurosci. 2012;15:39–47. doi: 10.1038/nn.2972. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. The control of dendrite development. Neuron. 2003;40:229–242. doi: 10.1016/s0896-6273(03)00631-7. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. Branching out: mechanisms of dendritic arborization. Nat Rev Neurosci. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M. Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin pathway. J Neurosci. 2005;25:11300–11312. doi: 10.1523/JNEUROSCI.2270-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong H, Boulter J, Weber JL, Lai C, Chao MV. An evolutionarily conserved transmembrane protein that is a novel downstream target of neurotrophin and ephrin receptors. J Neurosci. 2001;21:176–185. doi: 10.1523/JNEUROSCI.21-01-00176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konur S, Ghosh A. Calcium signaling and the control of dendritic development. Neuron. 2005;46:401–405. doi: 10.1016/j.neuron.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Kwon M, Fernández JR, Zegarek GF, Lo SB, Firestein BL. BDNF-promoted increases in proximal dendrites occur via CREB-dependent transcriptional regulation of cypin. J Neurosci. 2011;31:9735–9745. doi: 10.1523/JNEUROSCI.6785-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PC, Chan PM, Hall C, Manser E. Collapsin response mediator proteins (CRMPs) are a new class of microtubule-associated protein (MAP) that selectively interacts with assembled microtubules via a taxol-sensitive binding interaction. J Biol Chem. 2011;286:41466–41478. doi: 10.1074/jbc.M111.283580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Menéndez C, Gascón S, Sobrado M, Vidaurre OG, Higuero AM, Rodríguez-Peña A, Iglesias T, Díaz-Guerra M. Kidins220/ARMS downregulation by excitotoxic activation of NMDARs reveals its involvement in neuronal survival and death pathways. J Cell Sci. 2009;122:3554–3565. doi: 10.1242/jcs.056473. [DOI] [PubMed] [Google Scholar]
- Luo S, Chen Y, Lai KO, Arévalo JC, Froehner SC, Adams ME, Chao MV, Ip NY. α-Syntrophin regulates ARMS localization at the neuromuscular junction and enhances EphA4 signaling in an ARMS-dependent manner. J Cell Biol. 2005;169:813–824. doi: 10.1083/jcb.200412008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister AK, Lo DC, Katz LC. Neurotrophins regulate dendritic growth in developing visual cortex. Neuron. 1995;15:791–803. doi: 10.1016/0896-6273(95)90171-x. [DOI] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Miller FD, Kaplan DR. Signaling mechanisms underlying dendrite formation. Curr Opin Neurobiol. 2003;13:391–398. doi: 10.1016/s0959-4388(03)00072-2. [DOI] [PubMed] [Google Scholar]
- Pasquale EB. Eph receptor signalling casts a wide net on cell behaviour. Nat Rev Mol Cell Biol. 2005;6:462–475. doi: 10.1038/nrm1662. [DOI] [PubMed] [Google Scholar]
- Sutachan JJ, Chao MV, Ninan I. Regulation of inhibitory neurotransmission by the scaffolding protein ankyrin repeat-rich membrane spanning/kinase D-interacting substrate of 220 kDa. J Neurosci Res. 2010;88:3447–3456. doi: 10.1002/jnr.22513. [DOI] [PubMed] [Google Scholar]
- Wu SH, Arévalo JC, Sarti F, Tessarollo L, Gan WB, Chao MV. Ankyrin Repeat-rich Membrane Spanning/Kidins220 protein regulates dendritic branching and spine stability in vivo. Dev Neurobiol. 2009;69:547–557. doi: 10.1002/dneu.20723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SH, Arévalo JC, Neubrand VE, Zhang H, Arancio O, Chao MV. The ankyrin repeat-rich membrane spanning (ARMS)/Kidins220 scaffold protein is regulated by activity-dependent calpain proteolysis and modulates synaptic plasticity. J Biol Chem. 2010;285:40472–40478. doi: 10.1074/jbc.M110.171371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N, Mosinger B, Roy A, Miyazaki M, Ugajin K, Nakamura F, Sasaki Y, Yamaguchi K, Kolattukudy P, Goshima Y. CRMP5 (collapsin response mediator protein 5) regulates dendritic development and synaptic plasticity in the cerebellar Purkinje cells. J Neurosci. 2011;31:1773–1779. doi: 10.1523/JNEUROSCI.5337-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]


