Abstract
During brain morphogenesis, the mechanisms through which the cell cycle machinery integrates with differentiation signals remain elusive. Here we show that the Rb/E2F pathway regulates key aspects of differentiation and migration through direct control of the Dlx1 and Dlx2 homeodomain proteins, required for interneuron specification. Rb deficiency results in a dramatic reduction of Dlx1 and Dlx2 gene expression manifested by loss of interneuron subtypes and severe migration defects in the mouse brain. The Rb/E2F pathway modulates Dlx1/Dlx2 regulation through direct interaction with a Dlx forebrain-specific enhancer, I12b, and the Dlx1/Dlx2 proximal promoter regions, through repressor E2F sites both in vitro and in vivo. In the absence of Rb, we demonstrate that repressor E2Fs inhibit Dlx transcription at the Dlx1/Dlx2 promoters and Dlx1/2-I12b enhancer to suppress differentiation. Our findings support a model whereby the cell cycle machinery not only controls cell division but also modulates neuronal differentiation and migration through direct regulation of the Dlx1/Dlx2 bigene cluster during embryonic development.
Introduction
During brain development, cell cycle regulation and differentiation are tightly coordinated developmental programs. Cross talk exists between the cell cycle machinery and differentiation pathways to ensure that progenitor populations are maintained and that differentiation is induced at the time of terminal mitosis (McConnell and Kaznowski, 1991; Nguyen et al., 2006; Farkas and Huttner, 2008; Frank and Tsai, 2009). Despite the importance of the precise coordination of these events, the mechanisms by which the cell cycle machinery integrates with differentiation signals remain poorly understood.
The retinoblastoma protein, pRb, is a tumor suppressor gene that controls the G1-S phase checkpoint during cell cycle regulation (McClellan and Slack, 2006; Chen et al., 2009; Freedman et al., 2009). Rb regulates the transcription of genes that are required for DNA replication and cell cycle progression by binding and inhibiting E2F transcription factors (Burkhart and Sage, 2008). There are eight E2Fs, five of which can bind Rb (E2F1–5) and are considered among the classical Rb partners while E2F6–8 are Rb-independent repressors (Dick and Dyson, 2006; Chen et al., 2009; Lammens et al., 2009). E2F1, 2, and 3 are primarily transcriptional activators while E2F4 and 5 repress transcription and induce gene silencing through pocket protein binding (Dick and Dyson, 2006). E2F7 and E2F8, two atypical E2Fs (Lammens et al., 2009), can form homo and heterodimers which, in the absence of pocket proteins, bind and repress E2F target genes. The expression of E2F7 and 8 is induced by activating E2Fs and are believed to serve as a fine tuning mechanism to modulate E2F target gene regulation (Di Stefano et al., 2003; Christensen et al., 2005; Lammens et al., 2009).
It has been proposed that the Rb pathway may have novel function(s) that extend beyond cell cycle control (McClellan and Slack, 2006, 2007). Conditional knock-out studies have suggested that Rb may have a role in regulating differentiation and migration (Takahashi et al., 2003; Ferguson et al., 2005; Chen et al., 2007; McClellan et al., 2007); however, the underlying mechanisms remain unknown. Clearly, indirect cross talk between the cell cycle machinery and differentiation pathways is essential to prevent premature differentiation of proliferating progenitors while promoting differentiation as cell division ceases. If the cell cycle proteins themselves could directly regulate genes required for differentiation, these two processes could become intimately coordinated.
Here we have uncovered a more direct role for cell cycle proteins in neuronal differentiation through the control of Dlx1 and Dlx2 homeodomain protein regulation, two key proteins that specify GABAergic neurons in the brain. Consistent with a deficit in Dlx1/Dlx2 gene expression, mice lacking Rb exhibited a pronounced reduction of specific GABAergic subtypes and migration defects in the developing brain. We demonstrate that: (1) Rb directly modulates Dlx gene regulation through E2F repressor sites found in the Dlx1/2-I12b enhancer and through the Dlx1 and Dlx2 proximal promoters, (2) repressor E2Fs, the expression of which is deregulated in the absence of Rb, can repress Dlx1 and Dlx2 gene transcription.
Materials and Methods
Conditional Rb-null mice, transgenic mice, and β-galactosidase staining.
Rb conditional knock-out mice of either sex were generated by crossing Rbfloxed/floxed animals (Marino et al., 2000) with BF1-Cre (brain factor 1 or Foxg1) (Hébert and McConnell, 2000). To generate transgenic mice, the enhancer sequences of wild-type-I12b and the mutant enhancers (I12b-mutant A, I12b-mutant B and I12b-mutant C) were subcloned into the p1230 vector (Yee and Rigby, 1993) that contain a human β-globin minimal promoter and the lacZ reporter gene. Subclonings were done using a PCR-based approach or convenient restriction sites. β-Galactosidase staining was performed on sections overnight at 28°C in a solution of 1 mg/ml X-gal, 5 mm K3Fe(CN)6, 5 mm K4Fe(CN)6, 2 mm MgCl2, and 0.02% NP-40 in 1× PBS.
Histology.
Pregnant mice (n > 3) were killed at different embryonic ages and embryo heads were cut and fixed in 4% PFA overnight in phosphate buffer (PB) (1× PBS), pH 7.4. Cryoprotection was performed in a sucrose gradient by overnight incubation in 12%, 18% and 22% sucrose in 1× PBS, respectively. Embryonic heads were embedded in Tissue Tek (Sakura Finetek USA) and frozen using liquid CO2. Coronal and sagittal sections (10–12 μm) were cut on a cryostat and mounted on Superfrost slides (Fisher Scientific).
Cell counts.
Cell counts were performed on three to four consecutive sections from three embryos for the regions indicated in the text. SST (somatostatin-positive interneuron) counts were done in a 0.6 mm2 region spanning the mantle zone of the lateral ganglionic eminence (LGE) and medial ganglionic eminence (MGE), the piriform cortex and part of the marginal zone. TH-positive cells and GABA-positive cells were counted in the granule cell layer (GCL) and glomerular layer (GL) of the olfactory bulb (OB). BrdU-expressing cells, Active-Caspase 3 (AC-3)-positive cells, I12b-LacZ-positive cells, and GFP/Dlx2 double-positive cells were counted in the regions indicated in the corresponding figures. Averages and SEs as well as the statistical analyses were calculated as shown in the figure legends.
BrdU labeling.
Pregnant mouse dams received an intraperitoneal injection of 50 mg/kg bromodeoxyuridine (BrdU) (Sigma-Aldrich) in 0.9% NaCl at E13.5, E15.5, and E16.5, separately, and killed before giving birth. BrdU detection was performed according to the method of Ferguson et al. (2002).
Immunohistochemistry.
Frozen sections were incubated in blocking solution containing 1% bovine serum albumin, 5% goat serum and 0.3% Triton X-100 in 0.1 m PB for at least 1 h at room temperature. Incubation with the primary antibody(ies) was performed overnight at 4°C. The next day 3 × 10 min washes in 0.1 m PB was followed by incubation with the secondary antibody(ies) for 2 h at room temperature. Sections were washed and mounted in PBS/glycerol. (3:1). The following antibodies were used: mouse anti-Reelin (1:750, Calbiochem), mouse anti-Tuj1 (1:50, Developmental Studies Hybridoma Bank), rabbit anti-MAP2 (microtubule-associated protein 2; 1:200, Santa Cruz Biotechnology), mouse anti-BrdU (1:50, AbD Serotec), rabbit anti-β-galactosidase (1:5000, MP Biomedicals), rat anti-SST (1:100, Millipore Bioscience Research Reagents), rabbit anti-TH (1:350, Millipore Bioscience Research Reagents), rabbit anti-GABA (1:4000, Sigma), anti-AC-3 (1:500, Cell Signaling Technology); guinea-pig anti-Dlx2 (1:200, generous gift from Kazuaki Yoshikawa, Institute for Protein Research, Osaka University, Japan). Alexa Fluor secondary antibodies were purchased from Invitrogen.
In situ hybridization.
In situ hybridization on frozen tissue sections and digoxigenin RNA probe labeling were performed according to the procedures described in (Wallace and Raff, 1999). Hybridized probes were detected with an AP-conjugated anti-digoxigenin Fab fragment antibody (1:2000, Roche) and visualized with the NBT/BCIP substrate system. Antisense riboprobes for Dlx2 (Porteus et al., 1991), Dlx1, Dlx5, GAD67 (generous gifts from J. L. Rubenstein, University of California, San Francisco, San Francisco, CA), Dlx6 (generous gift from T. Lufkin, National University of Singapore), and Hes5 (Tomita et al., 1996) were prepared from plasmids.
