Abstract
The astroglial lineage consists of heterogeneous cells instrumental for normal brain development, function and repair. Unfortunately, this heterogeneity complicates research in the field, which suffers from lack of truly specific and sensitive astroglial markers. Nevertheless, single astroglial markers are often used to describe astrocytes in different settings. We therefore investigated and compared spatiotemporal patterns of immunoreactivity in developing human brain from 12 to 21 weeks post conception and publicly available RNA expression data for four established and potential astroglial markers – GFAP, S100, AQP4 and YKL‐40. In the hippocampal region, we also screened for C3, a complement component highly expressed in A1‐reactive astrocytes. We found diverging partly overlapping patterns of the established astroglial markers GFAP, S100 and AQP4, confirming that none of these markers can fully describe and discriminate different developmental forms and subpopulations of astrocytes in human developing brain, although AQP4 seems to be the most sensitive and specific marker for the astroglial lineage at midgestation. AQP4 characterizes a brain‐wide water transport system in cerebral cortex with regional differences in immunoreactivity at midgestation. AQP4 distinguishes a vast proportion of astrocytes and subpopulations of radial glial cells destined for the astroglial lineage, including astrocytes determined for the future glia limitans and apical truncated radial glial cells in ganglionic eminences, devoid of GFAP and S100. YKL‐40 and C3d, previously found in reactive astrocytes, stain different subpopulations of astrocytes/astroglial progenitors in developing hippocampus at midgestation and may characterize specific subpopulations of ‘developmental astrocytes’. Our results clearly reflect that lack of pan‐astrocytic markers necessitates the consideration of time, region, context and aim when choosing appropriate astroglial markers.
Keywords: astrocytes, astrogliogenesis, glial markers, human brain development, radial glial cells
Introduction
For decades, the study of astrogliogenesis was neglected in favor of unraveling neuronal development, dismissing astroglial cells as mere glue. Time revealed these cells to be multifunctional heterogeneous cells regulating neurogenesis, neural function and many other aspects of human brain development. Nevertheless, the important study of astrogliogenesis is persistently obstructed due to incomplete and miscellaneous definitions and lack of pan‐astrocytic markers (Molofsky et al. 2012; Molofsky & Deneen, 2015; Ben Haim & Rowitch, 2017). Through review of existing evidence and new findings based on RNA‐sequencing data, immunofluorescence and immunohistochemistry with a panel of structural and functional markers, we will discuss the complexity of astrogliogenesis in early developing human brain.
Astrogliogenesis
Creating a timeline for human astrogliogenesis is impeded by differences in ontogeny, difficulties in converting results on glial development across species (Smart et al. 2002; Molofsky et al. 2012; Molofsky & Deneen, 2015; Zhang et al. 2016), the small number of applicable experimental human models, spatiotemporal heterogeneity, lack of sensitive and specific markers, lack of clear definitions for particularly astrocytes and their precursors and no well‐defined starting or endpoint for astrocyte development.
From the time of Wilhelm His, neurons and macroglial cells were believed to originate from two separate progenitor populations (see Alvarez‐Buylla et al. 2001). This belief was slowly rejected, when radial glial cells were found initially to produce neurons followed by production of astrocytes (Malatesta et al. 2000; Alvarez‐Buylla et al. 2001; Rowitch & Kriegstein, 2010). The concept of adult neurogenesis from radial glia‐like cells with astrocytic features (Gotz et al. 2015; Falk & Gotz, 2017; Berg et al. 2018), bipotent gliogenic precursors (Miller & Gauthier, 2007) and changes in lineage restriction for both astrocytes and oligodendrocyte precursors based on culture conditions (Alvarez‐Buylla et al. 2001; Gotz et al. 2015) further challenged our understanding of lineage relationships. It should be noted that the idea of adult human neurogenesis is currently under revision (Boldrini et al. 2018; Kempermann et al. 2018; Sorrells et al. 2018). Cell–cell interaction, epigenetic regulation and the ultimate environment also influence cell fate and function (Rowitch & Kriegstein, 2010; Jiang & Nardelli, 2016).
