ABSTRACT
The ability to monitor changes in the expression and localization of integrins is essential for understanding their contribution to development, tissue homeostasis and disease. Here, we pioneered the use of Crispr/Cas9 genome editing to tag an allele of the β4 subunit of the α6β4 integrin. A tdTomato tag was inserted with a linker at the C-terminus of integrin β4 in mouse mammary epithelial cells. Cells harboring this tagged allele were similar to wild-type cells with respect to integrin β4 surface expression, association with the α6 subunit, adhesion to laminin and consequent signaling. These integrin β4 reporter cells were transformed with YAP (also known as YAP1), which enabled us to obtain novel insight into integrin β4 dynamics in response to a migratory stimulus (scratch wound) by live-cell video microscopy. An increase in integrin β4 expression in cells proximal to the wound edge was evident, and a population of integrin β4-expressing cells that exhibited unusually rapid migration was identified. These findings could shed insight into integrin β4 dynamics during invasion and metastasis. Moreover, these integrin β4 reporter cells should facilitate studies on the contribution of this integrin to mammary gland biology and cancer.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Integrin β4, Crispr/Cas9, Migration, YAP
Summary: Crispr/Cas9 technology was used to tag an allele of the β4 integrin; reporter cells harboring this allele are useful for obtaining new insight into integrin β4 dynamics during cell migration.
INTRODUCTION
Changes in the expression and localization of specific integrins underlie the contribution of these receptors to a wide range of biological and pathological processes (Bridgewater et al., 2012; Desgrosellier and Cheresh, 2010; Hynes, 2002; Longmate and DiPersio, 2017). The monitoring of these changes in real-time facilitates rigorous evaluation of their significance and functional contribution. The challenge to this approach, however, is preserving integrin function and cell homeostasis. For this reason, genome editing of integrins with Crispr/Cas9 technology has considerable potential. Although techniques describing integrin tagging have been previously described (De Franceschi et al., 2016; Huet-Calderwood et al., 2017; Nader et al., 2016; Wang et al., 2015), Crispr/Cas9 genomic engineering to knock-in fluorescent tags has not been used for this purpose. This approach is ripe for employment because it allows direct visualization of the integrin transcribed from the endogenous gene and it circumvents the use of ectopic expression systems that have the potential for artifacts. Given the challenges of monitoring integrins in real-time, the use of this technology to study their plasticity is timely and could be a useful resource for cell biologists.
A prime candidate for genome editing is the α6β4 integrin (referred to as ‘integrin β4’ because this is the only one β4-subunit-containing integrin). This integrin functions as a receptor for most laminins, and it is expressed at the basal surface of many epithelial tissues (Giancotti, 2007; Mercurio, 1995). The distinguishing structural feature of integrin β4 is the atypical intracellular domain of the integrin β4 subunit, which is distinct both in size (∼1000 amino acids) and structure from any other integrin subunit (Tamura et al., 1990). A major function of this intracellular domain is to link integrin β4 to intermediate filaments in stable adhesive structures, termed hemidesmosomes (HDs) (Borradori and Sonnenberg, 1999; Green and Jones, 1996). Our lab pioneered studies demonstrating that integrin β4 has a more dynamic role in promoting cell migration and invasion (Lipscomb and Mercurio, 2005; Mercurio and Rabinovitz, 2001; Mercurio et al., 2001). Specifically, we discovered that integrin β4 is mobilized from HDs in response to epithelial wounds or as a consequence of carcinoma progression, and that it localizes in F-actin protrusions where it facilitates migration and invasion (Lotz et al., 1997; O'Connor et al., 2000, 1998; Rabinovitz and Mercurio, 1997; Rabinovitz et al., 1999; Shaw et al., 1997). Subsequent studies have confirmed and extended these findings (e.g. Colburn and Jones, 2018; Santoro et al., 2003). These conclusions, however, were based primarily on immunofluorescence microscopy of fixed cells using β4-specific antibodies and not real-time imaging of integrin β4 in live cells. This consideration is significant because the published data suggest that rapid changes in integrin β4 expression and localization may occur as cells acquire the ability to migrate.
In this Tools and Resources study, we used Crispr/Cas9-mediated homologous donor recombination (HDR) to knock-in a tdTomato tag to the cytoplasmic domain of the β4 integrin in mouse mammary epithelial cells. Oncogenic transformation of these cells with YAP (also known as YAP1) enabled real-time monitoring and comparison of integrin β4 expression and localization in live, normal and transformed epithelial cells in response to a scratch wound.
RESULTS
Design and approach for Crispr/Cas9-mediated integrin β4 tagging
We designed a strategy to knock-in a tdTomato tag connected by an eight-amino-acid linker to the C-terminus of mouse integrin β4 by using Crispr/Cas9-mediated HDR. Our approach was based on limiting the likelihood of the endogenous tag altering integrin β4 function, which includes laminin binding extracellularly and integrin β4 signaling intracellularly. To accomplish this goal, we inserted the tdTomato tag connected by a linker at the C-terminus (cytoplasmic domain) near the last exon of integrin β4 because it ensures that the tag will not interfere with laminin binding extracellularly. The purpose of the linker was to provide space between integrin β4 and the fluorescent tag to limit disruption of integrin β4 interactions with cytoplasmic molecules.
