Abstract
Purpose:
Cancer care professionals are confronted with interpreting results from multiplexed gene sequencing of patients at hereditary risk for cancer. Assessments for variant classification now require orthogonal data searches and aggregation of multiple lines of evidence from diverse resources. The clinical genetics community needs a fast algorithm that automates ACMG based variant classification and provides uniform results.
Methods:
Pathogenicity of Mutation Analyzer (PathoMAN) automates germline genomic variant curation from clinical sequencing based on ACMG guidelines. PathoMAN aggregates multiple tracks of genomic, protein and disease specific information from public sources. We compared expertly curated variant data from clinical laboratories to assess performance.
Results:
PathoMAN achieved a high overall concordance of 94.4% for pathogenic and 81.1% for benign variants. We observed negligible discordance (0.3% pathogenic, 0% benign) when contrasted against expert curated variants. Some loss of resolution (5.3% pathogenic, 18.9% benign) and gain of resolution (1.6% pathogenic, 3.8% benign) was also observed.
Conclusion:
Automation of variant curation enables unbiased, fast, efficient delivery of results in both clinical and laboratory research. We highlight the advantages and weaknesses related to the programmable automation of variant classification. PathoMAN will aid in rapid variant classification by generating robust models using a knowledge-base of diverse genetic data. https://pathoman.mskcc.org
Keywords: Pathogenicity, ACMG, germline, cancer, curation
INTRODUCTION
Genetic testing and targeted resequencing of cancer susceptibility genes to facilitate precision cancer prevention and early diagnosis, has grown exponentially because of the decreasing costs of next generation sequencing (NGS)1,2. A major challenge in clinical sequencing is interpreting sequence variants. The American College of Medical Genetics and Genomics (ACMG) and the Association of Molecular Pathology (AMP) have published guidelines on interpretation of germline variants, considering both their pathogenicity and clinical actionability3. Yet, germline variant classification continues to pose an immense burden on the time and resources of diagnostic molecular laboratories and cancer care professionals. The ACMG classification schema requires manually exploring multiple lines of public data, other orthogonal data sources and literature; then aggregation and scoring to provide evidence for classifying variants4. Currently, there is no widely available automated computational framework for classifying genetic variants based on ACMG criteria. We developed PathoMAN, a computational resource that automates germline variant classification with uniformity, transparency and speed, to facilitate variant curation for the cancer genetics community.
PathoMAN’s variant curation algorithm classifies germline genomic variants. The schema is inspired by ACMG/AMP classification3. It aggregates multiple tracks of genetic and molecular evidences using variant annotators and from public repositories containing evidence necessary for pathogenicity assertion. The compiled data is then used in 28 distinct categories which are grouped as: variant type, biological impact, in silico predictions, presence in the control cohort, familial information, and inheritance mode. The aggregate score resulting from evaluation of these categories is used in generating the assertion for a variant as Pathogenic (P), Likely-Pathogenic (LP), Benign (B), Likely-Benign (LB) or Variant of Uncertain Significance (VUS).
PathoMAN’s performance was measured by re-evaluating expertly curated germline cancer variants from three clinical testing laboratories – Ambry, Invitae and GeneDx. We selected the commonly tested cancer susceptible genes in multiplex panels, many of which are in the ACMG recommended gene list. We also tested the algorithm on reported P/LP variants from four published cancer studies on non-ACMG heritable cancer risk and putative risk genes. In this study, we also assessed the frequency of clinically actionable variants present in general population using ExAC (noTCGA) data in cancer susceptibility and predisposition genes. We tested the application of ACMG criteria for germline cancer variants and addressed the bottleneck of variant curation in using automated algorithms in variant classification.
MATERIALS AND METHODS
Test datasets
To test the performance of PathoMAN against manual curation, we selected variants that were commonly reported by CLIA certified clinical testing laboratories - Ambry, Invitae and GeneDx (3,513 variants in 27 genes) with identical assertions in ClinVar (version Sept 2018). Many variants in this test dataset have also been reported by multiple other submitters in ClinVar. The Invitae dataset consisted of 1,494 B/LB, 608 P/LP and 1,412 VUS variants, the Ambry dataset 1,517 B/LB, 646 P/LP and 1,351 VUS variants and the GeneDx dataset 1,512 B/LB, 633 P/LP and 1,369 VUS variants (Table S1). We used these datasets to test the assertions of pathogenic and benign classifications by ACMG.
We also tested PathoMAN on 300 P/LP variants in 55 genes from four published reports on multiple cancers5–8, where ACMG criteria and primary assertions were available. We chose these datasets to test lesser known cancer predisposition genes that are not part of the ACMG list.
ACMG/AMP guidelines
The variant classification criteria used by PathoMAN utilizes ACMG/AMP guidelines3. The ACMG/AMP guidelines constitute 16 criteria that aid in classifying pathogenicity and 12 criteria that aid for classifying benignity. This classification system resolves a variant as pathogenic or benign based on eight major components – population frequency data, genomic annotation and computational predictive data, functional data, segregation data, de novo data, allelic/genotypic data, public databases and literature and other data (Table 1). The detailed usage is described in the Determination of ACMG criteria for variant classification section of the supplemental materials.