To define astrogliogenesis it may be necessary to distinguish between an initial neuron to glial cell switch (Rowitch & Kriegstein, 2010), primary gliogenesis and adult reactivation of glial cells. The neuron to glial cell switch is subjected to complex regulation mediated by both intrinsic and extrinsic factors, epigenetics, neuronal feedback and spatiotemporal interplay (Morrow et al. 2001; Miller & Gauthier, 2007; Rowitch & Kriegstein, 2010; Jiang & Nardelli, 2016). It also seems that single precursors can contribute to both neuronal and glial populations or produce only neurons or only glial cells depending on the time‐point (Miller & Gauthier, 2007). The above‐mentioned findings and considerations question the concept of differentiated cells and whether morphology and structural cell‐specific markers fully reflect cell function and potential. It also highlights the complexity and misconceptions of gliogenesis in the developing and adult brain.
Radial glial cells
Establishment of the human cerebral cortex follows induction of the neural tube, which at the onset is lined with a founder population of neuroepithelial cells. This pseudostratified layer of epithelial cells with characteristic apico‐basal radial processes forms the initial proliferative ventricular zone (Rakic, 1988; Bystron et al. 2008). At the onset of neurogenesis, neuroepithelial cells transform through asymmetric divisions into classical radial glial cells, the processes of which span the entire cerebral wall with apical ventricular connections and basal pial‐facing end feet (Rakic, 1972). In the developing central nervous system (CNS) there are several populations of RGCs with morphological and to some extent molecular similarities, specialized to accommodate regional needs (Rakic, 2003). They include RGCs in spinal cord and telencephalon, tanycytes in hypothalamus, Bergmann glia in cerebellum and Müller glia in the retina (Edwards et al. 1990; Rakic, 2003; Hamilton et al. 2009; Barry et al. 2014).
In cerebral cortex we classify radial glial cells as: (1) classical radial glial cells (cRGCs) spanning the entire cerebral wall, (2) apical truncated radial glial cells (atRGCs) in contact with the ventricular but not the pial surface and (3) basal radial glial cells (bRGCs), also referred to as outer radial glial cells (oRGs) (Rakic, 1988; Fietz et al. 2010; Hansen et al. 2010; Reillo et al. 2011; Nowakowski et al. 2016), situated primarily in the oSVZ with end feet at the pial surface but detached from the ventricular surface. Classical methods for demonstrating radial glia involve different Golgi impregnation methods and electron microscopy (see Sidman & Rakic, 1973; Choi & Lapham, 1978). Later, GLAST (Shibata et al. 1997), BLBP (Feng et al. 1994), GFAP, vimentin (Levitt & Rakic, 1980; Dahl et al. 1981; Stagaard & Møllgård, 1989) and other markers have been used to describe the radial glial cell population. Basal RGCs and atRGCs can be distinguished by expression of SOX2 and HOPX in bRGs only. Through cell divisions, cRGCs give rise to Tbr2‐positive intermediate progenitor cells (IPCs) of the SVZ, which in turn give rise to post‐mitotic neurons (Nowakowski et al. 2016). The radial glia scaffold in the developing human cerebral cortex becomes discontinuous at mid‐neurogenesis [around 16 weeks post conception (wpc)] but before midgestation (19 wpc), which leads to transformation of cRGCs to atRGCs (or truncated RGCs) (Rakic, 2003; Nowakowski et al. 2016). These often terminate on blood vessels in the oSVZ. This discontinuity of the radial glia scaffold heralds the transition from infragranular to supragranular neuron production, which from this stage in human cortical development is controlled by bRG/oRG (Nowakowski et al. 2016). Following and perhaps overlapping with neurogenesis, subpopulations of all three RGC subtypes initiate gliogenesis, and give rise to astrocyte precursor cells, astrocytes and oligodendrocyte precursor cells.
