Summary
σV is an Extracytoplasmic Function (ECF) σ factor that is found exclusively in Firmicutes including Bacillus subtilis and the opportunistic pathogens Clostridioides difficile and Enterococcus faecalis. σV is activated by lysozyme and is required for lysozyme resistance. The activity of σV is normally inhibited by the anti-σ factor RsiV, a transmembrane protein. RsiV acts as a receptor for lysozyme. The binding of lysozyme to RsiV triggers a signal transduction cascade which results in degradation of RsiV and activation of σV. Like the anti-σ factors for several other ECF σ factors, RsiV is degraded by a multi-step proteolytic cascade that is regulated at the step of site-1 cleavage. Unlike other anti-σ factors, site-1 cleavage of RsiV is not dependent upon a site-1 protease whose activity is regulated. Instead constitutively active signal peptidase cleaves RsiV at site-1 in a lysozyme-dependent manner. The activation of σV leads to the transcription of genes, which encode proteins required for lysozyme resistance.
Keywords: σ factors, cell envelope, stress response, signal transduction, gene expression
Graphical Abstract
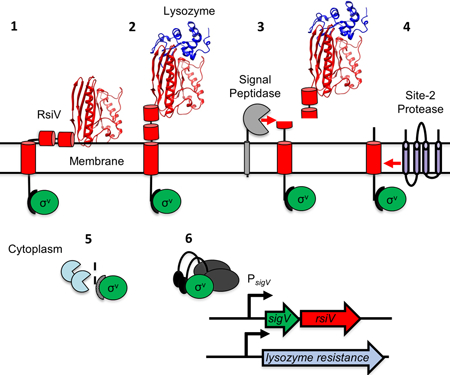
Activation of σV by lysozyme.
In the absence of lysozyme, RsiV (red) and inhibits σV activity (green). Lysozyme (blue) binds RsiV inducing σV activation. RsiV binds to lysozyme exposing the membrane-embedded site-1 cleavage site of RsiV to cleavage by signal peptidase. This leads to cleavage of RsiV by the site-2 protease and further degradation of the cytosolic portion of RsiV. Thus, freeing σV to interact with RNA polymerase and σV promoters to induce lysozyme resistance.
ECF σ factor background
ECF σ factors belong to the σ70 family of σ factors and contain only the σ2 and σ4.2 domains. These are homologous to the σ70 σ2 and σ4.2 domains and are required for binding to the −35 and −10 regions of target promoters (Helmann, 2002; Staroń et al., 2009; Helmann, 2016). In addition, most ECF σ factors are required for their own expression. The activity of most ECF σ factors is inhibited by a cognate anti-σ factor which is often encoded within the σ factor operon. Many but not all anti-σ factors are membrane proteins (Staroń et al., 2009). The σ factor activity is induced by inhibiting the function of the anti-σ factor. Activation of the σ factor can occur by one of several mechanisms; a conformational change in the anti-σ factor releases the σ factor, an anti-anti-σ factor binds to the anti-σ factor inducing σ factor release, or destruction of the anti-σ factor via Regulated Intramembrane Proteolysis (RIP) (Ho and Ellermeier, 2012; Helmann, 2016). Here we will focus on activation of ECF σ factors by degradation of the anti-σ factor. In the presence of an inducing signal, a site-1 protease cleaves the extracellular domain of an anti-σ factor. Following site-1 protease cleavage, the truncated anti-σ factor is cleaved by a site-2 protease within the transmembrane region of the anti-σ factor. The remainder of the anti-σ factor is then degraded by cytosolic proteases. This frees the σ factor to interact with RNA polymerase and transcribe target genes (Brown et al., 2000; Ho and Ellermeier, 2012; Helmann, 2016).
The ECF σ factor σV is found exclusively in Firmicutes or low GC Gram-positive bacteria and belongs to the ECF30 family of ECF σ factors (Staroń et al., 2009). The σV system is encoded by the model organism Bacillus subtilis, as well as the opportunistic pathogens Enterococcus faecalis and Clostridioides difficile (Fig. 1). σV is not present in all Firmicutes or even closely related species. While present in B. subtilis, σV is not present in B. anthracis or related organisms. Similarly, σV is present in C. difficile but not in C. perfringens or C. botulinum suggesting possible horizontal transfer of the sigV operon. While σV and RsiV are well-conserved, the compositions of the sigV operons vary (Fig 1.). In E. faecalis, the sigV operon contains only the sigV and rsiV genes (Benachour et al., 2005). In B. subtilis, the genes of the sigV operon encode the σV (SigV), the anti-σV (RsiV), a peptidoglycan O-acetyltransferase (OatA) and an uncharacterized protein YrhK (Guariglia-Oropeza and Helmann, 2011; Ho et al., 2011). In C. difficile, the sigV operon contains seven genes which encode, a polysaccharide deacetylase (PdaV), a putative peptidyl-prolyl cis-trans isomerase (PrsA2), σV (SigV or CsfV), the anti-σV (RsiV), a second RsiV-like protein, and two hypothetical proteins (a putative XdhC-like Xanthine dehydrogenase maturation factor and a putative zinc metalloprotease) (Ho and Ellermeier, 2011; Ho et al., 2014). The σV regulons have been experimentally determined in B. subtilis and C. difficile using microarrays to perform whole genome transcriptomics while the E. faecalis regulon has been partially determined using bioinformatic approaches (Benachour et al., 2005; Guariglia-Oropeza and Helmann, 2011; Ho et al., 2014) (Table 1). The σV regulons are summarized in Table 1 and discussed later in this review.
Figure 1. Alignment of sigV regions from B. subtilis, C. difficile and E. faecalis.