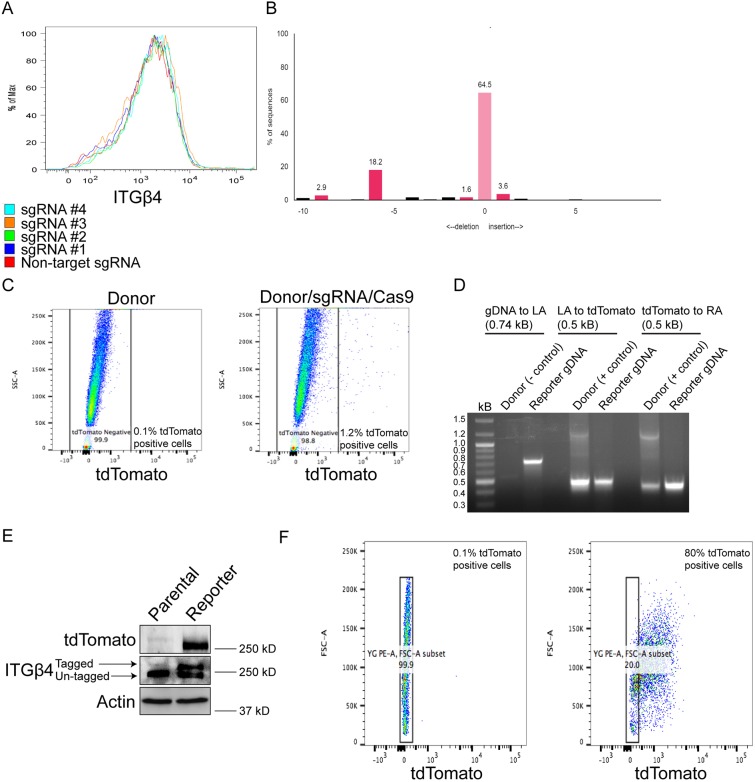
Our initial step was to design, and test the cutting efficiency and the effects on integrin β4 expression of four single guide (sg)RNAs targeting the region of the last exon of mouse integrin β4. Our goal in choosing a sgRNA and designing a corresponding donor plasmid was to identify a sgRNA that exhibits the greatest cutting efficiency and does not alter integrin β4 expression. As shown in Fig. 1A, none of the four sgRNAs we tested reduced integrin β4 surface abundance compared to a non-target sgRNA when co-transfected with Cas9. This result can be explained because the sgRNAs we designed correspond to the region of the last exon of integrin β4, as opposed to what is undertaken for Crispr/Cas9-mediated gene knockout, which typically targets one of the first exons of a gene. We ultimately chose sgRNA #2 because it exhibited the greatest cutting efficiency as determined by Tracking of Indels by Decomposition (TIDE) analysis (Fig. 1B), which is a quantitative Sanger sequencing assessment that uses a decomposition algorithm to quantify the frequency of insertions and deletions to determine the degree of sgRNA cutting (Brinkman et al., 2014). sgRNA #2 targets a component of the TGA stop codon and cuts three base pairs after the stop codon. Therefore, one consideration in the design of the donor plasmid that corresponds to sgRNA #2 is to avoid Cas9 cutting the endogenous locus after HDR has occurred. To accomplish this goal, we introduced silent mutations in the component of the stop codon of the sgRNA (TGA to TAA) and the protospacer adjacent motif (PAM), which also consists of a component of the last codon (threonine) of integrin β4 (ACC to ACG). We designed 900 base pair left and right homology arms in the donor plasmid based on a high recombination efficiency observed in prior studies (Zhang et al., 2017). The linker sequence (GGSGGSGS) was placed directly upstream of tdTomato, and we inserted a blasticidin-resistance gene in the donor plasmid because the sgRNA #2/Cas9 plasmid contains a puromycin-resistance gene.
Fig. 1.
Design, approach and validation for Crispr/Cas9-mediated integrin β4 tagging. (A) Comma-d1 cells expressing Cas9 and either a non-target sgRNA or one of four sgRNAs targeting the region of the last exon of mouse integrin β4 were processed for flow cytometry to determine the effects on integrin β4 surface abundance. (B) Genomic DNA was isolated from cells sorted as in A and was processed for TIDE analysis to quantify the cutting efficiency of each sgRNA. The y-axis indicates the percentage of sequences altered. The predicted Cas9 cut-site is indicated by ‘0’ in the x-axis, which also depicts other sites away from the predicted Cas9 cut-site that have insertions or deletions. (C) Comma-d1 cells were transfected with donor plasmid alone, or donor plasmid, sgRNA #2 and Cas9 plasmid, and processed for flow cytometry to quantify the proportion of tdTomato-positive cells, which were subsequently processed for single-cell sorting. (D) Genomic DNA from clones as described in C was isolated and processed for PCR to determine the correct genomic insert of tdTomato. (E) Immunoblotting was performed to confirm expression of tdTomato and the heterozygous tdTomato-tagged integrin β4 allele. (F) Expression of tdTomato was quantified by flow cytometry to determine the percentage of tdTomato-positive cells.
Validation of the correct genomic insert in mammary epithelial cells
Following the design of the donor plasmid that corresponds to sgRNA #2, we co-transfected circular donor, sgRNA and Cas9 plasmids into comma-d1 cells, which are mouse mammary epithelial cells (Danielson et al., 1984; Deugnier et al., 2002) that express integrin β4. After 72 h, we analyzed tdTomato expression by flow cytometry, and observed that cells transfected with donor, sgRNA and Cas9 exhibited 1.2% tdTomato-positive cells compared to 0.1% for control cells transfected with donor only (Fig. 1C), which is a similar number of fluorescent cells to what is observed for other Crispr/Cas9 knock-in strategies (Lackner et al., 2015). Of note, we also transfected linearized donor plasmid with sgRNA and Cas9 but did not obtain any positive clones.
We subsequently performed a single-cell sort for tdTomato-positive cells and grew surviving clones for the next 2–3 weeks to screen for the correct genomic insert. Clones were initially screened by PCR amplification of the junction between integrin β4 genomic DNA and the left homology arm using the donor plasmid as a negative control (Fig. 1D). Amplification of the regions between the left homology arm and tdTomato, and the right homology arm and tdTomato was performed using the donor plasmid as a positive control (Fig. 1D). The heterozygous tdTomato-tagged integrin β4 allele was verified by immunoblotting (Fig. 1E). Integrin β4 reporter comma-d1 cells were 80% positive for tdTomato (Fig. 1F), which is greater than other studies, reporting between 22% and 76% positive cells when using alternative Crispr/Cas9 knock-in strategies (Lackner et al., 2015). Subsequently, we validated our knock-in approach in 4T1 cells, a mouse mammary cancer cell line that expresses integrin β4 (Fig. S1A,B). This result demonstrates that our strategy to knock-in a tdTomato tag to integrin β4 can be applied to other experimental systems.
Integrin β4 reporter cells retain properties of parental cells
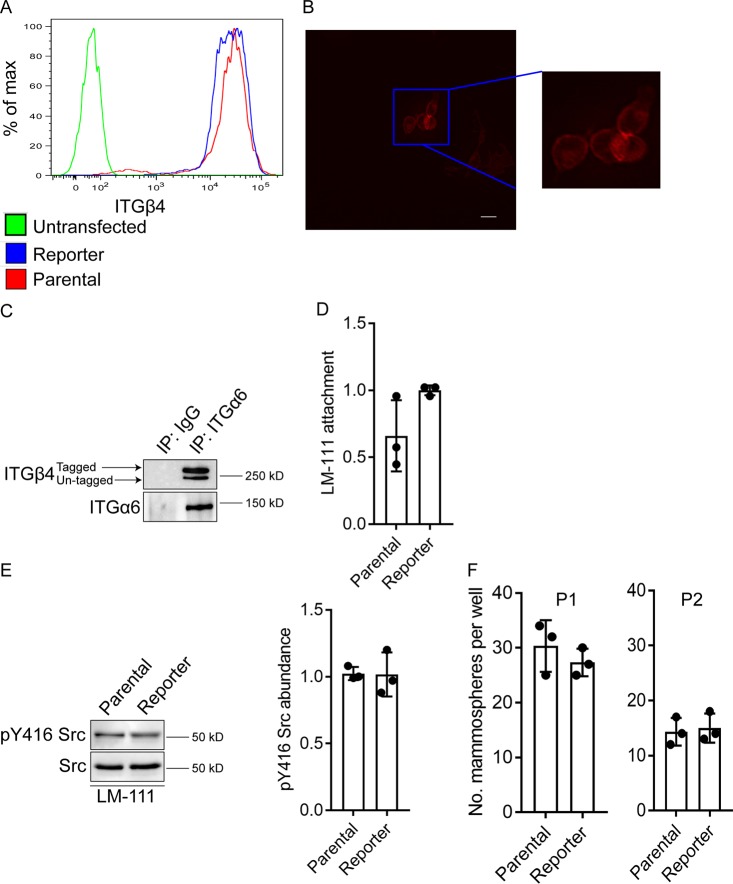
After validation of the correct genomic insert, we evaluated the potential effect of the tdTomato tag on the function of integrin β4. The comma-d1 reporter and parental cells exhibited similar levels of integrin β4 at the cell surface indicating that the C-terminal tdTomato tag did not alter integrin β4 surface localization (Fig. 2A). Immunofluorescence microscopy substantiated the localization of the tagged integrin on the cell surface (Fig. 2B). Also, analysis of Z stacks of confocal images revealed that the tdTomato signal is enriched on the basal surface of live adherent cells (Fig. S2). The tdTomato tag also did not interfere with integrin α6 pairing (Fig. 2C). Importantly, the reporter and parental cells did not differ significantly in their ability to adhere to laminin111 (Fig. 2D) and, consequently, activate Src (Fig. 2E), which is an effector of integrin β4-mediated signaling (Brown et al., 2017; Merdek et al., 2007).
