Abstract
Two-dimensional monolayer cell cultures are routinely utilized for preclinical cancer drug screening, but the results often do not translate well when drugs are tested in vivo. To address this limitation, a biocompatible chitosan-PEG hydrogel (CSPG gel) was synthesized to create a gel that can be easily dispensed into 96-well plates at room temperature and neutral pH. The stiffness of this gel was tailored to be within the stiffness range of human glioblastoma tissue to promote the formation of tumor spheroids. Differences in cell morphology, proliferation rate, and dose-dependent drug cytotoxicity were compared among cell spheroids grown on CSPG gels, cells in monolayer culture on tissue culture polystyrene and cells cultured on Matrigel. Tumor spheroids on CSPG gels displayed statistically significantly greater resistance to chemotherapeutics than in the conditions where cells did not form spheroids. Gene expression analysis suggests that resistance of cells on CSPG gels to the therapy may be partially attributed to upregulation of ATP-binding cassette transporters and downregulation of DNA mismatch repair genes, which was stimulated by spheroid formation. These findings suggest CSPG gel generates tumor spheroids that better reflect the malignant behavior of GBM and provides a cost-effective substrate for preclinical, high-throughput screening of potential cancer therapeutics.
Keywords: chitosan hydrogel, genipin, tumor spheroids, high-throughput drug screening, HTS
1. Introduction
Recently high-throughput screening (HTS) technologies have accelerated the discovery and optimization of chemotherapeutics.[1,2] Yet a major limitation of HTS systems is that screening results often do not translate well when the targeted drugs are tested using pre-clinical animal models. A significant challenge in the design of effective platforms for high-throughput, cell-based assays is that the cell culture substrates rarely, if ever, present a physiologically relevant environment to model the tumor microenvironment. Preliminary screens performed in conventional, two-dimensional (2D) culture systems are convenient, but fail to reflect critical microenvironmental cues found in vivo. This contributes to poorly predictive drug response in animal models and humans.[2,3] Three-dimensional (3D) culture platforms are advantageous for cancer cell line cultures as they promote more extensive cell-cell interactions and cell-extracellular matrix (ECM) interactions. Specifically, 3D multicellular tumor spheroid models recapitulate several important characteristics of tumors, including growth kinetics, cellular heterogeneity, signaling pathway activity, and gene expression.[4,5] These features represent more relevant microenvironments and cell behaviors that are expected to provide more predictive screening results.[3,4,6,7]
Various tumor spheroid culture-based screening platforms utilize commercially available hydrogel substrates including Matrigel™, Cultrex™, HyStem™, Geltrex™, etc. [8–10] Some of these are composed of basement membrane extracts making them impractical for HTS assays due to their complex, undefined nature and prohibitive cost. [8,9,11] Naturally-derived ECM constituents such as collagen and gelatin are animal-derived proteins with inconsistencies in stability and antigenicity or immunogenicity[12,13] and hyaluronic acid is very costly.[14–17] Many tumor spheroid-based platforms associated with the aforementioned materials present promising results, but most are difficult to automate.[18–20]
Here we report the development of a 3D cell culture gel matrix composed of a polyethylene glycol (PEG)-modified chitosan hydrogel crosslinked with genipin. Chitosan is widely used in many biomedical applications because of its excellent biocompatibility and the similarity of its structure to glycosaminoglycans (GAGs), which present cell-binding sites. Yet its applicability as a 3D cell culture matrix for HTS platforms has been limited due to the low solubility of high molecular weight chitosan in neutral, aqueous solvents. Previously, we presented a neutral, water-soluble chitosan-PEG hydrogel with stiffness in the range of 1 to tens of Pascals. [21,22] However, the stiffness of malignant tissues is generally thousands of Pascals and matrix stiffness is critical to regulating cell behavior.[23–25] The gels with stiffness that mimics the stiffness of physiologically relevant matrices are commonly obtained by either covalent crosslinking or noncovalent bonding of supramolecular assembly.[26,27] Here, the physicochemical properties of the chitosan gel were optimized by crosslinking with genipin. Genipin is considered the most biocompatible, natural crosslinking agent for naturally-derived polymers, yielding complexes with no cytotoxicity to human and animal cells.[28] This chitosan-PEG solution can be easily dispensed at room temperature into arrays of wells after mixing with genipin, which is especially suitable for large-scale automated liquid handling systems, and thus high-throughput drug screening.
The chitosan-PEG-genipin (CSPG) gel was engineered to present similar mechanical properties to human glioblastoma tissue. Glioblastomas (GBM) are highly malignant, primary brain tumors, yet effective methods for screening potential drugs efficiently and effectively are lacking. [29–32] GBM tumor spheroids formed on CSPG gels can serve as a practical tumor model for HTS drug screening. The evolution of mechanical properties during gelation and the strength of the hydrogel network were monitored by rheological analysis. The viability of GBM spheroids on CSPG gels treated with chemotherapeutics temozolomide (TMZ) and carmustine (BCNU) was compared to cells grown in conventional 2D culture or Matrigel (the most widely used commercial gel) cultures. Gene expression analysis was performed on several markers associated with drug efflux and DNA repair, including ATP-binding cassette (ABC) transporters and DNA mismatch repair (MMR) systems to elucidate the source of chemotherapy resistance by glioblastoma cells. This work demonstrates that this engineered CSPG gel represents an economical, physiologically-relevant synthetic matrix that is useful for 3D cell-based assays in high-throughput drug screening systems.
2. Results and discussion
2.1. Hydrogel formation and rheological analysis
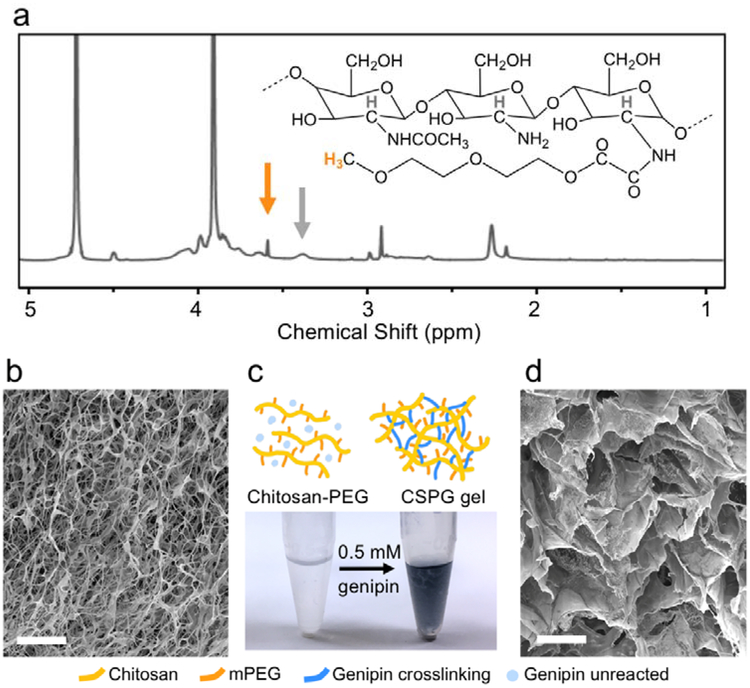
To generate a practical, 3D hydrogel cell culture matrix that can be utilized within multiwell plates for high-throughput drug screening, a dispensable gel solution is preferable to allow for efficient processing and is critical to automated HTS applications. To render chitosan soluble in a neutral, aqueous, injectable solution, chitosan was modified with mPEG. The degree of mPEG modification was characterized by 1H NMR spectroscopy as shown in Figure 1a and peak assignments are referenced from previous works.[21,22] By comparing the integration of the peaks from the methylene group at the end of mPEG (black arrow) and a hydrogen presenting on all chitosan backbone units (grey arrow), this analysis indicated 14 ± 0.4% of the chitosan monomers were grafted with mPEG. The sample was then lyophilized (Figure 1b) and reconstituted in PBS to form a dispensable, neutral solution making it amenable for cell culture. The hydrogel formed at this stage was relatively weak, therefore crosslinking was introduced by adding genipin, a naturally-derived, biocompatible crosslinker, which strengthened and stabilized the chitosan gels. The blended solutions were incubated overnight resulting in a crosslinked network. Gelation was characterized by the appearance of blue color (Figure 1c) due to a simultaneous side reaction that generates highly delocalized carbon double bonds resulting from genipin copolymer bridges.[33–35]
Figure 1.
