Abstract
Electrospray ionization mass spectrometry (ESI-MS) is a ubiquitously used analytical method applied across multiple departments in biopharma, ranging from early Research Discovery to Process Development. Accurate, efficient and consistent protein MS spectral deconvolution across multiple instrument and detector platforms (ToF, Orbitrap, FT-ICR) is essential. When proteins are ionized during the ESI process, a distribution of consecutive multiply charged ions are observed on the m/z scale, either positive [M+nH]n+ or negative [M-nH]n− depending on the ionization polarity. The manual calculation of the neutral molecular weight (MW) of single proteins measured by ESI-MS is simple, however algorithmic deconvolution is required for more complex protein mixtures to derive accurate MWs. Multiple deconvolution algorithms have evolved over the past two decades, all of which have their advantages and disadvantages, in terms of speed, user-input parameters (or ideally lack thereof) and whether they perform optimally on proteins analyzed under denatured or native solution and MS conditions. Herein we describe the utility of a parsimonious deconvolution algorithm (explaining the observed spectra with a minimum number of masses) over of a wide range of highly diverse biopharma relevant and research grade proteins and complexes (PEG-GCSF; an IgG1k; IgG1 and IgG2-biotin covalent conjugates; the membrane protein complex AqpZ; a highly polydisperse empty nanodisc, MSP1D1 and the tetradecameric chaperone protein complex GroEL) analysed under native MS, denaturing LC-MS, positive and negative modes of ionization, using multiple instruments and therefore multiple data formats. The implementation of a comb filter and peak sharpening option are also demonstrated to be highly effective for deconvolution of highly polydisperse and enhanced separation of a low level lysine glycation post translational modification (+162.1 Da), partially processed heavy chain lysine resides (+128.1 Da) and loss of N-Acetylglucosamine (GlcNAc; −203.1 Da) respectively.
Graphical Abstract

Introduction
Mass spectrometry plays a critical role in multiple stages of pharmaceutical research. From small molecule medicinal chemistry research efforts1 to high throughput screening efforts of drug targets2 to monoclonal antibody separation and accurate molecular weight determination3, mass spectrometry (MS) is a ubiquitous analytical method throughout biopharma. All the aforementioned examples rely on either liquid chromatographic (LC) separation or solid phase extraction (SPE) prior to MS analysis. A far less routine MS analytical method, in pharma at least, is native MS, where non-covalent protein-drug or protein-protein4 interactions remain intact within the gas-phase of the MS. In both cases, algorithmic spectral deconvolution is routinely performed within pharma, for routine accurate and rapid MW determination, on data derived from multiple instrument platforms (time-of-flight, Orbitrap and Fourier transform ion cyclotron resonance MS systems).
Since the initial demonstration of native MS experiments on proteins and complexes by Chait5, Loo6 and others7, 8, this unique area of MS has steadily grown from what was initially a niche area, to a fully established research field, described as gas-phase structural biology9, 10. The proteins investigated using native MS and solution conditions (typically 200 mM ammonium acetate, pH 6–711, 12) have ranged from the original holo-myoglobin5 and the alcohol dehydrogenase homotetrameric complex13, to multi-subunit complexes such as GroEL14 and valinyl-oxidase15 described in the late 1990s and early 2000s, to the present day, highly polydisperse nanodisc molecules16, 17, membrane proteins18 and mega-Dalton virus capsids19. Denaturing LC-MS has also proven itself to be a highly enabling platform for the rapid determination of accurate MWs of denatured proteins 20 and is routinely used within pharmaceutical research for monoclonal antibody characterization, bispecific antibodies and proteins of therapeutic interest2, 3, 21,22.
MW determination of a protein or complex can be performed either manually or through software, by performing data smoothing and centroid processing, followed by adjacent peak assignment (Figure S1, Supporting Information) based on the following formulae reproduced from Fenn23:
| (1) |
where z is the calculated charge for m1; m2 is the ion with x less charge, therefore in this case x=1, appearing higher in the m/z scale; x can also be 2, 3, 4, 5 etc, as long as the correct m/z value for m2 is chosen; mp is the mass of the proton (1.00728 Da). The numerical values for m1 and m2 are based on MS derived m/z values. Once the value for z is determined, the intact MW can easily be calculated:
| (2) |
For the more complex MS spectra, such as those derived from tandem MS experiments of poly disperse molecules, such as alpha-Crystallin B24, choatropic partial disruption of transcription factor iEF325 and the highly polydisperse empty MSP1D1 nanodisc16, 17 manual peak picking is challenging, if not impossible. In these cases, algorithmic deconvolution is a prerequisite for accurate MW determination. However recently, effective manual MW determination of an empty MSP1D1 nanodisc has been described17.
One of the first and arguably the most heavily used protein deconvolution algorithms is the Bayesian probability based Maximum Entropy26. Maximum Entropy was originally designed to deconvolve MS data of low MW, denatured, multiply charged protein spectra, acquired on low resolution quadrupole-based instruments20. On MS instruments where the proteins are analysed in near neutral pH aqueous solutions11, 12 the measured charge states are typically wider than the expected isotopic peak width distribution and also asymmetric due to solution, buffer and salt adducting11.
To date there are multiple algorithms available for protein ESI MS spectral deconvolution that have evolved over the last twenty years, some of which have focused on denaturing protein spectral data23, 26, 27. However, recently with the advent of native MS9, 10, there has been a renewal of interest in charge deconvolution algorithms28–33. UniDec29 and FFT-based deconvolution32 represent a significant step forward in protein spectral deconvolution. UniDec has the ability to efficiently deconvolve ion mobility based and highly polydisperse native MS data, such as those generated on empty MSP1D1 nanodiscs. UniDec also incorporates a comb filter which allows the user to explicitly define the MW repeat of the incorporated phospholipid. The FFT-based deconvolution method developed in the Prell lab32 does not require any prior known charge or repeat unit values, but solely relies on the fundamental frequencies and higher harmonics for MW determination of highly polydisperse and polymeric ions such as nanodiscs and polyethelene glycol. The latest algorithm development called PMI Intact (Protein Metrics) enables rapid and efficient deconvolution of native MS and denaturing LC-MS spectral data.
Herein, we present the deconvolution of native and denatured MS spectral data for a monoclonal antibody (NIST IgG1k); an antibody drug conjugate-like molecule (IgG1 and IgG2 conjugated to biotin); the PEG-GCSF protein; a membrane protein (AqpZ); an empty nanodisc (MSP1D1) and a chaperone protein complex (GroEL), acquired on a quadrupole time-of-flight (Q-ToF), an LC-ToF, a Fourier transform ion cyclotron resonance (FT-ICR) and an Orbitrap MS instrument using both positive and negative modes of ionization; all of which are discussed in the context of a biopharmaceutical relevant universal deconvolution algorithm. The deconvolution algorithm described is the PMI Intact Mass algorithm33, 34 which uses both forward (m to m/z) and backward (m/z to m) mappings. The Intact Mass algorithm also includes a step to bias the deconvoluted neutral mass spectrum to a “parsimonious” solution with minimal mass peaks as necessary to explain the observed m/z spectrum.
