Abstract
Background
Poly ADP-ribose polymerase (PARP) inhibitors, such as olaparib, are being explored as a treatment option for metastatic castration-resistant prostate cancer (mCRPC) in men harboring mutations in homologous recombination DNA-repair genes. Whether responses to PARP inhibitors differ according to the affected gene is currently unknown.
Objective
To determine whether responses to PARP inhibitors differ between men with BRCA1/2 and those with ATM mutations.
Design, setting, and participants
This was a multicenter retrospective review of 23 consecutive men with mCRPC and pathogenic germline and/or somatic BRCA1/2 or ATM mutations treated with olaparib at three academic sites in the USA.
Outcome measurements and statistical analysis
The proportion of patients achieving a >50% decline in prostate-specific antigen (PSA50 response) was compared using Fisher’s exact test. Clinical and radiographic progression-free survival (PFS) and overall survival were estimated using Kaplan-Meier analyses and compared using the log-rank test.
Results and limitations
The study included two men with BRCA1 mutations, 15 with BRCA2 mutations, and six with ATM mutations. PSA50 responses to olaparib were achieved in 76% (13/17) of men with BRCA1/2 versus 0% (0/6) of men with ATM mutations (Fisher’s exact test; p = 0.002). Patients with BRCA1/2 mutations had median PFS of 12.3 mo versus 2.4 mo for those with ATM mutations (hazard ratio 0.17, 95% confidence interval 0.05–0.57; p = 0.004). Limitations include the retrospective design and relatively small sample size.
Conclusions
Men with mCRPC harboring ATM mutations experienced inferior outcomes to PARP inhibitor therapy compared to those harboring BRCA1/2 mutations. Alternative therapies should be explored for patients with ATM mutations.
Patient summary
Mutations in BRCA1/2 and ATM genes are common in metastatic prostate cancer. In this study we compared outcomes for men with BRCA1/2 mutations to those for men with ATM mutations being treated with olaparib. We found that men with ATM mutations do not respond as well as men with BRCA1/2 mutations do.
Keywords: ATM, BRCA2, Olaparib, PARP inhibitor, Prostate cancer
1. Introduction
While there are currently seven therapies approved by the US Food and Drug Administration (FDA) for the treatment of metastatic castration-resistant prostate cancer (mCRPC), which is the lethal form of the disease, there are few genetic biomarkers to predict an individual’s response to therapy. In ovarian and breast cancer, poly ADP-ribose polymerase (PARP) inhibitors are used for patients harboring germline mutations in BRCA1 or BRCA2, supporting the concept of synthetic lethality [1]. Across all solid tumor types, the presence of mismatch repair (MMR) gene mutations predicts sensitivity to immune checkpoint blockade [2].
Although there are many molecular determinants of prostate cancer, few have given rise to genomically targeted therapies [3]. The FDA recently granted “breakthrough” designation status to the PARP inhibitor olaparib for treatment of mCRPC patients harboring germline and/or somatic mutations in the DNA-repair genes BRCA1 and BRCA2, as well as ATM [4]. This decision was based on earlier trials suggesting that men with mCRPC harboring mutations in homologous recombination DNA-repair genes are more likely to respond to olaparib than men without such mutations [5,6]. More recently, FDA “breakthrough” status was also granted to another PARP inhibitor, rucaparib, for mCRPC patients with BRCA1/2 mutations [7]. However, because ATM functions as a sensor of DNA damage rather than a mediator of DNA repair [8], we hypothesized that patients harboring ATM mutations might not show the same responses to PARP inhibitor therapy as those harboring BRCA1/2 mutations (which are bona fide homologous recombination genes) [9]. Here we describe the differential response to treatment with the PARP inhibitor olaparib among men with BRCA1/2 versus ATM mutations.
2. Patients and methods
This was a retrospective observational study of 46 consecutive patients with progressive mCRPC who were prescribed off-label single-agent olaparib at Johns Hopkins Hospital, University of Washington, and Mayo Clinic–Scottsdale from December 2014 (the date of olaparib FDA approval for ovarian cancer [10]) through October 2018. Patients who were deemed fit for therapy and were ineligible, declined, or did not have access to a clinical trial with PARP inhibitors were offered therapy. Those harboring pathogenic mutations (somatic or germline) in BRCA1, BRCA2, or ATM were included in this analysis. All centers participating in the study obtained local institutional review board approval before data abstraction.
