Summary
mRNAs are regulated by nucleotide modifications that influence their cellular fate. Two of the most abundant modified nucleotides are N6-methyladenosine (m6A), found within mRNAs, and N6,2′-O-dimethyladenosine (m6Am), which is found at the first-transcribed nucleotide. Distinguishing these modifications in mapping studies has been difficult. Here we identify and biochemically characterize, PCIF1, the methyltransferase that generates m6Am. We find that PCIF1 binds and is dependent on the m7G cap. By depleting PCIF1, we generated transcriptome-wide maps that distinguish m6Am and m6A. We find that m6A and m6Am misannotations arise from mRNA isoforms with alternate transcription-start sites (TSSs). These isoforms contain m6Am that map to “internal” sites, increasing the likelihood of misannotation. We find that depleting PCIF1 does not substantially affect mRNA translation but is associated with reduced stability of a subset of m6Am-annotated mRNAs. The discovery of PCIF1 and our accurate mapping technique will facilitate future studies to characterize m6Am’s function.
Keywords: m6Am, m6A, PCIF1, mRNA methylation, mRNA translation, mRNA stability
Graphical Abstract

eTOC Blurb
m6Am is a prevalent mRNA modifications occurring adjacent to the m7G cap. Boulias, Toczydlowska-Socha, Hawley et al. identify PCIF1 as the m6Am methyltransferase and perform transcriptome-wide mapping to distinguish m6Am from m6A and identify “internal” TSSs. m6Am increases stability of a subset of mRNAs but has minimal effects on translation.
Introduction
The most prevalent regulated methyl modifications in mRNA occur on two similar nucleotides: adenosine (A) and 2′-O-methyladenosine (Am) (Perry et al., 1975; Wei et al., 1975). METTL3 catalyzes the methylation on the N6 position of the adenine ring to form N6-methyladenosine (m6A) at internal sites in mRNA (Bokar et al., 1997). At least 25% of mRNAs contain at least one m6A (Dominissini et al., 2012; Meyer et al., 2012).
N6 methylation also occurs on Am to form a dimethylated adenosine: N6, 2′-O-dimethyladenosine (m6Am) (Keith et al., 1978; Wei et al., 1975). Am is primarily located at the first transcribed nucleotide position in mRNAs, adjacent to the m7G cap. Nucleotides located at the first transcribed nucleotide position in an mRNA are typically methylated on the ribose at the 2′-hydroxyl position. However, if this nucleotide is Am, it can undergo further N6 methylation to m6Am. Since m6Am is present at the first transcribed nucleotide in ~30% of all cellular mRNAs, m6Am can affect the fate of a large subset of the transcriptome (Wei et al., 1975).
Recent studies have started to reveal the functions of m6Am. m6Am is enriched in mRNAs with high stability and translation efficiency (Mauer et al., 2017a). Mechanistically, m6Am was shown to impair mRNA decapping mediated by DCP2, leading to increased stability of at least some m6Am-modified mRNAs (Mauer et al., 2017a). However, DCP2 does not regulate the stability of most mRNAs in the cell (Li et al., 2012; Li et al., 2011). Instead, DCP2 is important for specific mRNA degradation pathways, such as nonsense-mediated decay, microRNA-mediated mRNA degradation, and mRNA degradation in response to interferon (Li et al., 2012; Li et al., 2011). Therefore, it is not clear whether m6Am has a general role in promoting the high stability of m6Am-containing transcripts, or if m6Am confers mRNA stability to a subset of mRNAs that are degraded though selective decapping pathways.
Predicting the function of m6Am is complicated by the difficulty in determining whether an mRNA contains m6A or m6Am. Transcriptome-wide mapping of m6A and m6Am uses antibodies that bind the 6-methyladenine (6mA) nucleobase portion found in both of these methylated adenosine nucleotides. The two mapping methods, i.e., MeRIP-Seq (methyl RNA immunoprecipitation followed by sequencing) (Dominissini et al., 2012; Meyer et al., 2012) and miCLIP (m6A individual-nucleotide-resolution crosslinking and immunoprecipitation) (Linder et al., 2015) both map sites of 6mA, rather than m6A or m6Am. The 6mA “peaks” are then interpreted to be either m6A or m6Am using a variety of criteria. For example, if the 6mA peak is in the 5′UTR, this suggests that the 6mA peak is caused by m6Am since this nucleotide is exclusively found as the transcription-start nucleotides. Nevertheless, it can be difficult to distinguish m6Am from m6A located within the 5′UTR of mRNAs. As a result, previous maps of m6Am may have inaccuracies which may make it difficult for predicting its function.
To definitively distinguish m6A and m6Am in transcriptome-wide maps, depletion of either m6A or m6Am would be required. m6A depletion cannot be readily achieved as Mettl3 is essential for survival in nearly all of 341 cell lines that were screened (Tsherniak et al., 2017). The methyltransferase that generates m6Am is not known, but its depletion could enable the identification of the sites that are m6Am, since the remaining sites would be m6A.
Here we describe the identification of PCIF1 as the methyltransferase that is responsible for generating m6Am in mRNA. We show that PCIF1 methylates Am in the context of the m7G cap, and has negligible ability to methylate adenosine in mRNA outside this context. By mapping m6A in the transcriptome of PCIF1-deleted cells, we distinguish between m6Am and 5′UTR m6A. We find numerous examples where previously annotated m6Am sites reflect m6A and vice versa. We show that transcript isoforms with alternate TSSs account for many of these discrepancies facilitating the identification of these “internal” TSSs. Using this new high-confidence map of m6Am sites, we characterize the fate of m6Am-modified mRNAs in PCIF1 knockout cells, and show that m6Am has negligible effects on translation under basal conditions but is associated with increased stability of a subset of m6Am-initiated transcripts. Overall, our studies identify PCIF1 as the methyltransferase that generates m6Am in the transcriptome and provides revised transcriptome-wide maps that distinguish between m6A and m6Am.
RESULTS
Identification of PCIF1 as a candidate m6Am-forming methyltransferase
Studies in the 1970’s provided initial characterization of an enzymatic activity in HeLa cells that synthesizes m6Am (Keith et al., 1978). This enzyme selectively methylates Am adjacent to an m7G cap in synthetic RNA substrates (Keith et al., 1978).
In order to identify the m6Am-forming enzyme, we performed a comparative bioinformatic analysis of orphan adenosine methyltransferases. These enzymes contain the [DNSH]PP[YFW] motif which is present in all adenine N6-methylases (Iyer et al., 2016). Among these putative adenine methylases, PCIF1 is notable since it evolved at the same time that the 5′ cap emerged in mRNA (Iyer et al., 2016). It has been hypothesized that the 5′ cap emerged with eukaryotic evolution to replace the Shine-Dalgarno sequence and direct ribosomes to mRNAs and to protect from 5′ exoribonucleases to distinguish self-versus-foreign mRNAs (Furuichi et al., 1977; Shimotohno et al., 1977; Shuman, 2002). The PCIF1 methylase family is derived from the prokaryotic M.EcoKI/M.TaqI methylases of the bacterial restriction-modification systems (Iyer et al., 2016). All of these methylases contain helices before and after the conserved core strand-3 which display partial or complete degeneration into coil elements.
Another feature of these methylases is the addition of a conserved residue from a helix N-terminal to the core methylase catalytic domain. A PCIF1 crystal structure revealed that its putative methyltransferase domain indeed adopts the classical Rossmann fold of many RNA methylases (Akichika et al., 2019). PCIF1 also contains a WW domain which interacts with the C-terminal domain of RNA polymerase II (Fan et al., 2003) (Figure 1A), suggesting that its function is linked to transcription. Based on this, we asked whether PCIF1 is an adenine N6-methyltransferase in mRNA.
Figure 1. PCIF1 N6-methylates 2′-O-methyladenosine in vitro in an m7G cap-dependent manner.

A. Schematic of PCIF1 indicating the position of predicted functional domains. The location of the sites of mutations used in the study are shown. The catalytic domain includes a four amino acid motif, NPPF, which is predicted to be essential for mediating methylation (Iyer et al., 2016). The location of the site guide RNAs (gRNAs; 5′- CGGUUGAAAGACUCCCGUGG-3′ and 5′- ACUUAACAUAUCCUGCGGGG-3′) used in Figure 2 are indicated.
B. Oligonucleotide sequences used in methyltransferase assays.
C. PCIF1 methylates m7G-ppp-Am-N20 RNA. GST-PCIF1 (50 nM), but not the catalytically inactive mutants APPA or SPPG efficiently converts m7G-ppp-Am (4 μM) to m7G-ppp-m6Am as assessed by UHPLC-MS/MS. Under the same conditions (SAM, 160 μM, 10 min), PCIF1 does not convert any of the 5 internal adenosines to m6A. Each bar represents the mean ± s.e.m of 3 independent experiments. n.s: not significant, ***: p < 0.001, as assessed by unpaired Student’s t-tests.
D. PCIF1 methylates cap-adjacent adenosine regardless of 2′-O-ribose methylation. GST-PCIF1, but not the APPA or SPPG PCIF1 mutants efficiently converts m7G-ppp-A-N20 (4 μM) to m7G-ppp-m6A-N20. Assays were performed as in C. Each bar represents the mean ± s.e.m of 3 independent experiments. ***: p < 0.001, as assessed by unpaired t-tests.
E. PCIF1 enzyme kinetics. m7G-ppp-Am-N20 (at indicated concentration) was incubated with GST-PCIF1 (20 nM) for the indicated times in the presence of 1.33 μM 3H-SAM and 10 μM SAM. Methylation was determined by the presence of 3H in the RNA, as assessed by scintillation counting. Each point represents the mean ± s.e.m of 3 independent experiments.
F. Michaelis-Menten kinetics of PCIF1 methylytransferase activity toward m7G-ppp-Am and m7G-ppp-A. Each point represents the mean ± s.e.m of 3 independent experiments.
G. PCIF1 activity depends on the presence of the m7G cap. m7G-ppp-Am-N20 or ppp-Am-N20 (4 μM) was incubated with GST-PCIF1 as in C. PCIF1 converted Am to m6Am specifically in the m7G capped RNA. Each bar represents the mean ± s.e.m of 3 independent experiments. ***: p < 0.001, as assessed by unpaired t-tests.
H. PCIF1 directly binds the m7G cap. Anti-FLAG immunoblotting was used to detect binding of 3xFLAG-PCIF1 from HeLa cell extracts to m7GTP-conjugated beads. The beads were eluted with m7G-ppp-A or G-ppp-A. eIF4E and eIF4G were used to control for binding to m7G.
PCIF1 N6-methylates 2′-O-methyladenosine in an m7G cap-dependent manner in vitro
To identify potential PCIFI-dependent nucleotide methylase activity, we bacterially expressed and purified glutathione S-transferase (GST)-tagged PCIF1. To test whether PCIF1 can methylate the cap-adjacent adenosine of mRNAs, we performed in vitro methylase assays with an RNA oligonucleotide containing a 5′ m7G cap followed by 2′-O-methyladenosine (m7G-ppp-Am-N20) (Figure 1B). We found that PCIF1 methylates Am in this RNA to produce m6Am, as assessed by UHPLC-MS/MS (Figure 1C).
Interestingly, we did not detect m6A formation in these methylation reactions despite the presence of 5 internal adenosines in the RNA sequence (Figure 1C). Although PCIF1 may methylate an internal adenosine in a currently unknown sequence context, these findings suggest that PCIF1 preferentially N6-methylates 2′-O-methyladenosine rather than internal adenosines.
As a control, we generated predicted catalytically inactive PCIF1 by mutating both asparagine 553 and phenylalanine 556 to alanines (NPPF→PPA) or to a serine and a glycine (NPPF→PPG). The corresponding mutations inactivate the EcoKI and Dam N6-methylases (Guyot et al., 1993; Willcock et al., 1994). Neither the APPA nor SPPG mutant was able to methylate RNA (Figure 1C), suggesting that the PCIF1 catalytic domain is required for Am methylation in vitro.
We next asked if PCIF1-mediated N6 methylation of the m7G-adjacent A requires 2′-O-methyl modification on the A. To test this, we used an RNA substrate with a 5′ m7G cap followed by adenosine (m7G-ppp-A-N20) rather than Am. Using in vitro methylation assays, we found that wild-type PCIF1 but not the SPPG or APPA PCIF1 mutant was able to N6-methylate adenosine to m6A (Figure 1D). We next examined the rate and substrate preference of PCIF1 using a serial dilution of the m7G-ppp-Am- and the m7G-ppp-A-capped oligonucleotides and tritiated S-adenosyl methionine [3H]-SAM as the methyl donor (Figure 1E). Michaelis-Menten analysis yielded a KM = 82 +/− 18.2 nM for the capped 2′-O-methylated RNA and a KM = 630 +/− 84.2 nM for the unmethylated A RNA (Figure 1F), suggesting that PCIF1 has a ~7.6-fold higher preference for binding the 2′-O-methylated adenosine substrate.