Astrocytes
Astrocytes are present in all areas of the CNS (Molofsky & Deneen, 2015) with regional specialization (Ben Haim & Rowitch, 2017) and differences in density (Molofsky et al. 2012). They are typically grouped into protoplasmic (mainly in gray matter) and fibrous astrocytes (in white matter) (Andriezen, 1893). In the cerebral cortex, astrocytes are produced from radial glial cells in humans, and also from progenitors in the subventricular zone, locally proliferating glia and NG2 glia (Ge & Jia, 2016) in rodents. Transformation from radial glial cells to astrocytes occurs in rhesus monkeys in the middle prenatal stage (E60–E95) (Schmechel & Rakic, 1979). In humans this transformation is agreed by most authors to take place mainly throughout the second half of human fetal development (Kadhim et al. 1988; Marín‐Padilla, 1995), but this varies depending on location and method (e.g. 12–20 wpc in frontoparietal cortex found by Choi & Lapham, 1978, 15–35 wpc in frontal cortex found by Marín‐Padilla, 1995, or 21–40 gestational weeks (gw) in unspecified neocortical regions found by Kadhim et al. 1988) (see Table 2 and deAzevedo et al. 2003). This variability could reflect interregional differences in brain development or/and gliogenic potential, and lack of sensitive and specific methods to distinguish radial glial cells and their progeny. Adding to the complexity, transitional forms between RGCs and astrocytes exist (Choi & Lapham, 1978, 1980) and are difficult to define. RGCs are regionally specified to produce distinct subtypes of neuronal progeny in mice (Merkle et al. 2007; Bayraktar et al. 2014), which are not altered by culture manipulation or transplantation (Merkle et al. 2007). In vitro, astrocytes have also been shown to maintain region‐specific expression and functions (Molofsky et al. 2014; Ben Haim & Rowitch, 2017), suggesting that diversified regional astrocyte identity may already be determined at embryonic stages (Ben Haim & Rowitch, 2017). Astrocyte heterogeneity is also influenced by extrinsic signals from the microenvironment (Molofsky et al. 2012; Ben Haim & Rowitch, 2017) and morphology and function may change at different stages of development and maturation (Christopherson et al. 2005; Molofsky et al. 2012).
Astrocytes are involved in brain homeostasis, brain‐barrier integrity (Abbott et al. 2006; Saunders et al. 2018), neurogenesis, neuronal survival (Drukarch et al. 1998), synaptogenesis (Ullian et al. 2001, 2004; Allen, 2013), synaptic and axonal pruning (Chung et al. 2013; Reemst et al. 2016), synaptic transmission (Bayraktar et al. 2014; Molofsky & Deneen, 2015; Jiang & Nardelli, 2016; Reemst et al. 2016; Zhang et al. 2016; Ben Haim & Rowitch, 2017), axonal guidance (Reemst et al. 2016), myelination (Ishibashi et al. 2006; Watkins et al. 2008), postnatal angiogenesis (Ma et al. 2012; Reemst et al. 2016) and regulation of cerebral blood flow (Iadecola & Nedergaard, 2007; Ben Haim & Rowitch, 2017). Astrocytes may also function as neural stem cells (Bayraktar et al. 2014). This plethora of astrocyte functions may vary between species and depends on time and localization. Therefore, investigating developmental spatiotemporal patterning of human astrocytes and their progenitors seems crucial in understanding both developmental and mature astrocyte diversity.
Astroglial markers
GLAST, BLBP, FGFR3 and Sox 9 have been used to describe astrocyte precursors (Molofsky et al. 2012; Molofsky & Deneen, 2015), but they may not be specific to the astrocytic lineage (Molofsky et al. 2012) and to our knowledge there are no reliable validated markers describing specific stages in astroglial development (Molofsky et al. 2012; Reemst et al. 2016). For many years, GFAP was the most widely used marker for astrocytes and their differentiation, but other markers such as S100B, Aldh1L1, AldoC, Ascgb1, Glt1 and AQP4 are now also used to characterize mature astrocytes (Molofsky et al. 2012; Molofsky & Deneen, 2015), with caveats for each marker (Molofsky et al. 2012).
GFAP expression varies between different types of astrocytes (Tabata, 2015) and is seen in radial glial cells and transitional forms (Antanitus et al. 1976; Choi & Lapham, 1978; Choi, 1986; Stagaard Janas et al. 1991a; Middeldorp et al. 2010) and subpopulations of neural precursors (Miller & Gauthier, 2007; Silbereis et al. 2010). Therefore GFAP cannot be used to define astrogliogenesis or terminal astrocyte differentiation. GFAP is upregulated in reactive and cultured astrocytes, which are different from normal resting astrocytes (Molofsky et al. 2012), and whereas astrocyte end feet on penetrating blood vessels are often GFAP‐positive, end feet around capillaries are GFAP‐negative (Iadecola & Nedergaard, 2007). Furthermore, GFAP has been seen in ependymal cells in mice (Doetsch et al. 1997) and cells of proposed oligodendroglial lineage (Choi & Kim, 1984, 1985).