Demographic, clinical, and genomic data were recorded and reported. Proportions were compared using a χ2 or Fisher’s exact test, while means were compared using a Kruskal- Wallis test.
The primary efficacy endpoint was the percentage of men achieving a >50% decline in prostate-specific antigen level from baseline (PSA50 response). Response rates were compared between men with BRCA1/2 mutations and men with ATM mutations using Fisher’s exact test. Radiographic or clinical progression-free survival (PFS) and overall survival (OS) were estimated using Kaplan-Meier analysis and comparisons between mutational groups were carried out using log-rank testing. Clinical or radiographic progression was defined as either radiologic progression or unequivocal clinical progression (or death), whichever occurred first. Radiographic progression was determined at the discretion of the local radiologists, broadly consistent with the PCWG3 guidelines [11]. Clinical progression was defined as worsening bone pain, a need for additional systemic or radiation therapy, or bone complications including fracture or spinal cord compression.
Patients were followed from the time of olaparib initiation until the time of last clinical or radiographic assessment for PFS and were censored at the time of last contact with the health system for OS. Stata version 15 (StataCorp, College Station, TX, USA) was used for statistical analyses.
3. Results
3.1. Cohort characteristics
Forty-six men received off-label olaparib treatment (300 mg orally twice daily) for mCRPC during the study period and were included in this study (Fig. 1). Thirteen patients did not undergo any genetic testing, seven patients underwent genetic testing that revealed no known ATM, BRCA1, or BRCA2 gene mutations, two patients had nonpathogenic mutations (silent variants) in BRCA2, and one patient was prescribed but did not start olaparib. These patients were not included in this analysis, as those without pathogenic DNA repair gene mutations would not be expected to benefit from PARP inhibitor treatment given current knowledge. Twenty-three patients had pathogenic mutations in BRCA1/2 (n = 17) or ATM (n = 6) and were included in this analysis (Table 1). Thirteen (57%) of these mutations were of germline and ten (44%) were of somatic origin. Table 2 displays the baseline clinical and demographic characteristics of the 23 patients. Patients with an ATM mutation did not significantly differ from those with a BRCA1/2 mutation other than a trend towards more germline mutations in the BRCA1/2 cohort and more bone-only disease in the ATM cohort. At diagnosis, nine (39%), four (17%), and nine (39%) patients had Gleason sum 7, 8, and 9, respectively. The median age at time of olaparib initiation was 66 yr (interquartile range [IQR] 61–71). The median baseline PSA level was 37 ng/ml (IQR 6–281). The median time on olaparib treatment was 5.4 mo (IQR 2.6–11.2), with six BRCA2-positive patients continuing on therapy at the time of database lock. The median time on therapy for patients with ATM mutations was 2.6 mo (IQR 2.4–4.7) versus 8.4 mo (IQR 4.2–11.9) for those with BRCA1/2 mutations.
Fig. 1 –
Consort diagram.
mCRPC = metastatic castration-resistant prostate cancer.
Table 1 –
List of pathogenic mutations in BRCA1/2 or ATM observed in our cohort
| ID | Gene | Mutation |
Platform used for detection | ||
|---|---|---|---|---|---|
| Origin | Name | Mechanism | |||