Fig. 2.
Integrin β4 reporter cells exhibit properties of parental cells. (A) Analysis of integrin β4 surface expression by flow cytometry of untransfected (green line), integrin β4 reporter (blue line) and parental (red line) comma-d1 cells. (B) Live-cell image showing that the tdTomato signal is localized on the surface of adherent comma-d1 reporter cells. Scale bar: 25 µm. (C) Extracts of integrin β4 reporter cells were immunoprecipitated using an anti-integrin α6 antibody and then immunoblotted using an anti-integrin β4 antibody. Note that both the tagged and untagged integrin β4 alleles associate with integrin α6. (D) Cell culture dishes were coated with laminin-111 and integrin β4 reporter and parental comma-d1 cells were allowed to attach for 1 h in serum-free medium. Subsequently, Crystal Violet staining was performed to compare laminin-111 attachment. (E) Cells as in D were immunoblotted using an anti-pY416 Src antibody to assess Src activation. Densitometry was performed on these immunoblots using ImageJ (right graph). (F) Mammosphere-forming ability was assessed in integrin β4 reporter and parental comma-d1 cells. P1 indicates passage 1 and P2 indicates passage 2. Bar graphs in D–F are mean±s.d., with dots representing the results from three independent experiments. In D, E, results are represented relative to control (set at 1). There are no significant differences between samples.
Comma-d1 cells exhibit mammary progenitor potential (Deugnier et al., 2002, 2006; Taddei et al., 2008), and we did not observe differences in the number of mammospheres between the reporter and parental cells in serial passage assays (Fig. 2F). This result indicates that progenitor properties are not altered in the integrin β4 reporter cells. Together, these data suggest that cyto-tagging integrin β4 using Crispr/Cas9 does not alter its function. To determine if tdTomato was inserted in genomic loci other than integrin β4, we predicted the most likely sites that Cas9 may cut based on the sgRNA we chose to generate the reporter cells (sgRNA #2). We observed that tdTomato was not inserted into these sites and that our knock-in is highly specific (Fig. S3). Therefore, the resulting reporter cells are similar in nature to parental comma-d1 cells and our strategy limited potential off-target effects related to Crispr/Cas9 genomic alterations.
Real-time visualization of the expression and localization of the β4 integrin in migrating cells
The generation of an integrin β4 reporter cell line provided an opportunity to visualize integrin β4 expression and localization in real-time by immunofluorescence video microscopy. Given the established role of integrin β4 in cell migration, a scratch wound was made in the monolayer immediately before filming. A burst of integrin β4 expression was observed ∼6 h following wounding, especially in cells at the wound edge, concomitant with an increase in chemokinetic migration (Fig. 3; Movies 1, 2). However, there was little evidence of directional migration to heal the wound.
Fig. 3.
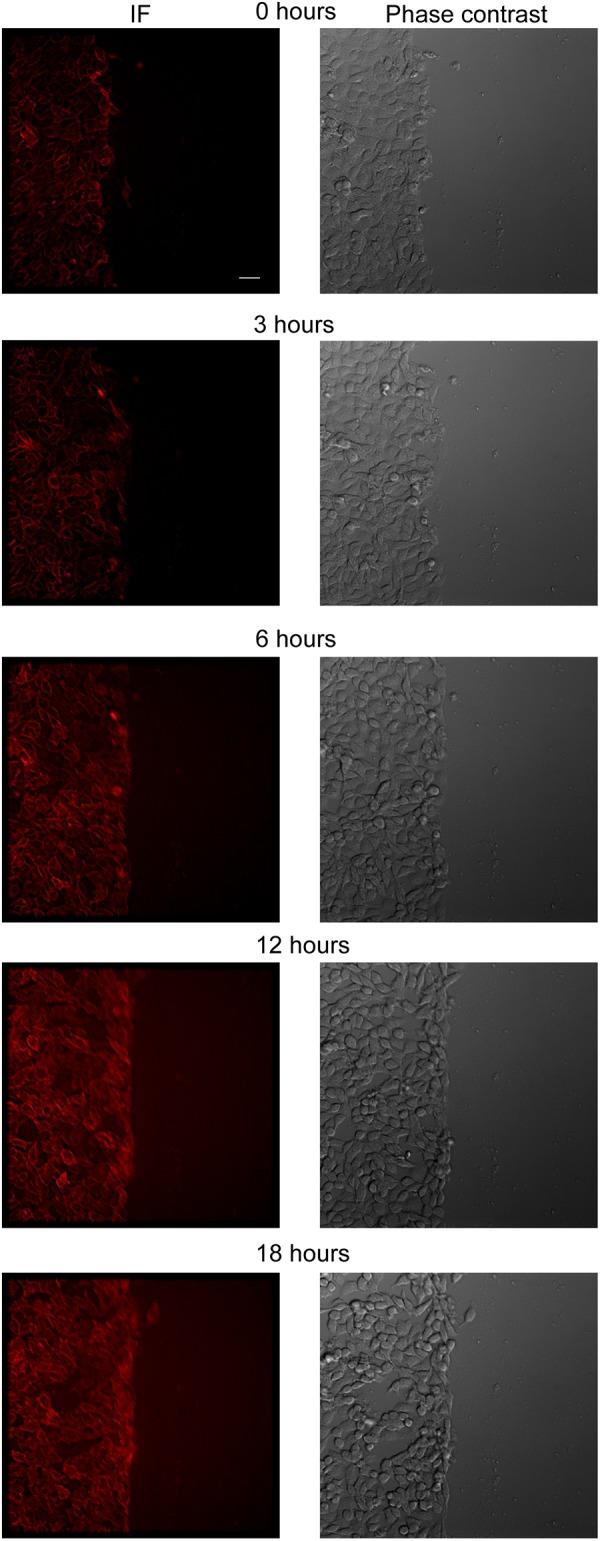
Monitoring integrin β4 dynamics in non-transformed reporter cells in response to a scratch wound. A scratch wound was introduced in integrin β4 reporter comma-d1 cells, which were monitored for 18 h. Representative still images are shown. See Movies 1 and 2 for real-time immunofluorescence video microscopy. Scale bar: 25 µm.