Mutagenesis experiments.
Three distinct mutations were generated in the I12b sequence by PCR using overlapping fragments and a proof-reading Taq polymerase as described by Poitras et al. (2007). The primers used to generate the mutations in the three E2F sites are: I12b-mutant A; 5′-GATTCCCTGGATGAAAAAATTGCTC-3′, I12b mutant B; 5′-CCAGAGAGATGATGACACTGCATTAGAATA-3′, and I12b-mutant C; 5′-CCTGCAAGCAATCTGATGGCTCCGGTACCG-3′.
Luciferase assays.
Transient cotransfection assays were performed in mouse neuroblastoma N2a cells and mouse embryonic carcinoma p19 cells using the dual luciferase reporter kit (Promega, E1910). The Dlx1/2-I12b enhancer sequence, and Dlx1 and Dlx2 proximal promoter regions (2 kb upstream of ATG for each gene) were PCR-amplified and subcloned into the pGL4.23 firefly reporter vector using the following primers: I12b forward (5′-CTGAAATAGGTTACACACAGC-3′) and I12b reverse (5′-CCTTTAGAGCTCGTGTGTGAGA-3′), Dlx1 promoter reverse (5′-TTGCCCGACACGGGGCTGTTGAGAC-3′), Dlx1 promoter forward (5′-GTGCTTGATTACAGAGGTCTCCCTG-3′), Dlx2 promoter reverse (5′-GGGACAGGAAAGAGCACGGGTG-3′), Dlx2 promoter forward (5′-GGCTGAGAACTAAATCCAGGCA-3′). The Dlx1/2-I12b carrying mutations in all three E2F sites, so-called (TM), was generated as described above. Expression vectors carrying the mouse cDNAs of Dlx2, E2F7, E2F4 and p107 were transfected as indicated in the text. All results were generated in triplicates and normalized to the expression of the Renilla luciferase used as internal control.
Quantitative real-time PCR.
Quantitative real-time PCR (qRT-PCR) assays were performed on total RNA isolated from embryonic forebrain from E13.5 and E15.5 Rbflox/± Foxg1-Cre and Rbflox/flox Foxg1-Cre animals using the TRIzol method (Invitrogen). A SuperScript III Platinum SYBR Green One-Step qRT-PCR kit (Invitrogen) was used to amplify E2f4, E2f5, E2f6, E2f7, E2f8, Dlx1, Dlx2 and GAPDH mRNAs from 50 ng of isolated total RNA. The primer sequences used were: E2f4 forward (5′-CGCTGACAAGCTGATTGAGCT-3′) and reverse (5′-AGTGACATTCCGGATGCTCTG-3′), E2f5 forward (5′-CGTGTCGTTGCTGCAGGAGGC-3′) and reverse (5′-AGCCAGCACCTACACCCTTCC-3′), E2f6 forward (5′-ACTGGGTGTTCGGAAGAGGCGA-3′) and reverse (5′-GGGGTGCGGCCCCAAAGTT-3′), E2f7 forward (5′-GTGCCTTGTGGCTGCTCCT-3′) and reverse (5′-GCACAGAGTGAACGGACCG-3′), E2f8 forward (5′-GAGAAATCCCAGCCGAGTC-3′) and reverse (5′-CATAAATCCGCCGACGTT-3′), Dlx1 forward (5′-ACTCACACAGACGCAGGTCA-3′) and reverse (5′-CTTCCCAGATGAGGAGTTCG-3′), Dlx2 forward (5′-TCACCCAAACTCAGGTCAAA-3′) and reverse (5′-GGAGGCACAAGGAGGAGAAG-3′), and GAPDH forward (5′-GGTGAAGGTCCGTGTGAACG-3′) and reverse (5′-CTCGCTCCTGGAAGATGGTG-3′). RT-PCR was done using a Qiagen Rotor Gene Q. Values obtained for E2f and Dlx expression levels were normalized to the GAPDH level as an internal control. At least three embryos from each genotype were analyzed by two-tailed Student's t test, with p values <0.05 and <0.01.
In utero electroporation.
E2F7 cDNA tagged with HA was subcloned into the pCig2 vector which contains GFP on a separate IRES promoter. CD1 embryos were electroporated at E13.5 with either an empty pCig2 construct or pCig2-E2F7 construct, expressing both GFP and E2F7. Plasmids were diluted to 2 μg/μl and mixed with trace amounts of Trypan Blue dye. The plasmid was injected into the lateral ventricle of the brain using an Eppendorf Femtojet Microinjector and electroporated into the ventral cortex using a BTX ECM 830 Square Wave Electroporator. Animals were killed 2 d later at E15.5.
Results
Rb is required for Dlx1/Dlx2 gene expression and development of interneuron subtypes
Previous studies have demonstrated a requirement for the Rb/E2F family of cell cycle proteins in regulating the differentiation and migration of ventrally derived interneurons (Ferguson et al., 2005; McClellan et al., 2007). Despite the compelling evidence suggesting cross talk between cell cycle and differentiation pathways, the mechanisms by which this occurs remains poorly understood. Here, we asked whether the Rb/E2F pathway may serve as a feedback mechanism to coordinate cell cycle regulation with neurogenesis and sought to identify whether such a link exists in the ventral telencephalon, a region severely affected by deregulation of the Rb/E2F pathway.
Given that the Dlx family of homeodomain proteins are master regulators of GABAergic neuron development, we first asked whether Rb deficiency results in a defect in the regulation of the Dlx1/Dlx2 bigene cluster, essential for interneuron development (Anderson et al., 1997, 1999, 2001; Pleasure et al., 2000; Panganiban and Rubenstein, 2002; Stühmer et al., 2002; Long et al., 2007, 2009; Petryniak et al., 2007). Using mice carrying a conditional telencephalic mutation for Rb (BF1-Cre; Rbflox/flox mice) (Hébert and McConnell, 2000; Marino et al., 2000), we examined the regulation of Dlx1 and Dlx2 in the developing telencephalon. Relative to control littermates, the expression patterns of these homeobox genes were similar to littermate controls at E13.5 (Fig. 1A–B′); however, a marked reduction in the levels of Dlx1 and Dlx2 mRNA was detected in the LGE and MGE and the septum in Rb conditional mutant mice starting at E15.5 (Fig. 1C,C′,F– G′; arrowheads in C′,F′,G′). Little, if any, Dlx1 and Dlx2-positive cells were found in the cortical layers, and the GCL and GL inside the OB in Rb conditional mutants around birth (Fig. 1E,E′,H,H′, arrowheads in G′,H′; data not shown). The reduction in Dlx1 and Dlx2 expression was confirmed by quantitative RT-PCR performed on total RNA extracted from ventral forebrain lysates (Fig. 1K) and by immunostaining for Dlx2 (Fig. 1I–J′). Together, these results demonstrate that Rb is required for proper expression of Dlx1 and Dlx2, two key members of the Dlx family of transcription factors that are expressed early on in uncommitted progenitor cells (Bulfone et al., 1993; Liu et al., 1997; Eisenstat et al., 1999) and play an essential role in the differentiation and migration of developing neurons during ventral telencephalic development.
Figure 1.
Reduced Dlx1 and Dlx2 gene expression in Rb conditional mutant brain. In situ hybridization with Dlx1 and Dlx2 antisense RNA probes in the ventral forebrain at E13.5 (A–B′, E15.5 (C, C′; F, F′) and P0 (E, E′; G–H′). Dlx1 and Dlx2 mRNA levels were severely reduced in Rb conditional mutants compared with controls between E15.5 and P0 in the LGE, MGE, Sp, RMS and inside the OB (arrowheads in A′–C′, E′–H′). Dlx2 protein level was also reduced in these regions (I–J′; arrowheads). D, D′, GAD67 mRNA (glutamic acid decarboxylase enzyme), a downstream target of Dlx gene, was also decreased in the ventral forebrain of Rb mutant mice at E15.5. K, Quantitative real-time PCR performed on ventral forebrain lysates confirmed the significant reduction in Dlx1 and Dlx2 gene expression at E15.5 but not earlier. Cx, Cortex; Sp, septum; SVZa, anterior subventricular zone; Pi, piriform cortex; MCL, mitral cell layer; SEL, subependymal layer. Scale bar, 100 μm.
