Summary
High affinity B cell selection in the germinal center (GC) is governed by signals delivered by follicular helper T cells (Tfh) to B cells. Selected B cells undergo clonal expansion and affinity maturation in the GC dark zone in direct proportion to the amount of antigen they capture and present to Tfh cells in the light zone. Here we examined the mechanisms whereby Tfh cells program the number of GC B cell divisions. Gene expression analysis revealed that Tfh induce MYC expression in light zone B cells in direct proportion to antigen capture. Conditional Myc haplo-insufficiency or over-expression combined with cell division tracking showed that MYC expression produces a metabolic reservoir in selected light zone B cells that is proportional to the number of cell divisions in the dark zone. Thus, MYC constitutes the germinal center B cell division timer that when deregulated leads to emergence of B cell lymphoma.
Graphical Abstract
eTOC Blurb:
B cell clonal expansion is regulated by Tfh cells in the germinal center. Finkin et al. report that Tfh induce Myc expression by light zone B cells in direct proportion to the amount of antigen presented, and that MYC proportionally regulates B cell size and division in the dark zone.
Introduction
Affinity maturation in GCs, is a product of sequential rounds of cell division, antibody gene mutation and selection (Bannard and Cyster, 2017; Rajewsky, 1996; Victora and Nussenzweig, 2012; Vinuesa et al., 2016). B cells divide and mutate their antibody genes in the GC dark zone (DZ), and test their newly mutated receptors for affinity to antigen in the light zone (LZ) (Allen et al., 2007; Oprea and Perelson, 1997; Victora et al., 2010). Movement between the zones is regulated such that B cells in the DZ turn off expression of CXCR4 and migrate to the LZ once they stop dividing (Allen et al., 2004; Allen et al., 2007; Victora and Nussenzweig, 2012; Victora et al., 2010). In the LZ, B cells with higher affinity receptors capture, process and present a greater proportion of antigen than lower affinity B cells, which leads to prolonged interactions with Tfh (Depoil et al., 2005; Shulman et al., 2014; Victora and Nussenzweig, 2012; Vinuesa et al., 2005) and preferential selection of high affinity cells for return to the DZ where they undergo additional rounds of division and mutation (Bannard and Cyster, 2017; Rajewsky, 1996; Victora and Nussenzweig, 2012; Vinuesa et al., 2016). The number of divisions and the amount of time a B cell spends in the DZ is directly proportional to the amount of antigen a LZ B cell presents to Tfh (Gitlin et al., 2014; Victora et al., 2010). Thus, T cells in the LZ appear to set a timer that controls affinity maturation by directing the number of B cell divisions in the DZ.
The transition to the DZ by selected B cells is associated with expression of the transcription factor MYC (Dominguez-Sola et al., 2012; Victora et al., 2012), which is also required for the GC reaction (Calado et al., 2012; Dominguez-Sola et al., 2012). In addition to MYC other key regulators that have been implicated in interzonal migration include the AP4 transcription factor, mTOR and FOXO1 (Chou et al., 2016; Dominguez-Sola et al., 2015; Ersching et al., 2017; Sander et al., 2015). How these or other factors or their combinations regulate the B cell division timer in the DZ has not been defined.
Here we measured MYC expression in LZ B cells undergoing positive selection in response to graded doses of antigen and found that MYC and cell growth are directly proportional to the amount of antigen captured. Conditional haploinsufficiency or overexpression experiments revealed that the amount of MYC expressed by selected LZ B cells dictates the number of B cell divisions in the DZ. Thus, MYC constitutes the GC B cell division timer.
Results
Relationship between antigen presentation, MYC expression and GC B cell expansion
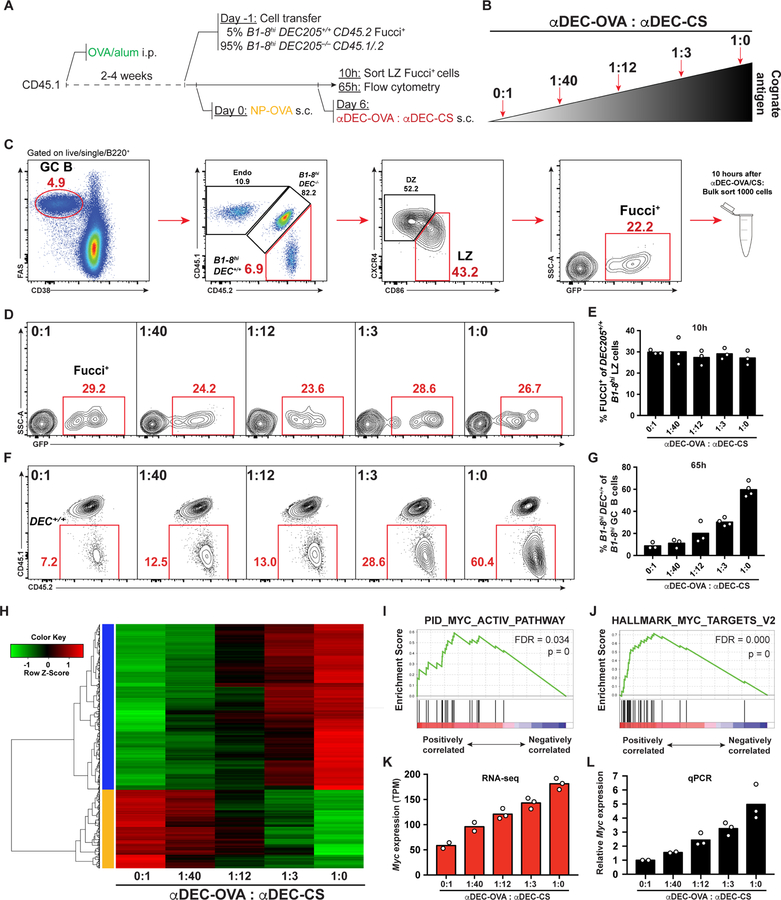
To determine whether there are molecular signatures of affinity selection that are directly proportional to the amount of antigen captured by B cells in the LZ we isolated GC B cells that had been positively selected after receiving graded amounts of antigen (Gitlin et al., 2014). In brief, mice were immunized with ovalbumin (OVA) to prime Tfh cells, and adoptively transferred with a mixture of DEC205+/+ and DEC205−/− NP-specific B1–8hi cells at a 5:95 ratio to limit the number of selected cells and facilitate interaction with Tfh. The B1–8hiDEC205+/+ also carried a fluorescent, ubiquitination-based cell cycle indicator (Fucci) (Aiba et al., 2010; Sakaue-Sawano et al., 2008) (B1–8hiDEC205+/+Fucci+ cells). GC reactions were initiated by injection with 4-hydroxy-3-nitro-phenylacetyl hapten coupled with OVA (NP-OVA). Graded doses of antigen were delivered to DEC205+/+ GC B cells by titrating mixtures of αDEC205 chimeric antibodies that carried the cognate antigen, OVA (αDEC-OVA), and an irrelevant antigen from Plasmodium falciparum (αDEC-CS) (Boscardin et al., 2006). Positively selected LZ B cells were identified by expression of the Fucci marker indicating that they had entered the S phase of the cell cycle and were isolated by cell sorting 10 hours after DEC205 targeting (Figures 1A–1C).
Figure 1. MYC pathway activation in GC B cells is proportional to T cell help.
(A-C) Schematic representation and gating strategy. Endo = Endogenous. (D) Representative GC B cells 10 hours after αDEC-OVA/CS treatment. (E) Quantification of (D) Each symbol represents 12 mice pooled to one group in three independent experiments. (F) Representative flow cytometry results showing percentage of B1–8hiDEC205+/+ among total B1–8hi GC B cells 65 hours after treatment with different amounts of αDEC-OVA (top left). Numbers in red indicate percentage of events in that gate. (G) Quantification of (F). Each symbol represents one mouse. Graph summarizes the results from 3–4 mice per group pooled from 3 independent experiments. (H) Heatmap and hierarchical clustering of proportionally increased or decreased genes expressed by LZ B1–8hiDEC205+/+Fucci+ GC B cells that were used for Gene Set Enrichment Analysis. (I and J) Gene Set Enrichment Plots for MYC pathway activated genes from pathway gene sets (I) and hallmark gene sets (J). FDR = False Discovery Rate. p = p-value. (K) Myc expression determined by RNA sequencing shown as Transcripts Per Million (TPM). (L) Real time PCR quantification of Myc expression performed on RNA from the same samples used for RNA sequencing. Results show the combined data from three independent experiments (see STAR Methods). See also Figure S1.
In agreement with the finding that T cell help is limited (Crotty, 2014; Victora et al., 2010), similar numbers of B cells were selected in the LZ 10 hours after DEC205 targeting irrespective of the antigen dose (Figures 1D and 1E). To verify that B1–8hiDEC205+/+ GC B cells received graded amount of antigen, the relative expansion of these cells was quantitated 65 hours after DEC205 targeting. As expected, the amount of subsequent B1–8hiDEC205+/+ GC B cell expansion was directly proportional to the dose of αDEC-OVA injected (Gitlin et al., 2014) (Figures 1F and 1G).
