Beyond mutualism, viruses manipulate host plant immunity not only to benefit vector allies but also to suppress their non-allies.
Abstract
A parasite-infected host may promote performance of associated insect vectors; but possible parasite effects on nonvector insects have been largely unexplored. Here, we show that Begomovirus, the largest genus of plant viruses and transmitted exclusively by whitefly, reprogram plant immunity to promote the fitness of the vector and suppress performance of nonvector insects (i.e., cotton bollworm and aphid). Infected plants accumulated begomoviral βC1 proteins in the phloem where they were bound to the plant transcription factor WRKY20. This viral hijacking of WRKY20 spatiotemporally redeployed plant chemical immunity within the leaf and had the asymmetrical benefiting effects on the begomoviruses and its whitefly vectors while negatively affecting two nonvector competitors. This type of interaction between a parasite and two types of herbivores, i.e., vectors and nonvectors, occurs widely in various natural and agricultural ecosystems; thus, our results have broad implications for the ecological significance of parasite-vector-host tripartite interactions.
INTRODUCTION
In natural ecosystems, humans and plants simultaneously face combinations of biotic stresses, especially in the cases of arthropod-borne diseases of humans, animals, crops, and wildlife, caused by Plasmodium, Zika virus, cassava mosaic begomovirus, and other parasites (1, 2). These diseases have disrupted human society and ecosystems and have reduced agricultural productivity. However, arthropod-borne diseases rarely occur, owing to the hosts’ efficient immune systems. As both sessile organisms and the trophic base of most ecosystems, plants have evolved immune systems that involve multiple defensive traits and can affect the whole plant community, e.g., through production of hundreds of thousands of secondary or specialized metabolites (3). Some plant-derived volatile blends releasing into the plant community function as long-distance signals that mediate plant interactions, such as repellent olfactory cues that attract (e.g., by mimicking sex pheromones) or repel herbivorous insects (4, 5). Pathogens and pests that come into contact with the plant surface can be perceived by plant receptor systems. Subsequently, several immune responses are triggered, including the emissions of various volatile organic compounds (e.g., terpenoids), phytoalexin [e.g., glucosinolates (GSs)], phytohormones (e.g., ethylene), and toxic polypeptides (e.g., defensin and pathogenesis-related protein 1).
Tripartite interactions within and between pathogens, pests, and hosts of arthropod-borne diseases are of fundamental importance for understanding many biological invasions and emerging infectious diseases. Arthropod-borne viruses (arboviruses), including begomoviruses, have evolved as masters at redirecting and reprogramming defensive traits and other host processes (6). Accumulating evidences support the wide occurrence of vector manipulation by plant arboviruses, which can either directly or indirectly influence vector behaviors and/or host-herbivore interactions (7). Pathogens manipulate their vector to facilitate their own transmission and spread. Some arboviruses, e.g., begomovirus and bunyavirus, are capable of achieving indirect mutualistic relationships with vectors via their shared host plant (8, 9). These arboviruses manipulate the host plant to attract their corresponding vector insects by inhibiting phytohormone-mediated and terpenoid-based communication between the plant and the vector, in turn promoting pathogen transmission among host plants. Some vector-borne plant viruses benefit their vectors by nutritionally deterring or physiologically impairing other herbivore competitors (1, 10). These negative effects on other herbivores have implications for enhancing the competitive strength of vectors and the spread of pathogens. However, it is unclear how a virus discriminates host-mediated interactions with various herbivores to favor its vector and facilitate disease spread.
Begomovirus consists of more than 320 species and infects dicotyledonous plants. It is exclusively transmitted by the whitefly Bemisia tabaci (Gennadius) species complex and can be found in all over the world. Most monopartite begomoviruses are associated with β-satellite DNA. In Asia and Africa, cotton leaf curl virus, one species of the begomovirus genus, causes the major viral disease in cotton. Cotton leaf curl Multan virus (CLCuMuV) is an important monopartite begomovirus, associated with β-satellite, endemic to the Indian subcontinent. CLCuMuV recently invaded China via infected Malvaceae horticulture plants. It has infected cotton and since then rapidly established itself in the south of China during the past decade (11). We have previously shown behavioral mechanisms for the worldwide invasion of whitefly species and begomoviruses (12). The β-satellite of begomoviruses encodes the βC1 protein, which can reprogram cellular processes in the host plant and facilitate begomovirus infection and transmission (13). The βC1 is also a suppressor on host immunities, including transcriptional and posttranscriptional gene silencing, phytohormone signaling, and the terpenoid-mediated plant chemical repellence. The βC1 protein attenuates jasmonate (JA) signaling and achieves indirect begomovirus-whitefly mutualism by targeting and interfering with at least three host factors, two transcription factors AS1 (asymmetric leaves 1) and MYC2 and a ubiquitination ligase component SKP1 (S-phase kinase-associated protein 1) (8, 14, 15). Studies of plant gene expression induced by βC1-AS1, βC1-MYC2, or βC1-SKP1 interaction have focused on viral infection and effects on the whitefly vector. However, little work has yet explored how arboviruses such as begomoviruses affect nonvector insect herbivores.
Here, we report that begomoviruses alter plant immunity in ways that promote vector performance and inhibit nonvector herbivores. Infected plants accumulated begomoviral βC1 proteins in the phloem where they were bound to the plant transcription factor WRKY20, inhibiting nonvector insects but benefiting whiteflies. These findings establish how begomoviruses influence plant-herbivore interactions that deter nonvectors by interfering with a host WRKY transcription factor.
RESULTS
Whitefly-transmitted begomoviruses enhance plant resistance to a nonvector insect, cotton bollworm
To examine whether and how CLCuMuV alters host plant interactions with the vector whitefly and local nonvector herbivores during an invasion, we first examined the interaction between whitefly and cotton bollworm (CBM; Helicoverpa armigera), a nonvector herbivore on cotton plants. CBM is a major generalist lepidopteran leaf-chewing pest that can live on a wide range of plants including cotton, Solanum lycopersicum, Nicotiana benthamiana (Nb), and Arabidopsis thaliana. When we placed CBM and whitefly together on noninfected control cotton plants, both herbivores had reduced performance on cotton compared to when either insect was present alone, suggesting that CBM is a native competitor of whitefly on cotton (fig. S1, A and B).
Next, to assess whether the begomovirus alters competition between whitefly and CBM, we tested the performance of whitefly and CBM on cotton infected with CLCuMuV and its associated β-satellite (hereafter referred to as CA + β) (fig. S1, C and D). Since begomoviruses can disable the JA signaling pathway to promote whitefly performance, we expected begomoviruses to benefit herbivores including CBM. The whiteflies indeed performed better on plants infected with the CLCuMuV complex, as indicated that CA + β–infected cotton showed significantly higher densities of whitefly than noninfected cotton (Fig. 1, A and B). The virus complex conferred strong resistance to CBM, as evident from the reduced feeding, reduced body weight, and higher mortality (Fig. 1, A, C, and D, and fig. S1, E and F). The superior inhibitory effects on CBM by begomovirus infection were as good as that of a widely used commercial transgenic cotton cultivar engineered to produce insecticidal toxins from Bacillus thuringiensis (Bt) (Fig. 1, C and D) (16). As most monopartite begomoviruses are associated with β-satellite, we next tested whether this asymmetric tripartite interaction also occurs in other β-satellite–associated begomoviruses. Nb plants infected with tomato yellow leaf curl China virus (TYLCCNV) and its associated β-satellite (TA + β), another begomovirus, had a similar asymmetric effect on the vector whitefly and the nonvector herbivore CBM (Fig. 1E) (8).
Fig. 1. Whitefly-transmitted begomoviruses enhance resistance to CBM on infected plants.
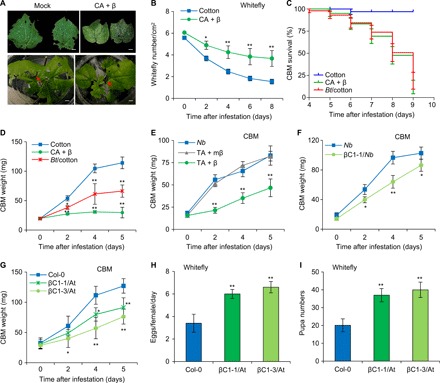
(A) Phenotype of cotton leaves infested with whiteflies or CBM after 5 days. Red arrows indicate CBM. Scale bars, 4 mm. (B) Numbers of live whiteflies infested on healthy cotton and the CLCuMuV complex (CA + β)–infected cotton. (C) Survival rates of CBM larvae infested on healthy cotton, CA + β–infected cotton, and Bt transgenic cotton (BD18). (D) Larval weight of CBM infested on healthy cotton, CA + β–infected cotton, and Bt transgenic cotton. (E) Larval weight of CBM infested on healthy Nicotiana benthamiana (Nb) plants and Nb plants infected with the TYLCCNV complex (TA + β) or with the TYLCCNV complex harboring mutant βC1 (TA + mβ). (F and G) Larval weight of CBM infested on wild-type Nb and transgenic Nb plants ectopically expressing βC1 (F) or Col-0 plants and Arabidopsis plants ectopically expressing βC1 (βC1-1/At and βC1-3/At) (G). (H) Daily number of eggs laid per female whitefly on wild-type Arabidopsis and the transgenic βC1 expression lines. (I) Pupa numbers of whiteflies present on wild-type Arabidopsis and the transgenic βC1 expression lines. (B to G) Bars represent means ± SD (n = 10). (H and I) Bars represent means ± SD (n = 8) (*P < 0.05 and **P < 0.01, Student’s t test). Photo credit: Pingzhi Zhao, Chinese Academy of Sciences.