Evaluation of hydrogel chemical structure and microstructure. (a) 1H NMR spectrum of chitosan-PEG in D2O at 50°C utilized to determine the percentage of chitosan monomers modified with mPEG. The peak at 3.6 ppm (orange arrow) indicates the methylene group on mPEG. The peak at 3.4 ppm (grey arrow) is associated with a hydrogen present on all chitosan monomers. (b) SEM reveals the microstructure of lyophilized chitosan-PEG samples as synthesized. Scale bar represents 20 μm. (c) The photo shows translucent reconstituted chitosan-PEG in 1× PBS versus the blue hydrogel (CSPG gel) formed after addition and mixing of 0.5 mM genipin and incubation at 37°C overnight. (d) The crosslinked hydrogel was lyophilized and examined under SEM. Scale bar represents 20 μm. The image shows significant structural changes compared to that in (b).
SEM images were taken to examine the structural changes in the lyophilized sample before and after crosslinking with genipin. Both structures were highly porous, but differed in architecture. Figure 1b shows the fine, fibrous microstructure of lyophilized, as-synthesized chitosan-PEG, while Figure 1d shows that the microstructure of genipin-crosslinked gel includes larger pores with interconnected walls.
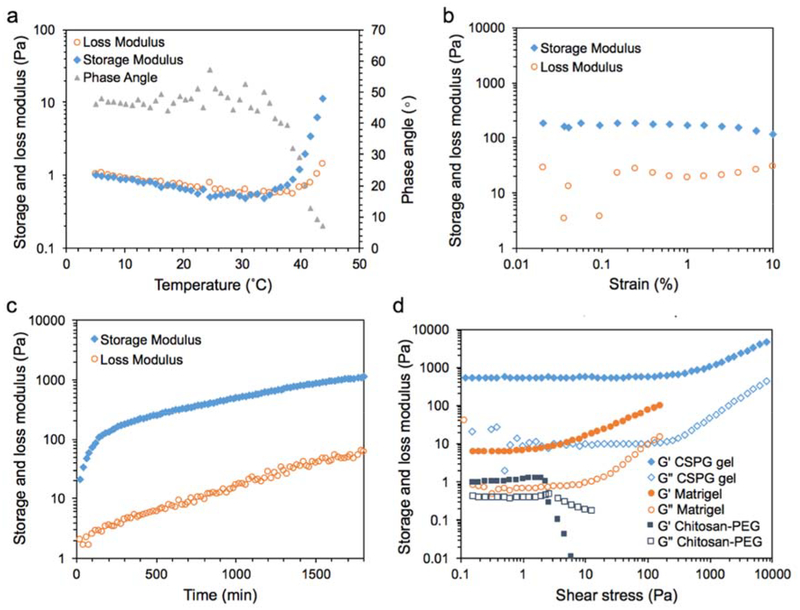
Rheological analyses were conducted on samples before and after crosslinking to understand changes in mechanical properties from a relatively soft gel to a stiffer gel (Figure 2). Prior to crosslinking, the chitosan-PEG solution possessed thermogelling properties.[21,36] The sol-gel transition was promoted with increasing temperature by disruption of hydrogen bonds between water and PEG, resulting in the domination of hydrophobic interactions from chitosan thus transforming the solution into a gel state. Figure 2a shows a typical thermogelling curve for chitosan-PEG where the sol-gel transition temperature was 32.7 ± 2.5°C.
Figure 2.
Rheological analysis of the chitosan gel. a) The storage modulus, loss modulus and phase angle changed with increasing temperature for uncrosslinked chitosan-PEG samples; the sol-gel transition was identified at 35°C. b) The strain sweep at 37°C and 1 Hz for CSPG gel showed that strain at 0.5% was within the lower end of LVR with less noise. c) The evolution of storage and loss moduli of 1.3 %w/v chitosan-PEG in the presence of 0.5 mM genipin while curing at 37°C. d) The storage (G’, filled symbols) and loss moduli (G”, empty symbols) were recorded under increasing shear stress for Matrigel, uncrosslinked chitosan-PEG and crosslinked CSPG gel at 37°C to compare the strength of different gels.
Next, we examined the rheological properties of crosslinked CSPG gel. The evolution of storage and loss moduli during gel formation helps explain the mechanical properties of the crosslinked hydrogel. In order to apply proper strain and frequency in time sweep experiments, gelled samples were first measured at increasing strain at 1 Hz and 37°C as shown in Figure 2b. Results show that strain at 0.2–5% lies in the linear viscoelastic regime (LVR) where the storage modulus is independent of the applied strain. Therefore, 0.5% strain at 1 Hz was chosen as the test condition to minimize damage to the gel as well as noise during the time sweep.
Figure 2c shows the typical time sweep profile of storage and loss moduli for freshly prepared samples. Unlike a few recently-reported chitosan-genipin hydrogel cell culture systems which require hundreds of minutes to gel,[34,37,38] the elastic property of CSPG gel was dominant at the beginning of the measurement period. The early gelation was due to the thermogelling property of chitosan-PEG, which is responsive within a minute at an elevated temperature at approximately 32°C as shown in Figure 2a. The storage modulus of the hydrogel developed from chitosan-PEG could be controlled within 1–10 Pa,[21,22,39] while the modulus of CSPG gel increased gradually with time from tens to almost 1000 Pa after 24 hr. The mechanical properties of the hydrogel changed from a relatively low stiffness value to a range that is relevant to glioblastoma tissue typically between 100–5000 Pa.[40,41]
The strength of the CSPG gel structure was further examined by stress sweep testing and compared with uncrosslinked chitosan-PEG hydrogel and Matrigel at 37°C (Figure 2d). In the case of the chitosan-PEG sample, within the LVR, the storage (G’, filled symbols) and loss moduli (G”, empty symbols) were independent of the applied stress up to approximately 2 Pa, which is also defined as yield/critical shear stress.[42] When the stress is higher than 3 Pa, the sample is subjected to significant structural destruction resulting in loss of elastic properties and the domination of viscous properties (the crossover of G’ and G” in Figure 2d for chitosan-PEG). With genipin crosslinking within the CSPG gel, the limit of the LVR was increased to approximately 100 Pa because of a higher degree of interconnection between polymer chains. These results show that crosslinking allowed the gel to maintain structural integrity against higher stresses as compared to the uncrosslinked samples.
The response of Matrigel to increasing shear stress was also shown in Figure 2d. The limit of LVR was similar to chitosan-PEG. At stresses above 2 Pa, the storage and loss moduli of Matrigel increased with stress, which was different from the transformation found with chitosan-PEG but similar to the tendency found in CSPG gel. This phenomenon may suggest that the deformation of Matrigel promotes interaction among the basement membrane components resulting in a more rigid network. However, the network broke down instantly at 160 Pa, resulting in the sudden boost of phase angel staying to 90°. The Matrigel lost its resistance to the loaded stress and deformation so no data points were collected above 160 Pa. A similar limitation was found for CSPG gel where the network broke down at approximately 7,000 Pa.