Genes that were proportionally increased or decreased transcriptionally were selected and subjected to unsupervised clustering (Figure 1H). Gene set enrichment analysis for metabolic pathways using curated data sets was used to determine whether the selected genes correspond to specific cellular programs (Liberzon et al., 2015; Subramanian et al., 2005). MYC, a transcription factor that is essential for the GC reaction, was at the apex of the only statistically significant pathway that was activated in a manner that was directly proportional to antigen dose (Figure 1I). Additional analysis using hallmark gene sets confirmed that genes downstream of MYC were the most significantly enriched genes (Figure 1J and Figure S1A), and revealed enrichment of STAT5, NF-kB signaling, the unfolded protein response, and mTOR signaling in that order (Figure S1B). Consistent with the gene set enrichment analysis Myc expression was directly proportional to antigen dose by RNA sequencing and quantitative PCR (Figures 1K and 1L). Thus, MYC and metabolic pathways activated by this transcription factor are induced in direct proportion to the amount of antigen captured by selected B cells in the LZ.
GC B cell size is proportional to MYC expression in the LZ
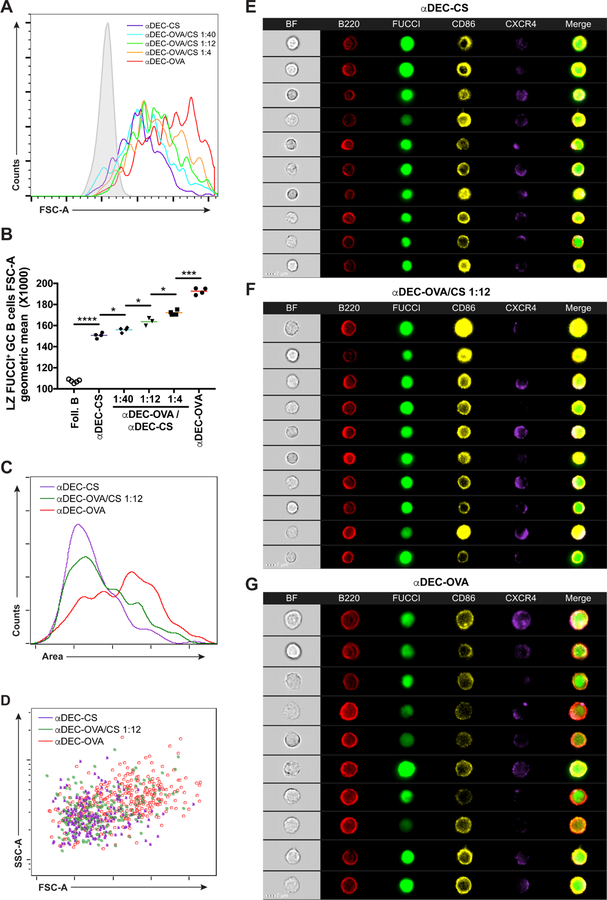
MYC is an important regulator of biogenic pathways (Preston et al., 2015; Wang et al., 2011). Its expression is associated with increased cell size and accumulation of metabolic intermediates required for cell division (Iritani and Eisenman, 1999; Lloyd, 2013). To determine whether the size of selected GC B cells in the light zone is proportional to the amount of antigen they capture, we delivered graded doses of antigen to B1–8hiDEC205+/+Fucci+ GC cells and measured the size of recently selected cells entering the S phase in the LZ by flow cytometry and ImageStream analysis (Figures S2A–S2C). In agreement with directly proportional increases in MYC expression, the size of selected LZ GC B cells was also directly proportional to the amount of antigen captured, both 10 hours and 30 hours following DEC205 targeting (Figures 2A–2G and not shown). Thus, the relative amount of MYC expression in LZ GC B cells is proportional to the increase in the size of selected cells.
Figure 2. GC B cell size is proportional to MYC expression.
(A) Histogram shows representative forward scatter of LZ Fucci+ GC B cells 30 hours after graded αDEC-OVA administration. Solid grey represents follicular B cells. (B) Quantification of (A). Each symbol represents one mouse. Graph summarizes the results from 3 – 4 mice per treatment in two independent experiments. (C and D) Histogram (C) and dot plot (D) representation of LZ B1–8hiDEC205+/+Fucci+ GC B cells size by ImageStream analysis. Graph summarizes the results from 5 combined mice per treatment in two independent experiments. (E-G) ImageStream galleries showing LZ B1–8hiDEC205+/+Fucci+ GC B cells analyzed 30 hours after αDEC-CS (E), αDEC-OVA/CS 1:12 (F) or αDEC-OVA (G) injection. Scale bar = 7 μm. Two-tailed unpaired student’s t test was used in (B). *p < 0.05. ***p < 0.001 ****p < 0.0001. See also Figure S2.
MYC proportionally regulates B cell size and division in vitro
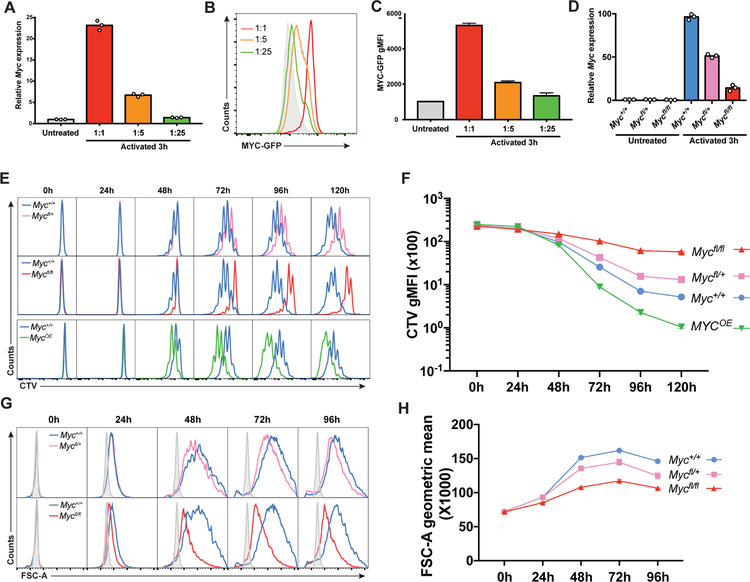
B cell selection is limited by CD40 and cytokine signals delivered by Tfh that induce MYC expression (Crotty, 2011; Vinuesa and Cyster, 2011). Moreover, the amount of CD40 signaling is proportional to MYC induction and B cell division in vitro (Heinzel et al., 2017; Luo et al., 2018). To confirm that MYC expression is proportional to the magnitude of Tfh signals in vitro, we cultured B cells with graded amounts of aCD40, IL-4, IL-21 and aCD180 and measured Myc mRNA expression. As expected, Myc mRNA and MYC-GFP fusion protein expression were directly proportional to the amount of the activating cocktail present in the culture medium in vitro (Figures 3A–3C).
Figure 3. MYC expression is proportional to the magnitude of Tfh signals in vitro.
(A to C) Quantification of Myc mRNA by real time PCR (A) or of MYC-GFP protein by flow cytometry (B and C) after graded activation of wild type B cells with IL-4, IL-21, αCD40 and αCD180 either undiluted or diluted 1:5 or 1:25. Solid grey represents MYC-GFP− cells. Graph shows one representative experiment with three technical repeats of two independent experiments. (D) Quantification of Myc mRNA by real time PCR in B cells extracted from spleens of the indicated R26ERT2-Cre mice, cultured in the presence of 4-OHT for 12 hours and activated for 3 hours with IL-4, αCD40 and αCD180. (E) Histograms show CTV dye dilution by spleen B cells cultured for the indicated times with IL-4, IL-21, αCD40 and αIgM and 4-OHT. Blue = Myc+/+, pink = Mycfl/+, red = Mycfl/fl and green = R26StopFLMYC (MYCOE/+). (F) Quantification of (E). (G) Histograms show size for cells treated as in (E). Solid grey represents unactivated B cells. (H) Quantification of (G). Histograms and graphs in (E) to (H) show one representative experiment with three technical repeats of each time point and condition in two independent experiments. Error bars represent SEM. See also Figure S3.
To determine whether MYC expression governs the number of B cell divisions we compared B cells that were Myc-sufficient, haploinsufficient and insufficient by combining a loxP-flanked allele of Myc (Mycfl) (de Alboran et al., 2001) with an inducible ROSA26-CreERT2 allele (R26ERT2-Cre) (Ventura et al., 2007). In addition, MYC was over expressed in B cells carrying a loxP-flanked STOP human MYC cDNA in the ROSA26 locus (R26StopFLMYC) (Sander et al., 2012) by combining it with inducible CreERT2 (UbcERT2-Cre) (Ruzankina et al., 2007). When activated B cells were treated with the active metabolite of tamoxifen, 4-OHT, Myc expression decreased to 55% and 14% of the wild type in R26ERT2-CreMycfl/+ and R26ERT2-CreMycfl/fl B cells respectively (Figure 3D). Cell division, which was monitored using CellTrace Violet (CTV), and cell size were directly proportional to the amount of MYC expressed, while no difference was found in cell death (Figures 3E–3H and Figures S3A–S3C). Whereas R26ERT2-CreMycfl/+ and R26ERT2-CreMycfl/fl B cells divided less than wild type, R26StopFLMYC B cells (MycOE) underwent additional divisions (Figure 3E and 3F). Thus, Myc haploinsufficiency leads to a proportional decrease in MYC expression, cell size and the amount of B cell division in response to activation in vitro.