S100 proteins are multifunctional proteins involved in calcium signal transduction (Tiu et al. 2000). S100A and S100B are most abundant in the nervous system (Astrand et al. 2013). S100 appear in radial glial cells (Stagaard Janas et al. 1991a) and astrocytes in developing and adult human brain (Stagaard Janas et al. 1991a,1991b; Tiu et al. 2000; Steiner et al. 2007) with the earliest appearance around 6.5 wpc in the brainstem followed by immunoreactivity in astrocytes in diencephalon at 8 wpc and in the hippocampal formation at 9.5 wpc (Stagaard Janas et al. 1991a). Most studies on S100 in the brain focus on S100B (Tiu et al. 2000; Deloulme et al. 2004; Steiner et al. 2007; Gos et al. 2013), which has also been found in reactive astrocytes (Mrak & Griffinbc, 2001; Steiner et al. 2007), myelinated fibers, oligodendrocytes, ependyma, choroid plexus epithelium, lymphocytes and a few neurons in adult human brain. In white matter, S100B preferentially stain oligodendrocyte resembling cells according to Steiner et al. (2007). The finding of S100B in oligodendrocytes is in accordance with results from adult patients with depressive/bipolar disorders (Gos et al. 2013) and expression of S100B in oligodendrocyte progenitor cells in mice (Deloulme et al. 2004; Hachem et al. 2005).
Aquaporins (AQP) are specialized water channels, of which nine different subtypes have been found within the CNS (Xu et al. 2017). AQP4 is the predominant AQP in the CNS and is mainly expressed in perivascular and subpial astrocyte end feet, and in ependymal cells (Assentoft et al. 2015). Activated astrocytes also express AQP4, but with a different subcellular distribution (Xu et al. 2017). AQP4 controls water movements in the CNS and is a key constituent of the glymphatic clearance system (Iliff et al. 2012; Xie et al. 2013). In developing human brain, only little evidence exists concerning AQP4. Gömöri et al. (2006) found increasing AQP4 immunoreaction in astrocyte processes and end feet at the developing ependymal lining, at the pial surface and around capillaries, which is consistent with localization in adult brain. There was a clear spatiotemporal difference in expression patterns, with delayed onset in neocortex (21–22 gw), compared with archicortex (14 gw), which could reflect differences in maturation tempi (Gömöri et al. 2006) or/and function. In mouse brain, AQP4 immunoreactivity has also been demonstrated in radial glial cells (Fallier‐Becker et al. 2014) and emerging astrocytes, and Fallier‐Becker et al. (2014) propose that differences in onset of AQP4 immunoreactivity follow neuronal differentiation.
GFAP, S100, AQP4 and YKL‐40 as astroglial markers
To our knowledge, only one study has evaluated and compared expression of GFAP, S100B and AQP4 (El‐Khoury et al. 2006). The study focused on astrocyte end feet coverage of blood vessels in white matter, germinal matrix and gray matter from 16 to 40 weeks of gestation and found both interregional and subcellular differences in staining of the astroglial lineage (El‐Khoury et al. 2006). To explore marker distribution and spatiotemporal patterns of early‐stage human astrogliogenesis, we compared immunoexpression of the diverse astroglial markers GFAP, S100 and AQP4 in human developing fetal brain and further included YKL‐40, a glycoprotein, which has been found in possible fetal astrocytes and astroglial progenitors in human developing brain (Bjørnbak et al. 2014; Brøchner & Møllgård, 2016), primary cultures of human embryonic astrocytes (Singh et al. 2011), gliomas (Qin et al. 2017) and reactive astrocytes (Bonneh‐Barkay et al. 2010, 2012) and may influence astroglial differentiation and migration (Singh et al. 2011).