Given (1) the importance of Dlx homeodomain proteins in interneuron development, and (2) the deficit in Dlx gene expression in the absence of Rb, we asked whether there was a loss of interneuron subtypes in Rb-deficient brains. Immunostaining revealed a 23% reduction in the number of SSTs in the basal ganglia and the piriform cortex at E15.5 [control (Ct), 322 ± 19.2 vs mutant (mut), 247 ± 4.3; Fig. 2A–B′,H]. More strikingly, we detected a gradual loss of GABAergic neurons in the olfactory bulb between E15.5 (Fig. 2D,D′,H; Ct, 167 ± 4.25 vs mut, 125 ± 3.5; 25% reduction at E17.5) and P0 (Fig. 2H, Ct, 270 ± 5.7 vs mut, 40 ± 2.5; 85% reduction), and this was also true for a subpopulation of dopaminergic neurons as detected by staining for TH (Fig. 2C,C′,H; Ct, 110 ± 3.5 vs mut, 65 ± 5.3; 41% reduction at E17.5, and Ct, 117 ± 3.7 vs mut, 38 ± 2.7; 67% reduction at P0). This neuronal loss affected specific GABAergic subpopulations and not all subtypes (data not shown). Further, the loss is not attributed to accelerated apoptosis as we did not detect an increase in activated caspase-3-positive cells in the SVZ of LGE and MGE of Rb conditional mutant between E13.5 and E18.5 (Fig. 3E,E′,I; Ct, 9.5 ± 5 vs mut, 12.3 ± 3 at E13.5 and Ct, 21.5 ± 1.2 vs mut, 18 ± 2.8 at E17.5). Of note, loss of GABAergic neurons was paralleled by a reduction in the expression of the GABA synthesizing enzyme, glutamic acid decarboxylase (GAD67), at E15.5 (Fig. 1D,D′; arrowheads in D′). In contrast, the number of OB projection neurons was unaffected by Rb deficiency, as shown by Reelin staining in the mitral cell layer of the OB (Fig. 2E,E′). This phenotype provides support of the hypothesis that the cell cycle regulator, Rb, is required for the development of ventrally derived GABAergic neurons.
Figure 2.
Loss of specific interneuron subtypes and migration defects in Rb conditional mutant brain. A–D′, Immunostaining using antibodies against SST (A–B′; coronal sections), TH (C, C′; sagittal sections), and GABA (D, D′; sagittal sections) in Rb control (A–D) and mutant (A′–D′) animals. B and B′ are higher-magnification images of boxes shown in A and A′, respectively. The number of SST-positive interneurons is reduced in the LGE, MZ, and Pi in Rb-null animals compared with controls at E15.5 (arrowheads in B′). A gradual loss of dopaminergic neurons (C′; arrowheads) and GABAergic neurons (D′; arrowheads) is detected in Rb-null animals compared with controls between E17.5 (C–D′) and P0 (H). E, E′, Immunostaining against Reelin, a projection neuron marker, shows no difference in expression between Rb control and mutant mice in the OB (coronal sections). F, F′, Immunostaining against Tuj1 (β-tubulin), an early differentiation marker, showing accumulation of neuroblasts in the anterior SVZ and rostral RMS in the Rb mutant brain (arrowheads in F′) compared with controls (F) at E18.5 (sagittal sections). The RMS is delineated with yellow dashes. G, G′, Immunostaining with anti-MAP2, a late differentiation marker, showed mislocalized expression in LGE, Cx, RMS, and SEL in the mutant bulb at E19.5 (arrowheads in G′) compared with controls (G) (sagittal sections) suggesting the presence of radial migration defects in Rb mutant mice. H, Quantification of the distinct subtypes of GABAergic neurons affected in the absence of Rb. Error bars represent SD of measurements from n = 3, and asterisks indicate a significant difference between control and mutant genotypes: *p < 0.05; **p < 0.01; t test. Legend as in Figure 1. Scale bar, 100 μm.
Figure 3.

LGE progenitors fail to migrate properly to the OB in Rb mutant mice during late development. E16.5 females were injected with BrdU and killed 3 d later. Brains were removed and assayed for BrdU incorporation by immunohistochemistry. In control animals, BrdU staining was mainly detected along the medial and caudal RMS and inside the OB (C, D; sagittal sections) with few BrdU-positive cells in the anterior subventricular zone (SVZa) (A, B) at E19.5. In contrast, in Rb-null animals, many BrdU-positive cells were still lining the VZ and SVZa of the LGE (A′–B′; arrows in B′) and failed to migrate along the RMS to the OB where only a few BrdU-positive cells were detected (C′–D′; arrows), suggesting the presence of migration defects in Rb-null mice. (E, E′) No increase in apoptosis is detected in the mutant LGE compared with controls before E19.5 as indicated by anti-Active-Caspase 3 staining (sagittal sections). However, a dramatic increase in cell death was observed in the LGE and the RMS at birth (F, F′; arrows in F′). G, G′, In situ hybridization with Hes5, showing abnormal migration of neuroblasts in the rostral RMS in the Rb mutant brain (G′; red arrowheads) compared with control (G). H, Quantification of BrdU-positive in the SVZa, medial RMS and OB at E19.5, n = 3. I, Quantification of AC-3-positive cells in the SVZ at different embryonic ages, n = 3. Legend as in Figure 1. Scale bar; 100 μm.
Rb is required for the proper maturation and migration of OB interneurons
Considering that both Dlx1 and Dlx2 are required for migration of GABAergic neurons to the cortex and OB (Anderson et al., 1997, 2001), we examined whether Rb, through the control of Dlx gene expression, is also needed for interneuron migration. Thus, we examined the distribution of Tuj1, an early differentiation marker, in the brain between E17.5 and birth. At E18.5, Rb mutant mice displayed a stronger Tuj1 staining in the LGE including the anterior SVZ (SVZa) as well as the rostral RMS (rostral migratory stream) compared with control mice (Fig. 2F,F′, arrowheads). This phenotype is consistent with increased proliferation of committed neuroblasts as shown previously (Herrup and Silver, 1994; Ferguson et al., 2002) and suggests the presence of migration defects in the absence of Rb. To further investigate this possibility, we assessed the pattern of MAP2 expression, a late differentiation marker which labels the dendritic arbor of mature neurons. While Rb mutant and control mice displayed similar patterns of MAP2 staining during midgestation (E16.5–E17.5), a severe alteration of MAP2 distribution was evident in the mutant telencephalon at E19.5 (Fig. 2G,G′). Unlike control animals where MAP2 was primarily detected in the cortical plate with strong intensity in the superficial layers, this dendritic marker was scattered throughout the cortex in Rb mutant mice (Fig. 2G,G′; arrowheads in G′). Moreover, an abnormally intense MAP2 staining was observed in the RMS in Rb mutant brains compared with controls and instead of being localized to the GCL and GL as in wild-type animals, MAP2 was highly expressed in the subependymal layer of the mutant OB (Fig. 2G,G′; arrowheads in G′). These results demonstrate a requirement for Rb in radial migration in both the cerebral cortex and the olfactory bulb.
Next, we questioned whether the generation of neuroblasts that give rise to OB GABAergic interneurons may be altered in the LGE or whether the migration phenotype described earlier may result from defective neuroblast maturation. To address this question, we performed a series of single BrdU injections (birth-dating studies) at E13.5, E15.5, and E16.5, separately, and killed the animals at E19.5 and immunostained for the detection of BrdU. In control animals injected at E16.5, BrdU-positive cells were primarily distributed along the medial and caudal RMS and inside the OB, with a few labeled cells in the SVZa at E19.5 (Fig. 3A–D,H). Thus, in wild-type animals, most of these cells have exited the cell cycle, induced differentiation and migrated to their final destination. In contrast, in the Rb mutant animals, BrdU-expressing cells were still found in the VZ and SVZa of the LGE 3 d post-BrdU injection (Fig. 3A′–B′; arrows in B′,H). Furthermore, the RMS and OB were disorganized and contained 3- to 4-fold less BrdU labeled cells compared with controls at this age (Fig. 3C′,D′, arrows, H). These migration defects were also evident by the expression of Hes5 showing abnormal cell migration in the rostral RMS in Rb mutant mice (Fig. 3G,G′; red arrowheads in G′). These results revealed that neuronal progenitors are generated normally in the Rb mutant LGE but fail to differentiate and/or migrate appropriately to their final destination and, hence, many cells die by apoptosis as shown by the increased AC-3 staining in the LGE, RMS and OB around birth (Fig. 3F,F′; arrows, I; data not shown).