2.2. Proliferation and morphology of glioblastoma cells
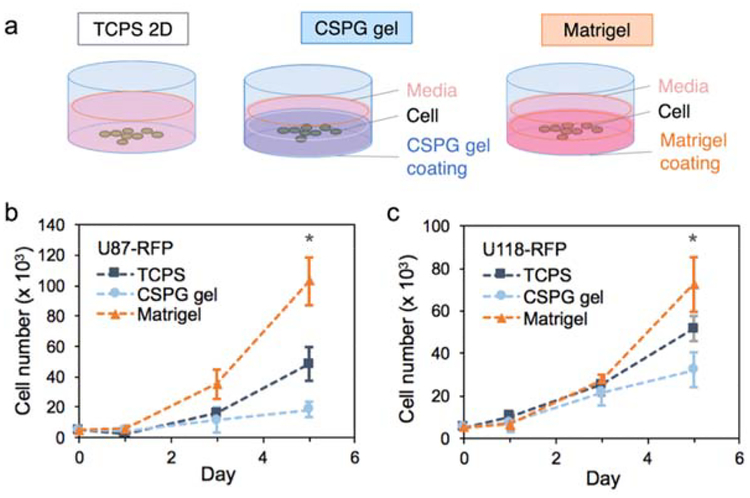
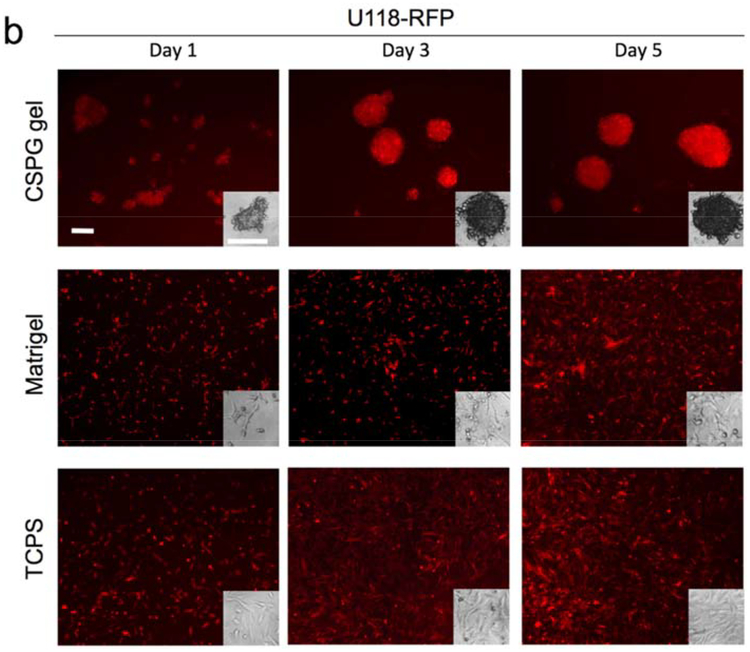
Multicellular tumor spheroid models are the most widely-used 3D tumor models in cancer research and here we examine if the CSPG gel culture platform promotes formation of glioblastoma multicellular spheroids. The initial attachment and subsequent proliferation of human glioblastoma cell lines were evaluated using an alamarBlue assay while the morphology was monitored using fluorescence imaging. Figure 3 shows the growth of U118-RFP and U87-RFP cells in TCPS, Matrigel, and CSPG gel, respectively, over 5 days. On day 1, cells attached to the surfaces of all substrates and the number of the cells remained similar or slightly higher than the number of cells seeded. After 5 days, regardless of the cell line, cells cultured on TCPS and on Matrigel had a much higher proliferation rate then cells cultured on the CSPG hydrogel.
Figure 3.
Effect of culture substrate on cell proliferation. a) 5,000 cells per well were seeded in 96-well plates at Day 0 on 2D TCPS, Matrigel, or CSPG gel. The number of cells was determined by the alamarBlue assay on Day 1, 3, and 5 for b) U87-RFP and (c) U118-RFP cell lines (n = 5). (* p < 0.05 compared to others)
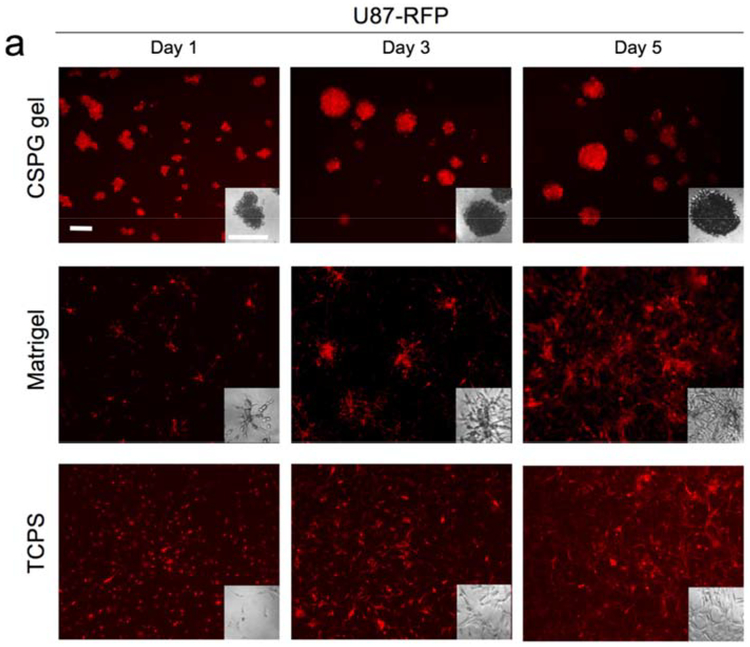
The relatively higher proliferation rate for cells cultured on Matrigel could be due to the presence of defined and undefined residual growth factors associated with the mouse Engelbreth-Holm-Swarm sarcoma source of Matri0.gel. The slower proliferation of cells on CSPG gel relative to cells on the other substrates may be better understood by examining cell morphology associated with each substrate (Figure 4). Cells attached on CSPG gel initially formed aggregates. The cells within the aggregates may have migrated toward each other and/or proliferated, resulting in cell spheroids with diameters much larger than that of the clusters associated with Matrigel culture. The cell clusters formed on Matrigel were not uniform and possessed a different morphology compared to typical spheroid cultures; some cells cultured on Matrigel had a polarized morphology similar to cells cultured on TCPS whereas the remaining cells formed small clusters of variable sizes with some elongated cells protruding from the clusters. CSPG hydrogels are thus a more consistent platform for generation of multicellular spheroids. Several studies reported similar findings of restricted cell proliferation within spheroids, due to physiological constraints, such as limited diffusion.[43–45]
Figure 4.
The morphology of cells a) U87-RFP and b) U118-RFP cultured on TCPS (2D), Matrigel, and CSPG gel. All scale bars represent 200 μm.
It also appears that the density of spheroids on CSPG gel increased over time with the spheroids appearing darker in phase contrast images and brighter in fluorescence images. Thus the tumor-like spheroids promote extensive cell-cell contact through the close proximity of neighboring cells. Intercellular interactions are essential to normal physiological and pathological processes in vivo and should be modeled, if possible, when using in vitro culture platforms for drug screening. Cell-cell communication modulated by proteins, including intercellular adhesion molecules, cadherins, gap junctions, and selectins, induces complex signal pathways that affect cell morphology, migration, differentiation, and sensitivity to environment assaults such as drug toxicity.[46] This high-density growth condition on CSPG gel is unlike those in the cell monolayer on TCPS or the seemingly few layers with undefined clusters on Matrigel, both of which showed relatively even cell distribution on the culture surfaces with fewer cell-cell contacts.