Table 1:
Utilization of ACMG criteria and knowledgebase in PathoMAN variant curation
| Category | ACMG criteria | Utilization of ACMG criteria in PathoMAN’s variant classification |
|---|---|---|
| literature and other sources | PVS1 | Tier 1 variants (Frameshift, truncating, essential splice and initiation codon) in curated list of OG and TSG and not in last exon |
| public databases | Missense variant (hgvs protein change) in ClinVar with review status >= 2 irrespective of the genomic alternative allele | |
| de novo data | PS2 | User Input |
| public databases; functional data | PS3 | Variant in list of pathogenic variants knowledgebase aggregated from literature with functional evidence, public database with loss of function reports, Tier 1 variants in ClinVar reported with review status >=2 and missense variants reported by ENIGMA |
| population frequency data | PS4 | Fisher’s case-control test odds ratio > 3 and Pval < 0.05 against ExAC-noTCGA and gnomAD population of interest (Applies to variants with BA1, BS1, BS2 and PM2 equal to 0) |
| public databases | PM1 | Amino acid residue in protein’s domain or residue for signalling or protein-protein interaction or in active site from Uniprot |
| population frequency data | PM2 | Variant absent from ExAC-noTCGA or gnomAD |
| allelic/genotypic data | PM3 | Not used in the current version |
| public databases | PM4 | Inframe ins/del or stop loss in a non-repetitive region from UCSC genome browser |
| public databases | PM5 | Missense variant in ClinVar with review status >=2 irrespective of the alternative amino acid change at the same position as that of the reported pathogenic variant in clinvar |
| de novo data | PM6 | User Input |
| segregation data | PP1 | User Input |
| public databases | PP2 | Variant in gene with significant pathogenic missense burden in ClinVar |
| genomic annotation and computational predictive data | PP3 | in-silico predictors agree on pathogenicity or deleteriousness of the variant |
| Other (disease specific) | PP4 | Not used in the current version |
| public databases | PP5 | Variant in ClinVar with review status <2 and pathogenic without conflicts |
| population frequency data | BA1 | Variant seen in ExAC-noTCGA or gnomAD with AF > 5% |
| population frequency data | BS1 | Variant seen in ExAC-noTCGA or gnomAD with AF between 1%–5% |
| population frequency data | BS2 | Variant seen in ExAC-noTCGA or gnomAD general population in homozygous form |
| public databases; functional data | BS3 | Variant in list of benign variants knowledgebase aggregated from literature with functional evidence, public database with loss of function reports, Tier 1 variants in ClinVar reported with review status >=2 and missense variants reported by ENIGMA |
| de novo data | BS4 | User Input |
| public databases | BP1 | Variant in gene with significant benign missense burden in ClinVar |
| allelic/genotypic data | BP2 | Not used in the current version |
| public databases | BP3 | Inframe ins/del or stop loss in a repetitive region from UCSC genome browser |
| genomic annotation and computational predictive data | BP4 | in-silico predictors agree on benignity or tolerance of the variant |
| Other (disease specific) | BP5 | Not used in the current version |
| public databases | BP6 | Variant in ClinVar with review status <2 and benign without conflicts |
| genomic annotation and computational predictive data | BP7 | synonymous variant with dbscSNV adaptive boosting and random forest score < 0.6 |
The results of PathoMAN were compared against the reported clinical assertion. We report them here in four categories: concordance, discordance, loss of resolution (LOR) and gain of resolution (GOR). When the reported P/LP and B/LB variants are re-classified as P/LP and B/LB respectively by PathoMAN, then the results are considered concordant. Similarly, when reported P/LP and B/LB variants are re-classified as B/LB and P/LP by PathoMAN respectively, then the variants are considered discordant. When reported P/LP or B/LB variants are re-classified as VUS by PathoMAN, then they are placed in the LOR category as PathoMAN cannot definitively classify these variants as either pathogenic or benign, thus losing resolution. Similarly, when the reported VUS are re-classified as P/LP or B/LB, they are considered as GOR as PathoMAN can resolve these variants as pathogenic or benign (Table S2). These evaluations aid in understanding the usage of the eight ACMG categories of evidence in the context of cancer genetics, and in their ability to differentiate between P/LP, B/LB and VUS.
ExAC subset of cancer predisposition genes
The Exome Aggregation Consortium9 (ExAC) is a joint effort to aggregate exome sequencing data from fourteen large sequencing projects to provide summary data such as ethnicity specific allele frequency for a wider scientific community. The ExAC-noTCGA data is a subset of 53,105 samples and it excludes The Cancer Genome Atlas (TCGA) cancer germline samples (n=7601). ExAC is often utilized as convenience controls for several cancers6,10,11 in case-control design. We wanted to estimate the burden of variants in ExAC-noTCGA as classified by PathoMAN and contrast against known information in ClinVar. We selected 55,566 variants from 76 known and putative cancer risk genes (Table S3) which were in exonic or essential splice site regions. This is not considered part of the test datasets described earlier, as ExAC data is used as part of the ACMG criteria PS4, PM2, BA1, BS1 and BS2.
Results
We developed PathoMAN, a germline variant classification algorithm, provided freely as a web-based service that allows user to either query single variants or batch upload a CSV file. Single variant query works on chromosome, position, reference allele, alternative allele, allele count, allele number, de novo status, co-segregation status and preferred control population sub-group. Batch upload requires six columns [chr, pos, ref, alt, ac, an] in a comma separated values (CSV) file. User can select de novo status, co-segregation status and preferred control population sub group. The program converts the CSV file to a minimal VCF4.2 file, annotates the minimal VCF and prepares it for PathoMAN variant classification. The result of a single variant query is displayed back on to the web-page immediately while the batch upload results will be e-mailed back to the submitter. For an annotated VCF file containing 100 variants, PathoMAN takes 6 minutes, which is 3.6 seconds per variant. This provides a massive advantage in terms of speed, uniformity, efficiency and service assurance compared to manual curation. In contrast, an expert reviewer may take 20–30 minutes to classify a novel variant.
PathoMAN versus manual curation for test datasets
To evaluate the performance of PathoMAN, we compared its results against clinical assertions from three clinical laboratories. The test dataset contained 3,513 variants with prior reported curation from the three clinical laboratories – Ambry Genetics, Invitae and GeneDx. Inter-lab agreement across the three laboratories was 84%. The 16% of the inter-lab disagreement variants were marked as VUS when comparing against PathoMAN results. Amongst 3,513 variants, missense variants accounted for 54.2%, synonymous variants for 24.7%, frameshift variants for 7.5%, stopgain variants for 4.7% and the rest distributed among splice variants, in-frame insertion/deletion and stop loss in the test dataset (Figure 1A, Table S4). Variants were annotated with CAVA, Annovar, ExAC noTCGA, gnomAD and ClinVar and ran through PathoMAN.