Together, our data demonstrate an essential role for Rb in the differentiation and migration of neuronal progenitors born in the LGE during mid-to-late gestation. Due to the profound defects in both of these processes in Rb-deficient brains, we next questioned whether Rb may be playing a direct role by controlling the Dlx genes that regulate these processes.
The Dlx1/Dlx2 enhancer I12b contains putative E2F binding sites acting as repressor sites in vivo
The transcription of Dlx genes in the forebrain is regulated primarily by highly conserved tissue-specific enhancers that act as cis-regulatory elements shared between the two Dlx genes belonging to one cluster (Zerucha et al., 2000; Ghanem et al., 2003, 2007). Given the significant reduction in Dlx1 and Dlx2 expressions in the absence of Rb, we examined the mechanism by which Rb may mediate the regulation of the Dlx1/Dlx2 bigene cluster. We hypothesized that Rb may regulate the expression of Dlx1 and Dlx2 by modulating the activity of their shared enhancer, I12b. This enhancer is particularly important because it controls Dlx expression in subpopulation(s) of interneurons that migrate to the dorsal cortex and OB. Furthermore, the enhancer activity is known to be high specifically in the ventral telencephalic regions severely affected by the absence of Rb (Ferguson et al., 2002; Ghanem et al., 2007; Batista-Brito et al., 2008). To test this possibility in vivo, we first generated Rb conditional mutants carrying LacZ reporter genes under the control of the human β-globin minimal promoter and the Dlx1/2-I12b enhancer (I12b-LacZ). As a consequence, the activity of the I12b-LacZ reporter gene was almost completely abolished in the LGE, the Cx and the OB of Rb conditional mutants between E15.5 and P0 (Fig. 4A–D′; arrowheads in A′–D′,E), suggesting that the activation of the Dlx1/2-I12b enhancer is dependent on the presence of Rb. These studies demonstrate that Rb is required for the activation of the Dlx1/2 intergenic enhancer which is essential for Dlx1/Dlx2 gene regulation. As Rb regulates gene transcription through interactions with its E2F binding partners, we next asked whether the Dlx-I12b enhancer and Dlx1/2 promoters contained E2F consensus sites.
Figure 4.

Loss of Dlx1/2-I12b enhancer activity in Rb-deficient brain. A–D′, Immunostaining with LacZ antibody performed on sagittal sections of Dlx transgenic animals carrying the LacZ reporter gene under the control of the Dlx1/2 enhancer, I12b, in the absence of Rb at E15.5 (A–B′) and P0 (C–D′). The expression of LacZ-I12b reporter gene is greatly reduced in the Cx, LGE, RMS, and OB in Rb-null animals (A′–D′; arrowheads) compared with controls (A–D), suggesting that the activity of I12b is Rb-dependent in the ventral forebrain. E, Quantification of I12b-LacZ-positive cells in indicated areas in Rb control versus mutant mice at E15.5 (n = 3) and P0 (n = 3). Legend as in Figure 1. Scale bar; 100 μm.
Since Dlx gene transcription is partially controlled by the forebrain-specific enhancer, I12b (Zerucha et al., 2000; Ghanem et al., 2003, 2007), we searched I12b for classical E2F consensus sites. Three putative E2F sites were identified in the enhancer, which we called A, B, and C (Fig. 5I; shown in red); the first two sites were intercalated between two Dlx binding sites (in blue), while the third site was adjacent to a Mash-1 site (in green) (Fig. 5I) (Poitras et al., 2007). To test whether these E2F sites function as repressors or activators in vivo, we mutated each site separately using a PCR-based approach (see Materials and Methods for details) and generated individual LacZ transgenic mice under the control of β-globin minimal promoter and I12b carrying each mutation (Fig. 5II; mutants A, B, and C). As a consequence of E2F consensus site deletion, the Dlx1/2-I12b enhancer activity was remarkably increased in mice carrying any of the three mutations, in particular, mutants B and C (Fig. 5; compare A–C and A″–C‴). Thus, stronger LacZ reporter expression was detected in the Cx, LGE and OB as well as in the diencephalon where I12b (and the Dlx genes) is also active (Fig. 5B′–C‴, arrowheads; data not shown). The increase in reporter expression was also confirmed by β-galactosidase staining in Rb control and mutant tissues simultaneously (Fig. 5a–a‴). All our results were reproducible and consistent among at least three primary lines that were generated independently for each mutation thus ruling out the possibility of an integration effect or a difference in copy number between the transgenic lines. Our mutagenesis study coupled to transgenesis in mice provides in vivo evidence that at least two of three putative E2F binding sites identified in Dlx1/2-I12b are functional repressor sites. We therefore asked which of the Rb target E2F transcription factors could be mediating the repression of the Dlx1/2-I12b enhancer activity in the absence of Rb.
Figure 5.
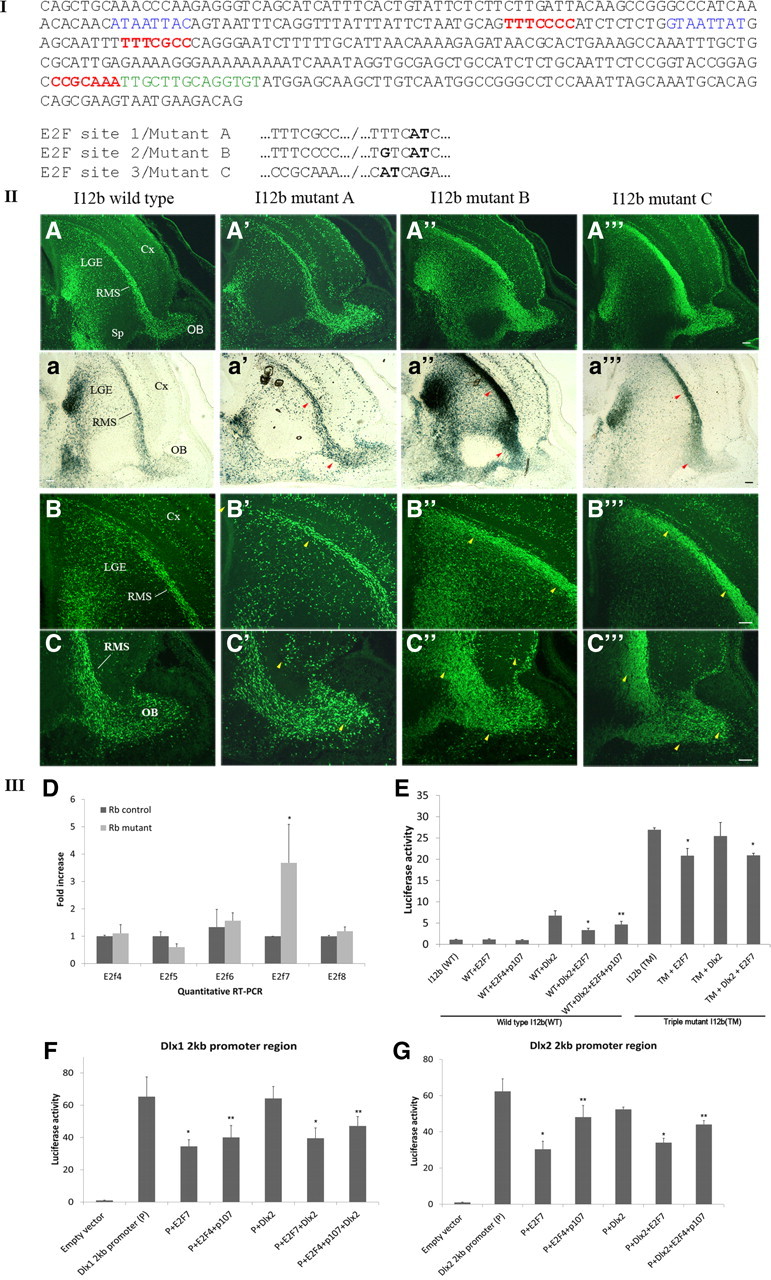
Repressor E2Fs such as E2F7 repress the activities of the Dlx1/2-I12b enhancer and the Dlx1 and Dlx2 proximal promoter regions. I, The sequence of the mouse Dlx1/2-I12b enhancer is shown and comprises three putative E2F binding sites (marked in red), two Dlx binding sites (in blue) and a Mash-1 site (in green) (Poitras et al., 2007). The three E2F sites were mutated separately as highlighted in bold and the corresponding mutants were called A, B, and C. II, The three consensus E2F sites found in I12b act as repressor sites in vivo. LacZ reporter mice under the control of I12b-wild-type (A–C), I12b-mutant A (A′–C′), I12b-mutant B (A″–C″) and I12b-mutant C (A‴–C‴) were generated and LacZ expression compared among these lines at E15.5. A marked increase in LacZ intensity was observed in the telencephalon of the I12b-mutant lines compared with I12b-wild-type control as shown by immunostaining against LacZ (A–A‴ and β-galactosidase staining (a–a‴) on sagittal sections. B–B‴ and C–C‴ are high-magnification images of LacZ staining in the LGE and RMS/OB, respectively. Yellow arrowheads indicate regions with increased LacZ intensity in the mutant lines. Legend as in Figure 1. III, Repressor E2Fs such as E2F7 repress the Dlx1/2-I12b enhancer activity and Dlx1/Dlx2 proximal promoter regions in vitro. D, Quantitative real-time PCR analysis of repressor E2Fs, E2F4–8, in Rb control versus Rb mutant brain showing a significant threefold increase in E2F7 expression but not the other E2Fs. *p < 0.01, t test. E, Luciferase reporter assays were performed in N2a cells. Firefly constructs under the control of Dlx1/2-I12b WT and Dlx1/2-I12b carrying three mutated E2F sites [Triple Mutant (TM)] were transfected alone or in combination with a Dlx2 expression vector to activate the enhancer. The effects of E2F7 and E2F4/p107 overexpression were assessed in each case, separately. F, Luciferase reporter assays were performed with firefly constructs carrying the 2 kb proximal promoter regions of Dlx1 and Dlx2 (G) and showing E2F repression of both Dlx promoter regions. Error bars represent SD of measurements from n = 3, and asterisks indicate a significant difference between control and mutant genotypes *p < 0.01; **p < 0.05; ANOVA and Tukey HSD tests.