| 1 | BRCA2 | Somatic | p.N899I fs*5; p.K1691N fs*15 | Frameshift deletions | PGDx-Cancer Select |
| 2 | BRCA2 | Germline | p.Y2215S fs*13 | Frameshift deletion | Color Genomics |
| 3 | ATM | Germline | p.S1905I fs*25 | Frameshift insertion | Color Genomics |
| 4 | BRCA2 | Germline | p.N319K fs*8 | Frameshift insertion | Color Genomics |
| 5 | BRCA2 | Germline | p.D3095E | Missense | Color Genomics |
| 6 | ATM | Somatic | p.Q284X* | Nonsense | PGDx-Plasma Select |
| 7 | BRCA2 | Germline | p.D156X* | Nonsense | Color Genomics |
| 8 | BRCA2 | Germline | p.W1692M fs*3 | Frameshift insertion | Color Genomics |
| 9 | BRCA2 | Somatic | p.S1982R fs*22 | Frameshift deletion | Foundation One |
| 10 | BRCA1 | Germline | p.Y1463X* | Nonsense | Color Genomics |
| 11 | BRCA1 | Germline | c.4357+1G>A | Splicing | Color Genomics |
| 12 | ATM | Somatic | Loss of exons 30–34 | Rearrangement | Foundation One |
| 13 | BRCA2 | Germline | p.S3147C fs*2 | Frameshift deletion | Myriad genetics |
| 14 | BRCA2 | Germline | p.Q2858A fs*5 | Frameshift insertion | Caris Genetics |
| 15 | BRCA2 | Somatic | p.F1546L fs*22 | Frameshift deletion | Foundation One |
| 16 | ATM | Somatic | p.M1V | Missense | Foundation One |
| 17 | ATM | Somatic | p.N405K fs*15 | Frameshift deletion | Foundation One |
| 18 | ATM | Somatic | 1.7 kbp deletion | Rearrangement | UW-Oncoplex |
| 19 | BRCA2 | Germline | p.L1908R fs*2 | Frameshift deletion | Myriad genetics |
| 20 | BRCA2 | Germline | p.T2125P fs*12 | Frameshift deletion | Color Genomics |
| 21 | BRCA2 | Germline | Exon loss, with hemizygous deletion of somatic allele | Rearrangement | Stand Up 2 Cancer (Ml-OncoSeq) |
| 22 | BRCA2 | Somatic | Homozygous deletion | Rearrangement | Stand Up 2 Cancer (MI-OncoSeq) |
| 23 | BRCA2 | Somatic | Homozygous deletion | Rearrangement | UW-Oncoplex |
Table 2 –
Baseline demographic and clinical data overall and by mutation type
| Overall | ATM | BRCA1/2 | p value | |
|---|---|---|---|---|
| Patients, n (%) | 23 | 6 (26) | 17 (74) | |
| Gleason sum at diagnosis, n (%) | 0.8 | |||
| 7 | 9 (39) | 2 (33) | 7 (41) | |
| 8 | 4 (17) | 2 (33) | 2 (12) | |
| 9 | 9 (39) | 2 (33) | 7 (41) | |
| Unavailable | 1 (4) | 0 (0) | 1 (6) | |
| Median age at start of therapy, yr (IQR) | 66 (61–71) | 71 (70–76) | 65 (61–70) | 0.07 |
| Median baseline PSA, ng/mi (IQR) | 37 (6.2–281) | 278 (95.2–365) | 22 (5.4–53.9) | 0.06 |
| Prior chemotherapy, n (%) | 1 | |||
| Yes | 15 (65) | 4 (67) | 11 (65) | |
| No | 8 (35) | 2 (33) | 6 (35) | |
| Prior enzalutamide/abiraterone, n (%) | 0.5 | |||
| Yes | 21 (91) | 5 (83) | 16 (94) | |
| No | 2 (9) | 1 (17) | 1 (6) | |
| Type of mutation, n (%) | 0.052 | |||
| Germline | 13 (57) | 1 (17) | 12 (71) | |
| Somatic | 10 (43) | 5 (83) | 5 (29) | |
| Presence of bone metastases, n (%) | 0.5 | |||
| Yes | 20 (87) | 6 (100) | 14 (82) | |
| No | 3 (13) | 0 (0) | 3 (18) | |
| Presence of visceral metastases, n (%) | 0.1 | |||
| Yes | 6 (26) | 0 (0) | 6 (35) | |
| No | 17 (74) | 6 (100) | 11 (65) | |
| Presence of nodal metastases, n (%) | 1 | |||
| Yes | 13 (52) | 3 (50) | 10 (59) | |
| No | 10 (48) | 3 (50) | 7 (41) | |
| Mean prior TLs for mCRPC, n (range) | 2.9 (0–5) | 2.8 (0–4) | 2.9 (1–5) | 0.7 |
| Mean TLs after olaparib, n (range) | 0.70 (0–4) | 0.83(0–2) | 0.65 (0–4) | 0.4 |
IQR = interquartile range; mCRPC = metastatic castrate-resistant prostate cancer; PSA = prostate-specific antigen; TL = treatment line.