DZ residence time and expansion are proportional to MYC in GC LZ B cells
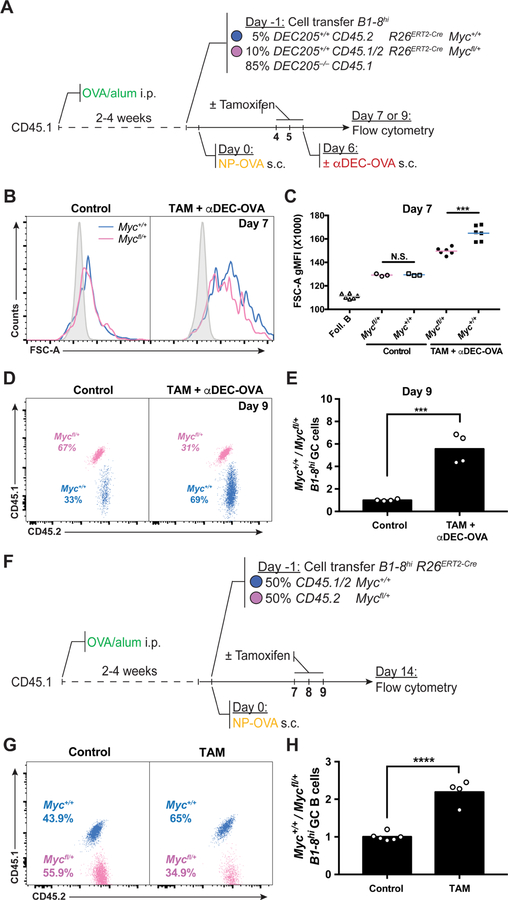
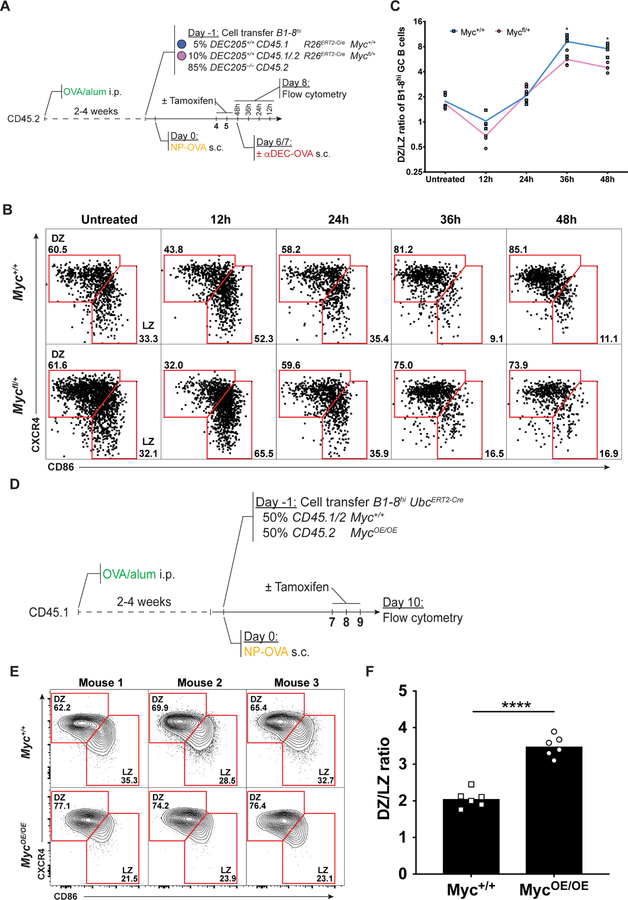
To directly measure the zonal distribution of MYC-expressing GC B cells in vivo, we sorted LZ and DZ DEC205+/+ NP-specific B1–8hi GC B cells 10 hours after injection of DEC-CS or DEC-OVA and quantified Myc mRNA expression in each zone by quantitative real-time PCR (Figure S4A). In agreement with previous reports (Calado et al., 2012; Dominguez-Sola et al., 2012), Myc expression was 8-fold higher in the LZ after DEC-CS treatment. In contrast, following DEC-OVA treatment Myc was drastically induced in the LZ, with only a subtle increase of its expression in the DZ, resulting in 20-fold difference in Myc expression between the zones (Figure S4B and S4C). To determine whether MYC amount regulates GC B cell size and expansion in vivo, we performed adoptive transfer experiments using allotypically marked B1–8hiR26ERT2-CreMyc+/+ and B1–8hiR26ERT2-CreMycfl/+ B cells. Recipient mice were primed with OVA and boosted with NP-OVA to initiate GC responses as described above and injected with tamoxifen before αDEC-OVA administration (Figure 4A). Whereas the size of GC B cells of the two genotypes was similar in control mice that did not receive tamoxifen, selected Myc-sufficient LZ GC B cells were larger than their haploinsufficient and insufficient counterparts (Figures 4B and 4C and Figures S4D–S4F). There was a 4.5-fold relative expansion of Myc-proficient (B1–8hiR26ERT2-CreMyc+/+) over haploinsufficient (B1–8hiR26ERT2-CreMycfl/+) GC B cells 72 hours after αDEC-OVA treatment, and Myc-insufficient GC B cells failed to expand (Figure 4D and 4E and Figures S4G–S4L). In addition, there was a 2-fold relative expansion of Myc-proficient (B1–8hiR26ERT2-CreMyc+/+) over haploinsufficient (B1–8hiR26ERT2-CreMycfl/+) GC B cells 7 days after tamoxifen injection in the absence of DEC-OVA injection (Figures 4F–4H). We conclude that the amount of MYC expressed by selected GC LZ cells is directly proportional to their cell size and amount of clonal expansion.
Figure 4. MYC proportionally regulates GC B cell expansion.
(A) Schematic representation of the experimental protocol for (B-E). (B) Representative histograms showing forward scatter of B1–8hiR26ERT2-CreDEC205+/+ GC B cells from Myc+/+ (blue) and Mycfl/+ (pink) mice without (left panel) or with (right panel) αDEC-OVA and tamoxifen administration 7 days after NP-OVA boost. Solid grey represents follicular B cells. (C) Quantification of (B). Each symbol represents one mouse. Summary of results from 3–6 mice in three independent experiments. (D) Representative dot plot showing the percentage of adoptively transferred B1–8hiR26ERT2-CreMyc+/+ and B1–8hiR26ERT2-CreMycfl/+ GC B cells in untreated control (left panel) or tamoxifen and αDEC-OVA (right panel) treated mice 9 days after NP-OVA boost. (E) Quantification of (D). Graph shows fold change of B1–8hiR26ERT2-CreMyc+/+ divided by B1–8hiR26ERT2-CreMycfl/+ in individual mice normalized to the average of the same in the control group. Summary of results from 4 – 6 mice in three independent experiments. (F) Schematic representation of the experimental protocol for (G) and (H). (G and H) Representative dot plots showing Percentage (G) and ratio (H, fold change over input) of GC B cells from the indicated mice 7 days after the first tamoxifen treatment. Each symbol represents one mouse. Data represents 3–6 mice in three independent experiments. Unpaired two-tailed student’s t test. N.S.: p > 0.05 (not statistically significant); ***p < 0.001 ****p < 0.0001. See also Figure S4 and Figure S5.
The difference in the number of Myc-sufficient and haploinsufficient B cells accumulating in GCs could be due to differential cell death or cell division (Mayer et al., 2017). However, we found no difference in cell death as measured by Caspase-3 activation (Figures S5A–S5F). The number of cell divisions per GC re-entry cycle is directly proportional to the amount of time a cell spends in the DZ (Gitlin et al., 2014; Victora et al., 2010). To determine whether MYC is proportional to the kinetics of GC cycling between LZ and DZ we monitored the number of Myc-sufficient and haploinsufficient GC B cells in each zone after αDEC-OVA injection (Figure 5A). The initial distribution of the two adoptively transferred B cell types and their initial accumulation in the DZ was similar. However, 36 hours after selection, there were more Myc-sufficient than haploinsufficient cells in the DZ suggesting that higher amounts of MYC expression during selection led to longer DZ residence times and more division (Figures 5B and 5C). Moreover, we found that MYC expression plays a role in LZ to DZ transition by selected cells since complete loss of its expression is associated with significantly reduced ability of LZ cells to transit to the DZ after receiving a selection signal (Figures S6A–S6C). To further explore how MYC influences the zonal distribution of GC B cells we monitored the number of co-transferred Myc-sufficient and MYC-overexpressing GC B cells in each zone (Figure 5D). MYC-overexpressing GC B cells showed a significant bias toward the DZ (Figures 5E and 5F). These results suggest that MYC regulates selected GC B cell division capacity.
Figure 5. MYC expression correlates with DZ residence time.
(A) Schematic representation of the experimental protocol for (B) and (C). (B) Representative Dot plots show DZ/LZ distribution of Myc+/+ (top) and Mycfl/+ (bottom) B1–8hiR26ERT2-CreDEC205+/+ GC B cells in mice treated with tamoxifen at the indicated time points after αDEC-OVA injection. (C) Quantification of (B). Summary of results from 4 mice in two experiments. (D) Schematic representation of the experimental protocol for (E) and (F). (E) Representative contour plots of DZ/LZ distribution of Myc+/+ (top) and MycOE/OE (bottom) B1–8hiUbcERT2-CreDEC205+/+ GC B cells. (F) Quantification of (E). Summary of results from 3–6 mice in two experiments. Unpaired two-tailed student’s t test. *p < 0.05, ****p < 0.0001. See also Figure S6.