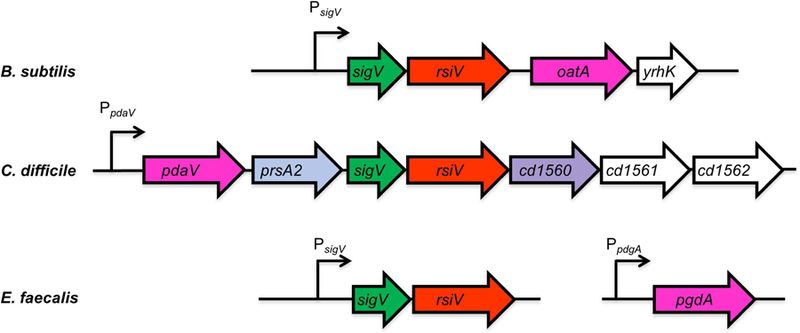
Shown in green is sigV (σV), rsiV (anti-σ factor RsiV) is red, the genes encoding peptidoglycan modifying enzymes are in pink (pdaV, pgdA and oatA), prsA2 (peptidylprolyl isomerase) is light blue and an rsiV ortholog cd1560 which lacks the σ factor binding domain is purple, the genes in white have no known function. The C. difficile genes are numbered based on C. difficile 630 numbering.
Table 1.
Comparison of the σV regulon in B. subtilis, C. difficile and E. faecalis
| Gene1 | Locus2 | Protein Function3 | Operon4 |
| B. subtilis | |||
| sigV | BSU27120 | RNA polymerase ECF-type sigma factor σV* | sigVrsiVoatAyrhK |
| rsiV | BSU27130 | Anti-σ factor for σV* | sigVrsiVoatAyrhK |
| oatA | BSU27140 | O-acetyl transferase* | sigVrsiVoatAyrhK |
| yrhK | BSU27150 | Hypothetical protein | sigVrsiVoatAyrhK |
| dltA | BSU27150 | D-alanyl-D-alanine carrier protein ligase | dltABCDE |
| dltB | BSU38510 | D-alanine transfer from DltC to undecaprenol-phosphate | dltABCDE |
| dltC | BSU38520 | D-alanine carrier protein | dltABCDE |
| dltD | BSU38530 | D-alanine transfer from undecaprenol-phosphate to the poly(glycerophosphate) chain | dltABCDE |
| dltE | BSU38540 | D-alanine transfer from undecaprenol-phosphate to the poly(glycerophosphate) chain | dltABCDE |
| pbpX | BSU16950 | penicillin-binding protein X endopeptidase | pbpX |
| bcrC | BSU36530 | Undecaprenyl pyrophosphate phosphatase | bcrC |
| sasA (ywaC) | BSU38480 | ppGpp synthase* | sasA |
| abh | BSU14480 | Transition state regulator similar to AbrB* | abh |
| C. difficile | |||
| pdaV | CD630_15560 | N-acetylglucosamine deacetylase* | pdaVprsA2sigVrsiV1560–1562 |
| prsA2 | CD630_15570 | Peptidyl isomerase | pdaVprsA2sigVrsiV1560–1562 |
| sigV (csfV) | CD630_15580 | σ factor* | pdaVprsA2sigVrsiV1560–1562 |
| rsiV | CD630_15590 | Anti-σ factor* | pdaVprsA2sigVrsiV1560–1562 |
| cd1560 | CD630_15600 | Lysozyme binding protein | pdaVprsA2sigVrsiV1560–1562 |
| cd1561 | CD630_15561 | XdhC-like Xanthine dehydrogenase maturation factor | pdaVprsA2sigVrsiV1560–1562 |
| cd1562 | CD630_15562 | cytoplasmic zinc metalloprotease | pdaVprsA2sigVrsiV1560–1562 |
| dltD | CD630_28540 | D-alanine transfer from undecaprenol-phosphate to the poly(glycerophosphate) chain | dltDABC |
| dltA | CD630_28530 | D-alanyl-D-alanine carrier protein ligase | dltDABC |
| dltB | CD630_28520 | D-alanine transfer from DltC to undecaprenol-phosphate | dltDABC |
| dltC | CD630_28510 | D-alanine carrier protein | dltDABC |
| cd0739 | CD630_07390 | Unknown transmembrane protein | cd0739 |
| cd1606 | CD630_16060 | GntR regulator | cd1606-cd1611 |
| cd1607 | CD630_16070 | ABC transport ATP binding protein | cd1606-cd1611 |
| cd1608 | CD630_16080 | Multidrug ABC family transport Permease | cd1606-cd1611 |
| cd1609 | CD630_16090 | Hypothetical protein | cd1606-cd1611 |
| cd1610 | CD630_16100 | Hypothetical protein | cd1606-cd1611 |
| cd1611 | CD630_16110 | Hypothetical protein | cd1606-cd1611 |
| E. faecalis | |||
| sigV | EF_3180 | σ factor* | sigVrsiV |
| rsiV | EF_3179 | Anti-σ factor* | sigVrsiV |
| pgdA | EF_1843 | N-acetylglucosamine deacetylase* | pdgA |
| ef0159 | EF_0159 | Hypothetical protein | ef0159 |
| ef1934 | EF_1934 | Hypothetical protein | ef1933–1934 |
| ef0315 | EF_0315 | Hypothetical protein | ef0315 |
Common gene name
Locus tags for B. subtilis 168, C. difficile CD630, and E. faecalis V583
Functions are based on homology unless marked by * where activity was determined experimentally.
Operon
σV Activation by Lysozyme
In B. subtilis, E. faecalis and C. difficile, σV is activated by lysozyme and σV is required for transcription of lysozyme resistance genes (Benachour et al., 2005; Guariglia-Oropeza and Helmann, 2011; Ho et al., 2011; Ho et al., 2014). Lysozymes are a large group of muramidases which are ubiquitous in nature and found in organisms from phage to bacteria to humans (Callewaert and Michiels, 2010; Callewaert et al., 2012). Lysozymes can be divided into six categories or types; chicken, goose, invertebrate, phage, plant, and bacterial (Bilej, 2015). Lysozymes are also an important component of the innate immune system of many organisms (Callewaert and Michiels, 2010; Ragland and Criss, 2017). Interestingly, only the c-type lysozymes human or hen egg white lysozyme activate σV (Hastie et al., 2014). Other muramidases like mutanolysin, a bacterial muramidase, (Yokogawa et al., 1974) fail to activate σV (Hastie et al., 2014). Additionally, the phage-type lysozyme from phage T4 also fails to induce σV activity (Lewerke and Ellermeier unpublished data). Thus, the enzymatic activity of lysozyme is not sufficient to induce σV activation. Hastie et al. demonstrated that a mutant of human lysozyme which lacks enzymatic activity still activates σV (Hastie et al., 2014). Thus, it is not the enzymatic activity of lysozyme but the structure of c-type lysozyme that triggers σV activation.