Notably, the m7G moiety was required for methylation as PCIF1 efficiently methylated Am to m6Am in an m7G capped RNA (m7G-ppp-Am-N20) but was unable to methylate Am in an RNA that lacked the m7G cap (ppp-Am-N20) (Figure 1G). Overall, these biochemical assays suggest that PCIF1 methyltransferase activity towards Am depends on the presence of the m7G cap, but does not require 2′-O-methylation on the adenosine.
We next asked if the PCIF1 preference for m7G-capped RNA was due to an ability to bind the m7G cap. We performed cap-binding assays with PCIF1 using 7-methylguanosine-5-triphosphate (m7G-ppp)-coupled Sepharose beads. In these experiments, we used lysates from HeLa cells expressing FLAG-tagged wild-type PCIF1. As expected, cap-binding proteins eIF4E and eIF4G were bound to m7G-ppp beads and were efficiently eluted using m7G-ppp-A but not G-ppp-A (Figure 1H). Similarly, PCIF1 bound to the m7G-ppp beads and was eluted with m7G-ppp-A but not G-ppp-A (Figure 1H). Together, these data suggest that PCIF1 binds directly to the m7G cap, which may account for its specificity towards adenosine adjacent to the m7G. These results are consistent with the crystal structure of PCIF1 which shows specific interactions with m7G (Akichika et al., 2019).
PCIF1 knockout abolishes m6Am levels without affecting m6A in RNA
To determine the ability of PCIF1 to generate m6Am in cells, we used CRISPR to delete PCIF1 in various cell lines and examined levels of m6Am and m6A in RNA (Figures 2A and S1A). To measure m6Am, we used a two-dimensional thin-layer chromatography (2D-TLC)-based method that can measure both m6Am and Am, allowing the ratio of these modified forms of adenosine to be calculated in mRNA (Kruse et al., 2011). In this assay, mRNA is decapped, and the 5′ nucleotide is selectively radiolabeled with [32P]-ATP by polynucleotide kinase (PNK). Thus, the first transcribed nucleotide in RNA samples can be selectively detected and quantified. As expected, all the known nucleotides located at the first transcribed nucleotide in mRNA were detected, i.e., m6Am, Am, Gm, Cm, and Um. However, in PCIF1 knockout cells, a selective and complete loss of m6Am was detected (Figures 2B and S1B). A similar effect was seen using UHPLC-MS/MS to quantify m6Am (Figure 2C). Thus, PCIF1 is required for the presence of m6Am at the first transcribed nucleotide in mRNA.
Figure 2. PCIF1 N6-methylates 2′-O-methyladenosine in cells.

A. CRISPR-mediated PCIF1 knockout (KO) in HEK293T cells was assessed by anti-PCIF1 immunoblotting. The upper band represents endogenous PCIF1, whereas the lower band is a non-specific band. β-actin, loading control.
B. PCIF1 is required for formation of m6Am in mRNA in cells. m6Am and Am levels in poly(A) RNA was detected by radiolabeling the 5′ nucleotide after decapping. RNA hydrolysates were resolved by 2D-TLC. Representative images are shown from 3 biological replicates. The bar graph on the right represents the mean ± s.e.m of 3 independent experiments. *** p < 0.001, Student’s t-test.
C. m6Am is depleted in PCIF1 KO HEK293T cells as assessed by UHPLC-MS/MS. Each bar represents the mean ± s.e.m of 3 independent experiments. ****: p <0.0001 as assessed by paired t-tests.
D. PCIF1 does not affect the level of internal m6A. 2D-TLC analysis of poly(A) RNA from HEK293T (WT) and PCIF1 KO HEK293T show no effect on the level of m6A.
E. Internal m6A is not affected in PCIF1 KO HEK293T cells as assessed by UHPLC-MS/MS. Each bar represents the mean ± s.e.m of 3 independent experiments. ns: not significant, as assessed by paired t-tests.
F. Wild-type but not a catalytically inactive PCIF1 mutant restores m6Am levels in PCIF1 knockout cells as assessed by 2D-TLC. Each bar represents the mean ± s.e.m of two independent experiments.
G. Wild-type but not catalytically inactive PCIF1 mutant restores m6Am levels in PCIF1 KO cells as assessed by UHPLC-MS/mS. Each bar represents the mean ± s.e.m of two independent experiments. Ns: not significant, * p < 0.05 as assessed by paired t-tests.
H. Western blot analysis demonstrates equivalent expression of wild-type and catalytically inactive FLAG-tagged PCIF1. β-actin, loading control.
I. Overexpression of wild-type but not catalytically inactive PCIF1 increases m6Am levels in HEK293T cells as assessed by 2D-TLC. Upper and left panels show representative images of 3 independent experiments. The bar graph represents the mean ± s.e.m of 3 independent experiments. ns: not significant, ****P ≤ 0.0001, unpaired t-tests.
See also Figure S1.
We next asked if PCIF1 deletion affects m6A levels in mRNA. To test this, we used a 2D-TLC-based method that selectively detects m6A in the G-A-C context (Zhong et al., 2008) and complemented this with UHPLC-MS/MS to quantify all m6A in mRNA. m6A was readily detected in mRNA in control cells, and no reduction was seen in PCIF1 knockout cells (Figures 2D and 2E).
To confirm that the loss of m6Am in the PCIF1 knockout cells was due to a loss of PCIF1 itself, we performed rescue experiments. In these experiments, we used wild-type or the SPPG catalytically inactive PCIF1 mutant (Figures S1C and S1D). We found that re-expression of the wild-type but not the catalytically inactive PCIF1 restored m6Am levels in mRNA of HEK293T PCIF1 knockout cells as assessed by 2D-TLC (Figure 2F) and by UHPLC-MS/MS (Figure 2G).
We next asked if PCIF1 was sufficient to increase m6Am levels in cells. We found that PCIF1 overexpression in HEK293T cells (Figure 2H) led to a ~3-fold increase in the m6Am to Am ratio (Figure 2I). This increase in m6Am levels was dependent on the catalytic activity of PCIF1, as overexpression of a catalytically inactive PCIF1 mutant had no effect on m6Am levels (Figure 2I). Together this data suggests that PCIF1 is both necessary and sufficient to generate m6Am in mRNA in cells.
miCLIP analysis of PCIF1 knockout cells distinguishes m6Am from 5′UTR m6A residues
Next, we used the PCIF1 knockout cells to distinguish m6Am and m6A in transcriptome-wide 6mA maps. We performed miCLIP, a method that produces narrow peaks, and nucleotide transitions at and adjacent to the m6A (Linder et al., 2015). m6A is nearly universally followed by cytosine in mRNA (Wei et al., 1976). This C is frequently observed to undergo a C to T transition as a result of antibody crosslinking in miCLIP, which can then be used to identify m6A (Linder et al., 2015). Because m6Am can also be followed by cytosine, C to T transitions alone are not sufficient to distinguish m6A from m6Am. Peaks caused by m6Am display a unique shape that exhibits a marked drop off of reads at an annotated A-starting TSS, and this feature can be used to identify m6Am (Linder et al., 2015). However, because m6A occurring near the TSS would also produce a similar shaped peak, this approach may result in false positive m6Am identifications.
Furthermore, these approaches are highly dependent on transcript annotations that may not have accurate TSS information for the cell type investigated. For example, annotated TSSs produced by RefSeq and ENSEMBL differ frequently for the same gene (Zhao and Zhang, 2015). Therefore, true m6Am peaks may have been discarded or thought to be m6A based on their location away from a TSS.
We therefore performed miCLIP in control and PCIF1 knockout cells to distinguish m6Am and m6A. In control cells, reads were enriched in the vicinity of the stop codon as well as the TSS, which is generally assumed to reflect m6A and m6Am, respectively (Figure 3A). PCIF1 knockout cells exhibited fewer reads mapping near the annotated TSS (Figure 3A, ~57.5% decrease in 5′UTR), suggesting these reads derive from an m6Am residue.
Figure 3. Depletion of PCIF1 distinguishes m6A and m6Am in transcriptome-wide 6mA maps.

A. Metagene of miCLIP reads in wild-type and PCIF1 knockout (KO) HEK293T cells. Shown is a metagene analysis of reads from the wild-type or PCIF1 KO miCLIP dataset. The first nucleotide of each read (with respect to the RNA strand) was extracted and plotted. Reads in the 5′UTR were lost in the PCIF1 knockout, suggesting a complete loss of m6Am in the PCIF1 knockout cells.
B. DREME motif search within called peaks show the DRACH motif as the most enriched in all datasets, consistent m6A as the most abundant 6mA-containing nucleotide mapped by miCLIP.
C. miCLIP peaks can be identified as m6Am or m6A based on their decrease in PCIF1 KO cells. Genome tracks were plotted for RPL35 and KDELR2 with called m6A sites (FDR<0.1) and m6Am sites indicated by red circles and blue triangle, respectively. Zoomed insets show m6Am peaks can be distinguished from nearby m6A sites.
D. The previously annotated m6Am site in RACK1 is actually a 5′UTR m6A. The TSS-proximal peak in RACK1 is not affected in the PCIF1 knockout and overlaps with a DRACH motif.
E. Metagene analysis of PCIF1 KO-validated m6Am sites shows m6Am sites throughout the 5′UTR and in the transcript body. Shown is a metagene of the exact sites of m6Am within the PCIF1-dependent peaks as determined by A to T transitions and the read drop-off method. The metagene reveals an overall enrichment at the TSS, with some sites that appear to be within the CDS and 3′UTR.
F. DREME motif search of the nucleotides surrounding each m6Am was performed confirms the previously reported BCA motif, and shows that the promoter sequence upstream of the m6Am is GC-enriched.
See also Figure S2.
A motif analysis of significant peaks showed the DRACH m6A consensus (D = A, G, U; R = A, G; H = A, C, U) as the most common motif in each data set (Figure 3B). This suggests that m6A is the most common modification mapped in both datasets, as expected.
To identify m6Am marked transcripts we next examined the 6mA peaks that showed differences in the control and PCIF1 knockout miCLIP datasets. As expected, we detected a loss of peaks near the TSS of certain genes in the PCIF1 knockout. For example, RPL35 and KDELR2 show peaks near the annotated TSS as well as at internal sites (Figure 3C). The TSS-proximal peaks were absent in the PCIF1 knockout miCLIP dataset. These data are consistent with the idea that the TSS peaks predominantly reflect m6Am.
However, in some cases, the peaks near the TSS were not affected in the PCIF1-knockout dataset. For example, peaks near the TSSs of RACK1 and RPS5, which were previously annotated as m6Am in HEK293T cells based on their location, peak shape and lack of C to T transitions (Mauer et al., 2017a), persist in the PCIF1 knockout dataset (Figure 3D and Figure S2A). These peaks contain a canonical DRACH m6A consensus motif, and C to T transitions are detected for RACK1 (Figure 3D), suggesting that these sites are actually m6A.
The variability in C to T transitions reflects the low transition rate induced by the antibody adduct on this transcript. Altogether, these data indicate that PCIF1 depletion can be used to determine the identity of an m6A peak.
Overall, only 60.2% of genes that had previously been annotated as m6Am (Mauer et al., 2017a) were validated as m6Am based on their loss in PCIF1 knockout cells. In some cases, this could be explained by peaks being below the threshold for detection in one or both replicates. Nevertheless, this difference highlights the importance of depleting cells of PCIF1 to reduce false positive m6Am identification.
A high-confidence transcriptome-wide map of m6A and m6Am based on PCIF1 depletion
To create a high confidence map of all m6Am sites in the transcriptome, we searched for all peaks that exhibit a marked reduction in miCLIP signal in the PCIF1 knockout dataset. The majority of peaks showed no substantial difference between control and PCIF1 knockout miCLIP datasets, suggesting that they are m6A (Figure S2B). However, 2360 peaks exhibited a significant reduction in both PCIF1 knockout datasets (Figure S2B). In contrast, only 11 sites appeared to increase, suggesting a very low incidence of false positives.