Materials and Methods
Mass Spectrometry
Nano-electrospray ionization (nESI) native MS was performed using the following source voltages and pressures: Q-ToF Synapt G1 (Waters Corporation): source temperature 25 °C, source backing pressure 6.0 mbar, sample cone 25–200 V, trap collision voltage 75–125 V, in cC4F8; OrbitrapEMR (ThermoScientific): source transfer capillary temperature 250 °C, source collision induced dissociation 80 V, higher-energy collision induced dissociation (HCD) 20 V, N2; 15 Tesla solariX FT-ICR (Bruker Daltonics): source transfer capillary temperature 100 °C, Skimmer 1 50 V , collision cell voltage 30 V, in SF6. A more detailed discussion of all instrument parameters can be found in the Supporting Information. Protein samples were all in the concentration range of 10–20 μM, in 200 mM ammonium acetate and introduced in to the MS systems using a gold coated glass nESI needles (Long thin wall, M956232AD1-S; Waters Corporation) in positive and negative ionization mode. High m/z calibration was performed in under both positive and negative nESI modes of acquisition using a 100 μg/μL solution of CsI (Figure S2, Supporting Information). Denaturing LC-MS was performed on an open access enabled 6230 ToF MS (Agilent) connected to an Infinity 1290 LC (Agilent) system, operated with a Zorbax SB300, C8 50 × 2.1 mm, 3.5 μm analytical column. More detailed native-MS and LC-MS conditions are noted in the Supporting Information.
Materials
The following proteins and complexes were used in this study: a homotetrameric membrane protein AqpZ35; an IgG1 mAb biotin covalent conjugate34 (deglycosylated using PNGaseF, QA Bio, E-PNG01); an IgG2 conjugated with NHS-PEG12-Biotin (ThermoFisher Scientific, 21312; 2.5 molar equivalents prepared in an identical manner to those described in34); an empty MSP1D1 nanodisc17; the PEG-GCSF protein36; a tetradecameric chaperone complex GroEL12 and the NIST IgGk mAb37. All proteins and complexes were buffer exchanged in to aqueous 200 mM ammonium acetate (diluted from Sigma-Aldrich 7.5 M stock, A2705–500ML) using a P6 micro bio-spin filter (BioRad, 7326221). The AqpZ 200mM ammonium acetate solution also contained 1.1% w/v octyglucoside35.
Computation
A detailed algorithm description can be found in the Supporting Information. Briefly, PMI Intact uses an iterative algorithm to deduce the mix of charges in each small interval of the m/z spectrum. All charge values are set equally likely for the first deconvolved mass spectrum; new charge values are then computed from the previous deconvolved mass spectrum and the process is repeated. PMI Intact applies a small “parsimony” bias against m/z intervals with many different charges, because multiple true masses mapping to the same m/z bin are less common than deconvolution artifacts caused by charge uncertainty. On each iteration, the algorithm updates the charge vectors, which provide probabilities for each charge at each point of the observed m/z spectrum. New charge vectors are determined by the last deconvolved mass spectrum along with a-priori assumptions about smoothness of charging and likelihood of mass coincidences. The new charge vectors give a new deconvolved mass spectrum, and each iteration reduces the sum of the squares of the differences between the observed m/z spectrum and the m/z spectrum computed from the last set of charge vectors and deconvolved mass spectrum. For polydisperse targets such as nanodiscs, the algorithm can incorporate a user defined comb filter. For example, 677.5 Da would be used to describe the delta mass for a nanodisc lipid containing dimyristoylphosphocholine (DMPC; Figure S3, Supporting Information). Native and denaturing MS deconvolution was performed using PMI Intact (w2.15–584 develop; Protein Metrics Inc). Raw unprocessed MS data files are dragged directly into the Create Project User Interface (Figures S4–S5, Supporting Information). More detailed discussions of the “Advanced” deconvolution parameters can be found in Figure S6 (Supporting Information). For additional algorithm information please refer to the Supporting Information or Bern et. al.33.
Results and Discussion
Native-MS Deconvolution
It is important to note that the PMI software is vendor neutral and accepts spectral data directly from the raw, unprocessed data files, therefore does not need to be converted to text format (typically m/z versus intensity) prior to deconvolution. The full deconvolution process ranges from approximately 0.5–2.0 min per data file (nESI infusion and LC-MS; and number of iteration and processor dependent) based on a HP Z620 Workstation (Intel Xeon 3.7 GHz, 16GB RAM, 12 cores) and the files described herein were processed across a network (data files not stored locally on the processing PC).
Figures 1a–d display the deconvoluted spectral data for multiple proteins and complexes ranging in MW (97.1 kDa to 802.4 kDa), stoichiometry (up to a tetradecamer) and polydispersity, all measured under native MS and solution conditions (200 mM ammonium acetate) by nESI using different MS instruments. See Supporting Information, Figures S7 and S8 for a discussion regarding denatured and native MS theoretical versus instrument derived peak widths.
Figure 1.
Native MS analyses using multiple MS instrumentation and subsequent algorithmic deconvolution of a diverse range of pharmaceutically relevant and research grade protein constructs: a) the NIST IgG1k mAb standard analyzed by nESI native-MS mode by Orbitrap-EMR MS; b) an IgG1 lysine-biotin (10 molar biotin equivalent;(34) PNGaseF treated) conjugate, analyzed by nESI native-MS by Q-ToF MS; c) aquaporin-Z analyzed by nESI native-MS by FTICR MS;(35) d) GroEL analyzed by negative native-MS nESI by Q-ToF MS. Insets display the unprocessed data. The most intense charge state, the formylation stoichiometry, and the drug-to-antibody ratio (DAR) values are annotated.