The anti-σ factor RsiV is a single pass transmembrane protein that inhibits σV activity (Fig. 2A) (Asai et al., 2003; Zellmeier et al., 2005). RsiV binds to hen egg white lysozyme at a 1:1 ratio and with ~70 nm affinity (Fig. 2B) (Hastie et al., 2014; Hastie et al., 2016). It was suggested that RsiV senses c-type lysozymes since both hen egg white lysozyme and human lysozyme directly bind RsiV while mutanolysin fails to bind RsiV (Hastie et al., 2014). Further evidence supporting RsiV as a lysozyme receptor came when the co-structure of RsiV and hen egg white lysozyme was solved and revealed a large number of contacts between RsiV and lysozyme (Hastie et al., 2016). Site-directed mutagenesis revealed that no single residue of RsiV was absolutely required for binding to hen egg white lysozyme (Hastie et al., 2016). Instead multiple contacts had to be disrupted (RsiVS169W P259A Y261A) to block binding to hen egg white lysozyme in vitro and activation of σV in response to lysozyme in vivo (Hastie et al., 2016). This demonstrated that binding of RsiV to hen egg white lysozyme is required for σV activation. As is the case with B. subtilis, c-type lysozyme activates σV homologs in multiple organisms including E. faecalis and C. difficile (Benachour et al., 2005; Ho and Ellermeier, 2011; Ho et al., 2011). The RsiV homologs from C. difficile and E. faecalis also bind to hen egg white lysozyme suggesting that the role of RsiV as a receptor for lysozyme is conserved across multiple species (Hastie et al., 2014).
Figure 2. Model of lysozyme-mediated σV activation.
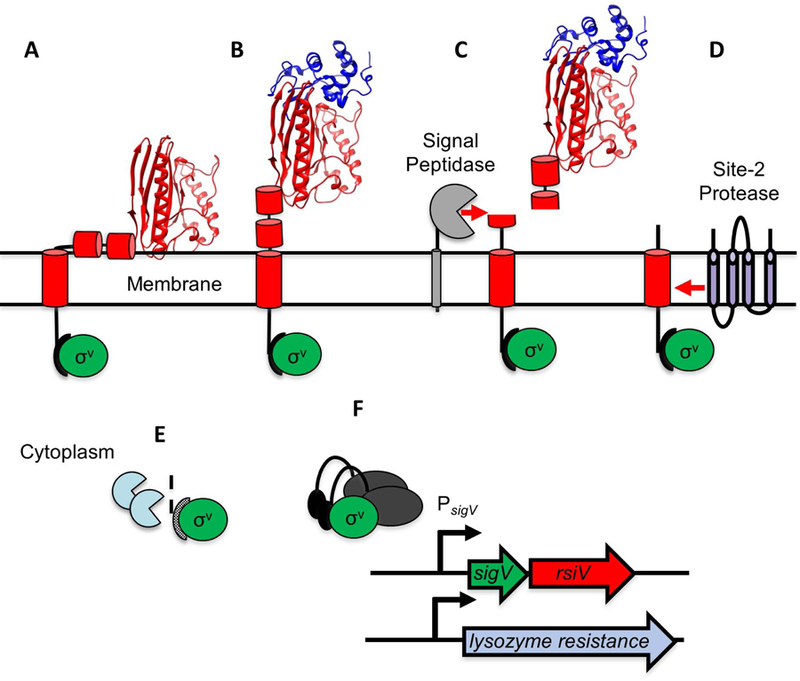
A. In the absence of lysozyme, RsiV (red) binds σV (green). Two amphipathic helices of RsiV (short red cylinders) are embedded in the membrane. B. Lysozyme (blue ribbon diagram) binds the C-terminal extracytoplasmic portion of RsiV (red ribbon diagram) and alters the structure of RsiV such that the amphipathic helices are no longer membrane-embedded. C. Signal peptidase (grey) can recognize and cleave RsiV at site-1 within the first amphipathic helix. D. The site-2 protease (RasP/Eep in purple) cleaves the site-1 cleavage product within the transmembrane domain. E. Cytoplasmic proteases (light blue) further degrade the RsiV site-2 cleavage product. F. σV (green), no longer bound to RsiV, is free to interact with RNA polymerase and σV promoters.
Some studies have suggested that other conditions can activate σV. For example, in B. subtilis, σV, σM and σX which have partially overlapping regulons (Mascher et al., 2007; Helmann, 2016) have a two- to three-fold increases in σ factor activity in strains lacking glycolipids (ugtP null mutants) (Hashimoto et al., 2013). In E. faecalis exposure to heat and SDS stress resulted in two to three-fold increase in σV activity (Benachour et al., 2005). While these levels of σV activation are modest, lysozyme induced σV activation is greater than 50-fold in B. subtilis (Guariglia-Oropeza and Helmann, 2011; Ho et al., 2011) and 300-fold in E. faecalis (Le Jeune et al., 2010). Thus, while some perturbations of the cell envelope may trigger a small degree of σV activation, lysozyme is the most potent inducer of σV activity.