Figure 1.
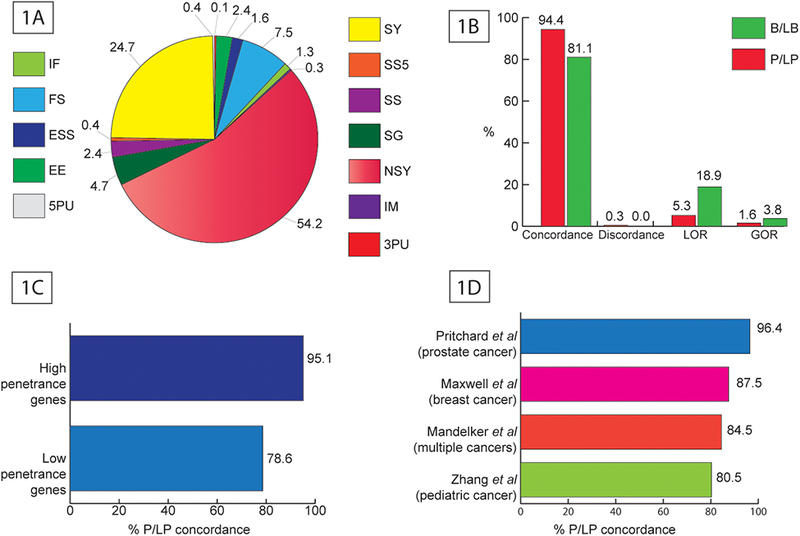
A: Distribution of 3513 variants in the test dataset by variant class (IF - Inframe insertion and/or deletion. It alters length but not frame of coding sequence, FS - Frameshifting insertion and/or deletion. It alters length and frame of coding sequence, ESS - Any variant that alters essential splice-site base (+1, +2, −1, −2), EE - Variant that alters the first or last three bases of an exon (i.e., the exon end), but not the frame of the coding sequence, 5PU - Any variant in 5′ untranslated region, SY - Synonymous variant. It does not alter amino acid or coding sequence length, SS5 - Any variant that alters +5 splice-site base but not an ESS base, SS - Any variant that alters splice-site base within the first eight intronic bases flanking exon (i.e., +8 to −8) but not an ESS or SS5 base, SG - Stop-gain (nonsense) variant caused by base substitution, NSY - Nonsynonymous variant. It alters amino acid(s) but not coding sequence length, IM - Variant that alters initiating methionine start codon, 3PU - Any variant in 3′ untranslated region). Figure 1B: Performance of PathoMAN’s variant classification against variant classification from three clinical laboratories – Ambry Genetics, Invitae and GeneDx. Figure 1C: Concordance of PathoMAN and clinical lab results for reported P/LP variants group by penetrance of the gene (1257 variants in 27 genes Supp table S5). Figure 1D: Concordance of PathoMAN and published reports for reported P/LP variants from four cancer studies (300 variants in 55 genes; Mandelker et al – 97 variants, Maxwell et al 40 variants, Pritchard et al – 56 variants and Zhang et al – 107 variants Supp table S7)
Upon comparing the clinical assertions from the three laboratories with PathoMAN’s results, the number of observed agreements was 90% (3,153 of 3,513 variants) and the Cohen’s Kappa coefficient for inter-rater agreement (к) was 0.83 (CI 95% 0.82–0.85).
PathoMAN achieved a concordance of 94.4% for P/LP and 81.1% for B/LB variants (Figure 1B). It showed 100% concordance for P/LP frameshift, splice sites and truncating variants. Similarly, there was 100% concordance for B/LB synonymous variants. There was a minimal discordance seen at 0.3% for P/LP variants (n=2). PathoMAN failed to resolve 22 P/LP missense variants and re-classified 2 P/LP variants as B/LB (a synonymous variant and an extended splice site variant) (Table 2). The synonymous PMS2 variant (c.825A>G p.Gln275Gln) was functionally shown to have aberrant splicing by experimental methods in literature. The MSH2 splice variant (c.942+3A>T) has been demonstrated to disrupt mRNA splicing and result in skipping of exon 5. PathoMAN assertions are made in real-time by the algorithm. In future versions of the program, we intend to incorporate a consensus splice prediction module and a literature-based evidence module, which would aid in correct classification of these edge case variants. No B/LB variants were re-classified as P/LP.