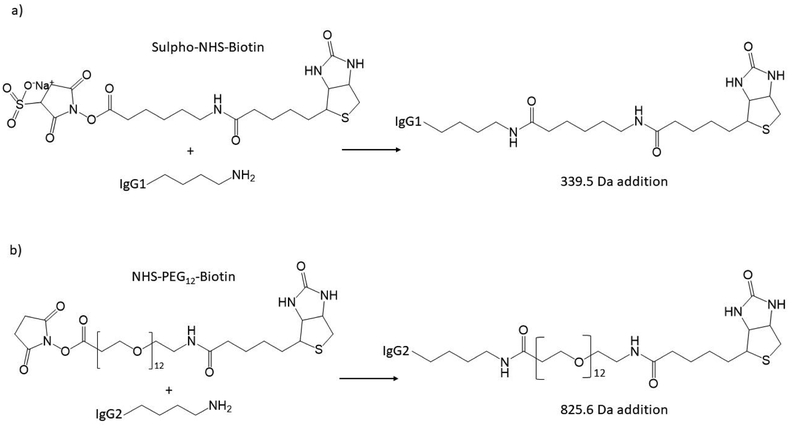
Figures 1a and 1b represent typical monoclonal antibodies (mAbs) and antibody drug conjugates (ADCs) which are highly specific and potent modalities used to treat multiple disease indications38, 39. Figure 1a displays the NIST IgG1k mAb standard analysed under positive ion native nESI mode on an Orbitrap-EMR MS. As can be seen, the main glycoforms are easily identified (G0F/G0F, G0F/G1F, G1F/G1F and G1F/G2F). A lower intensity and lower MW species are also identified as the aglycosylated G0F, G1F and G2F (146.5 kDa to 146.9 kDa40). Figure 1b represents a biotin conjugated IgG1 (Scheme 1a; 10 molar biotin equivalents) of relatively simple spectral complexity and MWs ranging from 145.9 kDa to 147.7 kDa, analysed under positive ion native nESI mode on a Q-ToF MS Adjacent mass differences correspond to 338.8 +/−10.1 Da (theoretical difference is 339.5 Da) representing biotin covalent conjugation to native lysine residues consistent to those previously described by FT-ICR34.
Scheme 1.
The synthetic scheme of lysine conjugation used to covalently modify the IgG1 and IgG2 mAbs described herein; a) sulfo-NHS-LC-biotin lysine conjugation34; b) NHS-PEG12-biotin lysine conjugation. A single native lysine residue is represented as only the primary amine side chain. Observed MW additions for a single conjugation are annotated (Da, average MW).
Membrane proteins constitute over 50% of current druggable targets41, 42 therefore their characterization by pharma using MS is of high importance17, 35, 43. The analysis of AqpZ, acquired on an FT-ICR (Figure 1c) represents a membrane protein homotetrameric complex with a low level of polydispersity (n = 0 to 4). For AqpZ, the observed MW differences in the deconvolved spectrum are small, ranging from 97.1 to 97.2 kDa, representing a previously described N-terminal formylation (theoretical MW addition of a formylation is 28.01 Da35). Based on the observed MW differences of adjacent formylation proteoforms (32.8 +/−1.9 Da) it would be challenging to positively determine which post translational modification is present. One would require either ultrahigh resolution mass measurements44 and/or proteolytic digestion35. An additional larger MW is observed corresponding to an approximate 182 Da increase, which is not observed when AqpZ is analysed under denaturing LC-MS conditions (Figure S9, Supporting Information), therefore we attribute this species to an unidentified non-covalent adduct.
Figure 1d represents the deconvolved spectrum of GroEL; the simplest in terms of spectral complexity presented herein. However, the measured charge states (negative nESI; z = 50- to 58-) are detected far higher in the m/z scale (>14,000) and represent a higher level of salt and buffer adduction than NIST IgG1k sample, the IgG1-biotin conjugate or the AqpZ complex, therefore representing a different challenge for spectral deconvolution. Upon deconvolution a major species of MW of 802.4 kDa is detected under negative nESI mode (consistent with the positive nESI mode data; Figure S10, Supporting Information). The raised baseline and partially resolved charge states in Figure 1d are indicative of additional species, close in MW. In both deconvolved spectra (positive and negative nESI) there is evidence of a lower MW species (791 kDa). Previously described GroEL spectra45, 46 have also displayed additional low level species present at similar m/z values to that of GroEL. The characterization of these species are beyond the scope of this manuscript, however are likely to be either truncated constructs of GroEL or additional protein complexes not removed during the purification procedure12. A denatured LC-MS GroEL spectrum is displayed in Figure S11 (Supporting Information) showing detection of additional lower MW species. Also note that the separation of this low intensity species from adjacent charge states is improved under negative nESI due to the charge states appearing higher in the m/z scale emphasizing the importance of the rarely utilized negative ion nESI in native MS47. In all the aforementioned cases (Figure 1a, b, c & d) highly comparable Basic and Advanced deconvolution parameters were used (Figure S6, Supporting Information).
Figure S12 (Supporting Information) displays a range of proteins (ubiquitin, myoglobin, NIST light and heavy chain and BSA) of varying MW (8.1 to 66.7 kDa) analysed by denaturing LC-MS, which have been deconvolved using the comparable settings to those used for native MS spectral processing described in Figures 1, 2 and 3. All spectra are artifact-free and are measured to an overall RMS error of 4.7 ppm to the expected theoretical values (Table S1, Supporting Information). In all processed examples described herein, an estimate of expected charge state distribution must be input and is typically set to a wide value (z = 10+ to 100+; Figure S6, Supporting Information). This is not the case for the original Maximum Entropy algorithm26 or the FFT-based deconvolution algorithm32 but is required for UniDec29. In rare cases such as the nanodisc16, 17, a charge state distribution can be challenging to predict and interpret. However, based on the MW versus z relationship established by de la Mora48, an approximate charge distribution can be predicted.. Additionally, it is also common practice to perform a “scouting” deconvolution over a wide m/z, MW and z range, then perform a narrow “focused” deconvolution. Ideally, a spectral deconvolution algorithm with minimal input parameters is preferred.
Figure 2.
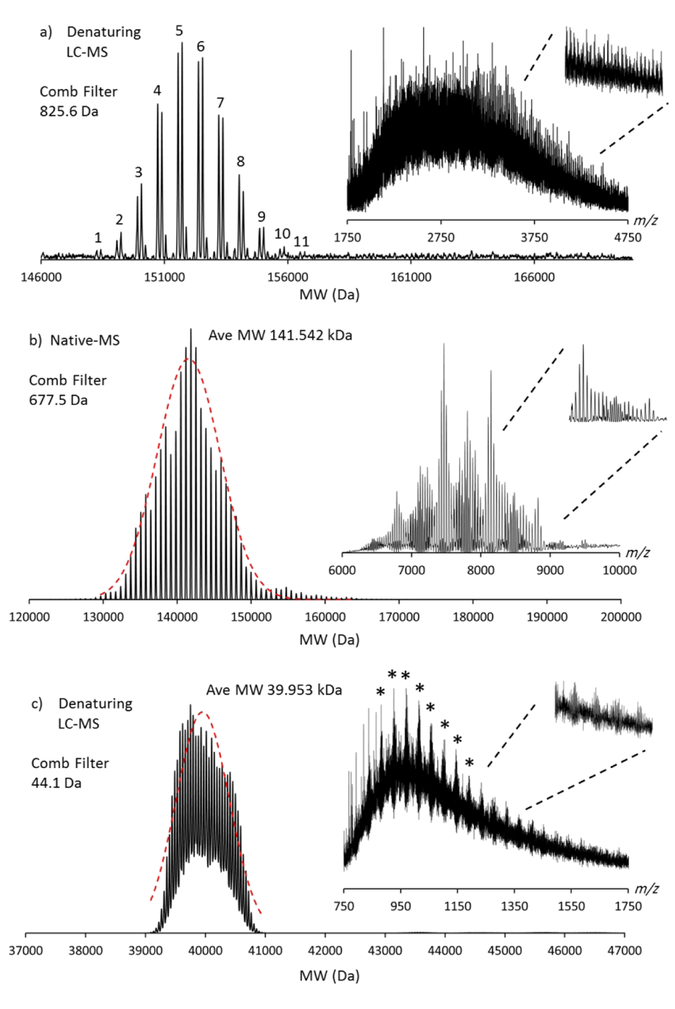
The effect of the comb filter (comb filter = 1) on the deconvolution of highly polydisperse MS spectral data: a) an IgG2 PEG12-biotin conjugate analyzed by denaturing LC-ToF MS; b) an empty MSP1D1 nanodisc(17) containing the phospholipid DMPC analyzed by nESI native-MS (Orbitrap-EMR); c) PEGylated protein analyzed by denaturing LC-ToF MS; * indicates free PEG differing by 44.1 Da. The MW (ave) is calculated from the superimposed (red hashed line) normal distribution. All insets display the unprocessed m/z data. The comb filter delta mass is also annotated. Figure S15a–c display the deconvolved spectra without the use of the comb filter.