Degradation of RsiV
In the presence of lysozyme RsiV is proteolytically degraded (Hastie et al., 2013). This degradation occurs in at least three steps of which the first two have been well-characterized (Fig. 2). Like other ECF anti-σ factors, degradation of RsiV is initiated by cleavage at site-1 (Fig. 2C) (Hastie et al., 2013; Hastie et al., 2014). The location of site-1 cleavage in vivo was determined to occur immediately after the canonical type 1 signal peptide motif (AXA) (Hastie et al., 2014). Mutation of the cleavage site blocks RsiV degradation and σV activation (Hastie et al., 2014). In contrast to other ECF anti-σ factors, which have dedicated site-1 proteases, cleavage of RsiV at site-1 requires signal peptidase (Hastie et al., 2014; Castro et al., 2018). B. subtilis encodes five type 1 signal peptidases, four prokaryotic type 1 signal peptidase (SipS, SipT, SipU and SipV) and one eukaryotic type 1 signal peptidase (SipW) (Tjalsma et al., 1997; Chu et al., 2002). SipS and SipT are the major signal peptidases in B. subtilis and the activity of either SipS or SipT is essential for viability (van Roosmalen et al., 2001). RsiV is cleaved at site-1 in either a sipS or a sipT null mutant (Hastie et al., 2014; Castro et al., 2018). However using a B. subtilis sipT sipSts double mutant strain which lacks SipT and produces a temperature sensitive SipS protein Castro et al demonstrated signal peptidase activity was required for site-1 cleavage of RsiV (Castro et al., 2018). Purified SipS or SipT both cleave RsiV in vitro (Castro et al., 2018). Importantly the addition of lysozyme to purified RsiV and SipS or SipT induces cleavage of RsiV (Castro et al., 2018). Thus SipS or SipT are sufficient for site-1 cleavage of RsiV in vitro only in the presence of lysozyme (Castro et al., 2018). To date it has not been reported if signal peptidase is responsible for site-1 cleavage of RsiV in C. difficile or E. faecalis; however, it seems likely as both have predicted signal peptidase cleavage sites.
Signal peptidases cleave proteins that are secreted via either the general secretory pathway or the twin arginine (TAT) secretion system (Paetzel et al., 2002; Auclair et al., 2012). This process is not thought to be regulated. Thus, an important question is how does RsiV avoid signal peptidase cleavage in the absence of lysozyme. Lewerke et al. determined that the region surrounding the signal peptidase cleavage site contains two predicted amphipathic helices separated by a turn (Lewerke et al., 2018). Disruption of these helices either by deletion or introduction of a charged residue on the hydrophobic face of the amphipathic helix leads to constitutive RsiV degradation and σV activation (Lewerke et al., 2018). Thus, in the absence of lysozyme, the amphipathic helices protect RsiV from cleavage by signal peptidase.
To date the amphipathic helices in RsiV have not been observed by X-ray crystallography. However, the presence of the amphipathic helices is supported by substituted cysteine scanning mutagenesis and labeling experiments (Lewerke et al., 2018). In these experiments, cysteines were placed on either the hydrophobic or hydrophilic surface of the amphipathic helix and then labeled with a membrane impermeable reagent Na-(3-maleimidylpropionyl) biocytin. It was found that the cysteines on the hydrophilic face of the helices were labeled in the presence or absence of lysozyme. In contrast, cysteines on the hydrophobic face were not labeled in the absence of lysozyme but could be labeled in the presence of lysozyme. This suggests that the predicted helices are membrane-embedded and binding of RsiV to lysozyme displaces the helices from the membrane resulting in cleavage by signal peptidase (Fig. 2B).
The RsiV-hen egg white lysozyme structure was solved using the extracellular domain of RsiV (Hastie et al., 2016). This included a portion of the first amphipathic helix and the entire second amphipathic helix. However, in this structure most of the residues of the amphipathic helices were unstructured (Hastie et al., 2016). Interestingly although the cysteine labelling experiments suggest that Ile80 is part of the amphipathic helix, since it can only be labeled in the presence of lysozyme, in the RsiV-hen egg white lysozyme structure it forms part of a β-sheet (Hastie et al., 2016; Lewerke et al., 2018). This suggests a possible mechanism by which RsiV becomes sensitive to signal peptidase in the presence of lysozyme. We hypothesize that upon binding lysozyme part of the amphipathic helix is pulled into a β-sheet conformation which results in the amphipathic helices being disrupted and the cleavage site rendered accessible to signal peptidase.
RsiV shares a domain of unknown function (DUF4179) with BAS1627 the anti-σ factor for another ECF30 family σ factor BAS1626 from B. anthracis (Fig. 3). Except for the DUF4179 domain, RsiV and BAS1627 are not homologous. To date the domain DUF4179 has only been defined in the anti-σ factors associated with the ECF30 σ factor family specifically those in the Firmicutes (1197 sequences) and Actinobacteria (20 sequences). The DUF4179 domain encompasses the transmembrane domain and two amphipathic helices separated by a turn (Fig. 3A). The X-ray crystal structure of BAS1627 (3FBQ) was determined and includes a portion of DUF4179 including the two amphipathic helices (Zhang et al.). Figure 3B and 3C show the 3FBQ structure with the two alpha helices separated by a turn highlighted in red (Zhang et al.). We hypothesize that the amphipathic helices separated by a turn may be a conserved mechanism for obscuring the proteolytic cleavage site. Solving the structure of RsiV alone will be critical to understand how lysozyme induces a conformational change making RsiV sensitive to signal peptidase.
Figure 3. Structural comparison of two ECF30 anti-σ factors.
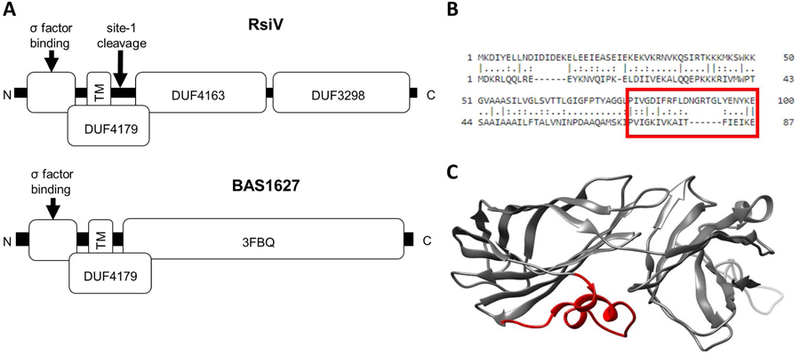
A. Domain structure of two ECF30 family anti-σ factors. RsiV (top) consists of 3 domains of unknown function. DUF4163 and DUF3298 comprise the lysozyme binding domain of RsiV. DUF4179 includes the transmembrane domain, amphipathic helices and site-1 cleavage site. The bottom portion shows BAS1627 an anti-σ factor from B. anthracis. The 3FBQ portion represents the solved X-ray crystal structure of BAS1627 and contains the c-terminal portion of the DUF4179. B. Alignment of RsiV and BAS1627. The region boxed in red are the amphipathic helices of RsiV. C. The structure of BAS1627 (3FBQ). The regions in red are the amphipathic helices that correspond the boxed region in B.