We next identified the exact m6Am residue within each of these peaks. In our previous approach, we used a “pile up” of reads that drop off at the 5′ end of these read clusters in A-starting genes to predict the m6Am site (Linder et al., 2015). In some cases, the drop off is not easily detected or several of these were found in close proximity. This appears to occur when (1) the total reads are too few; or (2) the reads terminate before the TSS, possibly due to impaired reverse transcription through the 2′-O-methyl modifications (Maden et al., 1995) in the cap-proximal nucleotides, or due to non-templated nucleotide addition that occurs at the ends of cDNAs generated by reverse transcriptases (Chen and Patton, 2001).
Therefore, we wanted to develop an alternative approach to identify m6Am within the PCIF1-dependent peaks. Previously we observed antibody-induced A to T transitions at the m6A site in miCLIP (Linder et al., 2015). We confirmed that A to T transitions are readily detected at known m6Am and m6A throughout the transcriptome (Figures S2C and S2D). Therefore, we used a 10% A to T transition rate to identify the m6Am within PCIF1-dependent peaks. The drop-off approach was used when the A to T transition rate did not meet these criteria (Figure S2E). There was high similarity in the m6Am sites that were called when using these methods separately (Figure S2F).
Overall, the m6Am sites mapped based on their dependence on PCIF1 (Table S1) were primarily located throughout the 5′UTR (~94%), with a prominent enrichment at the annotated TSS (Figure 3E). Motif analysis of the genomic context of the exact m6Am nucleotide revealed the BCA motif, with A representing the m6Am, and BC representing upstream genomic nucleotides (B = C, G, or T), as reported previously for m6Am (Linder et al., 2015). Additionally, motif analysis shows the upstream promoter sequence is GC-enriched (Figure 3F). Motifs downstream and including the transcription-start adenosine were also enriched, suggesting that m6Am occurs in specific sequence contexts within mRNA transcripts (Figure S2G).
Next, we mapped m6A in the 5′UTR. C to T transitions in a DRACH consensus were used to call m6A sites based on the miCLIP protocol (Linder et al., 2015). This identified 399 5′UTR m6A sites that were robustly called across all datasets (Table S2).
We next asked if mRNAs with 5′UTR m6A and mRNAs that contain m6Am are linked to different cellular processes, based on our updated m6A and m6Am sites. Functional annotation using DAVID shows that transcripts containing these distinct modified nucleotides are linked to different cellular processes, with 5′UTR m6A associated with processes such as transcription and cell division, while m6Am is primarily associated with splicing (Figures S2H, I and Table S3).
ATF4 contains a m6Am rather than m6A in its 5′UTR
We next wanted to understand if our revised map of m6Am and m6A can identify transcripts with misannotated modified nucleotides. A 5′UTR m6A site has been described as mediating the unusual stress-regulated translation of ATF4 (Zhou et al., 2018). ATF4 has two upstream open reading frames (uORFs) in its 5′UTR. In unstressed cells, the uORFs are translated, which prevents translation of the main open reading frame, which encodes the ATF4 protein (Vattem and Wek, 2004). However, during stress, the second uORF is skipped, and the ribosome scans to the main open read frame after translating the first uORF. This allows the ATF4 protein to be translated during stress. m6A was mapped to the second open reading frame and was described as disappearing in a stress-dependent manner, thus causing a stress-regulated switch in ATF4 translation (Zhou et al., 2018).
However, using miCLIP it is apparent that the 6mA peak in the 5′UTR of ATF4 is not located within the second open reading frame (Figure S3A). Instead the peak is located at the transcription-start nucleotide and does not overlap with the position of the putative m6A.
Based on the location of the peak, we asked if it instead reflects m6Am rather than m6A. To test this, we examined ATF4 in the PCIF1 knockout miCLIP dataset. Here, we observed a complete loss of this peak, further confirming that this site is m6Am (Figure S3A).
The role of m6A in controlling stress-induced Atf4 translation was described in mouse embryonic fibroblast cells (Zhou et al., 2018), rather than the HEK293T cells used here. Human cells appear to have lost the DRACH consensus sequence surrounding the putative m6A site (Figure S3B). Conceivably, human cells exhibit stress-induced regulation of ATF4 translation through an m6A-independent pathway and mouse cells utilize an m6A-dependent pathway. Therefore, we mapped 6mA in mouse embryonic fibroblasts using miCLIP (Figure S3C). Again, the 6mA peak was at the TSS, not at a position corresponding to the second uORF (Figure S3D). These data further show that this peak derives from a m6Am residue. In comparison, there were low levels of 6mA reads throughout the transcript body suggesting either background reads or low stoichiometry m6A sites (Figure S3D).
Overall, these data suggest that ATF4 contains a m6Am at the TSS, but no prominent m6A site within the 5′UTR as previously reported (Zhou et al., 2018). Thus, a role for a 5′UTR m6A in regulating ATF4 translation seems unlikely. Overall, these data demonstrate the ease with which m6A and m6Am sites can be confused for each other.
Identification of internal 6mA sites that reflect transcription-start m6Am
We noticed two unusual features in our mapping results. First, not all m6Am sites mapped to regions within annotated mRNA transcripts. Second, the m6Am metagene showed that while 94% of m6Am sites were located in the 5′UTR, many were not directly at the annotated TSS and, in some cases, further downstream within the transcript body (Figure 3E).
We considered that these findings could be due to m6Am that occurs in mRNA isoforms with alternate TSSs upstream or downstream of the TSS in the RefSeq-annotated transcript. To test this, we created an m6Am metaplot relative to RefSeq-annotated TSSs (Figure 4A). We observed 16.7% of m6Am sites mapping within 250 nucleotides upstream of annotated TSSs, suggesting that some m6Am occurs in isoforms with upstream TSSs. We similarly observed m6Am upstream of TSSs using GENCODE (Frankish et al., 2018) transcript annotations (Figure 4B). The FANTOM5 promoter-level expression atlas (Abugessaisa et al., 2017) uses a set of TSSs specifically mapped across multiple tissues using the cap analysis gene expression (CAGE) approach. Using FANTOM5 we observed a marked overlap with our m6Am sites, supporting the idea that these m6Am sites are indeed TSSs (Figure 4C).
Figure 4. Internally mapped m6Am sites reflect m6Am in mRNA isoforms with alternative TSSs.
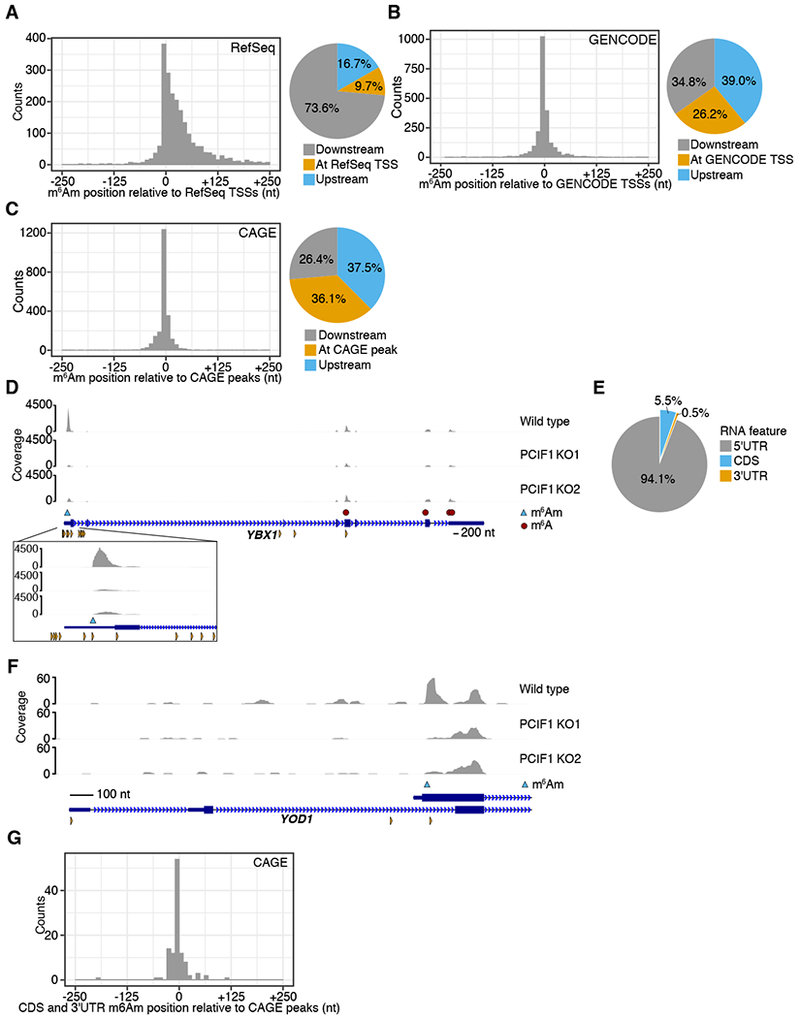
A. A metaplot centered on the closest RefSeq TSS for each called m6Am site shows most m6Am sites are found downstream of the annotated start site, but not at the annotated start site. The proportion of m6Am directly at the annotated TSS, or up- or downstream is shown.
B. A metaplot analysis of m6Am locations using GENCODE TSS annotations shows higher overlap with TSSs. GENCODE annotations include more transcript isoforms and TSSs than RefSeq.
C. A metaplot of the distance from each m6Am site to the closest CAGE peak in the FANTOM5 database shows that m6Am sites are indeed TSSs. Here, the overlap of m6Am was highest, suggesting that m6Am sites are selectively localized to TSSs and not internal nucleotides within mRNA.
D. The m6Am mapping to the annotated 5′UTR of the YBX1 transcript reflects a transcript isoform. The PCIF1-dependent 6mA peak in YBX1 maps within the annotated 5′UTR of YBX1. However, this peak overlaps with a CAGE site (orange triangles), indicating the existence of a transcript isoform that initiates at this 6mA site. m6Am peaks that appear within the 5′UTR reflect m6Am in transcript isoforms with alternative TSSs. The exact m6Am site (blue triangle) was determined using the A to T transition within the PCIF1-dependent peak.
E. Most m6Am are found in the annotated 5′UTR of transcripts.
F. The internally mapping m6Am in YOD1 derives from a TSS of a YOD1 transcript isoform. The m6Am peak in YOD1 begins beyond the start codon of both annotated isoforms. CAGE peaks (orange triangles) suggest this is indeed a TSS.
G. A metaplot analysis of CDS and 3′UTR mapping m6Am sites show overlap with CAGE data, indicating that m6Am occurs at TSSs. The closest CAGE peak to each of the 6% of sites that appeared to not map to the 5′UTR (E) was calculated and plotted.
See also Figure S3.
TSS heterogeneity likely explains why some m6Am sites map within the 5′UTR, rather than being solely located at the annotated TSS. In the case of YBX1, a 6mA peak is mapped to the 5′UTR and is lost in the PCIF1 knockout miCLIP dataset, suggesting that this peak is due to m6Am (Figure 4D). This m6Am likely reflects an isoform with a TSS located at this m6Am site, based on its overlap with a CAGE peak (Figure 4D). Thus, the presence of m6Am within the 5′UTR likely reflects TSS heterogeneity rather than “internal” m6Am nucleotides.
We next wanted to understand why ~6% of m6Am sites (121 sites) appear to map to coding sequences or 3′UTR regions (Figure 4E). For example, YOD1 shows an internal m6A peak in the first exon that is lost in the PCIF1 knockout miCLIP dataset (Figure 4F). As with YBX1, we observed a TSS that overlapped with the m6Am site. Thus, this internal site, which would normally have been assumed to be m6A using MeRIP-Seq and possibly miCLIP, derives from an isoform starting with m6Am.
To test this idea further, we performed a metagene analysis on m6Am sites mapping to coding sequences or the 3′UTR. Here we plotted the distance to the nearest CAGE sites (Figure 4G). This analysis shows that many m6Am sites in the coding sequence and 3′UTR are located at or near CAGE sites. Additionally, these m6Am sites show the BCA motif, which resembles the transcription initiation site (initiator element; Inr) motif of RNA Polymerase II (Figure S2G) (Yang et al., 2007). This motif was also previously found for m6Am mapped to canonical TSSs (Linder et al., 2015). 5′ RACE confirmed that our called m6Am sites are indeed transcription start nucleotides (Figure S3E). Overall, these data further suggest that m6Am is not internally located within transcripts but is instead found at the TSSs.
