Abstract
Evidence suggesting the involvement of P2X2 and P2X3 in chronic pain has been obtained mostly from rodent models. Here we show that rodents may be poor predictors of P2X3 pharmacology in human. We demonstrate that monkey and human dorsal root ganglion (DRG) neurons do not express appreciable levels of P2X2 subunit, contrary to rat sensory neurons. Additionally, we report functional P2X3 activity in monkey DRG neurons and confirm the absence of functional P2X2/3 receptors. Interestingly, native P2X3 receptors in rat and monkey DRGs show similar agonist potency, but different antagonist potencies for TNP-ATP [2-O-(2,4,6-trinitrophenyl)-ATP] and RO51. This unexpected difference in antagonist potency was confirmed by comparing rat and human P2X3 receptors in HEK293 cells. Mutagenesis studies reveal that two extracellular residues, A197 and T202, are synergistically responsible for the potency drop in primate P2X3 receptors. These results uncover species-specific P2X3 pharmacology and identify key mechanisms impacting the translatability of potential analgesics targeting P2X3 receptors.
Introduction
ATP and ionotropic P2X3 receptors have been proposed to be important mediators of chronic pain signaling. Animal models of neuropathic pain are associated with the release of high levels of ATP and increased purinergic signaling in persistent pain conditions (Matsuka et al., 2008). P2X3 receptors are predominantly expressed in a subset of small-diameter dorsal root ganglion (DRG) neurons (Lewis et al., 1995). Chronic pain conditions alter the expression of P2X3 receptors in nociceptive sensory neurons (Tsuzuki et al., 2001) and increase P2X3-mediated responses (Chen et al., 2005; Mo et al., 2011). Finally, transient P2X3 gene disruption (Barclay et al., 2002), P2X3 gene knockout (Cockayne et al., 2000), and P2X2/P2X3 double knockout (Cockayne et al., 2005) have all been shown to decrease pain-related behavior in nerve injury or inflammatory pain models, suggesting that ATP, P2X2, and P2X3 are essential for chronic pain signaling.
The evidence linking P2X3 to pain has supported the discovery of novel antagonists blocking homomeric P2X3 and heteromeric P2X2/3 receptors. TNP-ATP [2-O-(2,4,6-trinitrophenyl)-ATP] (Jarvis et al., 2001), A-317491 (McGaraughty et al., 2003), and RO51 (Jahangir et al., 2009) are P2X3/P2X2/3 antagonists that have shown reduced hyperalgesia and mechanical allodynia in inflammatory and neuropathic pain models. P2X3/P2X2/3 antagonists are currently being tested in the clinic, so their analgesic efficacy has yet to be established in patients (Ford, 2012). Progress in the discovery of novel analgesics has been hampered by limited predictability of efficacy from preclinical animal studies to validation in humans (Mogil et al., 2010). Part of this limitation can be attributed to the use of rodent models in pain research and the pharmacology of many receptors, including various P2X receptors (Gever et al., 2006), which exhibit limited translatability across species. To investigate some of these caveats, we used monkey native sensory neurons to identify species differences in P2X2 and P2X3 function that could prevent translatability of P2X pharmacology from rodents to primates.
Materials and Methods
Tissue acquisition.
This study was conducted under a protocol approved by an ethics committee. The animals were kept and experiments were performed at our main site (AstraZeneca R&D Montréal) or at a site that has accreditation from Canadian Council on Animal Care or Association for the Assessment and Accreditation of Laboratory Animal Care, and/or was approved by AstraZeneca Global Veterinary Council for study conduct. Rat DRG tissue samples were obtained from male Sprague Dawley rats (n = 47; Harlan) weighing between 200 and 250 g. Monkey DRGs were obtained from three male Macaca fascicularis aged 2 years 8 months (n = 2) and 2 years 7 months (n = 1). DRG levels included cervical, thoracic, and lumbar for both rat (L4–L6) and monkey (T12–L6). DRGs were rapidly dissected, snap frozen, and stored at −80°C until cryosectioned.
Frozen human (7 h postmortem) DRG tissues (n = 8) were acquired from the NICHD Brain and Tissue Bank for Developmental Disorders (University of Maryland School of Medicine, Baltimore, MD). The individuals were 10, 20 (n = 2), 22 (n = 3), 29, 55, and 58 years old at the time of death. The cause of death was accidental, mainly due to motor vehicle accidents, and the tissues are considered normal. DRG levels included cervical, thoracic, and lumbar. Human trigeminal ganglia and brain were used as positive control tissues.
Frozen tissue was cryosectioned at 14 μm and thaw mounted onto Superfrost Plus slides (VWR). Slides were stored at −80°C until processing for in situ hybridization.
Compounds and reagents.
RO51 was synthesized at AstraZeneca, prepared as previously described (Jahangir et al., 2009), and selected over other compounds based on its in vitro potency, bioavailability, and half-life. All reagents were purchased from Wisent Bioproducts or Sigma unless specified.
In situ hybridization.
Probes were designed to recognize all reported transcripts for rat or primate P2X2 or P2X3. The full-length probes used corresponded to human P2X2b, rat P2X2b, human P2X3, and rat P2X3. Partial probes used were as follows: human P2X2b probe from nucleotide 586 to 957 (from ATG start codon); and monkey P2X3 probe from nucleotide 586 to 957 (from ATG start codon). P2X2 and P2X3 antisense riboprobes were transcribed in vitro using RNA polymerase (Promega) in the presence of [35S]-UTP and [35S]-CTP (GE Healthcare). The quality of labeled riboprobes (and their hydrolysis) was verified by electrophoresis and scintillation counting. In situ hybridization was performed as previously described (Mennicken et al., 2003). Briefly, tissue sections were fixed in PFA, rinsed in standard buffer, and incubated overnight in hybridization buffer containing the [35S]-labeled cRNA probe. After treatment, sections were dehydrated and exposed to film for 2 weeks. The slides were dipped in emulsion and exposed before development and counterstaining. Bright-field and dark-field photomicrographs of sections were captured using a Leica microscope equipped with a Leica DFC490 camera.