3.2. PSA response rates
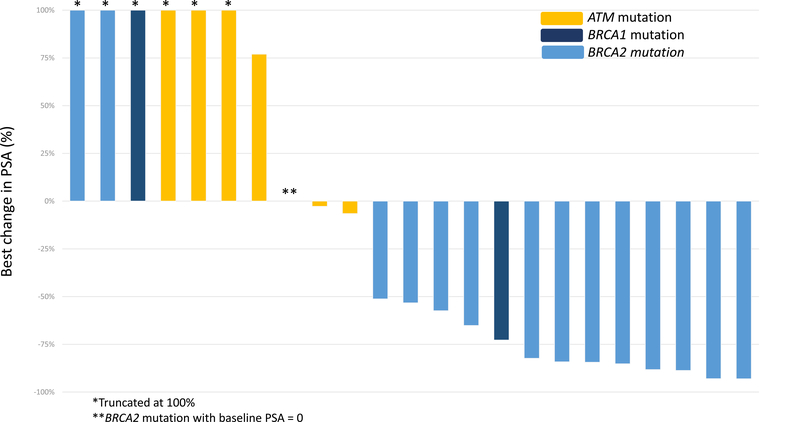
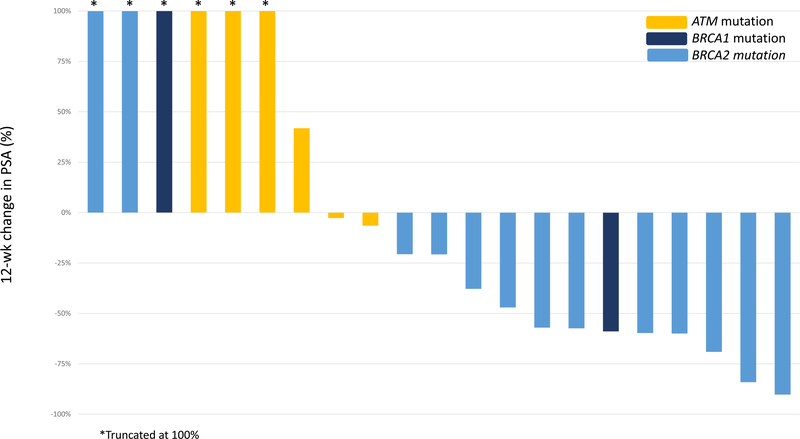
Figure 2A shows the best PSA response for each patient at any time point. Overall, 13/23 patients (57%) achieved a PSA50 response. No PSA responses (0/6) were observed among patients harboring ATM mutations, while 76% (13/17) of those with BRCA1/2 mutations achieved a PSA50 response (p = 0.002). The median time to best PSA response was 20 wk (IQR 7–35). The 12-wk PSA response is reported in Figure 2B; two patients could not be evaluated as they did not have PSA measured at the 12-wk time point (±4 wk). At 12 wk, 53% of the BRCA1/2 cohort (eight/15) achieved a PSA50 response versus 0% of ATM cohort (zero/six; p = 0.046).
Fig. 2 –
Best prostate-specific antigen (PSA) response by mutation status (A) at any time and (B) at 12 wk.
3.3. PFS and OS
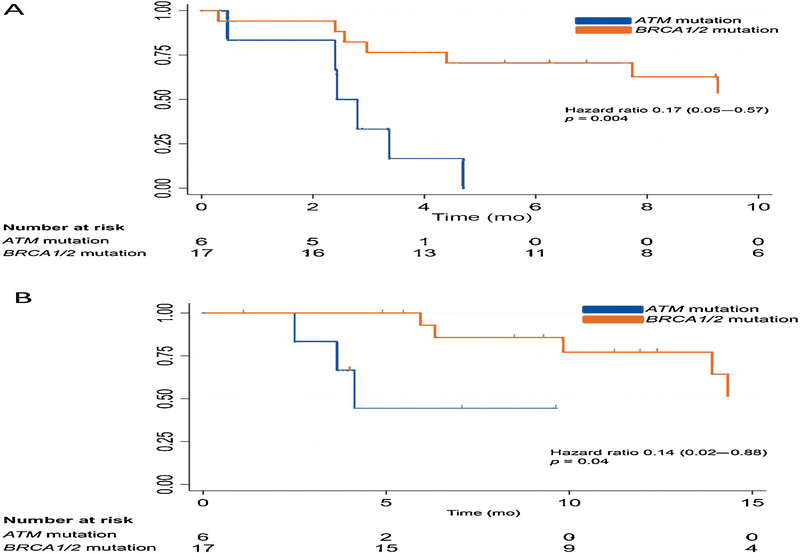
Overall, 17/23 patients had clinical or radiographic progression, all six of those with ATM mutations and 11/17 patients with BRCA1/2 mutations. Clinical or radiographic PFS with olaparib was significantly longer among patients with BRCA1/2 mutations than among those with ATM mutations (Fig. 3A). Men with BRCA1/2 mutations had median PFS of 12.3 mo, compared to 2.4 mo in the ATM group (hazard ratio [HR] 0.17, 95% confidence interval [CI] 0.05–0.57; p = 0.004). The median time to last assessment of progression for those who did not experience progression was 8 mo.