Approximately 8% of m6Am sites that mapped to the coding sequence or 3′UTR also contained an adjacent C to T transition. As a result, these peaks would likely have been called as an m6A. These data highlight the value of using PCIF1 depletion to validate the transcriptome-wide m6Am and m6A maps.
m6Am correlates with enhanced translation, expression, and stability of mRNAs
In our previous studies, we found that m6Am is correlated with transcripts that are highly expressed and have long half-lives in cells (Mauer et al., 2017a). We therefore wanted to reexamine this correlation based on the high-confidence m6Am annotation based on peaks that were depleted in the PCIF1 knockout miCLIP dataset. In some cases, mRNAs that had been previously annotated as beginning with Am, Cm, Gm, or Um were re-annotated as m6Am for this analysis, and mRNAs previously annotated as m6Am were re-annotated as Am based on our revised mapping data. Analysis of mRNA expression and half-lives showed that transcripts that begin with m6Am are indeed more highly expressed and stable than mRNAs with other start nucleotides. Notably, m6Am appears to be the predominant start nucleotide of the mRNAs that are the most abundant and have annotated half-lives greater than 24 hours (Figure 5A–D).
Figure 5. mRNA expression level and mRNA half-life depending on TSS.
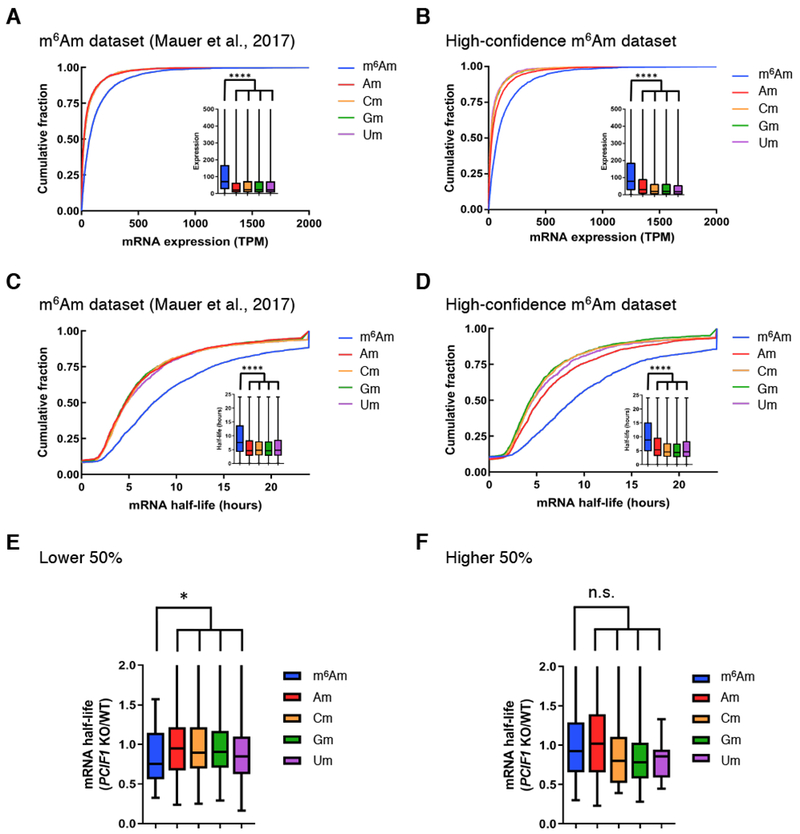
A. mRNAs with an annotated m6Am start nucleotide show higher mRNA expression than other mRNAs. mRNA expression level in wild-type HEK293T cells was based on the first annotated nucleotide and an earlier m6Am map (Mauer et al., 2017a) . Transcripts that start with m6Am are significantly upregulated. ****, P < 2.2 × 10-16, Student’s t-test. Cumulative distribution plot and boxplot represent the expression for mRNAs starting with m6Am, Am, Cm, Gm and Um. Data shown are the average gene expression measured from two replicates for HEK293T cells.
B. m6Am mRNAs annotated using the PCIF1 knockout (KO) miCLIP dataset show increased expression compared to mRNAs with other start nucleotides. Cumulative distribution plots were prepared as in A using the high-confidence m6Am dataset. The transcripts start with m6Am are significantly upregulated as in A. ****, P < 2.2 × 10-16, Student’s t-test.
C. mRNAs with an annotated m6Am start nucleotide show higher mRNA half-life than other mRNAs. Annotated mRNA half-lives were based on the first annotated nucleotide and an earlier m6Am map (Mauer et al., 2017a). mRNAs with an annotated m6Am exhibit a significantly elevated mRNA half-life than mRNAs with other annotated start nucleotides. ****, P < 2.2 × 10-16, Student’s t-test.
D. m6Am mRNAs annotated using the PCIF1 KO miCLIP dataset show increased expression compared to mRNAs with other start nucleotides. Transcripts with m6Am have significantly longer half-life with similar P-value. ****, P < 2.2 × 10-16, Student’s t-test.
E. Influence of PCIF1 depletion on mRNA half-life for transcripts in the lower half of gene expression. Transcripts with m6Am have significantly shorter half-life in comparison to mRNAs with other annotated start nucleotides. *, P = 0.0258 by Student’s t-test.
F. Influence of PCIF1 depletion on mRNA half-life for highly expressed transcripts. Transcripts with m6Am show no significant decrease in mRNA half-life in comparison to mRNAs with other annotated start nucleotides. n.s., Student’s t-test.
See also Figure S4.
Overall, these data suggest that the presence of m6Am correlates with an overall increase in mRNA stability, and that m6Am is the predominant starting nucleotide on “outlier” mRNAs with unusually high stability and expression. To determine whether the N6-methyl in m6Am was required for the unique properties of these outlier mRNAs, we examined mRNA stability in PCIF1 knockout HEK293T cells. mRNA stability was measured using SLAM-Seq (thiol(SH)-linked alkylation for the metabolic sequencing of RNA) (Herzog et al., 2017) (Figure S4A).
To examine the outlier mRNAs, which are highly expressed, we separately examined mRNAs in the lower and upper half of gene expression. We only used transcripts that exhibited a minimum threshold of transitions required for mRNA half-life quantification. For mRNAs in the lower half of gene expression, we observed a marked decrease in mRNA half-life upon PCIF1 depletion (Figure 5E). We confirmed this effect by examining the stability of individual mRNAs after treatment of control and PCIF1 knockout HEK293T cells with actinomycin D. Both NBR1 and AKAP12 transcripts exhibited decreased expression after 8 hours of actinomycin D treatment (Figure S4B). This effect was more prominent in PCIF1 knockout cells, consistent with a stabilizing effect of m6Am (Figure S4B).
However, when we examined the more abundant mRNAs, which are enriched in the outlier transcripts, these transcripts did not show a substantial change in mRNA half-life (Figures 5F and S4C). We observed a slight reduction in stability relative to Am-annotated transcripts, but compared to all mRNAs (Am, Cm, Gm, and Um), these mRNAs appeared to show small, but nonsignificant increase in mRNA stability in PCIF1 knockout cells.
Thus, although m6Am is highly enriched in these outlier transcripts, the N6 methyl does not appear to account for their unusual stability. In contrast, mRNAs in the lower half of gene expression appear to utilize m6Am for transcript stability.
Previously we found that m6Am-containing transcripts exhibit a subtle increase in translation relative to mRNAs with other start nucleotides (Mauer et al., 2017a). To more directly test the role of m6Am on translation, we compared the translation efficiency of transcripts in control and PCIF1 knockout cells by ribosome profiling (Figures S4D and S4E). Here, we found that transcripts that contained m6Am as the transcription-start nucleotide did not show a substantial change in translation efficiency upon PCIF1 depletion (Figure S4F). Rather than showing a decrease in translation, we observed a slight increase in translation upon loss of m6Am compared to transcripts annotated to begin with other nucleotides (Figure S4F). In agreement with the modest effects of PCIF1 depletion on translation rates, we found that levels of proteins encoded by several m6Am-modified mRNAs remained largely unchanged in PCIF1 KO cells (Figure S4G).
Together, these experiments suggest that under the conditions used in these experiments, N6 methylation does not mediate the increase in translation efficiency of m6Am-initiated mRNAs in HEK293T cells.
DISCUSSION
A major challenge when mapping m6A and m6Am is that both nucleotides are recognized by 6mA-specific antibodies and both can produce peaks in the 5′UTR of mRNA transcripts. Here, by identifying PCIF1 as the m6Am-forming methyltransferase, and by depleting PCIF1 to definitively identify m6Am sites, we present a revised annotation of m6Am and m6A in the transcriptome. We find that previous annotations contain errors that reflect the existence of mRNA isoforms that differ by TSSs. In some cases, the isoforms contain TSSs that map to internal sites within the annotated transcripts, resulting in the appearance of peaks that would otherwise be attributed to m6A. The identification and characterization of PCIF1 coupled with a precise m6Am annotations generated by PCIF1 depletion will facilitate the identification of functions for m6Am.
Our studies provide insights into the function of m6Am. Using our new high-confidence m6Am map, we find that m6Am is found on unusually stable and highly abundant transcripts in cells. However, depletion of m6Am by PCIF1 knockout does not markedly impair the stability of these unusual transcripts under basal conditions. This suggests that m6Am does not account for the stability of these unusual mRNAs. m6Am therefore is likely to co-occur with other transcript features that confer these unusual properties to these mRNAs.
However, m6Am does promote the stability of other mRNAs. When we examined mRNAs in the lower half of gene expression, we found a marked drop in mRNA stability in PCIF1-depleted cells. Why might some mRNAs be stabilized by m6Am, while others may not be affected? First, m6Am is enriched in different sequence contexts. This contrasts with m6A, which is nearly always found a single sequence context. m6Am is therefore likely to have different sensitivities to decapping or bind to different m6Am readers in a context-dependent manner. Thus, unlike traditional approaches where mRNAs are binned and bioinformatically analyzed based on the presence or absence of a modification, m6Am functions are more likely to be revealed by analysis of mRNAs binned based on m6Am sequence contexts.
Another important factor that might affect whether m6Am has a destabilizing effect is whether the mRNA utilizes DCP2, or potentially other m6Am-sensitive decapping mechanisms. Previous studies showed that m6Am confers stability of mRNAs to DCP2-mediated decapping (Mauer et al., 2017b). DCP2 is not the major decapping enzyme in cells, but DCP2 targets mRNAs with specific 3′UTR features. Thus, m6Am may stabilize mRNAs when DCP2-dependent pathways are activated. Overall, our studies show that m6Am acts in a transcript-selective manner, rather than a general mRNA stabilizing modification.
Although our study focused on mRNA stability, another study examined m6Am mRNA abundance, and found no effects upon PCIF1 depletion (Akichika et al., 2019). This might reflect compensatory upregulation of m6Am mRNAs. Notably, Akichika et al. examined all mRNAs, rather than specific subsets of m6Am-annotated mRNAs.
Two recent studies also reported PCIF1 as the m6Am cap-dependent methyltransferase (Akichika et al., 2019; Sun et al., 2019). Akichika et al. found that m6Am slightly enhances translation relative to the Am form of the transcript in PCIF1 knockout cells, based on ribosome profiling (Akichika et al., 2019). Our ribosome profiling analysis of PCIF1 knockout cells showed a slight repressive effect of m6Am on translation. Therefore, mRNAs modified with m6Am are efficiently translated but Am-modified mRNAs are even slightly more efficiently translated. Regardless, both our study and the Akichika et al. study are consistent in finding a very minor effect on translation of m6Am-annotated mRNAs upon PCIF1 depletion. It should be noted that effects of m6Am on both mRNA translation and mRNA stability are likely to depend on the sequence motif following m6Am, and may be different during signaling or stress conditions that were not examined in our study.
STAR METHODS
Synthesis and characterization of synthetic oligonucleotides
The sequences of all the oligonucleotides used in this study are shown in Figure 1B.
The synthetic RNA oligonucleotides, used in Figure 1E, were chemically assembled on an ABI 394 DNA synthesizer (Applied Biosystems) from commercially available long chain alkylamine controlled-pore glass (LCAA-CPG) solid support with a pore size of 1000 Å derivatized through the succinyl linker with 5′-O-dimethoxytrityl-2′-O-Ac-uridine (Link Technologies). All RNA sequences were prepared using phosphoramidite chemistry at 1-μmol scale in Twist oligonucleotide synthesis columns (Glen Research) from commercially available 2′-O-pivaloyloxymethyl amidites (5′-O-DMTr-2′-O-PivOM-[U, CAc, APac or GPac]-3′-O-(O-cyanoethyl-N,N-diisopropylphosphoramidite)(Lavergne et al., 2010) (Chemgenes). The 5′-terminal adenosine was methylated in 2′-OH (Am). The 5′-O-DMTr-2′-O-Me-APac-3′-O-(O-cyanoethyl-N,N-diisopropylphosphoramidite) (Chemgenes) was used to introduce Am at the 5′-end of RNA. All oligoribonucleotides were synthesized using standard protocols for solid-phase RNA synthesis with the PivOM methodology (Lavergne et al., 2008).