Increased level of E2F7, an Rb-independent repressor, in Rb conditional mutants
Deregulation of E2F activity is a common consequence for the loss of Rb both in vitro and in vivo. Hence, depending on the tissue or the cell line in question, one or more E2F genes become deregulated in the absence of Rb and may be directly associated with the aberrant phenotype (Callaghan et al., 1999; Chen et al., 2007; McClellan et al., 2007). Since the Dlx1/2-I12b enhancer activity and Dlx1/2 expressions are largely decreased in Rb-deficient mice, we hypothesized that these defects resulted from deregulation in the level(s) of one or more repressor E2Fs that may inhibit Dlx transcription via the Dlx1/2-I12b enhancer and/or the Dlx1/Dlx2 promoters. To address this question, we first compared the levels of repressor E2Fs, E2F4, 5, 6 in Rb control and mutant brain tissues using quantitative RT-PCR. We also included Rb-independent repressors, E2F7 and 8 since both have been shown to be activated by E2F1 which is elevated in the absence of Rb (Di Stefano et al., 2003; Christensen et al., 2005). No significant difference was observed in the mRNA levels of E2F4, 5, 6 and 8; however, a threefold increase in E2F7 mRNA was consistently detected (Fig. 5D, n = 3). As E2F7 is a downstream target of the Rb/E2F pathway (Di Stefano et al., 2003) and is upregulated in the absence of Rb, we next investigated whether E2F7 could suppress Dlx1 and Dlx2, two key genes in the regulation of neurogenesis.
Repressor E2Fs such as E2F7 negatively regulate Dlx1/Dlx2 gene expression in vitro
Dlx genes have been to shown to positively auto-regulate their transcription level by binding to their own enhancers such as I12b both in vivo and in vitro (Zerucha et al., 2000; Poitras et al., 2007; Potter et al., 2009). Given the upregulation of E2F7 in the Rb mutant brain, we sought to determine whether E2F7 upregulation itself can repress the enhancer activity of I12b and/or the endogenous activities of the Dlx1 and Dlx2 promoters. To address this question in vitro, we performed transient cotransfection experiments in two immortalized cell lines, the mouse neuroblastoma N2a cells and the mouse embryonic carcinoma p19 cells, independently. A Firefly reporter construct under the control of the Dlx1/2-I12b enhancer was transfected either alone or with a Dlx2 expression vector to activate the enhancer and thereby better visualize any repressive effects of E2F7. All readings were normalized to the Renilla luciferase used as internal control. Our results revealed that E2F7 repressed the reporter activity of I12b when activated by Dlx2 by twofold in both N2a cells and p19 cells lines [Fig. 5E; 6.74 (Dlx2) vs 3.34 (Dlx2 + E2F7); data not shown; n = 3]. We also examined E2F4, another repressor E2F, which mediates repression by recruitment of pocket proteins such as p107 to the site. To test this, we expressed E2F4 together with p107, and found that it was also able to repress the activity of I12b by 1.44-fold, although less efficiently than E2F7 [Fig. 5E; 6.74 (Dlx2) vs 4.66 (Dlx2 + E2F4), n = 3]. We then tested the responsiveness of a modified version of the Dlx1/2-I12b enhancer carrying three mutated E2F sites (Triple mutant of A, B, and C sites called TM-I12b). The mutant enhancer (TM) was transfected along with Dlx2 (to activate) and/or E2F7. Our results revealed the following: (1) the E2F7 repression of I12b was reduced from 2-fold (with WT-I12b) to 0.77-fold, suggesting that this regulation is indeed mediated by one or more of these E2F sites [Fig. 5E; 26.94 ± 0.43 (TM-I12b) vs 20.85 ± 1.7 (TM-I12b+E2F7), n = 3]. (2) The basal reporter activity of the triple mutant enhancer TM-I12b was ∼20-fold higher than the WT-I12b [Fig. 5E; 26.94 ± 0.43 (TM-I12b) vs 1.38 ± 0.13 (WT-I12b), n = 3] and ∼4-fold stronger than the activity of WT-I12b when induced by Dlx2 activation [26.94 ± 0.43 (TM-I12b) vs 6.74 ± 1.15 (Dlx2 + WT-I12b); Fig. 5E]. Given that the mutated E2F sites are in close proximity to the Dlx and Mash1 binding sites and function as repressor sites (Fig. 5I,II), this could suggest that E2Fs may compete with the activators of this enhancer in vivo.
We conducted a second set of luciferase assays to test the effect of E2F7 on the activity of two firefly reporter constructs each carrying a 2 kb fragment corresponding to the Dlx1 and Dlx2 proximal promoter regions located upstream of the ATG start site of each gene, respectively. Consequently, we detected a robust endogenous reporter activity associated with both promoter regions and corresponding to ∼60-fold activation compared with the empty vector [(1.03 ± 0.2 (empty vector) vs 62.36 ± 6.91(Dlx2 2 kb promoter); Fig. 5F,G]. This reporter activity was reduced by twofold following E2F7 expression [Fig. 5G; 62.36 ± 6.91(Dlx2 2 kb promoter) vs 30.39 ± 4.43 (Dlx2 2 kb promoter + E2F7)] and only by 0.77-fold after the combined expression of E2F4 and p107 [Fig. 5G; 62.36 ± 6.91 (Dlx2 2 kb promoter) vs 48.12 ± 6.47 (Dlx2 2 kb promoter + E2F4)]. E2F7 and E2F4/p107 repressed in a similar manner the Dlx1 promoter (Fig. 5F). Hence, the increased levels of E2F7 and, to a lesser extent, E2F4, could repress the endogenous activity of the Dlx1/2 proximal promoter regions. Together, our transfection experiments demonstrate that repressor E2Fs can directly repress Dlx expression via putative E2F sites found in the shared Dlx1/2-I12b enhancer and/or through the Dlx1/2 proximal promoter regions.