MYC determines the number of GC B cell divisions
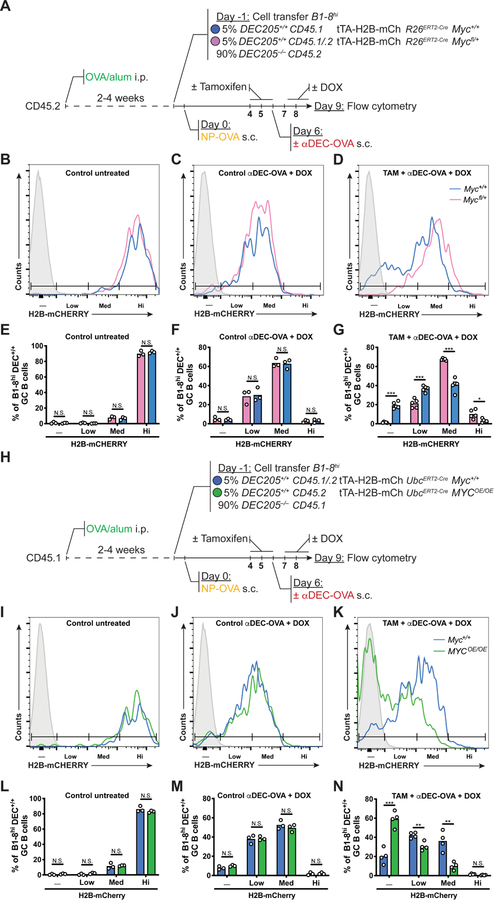
To determine whether the amount of MYC directly regulates the number of GC B cell divisions in vivo, we measured cell division by combining a transgene encoding the tetracycline transactivator (tTA) expressed under the Vav promoter and a histone H2B– mCherry fusion protein driven by a doxycycline (DOX)-regulated promoter (tTA– H2B– mCh) (Egli et al., 2007; Wiesner et al., 2005). Under homeostatic conditions H2B– mCherry is constitutively expressed; DOX administration represses its expression resulting in H2B– mCherry dilution that is directly proportional to cell division (Gitlin et al., 2014; Tumbar et al., 2004).
OVA primed recipient mice received B1–8hiR26ERT2-CreMyc+/+ tTA-H2B-mCh and B1–8hiMycfl/+ tTA-H2B-mCh B cells before boost with NP-OVA. Tamoxifen was administered on days 4–6 after immunization to produce Myc haploinsufficiency followed by αDEC-OVA and DOX to induce selection and to measure cell division, respectively (Figure 6A). Control untreated mice and controls that received αDEC-OVA and DOX showed comparable amounts of H2B-mCh indicator dilution among Myc+/+ and Mycfl/+ GC B cells (Figures 6B, 6C, 6E and 6F). In contrast, in mice that received tamoxifen, αDEC-OVA and DOX Myc-sufficient cells had undergone more rounds of cell division than their haploinsufficient counterparts (Figures 6D and 6G). Moreover, increasing the amount of MYC expression using B1–8hiR26StopFLMYC tTA-H2B-mCh B cells revealed that MYC overexpression was sufficient to increase selected B cell division (Figures 6H–6N). Thus, the amount of MYC expressed in the LZ determines the number of GC B cell divisions in the DZ.
Figure 6. MYC is both necessary and sufficient for GC B cell division.
(A) Schematic representation of the experimental protocol for (B-G). (B-D) Representative histograms show H2B-mCherry fluorescence among B1–8hiR26ERT2-CreMyc+/+tTA–H2B– mCh (blue) and B1–8hR26ERT2-CreMycfl/+tTA–H2B–mCh (pink) GC B cells in mice treated as indicated. (E-G) Quantification of (B-D). Control mice in (B) and (E) are untreated with TAM, αDEC-OVA and DOX. Summary of results from 3–6 mice in two independent experiments. Mean percentage of H2B-mCherry negative, low, medium or high B1–8hiR26ERT2-CreMyc+/+tTA–H2B–mCh and B1–8hiR26ERT2-CreMycfl/+tTA–H2B–mCh GC B cells. (H) Schematic representation of the experimental protocol for (I-N). (I-K) Representative histograms show H2B-mCherry fluorescence among B1–8hiUbcERT2-CreMyc+/+tTA–H2B–mCh (blue) and B1–8hiUbcERT2-CreR26StopFLMYC tTA–H2B–mCh (MycOE/OE, green) GC B cells in mice treated as indicated. (L-N) Quantification of (I-K). Control mice in (I) and (L) are untreated with TAM, αDEC-OVA and DOX. Summary of results from 3–5 mice in two independent experiments. Mean percentage of H2B-mCherry negative, low, medium or high B1–8hiUbcERT2-CreMyc+/+tTA–H2B–mCh and B1–8hiUbcERT2-CreR26StopFLMYC tTA–H2B–mCh GC B cells. Solid grey represents non-fluorescent cells. DOX = Doxycyclin hyclate. Unpaired two-tailed student’s t test. N.S.: p > 0.05 (not statistically significant); *p < 0.05, **p < 0.01, ***p < 0.001.
Direct correlation between affinity and size of selected LZ B cells
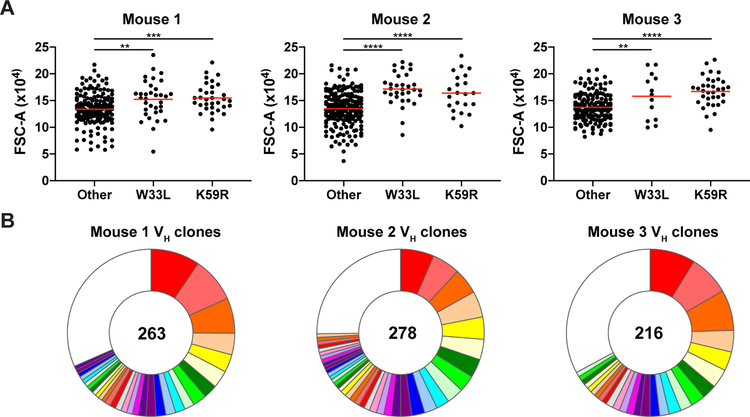
To determine whether a correlation exist between affinity and the size of selected GC B cells in a polyclonal immune response, we immunized otherwise wild type Fucci+ reporter mice with NP–OVA. Single Fucci+ LZ GC B cells were isolated from the draining lymph nodes by index sorting, their VH genes sequenced and the presence of characteristic high affinity mutations W33L and K59R documented (Allen et al., 1988; Furukawa et al., 1999) (Figure S7A and S7B). 168 of 757 sequences (22.2%) obtained from 3 separate mice carried the high affinity mutations. The high affinity cells were significantly larger than cells that did not carry the mutations (Figure 7A and 7B and Figure S7C). We conclude that MYC expression dictates selected cell size in the LZ and the number of divisions by these cells in the DZ.
Figure 7. Correlation between affinity and size of selected LZ B cells in polyclonal GCs.
(A) Forward scatter of individual l+ Fucci+ LZ GC B cells carrying the affinity-enhancing mutations VH186.2 W33L+ or K59R+ or those that do not 10 days after immunization with NP-OVA (see STAR Methods). Red bars indicate mean. Summary of three independent repeats of the experiment, with 1–2 mice each. **p < 0.01, ***p < 0.001, ****p < 0.0001, Unpaired two-tailed student’s t test. (B) Pie charts showing clonal composition of VH sequences for each mouse. Colored slices represent expanded clones, white slice represents singles. Total number of sequences analyzed for each mouse is shown in center. See also Figure S7.
Discussion
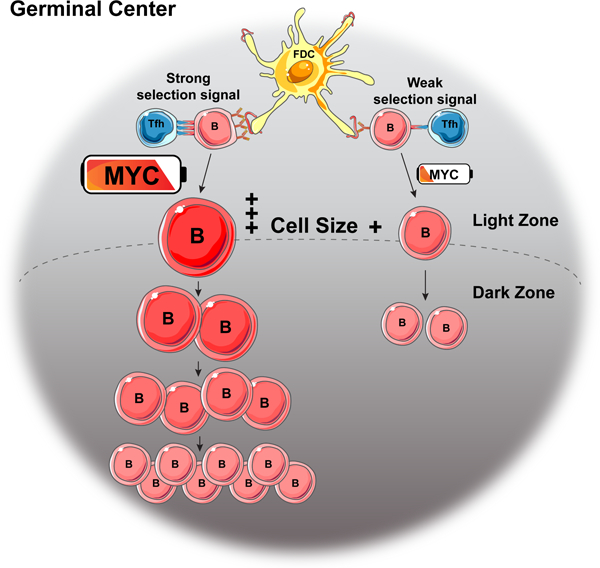
MYC is rapidly induced and only transiently expressed in selected GC B cells in the LZ (Calado et al., 2012; Dominguez-Sola et al., 2012). Our experiments show that the amount of MYC expression is directly related to the amount of antigen presented by the selected B cells and the strength of the Tfh signal. How does transient MYC expression in the LZ set the timer for DZ division? Cell division cycles in the DZ are among the fastest recorded in mammalian cells lasting 4–6 hours (Victora and Nussenzweig, 2012). Nearly all of that time is devoted to the S phase and G2/M with little or no time left for G (Gitlin et al., 2015). Therefore, there is little if any time to accumulate the metabolites required for additional cell division (Allen et al., 2007; Bannard and Cyster, 2017; Hauser et al., 2007). G1 cells in the DZ are likely to be in transition to the LZ, a process that requires several hours (Stewart et al., 2018; Victora et al., 2010). MYC expression in selected LZ B cells resolves this metabolic problem by regulating the amount of selected LZ B cell growth and accumulation of necessary metabolites in direct proportion to MYC expression. Thus, MYC expression produces a metabolic reservoir that is proportional to the strength of the Tfh signal. Since the metabolic reservoir created upon selection in the LZ cannot be fully replenished during rapid cycling in the DZ, its rate of exhaustion during division appears to dictate the number of cell cycles in the DZ.