Cell counts.
Three DRG tissue sections per donor were quantified using the following method. First, images of DRG were viewed under 10× magnification. The total number of neurons within a defined sampling square was counted. A neuron was defined as a pale pink cell showing a visible (light purple) nucleus present within the area inside the sampling square. Neurons touching the edges of the sampling square were excluded. P2X2- or P2X3-expressing neurons were considered “positive” if emulsion grains were present above the background level for that particular DRG section. Neurons were counted over 10 randomly selected sampling regions per tissue section per donor. Positive neuron counts were expressed as a percentage of the total number of neurons over a sampling of DRG sections. Because expression of P2X2 and P2X3 mRNA was consistent within a species, cell counts were performed on a subset of the samples (human n = 3; monkey n = 2; rat n = 2).
RT-PCR.
P2X1, P2X2, P2X3, and P2X4 mRNA were detected by distinct species-specific PCR primers designed to amplify all known variants of each receptor's mRNA within each species. PCR primer sequences were as follows: primate P2X1 (TAGGTGGTGCAAGAGTCAGG/AAGATGCCAATTCCAGAGCC); primate P2X2 (TCGTCAGGCTACAACTTCAGG/TGTTAGCAAGATCCAGTCGC); primate P2X3 (GTTGTGAAGAGCTGGACCATC/GGCAGAATCCTTGCATCTG); and primate P2X4 (ATCCTTCCCAACATCACCAC/GGCAAACCTGAAATTGTAGCC). All primate sequences were confirmed in monkey bladder reverse transcribed cDNA. The P2X2 sequence was also confirmed in a human placenta cDNA library. Total mRNA was extracted from monkey DRG homogenates, reverse transcription was performed using Superscript II reverse transcriptase (Invitrogen), and amplification was performed for 35 cycles.
Cell culture and mutagenesis studies.
DRGs were harvested (L4–L6 in 200–250 g rats and T12–L6 in 2.5–4.0 kg monkeys) and digested with collagenase-A (2 mg/ml) and papain (1 mg/ml) in DMEM. Cells were plated on poly-d-lysine-coated coverslips and maintained in F12 supplemented with 10% horse serum, 2 mm glutamine-penicillin-streptomycin, and 2 ng/ml NGF and GDNF. Culturing media for the HEK293 cells was supplemented with DMEM with 10% fetal bovine serum, 1% penicillin-streptomycin, 1% MEM, and 1% glutamine. HEK293 cells were transfected with EGFP and P2X3 using PolyFect (Qiagen). Residues S67, L69, L127, T162, A197, T202, and S267 were mutated using the Quikchange method (Stratagene), and mutations were confirmed by sequencing.
Electrophysiology.
HEK293 cells and DRG neurons were used in patch-clamp experiments. Briefly, cells were perfused with an extracellular solution containing the following (in mm): 137 NaCl; 8.1 Na2HPO4; 2.68 KCl; 1.47 KH2PO4; 0.9 CaCl2; 0.5 MgCl2 at pH 7. Whole-cell voltage or current-clamp recordings (filtered at 1 kHz, digitized at 10 kHz) were performed using an Axopatch 200B amplifier and PClamp software (Molecular Devices). Cells were recorded with borosilicate pipettes (2–6 MΩ) containing the following (in mm): 110 K-gluconate, 20 KCl, 10 EGTA, 8 NaCl, 10 HEPES, 1 MgCl2, and 4 Mg-ATP at pH 7.3. RO51 was applied through the bath perfusion system. The percentage of rescue was calculated as (Ih − Im)/(Ih − Ir) * 100%, where Ih, Im, and Ir are the inhibition produced by each concentration of RO51 on wild-type (WT) human, mutant human, or rat P2X3, respectively, in HEK293 cells. Data are presented as mean ± SEM. For concentration–response curves, current amplitude was normalized to the amplitude of the control current. Potency (EC50 and IC50) values are represented as the best-fit value ± 95% confidence interval. Statistical analyses for differences in mean were performed using either unpaired Student's t test or two-way ANOVA followed by Bonferroni's correction for multiple comparisons.
Results
P2X2 expression in DRG neurons differs across species
We examined cross-species cellular expression of P2X2 and P2X3 mRNA in rat, monkey, and human DRG sections. In rat DRG, P2X3 mRNA expression was readily detected in 70% of neurons (191/271, primarily small diameter neurons), consistent with previous in situ hybridization reports (Kobayashi et al., 2005). P2X3 expression was also readily detected in monkey and human DRGs, albeit at lower levels relative to rat, being detected in 51% (306/600) of human DRG neurons (Fig. 1A), in line with a previous report on human P2X3 protein (Yiangou et al., 2000). Human trigeminal ganglia were used as positive control tissue to validate our methods and riboprobes for P2X3. P2X3 mRNA expression was detected in trigeminal ganglia (data not shown) in line with previous immunohistochemical (Xiang et al., 1998) and in situ hybridization studies in rat (Collo et al., 1996).
Figure 1.