Figure 3.
Comparative deconvolved spectra of the NIST IgG1k mAb analyzed under: a) denaturing LC-ToF MS; b) native-MS and solution conditions by FTICR. The insets in all cases display a zoom-in of the glycoform G0F/G1F post-peak sharpened processing; * represents a species previously postulated to be a low level unprocessed heavy chain lysine residue (+128.1 Da).(40) Minor differences in charge state distributions are observed when analyzed under native and denaturing MS conditions, consistent with other groups observations.(34,57,64) c) Deconvolved MWs of denatured and native-MS analyzed NIST mAb glycoforms and their respective errors (in ppm) were calculated from n = 1 experiments. The major glycoform theoretical MWs are reproduced from Formolo et al.(40) MW measurements were derived directly from the software centered peak.
Comb Filter: Deconvolution of high polydisperse pharmaceutical relevant molecules:
A comb filter sums or averages evenly spaced points in a signal. PMI Intact allows the user to specify a comb filter to average peaks corresponding to MWs with anticipated mass deltas. The comb filter was added to the “backwards step”. A comb filter of width 1 is implemented as an averaging filter with weights 0.25, 0.5, 0.25 applied to points in the last neutral MW spectrum at masses:
| (3) |
where Δ is the delta mass (79.98 Da for phosphorylation, for example) and m is the neutral MW. The averaged value is then used to set the probability for charge k at m/z point mi = 1.00728 + m/k. A comb filter of width 2 uses a weighted average of:
| (4) |
An ADC is composed of an antibody with high affinity to a specific target and a covalently attached cytotoxic agent, via native lysine, reduced cysteine or engineered cysteine residue49–51. The resultant drug-to-antibody ratio (DAR) typically ranges from 1 to 8 covalent drug conjugations, using a cleavable or non-cleavable linker52. Depending on the level/heterogeneity of the glycosylation and the MW of the covalently attached moiety, the resultant MS spectrum can be polydisperse. Currently, there are four ADCs approved by the US FDA and more in clinical trials53, 54. If one considers the FDA approved chimeric IgG1-based ADC Brentuximab vedotin (ADCETRIS®), multiple overlaps between glycosylated charge states (G0F/G0F, G0F/G1F, G1F/G1F) and the cysteine conjugated mono-methyl-aurustatin E (MMAE; MW 1316.6 Da55) will occur at higher DAR values, resulting in a complex MS spectrum. To reduce the spectral complexity, groups have deglycosylated (PNGaseF treated) and analysed ADCs under native-MS conditions56 where charge states appear higher in the m/z scale, resulting in an improved level of separation. However, processing highly congested MS data with an effective deconvolution algorithm, regardless of where in the m/z scale the charge states appear, an accurate MW should be readily achieved. Figure 2a displays the deconvolved spectrum of an IgG2 mAb (glycans intact) covalently modified with NHS-PEG12-biotin (2.5-molar equivalents) at native lysine residues (Scheme 1b) resulting in multiple covalent MW additions of 825.6 Da. Within the lower m/z regions of the spectrum, the data is highly congested, however, higher regions of the spectrum (m/z 3750–4750) individual charge states begin to be resolved (Figure 2a, Inset). Including the comb filter (delta mass 825.6 Da) effective deconvolution was achieved. Utilizing the comb filter, the detection of lower S/N species, such as DAR 1, DAR2, DAR 10 and DAR 11 are now significantly improved (Figure S15a, Supporting Information). The average DAR value, with and without the use of the comb filter is also subtly different (5.63 vs 5.54 respectively). This has implications for not only which techniques (LC-MS, native MS, LC-UV, LC-HIC) are used to derive the DAR value34, 57, but also which deconvolution parameters are used to process the MS data; the use of consistent parameters and algorithms is key to optimized experimental process and consistent results.
Figure 2b represents an empty MSP1D1 nanodisc acquired on the Orbitrap-EMR instrument using intermediate activation energies (Supporting Information). Nanodiscs are enabling membrane mimetics and have been demonstrated as an effective means of immobilizing membrane proteins for further drug or fragment screening campaigns within pharma, using surface plasmon resonance58. The Inset displays a broad, polydisperse spectrum with clear areas of constructive overlap59. Effective deconvolution is achieved using a comb filter delta mass 677.5 Da (average MW of DMPC). An average MW of 141.542 kDa is derived of which there are approximately 143 ± 20–30 DMPC molecules (based on 2 x membrane scaffold proteins of MW 22.4 kDa) constituting this empty MSP1D1 nanodisc molecule, consistent with values previously described by native MS16, 17 and analytical ultracentrifugation58. The MW polydispersity determined using PMI (130 kDa to 160 kDa) using an applied Comb Filter setting of 1, is consistent with previously deconvolved MSP1D1 nanodisc spectral data17.