The CSPG gel-cell system mimics several key features of the in vivo glioblastoma environment in terms of composition and stiffness. Brain parenchyma ECM contains a large amount of proteoglycans, GAGs, hyaluronan, and tenascin, but lacks fibrous protein such as collagen and fibronectin,[47–49] resulting in the loss of gelatinous network with stiffness as low as hundreds of Pascals.[40,41] Chitosan has a proxy structure of GAGs, a major component of brain extracellular matrix (ECM). In terms of mechanical properties, the stiffness of the TCPS substrate exceeds 3 GPa, which is many orders of magnitude stiffer than brain tissue.[40] CSPG gel modulus (hundreds to thousands of Pascals) is designed to lie within the range of human brain and glioblastoma tissue stiffness. CSPG gel also displays cell-binding sites because of the similarity in chemical structure of chitosan to that of GAGs. Thus the CSPG gel matrix surrounding the cells likely promotes a morphology that was more amenable to cell-cell contact, leading to spheroid formation and proliferation within the spheroids. This leads to development of other important features of solid tumors, including the compact multicellular structure, enhanced cell-cell and cell-matrix interactions, and a nutrient/waste/oxygen gradient that more accurately represents the in vivo environment.
High-throughput screening is often utilized to efficiently determine drug candidate potency. HTS cell assays require a low-cost, simple and reliable cell culture platform that can be assessed in a reasonable amount of time. Conventional, 2D, monolayer culture is convenient, but fails to recapitulate several important aspects of the tumor microenvironment, such as cell-cell and cell-matrix interactions, physiologically-relevant nutrient and oxygen levels, and tissue-level architectural organization. Many proposed 3D culture systems possess some of these features, but require sophisticated preparation processes and 2–14 days of cell culture.[4] For example, Matrigel is typically mixed with cells to represent a 3D environment, but the process for generating cell-Matrigel cultures requires strict temperature control. In addition, the generally undefined basement membrane components, due to source material variability, may result in unpredictable cell proliferation rates. Here, we observed visible batch-to-batch variation and high cell proliferation rates.[50,51] On the other hand, CSPG gel provides a simpler, but better microenvironment to develop human glioblastoma spheroids in vitro for HTS systems.
2.3. Dose-dependent drug response
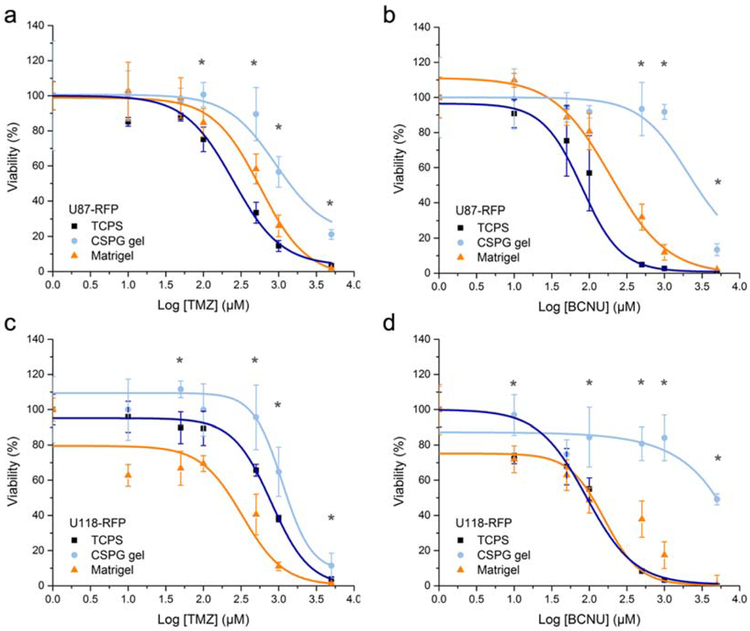
The The differences in cell proliferation rate and morphology for cell lines cultured with the different substrates indicate an impact of the culture substrate on cell behavior. Therefore, different cell responses to chemotherapeutic agents were anticipated and this was examined to further understand the impact of the culture microenvironment (Figure 5). Temozolomide (TMZ) represents a model chemotherapeutic because it is a component of the standard therapy regime for treatment of maglignant gliomas, which includes surgical resection and radiotherapy and/or chemotherapy.[52] A second representative chemotherapy drug, carmustine (BCNU), is also a commonly used treatment that is implanted (GLIADEL® Wafers) into the resection cavity for glioblastoma and malignant glioma. Table 1 summarizes the median lethal dose (LD50) of each drug with respct to cell line and culture condition. U87-RFP cells cultured using CSPG gel resulted in elevated cell viability at high drug concentrations as compared with those cultured on 2D TCPS and Matrigel (Figure 5a–b). In the case of U118-RFP cells, the Matrigel environment sensitized the cells to TMZ treatment compared to TCPS, whereas cells in CSPG gel culture demonstarted increased resistance to TMZ (Figure 5c). U118-RFP cell spheroids on CSPG gel showed significaly higher survival when exposed to BNCU relative to cells grown on TCPS and Matrigel, both of which presented similar viability (Figure 5d).
Figure 5.
a, b) Dose-dependent cytotoxicity of TMZ and BCNU on U87-RFP and c, d) U118-RFP cells cultured in different conditions. The viability was analyzed via alamarBlue assay 3 days post-treatment (n = 5) and shown as function of the dose. (* p < 0.05 for CSPG compared to either TCPs or Matrigel)
Table 1.
Estimated LD50 of TMZ and BCNU in treating human glioblastoma cell lines U87-RFP and U118-RFP cultured in 2D TCPS, Matrigel, and CSPG gel conditions. The values correspond to the 50% viability on the curves shown in Figure 5.
| Cell line | Drug | LD50 (μM) in each culture conditions | ||
|---|---|---|---|---|
| TCPS | Matrigel | CSPG gel | ||
| U87-RFP | TMZ | 260 | 535 | 1340 |
| BCNU | 80 | 230 | 2615 | |
| U118-RFP | TMZ | 745 | 235 | 1300 |
| BCNU | 90 | 110 | 4830 | |
Previous work by other researchers showed similar findings that cancer cells forming multicellular spheroids demonstrated greater chemoresistance.[4,18,19] However, matrix-based cutlure systems usaully takes 7–14 days to form tumor spheroids of ~100 μm in diameter.[18,54,55] Our gel system forms tumor spheroids of ~200 μm in diameter in 3–5 days. The tumor spheroid models were reported to introduce decreased drug penetration, cell cycle arrest, and altered gene expression induced by cell-cell interaction and cell-matrix adhesion, and have the oxygen and nutrient gradient constraints, which lead to increased. chemoresistance.[45,53] From the architectural point of view, in our CSPG gel system, the self-assembled, densely-packed spheroids (Figure 4) may establish biological barriers that limit the penetration of cytotoxic agents. More impotantly, the spheroid morphology induced by the CSPG gel microenvironment promoted significant cell-ECM and intercellular interactions that could alter the response to the treatment. Earlier studies showed that higly interactive spheroid models, recapitulating many features in vivo, may simulate signalling pathways and activate gene and protein expression thereby fostering treatment reistance.[4,18]
2.4. Expression of genes relevant to chemoresistance
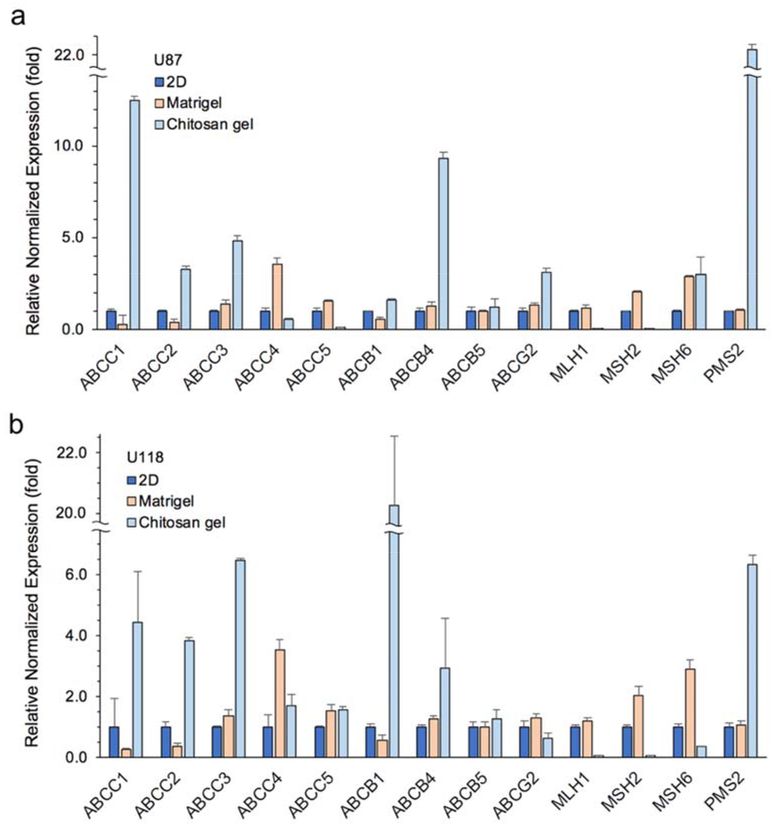