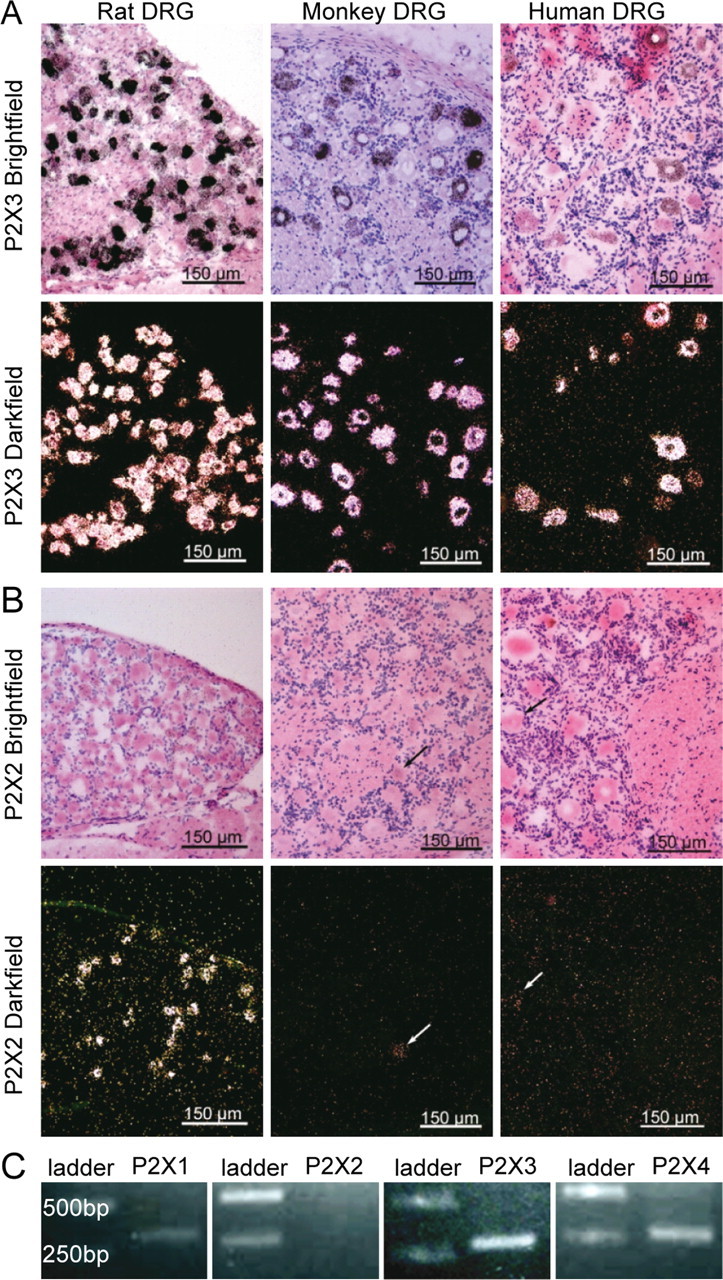
Species differences in the expression of P2X2 mRNA. A, In situ hybridization of P2X3 and P2X2 mRNA expression in rat, monkey, and human DRGs. P2X3 mRNA was predominantly detected in small-diameter neurons, and this pattern was consistent across species. B, P2X2 was found in small- and medium-diameter neurons in rat DRGs but was virtually absent in monkey and human DRGs. Neurons were considered positive when there was an accumulation of grains above the level of background (see Materials and Methods). Some human samples contained lipofuscin, which was clearly identified as such in microscopy. C, RT-PCR amplification of mRNA from monkey DRG homogenates reveals P2X1, P2X3, and P2X4 expression and confirms the absence of P2X2 mRNA.
P2X2 and P2X3 mRNA expression was assessed on adjacent tissue sections. In rat, 26% (49/183) of small- and medium-diameter DRG neurons expressed low or moderate levels of P2X2 mRNA, in agreement with a previous study (Kobayashi et al., 2005). A similar pattern of P2X2 mRNA expression was expected in monkey and human DRG samples (Vulchanova et al., 1997). Interestingly, <1% of P2X2-positive neurons were detected in monkey and human DRG tissue (Fig. 1B; monkey: 1/316 neurons, human: 2/750 neurons).
The relative absence of P2X2 mRNA in primate DRGs was confirmed using several approaches. First, human brain was used as positive control tissue to validate our methodology and our P2X2 riboprobes. P2X2 mRNA expression was consistently detected in several brain regions including hippocampus, amygdala, and cortex (data not shown), with or without hydrolyzing the full-length riboprobe to enhance hybridization (Cox et al., 1984). An additional shorter 364 bp riboprobe was generated targeting the exons 6–9 conserved by all nine reported transcripts of human P2X2 gene, and the low expression level of P2X2 mRNA in primate DRGs was confirmed (<1% of positive neurons; monkey: 1/197 neurons, human: 2/661 neurons). RT-PCR analysis of DRG homogenates also failed to detect expression of P2X2 mRNA in monkey sensory neurons, whereas primers specific for primate P2X1, P2X3, and P2X4 amplified a fragment corresponding to their respective cDNA (match confirmed by sequencing; Fig. 1C). Thus, although the distribution of P2X3 mRNA expression is conserved across rat, monkey, and human sensory neurons, P2X2 mRNA expression levels in DRGs appear to differ across these species.