PEGylation is used to enhance the half-life of therapeutically active molecules60, 61 however, MS analyses typically result in highly polydisperse spectra within which neutral MWs cannot be ascertained manually; algorithmic deconvolution is essential. Figure 2c shows a deconvolved average MW distribution of 39.953 kDa. This data represents a 18.8 kDa protein covalently modified with 21.9 kDa PEG similar to that described by Bagal36. The deconvolution was achieved using a comb filter delta mass of 44.1 Da resulting in an average MW of 39.953 kDa consistent with that previously published, without the need for spectral simplification by charge reduction36. The LC-MS deconvolved MW (Figure 2c) is highly consistent with that derived by linear MALDI-TOF-MS (40.050 kDa; Figure S13, Supporting Information) and contrary to recent opinion36, MALDI-MS is in fact an effective analytical method for MW determination of larger PEGylated protein constructs., with the caveat that linear MALDI-TOF will result in a larger MW spread (Figure S16, Supporting Information). Figure 2c inset shows the unprocessed data (from LC-MS) and highlighted (*) are a series of abundant ions differing by 44.1 Da superimposed on top of a highly polydisperse series of protein-PEG conjugate charge states and in the m/z scale where the two series overlap, constructive enhancement is observed. These PEG ions are likely to be a result of fragmentation within the MS instrument, as operating at a lower source fragmentor voltage reduced the intensity of these interferences and subtle differences in the deconvolved ion distribution are therefore also evident (Figure S14, Supporting Information). Figure S15 (Supportng Information) displays the deconvolved data the IgG2-PEG12-Biotin, the empty MSP1D1 nanodisc and the PEG-GCSF molecule, all processed without the use of the comb filter. In all cases, the implementation of the comb filter improves spectral S/N. Finally, a brief but useful comparison and discussion is made between PMI Intact and five of the most commonly used protein deconvolution algorithms to process the highly polydisperse PEG-GCSF LC-MS spectral data: MaxEnt (MassLynx, Waters), MaxEnt and PMod (MassHunter, Agilent), UniDec29 and iFAMS62 (Figure S16, Supporting Information). In brief, a full algorithm comparison is well beyond the scope of this manuscript, however, qualitatively, the more recently developed algorithms such as UniDec29, iFAMS62 and PMI33 appear to produce deconvolved spectra of superior quality.
Comparing Native and Denaturing MS Spectra
One must now consider whether deconvolution parameters can remain constant when processing denatured and native-MS spectral data and whether mass measurement parity is retained, for the same protein molecule. In this case, the NIST IgG1k mAb is compared. Figure 3a represents the NIST IgG1k analysed under denaturing LC-MS conditions (oa-ToF, C8 reversed phase using n- propanol, TFA 0.1% and 70 °C3; Supporting Information) and Figure 3b, the native-MS and solution condition spectrum (nESI, 15 T FT-ICR, 200 mM ammonium acetate, Supporting Information). The zero-charged deconvolved MW and mass measurement for the glycoforms G0F/G0F-GlcNAc, G0F/G0F, G0F/G1F, G1F/G1F, G1F/G2F are presented in Figure 3c. The denatured LC-MS and native-MS spectra display highly comparable MWs and respective mass measurement values. Only the glycoform G0F/G0F-GlcNAc displays a significant difference in measured mass accuracy. Less adduction is observed under LC-MS denaturing conditions, therefore, improved mass measurement is achieved (unprocessed NIST data are displayed in Figure S17; Supporting Information). In the denatured LC-MS data (Figure 3a) a lower m/z leading edge species is detected. This species can be further resolved (Figure 3a, Inset, zoom of glycoform G0F/G1F) by using the Peak Sharpening option under Advanced Settings (Figures S6 and S18, Supporting Information). This feature can also be detected as a leading edge in the native- MS FT-ICR spectrum (Figure 3b). Upon Peak Sharpening, this species is further resolved (Figure 3d Inset) and is highly consistent to that presented in Figure 3a. This minor species has previously been characterized as an uncleaved C-terminal heavy chain lysine residue (+128.1 Da40) superimposed (but partially resolved) over the adjacent glycoform (G0F/G1F) species (des-K form). However, the MW difference obtained using two separate acquisitions (denaturing LC- MS-ToF and native FT-ICR MS) are also consistent with a loss of a GlcNAc (−203.1 Da) from an adjacent glycoform. Figure S19a (Supporting Information), shows the deconvolved LC-MS for the NIST mAb heavy chain where multiple, well resolved −GlcNAc (−203.1 Da) species are detected. In Figure S19b (Supporting Information) a +128.1 Da addition is detected, representing a low level unprocessed heavy chain C-terminal lysine residue40. It is likely these leading-edge partially resolved species in Figures 3a and 3b, are in fact a mixture of +128.1 Da and −203.1 Da and relative ion intensity values appear to support this hypothesis (Figures S18 and S19, Supporting Information).
A similar comparison between the IgG1-biotin 5-molar equivalent analysed under denaturing LC- MS and native MS (Figure S20, Supporting Information). In both cases, the application of the peak-sharpening feature allows for the improved definition and mass measurement of the +162 Da glycation; a lysine PTM commonly observed in mAbs63. This improvement in glycation definition has also been demonstrated through FT-ICR transient apodization34. Additionally, the trailing edge shoulder is now partially resolved in the native-MS spectra. The mass difference is approximately 40 Da, therefore likely a noncovalent sodium, potassium, ammonium adduct, or multiples thereof. Minor differences in charge state distributions of mAb conjugates and ADCs analyzed under native and denaturing MS conditions and related analytical techniques have been previously addressed by multiple groups34, 57, 64. We assume this difference also holds true for mAb glycoforms described herein (Figure 3a & 3b; note the minor intensity differences between glycoforms G1F/G2F).
Conclusions
The use of a parsimonious deconvolution algorithm has been demonstrated to efficiently deconvolute spectral data acquired for proteins and complexes, both pharmaceutically relevant constructs and research grade standards, analyzed under native MS and denaturing conditions (LC- MS) under both positive and negative modes of ionization. MS data from three different analysers (oa-ToF, Orbitrap and FT-ICR) and four different instrument vendors (Waters, ThermoScientific, Agilent and Bruker) were successfully deconvoluted without any file format change.
The proteins and complexes analysed varied in MW, stoichiometry and m/z range; the NIST IgG1k (mAb, 148.3 kDa); an IgG1-biotin conjugate (ADC-like; 146.5 kDa), IgG1-PEG-Biotin (ADC- like; 147.5 kDa); a PEG-GCSF (39.9 kDa; up to 43 measurable PEG 20k units) an empty MSP1D1 nanodisc (141.5 kDa; two membrane scaffold proteins, approximately 124 to 170 measurable DMPC phospholipid molecules,), the membrane protein AqpZ (noncovalent homotetramer, 97.5 kDa) and the chaperone protein complex GroEL (homotetradecameric, 802.4 kDa). Highly comparable deconvolution parameters were used in all cases and the resultant zero-charged spectra are artifact free (zero harmonics; third, half, double, and triple multiples of the protein MW). Additionally, when processing denatured LC-MS or native MS spectral data (of the same constructs, NIST IgG1k and the IgG1-biotin conjugate), the deconvolution parameters remained constant and unchanged. In both cases, the deconvolved, zero-charged data peak widths consistently reflect those of the unprocessed data. Mass accuracy is also highly comparable.