The MYC pathway is parallel to and complements the PI3K/mTOR pathway that also regulates many of the important aspects of cell growth and metabolism (Ersching et al., 2017). FOXO1, a transcription factor that is directly downstream of PI3K/AKT is required to instruct the DZ program (Dominguez-Sola et al., 2015; Inoue et al., 2017; Luo et al., 2018). MYC differs from PI3K/mTOR in that in addition to cellular metabolism MYC is also a master regulator of cell division. Many of the genes that control cell division including CDKs, cyclins and E2F transcription factors, CDK activating kinase (CAK), and CDC25 phosphatases are downstream of and controlled by MYC (Bretones et al., 2015; Dang, 2012). In addition, MYC also induces a proportional increase in genes that dictate a cellular program that can sustain cell division in the DZ including the AP4 transcription factor that reinforces the program in absence of persistent MYC expression (Chou et al., 2016).
In conclusion, transient interaction between Tfh and LZ B cells results in activation of Myc and its downstream cellular program in direct proportion to the amount of B cell antigen presentation and this program constitutes the timer that governs the number of cell divisions that a selected B cell makes in the DZ. Consistent with this idea, de-regulation of the timer in GC B cells that carry IgH to MYC chromosome translocations is associated with the emergence of B cell lymphoma (Adams et al., 1983; Crews et al., 1982; Dalla-Favera et al., 1982; Leder et al., 1983).
STAR METHODS
Lead Contact and Materials Availability
Further information and requests for reagents and resources should be directed to and will be fulfilled by the Lead Contact Michel C. Nussenzweig (nussen@rockefeller.edu). This study did not generate new unique reagents. The mouse lines obtained from other laboratories are described below and may require a Material Transfer Agreement (MTA) with the providing scientists.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice.
C57BL/6J, B6.SJL, Myc-GFP, R26StopFLMYC (MYCOE), R26ERT2-Cre, UbcERT2-Cre and Col1A1-tetO-H2B–mCherry mice were purchased from Jackson Laboratories. Vav-tTA transgenic mice were crossed to Col1A1-tetO-H2B–mCherry mice to produce tTA–H2B–mCh mice as previously described (Gitlin et al., 2014). Myc-flox (Mycfl) mice were obtained from J.M. Sedivy and F.J. Gonzalez. Fucci transgenic mice were obtained from T. Kurosaki and A. Miyawaki (Sakaue-Sawano et al., 2008). B1–8hi knock-in (Shih et al., 2002) and DEC205−/− mice (Guo et al., 2000) were generated and maintained in our laboratory. See Star Methods for more information of mice used. All experiments were performed with authorization from the Institutional Review Board and the Rockefeller University IACUC.
METHOD DETAILS
B cell transfers.
Spleens were forced through a 70 µm mesh into FACS buffer (FB: PBS, 2 % v/v fetal bovine serum, 2 mM EDTA). Resting B cell suspensions were purified by magnetic cell sorting (MACS) using CD43 microbeads according to manufacturer’s protocol (Miltenyi Biotec). ~5 × 106 B1–8hi B cells (5 × 105 Igλ+, NP-specific B cells) composed of the indicated populations were transferred intravenously into recipient mice as previously reported (Pasqual et al., 2015).
Immunizations and treatments.
C57BL/6J or B6.SJL recipient mice (6 – 12 weeks old) were primed by intraperitoneal immunization with 50 µg of OVA (Albumin from chicken egg white, Grade V, Sigma) precipitated in alum at a 2:1 ratio in PBS. Two to six weeks after priming, mice received adoptive cell transfers of indicated B cells and were boosted the following day by injection of 25 µg of NP15–OVA (Biosearch Technologies) into footpads. αDEC–OVA and αDEC–CS chimeric antibodies were produced by transient transfection in HEK293E cells, as described (Boscardin et al., 2006). 5 µg of chimeric antibody in PBS was injected into footpads of the recipient mice at indicated time points. Deletion of loxP-flanked alleles was induced by intraperitoneal injection of tamoxifen (Sigma) dissolved in corn oil (Sigma) at indicated doses and time points. For tTA–H2B–mCherry dilution experiments, mice were administered DOX (doxycyclin hyclate, Sigma) by intraperitoneal injection of 2 mg DOX in PBS and footpad injection of 0.2 mg DOX in PBS. Mice were maintained on DOX by adding DOX (2 mg/mL) and sucrose (50 mg/mL) to the drinking water for the indicated time periods. Draining lymph nodes were collected for flow cytometric analysis. H2B-mCherry dilution rate was monitored by flow cytometry and showed a slight general increase in dye dilution rate after tamoxifen treatment.
B cell culture.
Resting B cells were obtained from mouse spleens by forcing tissue through a 70 µm mesh into PBS containing 3 % fetal bovine serum (FBS). B cells were then purified using anti-CD43 magnetic beads (MACS) according to manufacturer’s protocol (Miltenyi Biotec). B cells were cultured in RPMI-1640 culture medium, supplemented with 10 % FBS, 10 mM HEPES, antibiotic-antimycotic (1X), 1 mM sodium pyruvate, and 50 µM 2-mercaptoethanol (all from Gibco). B cells were stimulated with 10 µg/mL anti-CD40 (BioXcell, clone FGK45), 5 ng/mL IL-4 (Sigma), 10 ng/mL IL-21 (Sigma) and 10 µg/mL anti-IgM (Jackson ImmunoResearch) or 0.5 µg/mL anti-CD180 (BD Pharmingen). Experiments shown in Figure 3 and Figure S3 were repeated with either anti-IgM or anti-CD180, in addition to the other cytokines listed above, with similar results and a slightly higher division rate following stimulation with anti-CD180, which is a stronger MYC inducer. Temporal deletion of loxP-flanked alleles in vitro was induced by administration of 4-hydroxytamoxifen (4-OHT, Sigma) at a final concentration of 1 µM 12–18 hours before B cell activation. Cell division in vitro was measured by labeling resting B cells with 5 µM CellTrace Violet (CTV, Thermo Fisher) according to manufacturer’s instructions. B cells were cultured at 2–5 × 105 cells mL−1 for all in vitro experiments.
Flow cytometry and bulk GC B cell sorting.
Single cell suspensions were prepared by gently macerating draining lymph nodes against the wall of 1.5-mL microcentrifuge tube using a plastic pestle in 100 mL FB supplemented with 1 µg/mL anti-mouse CD16/CD32 (Fc block, BD Pharmingen). Cells were stained on ice for 45 minutes with biotinylated anti-CXCR4 antibody or 25 minutes with the conjugated antibodies listed in the Star Methods Key Resource Table. Before sorting or analysis, the cell suspensions were filtered through a 40µM cell strainer. For cell-death analysis, cells were fixed and permeabilized using the Cytofix/Cytoperm kit (BD) according to the manufacturer’s instructions and stained with anti-active Caspase 3 (BD) for 25 minutes. For MYC-GFP expression analysis cells were permeabilized using the Cytofix/Cytoperm kit (BD) according to the manufacturer’s instructions and stained with anti-GFP-AF488 (Thermo Fisher) for 25 minutes. Samples were analyzed on an LSRFortessa or Symphony flow cytometer (BD). GC B cells were gated as live/single/Dump−, B220+, CD38− FAS+. DZ and LZ GC B cells were gated as CXCR4hi CD86low and CXCR4low CD86hi, respectively. CD45.1 and CD45.2 allotypic markers were used to track adoptively transferred B cells within either C57BL/6J (CD45.2/.2) or B6.SJL (CD45.1/.1) recipient mice as indicated for each experiment. For cell sorting, lymph node cell suspensions were enriched for B cells by magnetic cell sorting (MACS) using CD43 microbeads according to manufacturer’s protocol (Miltenyi Biotec). Cells were stained as above and sorted directly into 200 µL extraction buffer from PicoPure RNA Isolation Kit (Thermo Fisher) for bulk RNA-Sequencing using a FACS Aria cell sorter (BD).
single GC B-cell index sorting.