Monkey DRG neurons express functional P2X3 but not P2X2/3 channels
Species differences in P2X2 expression suggest that rat DRGs may not be predictive of human (or monkey) P2X pharmacology and highlight the need for a better comparative model. Based on the conserved expression profile of P2X2 and P2X3 in monkey and human DRGs, we developed a monkey DRG assay as a surrogate for human DRGs. This involved harvesting, culturing, and recording monkey DRG neurons to test the pharmacology of native P2X receptors. Monkey DRG neurons in culture showed a similar developmental time course as rat DRG neurons, growing processes after 48 h of incubation (Fig. 2A). Monkey and rat DRG neurons shared similar membrane resting potentials (−58.6 ± 0.7 and −58.4 ± 0.6 mV, respectively; n = 50 cells each; p > 0.8, unpaired Student's t test) and firing upon depolarization (Fig. 2B). P2X3 currents were then compared between rat and monkey DRG neurons. Brief application of the P2X3 and P2X2/3 receptor agonist αβmeATP (10 μm, 2 s) evoked fast-activating and quickly desensitizing currents with P2X3-like kinetics in both rat and monkey neurons (exponential decay fit: rat: τfast = 172 ± 10 ms, τslow = 1.9 ± 0.1 s; monkey: τfast = 160 ± 5 ms, τslow = 1.8 ± 0.2 s; n = 20 cells each; Fig. 2C). Currents resensitized after 3 min of recovery, suggesting that the gating properties of P2X3 receptors in monkey DRG neurons are consistent with predictions from rodent studies (Sokolova et al., 2006).
Figure 2.
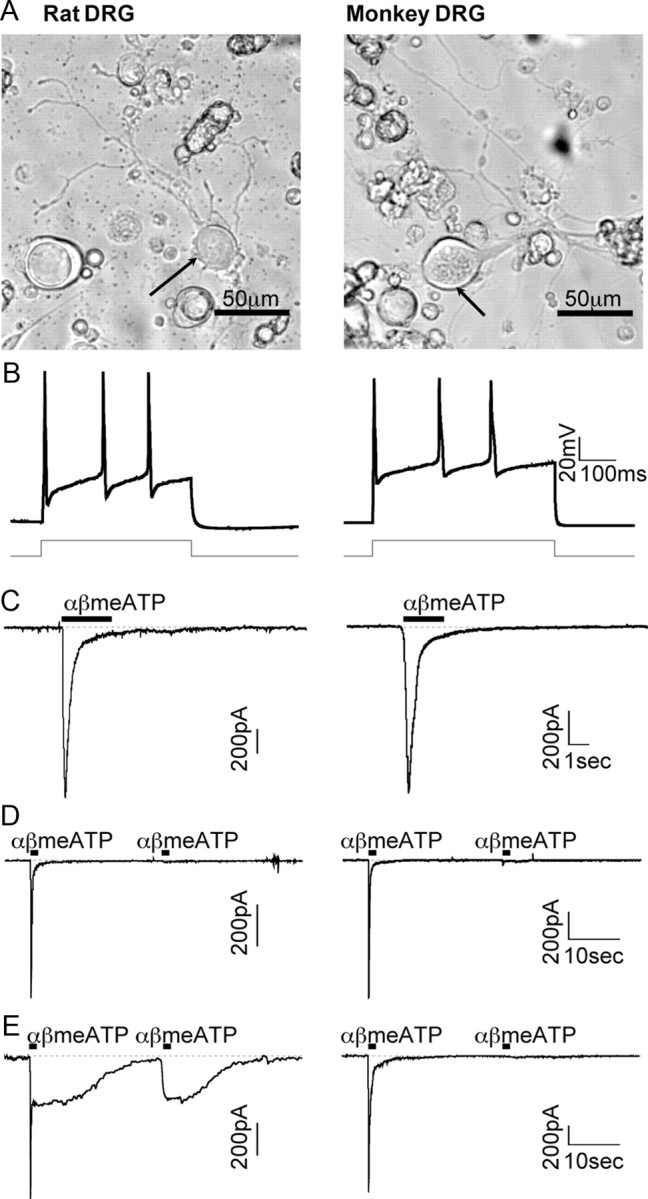
Functional recordings in monkey DRGs confirm the absence of P2X2/3 channels. A, Representative rat and monkey DRG neurons (arrow) in culture. B, C, Recordings from rat and monkey DRG neurons showing action potentials evoked upon depolarization (B) and P2X3 currents evoked by αβmeATP (10 μm, bar) (C). D, E, Currents generated by multiple applications of αβmeATP in cells with transient (D) or sustained (E) current profiles. Contrary to rat DRGs, none of the 256 monkey DRG neurons recorded displayed P2X2/3-like currents.
In rat DRG neurons, αβmeATP (10 μm, 2 s) evoked fast and slow currents based on their desensitizing properties. Fast responses attributed to homomeric P2X3 receptors rapidly desensitized and were not reactivated by a second application of αβmeATP within 30 s (Fig. 2D). Slowly desensitizing responses attributed to heteromeric P2X2/3 receptors (Pankratov et al., 2001) were observed in 27% of recorded neurons and were readily reactivated by multiple applications of αβmeATP (Fig. 2E). In contrast, none of the recorded monkey DRG neurons displayed P2X2/3-like currents (n = 256 neurons; Fig. 2D,E). Upon repeated applications (within a 30 s interval) of αβmeATP, P2X3 currents were completely desensitized in monkey DRG neurons. We also tested ATP (at 10 μm) in the majority of the DRG neurons we recorded. We did not observe any sustained currents or currents without rundown typical of P2X2 homomers (data not shown). Consistent with our in situ hybridization data, these findings confirmed the absence of functional PX2 and P2X2/3 receptors in monkey sensory neurons.