Table 2:
Distribution of PathoMAN re-classified of expertly curated germline cancer variants by variant class
| Variant Class | Description | Reported BLB | Reported PLP | Reported VUS | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| BLB | PLP | VUS | BLB | PLP | VUS | BLB | PLP | VUS | |||
| 3PU | Any variant in 3′ untranslated region | 0 | 0 | 3 | 0 | 0 | 3 | 0 | 0 | 7 | 13 |
| 5PU | Any variant in 5′ untranslated region | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 5 |
| EE | Variant that alters the first or last three bases of an exon (i.e., the exon end), but not the frame of the coding sequence | 6 | 0 | 13 | 0 | 17 | 0 | 2 | 2 | 46 | 86 |
| ESS | Any variant that alters essential splice-site base (+1, +2, −1, −2) | 0 | 0 | 0 | 0 | 51 | 0 | 1 | 3 | 2 | 57 |
| FS | Frameshifting insertion and/or deletion. It alters length and frame of coding sequence | 0 | 0 | 1 | 0 | 252 | 1 | 0 | 2 | 7 | 263 |
| IF | Inframe insertion and/or deletion. It alters length but not frame of coding sequence | 1 | 0 | 4 | 0 | 8 | 0 | 0 | 1 | 31 | 45 |
| IM | Variant that alters initiating methionine start codon | 0 | 0 | 0 | 0 | 3 | 2 | 0 | 0 | 4 | 9 |
| NSY | Nonsynonymous variant. It alters amino acid(s) but not coding sequence length | 169 | 0 | 211 | 0 | 71 | 22 | 19 | 17 | 1395 | 1904 |
| SG | Stop-gain (nonsense) variant caused by base substitution | 0 | 0 | 2 | 0 | 161 | 0 | 0 | 0 | 1 | 164 |
| SS | Any variant that alters splice-site base within the first eight intronic bases flanking exon (i.e., +8 to −8) but not an ESS or SS5 base | 17 | 0 | 3 | 1 | 4 | 3 | 1 | 0 | 56 | 85 |
| SS5 | Any variant that alters +5 splice-site base but not an ESS base | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 11 | 15 |
| SY | Synonymous variant. It does not alter amino acid or coding sequence length | 826 | 0 | 0 | 1 | 0 | 0 | 40 | 0 | 0 | 867 |
| Total | 1020 | 0 | 238 | 2 | 568 | 32 | 63 | 26 | 1565 | 3513 | |
PathoMAN re-classified 5.3% (n=32) reported P/LP variants and 18.9% (n=238) reported B/LB variants as VUS. Fifty three percent of these P/LP variants (17/32) and 74.4% B/LB variants (177/238) were classified as VUS in ClinVar by at least one submitter previously. This suggests that, for these variants, a consensus assertion has not been achieved due to insufficient clinical information. Similarly, PathoMAN re-classified 1.6% (n=26) and 3.8% (n=63) of VUS as P/LP and B/LB respectively. 62% of these reported VUS variants (55/89) had at least one submitter classify them as P/LP or B/LB among the three laboratories and in ClinVar. One of the major sources of gain of resolution is achieved by PathoMAN’s use of the saturation mutagenesis experiments on BRCA112. Other reasons for re-classification are version changes in ClinVar and updates on the public allele frequencies in ExAC and gnomAD.
When grouped as high and low penetrance genes (Figure 1C, Table S5), we calculated the absolute difference between classifications and observed Cohen’s Kappa coefficient for inter-rater agreement (к) of 0.82 (CI 95% 0.805 – 0.843) and 0.84 (CI 95% 0.77–0.92) respectively.
When compared against 300 P/LP variants in putative cancer predisposition candidate genes from four published cancer studies, PathoMAN showed 96.4% concordance for Pritchard et al6 (prostate cancer), 87.5% concordance for Maxwell et al5 (breast cancer), 84.5% concordance for Mandelker et al7 (multiple cancer types) and 80.3% for Zhang et al8 (pediatric cancer) (Table 1D).
PathoMAN results for ExAC dataset
PathoMAN classified < 1% of the heterozygous genotypes in ExAC-noTCGA dataset (55,566 exonic and essential splice variants from 76 cancer risk genes) as P/LP. We tabulated pathogenic variant burden by genes and compared them against ClinVar (Table 3). PathoMAN calls similar number of P/LP variants as reported in ClinVar for the high-risk cancer genes like BRCA1 and BRCA2. PathoMAN also predicts a few novel P/LP variants unreported in ClinVar. Investigators who intend to use the ExAC-noTCGA dataset as controls in a gene burden test against sequenced cancer cases, can use PathoMAN to get a list of P/LP variants across genes.
Table 3:
Comparison of PathoMAN P/LP burden vs ClinVar P/LP burden for ExAC (noTCGA) variants by cancer susceptibility genes
| GENE | ClinVar_PLP | PathoMAN_PLP | DIFF |
|---|---|---|---|
| BLM | 1 | 9 | 8 |
| TP53 | 15 | 7 | 8 |
| ATM | 80 | 74 | 6 |
| BRCA2 | 101 | 96 | 5 |
| BARD1 | 10 | 15 | 5 |
| SDHA | 5 | 0 | 5 |
| MLH1 | 6 | 11 | 5 |
| EGFR | 0 | 5 | 5 |
| RAD51B | 0 | 5 | 5 |
| PALB2 | 21 | 26 | 5 |
| RAD50 | 21 | 17 | 4 |
| PMS2 | 15 | 11 | 4 |
| CDH1 | 3 | 7 | 4 |
| MRE11A | 11 | 7 | 4 |
| EPCAM | 0 | 4 | 4 |
| BRCA1 | 68 | 65 | 3 |
| NF1 | 3 | 6 | 3 |
| PTEN | 2 | 5 | 3 |
| STK11 | 0 | 3 | 3 |
| KRAS | 2 | 0 | 2 |
| MUTYH | 26 | 24 | 2 |
| BRIP1 | 21 | 23 | 2 |
| RAD51C | 17 | 15 | 2 |
| FH | 6 | 4 | 2 |
| RET | 2 | 4 | 2 |
| BMPR1A | 1 | 3 | 2 |
| FAM175A | 1 | 0 | 1 |
| RAD51 | 1 | 0 | 1 |
| MSH6 | 13 | 12 | 1 |
| NBN | 10 | 9 | 1 |
| RAD51D | 6 | 7 | 1 |
| APC | 4 | 5 | 1 |
| CDKN2A | 5 | 4 | 1 |
| DICER1 | 3 | 2 | 1 |
| BAP1 | 0 | 1 | 1 |
Columns contain variant counts.