After cleavage at site-1 by signal peptidase, the conserved site-2 protease RasP is required for further processing of RsiV (Hastie et al., 2013). RasP cleaves RsiV within the transmembrane domain and is not regulated by lysozyme (Fig. 2D). RasP is a metalloprotease similar to RseP from E. coli (36% identity and 53% similarity), which is responsible for site-2 cleavage of the anti-σ factor RseA (Kanehara et al., 2002; Alba et al., 2002; Kanehara et al., 2003). RseP utilizes tandem PDZ domains to recognize substrates via a size-filtering rather than recognizing a specific amino acid sequence (Hizukuri et al., 2014). RasP contains a single PDZ domain that is also thought to function as a size exclusion filter for substrates (Parrell et al., 2017). In E. faecalis Eep a metalloprotease homologous to RasP (44% identity and 62% similarity) functions as the site-2 protease and is required for RsiV degradation and σV activation (Varahan et al., 2013). Upon cleavage by RasP the N-terminal portion of RsiV is released into the cytosol where it is presumably degraded by the cytosolic proteases (Fig. 2E). However, the specific proteases involved in degradation of the N-terminal portion of RsiV have not been identified in any organisms.
σV regulated modifications of peptidoglycan that mediate lysozyme resistance.
Lysozyme kills bacteria by cleaving the β−1,4-glycosidic bonds between the N-acetylglucosamine and N-acetylmuramic acid of peptidoglycan which can lead to cell lysis (Callewaert and Michiels, 2010). A common mechanism for lysozyme resistance in bacteria is altering the acetylation state of the peptidoglycan by either removing or adding acetyl groups to the N-acetylglucosamine or N-acetylmuramic acid backbone (Ragland and Criss, 2017). In B. subtilis σV is required for transcription of the sigV operon including oatA, which encodes for an O-acetyl transferase that is responsible for adding an acetyl-group to N-acetylmuramic acid (Ho et al., 2011; Laaberki et al., 2011; Guariglia-Oropeza and Helmann, 2011). Deletion of either sigV or oatA results in a ~2-fold increase in lysozyme sensitivity (Guariglia-Oropeza and Helmann, 2011; Ho et al., 2011).
Compared to B. subtilis, C. difficile is highly resistant to lysozyme (C. difficile MIC >16 mg/ml vs B. subtilis MIC ~3 µg/ml). The peptidoglycan of C. difficile is also highly deacetylated, >90% and σV is partially required for this high level of de-acetylation and lysozyme resistance (Peltier et al., 2011; Ho et al., 2014). In C. difficile σV is required for expression of pdaV, which encodes a peptidoglycan deacetylase (Ho and Ellermeier, 2011; Ho et al., 2014). PdaV has been shown to contribute to deacetylation of N-acetylglucosamine, which can increase lysozyme resistance (Ho et al., 2014). However, even in the absence of PdaV ~75% of the C. difficile peptidoglycan is deacetylated suggesting other deacetylases are involved in modifying the peptidoglycan for lysozyme resistance (Ho et al., 2014).
E. faecalis is also highly resistant to lysozyme (MIC ~32–64 mg/ml). This resistance is in part mediated by σV (Benachour et al., 2005; Le Jeune et al., 2010; Varahan et al., 2013). The peptidoglycan of E. faecalis is both O-acetylated and de-acetylated (Hébert et al., 2007). The expression of oatA, which encodes for a peptidoglycan O-acetylase, is σV-independent and contributes modestly to lysozyme resistance (Hébert et al., 2007; Le Jeune et al., 2010). σV is required for expression of pdgA, which encodes a homolog of S. pneumoniae N-acetylglucosamine deacetylase (Le Jeune et al., 2010). Although deletion of sigV results in a greater than 12.5-fold decrease in lysozyme resistance, a pdgA deletion has no effect on lysozyme resistance (Benachour et al., 2005; Hébert et al., 2007; Varahan et al., 2013). This suggest that σV must contribute at least partially to lysozyme resistance in a PdgA-independent manner.
Regulation of cell envelope modifications by σV
In B. subtilis σV also regulates expression of a number of genes outside its own operon (Table 1). One of the most notable operons is dltABCDE (Asai et al., 2003; Guariglia-Oropeza and Helmann, 2011). The dlt operon encodes proteins that contribute to D-alanylation of teichoic acids (Neuhaus and Baddiley, 2003; Nizet, 2006). Lysozyme is positively charged and the D-alanylation of teichoic acids has been associated with resistance to positively charged cationic antimicrobial peptides (CAMPs) by increasing the charge of the cell surface (Collins et al., 2002; Nizet, 2006). Although expression of dlt is modulated by σV, it is also controlled by other regulatory elements, including the ECF σ factor, σX (Cao and Helmann, 2004; Guariglia-Oropeza and Helmann, 2011; Helmann, 2016). Mutations in dlt result in ~2-fold decreases in lysozyme resistance (Guariglia-Oropeza and Helmann, 2011; Ho et al., 2011).
Like B. subtilis, expression of the C. difficile dltDABC operon is also modulated by lysozyme in a σV-dependent manner. The C. difficile dlt operon contributes to resistance to a number of CAMPs such as nisin, gallidermin, polymyxin B, and vancomycin in addition to lysozyme (McBride and Sonenshein, 2011; Woods et al., 2016). Although lysozyme and polymyxin B induce expression of dlt, σV only modulates expression upon exposure to only lysozyme and not polymyxin B (Woods et al., 2016).