We next examined whether the single protein βC1 encoded by β-satellite controls the asymmetric tripartite interactions among plants, viruses, and insects. A β-satellite mutant (mβ) of the βC1 protein was used as a control (17). The mutant virus with a nonfunctional βC1 protein did not deter CBM growth, as there was no significant difference in the larval weight of CBM feeding on TYLCCNV and nonfunctional βC1 mutant (TA + mβ)–infected Nb compared to healthy Nb plants (Fig. 1E). To further confirm this newly identified function for the βC1 protein, we measured the larval weights of CBM feeding on transgenic Nb or Arabidopsis lines ectopically expressing TYLCCNV βC1 (βC1-1/Nb, βC1-1/At, or βC1-3/At). CBM feeding on plants overexpressing βC1 weighed significantly less than those feeding on wild-type plants (Fig. 1, F and G). By contrast, whiteflies laid more eggs and exhibited faster pupa development on βC1-expressing lines than on the control Arabidopsis Col-0 plants (Fig. 1, H and I). These results demonstrate that βC1 protein triggers these asymmetric effects on the vector whitefly and nonvector CBM.
The interaction between βC1 and host transcription factor WRKY20 alters plant-mediated interactions with herbivores
Higher eudicot plants seem to have evolved at least two interconnecting antiherbivory immunity pathways, MYC2 branch and ERF (ethylene responsive factor) branch in JA signaling. We hypothesized that begomoviruses might hijack this interconnecting point to achieve asymmetrical effects on different types of herbivores. To identify how βC1 controls this asymmetric effect on the plant-herbivore community, we sought to identify βC1-targeted host factors by screening an Arabidopsis complementary DNA yeast two-hybrid library. We identified AtWRKY20, a transcription factor with unknown function that interacts with βC1. Arabidopsis WRKY20 belongs to the WRKY group I subfamily. Phylogenetic analysis revealed that WRKY20 is highly conserved in higher core eudicots, with likely orthologs from rosids and asterids species such as Brassica napus, Gossypium hirsutum, and S. lycopersicum (tomato), which are major hosts of begomoviruses (fig. S2A). We confirmed that CLCuMuV βC1 (βC1-C) interacts with GhWRKY20 homologs (GhWRKY20-1 and GhWRKY20-2) and that TYLCCNV βC1 interacts with AtWRKY20 by a yeast cotransformation assay. BD (binding domain)–βC1-C and AD (activation domain)–GhWRKY20 yeast transformants or BD-βC1 and AD-AtWRKY20 yeast transformants were able to grow on an SD-Leu-Trp-His selection plate with 2 mM 3-amino-1,2,4-triazole (3-AT), whereas yeast transformants carrying AD– and BD–βC1-C or AD- and BD-βC1 control constructs were unable to do so (Fig. 2A). Next, we performed pull-down assays to examine the interaction between βC1 and WRKY20 in vitro. GST (glutathione S-transferase)–βC1 could be pulled down by MBP (maltose binding protein)–AtWRKY20, but no signal was observed when the negative control protein GST replaced GST-βC1 (fig. S2B). To further confirm the interaction between plant WRKY20 and begomovirus-encoded βC1 in vivo, we performed a bimolecular fluorescence complementation (BiFC) assay on Nb leaves. Direct interactions between βC1 and WRKY20 proteins from tomato, cotton, and Arabidopsis were observed in nuclei and confirmed by colocalization with a signal from 4′,6-diamidino-2-phenylindole (DAPI) staining (Fig. 2B and fig. S2C). Together, these results consistently prove that βC1 interacts with plant WRKY20 proteins.
Fig. 2. Begomovirus-encoded βC1 proteins interact with plant WRKY20 proteins for differentiated resistance against herbivores.
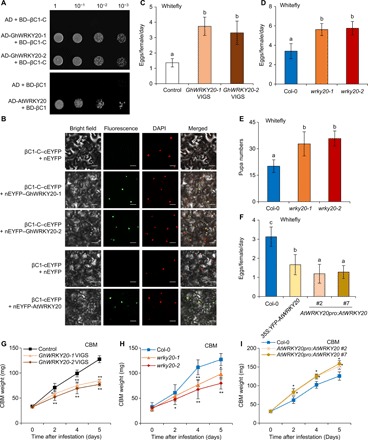
(A) Interaction of CLCuMuV βC1 (βC1-C) with GhWRKY20 homologs (GhWRKY20-1 and GhWRKY20-2) and TYLCCNV βC1 with AtWRKY20 in a yeast two-hybrid system. (B) BiFC analysis of βC1 proteins interaction with WRKY20 homologous proteins. Scale bars, 50 μm. (C and D) Daily number of eggs laid per female whitefly on vector control cotton and VIGS cotton silenced for GhWRKY20 (C) or on Arabidopsis Col-0 and wrky20 mutant plants (D). (E) Pupa numbers of whiteflies present on Arabidopsis Col-0 and wrky20 mutant plants. (F) Daily number of eggs laid per female whitefly on Arabidopsis Col-0 and overexpressing AtWRKY20 plants. (G to I) Larval weight of CBM reared on vector control cotton and GhWRKY20 VIGS cotton (G) on Col-0 and wrky20 mutant plants (H) or on Col-0 and plants overexpressing AtWRKY20 (I). (C to F) Bars represent means ± SD (n = 8). Means with different letters (a, b, and c) are significantly different [P < 0.05, one-way analysis of variance (ANOVA) along with Duncan’s multiple range test]. (G to I) Bars represent means ± SD (n = 10) (*P < 0.05 and **P < 0.01, Student’s t test).
To elucidate the possible roles of WRKY20 in plant-herbivore interactions, we performed whitefly and CBM bioassays on WRKY20–down-regulated plants and AtWRKY20-overexpressing plants. We obtained three species of WRKY20–down-regulated plants, Arabidopsis transferred DNA (T-DNA) insertion mutants, and virus-induced gene silencing (VIGS) tomato and cotton (fig. S3). Whiteflies laid more eggs on GhWRKY20 VIGS cotton and SlWRKY20-1 VIGS tomato than on vector control plants (Fig. 2C and fig. S3C). We further conducted bioassays on stable WRKY20 mutants of Arabidopsis, which is a host for both whitefly and begomoviruses. The whiteflies laid more eggs and exhibited faster pupa development on wrky20 mutants than on Arabidopsis Col-0 plants (Fig. 2, D and E). The complemented wrky20-1 mutant had the same effect on whitefly oviposition as wild-type Col-0 plants, whereas overexpressing AtWRKY20 in Col-0 plants resulted in resistance to whitefly (Fig. 2F and fig. S3H). However, CBM larvae feeding on GhWRKY20 VIGS cotton and Arabidopsis wrky20 mutants grew slower than those on control plants (Fig. 2, G and H), similar to the repressive effects of begomovirus infection and TYLCCNV βC1 transgenic plants on CBM growth traits (Fig. 1, D to G). CBM larvae feeding on plants overexpressing AtWRKY20 grew faster than those on wild-type Col-0 plants (Fig. 2I). Consistently, all of the lines with down-regulated WRKY20 expression had the same asymmetric effect as the begomovirus-infected and βC1-expressing plants (Fig. 2, C to E, G, and H, and fig. S3C). These results suggest that βC1 expression phenocopies plant mutants with down-regulated WRKY20 expression and asymmetrically regulates tripartite begomovirus-plant-herbivore interactions.