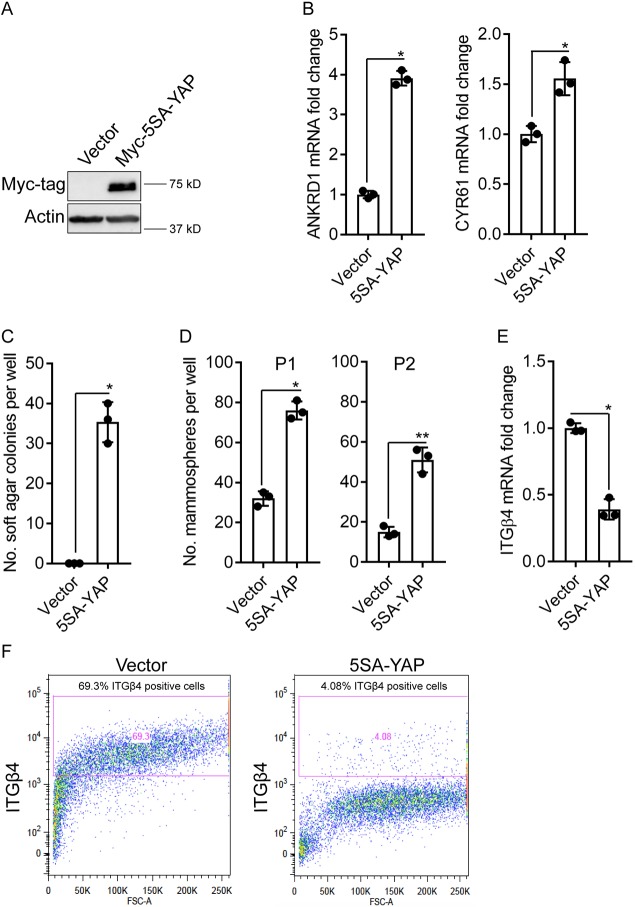
Based on the fact that the oncogenic transformation of epithelial cells stimulates their migration, we transformed our reporter cell line with YAP, which is a transcriptional effector of the Hippo tumor suppressor pathway that has been implicated in breast cancer progression, stemness, epithelial to mesenchymal transition and drug resistance (Di Agostino et al., 2016; Sorrentino et al., 2017; Zanconato et al., 2015, 2016, 2018; Zanconato and Piccolo, 2016). More specifically, we used a constitutively active form of YAP (5SA) to transform the comma-d1 reporter cells. This transformation increased the expression of YAP target genes ANKRD1 and Cyr61 (also known as CCN1) (Fig. 4A,B), promoted soft agar colony growth (Fig. 4C) and increased the number of mammospheres upon serial passage (Fig. 4D). Interestingly, YAP transformation decreased integrin β4 (ITGB4) mRNA expression (Fig. 4E), and it reduced integrin β4 surface expression abundance markedly (Fig. 4F).
Fig. 4.
YAP transformation of integrin β4 reporter cells. (A) Expression of Myc–5SA-YAP or empty vector in integrin β4 reporter comma-d1 cells was evaluated by immunoblotting. (B) Expression of the YAP target genes ANKRD1 and Cyr61 by means of qPCR in both populations was quantified and is presented relative to that in empty vector cells (set at 1). (C,D) Soft agar colony (C) and mammosphere formation (D) was assessed in the control and YAP-transformed integrin β4 reporter comma-d1 cells. P1 indicates passage 1 and P2 indicates passage 2. (E) integrin β4 mRNA expression was quantified in the control and YAP-transformed cells by means of qPCR and is presented relative to that in empty vector cells (set at 1). (F) Analysis of integrin β4 surface expression by flow cytometry of integrin β4 reporter cells expressing empty vector or 5SA-YAP. Bar graphs show mean±s.d., with dots representing the results from three independent experiments. *P≤0.05; **P<0.005 (two-tailed t-test).
Immunofluorescence video microscopy of YAP-transformed reporter cells in response to a scratch wound revealed directional migration of cells into the scratch by 3 h and restoration of the monolayer within 18 h of wounding (Fig. 5; Movies 3–6). We were able to detect several noteworthy aspects of integrin β4 expression and localization because of our ability to visualize endogenous integrin in real-time. The most striking observation was an induction of the integrin β4/tdTomato signal in cells proximal to the wound edge, which was evident within 3 h of wounding. Given that YAP transformation represses integrin β4 (Fig. 4E,F), this observation implies that that the migratory stimulus imposed by the wound is sufficient to induce expression of this integrin. It also appeared that the induction of integrin β4 expression was coincident with the acquisition of motility in multiple cells. As the wounds healed and the monolayer was restored, the integrin β4/tdTomato signal diminished.
Fig. 5.
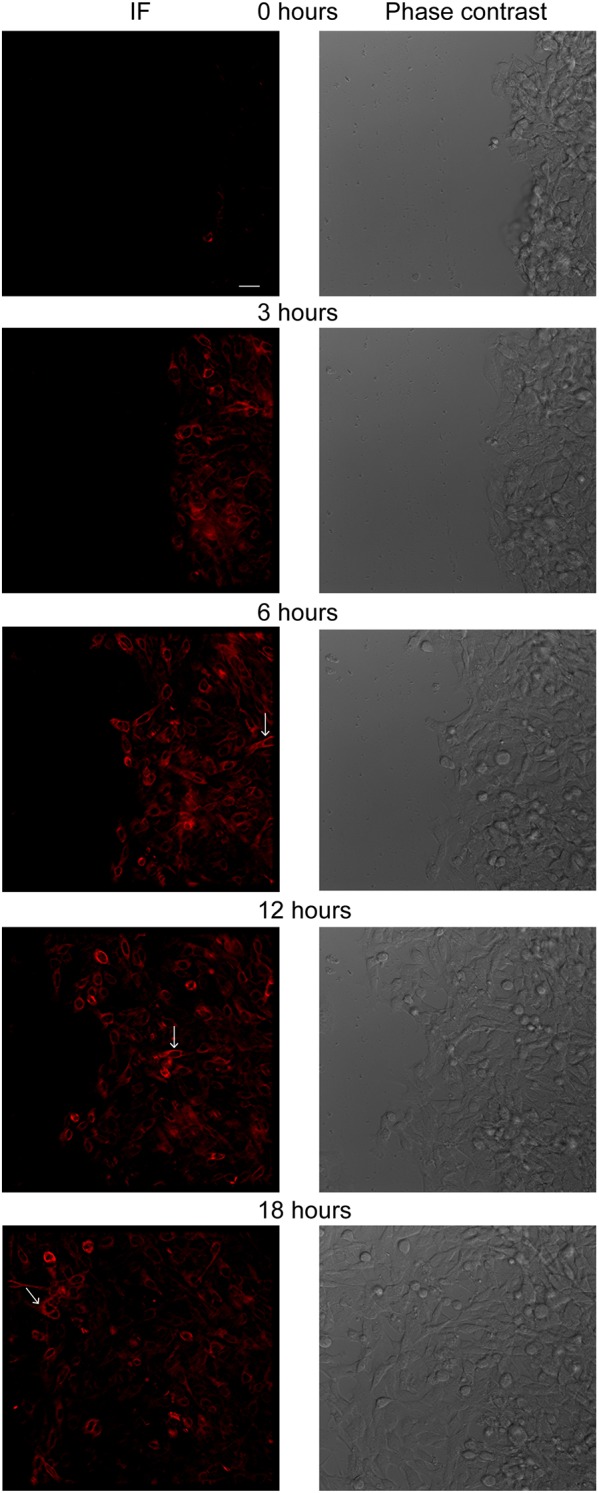
Monitoring integrin β4 dynamics in YAP-transformed reporter cells in response to a scratch wound. A scratch wound was introduced in YAP-transformed integrin β4 reporter comma-d1 cells, which were monitored for 18 h. Representative still images are shown. Arrows indicate an example of a integrin β4-positive cell that migrated more rapidly than neighboring cells. See Movies 3–6 for real-time immunofluorescence video microscopy. Scale bar: 25 µm.