After RNA assembly, the 5′-hydroxyl group of the 5′-terminal adenosine Am of RNA sequences, still anchored to solid support, was phosphorylated and the resulting H-phosphonate derivative was oxidized and activated into a phosphoroimidazolidate derivative to react with either pyrophosphate (for ppp(Am )-RNA synthesis) (Zlatev et al., 2010) or guanosine diphosphate (for G-ppp-Am-RNA synthesis) (Thillier et al., 2012).
After deprotection and release from the solid support upon basic conditions (DBU then aqueous ammonia treatment for 4h at 37°C), all RNA sequence s were purified by IEX-HPLC (Barral et al., 2013), they were obtained with high purity (>95 %) and they were unambiguously characterized by MALDI-TOF spectrometry.
N7-methylation of the purified G-ppp-Am-RNAs to give m7G-ppp-Am-RNAs was carried out quantitatively using human mRNA guanine-N7 methyltransferase and S-adenosylmethionine as previously described (Thillier et al., 2012). The oligonucleotides used in Figures 1C, 1D, 1G and 1H were synthesized by Trilink.
Cell culture
HEK293T and HeLa cells were maintained in DMEM (11995-065, ThermoFisher Scientific) with 10% FBS and antibiotics (100 units/ml penicillin and 100 μg/ml of streptomycin) under standard tissue culture conditions. Cells were split using TrypLE™ Express (Life Technologies) according to the manufacturer’s instructions. Mycoplasma contamination in cells were routinely tested by Hoechst staining.
Antibodies
Antibodies used for western blot analysis or immunostaining were as follows: mouse anti-FLAG M2 (F1804, Sigma), rabbit anti-PCIF1 (ab205016, Abcam), mouse anti-β actin (A5441, Sigma), anti-eIF4E (2067, Cell Signaling), anti-eIF4G (2498, Cell Signaling), rabbit anti-GAPDH (ab181602, Abcam), mouse anti-TRIM28 (ab22553, Abcam), rabbit anti-ATF5 (ab60126, Abcam), rabbit anti-EEF2 (ab33523), mouse anti-RACK1 (B-3, Santa Cruz), rabbit anti-PARP1 (9542, Cell Signaling), rabbit anti-HSPA8 (8444, Cell Signaling), mouse anti-HSP70/72 (C92F3A-5, Enzo Life Sciences). For m6A individual-nucleotide-resolution cross-linking and immunoprecipitation (miCLIP), rabbit anti-m6A (ab151230, Abcam) was used.
Generation of PCIF1 CRISPR knockout cells and overexpression cell lines
HEK293T and HeLa PCIFI-knockout cell lines were generated by CRISPR/Cas9 technology using two guide RNAs (gRNAs; 5′- CGGUUGAAAGACUCCCGUGG-3′ and 5′- ACUUAACAUAUCCUGCGGGG-3′) designed to target the PCIF1 genomic region between exon 8 and exon 17, that corresponds to the C terminal catalytic domain. Double-stranded DNA oligonucleotides corresponding to the gRNAs were inserted into the pSpCas9n(BB)-2A-Puro (PX459) V2.0 vector (62988, Addgene). Equal amounts of the two gRNA plasmids were mixed and transfected into HEK293T and HeLa cells using FuGENE 6 (Promega). The transfected cells were then subjected to puromycin selection for three days and viable cells were used for serial dilution to generate single-cell clones. The genomic deletion was screened by PCR and was confirmed by Sanger sequencing. HEK293T and HeLa PCIF1-knockout lines used in this study contained a 4655 or 4656 nt homozygous deletion that removed the region between exon 8 and exon 17, including the stop codon, resulting in the disruption of PCIF1 protein after P229 (aa 230-704). Loss of PCIF1 protein expression was confirmed by western blot with anti-PCIF1 antibody (Abcam).
Stable cell lines overexpressing PCIF1 WT or catalytically inactive mutant proteins were generated through retroviral infection. The coding sequence of human PCIF1 fused to a N-terminal 3X FLAG tag sequence that was cloned into the pBABE-puro retroviral vector (Addgene, 1764). Retroviral particles were generated in HEK293T cells through co-transfection of the packaging vectors pMD2.G (12259, Addgene) and pUMVC (8449, Addgene) with the appropriate pBABE-puro vectors. HEK293T and Hela cells were infected with retroviral particles of pBABE-puro-3X-FLAG-PCIF1 WT or pBABE-puro-3X-FLAG-PCIF1 SPPG or control pBABE-puro empty vector, followed by puromycin selection (1μg/ml).
Cells were maintained at 70-80% confluency before harvesting for mRNA purification. Two rounds of poly(A) mRNA isolation from mammalian cells was performed using oligo d(T)25 Magnetic mRNA isolation kit (NEB), according to the manufacturer’s instructions.
Protein expression and purification
The coding sequence of human PCIF1 was cloned as an in-frame fusion to the GST tagged vector pGEX-4T1. The catalytic site NPPF was mutated to APPA or SPPG thru site-directed mutagenesis using the Q5 mutagenesis kit (NEB), according to the manufacturer’s instructions. Recombinant GST-PCIF1 wild-type and catalytically inactive mutant proteins were expressed in E. coli T7 Express lysY. Overnight induction of protein expression was carried out with 0.5 mM IPTG at 18 °C. Bacteria were harvested at 4000 rpm, 4°C and the cell pellet was resuspended in protein purification lysis buffer (50 mM Tris-HCl pH 7.5, 0.25 M NaCl, 0.1% Triton-X, 1 mM PMSF, 1 mM DTT, and protease inhibitors). The lysate was sonicated 6 times in 30 seconds on/off cycles and then centrifuged at 12,000 rpm for 20 minutes. Lysates were incubated with glutathione Sepharose 4B beads (Sigma). Proteins and beads were washed 3 times with protein purification lysis buffer before incubating the beads with elution buffer (12 mg/ml Glutathione in protein purification lysis buffer, pH 8.0) for 30 minutes. Eluates were dialyzed overnight at 4 °C with enzyme storage buffer (40 mM Tris-HCl pH 8.0, 110 mM NaCl, 2.2 mM KCl, 1 mM DTT, 20% glycerol) and were subsequently stored at −80°C. Bradford assays and SDS-page gel electrophoresis followed by Coomassie staining was performed to determine integrity and quantity of purified proteins.
In Vitro methyltransferase assays
In vitro methylation reactions (50 μl) assaying PCIF1 activity against the m7G capped RNA oligonucleotides were performed in methylation reaction buffer (50 mM Tris pH 8.0, 1 mM EDTA, 1 mM DTT, 5% glycerol) supplemented with 160 μM SAM (NEB) using 50 nM GST-PCIF1 protein and 4 μM m7G capped oligonucleotide. Reactions were incubated for 10 minutes at 37°C, followed by heat inactivation for 20 minutes at 65°C and subsequent clean up and buffer exchange using Biospin P6 columns (Biorad). RNA oligonucleotides were decapped using 25 Units of RppH (NEB) in ThermoPol buffer for 3 hours at 37°C, followed by clean up and buffer exchange with Biospin P6 columns. Decapped RNA oligonucleotides were digested to nucleosides with 2 units of Nuclease P1 (Wako USA) at 37°C for 3 hours in a buffer containing 10 mM ammonium acetate pH 5.3, 2mM ZnCl2 followed by treatment with 2 units of Fast Alkaline Phosphatase (FastAP, Thermo Scientific) in FastAP reaction buffer for 1 hour at 37°C. After digestion the sample volume was brought to 100 μl with ddH20 followed by filtration using 0.22 μm Millex Syringe Filters (EMD Millipore). 5 μl of the filtered solution was analyzed by UHPLC-MS/MS.
Enzyme kinetics assaying PCIF1 activity against the m7G-Am and m7G-A RNA oligonucleotides were performed in methylation reaction buffer supplemented with 1.33 μM [3H]-SAM (Perkin Elmer) and 10 μM SAM (NEB), using 20 nM GST-PCIF1 protein and a range of concentrations of m7G-Am oligonucleotide for 2-4 min at 37°C in 50 μl reactions. The reactions were stopped with 0.1% TFA followed by removal of unincorporated [3H]-SAM with Biospin P30 columns (Biorad). The purified RNA oligonucleotide samples were then subjected to scintillation counting using a Perkin Elmer scintillation counter. The Michaelis-Menten curve and KM value were determined using Graphpad Prism software.
UHPLC-MS/MS analysis
For the detection and quantification of internal m6A in mRNA, 500 ng of poly(A) mRNA was denatured at 70°C for 5 minutes followed by digesti on to nucleotides using 20 units of S1 Nuclease (Thermo Scientific) in S1 Nuclease buffer for 2 hours at 37°C in 25 μl reactions. Nucleotides were then dephosphorylated to nucleosides by the addition of 2 units of Fast Alkaline Phosphatase (NEB) in FastAP reaction buffer for 1 hour at 37°C. After digestion the sample volume was brought to 100 μl with ddH20 followed by filtration using 0.22 μm Millex Syringe Filters (EMD Millipore). 5 μl of the filtered solution was analyzed by LC-MS/MS.
For the detection and quantification of cap-adjacent m6Am in mRNA, 500 ng of poly(A) mRNA was decapped using 25 Units of RppH (NEB) in ThermoPol buffer for 3 hours at 37 °C, followed by clean up and buffer exchange with Biospin P30 columns. Subsequently decapped RNA was denatured at 70 °C for 5 minutes followed by digest ion to nucleotides using 2 units of Nuclease P1 (Wako USA) in a buffer containing 10 mM ammonium acetate pH 5.3, 2mM ZnCl2 for 3 hours at 37G. Nucleotides were then dephosphorylated to nucleosides by the addition of 2 units of Fast Alkaline Phosphatase (NEB) in FastAP reaction buffer for 1 hour at 37°C. After digestion the sample volume was brought to 100 μl with ddH20 followed by filtration using 0.22 μm Millex Syringe Filters. 5 μl of the filtered solution was analyzed by LC-MS/MS.
The separation of nucleosides was performed using an Agilent 1290 UHPLC system with a C18 reversed-phase column (2.1 × 50 mm, 1.8 m). The mobile phase A was water with 0.1% (v/v) formic acid and mobile phase B was methanol with 0.1 % (v/v) formic acid. Online mass spectrometry detection was performed using an Agilent 6470 triple quadrupole mass spectrometer in positive electrospray ionization mode. Quantification of each nucleoside was accomplished in dynamic multiple reaction monitoring (dMRM) mode by monitoring the transitions of 268→136 (A), 282→136 (Am), 282→150 (m6A), 296→150 (m6Am), 244→112 (C). The amounts of A, C, Am, m6A and m6Am in the samples were quantified using corresponding calibration curves generated with pure standards. m6Am and m6A levels in the RNA oligonucleotides after in vitro methylation reactions were normalized by cytidine concentration.
Cap-binding assay
Cells were lysed in buffer B (20 mM HEPES-KOH pH 7.6, 100 mM KCl, 0.5 mM EDTA, 0.4% NP-40, 20% glycerol) supplemented with protease and phosphatase inhibitors (Roche), 1 mM dithiothreitol (DTT) and 80 units/ml RNasin (Promega). For pull down, 1-2.5 mg of total protein extract was first pre-cleared on Agarose beads (Jena Bioscience) followed by incubation with 25 μl m7GTP conjugated Agarose beads (Jena Bioscience) for 1 hour at 4°C degrees. Following pull-down the beads were washed three times and the supernatant was removed and replaced by lysis buffer. Beads were incubated with 0.25 mM cap analog, m7G-ppp-A, or G-ppp-A, or water (mock) for 1 hour at 4 °C. Supernatant (Eluate) was removed and diluted with Laemmli sample buffer. Beads were washed three times and resuspended in Laemmli sample buffer. Samples were resolved on a 4-15% Tris-HCl gradient gel (BioRad) and analyzed by western blotting using specific antibodies.