Usage of ACMG/AMP categories in PathoMAN
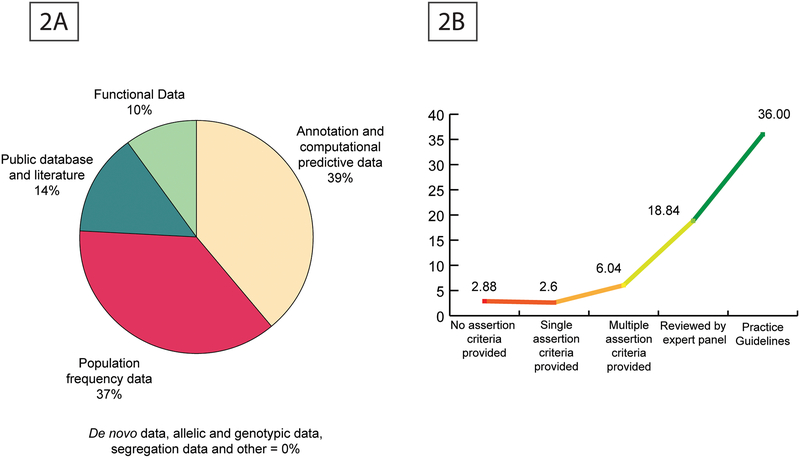
We analyzed the usability and frequency of use for the eight categories of evidence (population frequency, genomic annotation and computational prediction, functional evidence, co-segregation, de novo status, allelic/genotypic data, public databases, scientific literature and other data) described in the ACMG/AMP guidelines. Interestingly, we find that the categories: population frequency data, genomic annotation and computational predictions, databases and scientific literature (Figure 2A) are the most used. These are available due to generous data and tool-kit sharing policies in the genomics field. The categories that are rarely if ever used are familial co-segregation data or de novo status, allelic data, and functional data. We have also used ClinVar’s review status to upweight functional evidence in the current version, as we believe that the review status directly corresponds to the literature evidence reported for a variant (Figure 2B). The co-segregation data and de novo status data are limited to familial studies and are mostly unavailable in sporadic case- control settings since these are collected by investigators based on patient input. It was clear from ClinVar that, for a variant to be classified as pathogenic or likely pathogenic by ACMG criteria, one needs a maximum of 1 PVS1 or 2 PSs or 3 PMs or 4 PPs for which, the knowledge-base and resources used by PathoMAN were demonstrably sufficient. We describe below the bottlenecks in sharing this information and propose a novel framework to circumvent and ameliorate these issues.
Figure 2.
A: Utilization of knowledgebase components by PathoMAN during variant curation of test datasets. Figure 2B: Ratio of number of articles per variant across different review status reported in ClinVar.
Discussion
PathoMAN as a tool to aid variant curation
Traditionally, genetic variant curation has been performed manually by expert groups of individuals. However, this is a time intensive task that requires aggregation and interpretation of information from multiple sources. In the cancer realm, this was relatively easy when a single gene i.e BRCA1/2 was under investigation. In contemporary testing scenarios which routinely rely on multiplex gene-panels, this task is onerous. Large gene discovery efforts, as well as clinical reporting, could use a simplified, automated, method for prioritizing variants for a closer look or in the best case, be useful as the classification tool of choice. PathoMAN addresses this critical unmet need for an unbiased algorithmic approach towards classifying genetic variants of clinical interest in cancer predisposition. PathoMAN can be easily accessed through a web browser and results for individual variants are almost immediately available, while results of a batch query may take a few minutes.
Genetic testing laboratories are increasingly utilizing ACMG/AMP classification rules to classify variants for pathogenicity within cancer predisposition genes. However, results vary depending on availability of accessible data and interpretational differences13. Concordance between CLIA certified laboratories varied between 37 to 71% pre and post consultative processes using the ACMG guidelines. We observed 84% inter-laboratory agreement across all three clinical laboratories. Efforts are being made to narrow interpretational differences through initiatives underway such as ClinGen. In a recent report14, 13% of variants in ClinVar were re-analyzed, and were found to be unresolved, underscoring the difficulties even for expert curator groups. For manual or automated curation, the minimal set of information required to classify a variant as likely pathogenic or likely benign are: population frequency, in-silico predictors and prior reported evidence of pathogenicity from public databases. PathoMAN compiles this information uniformly in a machine accessible format which is used as a knowledge-base for variant classification. An additional advantage of using PathoMAN is that it can effortlessly identify benign variants based on public allele frequency and the genomic context information. In a typical multiplexed gene-panel variant list, after filtering for only rare high or moderate impact variants, PathoMAN will classify about one third of the variants as B/LB with high precision. This saves time and effort for the variant curators and helps them to focus on curating the remaining potentially actionable variants. PathoMAN will also identify known founder variants.
Cancer is a complex disease with multi-gene etiology. Some cancer genes confer high risk whereas some only moderately affect the carrier’s risk. Panel testing is currently used for active surveillance and intervention to lower disease risk. Large sequencing and genotyping efforts to discover new cancer predisposition genes are being carried out by several consortia like BCAC15, SIMPLEXO16; COMPLEXO17, CIMBA18, etc. As the cost for sequencing decreases, the number of genes tested is increasing. Automation allows for rapid processing, service assurance and reproducibility of results. Gold standard sets of curation pioneered by ClinGen19,20 would aid in refining these pathogenicity classifications further, while efforts such as the PROMPT21,22 registry enable accurate penetrance estimates of variants in susceptibility genes. The PROMPT registry has identified a 26% discordance rate among clinical laboratories and an 11% rate with conflicting interpretations, a discrepancy that has implications for altering medical management.
Many laboratories and certain programs such as CardioClassifier23 and InterVar24 use prior knowledge of disease-gene pair association. This is advantageous to reduce misclassifications leading in those genes that are not in a disease-gene pair. However, it also suffers from the disadvantage that it cannot be used for lesser known genes- disease pairs or for novel gene hunting. In a recent report7, we showed that, half of the cases, in a series consisting of selected advanced cancers at a single institution, were non-syndromic associations7. Probands or their close relatives had clinically actionable variants in cancer genes not directly associated with the specific cancers for which there were known syndromic associations. PathoMAN currently does not use the contextual syndromic association in deciphering pathogenicity of variants. This is a distinct advantage when searching for novel genetic association. However, in clinical sequencing, we acknowledge that limiting to known disease-gene pairs to identify pathogenic variants reduces false positives. In future versions, we hope to incorporate both clinical and gene discovery modes.