The glioblastoma cells cultured in each condition were characterized via quantitative real time PCR to investigate possible mechanisms of drug resistance developed in CSPG gel systems other than the barrier to drug diffusion created by the 3D, high density nature of the spheroids. There are multiple possible mechanisms for cancer cells to develop multidrug resistance (MDR), such as decreased drug uptake, increased drug efflux, activation of detoxifying systems, activation of DNA repair mechanisms, and evasion of drug-induced apoptosis.[56] qRT-PCR was used to examine the contribution of some of these factors. Figure 6 summarizes the relative expression of genes for each cell line associated with the ATP-binding cassette (ABC) transporter superfamily and DNA mismatch repair (MMR) system family. ABC transporters play a role in the ability of cells to expel cytotoxic molecules, while the MMR system is associated with cell apoptosis resulting from exposure to DNA-damaging agents. Figure 6a shows markedly higher expression of ABCC1, ABCC2, ABCC3, ABCB4, and ABCG2 for U87-RFP cells cultured on CSPG gel as compared to those in 2D culture, ranging from 3- to 12-fold upregulation. The MMR family genes, MLH1 and MSH2, were significantly downregulated for the CSPG gel condition, whereas MSH6 and PMS2 expression was upregulated. All ABC transporters examined for CSPG gel-cultured U118-RFP cells were upregulated with the exception of ABCG2, whereas all MMR markers were suppressed 2- to 16-fold with the exception of PMS2 (Figure 6b). Both cell lines in Matrigel condition showed upregulations on fewer ABC families and no downregulation was found in MMR systems compared to 2D controls. The GMB cells showed the mild resistance to TMZ found in Matrigel condition. These findings suggest that greater drug resistance by cells cultured on CSPG may be due to overexpression of several ABC transporters contributing to drug efflux, and the relative deficiency in MMR systems, leading to suppressed apoptosis, which should have been induced by genotoxic and cytotoxic chemotherapeutic agents like TMZ and BCNU.
Figure 6.
Relative expression of RNA content in a) U87-RFP and b) U118-PRF cells cultured on 3D CSPG gel, Matrigel and 2D TCPS. Gene expression was first normalized to GAPDH (reference gene) and then normalized to expression associated with 2D TCPS culture (set as 1-fold) (n = 3).
Both TMZ and BCNU are DNA alkylating drugs that cause alkylation lesions in genetic sequences. TMZ is a methylation agent; the breakdown of TMZ in the body delivers methyldiazonium cations, which methylates purine bases of DNA (O6-guanine; N7-guanine and N3-adenine).[57] BCNU is a chloroethylating agent, inducing cyclic N1O6ethanoguanine intermediates that eventually forms DNA interstrand crosslinks with the cytosine on the opposite strand.[58] Mismatch repair proteins, including MHS2, MHS6, MLH1, and PM2, recognize these repair mismatched bases.[59] For example, the carcinogenic O6-methylguanine mispair with thymine induced by TMZ, is recognized by the MMR proteins MSH2 and MSH6 (MutSα, a heterodimeric complex). Subsequently, MLH1 and PMS2 (MutLα complex) are recruited to excise the newly synthesized, yet miscoded strands; however, the miscoded template strand persists. The repeated attempts of MMR proteins to correct for mismatches in turn initiate futile MMR cycles of insertion and excision leading to cytogenetic changes, cell cycle arrest, and cell death.[60,61] Mismatch-repair deficient cells lose the sensitivity to the alkylation lesions via the MMR pathway, causing ineffective killing of tumor cells. Deficiency of MMR pathway could be characterized by one or more of the MMR proteins. It has been reported that knockdown of one or more MMR genes or proteins is a characteristic of glioblastoma recurrence following standard treatments, and frequently contributes to the development of therapy resistance.[62–64] Results in Figure 6 show significantly reduced expression up to 26-fold downregulation of MLH1 and MSH2 in CSPG gel culture. The CSPG gel-developed drug resistance to BCNU and TMZ may be due, in part, to the downregulation of these genes.
Mechanisms of drug resistance may also be attributed to ABC transmembrane protein transporters, such as ABCA, ABCB, ABCC, and ABCG transporters.[65] It is well recognized that MDR involves increased expression of ABC family genes where the transporters protect cells from endogenous or exogenous toxic molecules. The ABC transporter-mediated expulsion of drugs makes tumor cells resistant to the toxic effects of chemotherapeutics with various structures.[65] It was found that more than half of the ABC gene markers examined were upregulated in CSPG gel-cultured glioblastoma cells relative to 2D culture (Figure 6). The most significant increase in gene expression among the ABC transporters examined occurred for ABCC1 and ABCB1 for U87 and U118 cultures, respectively. It suggests that the heterogeneities in glioblastoma cell line established from different patients responded differently to the change in microenvironment via complex, undefined molecular mechanisms. These results, on some level, reflect some of the challenges faced when developing ABC inhibitor treatment.[66]
3. Conclusion
The CSPG gel developed, optimized and characterized in this study is an effective and chemically-defined chitosan-derived hydrogel system that promotes stable glioblastoma spheroid formation within 2–3 days of culture. This neutral, biocompatible, and genipincrosslinked chitosan hydrogel can be easily dispensed in multi-well plates making it highly applicable for high-throughput drug screening systems. The biophysical properties of the CSPG gel better mimic those of solid brain tumors than standard 2D TCPS substrates and Matrigel. The spheroids provide a more physiologically-relevant microenvironment in terms of cell morphology and organization by enhancing cell-cell and cell-ECM interactions. The glioblastoma spheroids on CSPG gels developed much higher resistance to the commonly used chemotherapeutics TMZ and BCNU as compared with Matrigel and 2D culture conditions. This resistance may be due to enhanced expression of ABC transporters promoting drug efflux and down-regulated expression of DNA mismatch repair genes which suppress cell death in tumor. This CSPG system is cost-effective for large-scale, high-throughput screening and could potentially be customizable, in terms of gel biophysical properties, by changing chitosan molecular weight, the degree of deacetylation, polymer concentration and/or degree of hydrogel crosslinking with genipin.
4. Experimental Section
4.1. Materials
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. Methoxypolyethylene glycol (mPEG, 750 Da), succinic anhydride, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), N-hydroxysuccinimide (NHS), and phosphate buffered saline (PBS) were used as received. Diethyl ether was purchased from J.T. Baker Chemical Company (Phillipsburg, NJ). Dulbecco’s modified Eagle media (DMEM), minimum essential medium (MEM), antibiotic-antimycotic (AA), Dulbecco’s phosphate-buffered saline (DPBS), and alamarBlue reagent were purchased from Invitrogen (Carlsbad, CA). Fetal bovine serum (FBS) was purchased from Atlanta Biologicals (Atlanta, GA). Growth factor-reduced Matrigel was purchased from Corning (Tewksbury, MA). iScript cDNA synthesis kit and iQ SYBR Green PCR Supermix were purchased from Bio-Rad (Hercules, CA).