Electroporation of E2F7 represses Dlx2 expression in vivo
Given that Dlx2 is the first Dlx gene to be induced during neurogenesis (Eisenstat et al., 1999), we asked whether Dlx2 gene expression can be repressed by E2F7 in vivo. Thus, we electroporated E2F7 in the developing brain at E13.5 using two constructs: a GFP-expressing vector (control vector) and a bicistronic vector expressing both GFP and E2F7, and animals were killed 2 d later. First, we confirmed by in situ hybridization that the brains electroporated with the GFP-E2F7 constructs expressed E2F7 at high levels and that the E2F7-expressing cells and GFP-positive cells were colocalized (data not shown). Second, we compared the colocalization of GFP and Dlx2 on adjacent slides in GFP-control tissue versus GFP-E2F7 electroporated tissue using immunostaining and in situ hybridization, respectively. A clear reduction in Dlx2 mRNA level was evident in electroporated regions that ectopically expressed GFP-E2F7 as indicated by the interrupted pattern of Dlx2 mRNA expression but not in electroporated regions expressing GFP alone (Fig. 6a–d,A–D′; arrows, n = 2 mutants, n = 4 controls). To confirm this result, we costained both GFP control tissue and GFP-E2F7-expressing tissue with GFP and Dlx2 antibodies and found little colocalization of the two proteins in the GFP-E2F7 electroporated brains in the SVZ (Fig. 6A″–B″, 8.7% overlap: 9 ± 2.12 GFP+/Dlx2+ of 109 ± 3.53 total GFP+ cells), whereas in GFP control brains, more than half of the GFP-positive cells coexpressed Dlx2 (Fig. 6C″–D″, 53.95% overlap: 92 ± 9.11 GFP+/Dlx2+ of 172 ± 22.42 total GFP+ cells). The absence of colocalization of Dlx2 and E2F7 in E2F7-electroporated regions could not result from tissue damage during electroporation as no difference was observed with DAPI staining in these regions in both control and E2F7 electroporated brains (Fig. 6A‴–D‴). In summary, our in vivo data demonstrates that repressor E2Fs such as E2F7, when upregulated in the absence of Rb, can repress endogenous Dlx2 gene expression and this is consistent with our in vitro luciferase reporter analyses with the Dlx1/2-I12b enhancer and the Dlx2 proximal promoter.
Figure 6.

E2F7 represses Dlx2 expression in vivo. In utero electroporation of a dual cassette containing both E2F7 and GFP, and an empty vector containing GFP alone in the ventral forebrain at E13.5. a–d, In situ hybridization performed with a Dlx2 probe on coronal sections at E15.5 and showing loss of Dlx2 expression in E2F7-electroporated brains (b; arrows) compared with control brains (c, d). Insets in a and c represent the non-electroporated contralateral side. A–D, Anti-GFP staining of brains electroporated with E2F7-GFP (A, B) and GFP alone (C, D). A′–D′ are merged images of a–d and A–D, respectively. A″–D″, Double immunostaining with anti-Dlx2 (red) and anti-GFP (green) showing little colocalization of the two proteins in E2F7 electroporated brains (B″; inset) compared with GFP controls where many cells are Dlx2+/GFP+ (D″; inset).
Together, our findings implicate a key role for the Rb/E2F proteins in the regulation of critical genes required for neuronal differentiation and migration, such as the Dlx1/Dlx2 bigene cluster. These studies identify a novel link between cell cycle regulation and homeobox genes that play a key role in neurogenesis. Furthermore, we provide new molecular evidence of a dual role of cell cycle proteins in proliferation control and regulating key aspects of differentiation during brain development.
Discussion
We investigated here the mechanisms by which cross talk may occur between cell cycle regulation and cellular differentiation in the developing brain. We demonstrated for the first time a requirement for cell cycle proteins in the direct regulation of key differentiation genes in the brain. We showed that E2Fs can regulate the expression of the Dlx1 and Dlx2 proteins through E2F functional sites found in the Dlx1/Dlx2 forebrain-specific enhancer, I12b, and through the Dlx1 and Dlx2 proximal promoter regions. In turn, Rb can modulate Dlx1/2 expression by controlling the activity of E2Fs during neurogenesis. Hence, in the absence of Rb, repressor E2Fs, such as E2F7, can repress the transcription of Dlx1 and Dlx2 in vitro and Dlx2 in vivo. This novel function of Rb/E2Fs in addition to their classical role in cell cycle control implicates these proteins in a dual role, which now includes the direct regulation of key genes required for neuronal differentiation and migration during brain development.
The Rb/E2F pathway regulates differentiation and migration through direct control of the Dlx1/2-I12b enhancer
Dlx1/Dlx2 expressions and the activity of a Dlx1/2 forebrain-specific enhancer, I12b, were drastically reduced in the absence of Rb (Figs. 1, 4). Considering that Dlx2 followed by Dlx1, are the earliest Dlx genes to be expressed in the SVZ, and that they activate the downstream Dlx5/Dlx6 genes, they are crucial regulators of interneuron differentiation and migration in the developing telencephalon (Bulfone et al., 1993; Liu et al., 1997; Eisenstat et al., 1999; Zerucha et al., 2000). I12b was previously identified as a cis-regulatory element controlling the expression of Dlx1 and/or Dlx2 (Ghanem et al., 2007; Poitras et al., 2007). Given that Dlx1 and Dlx2 arise from gene duplication and are regulated by a common enhancer I12b, it is logical that both of these homeobox genes are equally dependent on the presence of pRb for their regulation. The defects in interneuron differentiation and migration in the absence of pRb is consistent with a deficit in Dlx1/Dlx2 gene expressions during forebrain development.
We showed that Rb/E2F's regulation of Dlx1/2-I12b is directly mediated by three putative E2F binding sites on the enhancer acting as repressor sites in vivo (Fig. 5). Hence, the LacZ-I12b reporter activity was increased in the SVZ in transgenic animals carrying a single E2F mutant site (Fig. 5). Interestingly, site B (TTTCCCC) was adjacent to an homeobox binding motif (TAAT) that was previously shown to be a putative Meis2 repressor site (Poitras et al., 2007). Moreover, the E2Fs sites functioned as repressor sites in vitro; thus, the basal reporter activity of a firefly-I12b construct was ∼30-fold higher in the triple mutant enhancer TM-I12b compared with WT-I12b (Fig. 5E). Of note, the three E2F sites identified here were intercalated between two Dlx binding sites and one Mash1 site, all of which act as activator sites in vivo (Poitras et al., 2007).Together, these findings indicate that there is a dynamic interplay among activator and repressor transcription factors including Dlx, Mash1, E2Fs and Meis2 that may control the spatio-temporal regulation of Dlx gene expression.
Repressor E2Fs such as E2F7 can repress Dlx1/2 expression in the absence of Rb in vitro
We found that Rb regulation of Dlx1/2 is mediated by E2F repressors acting directly on the Dlx1/2-I12b enhancer. In fact, our results revealed a threefold increase in E2F7 mRNA, an Rb independent repressor E2F. Interestingly, upregulation of E2F7 was able to repress by twofold the activity of the Dlx1/2-I12b enhancer as well as the basal reporter activity of the Dlx1 and Dlx2 proximal promoter regions in vitro (Fig. 5E–G). Similarly, electroporation of E2F7 repressed Dlx2 expression in the ventral forebrain as indicated by the low number of E2F7-expressing cells (∼8.7%) that were colabeled with Dlx2 while more than half of the GFP+ control cells were Dlx2+ (∼54%; Fig. 6). In a wild-type brain, Rb normally represses the transcription of E2F1 which is an activator of E2F7 transcription (Di Stefano et al., 2003). Loss of Rb leads to upregulation of E2F7 which in turn can act as a potent transcriptional repressor interfering with developmental processes such as neuronal differentiation. Our in vitro studies also showed that other repressor E2Fs, for instance E2F4, in combination with p107, could also contribute to Dlx repression although to a lesser extent (Fig. 5E–G). Previous studies have clearly demonstrated a compensatory increase in p107 gene expression in Rb-deficient neural precursor cells (Callaghan et al., 1999). Therefore, the possibility exists that loss of pRb may also lead to an increase in p107 and/or p130 and a concomitant increase in nuclear E2F4 (Callaghan et al., 1999; Chen et al., 2007; McClellan et al., 2007), causing Dlx repression through repressive E2F sites on the Dlx1/Dlx2 enhancer/promoter regions.