Immunofluorescence
Cells were grown on poly-L-lysine pre-coated coverslips that were sterilized under UV light for 30 minutes - 1 hour. Cells were rinsed in 1X phosphate-buffered saline (PBS) solution followed by fixation in ice-cold methanol at −20 °C for 10 m inutes. Coverslips were then washed 3 times with 1X PBS before being blocked for 30 minutes in 1% BSA in 1X PBS. Primary antibody was diluted 1/200 in 1% BSA 1X PBS and incubated for 1 hour at room temperature in a humidified chamber. Slides were subsequently rinsed 3 times and washed 2 times for 15 minutes with 1% BSA in 1X PBS at room temperature before incubation with secondary antibody, diluted 1/200 in 1% BSA in 1X PBS, in a dark humidified chamber for 30 minutes at room temperature.
Coverslips were then rinsed 3 times and washed 3 times for 15 minutes with 1% BSA in 1X PBS in the dark before being rinsed 3 times with ddH2O. Coverslips were mounted using mounting medium containing DAPI. Image acquisition was carried out on a Nikon Eclipse Ti microscope (Nikon), using NIS-Elements AR software.
Determination of relative m6Am, Am, and m6A levels by thin layer chromatography
Levels of internal m6A in mRNA were determined by 2D-TLC essentially as previously described (Zhong et al., 2008). In brief, poly(A) RNA (100 ng) was digested with 2 units ribonuclease T1 (ThermoFisher Scientific) for 2h at 37°C in the presence of RNasin RNase Inhibitor (Promega). T1 cuts after every guanosine and exposes the 5′-hydroxyl of the following nucleotide, which can be A, C, U, or m6A. Thus, this method quantifies m6A in a GA sequence context. 5′ ends were subsequently labeled with 10 units T4 PNK (NEB) and 0.4 mBq [Y-32P] ATP at 37C for 30 min followed by removal of the γ-phosphate of ATP by incubation with 10 units Apyrase (NEB) at 30°C for 30 min. After phenol-chloroform extraction and ethanol precipitation, RNA samples were resuspended in 10 μl of DEPC-H2O and digested to single nucleotides with 2 units of P1 nuclease (Sigma) for 1h at 60°C. 1 μl of the releas ed 5′ monophosphates from this digest were then analyzed by 2D-TLC on glass-backed PEI-cellulose plates (MerckMillipore) as described previously (Kruse et al., 2011).
The protocol to detect the m6Am:Am ratio was based on the protocol developed by Fray and colleagues (Kruse et al., 2011), with some modifications. Poly(A) RNA (1 μg) was used for the assay. 300ng of poly(A) RNA was decapped with 15 units of RppH (NEB) for 3 h at 37°C. 5′ monophosphates in the resulting RNA were removed by addition of 5 units of rSAP phosphatase (NEB) for 1 h at 37°C. Up to this point, all enzymatic reactions were performed in the presence of SUPERase In RNase Inhibitor (ThermoFisher Scientific). After phenol-chloroform extraction and ethanol precipitation, RNA samples were resuspended in 10 μl of DEPC-H2O and 5′ ends were labeled using 30 units T4 PNK and 0.8 mBq [Y-32P] ATP at 37°C for 30 min. PNK was heat inactivated at 65°C for 20 min and the reaction was passed through a P-30 spin column (Bio-Rad) to remove unincorporated isotope. 8 μl of labeled RNA were then digested with 2 units of P1 nuclease (Sigma) for 1 h at 60°C. 2 μl of the released 5′ monophosphates from this digest were then analyzed by 2D-TLC on glass-backed PEI-cellulose plates (MerckMillipore) as described previously (Kruse et al., 2011).
Signal acquisition was carried out using a storage phosphor screen (GE Healthcare Life Sciences) at 200 μm resolution and ImageQuantTL software (GE Healthcare Life Sciences). Quantification was carried out with ImageJ (V2.0.0-rc-24/1.49m). For m6Am experiments, the m6Am:Am ratio was calculated. The use of this ratio has been described previously (Kruse et al., 2011). We confirmed that this assay is linear by spotting twice the sample material and confirming that the signal intensity doubles for the unmodified nucleotides (A, C, and U). Furthermore, exposure time of the TLC plates to the phosphor screen was chosen so the signal was not saturated. For m6A quantification, m6A was calculated as a percent of the total of the A, C, and U spots, as described previously (Jia et al., 2011). The use of relative ratios for each individual sample is important since it reduces the error derived from possible differences in loading. To minimize the effects of culturing conditions on the measured m6Am:Am ratios of each experimental group (e.g. control vs. knockout), all replicates were processed in parallel to minimize any source of variability between samples being compared.
miCLIP
Total RNA from wild-type and PCIF1 knockout HEK293T cells, and wild type mouse embryonic fibroblasts, was extracted using TRIzol following the manufacturer’s protocol. Any contaminating genomic DNA was degraded using DNase I and poly(A) RNA was isolated using two rounds of Dynabeads Oligo(dT) capture. 10 μg poly(A) RNA was then used as input for single nucleotide-resolution m6A mapping using the miCLIP protocol, as previously reported (Linder et al., 2015). Final libraries were amplified and subjected to 50-cycle paired-end sequencing on an Illumina HiSeq2500 at the Weill Cornell Medicine Epigenetic Core facility.
miCLIP bioinformatic analyses
The initial processing of raw FASTQ files was done as in the miCLIP protocol. Adapters and low quality nucleotides were first trimmed from paired reads using flexbar v2.5. The trimmed FASTQ file was then de-multiplexed using the pyBarcodeFilter.py script from the pyCRAC suite. The remainder of the random barcode was moved to the headers of the FASTQ reads using an awk script and PCR duplicates were removed using the pyCRAC pyDuplicateRemover.py script. Reads were aligned to hg38/mm10 using bwa v0.7.17 with the option “-n 0.06” as recommended in the CTK package. To identify m6A within the DRACH consensus, C to T transitions were extracted and the CIMS pipeline from the CTK package was used. Due to a high transition frequency in this dataset, putative m6A residues with an FDR<0.1 and in a DRACH consensus were used as the final list of m6A in this study.
To identify putative m6Am sites, coverage of wild type and PCIF1 knockout samples were compared genome-wide using the bamCompare tool from deeptools v3.1.3. In short, the genome was binned into 50 nt non-sliding windows and the coverage of reads in each was counted for each strand, discarding zero-coverage bins. This was normalized to the total number of reads in bins, per million (BPM) and the log2 ratio of BPM+1 for wild type to PCIF1 knockout was calculated. A log2 ratio threshold of 2 was chosen as the cutoff for each replicate. Adjacent bins passing threshold were merged using bedtools v2.27.1. The intersection of putative m6Am regions across replicates was taken using bedtools intersect, resulting in 2360 high-confidence m6Am peaks. To determine the precise m6Am nucleotide within these peaks, a combination of A to T transitions and a read pileup/drop-off method was used. In PCIF1-dependent peaks with an A to T transition occurring at a frequency of 10% or greater, this A was selected as the m6Am. For the remainder, a pileup/drop-off approach similar to the previous miCLIP criteria (Linder et al., 2015) was utilized. Here, the start nucleotide of each read (with respect to strand, i.e. the leftmost coordinate for + strand features and rightmost for – strand features) was extracted and piled up using the tag2cluster.pl script of the CTK package with the options “-s -v -maxgap −1”. Clusters of less than 5 reads were discarded, as were those that did not map to an A. When there was a single A-cluster in a PCIF1-dependent peak, this was selected as m6Am. When more than one occurred, the most piled-up cluster of the two closest to the beginning of the peak (with respect to strand) was selected.
To generate metagenes, MetaPlotR (Olarerin-George and Jaffrey, 2017) was used. In all cases, the longest GENCODE transcript isoform for each gene was selected. For metaplots centered on reference annotations, the closest m6Am to each feature was measured using bedtools closest and these distances were plotted as a histogram. Aligned reads in bigwig format and BED files with coordinates for m6A, m6Am, and CAGE peaks were used to generate genome tracks using pyGenomeTracks v1.0. Motif searches were performed using either DREME v5.0.2 or MEME v5.0.2. For functional annotation analyses of m6Am and 5′UTR m6A genes, DAVID v6.8 was used specifying a background of all genes covered with at least 20 reads.
Transcript 5′ end cloning
To validate called m6Am as transcription start nucleotides, an adapted 5′ RACE method was used. 10 μg poly(A) RNA was treated with Terminator exonuclease (Epicentre) and then CIP and rSAP (NEB) to remove 5′ -monophosphate RNA following the manufacturer’s recommendations. Half of this was then decapped using RppH (NEB) at a final concentration of 5 U per 100 ng RNA in 1X Thermopol buffer (NEB). The remaining half was incubated without RppH as a capped control, to control for any residual 5′ -monophosphates due to RNA degradation. 75 pmol of a biotinylated adapter (biotin-GTTCAGAGTTCTACAGTCCGACGATC) was then ligated onto the 5′ end of the processed RNA using T4 RNA ligase 1 (NEB) in a 30 μl reaction containing 1X T4 RNA ligase buffer, 1 mM ATP, and 20% PEG-8000 for 3 hours at 23°C. This was then diluted to 200 μl in streptavid in bead wash buffer (SA wash; 20 mM Tris-HCl pH 7.5, 0.5 M NaCl, 1 mM EDTA) and incubated with 0.5 mg hydrophilic streptavidin magnetic beads (NEB) for 15 minutes and room temperature. Beads were washed twice in SA wash buffer, then twice in annealing buffer (10 mM Tris-HCl pH 7.5, 50 mM NaCl, 1 mM EDTA). 12.5 pmol of each gene-specific reverse transcription primers (STAR Methods) were annealed to RNA in 150 μl annealing buffer by heating to 90°C for 5 minutes and cooled to room temperate over 20 minutes. Beads were then resuspended in a 50 μl Superscript III reverse transcription reaction (ThermoFisher Scientific) according to the manufacturer’s protocol. Beads were washed and resuspended in a 50 μl RNase H (NEB) reaction and incubated for 30 minutes at 37°C. All reactions on beads were performed in a thermoshaker (15 seconds on 1100 RPM, 30 seconds off) to ensure beads remained in suspension. 100 μl wash buffer was added, heated at 75°C for 2 minutes, placed on bead s and supernatant containing eluted cDNA immediately transferred to a fresh tube. The beads were resuspended in 50 μl and the elution repeated. Following an ethanol precipitation, 5% of the cDNA was used in 20 μl PCR reactions containing 1X Phusion master mix (NEB), 60% DMSO, 250 nM adapter primer, and 250 nM gene-specific primer (see STAR Methods for primer sequences). 5 μl of this was then loaded on a 6% TBE-PAGE gel and visualized. Bands of the correct size that were absent in the capped control were then identified as m6Am starting transcripts.
SLAM-seq
SLAM-seq was performed as described previously (Herzog et al., 2017) with minor modifications. HEK293T (WT and PCIF1 KO) cells (at 60% confluency) were incubated with cell culture growth medium supplemented with 25 μM 4-thiouridine (s4U) for 24 h (pulse phase). s4U incorporation was confirmed by HPLC analysis, as previously described (Herzog et al., 2017). The uridine chase was initiated by changing media containing 2.5 mM uridine (Sigma) and cells were collected for RNA extraction after 6 and 12 h. The 0 h sample were the cells that have completed the pulse with s4U, but without uridine-chase. Total RNA was extracted using RNAzol reagent (MRC) according to the manufacturer’s instructions, maintaining reducing conditions to prevent oxidation of s4U (0.1 mM DTT final concentration). For thiol alkylation, a master mix (10 mM iodoacetamide, 50 mM NaPO4 pH 8 and 50% DMSO) was prepared, centrifuged, and added to 20 μg of total RNA at 50°C for 15 min and then purified by ethanol precipitation. After that, two rounds of poly(A) mRNA enrichment was carried out with oligo d(T)25 Magnetic Beads (NEB). Standard RNA-seq libraries were prepared using NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB) following the instructions of the manufacturer. Sequencing was performed on a HiSeq2500 (Illumina) with 50 nucleotide reads.
Real-time PCR assay to determine transcript stability
Wild-type or PCIF1 knockout HEK293T cells were transfected with either empty vector or wild-type or SPPG mutant PCIF1 vectors for 48 hours and then treated with 5 μM actinomycin D or vehicle (DMSO) for 8 hours. Total RNA was extracted using Trizol and 2 μg of this reverse transcribed using random hexamers and Superscript IV (ThermoFisher Scientific) according to the manufacturer’s protocol. RT-PCR was performed in 20 μl reactions containing 250 nM forward and reverse primers and iQ SYBR Green supermix (Bio-Rad) on an Eppendorf RealPlex2 RT-PCR machine. A delta cycle threshold (Ct) was calculated using the average Ct values across technical triplicates, by subtracting the geometric mean of two control genes (RPS28 and ACTB). A delta-delta Ct was then calculated by subtracting the vehicle control delta Ct value for each sample and untransformed to obtain relative abundances. Fold changes were tested for P<0.1 by Student’s t-test.