Materials and methods
RNA‐sequencing of purified cells from healthy juvenile/adult brain, fetal astrocytes and astrocytes from glioblastoma core (www.brainrnaseq.org)
Gene‐level RNA‐seq quantification was generously shared by the Barres Lab (Dr. Steven Sloan). The RNA‐seq data on cells from fetal and human juvenile/adult healthy tissue is also available at www.brainrnaseq.org. Zhang et al. (2016) purified specific cell types from human brain samples by developing an immunopanning method using cell type‐specific antibodies. Anti‐CD45 antibodies were used to bind microglia (and macrophages), anti‐GalC hybridoma to bind oligodendrocytes and myelin debris, anti‐O4 hybridoma to bind oligodendrocyte precursor cells (OPCs), anti‐Thy1 antibody to bind neurons, anti‐HepaCAM antibody to bind astrocytes, and Bandeiraea simplicifolia lectin 1 (BSL‐1) to bind endothelial cells. For fetal samples, the protocol was shorter (further details regarding RNA‐seq data can be found in Zhang et al. 2016). ‘Normal’ juvenile/adult brain samples generally consisted of normal temporal lobe cortex resected during operations for epilepsy. Astrocytes from glioblastoma core were taken from regions with contrast enhancement and fetal astrocytes from fetal cortical tissue aged 18–18.5 gestational weeks corresponding to 16–16.5 wpc (Zhang et al. 2016). We extracted RNA‐seq quantification in fragments per kilobase of transcript sequence per million mapped (FPKM) for four genes of interest (GFAP, S100B, AQP4 and CHI3L1) for all available human cell types, which included fetal astrocytes, mature astrocytes, astrocytes from glioblastoma tumor core, oligodendrocytes, microglia/macrophages, neurons and endothelial cells.
RNA expression in brain regions from developing human brain (www.brainspan.org)
Data
Gene‐level RNA‐seq quantification was downloaded from BrainSpan (http://www.brainspan.org/static/download.html; 28 June 2018). Details about the RNA‐seq data can be found at http://help.brain-map.org/display/devhumanbrain/Documentation. Four genes of interest (GFAP, S100B, AQP4 and CHI3L1) were extracted for human samples with the following ages annotated in pcw: 8 (6), 9 (4), 12 (7,7,8), 13 (7,8,8), 16 (7,7,8), 17 (8), 19 (7), 21 (8), 24 (8), 26 (3), 35 (1), 37 (8) (the number of regions collected per sample is in brackets) for eight regions of interest: ‘anterior (rostral) cingulate (medial prefrontal) cortex’, ‘dorsolateral prefrontal cortex’, ‘hippocampus (hippocampal formation)’, ‘orbital frontal cortex’, ‘posterior (caudal) superior temporal cortex (area 22c)’, ‘posteroventral (inferior) parietal cortex’, ‘primary visual cortex (striate cortex, area V1/17)’ and ‘ventrolateral prefrontal cortex’. Regions were chosen based on data availability and cortical involvement. Neocortical regions collected from all prenatal brain samples included the subplate zone. Not all regions were collected from all fetuses. The samples were split into three groups: early prenatal (8–13 wpc), mid‐prenatal (16–23 wpc) and late prenatal (24–37 wpc) with at least two complete samples (containing all eight regions) in each group.
Deconvolutions of cell‐type proportions
The processed single‐cell expression data provided by Fan et al. (2018) was downloaded from GEO (accession number GSE103723) and the corresponding cell‐type classification was obtained from Fan et al. (2018) supplementary Table 3. The proportions of cell types were analyzed with the bioinformatic tool dTangle (https://www.biorxiv.org/content/early/2018/03/27/290262) following their single‐cell vignette (https://github.com/gjhunt/dtangle/blob/master/vign/sc_vignette.md). Specifically, all data from Fan et al. were used, except Pons and Medulla, because we mainly focus on cortical areas of developing human brain. The log2(FPKM/TPM) matrix of the two datasets (Fan et al. and BrainSpan) was reduced to genes analyzed in both cell types and combined into one big dataset using limmas normalizeBetweenArrays functionality (Smyth & Speed, 2003) (https://www.statsci.org/smyth/pubs/normalize.pdf). The cell‐type annotation from Fan et al. was simplified by combining sub‐cell types (e.g. the two sub‐groups of astrocytes, denoted _1 and _2, were combined). The combined matrix was then supplied to the dTangle function, along with the indexes of the cell‐type annotation from Fan et al., which estimated the cell‐type proportions using the ‘ratio’ method for identification of marker genes. The ‘ratio’ method refers to ranking marker genes by the ratio of the mean expression of each gene in each cell type to the mean of that gene in all other cell types. All other parameters were kept as default. Based on these estimated cell‐type proportions we defined a cell/sample as the cell type with the highest estimated proportion. To test the approach, we applied the method to the Fan et al. data and found that the cell‐type classification seemed very robust except for inhibitory (In) neurons, which included a proportion of excitatory (Ex) neurons and neural stem cells (NSC), including microglial cells (Micro) (see Box 2). Between 75 and 359 genes were used to describe each of the 14 cell types extracted from Fan et al. For the BrainSpan analysis we filtered the data to only contain cell types with an average estimated proportion larger than 10% in at least one sample group in at least one region resulting in six cell types. The top 10 marker genes used for the classification of cell types for the six most prevalent cell types are seen in Box 2.