Like B. subtilis and C. difficile, E. faecalis D-alanylation of teichoic acids contributes to lysozyme resistance as mutants of dlt show increase sensitivity to lysozyme (Le Jeune et al., 2010). Unlike B. subtilis and C. difficile, E. faecalis σV does not appear contribute significantly to dlt expression (Le Jeune et al., 2010). This suggests that control of dlt by σV is not conserved in all organisms.
Interestingly in several organisms it appears that deletion of dlt in combination with deletion of the genes for either de-aceytylase or O-acetylase result in a synergistic increase in lysozyme sensitivity. In S. aureus deletion of dlt and oatA results in a lysozyme-sensitized strain (Herbert et al., 2007). Similarly, in B. subtilis deletion of oatA and dlt is more sensitive to lysozyme than either mutant alone (Guariglia-Oropeza and Helmann, 2011). Thus σV-dependent genes function synergistically to increase lysozyme resistance.
Inhibition of Lysozyme Activity by RsiV
Many Gram-negative bacteria utilize lysozyme inhibitors to protect the cell wall however they appear less common in Gram-positive bacteria (Callewaert et al., 2012). Interestingly the co-structure of RsiV-hen egg white lysozyme revealed that RsiV binds to the active site of lysozyme (Hastie et al., 2016). Purified RsiV inhibits the activity of hen egg white lysozyme in vitro (Hastie et al., 2016). Thus, in addition to functioning as a sensor for lysozyme, RsiV also functions as a competitive inhibitor of lysozyme activity (Hastie et al., 2016). Production of RsiV in a ΔsigVrsiV mutant leads to a small (~1.5 fold) increase in lysozyme resistance (Hastie et al., 2016). Given that RsiV from C. difficile and E. faecalis bind lysozyme, it is reasonable to hypothesize they also inhibit lysozyme activity. In addition to RsiV, C. difficile encodes an RsiV-like protein (CD1560) that contains the lysozyme-binding and transmembrane domains but lacks the σV-binding portion. While lysozyme inhibition has been shown to be important for lysozyme resistance in a number of bacteria, it remains unclear if the inhibition of lysozyme by RsiV and the RsiV-like protein contribute to lysozyme resistance in vivo.
Other Genes Regulated by σV
Microarray analysis of C. difficile identified other genes induced by lysozyme in a σV-dependent manner (Table 1) (Ho et al., 2014). The role of many of these genes in lysozyme resistance and cell envelope maintenance remain unclear. PrsA2 is peptidyl prolyl isomerase and in other organisms has a role in proper folding of exported proteins (Kontinen and Sarvas, 1993; Hyyryläinen et al., 2001; Hyyryläinen et al., 2010; Alonzo et al., 2011). The cd0739 gene encodes a putative exported protein and is substantially induced by lysozyme in a σV-dependent manner. Similarly, expression of seven genes encoding a putative ABC transporter and a transcriptional regulator (cd1606-cd1611) are σV-regulated. Despite being induced by lysozyme, neither of these operons appear to be required for lysozyme resistance (Ho et al., 2014). It is possible these σV-regulated genes may be involved in protecting bacteria from other stresses that are encountered under similar conditions as lysozyme or their individual contribution to lysozyme resistance is masked by other σV regulated genes.
Evidence suggests that in E. faecalis, σV regulates expression of at least one other gene to increase lysozyme resistance. Deletion of sigV in an oatA pgdA dltA triple mutant shows a ~16-fold increase in lysozyme sensitivity compared to the triple mutant suggesting σV is required for expression of other genes involved in lysozyme resistance in E. faecalis (Smith et al., 2019). These other σV-regulated genes have not yet been identified.
It is clear in multiple organisms σV is induced by lysozyme and control lysozyme resistance mechanisms. First, most organisms utilize σV to control expression of peptidoglycan modifying enzymes which alter the structure of peptidoglycan to block lysozyme binding. The type acetylation or deacetylation of N-acetylmuramic acid or N-acetylglucosamine subunits and extent of modification vary between organisms. Second dlt is required for lysozyme resistance in each organism and multiple regulators contribute to dlt expression but the dependence on σV varies. Third in addition to its role in signal transduction, the anti-σV factor RsiV can also act as a competitive inhibitor of lysozyme by directly binding lysozyme to prevent it from recognizing peptidoglycan (Hastie et al., 2016).
σV Promoter Recognition
It is interesting to note that in both C. difficile and B. subtilis (where transcriptomics approaches have been used to identify the σV regulons) the sigV operons themselves are induced greater than 50-fold while most of the genes outside the sigV operon are induced to much lower levels two- to four-fold (Guariglia-Oropeza and Helmann, 2011; Ho et al., 2014). It remains unclear why induction of the sigV operon is significantly higher than other target genes presumably this promoter is closer to the σV consensus than the other σV-regulated genes. ECF30-dependent promoters, which includes σV, have a consensus sequence of −35 tgaAAC and −10 CGTC (Staroń et al., 2009). Recently, more detailed experimental analysis of the σV regulon in B. subtilis identified σV responsive and non-responsive promoters (Gaballa et al., 2017). The key feature of the σV responsive promoters was conservation of an extended run of tttt in the −35 region (tgaAACntttt) (Gaballa et al., 2017). Gaballa et al. demonstrated that mutating the run of tttt from a σV responsive promoter rendered it σV non-responsive (Gaballa et al., 2017). In contrast adding a run of tttt to an non-responsive promoter turned it into a σV responsive promoter (Gaballa et al., 2017). Further careful analysis of σV promoters from other organisms may help delineate strong versus weak σV promoters.
Future Directions
One of the key features of σV activation that is still not understood is the conformational change RsiV undergoes upon binding to lysozyme that allows it to be recognized by signal peptidase. To date the structure of RsiV alone has not been determined making understanding these changes difficult.
It will also be important to determine if other ECF σ factors utilize amphipathic helices in a similar manner to RsiV to control ECF σ factor activation. A clear example could be the ECF σ factor BAS1626 from B. anthracis. BAS1627 the putative anti-σ factor for BAS1626 contains both the amphipathic helices and a putative signal peptidase motif. Unfortunately, the signals required for inducing BAS1626 are not currently known.