4.2. Chitosan purification
Chitosan (85% deacetylated, medium molecular weight) was purified prior to modification. Chitosan powder was dissolved in 0.1 M acetic acid and filtered through 25 μm filter paper and centrifuged to discard insoluble impurities. The chitosan solution was dialyzed (MW 12,000–14,000 cutoff) in 0.1 M acetic acid and deionized (DI) water for three days. Chitosan precipitation was induced by adding 1M NH4OH solution to pH 9.0, collected by centrifugation, washed with DI water until pH 7.0 was reached, lyophilized and stored in −20°C for future use.
4.3. Synthesis and characterization of mPEG-chitosan
Chitosan Chitosan molecules modified with mPEG was prepared as described previously with minor modifications.[22,39,67] First, carboxylic acid-terminated mPEG (mPEG-acid) was synthesized. In brief, mPEG was dehydrated at 60°C under vacuum for 15 hr, followed by addition of three molar equivalents of succinic anhydride. The mixture was heated at 100°C for 30 minutes for dissolution of succinic anhydride. The reaction was heated under reflux at 120°C for 7 hr. After cooling to below 50°C, the product was mixed with diethyl ether in a separatory funnel and the top layer was collected following clear separation. The solution was mixed under vacuum at 50°C to remove the diethyl ether and pure mPEG-acid was obtained. The second part of the synthesis utilized amide bond coupling chemistry. 0.43 g mPEG-acid was reacted with 0.35 g purified chitosan dissolved in 30 mL 0.33% v/v acetic acid followed by addition of EDC (0.2 g) and NHS (0.12 g) in 10 mL DI water. The reaction was allowed to proceed for 4 h in a 45°C water bath. Following the reaction, 0.5 M NaOH solution was added to reach pH 7.0. The solution was dialyzed (MW 12,000–14,000 cutoff) six times against DI water and lyophilized.
The grafting efficiency of mPEG on chitosan was determined by 1H nuclear magnetic resonance spectroscopy (1H NMR, Bruker AV-500, MA) with the sample dissolved in 0.7 mL D2O with 7 μL acetic acid-d4. The % grafting was defined by the molar ratio of the methylene group at the end of mPEG and a hydrogen presenting on all chitosan backbone using the integral function in Topspin (Bruker, MA).[39,67]
Scanning electron microscopy (SEM, Model JSM-5600, JEOL Technics, Tokyo, Japan) was used to visualize lyophilized samples at 5 kV with spot size 3. The samples were mounted on double-sided carbon tape and sputter-coated with Au/Pd for 60 seconds at 9 mA.
4.4. Preparation and rheological analysis of crosslinked hydrogel
Dry chitosan-PEG was reconstituted in PBS at 1.3 % w/v to yield chitosan-mPEG hydrogel solutions. Genipin (Wako, Osaka, Japan) was dissolved in PBS at 21.5 mM and added to the chitosan-mPEG solution (0.5 mM genipin). The mixture was prepared immediately before use. The genipin concentration was optimized for spheroid formation in cell culture (Figure S1).
The viscoelastic properties and the modulus of uncrosslinked and crosslinked chitosan hydrogels were characterized using a stress-controlled rheometer (MCR 301, Anton Paar, Germany) with a cone and plate configuration of 24.982 mm diameter and 0.994° cone angle. The bottom plate was connected to a Peltier temperature controller and a circulating water bath. To prevent solvent evaporation during the measurements, a thin layer of light mineral oil and a customized solvent trap were applied.
First, the thermogelling properties of uncrosslinked chitosan-PEG solution were evaluated by measuring storage and loss moduli and phase angle with increasing temperature. Temperature was increased from 5°C to 45°C at 1°C/minute. Frequency (1 Hz) and strain (5%) were optimized previously[22] to conduct the measurement within the linear viscoelastic regime (LVR) with acceptable signal-to-noise ratio. The temperature at which a significant decrease of the phase angle by crossing 45° where the storage modulus and loss modulus cross over was determined as gelation temperature where the solution was transformed to a gel.
Secondly, the rheological properties of crosslinked chitosan-mPEG-genipin (CSPG) hydrogel were investigated. To acquire the proper strain for gelation measurements, CSPG gel was prepared 1 day prior to experiments for strain sweep conducted in a dynamic oscillatory mode with a constant 1 Hz frequency. Storage and loss moduli were recorded as a function of strain from 0.01% to 10% at 37°C. Strain at 0.5% and 1 Hz was chosen for subsequent time sweep measurements in a dynamic oscillatory mode where freshly blended chitosan-mPEG and genipin solutions were placed between plates within 3 minutes of mixing. Measurements began within 10 minutes of mixing. Storage and loss moduli were recorded as a function of time every 30 minutes for more than 24 h at 37°C to investigate the evolution during crosslinking formation.
The strength of the network of 50% Matrigel in DMEM, uncrosslinked CSPG gel and crosslinked CSPG gel were evaluated via stress sweep at 37°C and 1 Hz. Shear stress was applied on samples cured at 37°C after 24 hours for CSPG gel, and after 10 minutes for chitosan-PEG hydrogel and Matrigel, which for all conditions represents the time to ensure a stable gel. The storage modulus (G’) and loss modulus (G”) were recorded as a function of stress from 0.1–10,000 Pa.
4.5. Cell culture
U-118 MG and U-87 MG human glioblastoma cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA). U-118 MG and U-87 MG were previously transfected with red fluorescent protein (RFP) expression for visualization/imaging purposes.[68] For convenience, cell lines are abbreviated as U87-RFP and U118-RFP here. U118-RFP cells were maintained in DMEM with 10% FBS and 1%AA. U87-RFP cells were maintained in MEM with 10% FBS and 1%AA. All cell cultures were maintained in humidified incubator with 5% CO2 at 37°C.
4.6. Cell proliferation assay
The alamarBlue® assay was used to evaluate the cell proliferation rate on conventional 2D tissue culture polystyrene (TCPS), Matrigel, and CSPG hydrogel. Growth factor-reduced Matrigel was mixed with basal media at a 1:1 ratio on ice, dispensed into 96-well plates at 50 μL per well and incubated at 37°C for one hour. Freshly-mixed CSPG solution was dispensed into 96-well plates at 100 μL per well and incubated at 37°C for 24 hr. 200 μL of 1X PBS was added into each well and incubated overnight; before seeding, the wells were washed with 1× PBS one more time. Cells were seeded at 5,000 cells per well and cultured using previously described media formulations.
Cell proliferation was assessed by alamarBlue assay on Days 1, 3, and 5. Briefly, media containing 10% alamarBlue was added to each well, incubated with cells at 37°C for 2 h and then the reacted alamarBlue solution was transferred to black 96-well plates for measurement of fluorescence intensities using a microplate reader (Spectra Max M2, Molecular Devices, Union City, CA) at wavelength 560 nm for excitation and 590 nm for emission. A calibration curve (fluorescence intensity vs cell number) generated from a known number of cells reacting with the Alamar Blue was used to calculate the number of cells for each cell lines.
4.7. Dose-dependent drug response
The glioblastoma cell lines cultured on TCPS, Matrigel, and CSPG hydrogel for 2 days were treated with TMZ or BCNU, respectively. Drug concentrations were 0, 10, 50, 100, 500, 1000, 5000 μM for both drugs at n = 5 for all conditions. The viability of the cells was examined three days after the treatment using alamarBlue assay as described in previous section, relative to respective untreated conditions. The median lethal dose (LD50) of each drug was estimated via Dose-Respons Hill function curve fitting in OriginPro8. LD50 was defined by the dosage that caused death of 50% of the cell population. Note the almarBlue assay measures cell metabolic activity and thus a slight environment change would cause cell viability over or below 100%.