From an industrial and biopharmaceutical perspective, this deconvolution algorithm suite is highly advantageous, as most laboratories within a Research Discovery and Process Development setting will likely use multiple MS instruments from different vendors; the ability to drag-and-drop multiple MS data files of different format and subsequently process, is highly attractive. Also, in certain cases it may be required that both denaturing and native MS analyses be performed on the same protein construct. For example, one may want to derive an accurate mAb MW through LC- MS analysis3; levels of specific covalent modification from high throughput screening campaign2; drug-to-antibody ratio34, 57, 65; assess the levels of degradation66 of biotherapeutic molecules or assess the levels of aggregation (by SEC coupled to native MS67) present in the sample. Native- MS in biopharma is also used for assessing the correct assembly of a nanodisc; it is rapid (ca. 5 mins) and when combined with rapid and accurate deconvolution, one can accurately assess the level of DMPC incorporation and therefore ascertain its correct formation for further downstream manipulation of membrane proteins; for example SPR dose dependence experiments58. In summary, a single algorithm can now be used for protein deconvolution within the pharmaceutical Research Discovery environment, therefore removing much of the subjectivity that still exists in this most basic area of MS analytics.
Supplementary Material
Acknowledgments
Support from the US National Institutes of Health (R01GM103479 and S10RR028893 to J.A.L.), the US Department of Energy for the UCLA/DOE Institute for Genomics and Proteomics (DE-FC02–02ER63421 to J.A.L.) are gratefully acknowledged. We also thank Eric Carlson, Yong Joo Kil and Ilker Sen (Protein Metrics) for help with high MW and high m/z processing optimization. The authors would like to James Prell and Sean Cleary (University of Oregon) for processing the GCSF-PEG LC-MS data with iFAMS. We also thank Michael Marty (University of Arizona) for useful discussions regarding UniDec. The authors also wish to thank Ryan Holder and Nic Angell (Amgen) for their useful discussion on the GCSF-PEG LC-MS data. Peter Tieleman (University of Calgary) is thanked for the coarse-grain empty nanodisc image which is used in the TOC.
Footnotes
Supporting Information
Traditional peak-picking MW determination, detailed mass spectrometer instrument parameters, deconvolution algorithm details, external CsI m/z calibration in both positive and negative nESI mode, advanced deconvolution algorithm parameters, comb filter parameters, theoretical versus instrument derived peak widths, deconvolved denatured LC-MS AqpZ data, GroEL nESI positive ion mode data, deconvolved denatured LC-MS GroEL data; deconvolved denatured ubiquitin, myoglobin, BSA, NIST lysine and heavy chain data and accompanying mass measurement errors, IgG2-PEG12-Biotin, empty nanodisc and PEG-GCSF deconvolution without the implementation of the comb filter , PEG-GCSF intact linear MALDI analysis, denaturing LC-MS of GCSF-PEG analyzed at different source fragmentor voltages, a comparison of PMod, MaxEnt, iFAMS, UniDec and PMI Intact and the effects of algorithmic peak sharpening.
References
- 1.Wanner K; Hofner G; Mannhold R; Kubinyi H; Folkers G, Mass Spectrometry in Medicinal Chemistry: Applications in Drug Discovery. Methods and Principles in Medicinal Chemistry 2007, 36. [Google Scholar]
- 2.Campuzano ID; San Miguel T; Rowe T; Onea D; Cee VJ; Arvedson T; McCarter JD, High-Throughput Mass Spectrometric Analysis of Covalent Protein-Inhibitor Adducts for the Discovery of Irreversible Inhibitors: A Complete Workflow. Journal of biomolecular screening 2016, 21 (2), 136–44. [DOI] [PubMed] [Google Scholar]
- 3.Dillon TM; Bondarenko PV; Rehder DS; Pipes GD; Kleemann GR; Ricci MS, Optimization of a reversed-phase high-performance liquid chromatography/mass spectrometry method for characterizing recombinant antibody heterogeneity and stability. J Chromatogr A 2006, 1120 (1–2), 112–20. [DOI] [PubMed] [Google Scholar]
- 4.Marcoux J; Robinson CV, Twenty years of gas phase structural biology. Structure 2013, 21 (9), 1541–50. [DOI] [PubMed] [Google Scholar]
- 5.Katta V; Chait BT, Conformational changes in proteins probed by hydrogen-exchange electrospray-ionization mass spectrometry. Rapid Commun Mass Spectrom 1991, 5 (4), 214–7. [DOI] [PubMed] [Google Scholar]
- 6.Loo JA, Studying noncovalent protein complexes by electrospray ionisation mass spectrometry. Mass Spectrometry Reviews 1997, 16, 1–23. [DOI] [PubMed] [Google Scholar]
- 7.Ganem B; Li YT; Henion JD, Detection of noncovalent receptor–ligand complexes by mass spectrometry. Journal of the American Chemical Society 1991, 113, 6294–6296. [Google Scholar]
- 8.Robinson CV; Radford SE, Weighing the evidence for structure: electrospray ionization mass spectrometry of proteins. Structure 1995, 3 (9), 861–5. [DOI] [PubMed] [Google Scholar]
- 9.Benesch JL; Ruotolo BT; Simmons DA; Robinson CV, Protein complexes in the gas phase: technology for structural genomics and proteomics. Chem Rev 2007, 107 (8), 3544–67. [DOI] [PubMed] [Google Scholar]
- 10.Sharon M, Biochemistry. Structural MS pulls its weight. Science 2013, 340 (6136), 1059–60. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez H; Robinson CV, Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc 2007, 2 (3), 715–26. [DOI] [PubMed] [Google Scholar]
- 12.Campuzano I; Giles K, Nanospray Ion Mobility Mass Spectrometry of Selected High Mass Species. Nanoproteomics: Methods and Protocols, Methods in Molecular Biology, (Eds: Toms SA, Weil R), Humana Press, a part of Springer Science+Business Media, LLC, New York: 2011, 790, 57–70. [DOI] [PubMed] [Google Scholar]
- 13.Loo JA, Observation of large subunit protein complexes by electrospray ionisation mass spectrometry. Journal of Mass Spectrometry 1995, 30 (1), 180–183. [Google Scholar]
- 14.Rostom AA; Robinson CV, Detection of the Intact GroEL Chaperonin Assembly by Mass Spectrometry. Journal of the American Chemical Society 1999, 121, 4718–4719. [Google Scholar]