Another key observation was the dynamic polarization of integrin β4 signal in cell protrusions (Movies 3–6; Fig. 5; Fig. S4), which is consistent with its role in driving migration. Although it was not feasible to quantify the velocity of individual cells because of the heterogeneity of the population and the number of tdTomato-positive cells, it was evident from observing the videos that the tdTomato cells exhibited rapid directional migration and that some of these cells migrated across the monolayer extremely quickly relative to migrating neighboring cells (see arrows in Fig. 5; Movies 3–6).
DISCUSSION
The results of this study demonstrate the usefulness of Crispr/Cas9 genome editing to tag endogenous integrins for studies aimed at evaluating their expression, localization and function. We were able to engineer an allele of the β4 integrin subunit with a tdTomato tag at its C-terminus that retained the properties of the wild-type allele and generate reporter cells harboring this tagged allele. This resource, which enabled us to visualize integrin β4 in real-time, has distinct advantages over the use of immunofluorescence microscopy because it does not require fixation and is independent of issues related to antibody specificity. Moreover, although the use of expression plasmids containing an epitope tag has proven useful for integrin studies (e.g. De Franceschi et al., 2016; Huet-Calderwood et al., 2017; Nader et al., 2016; Wang et al., 2015), there are potential artifacts with this approach including the level of expression of the tagged integrin and competition with endogenous integrin that are obviated by the use of Crispr/Cas9 genome editing. Although it has been shown that expression of cyto-tagged integrin β1 plasmid may influence its function (Galbraith et al., 2018), we did not detect any deficiency in the ability of the cyto-tagged β4 integrin to heterodimerize with α6 or of the reporter cells to adhere to laminin and activate Src. Moreover, the data we generated from real-time imaging of migrating reporter cells is consistent with our previous studies on the function of this integrin. Although we cannot exclude the possibility that subtle differences may exist between the endogenous and tagged integrin, our strategy of adding a linker between the C-terminus of the integrin and the tdTomato tag appears to minimize adverse functional consequences.
The most exciting and informative data we obtained was from our analysis of integrin β4 expression and localization in response to a migratory stimulus (scratch wound). Although previous studies by our group and others had implicated integrin β4 in migration (Lotz et al., 1997; O'Connor et al., 2000; Rabinovitz and Mercurio, 1997; Rabinovitz et al., 1999; Sehgal et al., 2006; Shaw et al., 1997), the ability to visualize endogenous integrin in real-time proved to be quite powerful. This approach substantiated the dynamic nature of integrin β4 during migration that we and others had observed based on more indirect approaches (Mercurio et al., 2001), but it also revealed novel insights. Specifically, we note the relatively rapid increase in integrin β4 expression that appeared to be coincident with the acquisition of motility in YAP-transformed reporter cells at the wound edge. This observation is consistent with our previous work demonstrating that integrin β4 expression is induced at the edge of epithelial wounds (Lotz et al., 1997). Our new ability to monitor integrin β4 localization in real-time, however, reveals the dynamic nature of integrin β4 expression and localization in response to a wound. In this regard, we also found that integrin β4 expression diminished rapidly as the wounds healed, which is reminiscent of work on the regulation of integrin β4 half-life in carcinomas (Witkowski et al., 2000). We were also struck by the extremely rapid migration of some of the YAP-transformed integrin β4-expressing cells, which was unexpected. These cells would not have been detected without their ability to be imaged in real-time, and their nature clearly merits further investigation. Taken together, our data underscore the notion that integrin β4 expression and localization can change rapidly in response to alterations in the microenvironment. This consideration should be a note of caution, for example, when interpreting data on the expression and localization of integrin β4 in tumors based on the analysis of static images.
The integrin β4 reporter cells that we have generated should be quite useful for studying other aspects of the contribution of this integrin to mammary gland biology and cancer. Comma-d1 cells are progenitor cells and capable of populating a mammary gland upon injection into the mammary fat pad of mice (Deugnier et al., 2002, 2006; Taddei et al., 2008). Long-term intravital microscopy (Ewald et al., 2011) of the integrin β4-tagged comma-d1 cells could provide novel insight into integrin β4 dynamics during mammary gland development. The same approach could be used to visualize integrin β4 during tumor formation in vivo using the YAP-transformed reporter cells. Given that integrin β4 has been implicated in metastasis (Falcioni et al., 1986; Hoshino et al., 2015; Lipscomb and Mercurio, 2005), the presence of the tdTomato tag could facilitate the isolation and characterization of circulating tumor cells from mice harboring tumors generated by orthotopic injection of the YAP-transformed reporter cells.
MATERIALS AND METHODS
Antibodies, flow cytometry and cell culture
Antibodies for immunoblotting (used at 1:1000) were against the following proteins: actin (4970S, Cell Signaling Technologies), integrin β4 (ab29042, Abcam), red fluorescent protein that cross-reacts with tdTomato (ab62341, Abcam), Src (2108S, Cell Signaling Technologies), pY416 Src (2101S, Cell Signaling Technologies), α6 (provided by Dr Anne Cress, University of Arizona, Tucson, AZ) and the Myc tag (2278S, Cell Signaling Technologies). Anti-integrin β4 antibody 34611A (ab25254, Abcam) was used for flow cytometry. Comma-d1 cells were provided by Dr Nicholas Tonks (Cold Spring Harbor, NY) and were cultured in Dulbecco's Modified Eagle Medium (DMEM)/F12 medium supplemented with 5% fetal bovine serum, insulin, epidermal growth factor (EGF) and HEPES. 4T1 cells were provided by Dr Leslie Shaw (University of Massachusetts Medical School, Boston, MA) and were cultured in F12 medium supplemented with 5% fetal bovine serum.