αβmeATP is equipotent on rat and monkey P2X3
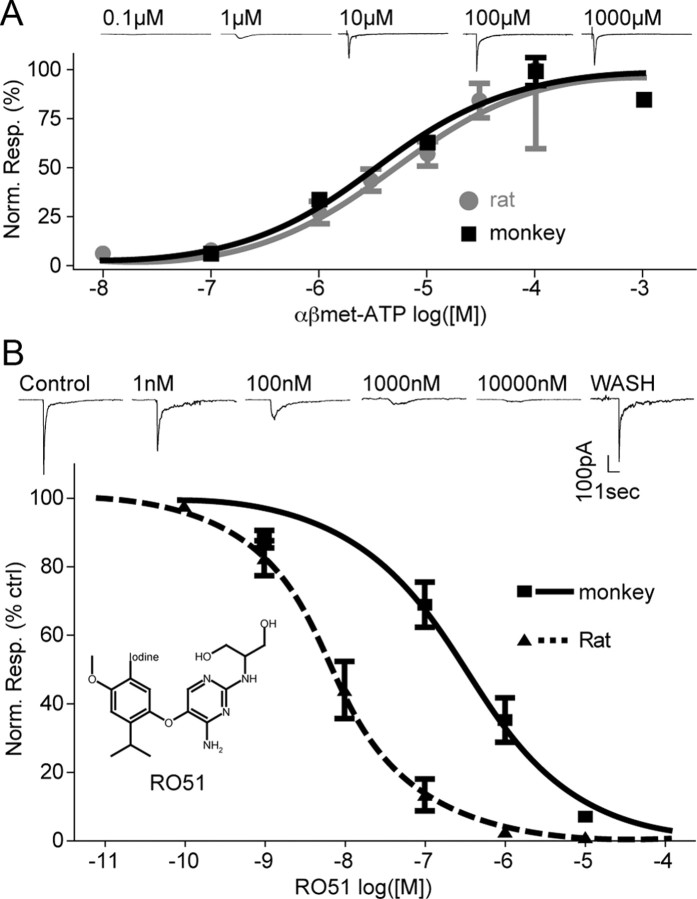
The similarity of P2X3 pharmacology between rodents and primates was investigated by comparing concentration–response curves of the agonist αβmeATP on rat and monkey DRG neurons. αβmeATP was found equipotent in activating P2X3 in monkey and rat DRG neurons (monkey DRG EC50 = 5.1 ± 1.9 μm, n = 3–11 neurons/point; rat DRG EC50 = 3.5 ± 1.6 μm, n = 3–20 neurons/point; p > 0.05, unpaired t test; Fig. 3A).
Figure 3.
Species differences in P2X3 pharmacology. A, Concentration–response curve generated by αβmeATP-evoked currents in rat and monkey neurons (EC50 in rat = 3.5 μm, Hill slope = 0.70, n = 20 neurons; EC50 in monkey = 5.1 μm, Hill slope = 0.70, n = 11 neurons). B, Concentration–response curve of RO51 on P2X3 currents in rat and monkey DRGs and shift in potency (rat Hill slope = −0.76; monkey Hill slope = −0.61).
RO51 and TNP-ATP are less potent antagonists on human than on rat P2X3
P2X3/P2X2/3 antagonists have been developed as a potential novel class of analgesics (McGaraughty et al., 2003; Kaan et al., 2010). Among them, RO51 and other diaminopyrimidines are bioavailable P2X3/P2X2/3 antagonists producing antinociception in rodent models of pain (Carter et al., 2009; Jahangir et al., 2009). We therefore generated concentration–response curves of RO51 by measuring the relative inhibition of P2X3 current amplitude at increasing antagonist concentrations and compared across species the potency of RO51. Interestingly, the potency of RO51 on P2X3 activity was significantly lower in monkey DRG neurons compared with rat (IC50 = 324.0 ± 2.1 and 7.3 ± 3.0 nm, respectively; n = 6–12 neurons/point; p < 0.001, unpaired t test; Fig. 3B). TNP-ATP, a structurally distinct P2X3 antagonist, was also 23-fold less potent on monkey than on rat DRGs (IC50 = 20.7 ± 3.4 and 0.9 ± 1.3 nm, respectively; n = 3–31 neurons/point, p < 0.005, unpaired t test, data not shown).
To evaluate whether the decreased potency observed in monkey DRGs also occurred on the human receptor, we compared the potency of RO51 on HEK293 cells expressing either rat or human P2X3. In this recombinant system ruling out cellular background differences, RO51 also showed a significant drop in potency on human P2X3 receptors (RO51 IC50 = 110.5 ± 14.8 and 0.04 ± 0.02 nm, respectively for human and rat P2X3; n = 5–9 cells/point; p < 0.05, unpaired t test; Fig. 4A). Similar studies with TNP-ATP confirmed that human and rat P2X3 receptors differed in their sensitivity to antagonists (TNP-ATP IC50 = 1.2 and 0.07 nm, respectively; data not shown).
Figure 4.
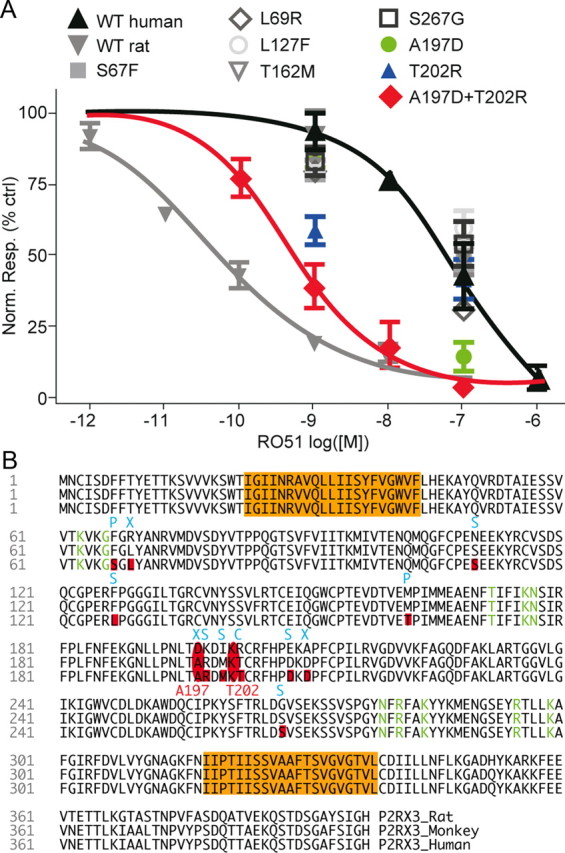
P2X3 structural determinants underlying species-dependent drop in RO51 potency. A, Concentration–response curve of RO51 on WT rat P2X3 (gray, Hill slope= −0.54), WT human P2X3 (black, Hill slope= −0.61), and mutants of human P2X3. A197D + T202R double mutant (red) increased the potency of RO51 on human P2X3 by 225-fold (Hill slope= −0.78). B, P2X3 protein sequence for rat, monkey, and human showing transmembrane domains (orange) and residues contributing to the agonist-binding site (green). Nonconserved residues between rat and human ectodomains are highlighted in red, resulting in a different charge (C), polarity (P), size (S), or charge and size (X).