The variants that are manually curated as P/LP or B/LB, however called as VUS by PathoMAN are grouped under loss of resolution (LOR) category. This loss of resolution due to lack of accessible supporting evidence could be due to several reasons - inability to programmatically parse inline texts from public databases, availability of updated proprietary databases like HGMD and LOVD, unavailability of in-house functional evidence25 or familial co-segregation information26, etc.
The upgrade for VUS to either LP or LB by PathoMAN is based on the three categories - lines of available evidence in public databases, population frequency and computational and in silico prediction on deleteriousness. These variants can be re-classified as either pathogenic or benign if additional functional or co-segregation data became available to the user.
Commercial testing laboratories have proprietary versions of interpretation pipelines such as Sherloc27 (Invitae Corporation) and MyVISION (Myriad Genetics). However, these are unavailable to the community at large. PathoMAN is designed to provide an optimized platform for clinical variant calling utilizing publically available data resources.
Using ACMG for variant classification in Cancer
Variants in tumor suppressors and oncogenes lead to tumorigenesis, and the Knudson two-hit hypothesis28 is seen to operate in many common cancers. Common examples include APC, TP53, BRCA1/2 genes etc. However, several of these genes, especially those that are part of the Fanconi complex (FANCS-BRCA1, FANCD1-BRCA2, FANCJ-BRIP1, FANCN-PALB2, FANCP-SLX4, RAD51C), neurofibromatosis (NF1), Ataxia-telangiectasia (ATM), Bloom syndrome (BLM), Nijmegen breakage syndrome (NBN), dyskeratosis congenita (TERT) that lead to autosomal recessive rare Mendelian disorders, are also found to be risk genes for autosomal dominant cancer predisposition. Heterozygous carriers of these gene mutations are reported to have increased risks for syndromic cancers29. Occasionally, gene disrupting heterozygous variants in these genes that are rare, absent in public controls such as ExAC and gnomAD may be observed in sequenced cancer cohorts. Their ClinVar record for pathogenicity is usually based on their Mendelian recessive syndrome and not to the cancer phenotypes. Hence, applying the ACMG rules to genes without membership in the ACMG list may be fraught with misclassification. However, we believe that continuing data streams for variants in these genes will lead to better classifications, especially when coupled with familial co-segregation and functional validations. While PathoMAN classifications for such genes are a useful starting point for identifying variants that may be pathogenic and discarding benign; we emphasize on expert manual curation to disentangle these issues.
Limitations of automation
Automating variant classification based on publicly available information has some pitfalls. Supporting evidences provided in ClinVar for variants by submitters are not computation friendly and requires manual curation to interpret free text. In several instances, the citations are not relevant to the specific records. Technologies such as natural language processing and tagging will eventually help to build a knowledge-base that can further be used for deep learning.
ACMG guidelines does not account for functional evidence provided in ClinVar (supporting observations), which leads to loss of information that could otherwise be used in variant classification. Due to this lack of data structure (free text), the bonafide variants in ClinVar are being coded only PP5 or BP6 and not PS3 or BS3. We employed the review status 2 or more status as a proxy for functional evidence. Not all submitters are equipped or do independent analyses to assess functional evidences for their clinical assertion. If the ClinVar evidence is informatically coded, it would be helpful for molecular geneticists and clinical curators to use this information for their pathogenicity estimation. For example, TP53 (R273H), BRCA1 (Y105C) and BRCA1 (V1688del) variants have overwhelming literature evidences (Figure S1); however, the evidence present in the description of the submissions within ClinVar, are computationally un-derivable. Similarly, there are many variants reported in the literature which may have some level of supporting evidence for pathogenicity or benignity in ClinVar. Currently all these data integration is done by manual curators on a case-by-case basis.
We propose a framework to report ClinVar data that can be structured and parsable for an automated algorithm in the context of cancer. This format consists of 6 important fields that compress the vast information that is present in literature or clinical reports.
Population/Ethnicity (NFE, AFR, SAS, AMR, ASJ, FIN, OTH, EAS, others)
Inheritance model (AD, AR, de novo, X-linked)
Allelic status (Hom, Het)
Family history/Co-segregation information (Yes-1; No-0)
Disease association (TCGA code/OncoTree code30)
Functional Evidence (Experiment type: NMC, LOH, etc.)
For example, ERCC3 (R109X) variant31 can be depicted as ASJ-AD-Het-1:1-BRCA, BLCA-NMD. This variant was seen in Ashkenazi Jewish individuals with an autosomal dominant inheritance for the heterozygous allele. This variant co-segregated in one family with cancer history. The variant was found in breast cancer and bladder cancer individuals and the functional evidence for pathogenicity was carried out by testing for non-sense mediated decay and other experiments.
Large sequencing studies and gene specific functional studies give curated list of variants with their pathogenic impacts like TP53 database32 and a functional study on PALB2 variants33,34. As a pilot project, we have collected a list of PALB2, TP53 variants from these literature as supporting the knowledge-base for PathoMAN (PS3/BS3 functional evidence) but there is a real need to create a publicly available well curated list of variants from the literature that is amenable to programmatic interpretation. Similarly, as standards evolve for the incorporation of somatic mutations into germline interpretation, we expect an integration of such events for well-established tumor suppressor and oncogenes. The roles played by the ENIGMA consortium35,36, G4GH37, BRCA-Share38 in this regard are meritorious. Though clinical laboratories collaborate to resolve the differences in variant interpretations submitted to ClinVar14, the fact remains however, that a unified framework for incorporation of supporting machine readable evidences in any variant database including ClinVar remains a critical bottleneck.