RsiV also represents an example of the anti-σ factor acting as a receptor for an inducing signal and initiating its own destruction. Given the diversity in the anti-σ factors it seems likely that other anti-σ factors may function as receptors for other signals. Identifying the inducers of ECF σ factors has often proven difficult. However, understanding the signals required for inducing uncharacterized ECF σ factors will likely reveal additional novel regulatory mechanisms.
References:
- Alba BM, Leeds J. a, Onufryk C, Lu CZ, and Gross CA (2002) DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev 16: 2156–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonzo F, Xayarath B, Whisstock JC, and Freitag NE (2011) Functional analysis of the Listeria monocytogenes secretion chaperone PrsA2 and its multiple contributions to bacterial virulence. Mol Microbiol 80: 1530–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai K, Yamaguchi H, Kang C, Yoshida K, Fujita Y, and Sadaie Y (2003) DNA microarray analysis of Bacillus subtilis sigma factors of extracytoplasmic function family. FEMS Microbiol Lett 220: 155–160. [DOI] [PubMed] [Google Scholar]
- Auclair SM, Bhanu MK, and Kendall DA (2012) Signal peptidase I: Cleaving the way to mature proteins. Protein Sci 21: 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benachour A, Muller C, Dabrowski-Coton M, Breton Y. Le, Giard J-C, Rincé A, et al. (2005) The Enterococcus faecalis SigV protein is an extracytoplasmic function sigma factor contributing to survival following heat, acid, and ethanol treatments. J Bacteriol 187: 1022–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilej M (2015) Mucosal Immunity in Invertebrates. Mucosal Immunol Fourth Ed 1–2: 135–144. [Google Scholar]
- Brown MS, Ye J, Rawson RB, and Goldstein JL (2000) Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100: 391–398. [DOI] [PubMed] [Google Scholar]
- Callewaert L, Herreweghe J.M. Van, Vanderkelen L, Leysen S, Voet A, and Michiels CW (2012) Guards of the great wall: bacterial lysozyme inhibitors. Trends Microbiol 20: 501–10. [DOI] [PubMed] [Google Scholar]
- Callewaert L, and Michiels CW (2010) Lysozymes in the animal kingdom. J Biosci 35: 127–160. [DOI] [PubMed] [Google Scholar]
- Cao M, and Helmann JD (2004) The Bacillus subtilis Extracytoplasmic-Function σX Factor Regulates Modification of the Cell Envelope and Resistance to Cationic Antimicrobial Peptides. J Bacteriol 186: 1136–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro AN, Lewerke LT, Hastie JL, and Ellermeier CD (2018) Signal Peptidase Is Necessary and Sufficient for Site 1 Cleavage of RsiV in Bacillus subtilis in Response to Lysozyme. J Bacteriol 200: e00663–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu HH, Hoang V, Kreutzmann P, Hofemeister B, Melzer M, and Hofemeister J (2002) Identification and properties of type I-signal peptidases of Bacillus amyloliquefaciens. Eur J Biochem 269: 458–469. [DOI] [PubMed] [Google Scholar]
- Collins LV, Kristian SA, Weidenmaier C, Faigle M, Kessel K.P.M. Van, Strijp J.A.G. Van, et al. (2002) Staphylococcus aureus strains lacking D-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J Infect Dis 186: 214–9. [DOI] [PubMed] [Google Scholar]
- Gaballa A, Guariglia-Oropeza V, Dürr F, Butcher BG, Chen AY, Chandrangsu P, and Helmann JD (2017) Modulation of extracytoplasmic function (ECF) sigma factor promoter selectivity by spacer region sequence. Nucleic Acids Res 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guariglia-Oropeza V, and Helmann JD (2011) Bacillus subtilis σV confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and D-alanylation of teichoic acids. J Bacteriol 193: 6223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Seki T, Matsuoka S, Hara H, Asai K, Sadaie Y, and Matsumoto K (2013) Induction of extracytoplasmic function sigma factors in Bacillus subtilis cells with defects in lipoteichoic acid synthesis. Microbiology 159: 23–35. [DOI] [PubMed] [Google Scholar]
- Hastie JL, Williams KB, Bohr LL, Houtman JC, Gakhar L, and Ellermeier CD (2016) The Anti-sigma Factor RsiV Is a Bacterial Receptor for Lysozyme: Co-crystal Structure Determination and Demonstration That Binding of Lysozyme to RsiV Is Required for σV Activation. PLOS Genet 12: e1006287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie JL, Williams KB, and Ellermeier CD (2013) The activity of σV, an Extra-Cytoplasmic Function σ factor of Bacillus subtilis, is controlled by a regulated proteolysis of the anti-σ factor RsiV. J Bacteriol 195: 3135–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie JL, Williams KB, Sepúlveda C, Houtman JC, Forest KT, and Ellermeier CD (2014) Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σV. PLoS Genet 10: e1004643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert L, Courtin P, Torelli R, Sanguinetti M, Chapot-Chartier MP, Auffray Y, and Benachour A (2007) Enterococcus faecalis constitutes an unusual bacterial model in lysozyme resistance. Infect Immun 75: 5390–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann JD (2002) The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol 46: 47–110. [DOI] [PubMed] [Google Scholar]
- Helmann John D. (2016) Bacillus subtilis extracytoplasmic function (ECF) sigma factors and defense of the cell envelope. Curr Opin Microbiol 30: 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert S, Bera A, Nerz C, Kraus D, Peschel A, Goerke C, et al. (2007) Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog 3: e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizukuri Y, Oda T, Tabata S, Tamura-Kawakami K, Oi R, Sato M, et al. (2014) A Structure-Based Model of Substrate Discrimination by a Noncanonical PDZ Tandem in the Intramembrane-Cleaving Protease RseP. Structure 1: 1–11. [DOI] [PubMed] [Google Scholar]