Ribosome profiling
Ribosome profiling was performed as described previously (McGlincy and Ingolia, 2017). In brief, wild-type and PCIF1 knockout HEK293T cells were grown to ~70% confluence, washed twice with ice cold PBS supplemented with 50 μg/ml of cycloheximide (CHX) and collected by scraping. After pelleting, cells were resuspended in 400 μl lysis buffer (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT and 100 μg/ml CHX) After incubation on ice for 10 min, lysate was triturated 5 times through a 25-gauge needle and then lysate was centrifuged at 20,000 x g for 10 min. 5 μl of lysate was flash frozen and saved as input. To generate ribosome-protected fragments the lysates (30 μg) were first mixed with 200 μl DEPC-H2O then incubated with 15 U RNase I for 45 min at room temperature. The reaction was stopped with 10 μl SUPERase*In RNase inhibitor. 0.9 ml of sucrose-supplemented polysome buffer was added to the digestion mixture and ultracentrifuged at 100,000 rpm, 4°C for 1 h. Pellets were resuspended in 300 μl of water and after phenol-chloroform extraction, precipitated with ethanol. The RNA was then run on a 15% 8 M urea TBE gel, stained with SYBR Gold, and a gel fragment between 17-34 nucleotides corresponding to ribosome-protected RNA was excised. RNA was eluted for 2 h at 37°C in 300 μl RNA extraction buffer (300 mM NaOAc pH 5.5, 1 mM EDTA, 0.25%v/v SDS) after crushing the gel fragment. RNA was ethanol precipitated and resuspended in 26 were resuspended inl water and treated with RiboZero Gold kit. Libraries from RNA-protected fragments were generated as previously described in the protocol (Linder et al., 2015). In brief, the RNA fragments were dephosphorylated with T4 PNK for 1 h at 37°C in dephosphorylation buffer (70 mM Tris, pH 6.5, 10 mM MgCl2, 1 mM DTT). The 3′ adaptor was ligated using T4 RNA Ligase 2, truncated K227Q ligase (New England BioLabs) for 3h at 22°C. Ligated sRNAs were purified by ethanol precipitation, and reverse transcribed using the primers complementary to the 3′ adaptor containing specific barcodes. After circularization with CircLigase II, cDNAs were relinearized by BamHI digestion and in the next step, PCR-amplified and subjected to Illumina HiSeq 2500 platform. Due to the similarity in size between ligated and unligated adapters, the libraries were gel purified.
RNA-Seq analysis was conducted using the ribosome profiling input material. Ribosomal RNAs were removed from the input RNA using the NEBNext rRNA Depletion Kit (NEB). Input RNA libraries were generated using the NEBNext Ultra Directional RNA library prep kit for Illumina (NEB). Libraries were sequenced using an Illumina HiSeq 2500 platform with 50 nt reads.
Ribosome footprint reads and corresponding RNA-Seq reads were processed essentially as described (Ingolia et al., 2012). Adaptors and short reads (<17nt) were trimmed using FLEXBAR v2.5, demultiplexed using pyBarcodeFilter.py (pyCRAC software). PCR duplicates were collapsed by pyFastqDuplicateRemover.py script. Ribosomal RNA reads were removed by STAR aligner38. Remaining reads were then aligned to the hg38 genome with STAR v2.5.2a in a splicing-aware manner and using UCSC refSeq as a transcript model database (version from June 02/2014 downloaded from Illumina iGenomes). Two mismatches were allowed and only unique alignments were reported. Aligned reads were then counted on transcript regions using custom R scripts considering only transcripts with annotated 5′ and 3′UTRs. Gene count tables generated from STAR were normalized using DESeq2 (R-Bioconductor). Translation efficiency was calculated using Riborex (Li et al., 2017), with pre-filtering for transcripts that had at least ten counted reads.
SLAM-seq bioinformatic analysis
Raw sequencing data were trimmed of adapter sequences and filtered of reads with uncalled bases and reads < 17 nucleotides in length using Flexbar. Duplicate reads were further removed using pyFastqDuplicateRemover.py script and remaining reads were aligned to the human genome (GrCh38) using the STAR aligner.
To identify T→C conversions, aligned reads were analyzed using Rsamtools Pileup (version 1.27.16). This program was used to determine the frequency of each of the four nucleotides present in mapped reads at every genomic position with read coverage. After summation of all nucleotide mapped to each transcript, we selected only those with at least 100 T→C conversions at time point 0 h. Additionally, to select for those transcripts with a longer half-life, transcripts were filtered for those with at least 50 T→C conversions at time point 6h. The mRNA half-life for each transcript was calculated based on the equation:
Statistics and software
P-values were calculated with a two-tailed unpaired Student’s t-test or, for the comparison of more than two groups, with a one- or two-way ANOVA followed by Bonferroni’s or Tukey’s post-test. Reproducibility of half-life and translation efficiency measurements was assessed by calculating the Spearman correlation coefficient between replicates. Significance of list overlaps was calculated using hypergeometric probability.
CONTACT FOR REAGENT AND RESOURCE SHARING
Please contact E.L.G. (Eric.Greer@childrens.harvard.edu) or S.R.J. (srj2003@med.cornell.edu) for reagents and resources generated in this study.
DATA AND SOFTWARE AVAILABILITY
The accession number for the RNA-sequencing and ribosome profiling data reported in this paper is NCBI GEO: GSE122948.
Reviewers may access these data deposited on GEO using the secure token qfmfaakyflkhvar.
Unprocessed and uncompressed imaging data is available at http://dx.doi.org/doi:10.17632/rnpfzjd7mj.1.
Supplementary Material
Table S1. Related to Figure 3. m6Am sites identified based on their dependence on PCIF1
Table S2. Related to Figure 3. m6A sites identified in the 5′ UTR
Table S3. Related to Figure 3. Functional annotations of cellular processes identified using DAVID of m6A and m6Am modified transcripts
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mouse anti-FLAG M2 | Sigma | F1804, RRID: AB_262044 |
| rabbit anti-PCIF1 | Abcam | ab205016, RRID: AB_2753142 |
| mouse anti-β actin | Sigma | A5441, RRID: AB_476744 |
| anti-eIF4E | Cell Signaling | 2067, RRID: AB_2097675 |
| anti-eIF4G | Cell Signaling | 2498, RRID: AB_2096025 |
| rabbit anti-GAPDH | Abcam | ab181602, RRID: AB_2630358 |
| mouse anti-KAP1 | Abcam | ab22553, RRID: AB_447151 |
| rabbit anti-EEF2 | Abcam | ab75748, RRID: AB_1310165 |
| mouse anti-RACK1 | Santa Cruz | B-3, RRID: AB_2247471 |
| mouse anti-HSP70/72 | Enzo Life Sciences | ADI-SPA-810-D, RRID: |
| rabbit anti-PARP1 | Cell Signaling | 9542, RRID: AB_10616513 |
| rabbit anti-HSPA8 | Cell Signaling | 8444, RRID: AB_10831837 |
| rabbit anti-m6A | Abcam | ab151230, RRID: AB_2753144 |
| rabbit anti-ATF5 | Abcam | ab60126, RRID: AB_940375 |
| rabbit anti-m6A | Abcam | ab151230. RRID:2753144 |
| Bacterial and Virus Strains | ||
| T7 Express lysY | New England Biolabs | C3010 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cycloheximide | Sigma Aldrich | C4859 |
| iodoacetamide | Sigma Aldrich | I1149 |
| 4-thiouridine (s4U) | Sigma Aldrich | T4509 |
| Actinomycin D | Sigma Aldrich | A1410 |
| Nuclease P1 | Sigma Aldrich | N8630 |
| Nuclease P1 | Wako USA | 145-08221 |
| RppH | New England Biolabs | M0356 |
| Fast AP | Thermo | EF0654 |
| T4 PNK | New England Biolabs | M0201 |
| rSAP | New England Biolabs | M0371 |
| Apyrase | New England Biolabs | M0398 |
| RNase I | Epicentre | N6901K |
| RNase T1 | Thermo | AM2283 |
| T4 ligase 2, truncated K227Q | New England BioLabs | M0351 |
| CircLigase™ II ssDNA Ligase | Lucigen | CL9021K |
| AccuPrime™ SuperMix I | Thermo | 12342010 |
| Superscript™ III Reverse Transcriptase | Thermo | 18080044 |
| S1 Nuclease | Thermo | EN 0321 |
| Terminator exonuclease | Epicentre | TER51020 |
| CIP | New England Biolabs | M0290 |
| T4 RNA ligase 1 | New England Biolabs | M0437 |
| RNase H | New England Biolabs | M0297 |
| Phusion Master Mix with HF buffer | New England Biolabs | M0531 |
| Actinomycin D | Sigma Aldrich | A1410 |
| SuperScript IV Reverse Transcriptase | Thermo | 18090010 |
| iQ SYBR Green supermix | Bio-Rad | 1708880 |
| Critical Commercial Assays | ||
| oligo d(T)25 Magnetic mRNA isolation kit | New England Biolabs | S1550 |
| Dynabeads oligo d(T)25 | Thermo | 61005 |
| Quant-iT™ RiboGreen™ RNA Assay Kit | Thermo | R11490 |
| Ribo-Zero Gold rRNA Removal Kit (Human, Mouse, Rat) | Illumina | MRZG12324 |
| NEBNext Ultra Directional RNA Library Prep Kit for Illumina | New England Biolabs | E7420 |
| Hydrophilic streptavidin magnetic beads | New England Biolabs | S1421 |
| Deposited Data | ||
| Raw and analyzed data | This Paper | GEO; GSE122948 |
| Unprocessed and uncompressed imaging data | This Paper | http://dx.doi.org/doi:10.17632/rnpfzjd7mj.1 |
| Experimental Models: Cell Lines | ||
| HEK293T | ATCC | CRL-3216 |
| HeLa | ATCC | CCL-2 |
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| m7GpppAmGUGGACUAACCACCAU | This Paper and Trilink | N/A |
| m7GpppAGUGGACUAACCACCAU | This Paper and Trilink | N/A |
| pppAmGUGGACUAACCACCAU | This Paper | N/A |
| sgRNAI; 5′- CGGUUGAAAGACUCCCGUGG-3′ | This Paper | N/A |
| sgRNA2; 5′- ACUUAACAUAUCCUGCGGGG-3′ | This Paper | N/A |
| Biotin 5’ adapter; biotin-GTTCAGAGTTCTACAGTCCGACGATC | This Paper | N/A |
| RT_ACOT7: cccgttctggctgttgcaatgc | This Paper | N/A |
| RT_BNIP2: agcctggattcaatgtcaactac | This Paper | N/A |
| RT_YOD1: cggctggacagcccctgcaaaac | This Paper | N/A |
| PCR_5’adapter: taatacgactcactatagggtctcGGATCCcgacGTTCAGAGTTCTACAGTCCGACGATC | This Paper | N/A |
| PCR_ACOT7: GTAAAACGACGGCCAGTaagaGAATTCggaacccgttctggctgttgcaatgc | This Paper | N/A |
| PCR_BNIP2: GTAAAACGACGGCCAGTaagaGAATTCggaaagcctggattcaatgtcaactac | This Paper | N/A |
| PCR_YOD1: GTAAAACGACGGCCAGTaagaGAATTCggaacggctggacagcccctgcaaaac | This Paper | N/A |
| qPCR_ACTB_Fw: GAAGATCAAGATCATTGCTCCTC | This Paper | N/A |
| qPCR_ACTB_Rv: ATCCACATCTGCTGGAAGG | This Paper | N/A |
| qPCR_RPS28_Fw: ATCAAGCTGGCTAGGGTAACC | This Paper | N/A |
| qPCR_RPS28_Rv: GGCCTTTGACATTTCGGATGA | This Paper | N/A |
| qPCR_AKAP12_Fw: CATTGTCACAGAGGTTGGA | This Paper | N/A |
| qPCR_AKAP12_Rv: GCTGACTTAGTAGCCATCTC | This Paper | N/A |
| qPCR_NBR1_Fw: GTCTAATACCCTGATGCTCCC | This Paper | N/A |
| qPCR_NBR1_Rv: GCAAATTCTCATCCACAAATGC | This Paper | N/A |
| Recombinant DNA | ||
| pSpCas9n(BB)-2A-Puro (PX459) V2.0 vector | Addgene | 62988 |
| pBABE-puro | Addgene | 1764 |
| pBABE-puro-PCIF1 WT | This Paper | N/A |
| pBABE-puro-PCIF1 SPPG | This Paper | N/A |
| pMD2.G | Addgene | 12259 |
| pUMVC | Addgene | 8449 |
| p3xFLAG-CMV-10 | Sigma | E7658 |
| p3xFLAG-CMV-10-PCIF1 WT | This Paper | N/A |
| p3xFLAG-CMV-10-PCIF1 SPPG | This Paper | N/A |
| Software and Algorithms | ||
| Prism 5 | Graphpad | N/A |
| MassHunter Suite | Agilent | N/A |
| NIS-Elements AR | Nikon | N/A |
| ImageQuantTL software | GE Healthcare | N/A |
| ImageJ (V2.0.0-rc-24/1.49m) | NIH | N/A |
| Flexbar v2.5 | Dodt, Roehr, Ahmed, and Dieterich, 2012 | https://github.com/seqan/flexbar |
| pyCRAC | Webb, Hector, Kudla, and Granneman, 2014 |
https://bitbucket.org/sgrann/pyc rac |
| bwa v0.7.17 | Li and Durbin, 2009 | http://bio-bwa.sourceforge.net/ |
| CTK v1.1.3 | Shah et al., 2017 | https://github.com/chaolinzhanglab/ctk |
| deeptools v3.1.3 | Ramírez et al., 2016 | https://deeptools.readthedocs.io/en/develop/ |
| bedtools v2.27.1 | Quinlan and Hall, 2010 | https://bedtools.readthedocs.io/en/latest/ |
| MetaPlotR | Olarerin-George and Jaffrey, 2017 | https://github.com/olarerin/metaPlotR |
| DREME/MEME v5.0.2 | Bailey et al., 2009 | http://meme-suite.org/ |
| Other | ||
Highlights.