Box 2.
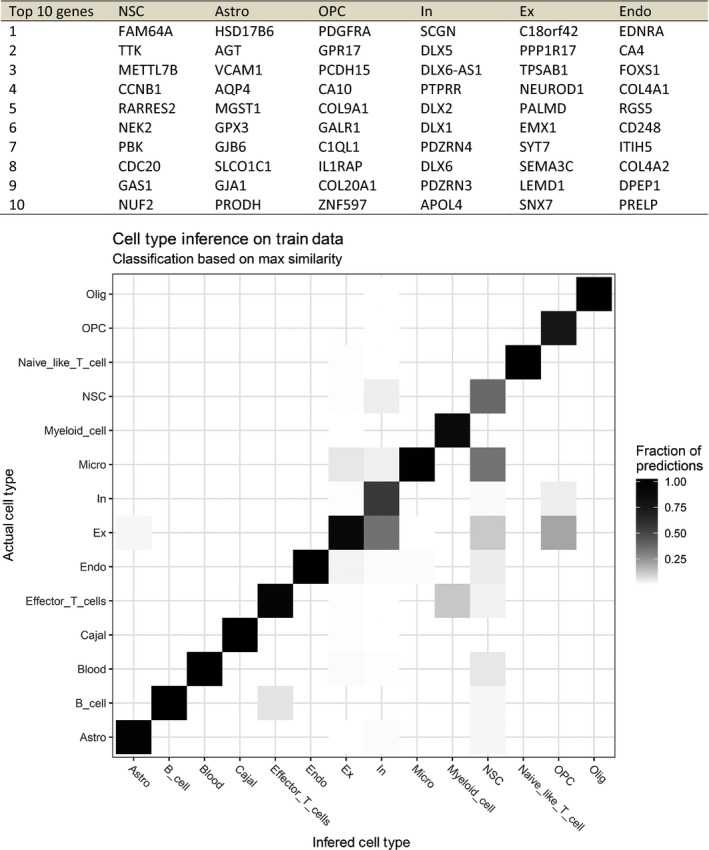
(A) Top 10 marker genes used for the classification of cell types for each of the six most prevalent BrainSpan cell types. Markers were defined based on single‐cell RNA expression data performed by Fan et al. (2018). (B) The fraction of the predicted cells (color) in each of the predicted cell type (x‐axis) compared with what they were originally classified as, in Fan et al. (y‐axis). See Material and methods for details. Astro, astrocytes; Cajal, Cajal‐Retzius cells; Endo, endothelial cells; Ex excitatory neurons; (Ex), In, inhibitory neurons; NSC, neural stem cells, OPC, oligodendrocyte progenitor cells; Olig, oligodendrocytes.