Fig. 3 –
Kaplan-Meier curve for (A) progression-free survival (the x-axis is truncated at 10 mo) and (B) overall survival (the x-axis is truncated at 15 mo) by mutation status.
Nine patients (three with ATM and six with BRCA1/2 mutations) died during follow-up. OS was longer in the BRCA1/2 cohort than in the ATM cohort (29.8 vs 4.1 mo; HR 0.14, 95% CI 0.02–0.88; p = 0.04; Fig. 3B). The median time to last assessment among those still living was 9.4 mo.
4. Discussion
Our results suggest that men with mCRPC harboring ATM mutations may not respond to PARP inhibitors as well as those with BRCA1/2 mutations. A similar pattern was recently observed in the preliminary results of the TRITON2 study investigating rucaparib, in which none of 18 patients with ATM mutations demonstrated a PSA50 response, compared to 51% (23/45) of those with BRCA1/2 mutations [12]. This is seemingly in contrast to the TOPARP- A study, in which four of six patients (67%) with ATM mutations responded to olaparib, although only two of the six patients achieved a PSA50 response at 12 wk, with the other two only achieving a decrease in circulating tumor cells [5]. It is also important to note that the time to best PSA response may be delayed (median of 20 wk in our study) and that five patients only achieved a PSA50 response after 12 wk. This should be taken into consideration when interpreting future studies, and may suggest that early PSA responses may not be the optimal measure of clinical benefit for PARP inhibitor therapy in prostate cancer.
One plausible biological mechanism explaining why men harboring BRCA1/2 mutations may respond differently to PARP inhibitors compared to those with ATM mutations is that the ATM protein functions primarily as a sensor of DNA damage rather than an effector of DNA repair like BRCA1 and BRCA2 [13]. Studies in breast cancer patients harboring germline ATM mutations have shown that ATM-mutant cancers are molecularly distinct from BRCA1/2-mutant cancers, lack characteristic genomic features reflective of homologous recombination deficiency, and exhibit significantly different immune infiltrates compared to BRCA1/2-mutant cancers [8]. Similar findings have been observed in prostate cancer: ATMmutant tumors showed a distinct transcriptional phenotype compared to tumors with mutations in true homologous repair genes [14]. A second potential explanation is that optimal response to PARP inhibitors probably requires biallelic gene inactivation for induction of synthetic lethality. In this context, BRCA1/2 mutation carriers may have a higher degree of biallelic loss than those with ATM mutations [15,16]. Furthermore, germline mutation carrier status, which was more prevalent in the BRCA1/2 group, may be relevant because every cancer cell will have at least monoallelic inactivation, whereas in patients with only somatic mutations, there may be only a tumor subclone that carries the mutation.
Taken together, our data suggest that alternative treatment strategies are needed for mCRPC patients harboring ATM mutations, as these men exhibit unfavorable responses to PARP inhibitors, progress relatively quickly, and are therefore not deriving significant benefit from therapy in comparison to those with BRCA1/2 mutations. Differing response rates to abiraterone and enzalutamide in ATM- and BRCA-deficient patients have also been observed in some studies [17,18]. Additional treatments such as ATR inhibitors have emerged as potential therapeutic approaches specifically for ATM-mutant cancers, including prostate cancers [19,20].
The limitations of this study include its retrospective nature with overall small sample size determined post hoc. In addition, this is a real-world experience with PARP inhibitors and does not control for many of the known and unknown baseline factors that may differ between the groups. Finally, we were not able to reliably assess biallelic mutations in most patients given the clinical-grade genomic assays used and were not able to perform immunohistochemical staining for BRCA1/2 or ATM protein loss.
5. Conclusions
Larger prospective studies are needed to confirm these preliminary findings. In addition, as these mechanisms are not unique to prostate cancer, similar findings should be explored in other cancer types for which PARP inhibitors are being used.