E2Fs bind specifically the Dlx2 locus in vivo
To identify gene promoters occupied by the E2F transcription factors, we performed ‘Chip-on-chip’ experiments involving chromatin immunoprecipitation (ChIP) with E2F3 and E2F4 antibodies followed by hybridization on microarrays (L. M. Julien and R. S. Slack, unpublished observation). These E2Fs are known targets for Rb in the context of brain development (McClellan et al., 2007). Interestingly, high E2F3 and E2F4 binding activities were detected on the Dlx2 locus (the 5′-UTR and 3′-UTR regions). These data were consistent with a previous report using similar techniques in five non-neuronal cell lines and showing recruitment of E2F1 and E2F4 mainly to the Dlx2 proximal promoter (Xu et al., 2007). In the present studies, however, we also found that Dlx1 regulation was dependent on pRb, where repressing E2Fs inhibited Dlx1 promoter activity. Indeed, we detected the presence of potential E2F binding sites in both Dlx1 and Dlx2 proximal promoter regions. Consistent with the presence of conserved E2F sites, luciferase reporter analysis revealed that both promoters were repressed by E2F4 and E2F7, the activities of which are deregulated in the absence of pRb. Unfortunately, we could not perform ChIP experiments with E2F7 since there were no effective antibodies available for ChIP experiments. Nevertheless, given that E2Fs share consensus binding sites, and given the ChIP studies described above, it is clearly evident that E2Fs can directly interact with the Dlx1 and Dlx2 loci in the context of native chromatin in vivo.
Context-dependent and time-dependent function for Rb in cell differentiation
Our study defines an important requirement for the Rb/E2Fs proteins in the generation of specific subtypes of GABAergic neurons during late gestation and through the direct regulation of the early differentiation genes, Dlx1 and Dlx2. Previous studies indicated that the requirement for Rb during cellular differentiation varies among different tissues and is time-dependent and context-dependent. Thomas et al. (2001) showed that Rb acts as a transcription coactivator required for late differentiation of osteoblasts. More recently, Rb was shown to maintain osteoblastic fate commitment and regulate fate choice between bone and brown adipose tissue in vivo (Calo et al., 2010). In contrast, Rb was required in the switch from proliferation to differentiation in skeletal muscle (Huh et al., 2004); however, its role in the maintenance of the terminally differentiated state in myotubes is still debated (Huh et al., 2004; Blais and Dynlacht, 2007). Furthermore, MacPherson et al. (2004) reported cell type-specific effects of Rb deletion during late embryogenesis and postnatal development in the murine retina. In other studies, Rb was shown to promote specifically cholinergic neurons differentiation in the developing retina (Chen et al., 2007) and the differentiation and survival of granule precursor cells during cerebellar development (Marino et al., 2003). Interestingly, this context-specific requirement for Rb seems to be evolutionary conserved as a study using the Drosophila model system has shown that the second Drosophila Retinoblastoma family gene (RBF2) and dE2F2 regulate different sets of target genes in different tissues (Stevaux et al., 2005). Together, our data and other studies indicate that the requirement for Rb during cellular differentiation is context-dependent and can vary among different tissues.
The Rb/E2F pathway coordinates the transition between proliferation and early differentiation during neurogenesis
We propose here a model whereby Rb/E2F is required to coordinate the transition between proliferation and differentiation by controlling key aspects of differentiation through regulation of the Dlx1/Dlx2 bigene cluster (Fig. 7). Hence, in normal brain development and when Rb is inactive, such as in uncommitted and rapidly proliferating cells, E2Fs inhibit Dlx gene expression to prevent premature differentiation and depletion of the progenitor pool. When activated, Rb represses E2F activity and thereby enables the full induction of the Dlx1/2 bigene cluster expression. This is essential to coordinate differentiation and neuronal migration with cell cycle regulation. In contrast, when Rb is absent, the level(s) of repressor E2Fs, such as E2F7 (an Rb-independent repressor) or E2F4/p107 become deregulated and interfere with the induction of differentiation regulators such as Dlx1/2, thereby resulting in impaired interneuron differentiation and migration.
Figure 7.
Proposed model for the role of the Rb/E2F pathway in the coordination of proliferation and differentiation during neurogenesis. I, In an uncommitted progenitor and in the absence of active Rb, high E2F levels control cell proliferation while repressing Dlx gene expression to prevent premature differentiation and depletion of the progenitor pool. When activated, Rb inhibits E2F activity to trigger cell cycle exist and simultaneously initiate differentiation of progenitor cells via the full induction of the Dlx1/Dlx2 genes. II, In Rb-null mice, the level(s) of repressor E2Fs, such as E2F7 (an Rb-independent repressor) or E2F4 along with other pocket proteins e.g., p107, are deregulated which could interfere with the transcription of key differentiation genes such as Dlx2 and Dlx1, thus impairing differentiation and migration in the developing brain.
In summary, our findings demonstrate a novel cell cycle independent function for the Rb/E2F pathway which involves the modulation of neurogenesis through direct control of the Dlx1/Dlx2 genes. By directly participating in the regulation of genes that specify neuronal subtypes, the Rb tumor suppressor coordinates cell cycle regulation with differentiation and neuronal migration, and thereby plays an essential function in modulating normal brain development.
Footnotes
This work was supported by a Canadian Institutes of Health Research (CIHR) grant to R.S.S., and grants from the Lebanese National Council for Scientific Research and the University Research Board at the American University of Beirut to N.G. N.G. was previously supported by a fellowship from the Heart and Stroke Foundation of Canada. M.G.A. is supported by a Heart and Stroke Foundation of Ontario studentship; L.M.J. and K.A.M. were supported by CIHR Canada Scholarships. We gratefully acknowledge equipment funding from the Centre for Stroke Recovery to R.S.S. and D.S.P. We thank Drs. Mireille Khacho, Renaud Vandenbosch, and Marc Germain for critical reading of the manuscript. We thank Jason MacLaurin, Rayan Naser, Carine Jaafar, and Maarouf Baghdadi for excellent technical assistance.
References
- Anderson S, Mione M, Yun K, Rubenstein JL. Differential origins of neocortical projection and local circuit neurons: role of Dlx genes in neocortical interneuronogenesis. Cereb Cortex. 1999;9:646–654. doi: 10.1093/cercor/9.6.646. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Marín O, Horn C, Jennings K, Rubenstein JL. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 2001;128:353–363. doi: 10.1242/dev.128.3.353. [DOI] [PubMed] [Google Scholar]
- Batista-Brito R, Close J, Machold R, Fishell G. The distinct temporal origins of olfactory bulb interneuron subtypes. J Neurosci. 2008;28:3966–3975. doi: 10.1523/JNEUROSCI.5625-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A, Dynlacht BD. E2F-associated chromatin modifiers and cell cycle control. Curr Opin Cell Biol. 2007;19:658–662. doi: 10.1016/j.ceb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulfone A, Kim HJ, Puelles L, Porteus MH, Grippo JF, Rubenstein JL. The mouse Dlx-2 (Tes-1) gene is expressed in spatially restricted domains of the forebrain, face and limbs in midgestation mouse embryos. Mech Dev. 1993;40:129–140. doi: 10.1016/0925-4773(93)90071-5. [DOI] [PubMed] [Google Scholar]
- Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan DA, Dong L, Callaghan SM, Hou YX, Dagnino L, Slack RS. Neural precursor cells differentiating in the absence of Rb exhibit delayed terminal mitosis and deregulated E2F 1 and 3 activity. Dev Biol. 1999;207:257–270. doi: 10.1006/dbio.1998.9162. [DOI] [PubMed] [Google Scholar]
- Calo E, Quintero-Estades JA, Danielian PS, Nedelcu S, Berman SD, Lees JA. Rb regulates fate choice and lineage commitment in vivo. Nature. 2010;466:1110–1114. doi: 10.1038/nature09264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Opavsky R, Pacal M, Tanimoto N, Wenzel P, Seeliger MW, Leone G, Bremner R. Rb-mediated neuronal differentiation through cell-cycle-independent regulation of E2f3a. PLoS Biol. 2007;5:e179. doi: 10.1371/journal.pbio.0050179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Cloos P, Toftegaard U, Klinkenberg D, Bracken AP, Trinh E, Heeran M, Di Stefano L, Helin K. Characterization of E2F8, a novel E2F-like cell-cycle regulated repressor of E2F-activated transcription. Nucleic Acids Res. 2005;33:5458–5470. doi: 10.1093/nar/gki855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick F, Dyson N. Diverse regulatory functions of the E2F family of transcription factors. In: Fanciulli M, editor. Rb and tumorigenesis. Austin, TX: Landes Bioscience; 2006. pp. 59–72. [Google Scholar]