The βC1 interaction partners WRKY20 and MYC2 form a negative regulation feedback loop
We next examined how WRKY20 integrates into the known signaling transduction networks controlling plant-herbivore interactions. MYC2 is a well-known early JA-responsive gene and a positive regulator of plant immunity against herbivores including CBM and whitefly (8, 18). We have previously shown that MYC2 interacts with TYLCCNV βC1 and affects host plant-mediated begomovirus-whitefly mutualism by regulating terpene synthase genes (8). Consistent with previous reports, more CBM feeding was observed on myc2-1 leaves than on wild-type Col-0 leaves (Fig. 3, A and B). Since both MYC2 and WRKY20 are targets of βC1, we next examined how Arabidopsis spatiotemporally fine tunes this antiherbivore network. First, we found that AtWRKY20 is a later JA-responsive gene (Fig. 3C). Second, we found that WRKY20 and MYC2 form a negative regulation feedback loop, as AtWRKY20 expression levels in the myc2-1 mutant were higher than those in Col-0 plants and there was higher expression of AtMYC2 and its homologs AtMYC3 and AtMYC4 in the wrky20-1 knockdown mutant (Fig. 3, D and E). It appears that WRKY20 and MYC2 are directly negatively transregulated vice versa by their transcription regulation activity.
Fig. 3. The βC1 interaction partners WRKY20 and MYC2 form a negative regulation feedback loop.
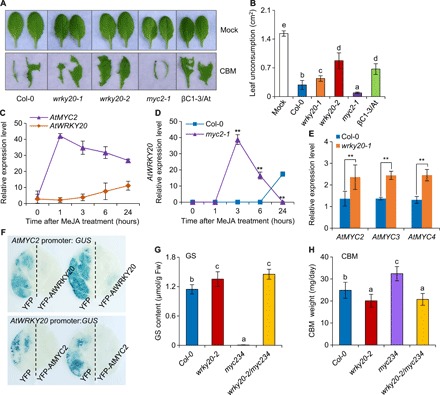
(A) Phenotypes of 6-week-old Arabidopsis leaves after CBM infestation for 12 hours. (B) Nonconsumed leaf areas were recorded after CBM infestation for 12 hours on Arabidopsis leaves. Bars represent means ± SD (n = 6). (C) Relative expression levels of AtMYC2 and AtWRKY20 in Col-0 treated with methyl JA (MeJA). (D) Relative expression level of AtWRKY20 in Col-0 and myc2-1 plants treated with MeJA. (E) Relative expression levels of AtMYC2, AtMYC3, and AtMYC4 in Col-0 and wrky20-1 plants after 6 hours of MeJA treatment. (F) AtMYC2 promoter activity in YFP-expressing and YFP-AtWRKY20–expressing leaves by β-glucuronidase (GUS) staining and AtWRKY20 promoter activity in YFP-expressing and YFP-AtMYC2–expressing leaves by GUS staining. (G) Total GSs content of 6-week-old Arabidopsis plants. Bars represent means ± SD (n = 4). Fw, fresh weight. (H) Growth rate of CBM reared on Arabidopsis plants. Bars represent means ± SD (n = 10). (B, G, and H) Means with different letters (a, b, c, d, and e) are significantly different (P < 0.05, one-way ANOVA along with Duncan’s multiple range test). (C to E) Bars represent means ± SD (n = 3) (**P < 0.01, Student’s t test). Photo credit: Pingzhi Zhao, Chinese Academy of Sciences.
WRKY20 belongs to the WRKY transcription factor family, which specifically binds to the W-box (TTGAC and TTGACC/T) or W-box–like (TGACC/T) elements containing the TGAC core sequence (19). Promoter analysis using the plant cis-acting regulatory DNA elements (PLACE) database (www.dna.affrc.go.jp/PLACE/?action=newplace) revealed that five W-box and three W-box–like elements are distributed in the 2.0-kb promoter of AtMYC2 (fig. S4A). One additional W-box–like element was in the 5′ untranslated region of AtMYC2. To test whether WRKY20 specifically binds to these elements in vivo, we performed a chromatin immunoprecipitation (ChIP) assay using a transgenic line expressing 35S:YFP-AtWRKY20 in the Col-0 background. Using a line expressing 35S:YFP as a negative control, we performed a ChIP-qPCR (quantitative polymerase chain reaction) analysis, which showed that 35S:YFP-AtWRKY20 lines significantly enriched immunoprecipitation on regions II, IV, and V of the AtMYC2 promoter, suggesting that AtWRKY20 directly binds to the AtMYC2 promoter (fig. S4B). Furthermore, we detected the transcriptional repression activity of AtWRKY20 by using two transient expression systems. Leaves expressing YFP-AtWRKY20 significantly suppressed the promoter activity of AtMYC2 compared to leaves expressing only YFP (Fig. 3F and fig. S4, C to E). Besides the direct repressive role of WRKY20 on the transcription of MYC2, we found that MYC2 also directly suppressed the transcription of the WRKY20 in a similar set of experiments (Fig. 3F and fig. S4, F to H). These results further support that WRKY20 and MYC2 form a negative regulation feedback loop.
To further understand the epistatic relationship between MYCs and WRKY20 in Arabidopsis resistant to CBM, we crossed the myc234 triple mutant with the wrky20-2 mutant. MYC2 and two homologs, MYC3 and MYC4, redundantly induce the production of toxic metabolites, such as GSs, which protect the plant against chewing herbivores (20). Unlike the myc234 triple mutant, which is almost completely devoid of GSs, the total GS content in the wrky20-2 mutant was significantly higher than that in Col-0 plants (Fig. 3G), which explains the enhanced resistance to CBM in plants lacking WRKY20. The wrky20-2/myc234 quadruple mutant has the same GS level and CBM insensitivity as the wrky20-2 mutant, in stark contrast to the near-null deficiency of GSs and CBM sensitivity in the myc234 mutant (Fig. 3, G and H). These results suggest that WRKY20 functions downstream of the MYCs in controlling the accumulation of GSs, which affect the Arabidopsis-CBM interaction.
βC1 alters WRKY20-regulated GS biosynthesis in a vascular-specific manner
As whitefly is a phloem-feeding insect and CBM is a leaf-chewing herbivore, each has its own unique ecological niche, although both feed on the leaf. Thus, we hypothesized that the asymmetric effect of begomoviruses on whitefly and CBM is due to WRKY20-mediated tissue-specific immunity. To test this, we generated AtWRKY20 promoter: GUS (β-glucuronidase) reporter lines to determine the spatiotemporal expression pattern of AtWRKY20. The reporter gene was expressed exclusively in Arabidopsis vascular tissue, which is the site of TYLCCNV βC1 expression and whitefly vector feeding (Fig. 4, A to H) (21, 22). After Arabidopsis had transitioned to the adult vegetative phase (6 weeks old), the expression of the reporter gene could not be detected in leaves under normal conditions, but methyl JA (MeJA) treatment could strongly induce its expression in a vascular-specific manner (Fig. 4, I and J). Therefore, we speculated that WRKY20 regulates the spatiotemporal production of plant secondary metabolites during begomovirus-plant-herbivore interactions in a manner similar to other transcription factors when exposed to biotic stresses (23, 24).
Fig. 4. βC1 regulates WRKY20-mediated GS biosynthesis in a vascular-specific manner.
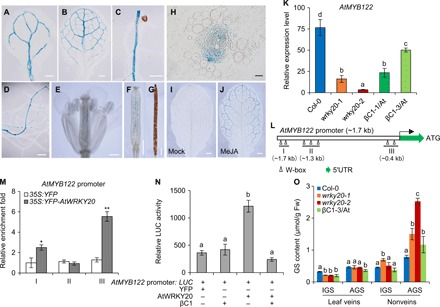
(A to G) AtWRKY20 promoter activity in a cotyledon (A), first rosette leaf (B), roots (C and D), inflorescence (E), and siliques (F and G). Scale bars, 1 mm. (H) Resin root slice of the AtWRKY20 promoter: GUS lines. Scale bars, 20 μm. (I and J) GUS staining of the leaves of 6-week-old plants before (I) and after (J) MeJA treatment in AtWRKY20 promoter: GUS lines. Scale bars, 2 mm. (K) Relative expression level of AtMYB122 in 6-week-old Arabidopsis plants after 6 hours MeJA treatment. Bars represent means ± SD (n = 3). (L) Schematic diagram of the AtMYB122 promoter. The small hollow triangles represent W-box motifs in the schematic diagram of the promoters. ATG indicates the coding initiation site of a gene. Three lines under triangles represent various DNA fragments were amplified in ChIP assay. (M) Fold enrichment of YFP-AtWRKY20 associated with DNA fragments of the AtMYB122 promoter in a ChIP assay. Enrichments are referred to the 35S:YFP-AtWRKY20 or 35S:YFP against wild-type Col-0 seedlings. Bars represent means ± SD (n = 4) (*P < 0.05 and **P < 0.01, Student’s t test). (N) Effects of βC1 on the transactivation activity of AtWRKY20 on the AtMYB122 promoter. Bars represent means ± SD (n = 8). (O) Contents of AGS and IGS in leaf veins and nonveins, respectively, of 6-week-old Arabidopsis plants after 6 hours of MeJA treatment. Bars represent means ± SD (n = 4). (K, N, and O) Means with different letters (a, b, c, and d) are significantly different (P < 0.05, one-way ANOVA along with Duncan’s multiple range test). Photo credit: Pingzhi Zhao, Chinese Academy of Sciences.