Functional data is rarely available for most genes. Exceptions are BRCA1/2 due to the concerted efforts of the ENIGMA consortium35,36. In single variant reports, data is usually buried within scientific jargon that is not compatible with genomic variant information. In many instances, functional data is dependent on the models used, e.g. over expression of a mutant construct, deletion of a region using a CRISPR endonuclease and sometimes, introduction of the specific nucleotide through homology directed DNA repair. It is also likely, that the results from these three methods do not agree. Novel methods to understand deleteriousness using saturation mutagenesis are also starting to emerge39,40 for e.g., for BRCA112, we have incorporated the loss of function information into PathoMAN’s algorithm. We hope these will add a uniform layer of functional data that can be used in determining pathogenicity in the coming years.
In conclusion, we performed pathogenicity assessment of 59,379 variants in germline cancer risk genes, the first and largest uniform classification using an unbiased computational tool. We demonstrate the high concordance and low discordance when compared with manual curation as a harbinger of how such programs will soon be able to help domain experts and manual curators. PathoMAN is a first step towards our goal of automating the complex process of variant classification and interpretation. A beta version of the web app is available at https://pathoman.mskcc.org/
Supplementary Material
Acknowledgements
We thank Sabine Topka PhD, Semanti Mukherjee PhD, Maria Carlo MD and Zoe Steinsnyder BS for helpful suggestions to improve the manuscript.
Funding/Support: Research reported in this pre-print was supported by National Cancer Institute of the National Institutes of Health under award number R21CA029533, P50CA221745 and as well as Cycle for Survival, the Breast Cancer Research Foundation and The V Foundation for Cancer Research. It is also supported by the Cancer Center core grant P30CA008748 and The Robert and Kate Niehaus Center for Inherited Cancer Genomics. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or other funding agencies.
Footnotes
- Annovar - https://annovar.openbioinformatics.org
- ExACnoTCGA - http://exac.broadinstitute.org
- gnomAD - http://gnomad.broadinstitute.org/
- ClinVar - https://www.ncbi.nlm.nih.gov/clinvar/
- IARC database - http://p53.iarc.fr/
- ClinVar parser tool - https://github.com/macarthur-lab/clinvar
- dbNSfP and dbscSNV - https://sites.google.com/site/jpopgen/dbNSFP
- Gene List - https://github.com/macarthur-lab/gene_lists
- Repeat masker - http://www.repeatmasker.org/
- UCSC Genome Browser - https://genome.ucsc.edu
- CardioClassifier - https://www.cardioclassifier.org/
- InterVar - https://github.com/WGLab/InterVar
- ACMG - https://www.acmg.net/
- ClinVar Miner - https://clinvarminer.genetics.utah.edu/
- PathoMAN- http://pathoman.mskcc.org/
- Ambry - https://ambrygen.com/
- Invitae - https://www.invitae.com/en/
- GeneDx - https://www.genedx.com/
Conflicts of Interest
Yelena Kemel: The author was a past employee of Bioreference Laboratories, a subisidiary of OPKO Health, with employment ending in January 2016
Karen Cadoo: The author declares institutional support for therapeutic clinical trial from Astra Zeneca and Syndax Pharmaceuticals outside the submitted work.
Liying Zhang: The author declares that she received compensation from Future Technology Research LLC (seminar on precision medicine), Roche Diagnostics Asia Pacific, BGI, Illumina (speaking activities at conferences/workshop). The author’s family member has a leadership position and ownership interest of Shanghai Genome Center.
Zsofia Stadler: The author declares her immediate family member works at the department of Ophthalmology at MSKCC. She also holds consulting/advisory role with Allergan, Adverum Biotechnologies, Alimera Sciences, Biomarin, Fortress Biotech, Genentech, Novartis, Optos, Regeneron, Regenxbio, Spark Therapeutics
Mark Robson: The author declares grants, personal fees and non-financial support from AstraZeneca, personal fees from McKesson, grants and personal fees from Pfizer, non-financial support from Myriad, non-financial support from Invitae, grants from AbbVie, grants from Tesaro, grants from Medivation outside the submitted work.
Vijai Joseph and Kenneth Offit : The authors declares that they holds patent on the “Diagnosis and Treatment of ERCC3 mutant cancer” PCT/US18/22588.
Vignesh Ravichandran, Zarina Shameer, Diana Mandelkar, Steve Lipkin and Michael F Walsh have no conflicts to disclose.