Single cell suspensions were obtained from the draining lymph nodes of NP15–OVA immunized C57BL/6J Fucci+ mice and stained as described above with the conjugated antibodies listed in Star Methods Key Resource Table. Sorted LZ selected GC B cells were gated as live/single/Dump−, B220+, Igλ+, CD38− FAS+, CXCR4low CD86hi, Fucci+. Single cells were index sorted into individual wells of a 96-well plate containing 5 µl of lysis buffer with 1% of 2-β-mercaptoethanol.Plates were immediately frozen on dry ice and stored at −80°C.
Antibody sequencing and cloning.
96-well plates containing single cell lysates were thawed on ice. Single cell RNA was purified in a designated clean area using magnetic beads (RNAClean XP) following the manufacturer instructions. In the final step of the purification protocol, RNA was eluted from the magnetic beads with 11µl of a solution containing (14.5 ng/µl of random primers (Invitrogen), 0.5% of tergitol, (Type NP-40, 70% in H2O, Sigma-Aldrich,), 0.6U/µl of RNAse inhibitor (Promega) in nuclease free water (Qiagen), and incubated at 65°C for 3 min. cDNA was subsequently synthesized by reverse transcription (SuperScript III Reverse Transcriptase, Invitrogen) as previously described (von Boehmer et al., 2016). cDNA was stored at −80°C or used for antibody gene amplification by nested Polymerase chain reaction (PCR). To amplify the antibody genes from single B-cells, 10 µl of nuclease free water was added to the solution containing cDNA. Mouse antibody genes were amplified by nested PCR as previously described (von Boehmer et al., 2016). PCR protocols: (annealing (°C)/ elongation (sec)/ number of cycles): 1st PCR (IgM and IgG VH): 46/55/50; 2nd PCR: 50/55/50 (see primer sequences in the Key Resource Table).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC anti-mouse/human CD45R/B220 (clone RA3–6B2) | BioLegend | Cat# 103212; RRID: AB_312997 |
| BUV395 anti-mouse CD45R/B220 (clone RA3–6B2) | BD Biosciences | Cat# 563793; RRID: AB_2738427 |
| BV605 anti-mouse CD45R/B220 (clone RA3–6B2) | BioLegend | Cat# 103244; RRID: AB_2563312 |
| PE anti-Mouse CD86 (B7–2) (clone GL1) | Thermo Fisher | Cat# 12–0862-82; RRID: AB_465768 |
| APC anti-mouse CD86 (clone GL1) | BioLegend | Cat# 105012; RRID: AB_493342 |
| Biotin anti-mouse CD184 (CXCR4) (clone 2B11/CXCR4) | BD Biosciences | Cat# 551968, RRID: AB_394307 |
| BV421 anti-mouse CD45.1 (clone A20) | BioLegend | Cat# 110732; RRID: AB_2562563 |
| FITC anti-mouse CD45.2 (clone104) | Thermo Fisher | Cat# 11–0454-85; RRID: AB_465062 |
| APC anti mouse CD45.2 (clone104) | Thermo Fisher | Cat# 17–0454-82; RRID: AB_469400 |
| PE anti-mouse CD45.2 (clone104) | BioLegend | Cat# 109808; RRID: AB_313445 |
| AF700 anti-mouse CD38 (clone 90) | Thermo Fisher | Cat# 56–0381-82; RRID: AB_657740 |
| APC eF780 anti mouse CD4 (clone RM4–5) | Thermo Fisher | Cat# 47–0042-82; RRID: AB_1272183 |
| APC eF780 anti mouse CD8a (clone 53–6.7) | Thermo Fisher | Cat# 47–0081-82; RRID: AB_1272185 |
| APC eF780 anti mouse NK1.1(clone PK136) | Thermo Fisher | Cat# 47–5941-82; RRID: AB_2735070 |
| APC eF780 anti mouse F4/80 (clone BM8) | Thermo Fisher | Cat# 47–4801-82; RRID: AB_2735036 |
| APC eF780 anti mouse LY6G (Gr1) (clone RB6–8C5) | Thermo Fisher | Cat# 47–5931-82; RRID: AB_1518804 |
| PE-Cy7 anti- mouse CD95 (FAS) (clone Jo2) | BD Biosciences | Cat# 557653; RRID: AB_396768 |
| Biotin anti mouse IgD (clone 11–26C) | Thermo Fisher | Cat# 13–5993-85; RRID: AB_466861 |
| Unconjugated anti-mouse CD16/32 (clone 2.4G2) | BD Biosciences | Cat# 553142; RRID: AB_394657 |
| InVivoMab anti-mouse CD40 (clone FKG4.5) | BioXCell | Cat# BE0016–2; RRID: AB_1107647 |
| Unconjugated anti-mouse CD180 (clone RP/14) | BD Biosciences | Cat# 552128; RRID: AB_394343 |
| AF488 polyclonal anti-GFP | Thermo Fisher | Cat# A-21311; RRID: AB_221477 |
| APC anti-mouse Igλ (clone RML-42) | BioLegend | Cat# 407306; RRID: AB_961363 |
| FITC anti-human CD2, (clone TS1/8) | BioLegend | Cat# 309206; RRID: AB_314756 |
| AF647 anti mouse Active Caspase-3 (clone C92–605) | BD Biosciences | Cat# 560626; RRID: AB_1727414) |
| Polyclonal Anti-mouse IgM | Jackson ImmunoResearch | Cat# 115–005-020; RRID: AB_2338450 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Mouse IL-4 | Sigma | I1020 |
| Mouse IL-21 | Sigma | I4032 |
| Streptavidin-BV510 | BD Biosciences | 563261 |
| Streptavidin-BV421 | BD Biosciences | 563259 |
| (Z)-4-Hydroxytamoxifen | Sigma | H7904–5MG |
| Zombie NIR | BioLegend | 423105 |
| Doxycyclin hyclate | Sigma | D9891–25G |
| Tamoxifen | Sigma | T5648–5G |
| Taqman Universal Master Mix | Thermo Fisher | 4304437 |
| CD43 (Ly-48) MicroBeads | Myltenyi Biotec | 130–049-801 |
| Streptavidin MicroBeads | Myltenyi Biotec | 130–048-101 |
| Albumin from chicken egg white | Sigma | A5503–25G |
| CellTrace Violet | Thermo Fisher | C34557 |
| NP-OVAL (Ovalbumin) | BioSearch Technologies | N-5051 |
| Random primers | Invitrogen | 48190–011 |
| Tergitol | Sigma-Aldrich | NP40S-100ML |
| RNAse inhibitor | Promega | N2615 |
| αDEC-CS and αDEC-OVA | Boscardin et al., 2006 | N/A |
| Critical Commercial Assays | ||
| BD Perm/Wash buffer | BD Biosciences | 554722 |
| BD Cytofix/Cytoperm | BD Biosciences | 554723 |
| PicoPure RNA Isolation Kit | Thermo Fisher | 12204–01 |
| RNeasy Plus Mini Kit | Qiagen | 74134 |
| SMART-Seq v4 Ultra Low Input RNA Kit | Clontech | 634888 |
| Nextera XT DNA sample preparation kit | Illumina | FC-131–1024 |
| SuperScript III First-Strand Synthesis kit | Invitrogen | 18080400 |
| Myc Taqman assay | Applied BioSystems | Mm00487804_m1 |
| Hprt Taqman assay | Applied BioSystems | Mm00446968_m1 |
| Ubc Taqman assay | Applied BioSystems | Mm02525934_g1 |
| Gapdh Taqman assay | Applied BioSystems | Mm99999915_g1 |
| Actb Taqman assay | Applied BioSystems | Mm01205647_g1 |
| RNAClean XP beads | Beckman Coulter | A63987 |
| TCL buffer | Qiagen | 1031576 |
| Deposited Data | ||
| RNA sequencing data | This paper | GEO: GSE132881 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664 |
| Mouse: B6.SJL | The Jackson Laboratory | JAX: 002014 |
| Mouse: Myc-GFP | The Jackson Laboratory | JAX: 021935 |
| Mouse: R26StopFLMYC (MycOE) | The Jackson Laboratory | JAX: 020458 |
| Mouse: Myc-flox (Mycfl) | The Jackson Laboratory | MMRRC#: 32046-JAX |
| Mouse: R26ERT2-Cre | The Jackson Laboratory | JAX: 008463 |
| Mouse: UbcERT2-Cre | The Jackson Laboratory | JAX: 008085 |
| Mouse: Col1A1-tetO-H2B–mCherry | The Jackson Laboratory | JAX: 014592 |
| Mouse: B1–8hi | The Jackson Laboratory | JAX: 007594 |
| Mouse: DEC205−/− | Shih et al., 2002 | N/A |
| Mouse: Vav-tTA | Egli et al., 2007 | N/A |
| Mouse: Fucci | Sakaue-Sawano et al., 2008 | N/A |
| Oligonucleotides | ||
| AGGAACTGCAGGTGTCC (Antibody sequencing 1st PCR forward) | von Boehmer et al., 2016 | N/A |
| CAGCTACAGGTGTCCACTCC (Antibody sequencing 1st PCR forward) | von Boehmer et al., 2016 | N/A |
| TGGCAGCAACAGCTACAGG (Antibody sequencing 1st PCR forward) | von Boehmer et al., 2016 | N/A |
| CTGCCTGGTGACATTCCCA (Antibody sequencing 1st PCR forward) | von Boehmer et al., 2016 | N/A |
| CCAAGCTGTGTCCTGTC (Antibody sequencing 1st PCR forward) | von Boehmer et al., 2016 | N/A |
| TTTTAAAAGGTGTCCAGTGT (Antibody sequencing 1st PCR forward) | von Boehmer et al., 2016 | N/A |
| CCTGTCAGTAACTACAGGTGTCC (Antibody sequencing 1st PCR forward) | von Boehmer et al., 2016 | N/A |
| CTTTWWMAAGGKRTSCAKTGT (Antibody sequencing 1st PCR forward) | von Boehmer et al., 2016 | N/A |
| GGCAGCTGCCCAAAGTRYCC (Antibody sequencing 1st PCR forward) | von Boehmer et al., 2016 | N/A |
| AGAAGGTGTGCACACCGCTGGAC (Antibody sequencing 1st PCR reverse) | von Boehmer et al., 2016 | N/A |
| AGGGGGCTCTCGCAGGAGACGAGG (Antibody sequencing 1st PCR reverse) | von Boehmer et al., 2016 | N/A |
| GGGAATTCGAGGTGCAGCTGCAGGAGTCTGG (Antibody sequencing 2st PCR forward) | von Boehmer et al., 2016 | N/A |
| AGCTGTATCATGCTCTTCTTGGCA (Antibody sequencing 2st PCR forward) | Dominguez-Sola et al., 2012 | N/A |
| GCTCAGGGAARTAGCCCTTGAC (Antibody sequencing 2st PCR reverse) | von Boehmer et al., 2016 | N/A |
| AGGGGGAAGACATTTGGGAAGGAC (Antibody sequencing 2st PCR reverse) | von Boehmer et al., 2016 | N/A |
| Software and Algorithms | ||
| FlowJo v10.3.5 | https://www.flowjo.com/ | RRID: SCR_008520 |
| Graphpad prism version 7 | https://www.graphpad.com/scientific-software/prism/ | RRID: SCR_002798 |
| Adobe Illustrator CC 2018 | https://www.adobe.com/uk/products/illustrator.html | RRID: SCR_014198 |
| IgBlast tool (NCBI) | https://www.ncbi.nlm.nih.gov/igblast/ | RRID: SCR_002873 |
| DataAssist 3.0 | https://www.thermofisher.com/us/en/home/technical-resources/software-downloads/dataassist-software.html | RRID: SCR_014969 |
| kallisto v0.43.0 | https://pachterlab.github.io/kallisto/about | RRID: SCR_016582 |
| R package tximport | https://github.com/mikelove/tximport | RRID: SCR_016752 |
| DESeq2 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html | RRID: SCR_015687 |
Gene expression analysis.