In Brassicaceae plants, some secondary metabolites such as indole and aliphatic GSs (IGS and AGS, respectively) are readily loaded into and transported through the vascular tissues (25). The AtWRKY20 expression pattern is highly similar to that reported for a number of transcription factor genes functioning in the GS pathway that gives rise to both IGS and AGS (26). Gene expression analysis showed that WRKY20 positively regulated IGS-related genes (AtMYB51, AtMYB122, AtCYP79B2, and AtCYP83B1) but negatively regulated AGS-related genes (AtMYB29, AtMYB76, and AtCYP79F1) (Fig. 4K and fig. S5). ChIP-qPCR analysis demonstrated that AtWRKY20 directly binds to the promoters of most IGS-related genes and one AGS-related gene (Fig. 4, L and M, and fig. S6). These results indicate that plant WRKY20 is a dual-function transcription factor controlling GS biosynthesis. The interaction between WRKY20 and βC1 blocked the ability of WRKY20 to bind the promoter of MYB122 (Fig. 4N), an IGS-related gene, and thereby possibly decreased the accumulation of IGS in vascular tissues of Arabidopsis.
Since AtWRKY20 expression is strongest in vascular tissue and is also JA responsive, we collected vein and nonvein sections of the leaf separately after MeJA treatment and performed GS analysis. At least 50% increase of AGS content in nonvein leaf tissues of wrky20 mutants and βC1-3/At plants could explain the bioassay results for these lines, which had enhanced resistance against CBM (Fig. 4O and fig. S7A). Conversely, a significant 35% reduction of IGS content in the leaf veins of wrky20 mutants and βC1-expressing plants could explain why begomovirus infection promotes whitefly performance (Fig. 4O and fig. S7B). Both the GS amounts and related gene expression results suggest that WRKY20 may act as a dual-function regulator of host defense mechanisms against two herbivores with different feeding patterns. WRKY20 deficiency would make the host sensitive to whitefly but resistant to CBM. If this hypothesis is correct, then WRKY20 overexpression should produce the opposite result. This was indeed the case in Arabidopsis (Fig. 2, F and I, and fig. S7C). Overexpressing WRKY20 in Arabidopsis plants increased whitefly resistance and reduced CBM resistance (Fig. 2, F and I). These findings are well explained by the enhanced GS levels in leaf veins and reduced GS levels in nonvein leaf tissue in plants overexpressing AtWRKY20 (fig. S7C). Together, the results demonstrate that βC1-WRKY20 interaction specifically promotes begomovirus-whitefly mutualism by modulating the tissue-specific biosynthesis of GSs in Arabidopsis.
βC1 suppresses WRKY20 activity by interfering with its dimerization
In addition to inducing the production of antiherbivory metabolites against CBM, begomovirus infection may influence other defensive mechanisms mediated by phytohormone-regulated biosynthesis and the release of toxic polypeptides, such as mechanisms involving the ethylene- and JA-responsive defensin gene PDF1.2 (plant defensin 1.2) and the salicylic acid (SA)–regulated gene PR1 (pathogenesis-related 1) (14, 27). Here, we demonstrated that AtWRKY20 is also involved in the ERF branch of JA signaling and that the JA responsiveness of both PDF1.2 and ORA59 (octadecanoid-responsive arabidopsis AP2/ERF 59), the major regulators of the ERF branch, was strongly suppressed in wrky20 mutants and βC1-3/At plants compared with wild-type Col-0 plants (Fig. 5, A and B). To understand the exact mechanism through which WRKY20 is involved in JA signaling, we next tested whether WRKY20 directly interacts with one or more key regulators in the JA pathway. In vitro pull-down and BiFC assays jointly demonstrated that AtWRKY20 directly interacts with ERF-AtORA59 (Fig. 5, C and D). In addition, there was a positive correlation between the expression of AtWRKY20 and AtORA59 in Arabidopsis (fig. S8A). These results suggest that WRKY20 is a key regulator in the ERF branch of JA signaling.
Fig. 5. βC1 interferes with the interaction between WRKY20 and ERF-ORA59.
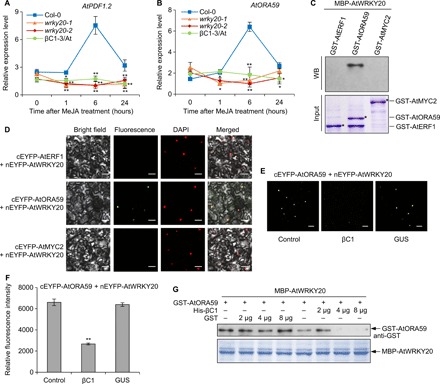
(A and B) Relative expression level of AtPDF1.2 (A) or AtORA59 (B) in Arabidopsis plants under MeJA treatment. Bars represent means ± SD (n = 3) (*P < 0.05 and **P < 0.01, Student’s t test). (C) GST pull-down assay. Two micrograms of MBP-AtWRKY20 fusion protein was used to pull down 2 μg of GST fusion proteins. WB, Western blot. (D) BiFC analysis of AtWRKY20 interaction only with AtORA59 but not with AtMYC2 or AtERF1. Scale bars, 50 μm. (E) Effects of βC1 on the interaction of WRKY20 with ORA59 by modified BiFC assay. The EYFP fluorescences were detected after coproduction of cEYFP-AtORA59 + nEYFP-AtWRKY20 (control), βC1 + cEYFP-AtORA59 + nEYFP-AtWRKY20 (βC1), or GUS + cEYFP-AtORA59 + nEYFP-AtWRKY20 (GUS). Scale bars, 50 μm. (F) Quantitative data of EYFP fluorescence intensity show effects of βC1 on the interaction of WRKY20 with ORA59. Bars represent means ± SD (n = 20) (**P < 0.01, Student’s t test). (G) Pull-down protein competition assays. The indicated protein amount of His-βC1 or GST was mixed with 2 μg of GST-AtORA59 and pulled down by 2 μg of MBP-AtWRKY20. Immunoblots were performed using anti-GST antibody to detect the associated proteins. (C and G) Membranes were stained with Coomassie brilliant blue to monitor input protein amount.
Because of WRKY20 interaction with either βC1 or ORA59, we raised a possibility that βC1 competes with ORA59 for interaction with WRKY20. To test this hypothesis, we performed a modified BiFC assay, as in a previous report (28). In the presence of coexpressing βC1, the interaction signal strength of AtWRKY20-AtORA59 indicated by enhanced yellow fluorescent protein (EYFP) fluorescence intensity markedly decreased (Fig. 5, E and F). A negative control of GUS coexpression did not affect the interaction between AtWRKY20 and AtORA59. Moreover, in vitro competitive pull-down assays also showed that βC1 interferes with the interaction between WRKY20 and ORA59 (Fig. 5G).
A previous study reported that ORA59 binds to GCC-box elements of the PDF1.2 promoter and transactivates the PDF1.2 promoter (29). In addition, there are three W-box–like elements in the AtPDF1.2 promoter (fig. S8B). Further ChIP analysis demonstrated that AtWRKY20 directly binds to the AtPDF1.2 promoter (fig. S8C). In transient assays, the cotransfection of AtWRKY20 and AtORA59 promoted activation of the AtPDF1.2 promoter compared with that observed in the presence of AtPDF1.2 reporter by AtWRKY20 or AtORA59 alone (fig. S8D). In addition, reduction of ORA59 expression or knockdown of PDF1.2 expression in Arabidopsis led to increased whitefly oviposition compared with wild-type Col-0 plants (fig. S8E). Moreover, coexpression of the βC1 protein with AtWRKY20-cEYFP and nEYFP-AtWRKY20 reduced the intensity of EYFP fluorescence in Nb leaves, indicating that βC1 and WRKY20 forms a complex that interferes with WRKY20 homodimerization (fig. S8, F to I). As expected, βC1 severely repressed the transactivation activity of WRKY20 on PDF1.2 transcription (fig. S8J).