References
- 1.Offit K Multigene Testing for Hereditary Cancer: When, Why, and How. J Natl Compr Canc Netw. 2017;15(5S):741–743. [DOI] [PubMed] [Google Scholar]
- 2.Desmond A, Kurian AW, Gabree M, et al. Clinical Actionability of Multigene Panel Testing for Hereditary Breast and Ovarian Cancer Risk Assessment. JAMA Oncol. 2015;1(7):943–951. doi: 10.1001/jamaoncol.2015.2690. [DOI] [PubMed] [Google Scholar]
- 3.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandey KR, Maden N, Poudel B, Pradhananga S, Sharma AK. The curation of genetic variants: difficulties and possible solutions. Genomics Proteomics Bioinformatics. 2012;10(6):317–325. doi: 10.1016/j.gpb.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maxwell KN, Hart SN, Vijai J, et al. Evaluation of ACMG-Guideline-Based Variant Classification of Cancer Susceptibility and Non-Cancer-Associated Genes in Families Affected by Breast Cancer. Am J Hum Genet. 2016;98(5):801–817. doi: 10.1016/j.ajhg.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med. 2016;375(5):443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandelker D, Zhang L, Kemel Y, et al. Mutation Detection in Patients With Advanced Cancer by Universal Sequencing of Cancer-Related Genes in Tumor and Normal DNA vs Guideline-Based Germline Testing. JAMA. 2017;318(9):825–835. doi: 10.1001/jama.2017.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Walsh MF, Wu G, et al. Germline Mutations in Predisposition Genes in Pediatric Cancer. N Engl J Med. 2015;373(24):2336–2346. doi: 10.1056/NEJMoa1508054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu C, Hart SN, Polley EC, et al. Association Between Inherited Germline Mutations in Cancer Predisposition Genes and Risk of Pancreatic Cancer. JAMA. 2018;319(23):2401–2409. doi: 10.1001/jama.2018.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson DR, Wu YM, Lonigro RJ, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548(7667):297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Findlay GM, Daza RM, Martin B, et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562(7726):217–222. doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amendola LM, Jarvik GP, Leo MC, et al. Performance of ACMG-AMP Variant-Interpretation Guidelines among Nine Laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet. 2016;99(1):247. doi: 10.1016/j.ajhg.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison SM, Dolinsky JS, Knight Johnson AE, et al. Clinical laboratories collaborate to resolve differences in variant interpretations submitted to ClinVar. Genet Med. 2017;19(10):1096–1104. doi: 10.1038/gim.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breast Cancer Association C. Commonly studied single-nucleotide polymorphisms and breast cancer: results from the Breast Cancer Association Consortium. J Natl Cancer Inst. 2006;98(19):1382–1396. doi: 10.1093/jnci/djj374. [DOI] [PubMed] [Google Scholar]
- 16.Hart SN, Maxwell KN, Thomas T, et al. Collaborative science in the next-generation sequencing era: a viewpoint on how to combine exome sequencing data across sites to identify novel disease susceptibility genes. Brief Bioinform. 2016;17(4):672–677. doi: 10.1093/bib/bbv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Complexo, Southey MC, Park DJ, et al. COMPLEXO: identifying the missing heritability of breast cancer via next generation collaboration. Breast Cancer Res. 2013;15(3):402. doi: 10.1186/bcr3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chenevix-Trench G, Milne RL, Antoniou AC, et al. An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA). Breast Cancer Res. 2007;9(2):104. doi: 10.1186/bcr1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel RY, Shah N, Jackson AR, et al. ClinGen Pathogenicity Calculator: a configurable system for assessing pathogenicity of genetic variants. Genome Med. 2017;9(1):3. doi: 10.1186/s13073-016-0391-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehm HL, Berg JS, Brooks LD, et al. ClinGen--the Clinical Genome Resource. N Engl J Med. 2015;372(23):2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balmana J, Digiovanni L, Gaddam P, et al. Conflicting Interpretation of Genetic Variants and Cancer Risk by Commercial Laboratories as Assessed by the Prospective Registry of Multiplex Testing. J Clin Oncol. 2016;34(34):4071–4078. doi: 10.1200/JCO.2016.68.4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh MF, Gaddam P, Digiovanni L, et al. Prospective registry of multiplex testing (PROMPT): A web-based platform to assess cancer risk of genetic variants. Journal of Clinical Oncology. 2016;34(15_suppl):1518–1518. doi: 10.1200/JCO.2016.34.15_suppl.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whiffin N, Walsh R, Govind R, et al. CardioClassifier: disease- and gene-specific computational decision support for clinical genome interpretation. Genet Med. 2018. doi: 10.1038/gim.2017.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q, Wang K. InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. Am J Hum Genet. 2017;100(2):267–280. doi: 10.1016/j.ajhg.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly MA, Caleshu C, Morales A, et al. Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: recommendations by ClinGen’s Inherited Cardiomyopathy Expert Panel. Genet Med. 2018. doi: 10.1038/gim.2017.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarvik GP, Browning BL. Consideration of Cosegregation in the Pathogenicity Classification of Genomic Variants. Am J Hum Genet. 2016;98(6):1077–1081. doi: 10.1016/j.ajhg.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nykamp K, Anderson M, Powers M, et al. Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet Med. 2017;19(10):1105–1117. doi: 10.1038/gim.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knudson AG, Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68(4):820–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olsen JH, Hahnemann JM, Borresen-Dale AL, et al. Cancer in patients with ataxia-telangiectasia and in their relatives in the nordic countries. J Natl Cancer Inst. 2001;93(2):121–127. [DOI] [PubMed] [Google Scholar]
- 30. http://oncotree.mskcc.org.
- 31.Vijai J, Topka S, Villano D, et al. A Recurrent ERCC3 Truncating Mutation Confers Moderate Risk for Breast Cancer. Cancer Discov. 2016;6(11):1267–1275. doi: 10.1158/2159-8290.CD-16-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouaoun L, Sonkin D, Ardin M, et al. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum Mutat. 2016;37(9):865–876. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 33.Janatova M, Kleibl Z, Stribrna J, et al. The PALB2 gene is a strong candidate for clinical testing in BRCA1- and BRCA2-negative hereditary breast cancer. Cancer Epidemiol Biomarkers Prev. 2013;22(12):2323–2332. doi: 10.1158/1055-9965.EPI-13-0745-T. [DOI] [PubMed] [Google Scholar]
- 34.Southey MC, Goldgar DE, Winqvist R, et al. PALB2, CHEK2 and ATM rare variants and cancer risk: data from COGS. J Med Genet. 2016;53(12):800–811. doi: 10.1136/jmedgenet-2016-103839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guidugli L, Carreira A, Caputo SM, et al. Functional assays for analysis of variants of uncertain significance in BRCA2. Hum Mutat. 2014;35(2):151–164. doi: 10.1002/humu.22478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spurdle AB, Healey S, Devereau A, et al. ENIGMA--evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum Mutat. 2012;33(1):2–7. doi: 10.1002/humu.21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. https://www.ga4gh.org/
- 38.Beroud C, Letovsky SI, Braastad CD, et al. BRCA Share: A Collection of Clinical BRCA Gene Variants. Hum Mutat. 2016;37(12):1318–1328. doi: 10.1002/humu.23113. [DOI] [PubMed] [Google Scholar]
- 39.Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513(7516):120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Starita LM, Ahituv N, Dunham MJ, et al. Variant Interpretation: Functional Assays to the Rescue. Am J Hum Genet. 2017;101(3):315–325. doi: 10.1016/j.ajhg.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.