Constructs, sgRNAs and transfection
We initially tested the cutting efficiency of four sgRNAs targeting the region of the last exon of mouse integrin β4 (sgRNA #1, 5′-TGACCCAGGAATTCGTGACC-3′; sgRNA #2 5′-CTGGGGCGCGGGGGAGGTTC-3′; sgRNA #3 5′-GAGGAAGAAGGCGCTAGGAG-3′; sgRNA #4 5′-GAGAGAGCCACTGGCCGTTA-3′ by cloning each sgRNA into lentiCrispr V2 vector (Dr Wen Xue, University of Massachusetts Medical School, MA). Cells were subsequently infected with lentivirus carrying each sgRNA and puromycin selected (2 µg/ml) for 2 days. TIDE analysis (https://tide.nki.nl/) was performed to assess the cutting efficiency of each sgRNA, as well as flow cytometry to determine the effects on integrin β4 surface abundance.
After selecting the sgRNA #2 sequence, VectorBuilder (https://en.vectorbuilder.com/) was used to construct puromycin resistant sgRNA #2/Cas9 plasmid (VectorBuilder ID: VB180312-1135bvn) and the corresponding blasticidin-resistance donor plasmid (VectorBuilder ID: VB180312-1325zpa) to be transfected into comma-d1 cells. Approximately 75,000 cells were seeded in six-well plates. The next day, cells were transfected with 800 ng sgRNA/Cas9 plasmid and 500 ng circular donor plasmid using Lipofectamine 3000 (Thermo Fisher Scientific) and processed for single-cell sorting 72 h later. Hygromycin-resistance Myc-5SA-YAP vector was purchased from Addgene (plasmid #33093; deposited by K. Guan) and was used to transform the integrin β4 reporter comma-d1 cells.
Crispr/Cas9 off-target analysis
Off-target genomic loci were identified and ranked as previously described (Bae et al., 2014; Stemmer et al., 2015). PCR using primer pairs that bound the locus of interest and the tdTomato reporter cassette was utilized to assess for non-specific integration at the top four predicted sites. These genomic loci and the primer sequences used to amplify them are described in Fig. S3.
PCR
For quantitative (q)PCR, RNA was isolated using an RNA extraction kit (BS88133, Bio Basic Inc) and cDNAs were produced using qScript cDNA kit (#95047, Quantabio). The qPCR master mix used was SYBR green (Applied Biosystems) and experiments were performed with three technical replicates and normalized to the levels of 18S RNAs. The Massachusetts General Hospital/Harvard Medical School PrimerBank (http://pga.mgh.harvard.edu/primerbank/) was used to obtain qPCR primer sequences.
To assess the correct insertion of tdTomato, genomic DNA was isolated using mammalian genomic DNA isolation kit (G1N70, Sigma) from comma-d1 cells that were grown from single-cell clones. Primers were designed to amplify the junction between integrin β4 genomic DNA and the left homology arm (forward, 5′-AGGACCCCTCCAAATCAGTT-3′; reverse, 5′-CAGGTCACCAGGTAGCCAAG-3′), the left homology arm and tdTomato (forward, 5′- GAGCTGGGACCTGTACTCCA-3′; reverse, 5′-GCTTCTTGTAATCGGGGATG-3′) and tdTomato and the right homology arm (forward, 5′- CCCGGCTACTACTACGTGGA-3′; reverse, 5′-AGAACAAAAGGCTGGGGACT-3′).
Mammosphere and soft agar assays
UltraLow attachment six-well plates were used for mammosphere experiments. Cells were plated in DMEM/F12 medium supplemented with B27, EGF and fibroblast growth factor as previously described (Elaimy et al., 2018; Goel et al., 2014). Serial passaging was performed by pelleting and dissociating mammospheres with 0.05% trypsin for 15 min at 37°C to obtain single cells, which were washed in 1× PBS, counted and re-plated in UltraLow attachment six-well plates. Soft agar colony formation was performed as previously described (Goel et al., 2012).
Immunoblotting and co-immunoprecipitation
Immunoblotting was accomplished by washing cells in 1× PBS and scraping them on ice in RIPA buffer with EDTA and EGTA (BP-115DG, Boston Bioproducts) supplemented with protease and phosphatase inhibitors (Roche, 04693132001). Laemmli buffer (BP-111R, Boston Bioproducts) was added to each sample and the lysate was boiled and separated by SDS-PAGE. NP-40 lysis buffer (BP-119, Boston Bioproducts) containing protease and phosphatase inhibitors (Roche, 04693132001) was used to extract protein for co-immunoprecipitation. Protein A agarose beads were added to the lysate and incubated for one hour at 4°C for pre-clearing. Lysates were subsequently incubated with anti-integrin α6 antibody (555736, BD Biosciences) or IgG overnight at 4°. The next morning lysates were incubated again with protein A–agarose beads at 4°C for 1 h to pull down the protein complexes, which were separated using SDS-PAGE and immunoblotted for integrins α6 and β4.
Laminin attachment assay
Cell culture dishes were coated with laminin-111 (23017015, Thermo Fisher Scientific) and blocked in 1% bovine serum album at for 1 h at 37°C. Cells were trypsinized, plated on the laminin-111-coated plates in serum-free medium and allowed to attach for 1 h. Subsequently, cells were lysed and immunoblotted for pY416 Src and Src, or stained with Crystal Violet to measure the optical density (OD) at 595 nm to determine the level of adhesion.
Scratch wound assay and microscopy
Cells were seeded on six-well plates and grown to confluency. Following the introduction of a scratch with a pipet tip, a DeltaVision wide-field deconvolution fluorescence microscope with temperature and CO2 control was used to capture images at 20× total magnification of migrating cells at 15-min intervals for 18 h or 72 h using an A594 filter. A Zeiss Axio Observer microscope with a 63× objective lens was used to obtain a Z-stack of live adherent reporter cells.
Supplementary Material
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.L.E., A.M.M.; Methodology: A.L.E., A.S., C.W.B., M.R.W., M.W., J.J.A., W.X., A.C., C.E.B., H.L.G., A.M.M.; Software: A.L.E., A.M.M.; Validation: A.L.E., A.M.M.; Formal analysis: A.L.E., A.M.M.; Investigation: A.L.E., A.M.M.; Resources: A.L.E., A.M.M.; Data curation: A.L.E., A.M.M.; Writing - original draft: A.L.E., A.M.M.; Writing - review & editing: A.L.E., A.M.M.; Visualization: A.L.E., A.M.M.; Supervision: A.L.E., A.M.M.; Project administration: A.L.E., A.M.M.; Funding acquisition: A.L.E., A.M.M.