Acknowledgments
Financial disclosures: Emmanuel S. Antonarakis certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following. Emmanuel S. Antonarakis is a paid consultant/advisor to Janssen, Astellas, Sanofi, Dendreon, Medivation, AstraZeneca, Clovis, and Merck; has received institutional research funding from Janssen, Johnson & Johnson, Sanofi, Dendreon, Genentech, Novartis, Tokai, Bristol Myers-Squibb, AstraZeneca, Clovis, and Merck; and is co-inventor of a biomarker technology that has been licensed to Qiagen. Heather H. Cheng has received institutional research funding from Celgene, Clovis, Inovio, Janssen, Medivation, and Sanofi. Michael T. Schweizer is a paid consultant/advisor to Janssen and has received institutional research funding from Janssen, AstraZeneca, Hoffmann La Roche, Zenith Epigenetics, and Pfizer. Catherine H. Marshall has received research funding via the Conquer Cancer Foundation from Bristol Myers-Squibb and travel support from Dava Oncology, and is a paid consultant to the McGraw-Hill publishing company. The remaining authors have nothing to disclose.
Funding/Support and role of the sponsor: This work was partly supported by National Institutes of Health Cancer Center Support Grants P30 CA006973 and P30 CA015704, Pacific Northwest Prostate Cancer SPORE CA097186, Department of Defense grant W81XWH-16-PCRP-CCRSA, the Patrick C. Walsh Research Fund, and Institute for Prostate Cancer Research and Prostate Cancer Foundation Awards. The sponsors played no direct role in the study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.US Food & Drug Administration. FDA approves olaparib for germline BRCA-mutated metastatic breast cancer. www.fda.gov/drugs/informationondrugs/approveddrugs/ucm592357.htm
- 2.Le DT, Durham JN, Smith KN, et al. Mismatch-repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357409–1l3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armenia J, Wankowicz SAM, Liu D, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet 2018;50:645–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.AstraZeneca. Press release. Lynparza™ (olaparib) granted breakthrough therapy designation by US FDA for treatment of BRCA1/2 or ATM gene mutated metastatic castration resistant prostate cancer; 2016.
- 5.Mateo J, Carreira S, Sandhu S, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med 2015;373:1697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 2015;33:244–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katz A FDA grants rucaparib breakthrough designation for mCRPC. OncLive; 2018. www.onclive.com/web-exclusives/fda-grants-rucaparib-breakthrough-designation-for-mcrpc [Google Scholar]
- 8.Weigelt B, Bi R, Kumar R, et al. The landscape of somatic genetic alterations in breast cancers from ATM germline mutation carriers. J Natl Cancer Inst 2018;110:1030–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tutt A, Ashworth A. The relationship between the roles of BRCA genes in DNA repair and cancer predisposition. Trends Mol Med 2002;8:571–6. [DOI] [PubMed] [Google Scholar]
- 10.Kim G, Ison G, McKee AE, et al. FDA approval summary: olaparib monotherapy in patients with deleterious germline BRCA-mutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin Cancer Res 2015;21:4257–61. [DOI] [PubMed] [Google Scholar]
- 11.Scher HI, Morris MJ, Stadler WM, et al. Trial design and objectives for castration resistant prostate cancer: updated recommendations from the Prostate Cancer Clinical Trials Working Group 3. J Clin Oncol 2016;34:1402–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abida W, Bryce AH, Vogelzang NJ, et al. Preliminary results from TRITON2: a phase 2 study of rucaparib in patients with metastatic castration-resistant prostate cancer associated with homologous recombination repair gene alterations. Paper presented at: ESMO 2018 Congress; October 21, 2018; Munich. [Google Scholar]
- 13.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell 2007;28:739–45. [DOI] [PubMed] [Google Scholar]
- 14.Wu Y-M, Cieślik M, Lonigro RJ, et al. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell 2018;173:1770–82.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Annala M, Struss WJ, Warner EW, et al. Treatment outcomes and tumor loss of heterozygosity in germline DNA repair-deficient prostate cancer. Eur Urol 2017;72:34–42. [DOI] [PubMed] [Google Scholar]
- 16.Decker B, Karyadi DM, Davis BW, et al. Biallelic BRCA2 mutations shape the somatic mutational landscape of aggressive prostate tumors. Am J Hum Genet 2016;98:818–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antonarakis ES, Lu C, Luber B, et al. Germline DNA-repair gene mutations and outcomes in men with metastatic castration-resistant prostate cancer receiving first-line abiraterone and enzalutamide. Eur Urol 2018;74:218–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Annala M, Vandekerkhove G, Khalaf D, et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov 2018;8:444–57. [DOI] [PubMed] [Google Scholar]
- 19.Hickson I, Zhao Y, Richardson CJ, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res 2004;64:9152–9. [DOI] [PubMed] [Google Scholar]
- 20.Reaper PM, Griffiths MR, Long JM, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol 2011;7:428–30 [DOI] [PubMed] [Google Scholar]