The preserved species differences in P2X3 pharmacology in recombinant systems suggest that this discrepancy may be caused by intrinsic structural dissimilarities in the two receptors. Despite their relatively high homology (94% identity, 372/397 identical residues; Fig. 4B), a small number of amino acids might underlie significant functional changes. We performed mutagenesis studies to identify residues on the human P2X3 subunit that may be critical for the interaction with RO51. Individual residues in the ectodomain of the human P2X3 receptor were mutated to their rat counterpart (S67F, L69R, L127F, T162M, A197D, T202R, S267G; Fig. 4B), and the rescue produced by each mutation relative to WT human P2X3 was statistically compared at two concentrations (1 and 100 nm) of RO51. RO51 inhibition of the mutated receptor at S67F, L69R, L127F, T162M, and S267G was not statistically different from values obtained on WT human receptors (p > 0.05 at both 1 and 100 nm RO51, two-way repeated-measures ANOVA with Bonferroni test vs WT human; n = 5 cells each; Fig. 4A). Of all mutants tested, only A197D and T202R were significantly different from WT human P2X3 (A197D produced 47% rescue at 1 nm and T202R produced 74% rescue at 100 nm; p < 0.001, two-way repeated-measures ANOVA with Bonferroni test vs WT human; n = 5 cells each; Fig. 4A) suggesting that these two residues play a major role in P2X3 antagonist action.
To further investigate the functional contribution of residues A197 and T202, a double-mutant human P2X3 receptor was created (A197D+T202R) that combined both amino acid changes. Inhibition of A197D+T202R by RO51 was statistically different from WT human. The potency of RO51 on A197D+T202R revealed an IC50 of 0.49 ± 0.48 nm representing a significant 225-fold increase in potency compared with WT human P2X3, suggesting that the combination of these two amino acids synergized their individual interactions with RO51 (p < 0.01, unpaired t test; n = 11 cells; Fig. 4A). The degree of rescue reached 74.3% and 102.1% at 1 and 100 nm RO51, respectively, confirming that A197 and T202 together are critically involved in shifting the potency of this P2X3/P2X2/3 antagonist.
Discussion
Translatability between rodent and human P2X3 physiology has not, to our knowledge, been reported. The present study uncovers significant species differences that may prevent preclinical studies from predicting analgesic efficacy of P2X3/P2X2/3 antagonists in humans. Although the pattern of P2X3 mRNA distribution was similar in rats and primates, there were unexpected species differences in P2X2 mRNA expression. Consistent with the lack of P2X2 mRNA expression in primate DRGs, P2X2/3 function was absent from native monkey sensory neurons. Furthermore, the potencies of P2X3/P2X2/3 antagonists were decreased on primate P2X3R relative to rodent. Site-directed mutagenesis identified two extracellular amino acid residues in P2X3 receptors (A197 and T202 in rodent) that are critically involved in the species-dependent interaction with RO51. Understanding these species differences could help select the most relevant model species/assays and thereby design antagonists that translate into effective analgesics in human.
Several lines of evidence demonstrate the functional homology between monkey and human P2X physiology. First, P2X2 expression is relatively absent from monkey and human DRGs compared with rat DRGs. Second, homology is higher between monkey and human P2X3 sequences (4 different amino acid residues) than between rat and human P2X3 sequences (25 differences). Third, the ATP binding domain is highly conserved (Bodnar et al., 2011) consistent with αβmeATP being equipotent across species. Fourth, RO51 and TNP-ATP are equipotent on monkey and human P2X3 as well as on mutants S67F, L127F, and T162M. Overall, these studies emphasize the value of monkey tissue for predicting human P2X3 pharmacology and, potentially, clinical analgesic efficacy.
Primate sensory neurons do not express P2X2
Finding that P2X2 receptors are virtually absent from primate sensory neurons was unexpected because this receptor was readily observed in rodent DRG tissue (Kobayashi et al., 2005) and its expression was previously reported in monkey DRGs using an antibody directed against rat P2X2 (Vulchanova et al., 1997). The reasons for this discrepancy in P2X2 localization are unclear but may be due to the different methodologies used (in situ hybridization vs immunohistochemistry). In this study, we used two different in situ hybridization probes spanning distinct regions of the monkey and human P2X2 gene to confirm mRNA expression differences. The near absence of P2X2 was not due to a loss of mRNA integrity because mRNAs for other receptors were detected on adjacent sections. Moreover, all our P2X2 probes consistently detected low levels of P2X2 mRNA in various brain regions, indicating that our method was sensitive enough to detect low levels of P2X2 mRNA in DRGs. In addition, P2X2 mRNA was not detected in monkey DRG using RT-PCR, a method known for its high sensitivity. Finally, the fact that we did not record any P2X2/3 current in patch-clamp electrophysiology from monkey DRG neurons confirmed our findings on significant species differences in the expression of P2X2 in pain pathways. Whether this finding only applies to DRG sensory neurons or whether these species differences in P2X2 function also occur in cranial or autonomic ganglia remains to be assessed. The near absence of P2X2 mRNA in primate DRGs may have direct implications for drug discovery as we currently do not understand the relative contribution of P2X3 homomeric versus heteromeric P2X2/3 blockade underlying the analgesic effects observed in rodents with a majority of disclosed nonselective antagonists. Therefore, blockade of P2X2/3 receptors in rat sensory neurons may produce analgesic effects that would lead to an overestimation of efficacy in human.