The hijacking of WRKY20 by viral βC1 promotes plant resistance to aphid by activating SA signaling
So far, we have demonstrated that WRKY20 may confer plant tissue-specific immunity against two different herbivores that have unique ecological niches on the same plant. Given these observations, we next expected that the reduction of indole GSs in both βC1-expressing plants and wrky20 mutant plants would also benefit other piercing insects such as aphids. Like whiteflies, aphids are another key insect vector for many types of plant viruses including potyvirus and cucumovirus (30). Unexpectedly, wrky20 mutants and βC1-expressing plants exhibited resistance to the green peach aphid (Myzus persicae) (Fig. 6A), suggesting that there are different mechanisms for WRKY20-mediated immunity against this species, which is not a begomovirus vector. Our above results demonstrated that WRKY20 physically and functionally interacts with ORA59 (Fig. 5, C and D), which has been shown to be a key intersection point of pathways involving defensive phytohormones, i.e., JA, ethylene, and SA. It has been established that plant SA-mediated resistance is the major phytohormone-mediated defense against aphids (31). SA signaling negatively regulates resistance against whitefly nymphal development, but it is a prerequisite for the plant to build immunity toward infestation by aphids, another phloem sap-feeding local insect (32). This led us to examine the role of WRKY20 in SA signaling. We found that increased expression of SA synthesis and SA-responsive genes (AtPAL1, AtSID2, AtPAD4, AtEDS5, and AtPR1) in Arabidopsis plants with down-regulated WRKY20 expression or transgenic βC1 expression (Fig. 6, B to F) promoted tolerance to green peach aphid. Meanwhile, overexpressing AtWRKY20 in Arabidopsis reduced resistance to aphids by suppressing SA signaling (fig. S9, A and B). Furthermore, we demonstrated that AtWRKY20 could bind to some of the W-boxes in the promoter of AtPR1, which is a marker gene for the SA signaling pathway (fig. S9, C and D). Therefore, the interaction of between βC1 and WRKY20 activates plant SA signaling and enhances plant resistance to a nonvector aphid.
Fig. 6. The βC1-WRKY20 interaction promotes plant resistance to aphid by activating SA signaling.
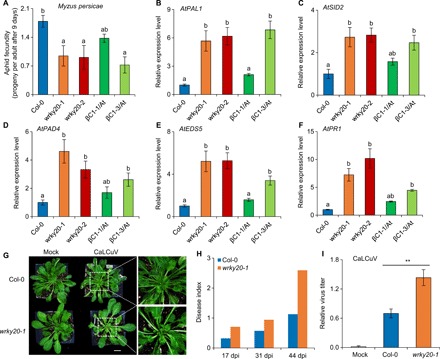
(A) Number of progeny produced by each aphid 9 days after infestation of 6-week-old Arabidopsis plants. Bars represent means ± SD (n = 8). (B to E) Relative expression levels of SA-related genes, AtPAL1 (B), AtSID2 (C), AtPAD4 (D), AtEDS5 (E), or AtPR1 (F) in 6-week-old Arabidopsis plants. Bars represent means ± SD (n = 3). (G) Symptoms of cabbage leaf curl virus (CaLCuV)–infected Arabidopsis plants at 31 days post infection (dpi). Scale bar, 1 cm. (H) Disease index of CaLCuV-infected Arabidopsis plants. Disease indexes is an indicator of the average disease severity on the survey plant samples, which were observed by level of viral symptomatic onset such as obvious yellow and curly leaves in infected plants. (I) Relative CaLCuV virus titer in Arabidopsis plants after 31 dpi. Values are means ± SD (n = 3) (**P < 0.01, Student’s t test). (A to F) Means with different letters (a and b) are significantly different (P < 0.05, one-way ANOVA along with Duncan’s multiple range test). Photo credit: Pingzhi Zhao, Chinese Academy of Sciences.
We also investigated how the begomovirus itself benefits from interfering with WRKY20. Because TYLCCNV replicates weakly in Arabidopsis, we used cabbage leaf curl virus (CaLCuV), a bipartite begomovirus, to infect Arabidopsis (8). The wrky20-1 mutant plants had more severe viral infection symptoms and higher virus accumulation than the wild-type Col-0 control (Fig. 6, G to I). These results suggest that WRKY20 mediates plant immunity against begomoviruses and that the suppression of WRKY20 activity by begomoviruses not only benefits the whitefly vector but also facilitates the multiplication of begomovirus in the host plant.
DISCUSSION
Within a few decades, Begomovirus has emerged as the largest genus of plant viruses and its members cause severe damage on almost every continent. Among their known adaptation advantages, the ability to suppress host immunity mechanisms such as RNA silencing and innate immunity is the best understood (13). We here report a new strategy interfering with the host plant WRKY20 to promote begomovirus multiplication and transmission by deterring nonvector competitors (fig. S9E). Host plants have evolved multiple immune mechanisms regulated by several transcription factors such as MYC2 and WRKY20 to circumvent against infections of arboviruses. Whitefly-transmitted begomovirus triggers several immune responses, including the emissions of terpenoids and the synthesis of GSs, phytohormones (e.g., JA, ethylene, and SA), and toxic polypeptides (e.g., PDF1.2 and PR1). The begomovirus-encoded βC1 interferes with multiple host defensive responses regulated by WRKY20 and MYC2 transcription factors. βC1 disrupts the dimerization of WRKY20-WRKY20 and WRKY20-ORA59, thereby mobilizing the biosynthesis and accumulation of GSs and several defensive compounds (e.g., PDF1.2 and PR1). This viral hijacking of both WRK20 and other host targets confers to benefit whitefly but deters nonvector CBM and aphids.
WRKY20 functions as a hub that integrates the signaling of multiple defensive phytohormones (JA, SA, and ethylene) in Arabidopsis and other higher plants to regulate specialized antiherbivore metabolites, such as GSs in Brassicaceae plants. WRKY20 is a vascular-specific gene that regulates two branches of GS biosynthesis (Fig. 4 and fig. S5 to S7). WRKY20 deficiency, caused by begomoviral infection, inhibits the biosynthesis of tryptophan-derived indolic GSs in plant vascular tissues, conferring to benefit both the begomovirus and the whitefly. Meanwhile, the promoted accumulation of methionine-derived aliphatic GSs in nonvein organs deters CBM, a nonvector leaf-chewing herbivore. Thus, the different modes of feeding by CBM and whitefly and the spatial distribution of GSs in wrky20 mutants may explain why begomovirus-infected or βC1-expressing plants deter CBM and benefit whitefly. Meanwhile, the activation of SA biosynthesis and signaling pathways confers that enhanced resistance to aphids due to the repressed SA signaling by WRKY20 is alleviated by the interference of begomoviral βC1 (Fig. 6). We speculate that cotton WRKY20 homologs may also regulate the biosynthesis of some uncharacterized and vascular-specialized metabolites that the cotton begomovirus used for against herbivores. It is still unknown whether this type of WRKY20-ORA59 interaction is conserved across plant species or limited to cruciferous plants. Notably, our results pinpointed the mechanisms underlying plant-dependent begomovirus-whitefly mutualism by targeting at least four host factors, such as AS1, MYC2, SKP1, and WRKY20.
Interactions among competitors, predators, and prey have traditionally been viewed as the foundation of community structure. Parasites including pathogens, which have long been ignored in community ecology, are now recognized as playing an important role in influencing many biological invasions and emerging infectious diseases, especially for vector-borne diseases (33). Here, we demonstrate how begomovirus promotes its vector and deters its nonvector competitors in the plant community, partially explaining why this virus is so successful across the world in places where agricultural products are exchanged. This type of spread is not limited to begomoviruses; some invasive animal pathogens have spread globally through similar processes (34, 35). For example, chytridiomycosis, a disease caused by the chytrid fungus Batrachochytrium dendrobatidis, which is associated with global amphibian declines and extinctions, is partially attributed to the introduction of non-native amphibians via international trade (32). Although the pathogenesis factors of the chytrid fungus are currently unclear, invasive pathogens, including viruses in general, have higher virulence in native species. Furthermore, parasites also have indirect effects on the species with which their hosts interact, these include density-mediated effects (resulting from parasite-induced reduction in host reproduction and survival) and trait-mediated indirect effects (resulting from parasite-induced changes in host phenotype, behavior, and life history).
This report describes studies of a whitefly-vectored begomovirus that has become an important pest of cotton in China. The data here are limited to laboratory observations; nevertheless, they provide evidence for how this event might have taken place. We here document a promising and previously unidentified broad-spectrum pest resistance strategy in crops by manipulating WRKY20. Ultimately, these studies will provide important ecological insights to predict pathogenic infections and provide effective methods of biological control for vector-borne diseases.
MATERIALS AND METHODS
Plant materials and growth conditions
Nb transgenic plants carrying 35S:βC1 have been reported previously (8). Nb, S. lycopersicum (Zhongza no.9; tomato), and Gossypium barbadense (Xinhai no. 21; cotton) plants were grown in a growth chamber at 25°C with a 12-hour light/12-hour dark cycle. G. hirsutum BD18 containing Bt gene was used as Bt/cotton plants.
A. thaliana ecotype Columbia (Col-0, At), βC1-1/At, and βC1-3/At (14), wrky20-1 (SALK_055904), wrky20-2 (SALK_116115), pdf1.2a (SALK_063966), myc2-1 (18), and myc234 (20) mutants were used in this study. We generated quadruple wrky20-2/myc234 mutants by crossing the corresponding parental single wrky20-2 and triple myc234 homozygous lines. After vernalization for 2 days at 4°C in the dark, sterilized seeds on Murashige and Skoog (MS) medium were transferred to a growth chamber at 22°C with a 12-hour light/12-hour dark cycle.