4.8. Quantitative real-time PCR (qRT-PCR)
qRT-PCR was performed on cDNA collected from cells grown in each condition after 5 days of culture to analyze relative gene expression. RNA was extracted from cells using the Qiagen RNeasy kit (Qiagen, Valencia, CA) following the manufacture’s protocol. cDNA was prepared using the iScript cDNA synthesis kit following the manufacturer’s instructions. SYBR Green PCR Mastermix was used for evaluating the amplification with a primer for each of the transcripts in a Bio-Rad CFX96 real-time PCR detection system. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the reference gene. Quantification was determined by the cycle number at threshold fluorescence intensity. Thermocycling for all samples was carried out in 10 μL solution containing 5 μL SYBR Mastermix, 300 nM primers (Integrated DNA Technologies, Coralville, IA), and cDNA at concentration 0.04–1 ng/μL optimized for each condition. The thermocycle was 95°C for 2 min, 40 cycles for denaturation at 95°C for 15 s, annealing at 58°C for 30 s, and extension at 72°C for 30 s. All qRT-PCR data was analyzed using the CFX Manager software (Bio-Rad). The primers are indicated in Table 2.
Table 2.
Primer sequences of the reference and target genes.
| Gene | Forward | Reverse |
|---|---|---|
| GAPDH | GGT GTG AAC CAT GAG AAG TAT GA | GAG TCC TTC CAC GAT ACC AAA G |
| ABCC1 | GCG CCC CCT GCA GGA ACA TT | ACA GCC GAG AGG GGG TCG TC |
| ABCC2 | GGT GGC TGT GGT GGG CAC TG | TCT TGC CAG CCA ACA GGC CG |
| ABCC3 | CAG AGA AGG TGC AGG TGA CA | CTA AAG CAG CAT AGA CGC CC |
| ABCC4 | ATG GTT CGC CGT GCG TCT GG | GGG TGG GCG CTT CCT GCA TT |
| ABCC5 | AGG GGC AAG AAA GAG AAG GTG AGG | GAG GGG GTC GTC CAG GAT GTA GAT |
| ABCB1 | CCC GCA TTC TGC CGA GCG TT | CCC AGC TGC CAG GCA CCA AA |
| ABCB4 | GGC CGG AAC AGT GCT CCT CG | ATG TTG GCC TCC TTG GCC GC |
| ABCB5 | CAG GCC CTC GAC AAA GCC CG | CCC GGC GCA CAC AAA GGC TA |
| ABCG2 | GTC GGT GTG CGA GTC AGG GC | CTT GCC TCC GCC TGT GGG TC |
| MLH1 | TTC GTG GCA GGG GTT ATT CG | GCC TCC CTC TTT AAC AAT CAC TT |
| MSH2 | GCT GGA AAT AAG GCA TCC AAG G | CAC CAA TGG AAG CTG ACA TAT CA |
| MSH6 | AGC TTA AAG GAT CAC GCC ATC | AAG CAC ACA ATA GGC TTT GCC |
| PMS2 | GAA GGT TGG AAC TCG ACT GAT | CGA ACA GGT AGT GTG GAA AA |
4.9. Statistical analysis
Data is represented as mean ± standard deviation. Statistical significance was evaluated with OriginPro8 software for CSPG gel condition versus TCPS and Matrigel conditions using an unpaired two-sample student’s t-test with p < 0.05 (*) considered as statistically significant.
Supplementary Material
Acknowledgement
The work was supported by National Institute of Health (NIH) grant R01CA172455 and Kyocera Endowment to Miqin Zhang. Sheeny Lan Levengood thanks the support from Ruth L. Kirschstein NIH Training grant T32CA138312. The authors acknowledge the use of the resources provided by Molecular Analysis Facility, Bindra Innovation Lab, and Pozzo research group at University of Washington.
References
- [1].Wilhelm S, Carter C, Lynch M, Lowinger T, Dumas J, Smith RA, Schwartz B, Simantov R, Kelley S, Nat Rev Drug Discov 2006, 5, 835. [DOI] [PubMed] [Google Scholar]
- [2].Macarron R, Banks MN, Bojanic D, Burns DJ, Cirovic DA, Garyantes T, Green DVS, Hertzberg RP, Janzen WP, Paslay JW, Schopfer U, Sittampalam GS, Nat Rev Drug Discov 2011, 10, 188. [DOI] [PubMed] [Google Scholar]
- [3].Gu L, Mooney DJ, Nature Reviews Cancer 2016, 16, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nath S, Devi GR, Pharmacology & Therapeutics 2016, 163, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Friedrich J, Seidel C, Ebner R, Kunz-Schughart LA, Nat Protoc 2009, 4, 309. [DOI] [PubMed] [Google Scholar]
- [6].Alemany-Ribes M, Semino CE, Advanced Drug Delivery Reviews 2014, 79–80, 40. [DOI] [PubMed] [Google Scholar]
- [7].Breslin S, O’Driscoll L, Drug Discov. Today 2013, 18, 240. [DOI] [PubMed] [Google Scholar]
- [8].Cox MC, Reese LM, Bickford LR, Verbridge SS, ACS Biomater. Sci. Eng 2015, 1, 877. [DOI] [PubMed] [Google Scholar]
- [9].Hayashi Y, Furue MK, Stem Cells Int 2016, 2016, 5380560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Engel BJ, Constantinou PE, Sablatura LK, Doty NJ, Carson DD, Farach-Carson MC, Harrington DA, Zarembinski TI, Advanced Healthcare Materials 2015, 4, 1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fridman R, Benton G, Aranoutova I, Kleinman HK, Bonfil RD, Nat Protoc 2012, 7, 1138. [DOI] [PubMed] [Google Scholar]
- [12].Gorgieva S, Kokol V, Collagen- vs. Gelatine-Based Biomaterials and Their Biocompatibility: Review and Perspectives, 2011. [Google Scholar]
- [13].Lynn AK, Yannas IV, Bonfield W, Journal of Biomedical Materials Research Part B: Applied Biomaterials 2004, 71B, 343. [DOI] [PubMed] [Google Scholar]
- [14].Infanger DW, Lynch ME, Fischbach C, Annu Rev Biomed Eng 2013, 15, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zeugolis DI, Paul RG, Attenburrow G, J. Biomed. Mater. Res 2008, 86, 892. [DOI] [PubMed] [Google Scholar]
- [16].Peng YY, Glattauer V, Ramshaw JAM, Werkmeister JA, J. Biomed. Mater. Res 2010, 93, 1235. [DOI] [PubMed] [Google Scholar]
- [17].Park S-H, Song T, Bae TS, Khang G, Choi BH, Park SR, Min B-H, Int. J. Precis. Eng. Manuf 2012, 13, 2059. [Google Scholar]
- [18].Sant S, Johnston PA, Drug Discov Today Technol 2017, 23, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gurski LA, Petrelli NJ, Jia X, Farach-Carson MC, Oncology Issues 2017. [Google Scholar]
- [20].Thoma CR, Zimmermann M, Agarkova I, Kelm JM, Krek W, Advanced Drug Delivery Reviews 2014, 69–70, 29. [DOI] [PubMed] [Google Scholar]
- [21].Tsao C-T, Hsiao MH, Zhang MY, Levengood SL, Zhang M, Macromol. Rapid Commun 2015, 36, 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chang F-C, Tsao C-T, Lin A, Zhang M, Levengood SL, Zhang M, Polymers 2016, 8, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nagelkerke A, Bussink J, Rowan AE, Span PN, Seminars in Cancer Biology 2015, 35, 62. [DOI] [PubMed] [Google Scholar]