Primate sensory neurons are less sensitive to P2X3 antagonists
In the present study, a species difference in sensitivity to antagonists between native rat and primate P2X3 receptors was revealed. This drop in antagonist sensitivity indicates that efficacious doses of RO51 used in rats could underestimate the clinically relevant dose required for efficacy in pain patients. It is unlikely that these shifts in pharmacology are caused by differences in cellular background between sensory neurons from primates and rodents since similar differences in antagonist potency between rat and human P2X3 are measured in the same recombinant cell line. Differences in potency between native tissue and recombinant systems are also noted and confirm the need for native tissue in drug discovery procedures. The mutagenesis data demonstrated that amino acid residues at positions 197 and 202 in the ectodomain of the P2X3 receptor, located on the “dorsal fin” of P2X subunits (Kawate et al., 2009), explain intrinsic species differences underlying this pharmacological shift. It shows that discrete structural differences in P2X3 receptors have a strong impact on the binding of antagonists and affect their potency. The discovery of critical species differences in P2X3 receptors will facilitate the development of a new generation of antagonists with similar cross-species potency that could translate into new analgesics.
Footnotes
We thank Dominique Blais (Montreal Neurological Institute) for help with mutagenesis, Mylene Gosselin (AstraZeneca R&D Montréal) for help with transfections and Manon Pelletier (AstraZeneca R&D Montréal) for designing riboprobes.
A.S., G.M., R.G., M.P., D.O., X.H.Y., M.J.T., M.N.P., and C.Q.C. are employed by AstraZeneca.
References
- Barclay J, Patel S, Dorn G, Wotherspoon G, Moffatt S, Eunson L, Abdel'al S, Natt F, Hall J, Winter J, Bevan S, Wishart W, Fox A, Ganju P. Functional downregulation of P2X3 receptor subunit in rat sensory neurons reveals a significant role in chronic neuropathic and inflammatory pain. J Neurosci. 2002;22:8139–8147. doi: 10.1523/JNEUROSCI.22-18-08139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar M, Wang H, Riedel T, Hintze S, Kato E, Fallah G, Gröger-Arndt H, Giniatullin R, Grohmann M, Hausmann R, Schmalzing G, Illes P, Rubini P. Amino acid residues constituting the agonist binding site of the human P2X3 receptor. J Biol Chem. 2011;286:2739–2749. doi: 10.1074/jbc.M110.167437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter DS, Alam M, Cai H, Dillon MP, Ford AP, Gever JR, Jahangir A, Lin C, Moore AG, Wagner PJ, Zhai Y. Identification and SAR of novel diaminopyrimidines. Part 1: the discovery of RO-4, a dual P2X(3)/P2X(2/3) antagonist for the treatment of pain. Bioorg Med Chem Lett. 2009;19:1628–1631. doi: 10.1016/j.bmcl.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Chen Y, Li GW, Wang C, Gu Y, Huang LY. Mechanisms underlying enhanced P2X receptor-mediated responses in the neuropathic pain state. Pain. 2005;119:38–48. doi: 10.1016/j.pain.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Cockayne DA, Dunn PM, Zhong Y, Rong W, Hamilton SG, Knight GE, Ruan HZ, Ma B, Yip P, Nunn P, McMahon SB, Burnstock G, Ford AP. P2X2 knockout mice and P2X2/P2X3 double knockout mice reveal a role for the P2X2 receptor subunit in mediating multiple sensory effects of ATP. J Physiol. 2005;567:621–639. doi: 10.1113/jphysiol.2005.088435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockayne DA, Hamilton SG, Zhu QM, Dunn PM, Zhong Y, Novakovic S, Malmberg AB, Cain G, Berson A, Kassotakis L, Hedley L, Lachnit WG, Burnstock G, McMahon SB, Ford AP. Urinary bladder hyporeflexia and reduced pain-related behaviour in P2X3-deficient mice. Nature. 2000;407:1011–1015. doi: 10.1038/35039519. [DOI] [PubMed] [Google Scholar]
- Collo G, North RA, Kawashima E, Merlo-Pich E, Neidhart S, Surprenant A, Buell G. Cloning of P2X5 and P2X6 receptors and the distribution and properties of an extended family of ATP-gated ion channels. J Neurosci. 1996;16:2495–2507. doi: 10.1523/JNEUROSCI.16-08-02495.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, DeLeon DV, Angerer LM, Angerer RC. Detection of mrnas in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol. 1984;101:485–502. doi: 10.1016/0012-1606(84)90162-3. [DOI] [PubMed] [Google Scholar]
- Ford AP. In pursuit of P2X3 antagonists: novel therapeutics for chronic pain and afferent sensitization. Purinergic Signal. 2012;8:3–26. doi: 10.1007/s11302-011-9271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gever JR, Cockayne DA, Dillon MP, Burnstock G, Ford AP. Pharmacology of P2X channels. Pflugers Arch. 2006;452:513–537. doi: 10.1007/s00424-006-0070-9. [DOI] [PubMed] [Google Scholar]