Virus inoculation and viral DNA measurement
For TYLCCNV infection, Nb plants at the sixth true leaf stage were inoculated with Agrobacterium tumefaciens carrying TYLCCNV and β-satellite (TA + β), as described previously (8, 14). Inoculation with TYLCCNV and a mutant βC1 (TA + mβ) was used as a control (17).
For CLCuMuV infection, cotton plants at the two-cotyledon stage were inoculated with A. tumefaciens carrying CLCuMuV and β-satellite (CA + β) mixed at 1:1 ratio, as described previously (15). We used a CLCuMuV-specific primer set (CLCuMuV V1-F and CLCuMuV V1-R) to quantify viral DNA by real-time PCR.
Insect bioassays
Whiteflies (Middle East-Asia Minor 1, formerly biotype B) were collected in a suburb of Beijing, China, and a culture of the whitefly was established and maintained on healthy cotton in a growth chamber. We performed the whitefly oviposition and development experiments according to previous method (8) by using plant leaves of similar size and a leaf-clip cage (diameter, 45 mm; height, 30 mm). For the whitefly oviposition experiment, each cohort of three male and female adult whiteflies was released into a leaf-clip cage. Whitefly eggs on the leaf area enclosed in a cage were counted after 10 days under a microscope. For whitefly development experiment, a cohort of 16 adult female whiteflies was released into a leaf-clip cage. All adult females were removed after 2 days of oviposition, and the eggs were left to develop in the leaf-clip cage. The numbers of whitefly pupae in each of the cages were recorded after 20 days. Eight plants of each line were used in the experiment. The experiment was repeated three times with similar results.
Short-term feeding experiments were performed to evaluate the effect of leaf quality on the growth rate of third instar larvae of CBM. CBM larvae were fed on living plants in a growth chamber (25°C, 8-hour light/16-hour dark cycle). Each larva was weighted for 1 or 2 days. Four larvae as one sample were weighed together to obtain one datum for average weight. Forty larvae (10 independent samples) for each genotype in each biological experiment were examined. The data in Fig. 1 (A to D) were obtained by rearing one CBM larva and 50 adult whiteflies on one healthy cotton or CA + β–infected cotton for 9 days. The experiment was repeated three times with similar results.
The experiment on aphid fecundity on different lines was performed, as described previously (36). One apterous adult aphid was placed on one plant. All aphids except three nymphs were removed after 24 hours. The nymphs were allowed to develop to adult, and their progeny were counted after 9 days. Eight biological replicates were conducted. The experiment was repeated two times with similar results.
MeJA treatment
Arabidopsis seeds were grown on MS medium. Three-week-old plants were transferred into the soil. Plants were grown on soil under a 12-hour light/12-hour dark cycle at 22°C. Six-week-old plants were treated with 100 μM MeJA using foliar sprays and then covered with a plastic cover. Plant samples were collected at various times or after 6 hours treatment for gene expressions, GS analysis, and GUS staining.
Plasmid constructs
For yeast two-hybrid experiments, DNA fragments encoding GhWRKY20 and AtWRKY20 were cloned into a pGADT7 vector to separately generate AD constructs. Full-length encoding CLCuMuV βC1-C and TYLCCNV βC1 were cloned into pGBKT7 vector to generate BD constructs. These constructs were transformed into yeast stain AH109.
For pull-down assay in vitro, full-length open-reading frames encoding AtWRKY20, TYLCCNV βC1, AtMYC2, AtORA59, and AtERF1 were cloned into a pGEX-DC or pMAL-DC vector to generate GST fusion or MBP fusion constructs. Full-length TYLCCNV βC1 was cloned into a pET-29a vector to generate a His-βC1 fusion construct. These protein expression constructs were transformed into Escherichia coli BL21 (DE3).
For BiFC, the DNA fragments of TYLCCNV βC1, CLCuMuV βC1-C, GhWRKY20, SlWRKY20, AtWRKY20, AtMYC2, AtORA59, and AtERF1 were cloned into pENTR-3C entry vector and then transferred into pBA3032 and pBA3036 destination vectors by attL× attR (LR) recombination reactions to generate fusion genes with cEYFP or nEYFP at the N or C terminus under the control of the CaMV 35S promoter. Constructs for BiFC were transferred into A. tumefaciens strain C58C1.
For transactivation repression assay, the promoter fragment of AtMYC2 (~2.0 kb, before the initiation codon) was cloned into a pKGWFS7 vector to generate AtMYC2 promoter: GUS fusion constructs. For transactivation assay, the promoter fragment of AtMYC2 (~2.0 kb), AtMYB122 (~1.7 kb), or AtPDF1.2 (~1.4 kb) was separately cloned into a pGWB435 vector to generate AtMYC2 promoter: LUC, AtMYB122 promoter: LUC, or AtPDF1.2 promoter: LUC fusion constructs. Full-length encoding AtWRKY20, YFP, or TYLCCNV βC1 was cloned into pBA-DC-3HA to generate hemagglutinin-tagged constructs with a CaMV 35S promoter. Full-length encoding AtWRKY20 was cloned into pH7YWG2 to generate YFP-tagged constructs with a CaMV 35S promoter. Plasmids were transferred into A. tumefaciens EHA105 strain.
The full-length encoding fragments of AtWRKY20 and AtORA59 were separately cloned into the pBA-YFP or pH7GWIGWII vector to generate 35S:YFP-AtWRKY20–fused plasmid and RNAi-AtORA59 plasmid for stable plant transformation. The promoter fragments of AtWRKY20 (~2 kb) were amplified and inserted into the pKGWFS7 vector to generate the AtWRKY20 promoter: GUS fusion constructs. To produce the construct used for functional complementation of the wrky20-1 mutant, the genomic DNA fragment of AtWRKY20 (AtWRKY20pro:AtWRKY20) including the 5′ promoter region, coding region, and Flag tag was cloned into a binary vector pBA002a with ClonExpress-II One Step Cloning Kit (Vazyme). Plasmids were transferred into the A. tumefaciens EHA105 strain. Arabidopsis transformations were performed using the floral-dip method.
Yeast two-hybrid analysis
The library of Arabidopsis Mate and Plate was used to screen with the Matchmaker Gold Yeast Two-Hybrid System according to the manufacturer’s protocol (Clontech). All constructs were transformed into yeast stain AH109 through the method of modified lithium acetate. Yeast cotransformants were screened on the selective dropout medium SD-Leu-Trp-His with 2 mM 3-AT.
Confirmation of the interactions between begomovirus-encoded βC1 proteins and WRKY20 proteins was performed according to the manufacturer’s protocol. The yeast strain AH109 was cotransformed with AD-GhWRKY20 and BD–βC1-C or AD-AtWRKY20 and BD-βC1 constructs. Yeast cotransformants were plated on SD-Leu-Trp–selective dropout medium. Colonies were transformed onto SD-Leu-Trp-His plates with 2 mM 3-AT.
In vitro pull-down and protein competitive interaction assay
The recombinant GST and MBP tag proteins were separately purified using glutathione sepharose (GE Healthcare) and amylose resin (New England Biolabs) beads according to the manufacturer’s instructions. The in vitro binding assay was performed as described (14). Two micrograms of bait MBP-AtWRKY20 fusion protein and 2 μg of prey protein (GST-fusion proteins) were added into 1 ml of binding buffer [50 mM tris-HCl (pH 7.5), 100 mM NaCl, 0.5% Triton X-100, 0.5 mM β-mercaptoethanol, and 2% proteinase inhibiter cocktail] and incubated with amylose resin beads at 25°C for 2 hours. For competitive pull-down assay, His-βC1 fusion proteins were purified using Ni-nitrilotriacetate (Ni-NTA) agarose (Qiagen) according to the manufacturer’s instructions. Indicated amounts of His-βC1 or GST were mixed with 2 μg of MBP-AtWRKY20 and 50 μl of amylose resin overnight. After two times of centrifugation and two washes with binding buffer [50 mM tris-HCl (pH 7.5), 100 mM NaCl, 0.25% Triton X-100, and 35 mM β-mercaptoethanol], 2 μg of GST-AtWRKY20 or GST-ORA59 was added and the mixture was incubated for 2 hours at 4°C. After incubation, beads were washed six times with fresh binding buffer. Pull-down proteins were separated on 10% SDS–polyacrylamide gels and detected through immunoblotting with anti-GST antibody (TransGen Biotech).