- [24].Chin L, Xia Y, Discher DE, Janmey PA, Curr Opin Chem Eng 2016, 11, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pickup MW, Mouw JK, Weaver VM, EMBO Rep 2014, 15, 1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Xing R, Liu K, Jiao T, Zhang N, Ma K, Zhang R, Zou Q, Ma G, Yan X, Adv. Mater. Weinheim 2016, 28, 3669. [DOI] [PubMed] [Google Scholar]
- [27].Xing R, Yuan C, Li S, Song J, Li J, Yan X, Angewandte Chemie International Edition 2018, 57, 1537. [DOI] [PubMed] [Google Scholar]
- [28].Muzzarelli RAA, El Mehtedi M, Bottegoni C, Aquili A, Gigante A, Marine Drugs 2015, 13, 7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A, Acta Neuropathol 2015, 129, 829. [DOI] [PubMed] [Google Scholar]
- [30].Omuro A, DeAngelis LM, JAMA 2013, 310, 1842. [DOI] [PubMed] [Google Scholar]
- [31].Tanaka S, Louis DN, Curry WT, Batchelor TT, Dietrich J, Nat Rev Clin Oncol 2013, 10, 14. [DOI] [PubMed] [Google Scholar]
- [32].Thomas AA, Brennan CW, DeAngelis LM, Omuro AM, JAMA Neurol 2014, 71, 1437. [DOI] [PubMed] [Google Scholar]
- [33].Dimida S, Demitri C, De Benedictis VM, Scalera F, Gervaso F, Sannino A, J. Appl. Polym. Sci 2015, 132. [Google Scholar]
- [34].Delmar K, Bianco-Peled H, Carbohydrate Polymers 2015, 127, 28. [DOI] [PubMed] [Google Scholar]
- [35].Mi F-L, Shyu SS, Peng CK, Journal of Polymer Science Part A: Polymer Chemistry 2005, 43, 1985. [Google Scholar]
- [36].Bhattarai N, Gunn J, Zhang M, Advanced Drug Delivery Reviews 2010, 62, 83. [DOI] [PubMed] [Google Scholar]
- [37].Xu J, Strandman S, Zhu JXX, Barralet J, Cerruti M, Biomaterials 2015, 37, 395. [DOI] [PubMed] [Google Scholar]
- [38].Zhang W, Ren G, Xu H, Zhang J, Liu H, Mu S, Cai X, Wu T, J Polym Res 2016, 23, 156. [Google Scholar]
- [39].Tsao C-T, Kievit FM, Wang K, Erickson AE, Ellenbogen RG, Zhang M, Mol. Pharmaceutics 2014, 11, 2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rape A, Ananthanarayanan B, Kumar S, Advanced Drug Delivery Reviews 2014, 79–80, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zuidema JM, Rivet CJ, Gilbert RJ, Morrison FA, Journal of Biomedical Materials Research Part B: Applied Biomaterials 2014, 102, 1063. [DOI] [PubMed] [Google Scholar]
- [42].Moura MJ, Figueiredo MM, Gil MH, Biomacromolecules 2007, 8, 3823. [DOI] [PubMed] [Google Scholar]
- [43].Theodoraki MA, Rezende CO, Chantarasriwong O, Corben AD, Theodorakis EA, Alpaugh ML, Oncotarget 2015, 6, 21255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Vinci M, Gowan S, Boxall F, Patterson L, Zimmermann M, Court W, Lomas C, Mendiola M, Hardisson D, Eccles SA, BMC Biol 2012, 10, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gong X, Lin C, Cheng J, Su J, Zhao H, Liu T, Wen X, PLoS ONE 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Astashkina A, Mann B, Grainger DW, Pharmacology & Therapeutics 2012, 134, 82. [DOI] [PubMed] [Google Scholar]
- [47].Koh I, Cha J, Park J, Choi J, Kang S-G, Kim P, Sci. Rep 2018, 8, 4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gritsenko PG, Ilina O, Friedl P, The Journal of Pathology 2011, 226, 185. [DOI] [PubMed] [Google Scholar]
- [49].Manini I, Caponnetto F, Bartolini A, Ius T, Mariuzzi L, Di Loreto C, Beltrami A, Cesselli D, International Journal of Molecular Sciences 2018, Vol. 19, Page 147 2018, 19, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nguyen EH, Daly WT, Le NNT, Farnoodian M, Belair DG, Schwartz MP, Lebakken CS, Ananiev GE, Saghiri MA, Knudsen TB, Sheibani N, Murphy WL, Nat Biomed Eng 2017, 1, 0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cruz-Acuña R, García AJ, Matrix Biol 2017, 57–58, 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Weller M, van den Bent M, Hopkins K, Tonn JC, Stupp R, Falini A, Cohen-Jonathan-Moyal E, Frappaz D, Henriksson R, Balana C, Chinot O, Ram Z, Reifenberger G, Soffietti R, Wick W, EANO EAN, Lancet Oncol 2014, 15, E395. [DOI] [PubMed] [Google Scholar]
- [53].Wang K, Kievit FM, Erickson AE, Silber JR, Ellenbogen RG, Zhang M, Advanced Healthcare Materials 2016, DOI 10.1002/adhm.201600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Li Y, Kumacheva E, Sci Adv 2018, 4, eaas8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Gonçalves DPN, Rodriguez RD, Kurth T, Bray LJ, Binner M, Jungnickel C, Gür FN, Poser SW, Schmidt TL, Zahn DRT, Androutsellis-Theotokis A, Schlierf M, Werner C, Acta Biomaterialia 2017, 58, 12. [DOI] [PubMed] [Google Scholar]
- [56].Gillet J-P, Gottesman MM, in Multi-Drug Resistance in Cancer (Ed.: Zhou J), Humana Press, Totowa, NJ, 2009, pp. 47–76. [Google Scholar]
- [57].Zhang J, Stevens MFG, Bradshaw TD, Curr Mol Pharmacol 2012, 5, 102. [DOI] [PubMed] [Google Scholar]
- [58].Gerson SL, Journal of Clinical Oncology 2016, 20, 2388. [DOI] [PubMed] [Google Scholar]
- [59].Bouwman P, Jonkers J, Nature Reviews Cancer 2012, 12, 587. [DOI] [PubMed] [Google Scholar]
- [60].Johannessen T-CA, Bjerkvig R, Tysnes BB, Cancer Treat. Rev 2008, 34, 558. [DOI] [PubMed] [Google Scholar]
- [61].Drabløs F, Feyzi E, Aas PA, Vaagbø CB, Kavli B, Bratlie MS, Peña-Diaz J, Otterlei M, Slupphaug G, Krokan HE, DNA Repair (Amst.) 2004, 3, 1389. [DOI] [PubMed] [Google Scholar]
- [62].Begum R, Martin SA, DNA Repair (Amst.) 2016, 38, 135. [DOI] [PubMed] [Google Scholar]
- [63].Shinsato Y, Furukawa T, Yunoue S, Yonezawa H, Minami K, Nishizawa Y, Ikeda R, Kawahara K, Yamamoto M, Hirano H, Tokimura H, Arita K, Oncotarget 2013, 4, 2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Felsberg J, Thon N, Eigenbrod S, Hentschel B, Sabel MC, Westphal M, Schackert G, Kreth FW, Pietsch T, Löffler M, Weller M, Reifenberger G, Tonn JC, German Glioma Network, Int. J. Cancer 2011, 129, 659. [DOI] [PubMed] [Google Scholar]
- [65].Fletcher JI, Haber M, Henderson MJ, Norris MD, Nature Reviews Cancer 2010, 10, 147. [DOI] [PubMed] [Google Scholar]
- [66].Li W, Zhang H, Assaraf YG, Zhao K, Xu X, Xie J, Yang D-H, Chen Z-S, Drug Resist. Updat 2016, 27, 14. [DOI] [PubMed] [Google Scholar]
- [67].Chen S-H, Tsao C-T, Chang C-H, Lai Y-T, Wu M-F, Liu Z-W, Chuang C-N, Chou H-C, Wang C-K, Hsieh K-H, Macromol. Mater. Eng 2013, 298, 429. [Google Scholar]
- [68].Tsao C-T, Kievit FM, Ravanpay A, Erickson AE, Jensen MC, Ellenbogen RG, Zhang M, Biomacromolecules 2014, 15, 2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.