PCIF1 is the N6-adenosine methylase that produces m6Am in an m7G cap-dependent manner
PCIF1 depletion allows transcriptome-wide mapping of m6A and m6Am
m6Am mapping identifies alternative “internal” transcription start sites
m6Am increases stability of a subset of mRNAs and has no effect on translation
ACKNOWLEDGMENTS
We thank J. Lipton for reagents and A. O. Olarerin-George for assistance with data analysis. This work was supported by NCN 2017/24/T/NZ1/00170 (D.T.S.), and NIH grants R00AG043550 and DP2AG055947 (E.L.G.), and R01DA037755 (S.R.J.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
S.R.J. is scientific founder, advisor to, and owns equity in Gotham Therapeutics.
REFERENCES
- Abugessaisa I, Noguchi S, Hasegawa A, Harshbarger J, Kondo A, Lizio M, Severin J, Carninci P, Kawaji H, and Kasukawa T (2017). FANTOM5 CAGE profiles of human and mouse reprocessed for GRCh38 and GRCm38 genome assemblies. Sci Data 4, 170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akichika S., Hirano S, Shichino Y, Suzuki T, Nishimasu H, Ishitani R, Sugita A, Hirose Y, Iwasaki S, Nureki O, et al. (2019). Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363, 6423, eaav0080, doi: 10.1126/science.aav0080. [DOI] [PubMed] [Google Scholar]
- Barral K, Sallamand C, Petzold C, Coutard B, Collet A, Thillier Y, Zimmermann J, Vasseur JJ, Canard B, Rohayem J, et al. (2013). Development of specific dengue virus 2′-O- and N7-methyltransferase assays for antiviral drug screening. Antiviral Res 99, 292–300. [DOI] [PubMed] [Google Scholar]
- Belanger F, Stepinski J, Darzynkiewicz E, and Pelletier J (2010). Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. J Biol Chem 285, 33037–33044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar JA, Shambaugh ME, Polayes D, Matera AG, and Rottman FM (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Chen D, and Patton JT (2001). Reverse transcriptase adds nontemplated nucleotides to cDNAs during 5′-RACE and primer extension. Biotechniques 30, 574–580, 582. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. [DOI] [PubMed] [Google Scholar]
- Fan H, Sakuraba K, Komuro A, Kato S, Harada F, and Hirose Y (2003). PCIF1, a novel human WW domain-containing protein, interacts with the phosphorylated RNA polymerase II. Biochem Biophys Res Commun 301, 378–385. [DOI] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, et al. (2018). GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y, LaFiandra A, and Shatkin AJ (1977). 5′-Terminal structure and mRNA stability. Nature 266, 235–239. [DOI] [PubMed] [Google Scholar]
- Guyot JB, Grassi J, Hahn U, and Guschlbauer W (1993). The role of the preserved sequences of Dam methylase. Nucleic Acids Res 21, 3183–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemeon I, Gutierrez JA, Ho MC, and Schramm VL (2011). Characterizing DNA methyltransferases with an ultrasensitive luciferase-linked continuous assay. Anal Chem 83, 4996–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog VA, Reichholf B, Neumann T, Rescheneder P, Bhat P, Burkard TR, Wlotzka W, von Haeseler A, Zuber J, and Ameres SL (2017). Thiol-linked alkylation of RNA to assess expression dynamics. Nat Methods 14, 1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Iwamoto Y, Sakuraba K, Yunokuchi I, Harada F, and Ohkuma Y (2008). Human phosphorylated CTD-interacting protein, PCIF1, negatively modulates gene expression by RNA polymerase II. Biochem Biophys Res Commun 369, 449–455. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, and Weissman JS (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7, 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Zhang D, and Aravind L (2016). Adenine methylation in eukaryotes: Apprehending the complex evolutionary history and functional potential of an epigenetic modification. Bioessays 38, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7, 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith JM, Ensinger MJ, and Moss B (1978). HeLa cell RNA (2′-O-methyladenosine-N6-)-methyltransferase specific for the capped 5′-end of messenger RNA. J Biol Chem 253, 5033–5039. [PubMed] [Google Scholar]
- Kruse S, Zhong S, Bodi Z, Button J, Alcocer MJ, Hayes CJ, and Fray R (2011). A novel synthesis and detection method for cap-associated adenosine modifications in mouse mRNA. Sci Rep 1, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavergne T, Bertrand JR, Vasseur JJ, and Debart F (2008). A base-labile group for 2′-OH protection of ribonucleosides: a major challenge for RNA synthesis. Chemistry 14, 9135–9138. [DOI] [PubMed] [Google Scholar]
- Lavergne T, Janin M, Dupouy C, Vasseur JJ, and Debart F (2010). Chemical synthesis of RNA with base-labile 2′-O-(pivaloyloxymethyl)-protected ribonucleoside phosphoramidites. Curr Protoc Nucleic Acid Chem Chapter 3, Unit 3 19. [DOI] [PubMed] [Google Scholar]
- Li F, Kennedy S, Hajian T, Gibson E, Seitova A, Xu C, Artrsmith CH, and Vedadi M (2016). A Radioactivity-Based Assay for Screening Human m6A-RNA Methyltransferase, METTL3-METTL14 Complex, and Demethylase ALKBH5. J Biomol Screen 21, 290–297. [DOI] [PubMed] [Google Scholar]
- Li W, Wang W, Uren PJ, Penalva LOF, and Smith AD (2017). Riborex: fast and flexible identification of differential translation from Ribo-seq data. Bioinformatics 33, 1735–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dai J, Song M, Fitzgerald-Bocarsly P, and Kiledjian M (2012). Dcp2 decapping protein modulates mRNA stability of the critical interferon regulatory factor (IRF) IRF-7. Mol Cell Biol 32, 1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Song M, and Kiledjian M (2011). Differential utilization of decapping enzymes in mammalian mRNA decay pathways. RNA 17, 419–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, and Jaffrey SR (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 12, 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden BE, Corbett ME, Heeney PA, Pugh K, and Ajuh PM (1995). Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie 77, 22–29. [DOI] [PubMed] [Google Scholar]
- Martin SA, and Moss B (1976). mRNA guanylyltransferase and mRNA (guanine-7-)-methyltransferase from vaccinia virions. Donor and acceptor substrate specificites. J Biol Chem 251, 7313–7321. [PubMed] [Google Scholar]
- Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, et al. (2017a). Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, et al. (2017b). Reversible methylation of m(6)Am in the 5′ cap controls mRNA stability. Nature 541, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy NJ, and Ingolia NT (2017). Transcriptome-wide measurement of translation by ribosome profiling. Methods 126, 112–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, and Jaffrey SR (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olarerin-George AO, and Jaffrey SR (2017). MetaPlotR: a Perl/R pipeline for plotting metagenes of nucleotide modifications and other transcriptomic sites. Bioinformatics 33, 1563–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RP, Kelley DE, Friderici K, and Rottman F (1975). The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5′ terminus. Cell 4, 387–394. [DOI] [PubMed] [Google Scholar]
- Robertson AK, Geiman TM, Sankpal UT, Hager GL, and Robertson KD (2004). Effects of chromatin structure on the enzymatic and DNA binding functions of DNA methyltransferases DNMT1 and Dnmt3a in vitro. Biochem Biophys Res Commun 322, 110–118. [DOI] [PubMed] [Google Scholar]
- Shimotohno K, Kodama Y, Hashimoto J, and Miura KI (1977). Importance of 5′-terminal blocking structure to stabilize mRNA in eukaryotic protein synthesis. Proc Natl Acad Sci U S A 74, 2734–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S (2002). What messenger RNA capping tells us about eukaryotic evolution. Nature Rev Mol Cell Biol 3, 619–625. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Morgan MA, Merrick WC, and Shatkin AJ (1978). A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc Natl Acad Sci U S A 75, 4843–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, and Shatkin AJ (1977). Reovirus mRNA can be covalently crosslinked via the 5′ cap to proteins in initiation complexes. Proc Natl Acad Sci U S A 74, 4288–4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Zhang M, Li K, Bai D, and Yi C (2019). Cap-specific, terminal N(6)-methylation by a mammalian m(6)Am methyltransferase. Cell Res 29, 80–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thillier Y, Decroly E, Morvan F, Canard B, Vasseur JJ, and Debart F (2012). Synthesis of 5′ cap-0 and cap-1 RNAs using solid-phase chemistry coupled with enzymatic methylation by human (guanine-N(7))-methyl transferase. RNA 18, 856–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, et al. (2017). Defining a Cancer Dependency Map. Cell 170, 564–576 e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem KM, and Wek RC (2004). Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A 101, 11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Gershowitz A, and Moss B (1975). N6, O2’-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature 257, 251–253. [DOI] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, and Moss B (1976). 5′-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry 15, 397–401. [DOI] [PubMed] [Google Scholar]
- Willcock DF, Dryden DT, and Murray NE (1994). A mutational analysis of the two motifs common to adenine methyltransferases. The EMBO journal 13, 3902–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Bolotin E, Jiang T, Sladek FM, and Martinez E (2007). Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene 389, 52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, and Zhang B (2015). A comprehensive evaluation of ensembl, RefSeq, and UCSC annotations in the context of RNA-seq read mapping and gene quantification. BMC Genomics 16, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, and Fray RG (2008). MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20, 1278–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Shu XE, Mao Y, Liu XM, Yuan X, Zhang X, Hess ME, Bruning JC, and Qian SB (2018). N(6)-Methyladenosine Guides mRNA Alternative Translation during Integrated Stress Response. Mol Cell 69, 636–647 e637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatev I, Lavergne T, Debart F, Vasseur JJ, Manoharan M, and Morvan F (2010). Efficient solid-phase chemical synthesis of 5′-triphosphates of DNA, RNA, and their analogues. Org Lett 12, 2190–2193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Related to Figure 3. m6Am sites identified based on their dependence on PCIF1
Table S2. Related to Figure 3. m6A sites identified in the 5′ UTR
Table S3. Related to Figure 3. Functional annotations of cellular processes identified using DAVID of m6A and m6Am modified transcripts
Data Availability Statement
The accession number for the RNA-sequencing and ribosome profiling data reported in this paper is NCBI GEO: GSE122948.
Reviewers may access these data deposited on GEO using the secure token qfmfaakyflkhvar.
Unprocessed and uncompressed imaging data is available at http://dx.doi.org/doi:10.17632/rnpfzjd7mj.1.