BiFC assay
To confirm protein-protein interactions in vivo, the BiFC experiments were performed using methods as described. Recombinant plasmids encoding cEYFP and nEYFP fusions were transformed into competent Agrobacterium (strain C58C1) cells, which were then cultured. Agrobacterial cells containing various constructs were collected and suspended in a solution containing 10 mM MgCl2 and 150 μM acetosyringone and then kept at 25°C for at least 3 hours without shaking. Agrobacterial suspension was used to infiltrate into leaves of Nb plants. Images of fluorescence and DAPI staining were taken by a confocal microscopy (Leica SP8) after 2 days of incubation. The experiment was repeated three times with similar results.
Virus-induced gene silencing
Leaves of 3-week-old Xinhai 21 cotton plants were agroinfiltrated with psTRV1 and psTRV2-GhWRKY20, and leaves of 3-week-old tomato plants were also agroinfiltrated with psTRV1 and psTRV2-SlWRKY20-1. Plants co-infiltrated with psTRV1 and psTRV2 were used as the control (37).
Antibody preparation
The DNA fragment of the N terminus of AtWRKY20 (1 to 300 base pair) was cloned into pET-28a (+) vector to generate His-AtWRKY201–300 fusion construct. His-AtWRKY201–300 protein was purified using Ni-NTA agarose (Qiagen) according to the manufacturer’s instructions. His-AtWRKY201–300 protein was then injected into mice, and the corresponding monoclonal antibody was prepared by the Animal Center of Institute of Genetics and Developmental Biology (Chinese Academy of Sciences, China).
ChIP assay
The 2-week-old transgenic Arabidopsis plant overexpressing 35S:YFP or 35S:YFP-AtWRKY20 and the wild-type Col-0 seedlings were used for ChIP assay. Expressing 35S:YFP line was used as a negative control. Three micrograms of seedlings was fixed in 1% formaldehyde for 10 min in a vacuum. Glycine was added to a final concentration of 0.125 M, and the reaction was terminated by incubation for 5 min in a vacuum. Seedlings were rinsed three times with distilled water and frozen in liquid nitrogen for ChIP experiments. ChIP experiments were performed, as described previously, using anti–green fluorescent protein agarose beads (ChromoTek) for immunoprecipitation (38). The resulting DNA samples were purified with a PCR Cleanup kit (Axygen). DNA fragments were analyzed by qPCR, with the Arabidopsis ACTIN2 promoter as a reference. Enrichments were referred to the 35S:YFP or 35S:YFP-AtWRKY20 against wild-type seedlings. Primers of ChIP assays are listed in table S1.The experiments were repeated with four independent biological replicates and were repeated twice with similar results.
Transcriptional repression assay
Leaves of Nb were co-infiltrated with AtMYC2 promoter: GUS and 35S:YFP or AtMYC2 promoter: GUS and 35S:YFP-AtWRKY20 at a ratio of 1:1. Two days after infiltration, leaves were harvested and stained with GUS staining buffer [0.1 M sodium phosphate buffer (pH 7.0), 0.1% Triton X-100, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, and 10 mM EDTA]. In addition, leaves were collected and frozen in liquid nitrogen, and GUS quantitative assay was performed, as described previously (39). Eight biological samples were used for each experiment, and similar results were found in two experiments.
Resin embedding method
A historesin embedding kit (Leica) was used to dissect resin slice of roots of AtWRKY20 promoter: GUS transgenic lines. The images were recorded by a stereoscopic microscope.
GS assay
Six-week-old Arabidopsis leaves were separately collected by dissecting with a scalpel into leaf veins and leaf nonveins (the rest of leaf veins) tissues, as described previously (40), after MeJA treatment for 6 hours. The same weight of each leaf section was used in the experiments. Four replicates were conducted for each genotype. GSs were extracted and analyzed as described. The high-performance liquid chromatography (HPLC) analysis was performed using an Agilent 1260 HPLC system. Data were quantified as micromole per gram fresh weight.
Quantitative reverse transcription polymerase chain reaction
Total RNA was isolated from 6-week-old Arabidopsis plants using the RNeasy Plant Mini Kit (Qiagen) including on-column deoxyribonuclease treatment. Reverse transcription was performed using 1 μg of RNA of each sample and oligo(dT)15 primers using Moloney-murine leukemia virus (M-MLV) reverse transcriptase (Promega). Three independent biological samples were collected and analyzed. Real-time PCR was performed on a Bio-Rad CFX96 real-time PCR system with Thunderbird SYBR qPCR mix (TOYOBO) and gene-specific primers listed in table S1. The Arabidopsis ACTIN2 (At3g18780) mRNA was used as internal control. These experiments of gene expressions were repeated three times with similar results.
Luciferase assays
Nb leaves were agroinfiltrated with A. tumefaciens EHA105 strains carrying different combinations of DNA constructs as indicated in the figures. Forty-eight hours after infiltration, leaves of Nb coexpressing different constructs were harvested and assayed for luciferase (LUC) activity with eight independent biological replicates. AtMYC2 promoter: LUC, AtMYB122 promoter: LUC, or AtPDF1.2 promoter: LUC was used as a reporter construct. The CaMV 35S promoter-driven AtWRKY20 or βC1 was used as effector constructs, and CaMV 35S promoter-driven YFP was used as the control. The experiment was repeated three times with similar results.
Statistical analyses
Significance of differences in insect performance, nonconsumed leaf area, gene expression levels and GS contents, relative fluorescence intensity, relative enrichment fold of DNA fragments in the promoter, GUS activity, and relative LUC activity was determined using Student’s t tests for comparing two treatments or two lines or using one-way analysis of variance (ANOVA), followed by Duncan’s multiple-range tests for more than two lines or treatments.
Supplementary Material
Acknowledgments
We thank Y. Liu (Tsinghua University, China) for providing CLCuMuV infectious clones pCAMBIA-2300-CA and pCAMBIA-2300-β. We thank J. Wu, J. Liu, L. Su, Y. Wu (Institute of Microbiology, Chinese Academy of Sciences), and S. Jie (Shihezi University, Xinjiang, China) for their invaluable assistance with research materials and experiments. Funding: The study was supported by the Chinese Academy of Sciences (Strategic Priority Research Program grant no. XDB11040300), the National Natural Science Foundation of China (grant nos. 31830073, 31522046, 31672001, 31701783, 31522046, and 31390421), and China Postdoctoral Science Foundation (grant no. 2016 M600142). Author contributions: J.Y. and P.Z. conceived and designed experiments, which were mainly carried out by P.Z. The levels of GSs were measured by C.C., M.W, and Q.W. Insect assays were performed with the help of X.Y., J.D., and Z.Z. Begomovirus infections were performed by Y.S. Screening T-DNA lines of WRKY20 and yeast works were performed by R.L. Data were analyzed by P.Z. and J.Y. The manuscript was written by P.Z. and J.Y. with contributions from D.J.K, S.-S.L., and R.-X.F. Competing interests: J.Y. and P.Z. are inventors on a patent related to this work, filed by the Chinese Academy of Sciences (no. 201811183208.6, filed 11 October 2018). The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/8/eaav9801/DC1
Fig. S1. Whitefly vector competes with nonvector CBM on cotton.
Fig. S2. TYLCCNV βC1 protein interacts with WRKY20 proteins.
Fig. S3. WRKY20 mediates plant immunity against whitefly.
Fig. S4. WRKY20 and MYC2 form a negative feedback loop.
Fig. S5. Plant WRKY20 is a dual-function transcription factor controlling GS biosynthesis.
Fig. S6. WRKY20 directly targets GS biosynthetic-related genes by binding to their promoters.
Fig. S7. WRKY20 regulates a JA-mediated GS accumulation in a vascular-specific pattern.
Fig. S8. βC1 suppresses the WRKY20 activity by interfering with its homodimerization.
Fig. S9. WRKY20 negatively regulates SA-mediated defense against the green peach aphid.
Table S1. DNA primers used in this study.