- Di Stefano L, Jensen MR, Helin K. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 2003;22:6289–6298. doi: 10.1093/emboj/cdg613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstat DD, Liu JK, Mione M, Zhong W, Yu G, Anderson SA, Ghattas I, Puelles L, Rubenstein JL. DLX-1, DLX-2, and DLX-5 expression define distinct stages of basal forebrain differentiation. J Comp Neurol. 1999;414:217–237. doi: 10.1002/(sici)1096-9861(19991115)414:2<217::aid-cne6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Farkas LM, Huttner WB. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr Opin Cell Biol. 2008;20:707–715. doi: 10.1016/j.ceb.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Ferguson KL, Vanderluit JL, Hébert JM, McIntosh WC, Tibbo E, MacLaurin JG, Park DS, Wallace VA, Vooijs M, McConnell SK, Slack RS. Telencephalon-specific Rb knockouts reveal enhanced neurogenesis, survival and abnormal cortical development. EMBO J. 2002;21:3337–3346. doi: 10.1093/emboj/cdf338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KL, McClellan KA, Vanderluit JL, McIntosh WC, Schuurmans C, Polleux F, Slack RS. A cell-autonomous requirement for the cell cycle regulatory protein, Rb, in neuronal migration. EMBO J. 2005;24:4381–4391. doi: 10.1038/sj.emboj.7600887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CL, Tsai LH. Alternative functions of core cell cycle regulators in neuronal migration, neuronal maturation, and synaptic plasticity. Neuron. 2009;62:312–326. doi: 10.1016/j.neuron.2009.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman JA, Chang JT, Jakoi L, Nevins JR. A combinatorial mechanism for determining the specificity of E2F activation and repression. Oncogene. 2009;28:2873–2881. doi: 10.1038/onc.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem N, Jarinova O, Amores A, Long Q, Hatch G, Park BK, Rubenstein JL, Ekker M. Regulatory roles of conserved intergenic domains in vertebrate Dlx bigene clusters. Genome Res. 2003;13:533–543. doi: 10.1101/gr.716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem N, Yu M, Long J, Hatch G, Rubenstein JL, Ekker M. Distinct cis-regulatory elements from the Dlx1/Dlx2 locus mark different progenitor cell populations in the ganglionic eminences and different subtypes of adult cortical interneurons. J Neurosci. 2007;27:5012–5022. doi: 10.1523/JNEUROSCI.4725-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev Biol. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- Herrup K, Silver J. Cortical development and topographic maps: patterns of cell dispersion in developing cerebral cortex. Curr Opin Neurobiol. 1994;4:108–111. doi: 10.1016/0959-4388(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Huh MS, Parker MH, Scimè A, Parks R, Rudnicki MA. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J Cell Biol. 2004;166:865–876. doi: 10.1083/jcb.200403004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens T, Li J, Leone G, De Veylder L. Atypical E2Fs: new players in the E2F transcription factor family. Trends Cell Biol. 2009;19:111–118. doi: 10.1016/j.tcb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JK, Ghattas I, Liu S, Chen S, Rubenstein JL. Dlx genes encode DNA-binding proteins that are expressed in an overlapping and sequential pattern during basal ganglia differentiation. Dev Dyn. 1997;210:498–512. doi: 10.1002/(SICI)1097-0177(199712)210:4<498::AID-AJA12>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Long JE, Garel S, Alvarez-Dolado M, Yoshikawa K, Osumi N, Alvarez-Buylla A, Rubenstein JL. Dlx-dependent and -independent regulation of olfactory bulb interneuron differentiation. J Neurosci. 2007;27:3230–3243. doi: 10.1523/JNEUROSCI.5265-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Cobos I, Potter GB, Rubenstein JL. Dlx1&2 and Mash1 transcription factors control MGE and CGE patterning and differentiation through parallel and overlapping pathways. Cereb Cortex. 2009;19(Suppl 1):i96–i106. doi: 10.1093/cercor/bhp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson D, Sage J, Kim T, Ho D, McLaughlin ME, Jacks T. Cell type-specific effects of Rb deletion in the murine retina. Genes Dev. 2004;18:1681–1694. doi: 10.1101/gad.1203304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- Marino S, Hoogervoorst D, Brandner S, Berns A. Rb and p107 are required for normal cerebellar development and granule cell survival but not for Purkinje cell persistence. Development. 2003;130:3359–3368. doi: 10.1242/dev.00553. [DOI] [PubMed] [Google Scholar]
- McClellan KA, Slack RS. Novel functions for cell cycle genes in nervous system development. Cell Cycle. 2006;5:1506–1513. doi: 10.4161/cc.5.14.2980. [DOI] [PubMed] [Google Scholar]
- McClellan KA, Slack RS. Specific in vivo roles for E2Fs in differentiation and development. Cell Cycle. 2007;6:2917–2927. doi: 10.4161/cc.6.23.4997. [DOI] [PubMed] [Google Scholar]
- McClellan KA, Ruzhynsky VA, Douda DN, Vanderluit JL, Ferguson KL, Chen D, Bremner R, Park DS, Leone G, Slack RS. Unique requirement for Rb/E2F3 in neuronal migration: evidence for cell cycle-independent functions. Mol Cell Biol. 2007;27:4825–4843. doi: 10.1128/MCB.02100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SK, Kaznowski CE. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- Nguyen L, Besson A, Roberts JM, Guillemot F. Coupling cell cycle exit, neuronal differentiation and migration in cortical neurogenesis. Cell Cycle. 2006;5:2314–2318. doi: 10.4161/cc.5.20.3381. [DOI] [PubMed] [Google Scholar]
- Panganiban G, Rubenstein JL. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- Petryniak MA, Potter GB, Rowitch DH, Rubenstein JL. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007;55:417–433. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasure SJ, Anderson S, Hevner R, Bagri A, Marin O, Lowenstein DH, Rubenstein JL. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron. 2000;28:727–740. doi: 10.1016/s0896-6273(00)00149-5. [DOI] [PubMed] [Google Scholar]
- Poitras L, Ghanem N, Hatch G, Ekker M. The proneural determinant MASH1 regulates forebrain Dlx1/2 expression through the I12b intergenic enhancer. Development. 2007;134:1755–1765. doi: 10.1242/dev.02845. [DOI] [PubMed] [Google Scholar]
- Porteus MH, Bulfone A, Ciaranello RD, Rubenstein JL. Isolation and characterization of a novel cDNA clone encoding a homeodomain that is developmentally regulated in the ventral forebrain. Neuron. 1991;7:221–229. doi: 10.1016/0896-6273(91)90260-7. [DOI] [PubMed] [Google Scholar]
- Potter GB, Petryniak MA, Shevchenko E, McKinsey GL, Ekker M, Rubenstein JL. Generation of Cre-transgenic mice using Dlx1/Dlx2 enhancers and their characterization in GABAergic interneurons. Mol Cell Neurosci. 2009;40:167–186. doi: 10.1016/j.mcn.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevaux O, Dimova DK, Ji JY, Moon NS, Frolov MV, Dyson NJ. Retinoblastoma family 2 is required in vivo for the tissue-specific repression of dE2F2 target genes. Cell Cycle. 2005;4:1272–1280. doi: 10.4161/cc.4.9.1982. [DOI] [PubMed] [Google Scholar]
- Stühmer T, Anderson SA, Ekker M, Rubenstein JL. Ectopic expression of the Dlx genes induces glutamic acid decarboxylase and Dlx expression. Development. 2002;129:245–252. doi: 10.1242/dev.129.1.245. [DOI] [PubMed] [Google Scholar]
- Takahashi C, Bronson RT, Socolovsky M, Contreras B, Lee KY, Jacks T, Noda M, Kucherlapati R, Ewen ME. Rb and N-ras function together to control differentiation in the mouse. Mol Cell Biol. 2003;23:5256–5268. doi: 10.1128/MCB.23.15.5256-5268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, Hinds PW. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell. 2001;8:303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- Tomita K, Ishibashi M, Nakahara K, Ang SL, Nakanishi S, Guillemot F, Kageyama R. Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron. 1996;16:723–734. doi: 10.1016/s0896-6273(00)80093-8. [DOI] [PubMed] [Google Scholar]
- Wallace VA, Raff MC. A role for Sonic hedgehog in axon-to-astrocyte signalling in the rodent optic nerve. Development. 1999;126:2901–2909. doi: 10.1242/dev.126.13.2901. [DOI] [PubMed] [Google Scholar]
- Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, Farnham PJ. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17:1550–1561. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee SP, Rigby PW. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 1993;7:1277–1289. doi: 10.1101/gad.7.7a.1277. [DOI] [PubMed] [Google Scholar]
- Zerucha T, Stühmer T, Hatch G, Park BK, Long Q, Yu G, Gambarotta A, Schultz JR, Rubenstein JL, Ekker M. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]