Funding
This work was supported by National Institutes of Health (NIH) grants CA168464, CA203439 (A.M.M), F30CA206271 (A.L.E.), F30CA232657 (A.S.) and CA221780 (H.L.G.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.231241.supplemental
References
- Bae S., Park J. and Kim J.-S. (2014). Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473-1475. 10.1093/bioinformatics/btu048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borradori L. and Sonnenberg A. (1999). Structure and function of hemidesmosomes: more than simple adhesion complexes. J. Invest. Dermatol. 112, 411-418. 10.1046/j.1523-1747.1999.00546.x [DOI] [PubMed] [Google Scholar]
- Bridgewater R. E., Norman J. C. and Caswell P. T. (2012). Integrin trafficking at a glance. J. Cell Sci. 125, 3695-3701. 10.1242/jcs.095810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman E. K., Chen T., Amendola M. and van Steensel B. (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 10.1093/nar/gku936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. W., Amante J. J., Goel H. L. and Mercurio A. M. (2017). The alpha6beta4 integrin promotes resistance to ferroptosis. J. Cell Biol. 216, 4287-4297. 10.1083/jcb.201701136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colburn Z. T. and Jones J. C. R. (2018). Complexes of alpha6beta4 integrin and vimentin act as signaling hubs to regulate epithelial cell migration. J. Cell Sci. 131, jcs214593 10.1242/jcs.214593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson K. G., Oborn C. J., Durban E. M., Butel J. S. and Medina D. (1984). Epithelial mouse mammary cell line exhibiting normal morphogenesis in vivo and functional differentiation in vitro. Proc. Natl. Acad. Sci. USA 81, 3756-3760. 10.1073/pnas.81.12.3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Franceschi N., Arjonen A., Elkhatib N., Denessiouk K., Wrobel A. G., Wilson T. A., Pouwels J., Montagnac G., Owen D. J. and Ivaska J. (2016). Selective integrin endocytosis is driven by interactions between the integrin alpha-chain and AP2. Nat. Struct. Mol. Biol. 23, 172-179. 10.1038/nsmb.3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desgrosellier J. S. and Cheresh D. A. (2010). Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer 10, 9-22. 10.1038/nrc2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deugnier M.-A., Faraldo M. M., Janji B., Rousselle P., Thiery J. P. and Glukhova M. A. (2002). EGF controls the in vivo developmental potential of a mammary epithelial cell line possessing progenitor properties. J. Cell Biol. 159, 453-463. 10.1083/jcb.200207138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deugnier M.-A., Faraldo M. M., Teulière J., Thiery J. P., Medina D. and Glukhova M. A. (2006). Isolation of mouse mammary epithelial progenitor cells with basal characteristics from the Comma-Dbeta cell line. Dev. Biol. 293, 414-425. 10.1016/j.ydbio.2006.02.007 [DOI] [PubMed] [Google Scholar]
- Di Agostino S., Sorrentino G., Ingallina E., Valenti F., Ferraiuolo M., Bicciato S., Piazza S., Strano S., Del Sal G. and Blandino G. (2016). YAP enhances the pro-proliferative transcriptional activity of mutant p53 proteins. EMBO Rep. 17, 188-201. 10.15252/embr.201540488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elaimy A. L., Guru S., Chang C., Ou J., Amante J. J., Zhu L. J., Goel H. L. and Mercurio A. M. (2018). VEGF-neuropilin-2 signaling promotes stem-like traits in breast cancer cells by TAZ-mediated repression of the Rac GAP beta2-chimaerin. Sci. Signal. 11, eaao6897 10.1126/scisignal.aao6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald A. J., Werb Z. and Egeblad M. (2011). Preparation of mice for long-term intravital imaging of the mammary gland. Cold Spring Harb. Protoc. 2011, pdb.prot5562 10.1101/pdb.prot5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcioni R., Kennel S. J., Giacomini P., Zupi G. and Sacchi A. (1986). Expression of tumor antigen correlated with metastatic potential of Lewis lung carcinoma and B16 melanoma clones in mice. Cancer Res. 46, 5772-5778. [PubMed] [Google Scholar]
- Galbraith C. G., Davidson M. W. and Galbraith J. A. (2018). Coupling integrin dynamics to cellular adhesion behaviors. Biol. Open 7, bio036806 10.1242/bio.036806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancotti F. G. (2007). Targeting integrin beta4 for cancer and anti-angiogenic therapy. Trends Pharmacol. Sci. 28, 506-511. 10.1016/j.tips.2007.08.004 [DOI] [PubMed] [Google Scholar]
- Goel H. L., Chang C., Pursell B., Leav I., Lyle S., Xi H. S., Hsieh C.-C., Adisetiyo H., Roy-Burman P., Coleman I. M. et al. (2012). VEGF/neuropilin-2 regulation of Bmi-1 and consequent repression of IGF-IR define a novel mechanism of aggressive prostate cancer. Cancer Discov. 2, 906-921. 10.1158/2159-8290.CD-12-0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel H. L., Gritsko T., Pursell B., Chang C., Shultz L. D., Greiner D. L., Norum J. H., Toftgard R., Shaw L. M. and Mercurio A. M. (2014). Regulated splicing of the alpha6 integrin cytoplasmic domain determines the fate of breast cancer stem cells. Cell Rep. 7, 747-761. 10.1016/j.celrep.2014.03.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K. J. and Jones J. C. (1996). Desmosomes and hemidesmosomes: structure and function of molecular components. FASEB J. 10, 871-881. 10.1096/fasebj.10.8.8666164 [DOI] [PubMed] [Google Scholar]
- Hoshino A., Costa-Silva B., Shen T. L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S. et al. (2015). Tumour exosome integrins determine organotropic metastasis. Nature 527, 329-335. 10.1038/nature15756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet-Calderwood C., Rivera-Molina F., Iwamoto D. V., Kromann E. B., Toomre D. and Calderwood D. A. (2017). Novel ecto-tagged integrins reveal their trafficking in live cells. Nat. Commun. 8, 570 10.1038/s41467-017-00646-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R. O. (2002). Integrins: bidirectional, allosteric signaling machines. Cell 110, 673-687. 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Lackner D. H., Carré A., Guzzardo P. M., Banning C., Mangena R., Henley T., Oberndorfer S., Gapp B. V., Nijman S. M. B., Brummelkamp T. R. et al. (2015). A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat. Commun. 6, 10237 10.1038/ncomms10237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb E. A. and Mercurio A. M. (2005). Mobilization and activation of a signaling competent alpha6beta4integrin underlies its contribution to carcinoma progression. Cancer Metastasis Rev. 24, 413-423. 10.1007/s10555-005-5133-4 [DOI] [PubMed] [Google Scholar]