- 15.van Berkel WJ; van den Heuvel RH; Versluis C; Heck AJ, Detection of intact megaDalton protein assemblies of vanillyl-alcohol oxidase by mass spectrometry. Protein Sci 2000, 9 (3), 435–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marty MT; Zhang H; Cui W; Gross ML; Sligar SG, Interpretation and deconvolution of nanodisc native mass spectra. J Am Soc Mass Spectrom 2014, 25 (2), 269–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campuzano ID; Li H; Bagal D; Lippens JL; Svitel J; Kurzeja RJ; Xu H; Schnier PD; Loo JA, Native MS Analysis of Bacteriorhodopsin and an Empty Nanodisc by Orthogonal Acceleration Time-of-Flight, Orbitrap and Ion Cyclotron Resonance. Anal Chem 2016, 88 (24), 12427–12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landreh M; Marty MT; Gault J; Robinson CV, A sliding selectivity scale for lipid binding to membrane proteins. Curr Opin Struct Biol 2016, 39, 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van de Waterbeemd M; Snijder J; Tsvetkova IB; Dragnea BG; Cornelissen JJ; Heck AJ, Examining the Heterogeneous Genome Content of Multipartite Viruses BMV and CCMV by Native Mass Spectrometry. J Am Soc Mass Spectrom 2016, 27 (6), 1000–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rai DK; Griffiths WJ; Landin B; Wild BJ; Alvelius G; Green BN, Accurate mass measurement by electrospray ionization quadrupole mass spectrometry: detection of variants differing by <6 Da from normal in human hemoglobin heterozygotes. Anal Chem 2003, 75 (9), 1978–82. [DOI] [PubMed] [Google Scholar]
- 21.Schachner L; Han G; Dillon M; Zhou J; McCarty L; Ellerman D; Yin Y; Spiess C; Lill JR; Carter PJ; Sandoval W, Characterization of Chain Pairing Variants of Bispecific IgG Expressed in a Single Host Cell by High-Resolution Native and Denaturing Mass Spectrometry. Anal Chem 2016, 88 (24), 12122–12127. [DOI] [PubMed] [Google Scholar]
- 22.He J; Su D; Ng C; Liu L; Yu SF; Pillow TH; Del Rosario G; Darwish M; Lee BC; Ohri R; Zhou H; Wang X; Lu J; Kaur S; Xu K, High-Resolution Accurate-Mass Mass Spectrometry Enabling In-Depth Characterization of in Vivo Biotransformations for Intact Antibody-Drug Conjugates. Anal Chem 2017, 89 (10), 5476–5483. [DOI] [PubMed] [Google Scholar]
- 23.Mann M; Meng CK; Fenn JB, Interpreting Mass Spectra of Multiply Charged Ions. Analytical Chemistry 1989, 61, 1702–1708. [Google Scholar]
- 24.Benesch JL; Aquilina JA; Ruotolo BT; Sobott F; Robinson CV, Tandem mass spectrometry reveals the quaternary organization of macromolecular assemblies. Chem Biol 2006, 13 (6), 597–605. [DOI] [PubMed] [Google Scholar]
- 25.Zhou M; Sandercock AM; Fraser CS; Ridlova G; Stephens E; Schenauer MR; Yokoi- Fong T; Barsky D; Leary JA; Hershey JW; Doudna JA; Robinson CV, Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc Natl Acad Sci U S A 2008, 105 (47), 18139–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrige AG; Seddon MJ; Green BN; Jarvis SA; Skilling J, Disentangling Electrospray Spectra with Maximum Entropy. Rapid Commun Mass Spectrom 1992, 6, 707–711. [Google Scholar]
- 27.Zhang Z; Marshall AG, A Universal Algorithm for Fast and Automated Charge State Deconvolution of Electrospray Mass-to-Charge Ratio Spectra. Journal of the American Society for Mass Spectrometry 1998, 9, 225–233. [DOI] [PubMed] [Google Scholar]
- 28.Morgner N; Robinson CV, Massign: an assignment strategy for maximizing information from the mass spectra of heterogeneous protein assemblies. Anal Chem 2012, 84 (6), 2939–48. [DOI] [PubMed] [Google Scholar]
- 29.Marty MT; Baldwin AJ; Marklund EG; Hochberg GK; Benesch JL; Robinson CV, Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal Chem 2015, 87 (8), 4370–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu J; Trnka MJ; Roh SH; Robinson PJ; Shiau C; Fujimori DG; Chiu W; Burlingame AL; Guan S, Improved Peak Detection and Deconvolution of Native Electrospray Mass Spectra from Large Protein Complexes. J Am Soc Mass Spectrom 2015, 26 (12), 2141–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guan S; Trnka MJ; Bushnell DA; Robinson PJ; Gestwicki JE; Burlingame AL, Deconvolution method for specific and nonspecific binding of ligand to multiprotein complex by native mass spectrometry. Anal Chem 2015, 87 (16), 8541–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cleary SP; Thompson AM; Prell JS, Fourier Analysis Method for Analyzing Highly Congested Mass Spectra of Ion Populations with Repeated Subunits. Anal Chem 2016, 88 (12), 6205–13. [DOI] [PubMed] [Google Scholar]
- 33.Bern M; Caval T; Kil YJ; Tang W; Becker C; Carlson E; Kletter D; Sen KI; Galy N; Hagemans D; Franc V; Heck AJR, Parsimonious Charge Deconvolution for Native Mass Spectrometry. J Proteome Res 2018, 17 (3), 1216–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campuzano IDG; Netirojjanakul C; Nshanian M; Lippens JL; Kilgour DPA; Van Orden S; Loo JA, Native-MS Analysis of Monoclonal Antibody Conjugates by Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Anal Chem 2018, 90 (1), 745–751. [DOI] [PubMed] [Google Scholar]
- 35.Lippens JL; Nshanian M; Spahr C; Egea PF; Loo JA; Campuzano IDG, Fourier Transform-Ion Cyclotron Resonance Mass Spectrometry as a Platform for Characterizing Multimeric Membrane Protein Complexes. J Am Soc Mass Spectrom 2018, 29 (1), 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bagal D; Zhang H; Schnier PD, Gas-phase proton-transfer chemistry coupled with TOF mass spectrometry and ion mobility-MS for the facile analysis of poly(ethylene glycols) and PEGylated polypeptide conjugates. Anal Chem 2008, 80 (7), 2408–18. [DOI] [PubMed] [Google Scholar]
- 37.Campuzano IDG; Larriba C; Bagal D; Schnier PD, Ion Mobility and Mass Spectrometry Measurements of the Humanized IgGk NIST Monoclonal Antibody. ACS Symposium Series 2015, 1202, 75–112. [Google Scholar]
- 38.Leavy O, Therapeutic antibodies: past, present and future. Nature reviews. Immunology 2010, 10 (5), 297. [DOI] [PubMed] [Google Scholar]
- 39.Alley SC; Okeley NM; Senter PD, Antibody-drug conjugates: targeted drug delivery for cancer. Curr Opin Chem Biol 2010, 14 (4), 529–37. [DOI] [PubMed] [Google Scholar]
- 40.Formolo T; Ly M; Levy M; Kilpatrick L; Lute S; Phinney K; Marzilli L; Brorson K; Boyne M; Davis D; Schiel J, Determination of the NISTmAb Primary Structure State-of-the-Art and Emerging Technologies for Therapeutic Monoclonal Antibody Characterization Volume 2 ACS Symposium Series; American Chemical Society: Washington, DC, doi: 10.1021/bk-2015-1201.ch001 2015. [DOI] [Google Scholar]