- Ho TD, and Ellermeier CD (2011) PrsW is required for colonization, resistance to antimicrobial peptides, and expression of extracytoplasmic function σ factors in Clostridium difficile. Infect Immun 79: 3229–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TD, and Ellermeier CD (2012) Extra cytoplasmic function σ factor activation. Curr Opin Microbiol 15: 182–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Theresa D, Hastie JL, Intile PJ, and Ellermeier CD (2011) The Bacillus subtilis extracytoplasmic function σ factor σ(V) is induced by lysozyme and provides resistance to lysozyme. J Bacteriol 193: 6215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TD, Williams KB, Chen Y, Helm RF, Popham DL, and Ellermeier CD (2014) Clostridium difficile extracytoplasmic function σ factor σV regulates lysozyme resistance and is necessary for pathogenesis in the hamster model of infection. Infect Immun 82: 2345–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyyryläinen H-L, Marciniak BC, Dahncke K, Pietiäinen M, Courtin P, Vitikainen M, et al. (2010) Penicillin-binding protein folding is dependent on the PrsA peptidyl-prolyl cis-trans isomerase in Bacillus subtilis. Mol Microbiol 77: 108–27. [DOI] [PubMed] [Google Scholar]
- Hyyryläinen HL, Bolhuis A, Darmon E, Muukkonen L, Koski P, Vitikainen M, et al. (2001) A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol Microbiol 41: 1159–1172. [DOI] [PubMed] [Google Scholar]
- Jeune A. Le, Torelli R, Sanguinetti M, Giard J-CC, Hartke A, Auffray Y, and Benachour A (2010) The extracytoplasmic function sigma factor SigV plays a key role in the original model of lysozyme resistance and virulence of Enterococcus faecalis. PLoS One 5: e9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehara K, Ito K, and Akiyama Y (2002) YaeL (EcfE) activates the σE pathway of stress response through a site-2 cleavage of anti-σE, RseA. Genes Dev 16: 2147–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehara K, Ito K, and Akiyama Y (2003) YaeL proteolysis of RseA is controlled by the PDZ domain of YaeL and a Gln-rich region of RseA. EMBO J 22: 6389–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontinen VP, and Sarvas M (1993) The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high‐level secretion. Mol Microbiol 8: 727–737. [DOI] [PubMed] [Google Scholar]
- Laaberki M-H, Pfeffer J, Clarke AJ, and Dworkin J (2011) O-Acetylation of peptidoglycan is required for proper cell separation and S-layer anchoring in Bacillus anthracis. J Biol Chem 286: 5278–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewerke LT, Kies PJ, Müh U, and Ellermeier CD (2018) Bacterial sensing: A putative amphipathic helix in RsiV is the switch for activating σV in response to lysozyme. PLoS Genet 14: e1007527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T, Hachmann A-B, and Helmann JD (2007) Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function sigma factors. J Bacteriol 189: 6919–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride SM, and Sonenshein AL (2011) Identification of a genetic locus responsible for antimicrobial peptide resistance in Clostridium difficile. Infect Immun 79: 167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus FC, and Baddiley J (2003) A Continuum of Anionic Charge: Structures and Functions of. Microbiology 67: 686–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V (2006) Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Curr Issues Mol Biol 8: 11–26. [PubMed] [Google Scholar]
- Paetzel M, Karla A, Strynadka NCJ, and Dalbey RE (2002) Signal peptidases. Chem Rev 102: 4549–4579. [DOI] [PubMed] [Google Scholar]
- Parrell D, Zhang Y, Olenic S, and Kroos L (2017) Bacillus subtilis Intramembrane Protease RasP Activity in Escherichia coli and in Vitro. J Bacteriol JB 00381–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J, Courtin P, Meouche I. El, Lemée L, Chapot-Chartier M-P, and Pons J-L (2011) Clostridium difficile has an original peptidoglycan structure with a high level of N-acetylglucosamine deacetylation and mainly 3–3 cross-links. J Biol Chem 286: 29053–29062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland SA, and Criss AK (2017) From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLOS Pathog 13: e1006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosmalen M.L. van, Jongbloed JDH, Dubois JYF, Venema G, Bron S, and Dijl J.M. Van (2001) Distinction between Major and Minor Bacillus Signal Peptidases Based on Phylogenetic and Structural Criteria. J Biol Chem 276: 25230–25235. [DOI] [PubMed] [Google Scholar]
- Smith RE, Salamaga B, Szkuta P, Hajdamowicz N, Prajsnar TK, Bulmer GS, et al. (2019) Decoration of the enterococcal polysaccharide antigen EPA is essential for virulence, cell surface charge and interaction with effectors of the innate immune system. PLOS Pathog 15: e1007730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staroń A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, and Mascher T (2009) The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol Microbiol 74: 557–81. [DOI] [PubMed] [Google Scholar]
- Tjalsma H, Noback MA, Bron S, Venema G, Yamane K, and Dijl J.M. Van (1997) Bacillus subtilis Contains Four Closely Related Type I Signal Peptidases with Overlapping Substrate Specificities. J Biol Chem 272: 25983–25992. [DOI] [PubMed] [Google Scholar]
- Varahan S, Iyer VS, Moore WT, and Hancock LE (2013) Eep Confers Lysozyme Resistance to Enterococcus faecalis via the Activation of the Extracytoplasmic Function Sigma Factor SigV. J Bacteriol 195: 3125–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods EC, Nawrocki KL, Suárez JM, and McBride SM (2016) The Clostridium difficile Dlt pathway is controlled by the extracytoplasmic function sigma factor σV in response to lysozyme. Infect Immun 84: 1902–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokogawa K, Kawata S, Nishimura S, Ikeda Y, and Yoshimura Y (1974) Mutanolysin, Bacteriolytic Agent for Cariogenic Streptococci: Partial Purification and Properties. Antimicrob Agents Chemother 6: 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellmeier S, Hofmann C, Thomas S, Wiegert T, and Schumann W (2005) Identification of sigmaV-dependent genes of Bacillus subtilis. FEMS Microbiol Lett 253: 221–9. [DOI] [PubMed] [Google Scholar]
- Zhang R, Joachimiak G, Kim Y, Gornicki P, and Joachimiak A The crystal structure of the conserved domain protein from Bacillus anthracis [Google Scholar]