- Longmate W. and DiPersio C. M. (2017). Beyond adhesion: emerging roles for integrins in control of the tumor microenvironment. F1000Res 6, 1612 10.12688/f1000research.11877.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotz M. M., Nusrat A., Madara J. L., Ezzell R., Wewer U. M. and Mercurio A. M. (1997). Intestinal epithelial restitution. Involvement of specific laminin isoforms and integrin laminin receptors in wound closure of a transformed model epithelium. Am. J. Pathol. 150, 747-760. [PMC free article] [PubMed] [Google Scholar]
- Mercurio A. M. (1995). Laminin receptors: achieving specificity through cooperation. Trends Cell Biol. 5, 419-423. 10.1016/S0962-8924(00)89100-X [DOI] [PubMed] [Google Scholar]
- Mercurio A. M. and Rabinovitz I. (2001). Towards a mechanistic understanding of tumor invasion--lessons from the alpha6beta 4 integrin. Semin. Cancer Biol. 11, 129-141. 10.1006/scbi.2000.0364 [DOI] [PubMed] [Google Scholar]
- Mercurio A. M., Rabinovitz I. and Shaw L. M. (2001). The alpha 6 beta 4 integrin and epithelial cell migration. Curr. Opin. Cell Biol. 13, 541-545. 10.1016/S0955-0674(00)00249-0 [DOI] [PubMed] [Google Scholar]
- Merdek K. D., Yang X., Taglienti C. A., Shaw L. M. and Mercurio A. M. (2007). Intrinsic signaling functions of the beta4 integrin intracellular domain. J. Biol. Chem. 282, 30322-30330. 10.1074/jbc.M703156200 [DOI] [PubMed] [Google Scholar]
- Nader G. P. F., Ezratty E. J. and Gundersen G. G. (2016). FAK, talin and PIPKIgamma regulate endocytosed integrin activation to polarize focal adhesion assembly. Nat. Cell Biol. 18, 491-503. 10.1038/ncb3333 [DOI] [PubMed] [Google Scholar]
- O'Connor K. L., Shaw L. M. and Mercurio A. M. (1998). Release of cAMP gating by the alpha6beta4 integrin stimulates lamellae formation and the chemotactic migration of invasive carcinoma cells. J. Cell Biol. 143, 1749-1760. 10.1083/jcb.143.6.1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. L., Nguyen B.-K. and Mercurio A. M. (2000). RhoA function in lamellae formation and migration is regulated by the alpha6beta4 integrin and cAMP metabolism. J. Cell Biol. 148, 253-258. 10.1083/jcb.148.2.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitz I. and Mercurio A. M. (1997). The integrin alpha6beta4 functions in carcinoma cell migration on laminin-1 by mediating the formation and stabilization of actin-containing motility structures. J. Cell Biol. 139, 1873-1884. 10.1083/jcb.139.7.1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitz I., Toker A. and Mercurio A. M. (1999). Protein kinase C-dependent mobilization of the alpha6beta4 integrin from hemidesmosomes and its association with actin-rich cell protrusions drive the chemotactic migration of carcinoma cells. J. Cell Biol. 146, 1147-1160. 10.1083/jcb.146.5.1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro M. M., Gaudino G. and Marchisio P. C. (2003). The MSP receptor regulates alpha6beta4 and alpha3beta1 integrins via 14-3-3 proteins in keratinocyte migration. Dev. Cell 5, 257-271. 10.1016/S1534-5807(03)00201-6 [DOI] [PubMed] [Google Scholar]
- Sehgal B. U., DeBiase P. J., Matzno S., Chew T.-L., Claiborne J. N., Hopkinson S. B., Russell A., Marinkovich M. P. and Jones J. C. (2006). Integrin beta4 regulates migratory behavior of keratinocytes by determining laminin-332 organization. J. Biol. Chem. 281, 35487-35498. 10.1074/jbc.M606317200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L. M., Rabinovitz I., Wang H. H.-F., Toker A. and Mercurio A. M. (1997). Activation of phosphoinositide 3-OH kinase by the alpha6beta4 integrin promotes carcinoma invasion. Cell 91, 949-960. 10.1016/S0092-8674(00)80486-9 [DOI] [PubMed] [Google Scholar]
- Sorrentino G., Ruggeri N., Zannini A., Ingallina E., Bertolio R., Marotta C., Neri C., Cappuzzello E., Forcato M., Rosato A. et al. (2017). Glucocorticoid receptor signalling activates YAP in breast cancer. Nat. Commun. 8, 14073 10.1038/ncomms14073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer M., Thumberger T., Del Sol Keyer M., Wittbrodt J. and Mateo J. L. (2015). CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE 10, e0124633 10.1371/journal.pone.0124633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei I., Deugnier M.-A., Faraldo M. M., Petit V., Bouvard D., Medina D., Fässler R., Thiery J. P. and Glukhova M. A. (2008). Beta1 integrin deletion from the basal compartment of the mammary epithelium affects stem cells. Nat. Cell Biol. 10, 716-722. 10.1038/ncb1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura R. N., Rozzo C., Starr L., Chambers J., Reichardt L. F., Cooper H. M. and Quaranta V. (1990). Epithelial integrin alpha 6 beta 4: complete primary structure of alpha 6 and variant forms of beta 4. J. Cell Biol. 111, 1593-1604. 10.1083/jcb.111.4.1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Arjonen A., Pouwels J., Ta H., Pausch P., Bange G., Engel U., Pan X., Fackler O. T., Ivaska J. et al. (2015). Formin-like 2 promotes beta1-integrin trafficking and invasive motility downstream of PKCalpha. Dev. Cell 34, 475-483. 10.1016/j.devcel.2015.06.015 [DOI] [PubMed] [Google Scholar]
- Witkowski C. M., Bowden G. T., Nagle R. B. and Cress A. E. (2000). Altered surface expression and increased turnover of the alpha6beta4 integrin in an undifferentiated carcinoma. Carcinogenesis 21, 325-330. 10.1093/carcin/21.2.325 [DOI] [PubMed] [Google Scholar]
- Zanconato F. and Piccolo S. (2016). Eradicating tumor drug resistance at its YAP-biomechanical roots. EMBO J. 35, 459-461. 10.15252/embj.201593584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F., Forcato M., Battilana G., Azzolin L., Quaranta E., Bodega B., Rosato A., Bicciato S., Cordenonsi M. and Piccolo S. (2015). Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 17, 1218-1227. 10.1038/ncb3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F., Cordenonsi M. and Piccolo S. (2016). YAP/TAZ at the roots of cancer. Cancer Cell 29, 783-803. 10.1016/j.ccell.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F., Battilana G., Forcato M., Filippi L., Azzolin L., Manfrin A., Quaranta E., Di Biagio D., Sigismondo G., Guzzardo V. et al. (2018). Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4. Nat. Med. 24, 1599-1610. 10.1038/s41591-018-0158-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-P., Li X.-L., Li G.-H., Chen W., Arakaki C., Botimer G. D., Baylink D., Zhang L., Wen W., Fu Y.-W. et al. (2017). Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 18, 35 10.1186/s13059-017-1164-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.