REFERENCES AND NOTES
- 1.Eigenbrode S. D., Bosque-Pérez N., Davis T. S., Insect-borne plant pathogens and their vectors: Ecology, evolution, and complex interactions. Annu. Rev. Entomol. 63, 169–191 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Weaver S. C., Charlier C., Vasilakis N., Lecuit M., Zika, chikungunya, and other emerging vector-borne viral diseases. Annu. Rev. Med. 69, 395–408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mithöfer A., Boland W., Plant defense against herbivores: Chemical aspects. Annu. Rev. Plant Biol. 63, 431–450 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Zhang P.-J., Wei J.-N., Zhao C., Zhang Y.-F., Li C.-Y., Liu S.-S., Dicke M., Yu X.-P., Turlings T. C. J., Airborne host–plant manipulation by whiteflies via an inducible blend of plant volatiles. Proc. Natl. Acad. Sci. U.S.A. 116, 7387–7396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurtovic A., Widmer A., Dickson B. J., A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature 446, 542–546 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Hanley-Bowdoin L., Bejarano E. R., Robertson D., Mansoor S., Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 11, 777–788 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Mauck K. E., Chesnais Q., Shapiro L. R., Evolutionary determinants of host and vector manipulation by plant viruses. Adv. Virus Res. 101, 189–250 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Li R., Weldegergis B. T., Li J., Jung C., Qu J., Sun Y., Qian H., Tee C., van Loon J. J. A., Dicke M., Chua N.-H., Liu S.-S., Ye J., Virulence factors of geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell 26, 4991–5008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X., Xu S., Zhao P., Zhang X., Yao X., Sun Y., Fang R., Ye J., The Orthotospovirus nonstructural protein NSs suppresses plant MYC-regulated jasmonate signaling leading to enhanced vector attraction and performance. PLOS Pathog. 15, e1007897 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saad K. A., Mohamad Roff M. N., Hallett R. H., Abd-Ghani I. B., Effects of cucumber mosaic virus-infected chilli plants on non-vector Bemisia tabaci (Hemiptera: Aleyrodidae). Insect Sci. 26, 76–85 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Du Z., Tang Y., He Z., She X., High genetic homogeneity points to a single introduction event responsible for invasion of Cotton leaf curl Multan virus and its associated betasatellite into China. Virol. J. 12, 163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S.-S., De Barro P. J., Xu J., Luan J.-B., Zang L.-S., Ruan Y.-M., Wan F.-H., Asymmetric mating interactions drive widespread invasion and displacement in a whitefly. Science 318, 1769–1772 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Yang X., Guo W., Li F., Sunter G., Zhou X., Geminivirus-associated betasatellites: Exploiting chinks in the antiviral arsenal of plants. Trends Plant Sci. 24, 519–529 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Yang J.-Y., Iwasaki M., Machida C., Machida Y., Zhou X., Chua N.-H., βC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev. 22, 2564–2577 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia Q., Liu N., Xie K., Dai Y., Han S., Zhao X., Qian L., Wang Y., Zhao J., Gorovits R., Xie D., Hong Y., Liu Y., CLCuMuB βC1 subverts ubiquitination by interacting with NbSKP1s to enhance geminivirus infection in Nicotiana benthamiana. PLOS Pathog. 12, e1005668 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu K.-M., Lu Y.-H., Feng H.-Q., Jiang Y.-Y., Zhao J.-Z., Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin–containing cotton. Science 321, 1676–1678 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Cui X., Tao X., Xie Y., Fauquet C. M., Zhou X., A DNA β associated with Tomato yellow leaf curl China virus is required for symptom induction. J. Virol. 78, 13966–13974 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dombrecht B., Xue G. P., Sprague S. J., Kirkegaard J. A., Ross J. J., Reid J. B., Fitt G. P., Sewelam N., Schenk P. M., Manners J. M., Kazan K., MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19, 2225–2245 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eulgem T., Rushton P. J., Robatzek S., Somssich I. E., The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Schweizer F., Fernández-Calvo P., Zander M., Diez-Diaz M., Fonseca S., Glauser G., Lewsey M. G., Ecker J. R., Solano R., Reymond P., Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25, 3117–3132 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guan C., Zhou X., Phloem specific promoter from a satellite associated with a DNA virus. Virus Res. 115, 150–157 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Wang X.-W., Li P., Liu S.-S., Whitefly interactions with plants. Curr. Opin. Insect Sci. 19, 70–75 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Züst T., Heichinger C., Grossniklaus U., Harrington R., Kliebenstein D. J., Turnbull L. A., Natural enemies drive geographic variation in plant defenses. Science 338, 116–119 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Li B., Tang M., Nelson A., Caligagan H., Zhou X., Clark-Wiest C., Ngo R., Brady S. M., Kliebenstein D. J., Network-guided discovery of extensive epistasis between transcription factors involved in aliphatic glucosinolate biosynthesis. Plant Cell 30, 178–195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S., Petersen B. L., Olsen C. E., Schulz A., Halkier B. A., Long-distance phloem transport of glucosinolates in Arabidopsis. Plant Physiol. 127, 194–201 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nintemann S. J., Hunziker P., Andersen T. G., Schulz A., Burow M., Halkier B. A., Localization of the glucosinolate biosynthetic enzymes reveals distinct spatial patterns for the biosynthesis of indole and aliphatic glucosinolates. Physiol. Plant. 163, 138–154 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Chen H., Zhang Z., Teng K., Lai J., Zhang Y., Huang Y., Li Y., Liang L., Wang Y., Chu C., Guo H., Xie Q., Up-regulation of LSB1/GDU3 affects geminivirus infection by activating the salicylic acid pathway. Plant J. 62, 12–23 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Qi T., Song S., Xie D., Modified bimolecular fluorescence complementation assay to study the inhibition of transcription complex formation by JAZ proteins. Methods Mol. Biol. 1011, 187–197 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Van der Does D., Leon-Reyes A., Koornneef A., Van Verk M. C., Rodenburg N., Pauwels L., Goossens A., Körbes A. P., Memelink J., Ritsema T., Van Wees S. C. M., Pieterse C. M. J., Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25, 744–761 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brault V., Uzest M., Monsion B., Jacquot E., Blanc S., Aphids as transport devices for plant viruses. C. R. Biol. 333, 524–538 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Züst T., Agrawal A. A., Mechanisms and evolution of plant resistance to aphids. Nat. Plants 2, 15206 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Zarate S. I., Kempema L. A., Walling L. L., Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol. 143, 866–875 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young H. S., Parker I. M., Gilbert G. S., Sofia Guerra A., Nunn C. L., Introduced species, disease ecology, and biodiversity-disease relationships. Trends Ecol. Evol. 32, 41–54 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Scheele B. C., Pasmans F., Skerratt L. F., Berger L., Martel A., Beukema W., Acevedo A. A., Burrowes P. A., Carvalho T., Catenazzi A., De la Riva I., Fisher M. C., Flechas S. V., Foster C. N., Frías-Álvarez P., Garner T. W. J., Gratwicke B., Guayasamin J. M., Hirschfeld M., Kolby J. E., Kosch T. A., La Marca E., Lindenmayer D. B., Lips K. R., Longo A. V., Maneyro R., McDonald C. A., Mendelson J. III, Palacios-Rodriguez P., Parra-Olea G., Richards-Zawacki C. L., Rödel M.-O., Rovito S. M., Soto-Azat C., Toledo L. F., Voyles J., Weldon C., Whitfield S. M., Wilkinson M., Zamudio K. R., Canessa S., Amphibian fungal panzootic causes catastrophic and ongoing loss of biodiversity. Science 363, 1459–1463 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Vilcinskas A., Stoecker K., Schmidtberg H., Röhrich C. R., Vogel H., Invasive harlequin ladybird carries biological weapons against native competitors. Science 340, 862–863 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Casteel C. L., Yang C., Nanduri A. C., De Jong H. N., Whitham S. A., Jander G., The NIa-Pro protein of Turnip mosaic virus improves growth and reproduction of the aphid vector, Myzus persicae (green peach aphid). Plant J. 77, 653–663 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Qu J., Ye J., Geng Y.-F., Sun Y.-W., Gao S.-Q., Zhang B.-P., Chen W., Chua N.-H., Dissecting functions of KATANIN and WRINKLED1 in cotton fiber development by virus-induced gene silencing. Plant Physiol. 160, 738–748 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye J., Yang J. Y., Sun Y., Zhao P., Gao S., Jung C., Qu J., Fang R., Chua N.-H., Geminivirus activates ASYMMETRIC LEAVES 2 to accelerate cytoplasmic DCP2-mediated mRNA turnover and weakens RNA silencing in Arabidopsis. PLOS Pathog. 11, e1005196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jefferson R. A., Kavanagh T. A., Bevan M. W., GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sønderby I. E., Burow M., Rowe H. C., Kliebenstein D. J., Halkier B. A., A complex interplay of three R2R3 MYB transcription factors determines the profile of aliphatic glucosinolates in Arabidopsis. Plant Physiol. 153, 348–363 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/8/eaav9801/DC1
Fig. S1. Whitefly vector competes with nonvector CBM on cotton.
Fig. S2. TYLCCNV βC1 protein interacts with WRKY20 proteins.
Fig. S3. WRKY20 mediates plant immunity against whitefly.
Fig. S4. WRKY20 and MYC2 form a negative feedback loop.
Fig. S5. Plant WRKY20 is a dual-function transcription factor controlling GS biosynthesis.
Fig. S6. WRKY20 directly targets GS biosynthetic-related genes by binding to their promoters.
Fig. S7. WRKY20 regulates a JA-mediated GS accumulation in a vascular-specific pattern.
Fig. S8. βC1 suppresses the WRKY20 activity by interfering with its homodimerization.
Fig. S9. WRKY20 negatively regulates SA-mediated defense against the green peach aphid.
Table S1. DNA primers used in this study.