Three independent biological replicates each consisting of pools of 11–12 mice were sorted at each time point for the experiment shown in Fig. 1H or 33–36 mice for each anti-DEC-CS/OVA titration group. Sorted cells were processed by extracting RNA using PicoPure RNA Isolation Kit (Thermo Fisher) according to manufacturer’s recommendations. Two of the 15 sorted samples used in the analysis shown in Fig. 2H produced RNA of poor quality and were not included in the analysis. 1 ng of total RNA was used to generate full length cDNA using SMART-Seq v4 Ultra Low Input RNA Kit (Clontech). 1 ng of cDNA was then used to prepare libraries using Nextera XT DNA sample preparation kit (Illumina). Libraries with unique barcodes were pooled at equal molar ratios and sequenced on Illumina NextSeq 500 sequencer to generate 75 bp single reads, following the manufacturer’s protocol (Cat# 15048776 Rev.E). Transcript abundance was quantified using kallisto v0.43.0 (Bray et al., 2016), and subsequently summarized to the gene-amount using the R package tximport (Soneson et al., 2015). Differential gene expression analysis was performed using DESeq2 (Love et al., 2014). Hierarchical clustering was done based on combined data from three experimental repeats. Hierarchical clustering done based on data from the individual repeats gave similar results.
qPCR.
RNA was extracted with RNeasy Plus Mini Kit (Qiagen) or using the SMART2-seq protocol modified for population, as previously described (Haber et al., 2017). RNA was reverse-transcribed using SuperScript III First-Strand Synthesis kit (Invitrogen) or Maxima H-minus RT (Thermo Fisher). Quantitative PCR was performed using the QuantStudio Flex 6 Real-Time PCR System (Applied BioSystems). Samples were run in triplicate using TaqMan assay for Myc and with at list two of the following TaqMan assays as reference genes: Hprt, Ubc, Actb and Gapdh (all from Applied BioSystems; See Key Resource Table for details). Results were analyzed with DataAssist 3.0 software.
ImageStream cell size analysis.
Lymph node single-cell suspensions were treated for 10 minutes with 1 µg/mL anti-mouse CD16/CD32 (BD Pharmingen). Enrichment for GC B cells was done by staining with anti-mouse IgD–biotin and magnetic cell sorting (MACS) using Streptavidin and CD43 microbeads according to manufacturer’s instructions (Miltenyi Biotec). Cells were stained on ice for 45 minutes with biotinylated anti-CXCR4 antibody and were then washed and stained with anti-CD86-PE, anti-Streptavidin-BV421 and anti-B220-APC and with the following reagents for live/dead and dump exclusion: Zombie-NIR, anti-CD4, anti-CD8, anti-F4/80, anti-NK1.1, anti-Gr1 (all conjugated to APC-eFluor780; See Key Resource Table for antibody details). Samples were analyzed on ImageStreamX equipped with 4 lasers using the following power set-up: 405 nm (90 mW), 488 nm (200 mW), 561 nm (200 mW) and 658 nm (120 mW). 60x magnification was used for image acquisition with pixel size of 0.3 × 0.3 μm and field of view 40 × 170 μm. Data were analyzed by IDEAS 6.2 software using the area feature for size estimation. Scale bar = 7 μm.
STATISTICAL ANALYSIS
The number of mice per group, number of replicates and the nature of error bars are indicated in the legend of each figure. Center bars always indicate mean. Statistical significance was determined with Graphpad Prism Version 7.0 using the tests indicated in each figure legend. Data were considered statistically significant at *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 and ****p ≤ 0.0001.
DATA AND SOFTWARE AVAILABILITY
Data Resources
Raw and processed data files for the RNA sequencing analysis have been deposited in the NCBI Gene Expression Omnibus under accession number GSE132881.
Supplementary Material
Highlights:
MYC expression in light zone B cells is proportional to the magnitude of T cell help
MYC proportionally regulates the size of light zone germinal center B cells
MYC amounts dictate division capacity and residence time in the dark zone
ACKNOWLEDGMENTS:
We thank A. Miyawaki, T. Kurosaki, J.M. Sedivy and F.J. Gonzalez for mice, T. Eisenreich for help with mouse colony management, K. Yao for technical help, A. Gazumyan and D. Yost for protein production, A. Gitlin and M. Biton for advice, B. Zhang and C. Zhao at The Rockefeller University Genomics Resource Center for assistance with high-throughput sequencing, K. Gordon for assistance with cell sorting, S. Semova and S. Mazel at The Rockefeller University Flow Cytometry Resource Center for assistance with ImageStream analysis and all members of the Nussenzweig laboratory for discussion. This work was supported by NIH grant 5R37 AI037526–24 and UM1-AI100663 to M.C.N. H.H. is a Cancer Research Institute Irvington fellow. E.E.K. is a National Health and Medical Research Council C.J. Martin Overseas Biomedical fellow. M.C.N. is an HHMI investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare no competing interests.