- 41.Hopkins AL; Groom CR, The druggable genome. Nature reviews. Drug discovery 2002, 1 (9), 727–30. [DOI] [PubMed] [Google Scholar]
- 42.Arinaminpathy Y; Khurana E; Engelman DM; Gerstein MB, Computational analysis of membrane proteins: the largest class of drug targets. Drug Discov Today 2009, 14 (23–24), 1130–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lippens JL; Egea PF; Spahr C; Vaish A; Keener JE; Marty MT; Loo JA; Campuzano IDG, Rapid LC-MS Method for Accurate Molecular Weight Determination of Membrane and Hydrophobic Proteins. Anal Chem 2018, 90 (22), 13616–13623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marshall AG; Hendrickson CL; Jackson GS, Fourier transform ion cyclotron resonance mass spectrometry: a primer. Mass Spectrom Rev 1998, 17 (1), 1–35. [DOI] [PubMed] [Google Scholar]
- 45.Rose RJ; Damoc E; Denisov E; Makarov A; Heck AJ, High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nature methods 2012, 9 (11), 1084–6. [DOI] [PubMed] [Google Scholar]
- 46.Belov ME; Damoc E; Denisov E; Compton PD; Horning S; Makarov AA; Kelleher NL, From protein complexes to subunit backbone fragments: a multi-stage approach to native mass spectrometry. Anal Chem 2013, 85 (23), 11163–73. [DOI] [PubMed] [Google Scholar]
- 47.Liko I; Hopper JT; Allison TM; Benesch JL; Robinson CV, Negative Ions Enhance Survival of Membrane Protein Complexes. J Am Soc Mass Spectrom 2016, 27 (6), 1099–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mora FJ, Electrospray ionization of large multiply charged species proceeds via Dole’s charged residue mechanism. Analytica Chimica Acta 2000, 406 (1), 93–104. [Google Scholar]
- 49.Kim MT; Chen Y; Marhoul J; Jacobson F, Statistical modeling of the drug load distribution on trastuzumab emtansine (Kadcyla), a lysine-linked antibody drug conjugate. Bioconjug Chem 2014, 25 (7), 1223–32. [DOI] [PubMed] [Google Scholar]
- 50.Panowski S; Bhakta S; Raab H; Polakis P; Junutula JR, Site-specific antibody drug conjugates for cancer therapy. MAbs 2014, 6 (1), 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valliere-Douglass JF; Hengel SM; Pan LY, Approaches to Interchain Cysteine-Linked ADC Characterization by Mass Spectrometry. Molecular pharmaceutics 2015, 12 (6), 1774–83. [DOI] [PubMed] [Google Scholar]
- 52.McCombs JR; Owen SC, Antibody drug conjugates: design and selection of linker, payload and conjugation chemistry. The AAPS journal 2015, 17 (2), 339–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mullard A, Maturing antibody-drug conjugate pipeline hits 30. Nature reviews. Drug discovery 2013, 12 (5), 329–32. [DOI] [PubMed] [Google Scholar]
- 54.Diamantis N; Banerji U, Antibody-drug conjugates--an emerging class of cancer treatment. British journal of cancer 2016, 114 (4), 362–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.D’Atri V; Fekete S; Stoll D; Lauber M; Beck A; Guillarme D, Characterization of an antibody-drug conjugate by hydrophilic interaction chromatography coupled to mass spectrometry. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences 2018, 1080, 37–41. [DOI] [PubMed] [Google Scholar]
- 56.Dyachenko A; Wang G; Belov M; Makarov A; de Jong RN; van den Bremer ET; Parren PW; Heck AJ, Tandem Native Mass-Spectrometry on Antibody-Drug Conjugates and Submillion Da Antibody-Antigen Protein Assemblies on an Orbitrap EMR Equipped with a High-Mass Quadrupole Mass Selector. Anal Chem 2015, 87 (12), 6095–102. [DOI] [PubMed] [Google Scholar]
- 57.Chen J; Yin S; Wu Y; Ouyang J, Development of a native nanoelectrospray mass spectrometry method for determination of the drug-to-antibody ratio of antibody-drug conjugates. Anal Chem 2013, 85 (3), 1699–704. [DOI] [PubMed] [Google Scholar]
- 58.Xu H; Hill JJ; Michelsen K; Yamane H; Kurzeja RJ; Tam T; Isaacs RJ; Shen F; Tagari P, Characterization of the direct interaction between KcsA-Kv1.3 and its inhibitors. Biochimica et biophysica acta 2015, 1848 (10 Pt A), 1974–80. [DOI] [PubMed] [Google Scholar]
- 59.Marty MT; Hoi KK; Gault J; Robinson CV, Probing the Lipid Annular Belt by Gas-Phase Dissociation of Membrane Proteins in Nanodiscs. Angew Chem Int Ed Engl 2016, 55 (2), 550–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Molineux G, Pegylation: engineering improved biopharmaceuticals for oncology. Pharmacotherapy 2003, 23 (8 Pt 2), 3S–8S. [DOI] [PubMed] [Google Scholar]
- 61.Molineux G, Pegfilgrastim: using pegylation technology to improve neutropenia support in cancer patients. Anti-cancer drugs 2003, 14 (4), 259–64. [DOI] [PubMed] [Google Scholar]
- 62.Cleary SP; Li H; Bagal D; Loo JA; Campuzano IDG; Prell JS, Extracting Charge and Mass Information from Highly Congested Mass Spectra Using Fourier-Domain Harmonics. J Am Soc Mass Spectrom 2018, 29 (10), 2067–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haberger M; Bomans K; Diepold K; Hook M; Gassner J; Schlothauer T; Zwick A; Spick C; Kepert JF; Hienz B; Wiedmann M; Beck H; Metzger P; Molhoj M; Knoblich C; Grauschopf U; Reusch D; Bulau P, Assessment of chemical modifications of sites in the CDRs of recombinant antibodies: Susceptibility vs. functionality of critical quality attributes. MAbs 2014, 6 (2), 327–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Debaene F; Boeuf A; Wagner-Rousset E; Colas O; Ayoub D; Corvaia N; Van Dorsselaer A; Beck A; Cianferani S, Innovative native MS methodologies for antibody drug conjugate characterization: High resolution native MS and IM-MS for average DAR and DAR distribution assessment. Anal Chem 2014, 86 (21), 10674–83. [DOI] [PubMed] [Google Scholar]
- 65.Luo Q; Chung HH; Borths C; Janson M; Wen J; Joubert MK; Wypych J, Structural Characterization of a Monoclonal Antibody-Maytansinoid Immunoconjugate. Anal Chem 2016, 88 (1), 695–702. [DOI] [PubMed] [Google Scholar]
- 66.Ren D; Pipes GD; Hambly DM; Bondarenko PV; Treuheit MJ; Brems DN; Gadgil HS, Reversed-phase liquid chromatography of immunoglobulin G molecules and their fragments with the diphenyl column. J Chromatogr A 2007, 1175 (1), 63–8. [DOI] [PubMed] [Google Scholar]
- 67.Haberger M; Leiss M; Heidenreich AK; Pester O; Hafenmair G; Hook M; Bonnington L; Wegele H; Haindl M; Reusch D; Bulau P, Rapid characterization of biotherapeutic proteins by size-exclusion chromatography coupled to native mass spectrometry. MAbs 2016, 8 (2), 331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.