- Jahangir A, Alam M, Carter DS, Dillon MP, Bois DJ, Ford AP, Gever JR, Lin C, Wagner PJ, Zhai Y, Zira J. Identification and SAR of novel diaminopyrimidines Part 2: the discovery of RO-51, a potent and selective, dual P2X(3)/P2X(2/3) antagonist for the treatment of pain. Bioorg Med Chem Lett. 2009;19:1632–1635. doi: 10.1016/j.bmcl.2009.01.097. [DOI] [PubMed] [Google Scholar]
- Jarvis MF, Wismer CT, Schweitzer E, Yu H, van Biesen T, Lynch KJ, Burgard EC, Kowaluk EA. Modulation of BzATP and formalin induced nociception: attenuation by the P2X receptor antagonist, TNP-ATP and enhancement by the P2X(3) allosteric modulator, cibacron blue. Br J Pharmacol. 2001;132:259–269. doi: 10.1038/sj.bjp.0703793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaan TK, Yip PK, Patel S, Davies M, Marchand F, Cockayne DA, Nunn PA, Dickenson AH, Ford AP, Zhong Y, Malcangio M, McMahon SB. Systemic blockade of P2X3 and P2X2/3 receptors attenuates bone cancer pain behaviour in rats. Brain. 2010;133:2549–2564. doi: 10.1093/brain/awq194. [DOI] [PubMed] [Google Scholar]
- Kawate T, Michel JC, Birdsong WT, Gouaux E. Crystal structure of the ATP-gated P2X(4) ion channel in the closed state. Nature. 2009;460:592–598. doi: 10.1038/nature08198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Fukuoka T, Yamanaka H, Dai Y, Obata K, Tokunaga A, Noguchi K. Differential expression patterns of mRNAs for P2X receptor subunits in neurochemically characterized dorsal root ganglion neurons in the rat. J Comp Neurol. 2005;481:377–390. doi: 10.1002/cne.20393. [DOI] [PubMed] [Google Scholar]
- Lewis C, Neidhart S, Holy C, North RA, Buell G, Surprenant A. Coexpression of P2X2 and P2X3 receptor subunits can account for ATP-gated currents in sensory neurons. Nature. 1995;377:432–435. doi: 10.1038/377432a0. [DOI] [PubMed] [Google Scholar]
- Matsuka Y, Ono T, Iwase H, Mitrirattanakul S, Omoto KS, Cho T, Lam YY, Snyder B, Spigelman I. Altered ATP release and metabolism in dorsal root ganglia of neuropathic rats. Mol Pain. 2008;4:66. doi: 10.1186/1744-8069-4-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaraughty S, Wismer CT, Zhu CZ, Mikusa J, Honore P, Chu KL, Lee CH, Faltynek CR, Jarvis MF. Effects of A-317491, a novel and selective P2X3/P2X2/3 receptor antagonist, on neuropathic, inflammatory and chemogenic nociception following intrathecal and intraplantar administration. Br J Pharmacol. 2003;140:1381–1388. doi: 10.1038/sj.bjp.0705574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennicken F, Zhang J, Hoffert C, Ahmad S, Beaudet A, O'Donnell D. Phylogenetic changes in the expression of delta opioid receptors in spinal cord and dorsal root ganglia. J Comp Neurol. 2003;465:349–360. doi: 10.1002/cne.10839. [DOI] [PubMed] [Google Scholar]
- Mo G, Grant R, O'Donnell D, Ragsdale DS, Cao CQ, Séguéla P. Neuropathic Nav1.3-mediated sensitization to P2X activation is regulated by protein kinase C. Mol Pain. 2011;7:14. doi: 10.1186/1744-8069-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogil JS, Davis KD, Derbyshire SW. The necessity of animal models in pain research. Pain. 2010;151:12–17. doi: 10.1016/j.pain.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Pankratov YuV, Lalo UV, Dashkin AN, Krishtal A. Heterogeneity of the functional expression of P2X3 and P2X2/3 receptors in the primary nociceptive neurons of rat. Neurochem Res. 2001;26:993–1000. doi: 10.1023/a:1012344803672. [DOI] [PubMed] [Google Scholar]
- Sokolova E, Skorinkin A, Moiseev I, Agrachev A, Nistri A, Giniatullin R. Experimental and modeling studies of desensitization of P2X3 receptors. Mol Pharmacol. 2006;70:373–382. doi: 10.1124/mol.106.023564. [DOI] [PubMed] [Google Scholar]
- Tsuzuki K, Kondo E, Fukuoka T, Yi D, Tsujino H, Sakagami M, Noguchi K. Differential regulation of P2X(3) mRNA expression by peripheral nerve injury in intact and injured neurons in the rat sensory ganglia. Pain. 2001;91:351–360. doi: 10.1016/S0304-3959(00)00456-5. [DOI] [PubMed] [Google Scholar]
- Vulchanova L, Riedl MS, Shuster SJ, Buell G, Surprenant A, North RA, Elde R. Immunohistochemical study of the P2X2 and P2X3 receptor subunits in rat and monkey sensory neurons and their central terminals. Neuropharmacology. 1997;36:1229–1242. doi: 10.1016/s0028-3908(97)00126-3. [DOI] [PubMed] [Google Scholar]
- Xiang Z, Bo X, Burnstock G. Localization of ATP-gated P2X receptor immunoreactivity in rat sensory and sympathetic ganglia. Neurosci Lett. 1998;256:105–108. doi: 10.1016/s0304-3940(98)00774-5. [DOI] [PubMed] [Google Scholar]
- Yiangou Y, Facer P, Birch R, Sangameswaran L, Eglen R, Anand P. P2X3 receptor in injured human sensory neurons. Neuroreport. 2000;11:993–996. doi: 10.1097/00001756-200004070-00019. [DOI] [PubMed] [Google Scholar]