References
- Adams JM, Gerondakis S, Webb E, Corcoran LM, and Cory S (1983). Cellular myc oncogene is altered by chromosome translocation to an immunoglobulin locus in murine plasmacytomas and is rearranged similarly in human Burkitt lymphomas. Proc Natl Acad Sci U S A 80, 1982–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiba Y, Kometani K, Hamadate M, Moriyama S, Sakaue-Sawano A, Tomura M, Luche H, Fehling HJ, Casellas R, Kanagawa O, et al. (2010). Preferential localization of IgG memory B cells adjacent to contracted germinal centers. Proc Natl Acad Sci U S A 107, 12192–12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen CD, Ansel KM, Low C, Lesley R, Tamamura H, Fujii N, and Cyster JG (2004). Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat Immunol 5, 943–952. [DOI] [PubMed] [Google Scholar]
- Allen CD, Okada T, Tang HL, and Cyster JG (2007). Imaging of germinal center selection events during affinity maturation. Science 315, 528–531. [DOI] [PubMed] [Google Scholar]
- Allen D, Simon T, Sablitzky F, Rajewsky K, and Cumano A (1988). Antibody engineering for the analysis of affinity maturation of an anti-hapten response. EMBO J 7, 1995–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannard O, and Cyster JG (2017). Germinal centers: programmed for affinity maturation and antibody diversification. Curr Opin Immunol 45, 21–30. [DOI] [PubMed] [Google Scholar]
- Boscardin SB, Hafalla JC, Masilamani RF, Kamphorst AO, Zebroski HA, Rai U, Morrot A, Zavala F, Steinman RM, Nussenzweig RS, and Nussenzweig MC (2006). Antigen targeting to dendritic cells elicits long-lived T cell help for antibody responses. J Exp Med 203, 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Bretones G, Delgado MD, and Leon J (2015). Myc and cell cycle control. Biochim Biophys Acta 1849, 506–516. [DOI] [PubMed] [Google Scholar]
- Calado DP, Sasaki Y, Godinho SA, Pellerin A, Kochert K, Sleckman BP, de Alboran IM, Janz M, Rodig S, and Rajewsky K (2012). The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat Immunol 13, 1092–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C, Verbaro DJ, Tonc E, Holmgren M, Cella M, Colonna M, Bhattacharya D, and Egawa T (2016). The Transcription Factor AP4 Mediates Resolution of Chronic Viral Infection through Amplification of Germinal Center B Cell Responses. Immunity 45, 570–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews S, Barth R, Hood L, Prehn J, and Calame K (1982). Mouse c-myc oncogene is located on chromosome 15 and translocated to chromosome 12 in plasmacytomas. Science 218, 1319–1321. [DOI] [PubMed] [Google Scholar]
- Crotty S (2011). Follicular helper CD4 T cells (TFH). Annu Rev Immunol 29, 621–663. [DOI] [PubMed] [Google Scholar]
- Crotty S (2014). T follicular helper cell differentiation, function, and roles in disease. Immunity 41, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, and Croce CM (1982). Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci U S A 79, 7824–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV (2012). MYC on the path to cancer. Cell 149, 22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alboran IM, O’Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, and Alt FW (2001). Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14, 45–55. [DOI] [PubMed] [Google Scholar]
- Depoil D, Zaru R, Guiraud M, Chauveau A, Harriague J, Bismuth G, Utzny C, Muller S, and Valitutti S (2005). Immunological synapses are versatile structures enabling selective T cell polarization. Immunity 22, 185–194. [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Kung J, Holmes AB, Wells VA, Mo T, Basso K, and Dalla-Favera R (2015). The FOXO1 Transcription Factor Instructs the Germinal Center Dark Zone Program. Immunity 43, 1064–1074. [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Victora GD, Ying CY, Phan RT, Saito M, Nussenzweig MC, and Dalla-Favera R (2012). The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat Immunol 13, 1083–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D, Rosains J, Birkhoff G, and Eggan K (2007). Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature 447, 679–685. [DOI] [PubMed] [Google Scholar]
- Ersching J, Efeyan A, Mesin L, Jacobsen JT, Pasqual G, Grabiner BC, Dominguez-Sola D, Sabatini DM, and Victora GD (2017). Germinal Center Selection and Affinity Maturation Require Dynamic Regulation of mTORC1 Kinase. Immunity 46, 1045–1058 e1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Akasako-Furukawa A, Shirai H, Nakamura H, and Azuma T (1999). Junctional amino acids determine the maturation pathway of an antibody. Immunity 11, 329–338. [DOI] [PubMed] [Google Scholar]
- Gitlin AD, Mayer CT, Oliveira TY, Shulman Z, Jones MJ, Koren A, and Nussenzweig MC (2015). HUMORAL IMMUNITY. T cell help controls the speed of the cell cycle in germinal center B cells. Science 349, 643–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin AD, Shulman Z, and Nussenzweig MC (2014). Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature 509, 637–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Gong S, Maric S, Misulovin Z, Pack M, Mahnke K, Nussenzweig MC, and Steinman RM (2000). A monoclonal antibody to the DEC-205 endocytosis receptor on human dendritic cells. Hum Immunol 61, 729–738. [DOI] [PubMed] [Google Scholar]
- Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, et al. (2017). A single-cell survey of the small intestinal epithelium. Nature 551, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser AE, Junt T, Mempel TR, Sneddon MW, Kleinstein SH, Henrickson SE, von Andrian UH, Shlomchik MJ, and Haberman AM (2007). Definition of germinal-center B cell migration in vivo reveals predominant intrazonal circulation patterns. Immunity 26, 655–667. [DOI] [PubMed] [Google Scholar]
- Heinzel S, Binh Giang T, Kan A, Marchingo JM, Lye BK, Corcoran LM, and Hodgkin PD (2017). A Myc-dependent division timer complements a cell-death timer to regulate T cell and B cell responses. Nat Immunol 18, 96–103. [DOI] [PubMed] [Google Scholar]
- Inoue T, Shinnakasu R, Ise W, Kawai C, Egawa T, and Kurosaki T (2017). The transcription factor Foxo1 controls germinal center B cell proliferation in response to T cell help. J Exp Med 214, 1181–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iritani BM, and Eisenman RN (1999). c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci U S A 96, 13180–13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder P, Battey J, Lenoir G, Moulding C, Murphy W, Potter H, Stewart T, and Taub R (1983). Translocations among antibody genes in human cancer. Science 222, 765–771. [DOI] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, and Tamayo P (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AC (2013). The regulation of cell size. Cell 154, 1194–1205. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Weisel F, and Shlomchik MJ (2018). B Cell Receptor and CD40 Signaling Are Rewired for Synergistic Induction of the c-Myc Transcription Factor in Germinal Center B Cells. Immunity 48, 313–326 e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer CT, Gazumyan A, Kara EE, Gitlin AD, Golijanin J, Viant C, Pai J, Oliveira TY, Wang Q, Escolano A, et al. (2017). The microanatomic segregation of selection by apoptosis in the germinal center. Science 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprea M, and Perelson AS (1997). Somatic mutation leads to efficient affinity maturation when centrocytes recycle back to centroblasts. J Immunol 158, 5155–5162. [PubMed] [Google Scholar]
- Pasqual G, Angelini A, and Victora GD (2015). Triggering positive selection of germinal center B cells by antigen targeting to DEC-205. Methods Mol Biol 1291, 125–134. [DOI] [PubMed] [Google Scholar]
- Preston GC, Sinclair LV, Kaskar A, Hukelmann JL, Navarro MN, Ferrero I, MacDonald HR, Cowling VH, and Cantrell DA (2015). Single cell tuning of Myc expression by antigen receptor signal strength and interleukin-2 in T lymphocytes. EMBO J 34, 2008–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky K (1996). Clonal selection and learning in the antibody system. Nature 381, 751–758. [DOI] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, and Brown EJ (2007). Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1, 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. (2008). Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498. [DOI] [PubMed] [Google Scholar]
- Sander S, Calado DP, Srinivasan L, Kochert K, Zhang B, Rosolowski M, Rodig SJ, Holzmann K, Stilgenbauer S, Siebert R, et al. (2012). Synergy between PI3K signaling and MYC in Burkitt lymphomagenesis. Cancer Cell 22, 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander S, Chu VT, Yasuda T, Franklin A, Graf R, Calado DP, Li S, Imami K, Selbach M, Di Virgilio M, et al. (2015). PI3 Kinase and FOXO1 Transcription Factor Activity Differentially Control B Cells in the Germinal Center Light and Dark Zones. Immunity 43, 1075–1086. [DOI] [PubMed] [Google Scholar]
- Shih TA, Roederer M, and Nussenzweig MC (2002). Role of antigen receptor affinity in T cell-independent antibody responses in vivo. Nat Immunol 3, 399–406. [DOI] [PubMed] [Google Scholar]
- Shulman Z, Gitlin AD, Weinstein JS, Lainez B, Esplugues E, Flavell RA, Craft JE, and Nussenzweig MC (2014). Dynamic signaling by T follicular helper cells during germinal center B cell selection. Science 345, 1058–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love MI, and Robinson MD (2015). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart I, Radtke D, Phillips B, McGowan SJ, and Bannard O (2018). Germinal Center B Cells Replace Their Antigen Receptors in Dark Zones and Fail Light Zone Entry when Immunoglobulin Gene Mutations are Damaging. Immunity 49, 477–489 e477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, and Fuchs E (2004). Defining the epithelial stem cell niche in skin. Science 303, 359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, and Jacks T (2007). Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665. [DOI] [PubMed] [Google Scholar]
- Victora GD, Dominguez-Sola D, Holmes AB, Deroubaix S, Dalla-Favera R, and Nussenzweig MC (2012). Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood 120, 2240–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, and Nussenzweig MC (2012). Germinal centers. Annu Rev Immunol 30, 429–457. [DOI] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, and Nussenzweig MC (2010). Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell 143, 592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa CG, and Cyster JG (2011). How T cells earn the follicular rite of passage. Immunity 35, 671–680. [DOI] [PubMed] [Google Scholar]
- Vinuesa CG, Linterman MA, Yu D, and MacLennan IC (2016). Follicular Helper T Cells. Annu Rev Immunol 34, 335–368. [DOI] [PubMed] [Google Scholar]
- Vinuesa CG, Tangye SG, Moser B, and Mackay CR (2005). Follicular B helper T cells in antibody responses and autoimmunity. Nat Rev Immunol 5, 853–865. [DOI] [PubMed] [Google Scholar]
- von Boehmer L, Liu C, Ackerman S, Gitlin AD, Wang Q, Gazumyan A, and Nussenzweig MC (2016). Sequencing and cloning of antigen-specific antibodies from mouse memory B cells. Nat Protoc 11, 1908–1923. [DOI] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, and Green DR (2011). The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner SM, Jones JM, Hasz DE, and Largaespada DA (2005). Repressible transgenic model of NRAS oncogene-driven mast cell disease in the mouse. Blood 106, 1054–1062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data Resources
Raw and processed data files for the RNA sequencing analysis have been deposited in the NCBI Gene Expression Omnibus under accession number GSE132881.