Tissue samples
Brains from 11 fetuses (70–200 mm CRL) corresponding to 12–21 wpc were examined. Eight of the fetal brains were from the Copenhagen University Human Embryonic/Fetal Biobank (humanGENESIS), Institute of Cellular and Molecular Medicine (ICMM), Faculty of Health and Medical Sciences, University of Copenhagen, Denmark. The hGENESIS collection of embryos and fetuses was obtained during more than 50 years from legal abortions after informed consent from all contributing women following oral and written information, in accordance with the Helsinki Declaration II, and was approved by the Danish Regional Committee on Health Research Ethics (KF–V.100.1735/90). Immediately following the abortion, brains were removed and promptly fixed for 12–24 h at 4 °C in either 10% neutral buffered formalin, 4% formol‐calcium, Lillie's or Bouin's fixatives. This procedure kept the time from delivery to fixation at a minimum, normally less than 2 h, in order to retain optimal tissue quality. Following 2–6 h fixation, larger fetal brains were dissected into well‐defined blocks and serial‐sectioned to provide full spatial developmental analysis (see Box 1). Smaller brains were dissected and paraffin‐embedded for coronal, sagittal and horizontal sections. In addition, three fetal brains were obtained from the general clinical biobank at the Department of Pathology, Copenhagen University Hospital, Copenhagen, Denmark, with approval from the Danish Regional Committee on Health Research Ethics (H‐17016416, project: Developing brain – a key to the adult). Brains from the clinical biobank were fixed for up to 21 days in 10% neutral buffered formalin preceded by addition of picric acid (see Box 1).
Box 1.
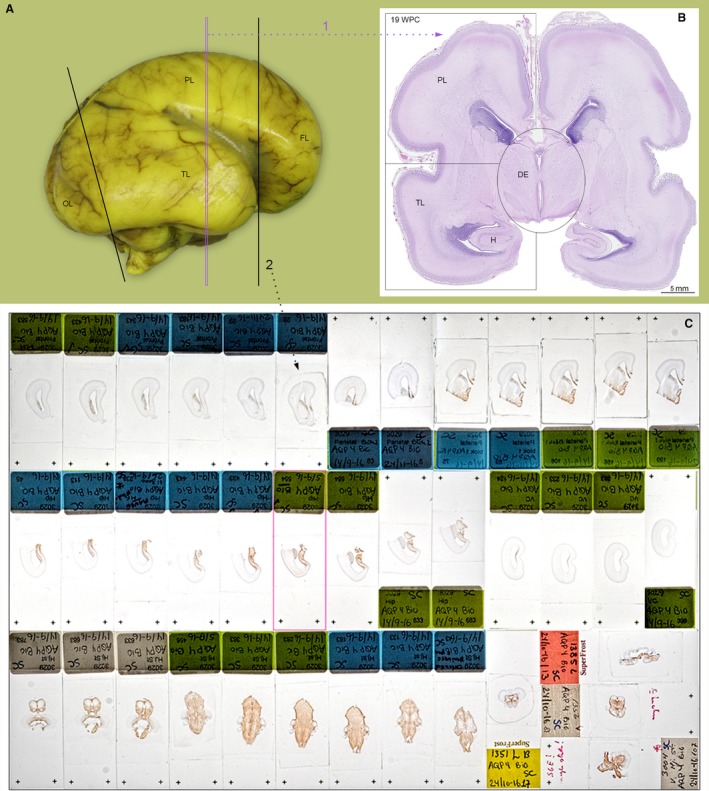
(A) A 19 wpc fetal brain from the clinical biobank fixed for 3 weeks. This part of the material was cut into 4‐mm coronal slices prior to paraffin embedding, 5‐μm‐thick microtome sectioning and staining. (B) A hematoxylin‐eosin (HE) stained section from a 5 × 5 cm paraffin block of the parieto‐temporal cortex indicated by the magenta dotted line (1) in (A). An alternative procedure (2) was used for material obtained from the hGENESIS, ICMM collection: After 2–6 hours initial fixation, the brain was dissected into frontal (FL) and occipital lobes (OL) corresponding to black lines in (A). From the remaining central tissue block, the diencephalon (DE) and brainstem was dissected out and the remaining telencephalon was subdivided into blocks containing parietal lobe (PL) and temporal lobe (TL) as indicated in (B). In the hGENESIS, ICMM collection samples were embedded for frontal sectioning apart from the brainstem, which was cut horizontally. For some fetuses, more than 4000 2‐μm‐thick serial sections are available from immediately fixed brains. Examples of hGENESIS sections stained for AQP4 are shown in (C), where the upper line of sections includes six sections from frontal lobe (dotted black line 2 in A) and seven sections from parietal lobe. The middle line shows seven sections from temporal lobe and hippocampus (H) and four from occipital lobe. The lower line depicts nine horizontal sections from the brainstem and four coronal and sagittal sections from other fetuses to demonstrate consistency in AQP4 staining patterns. FL, frontal lobe; H, hippocampus; OL, occipital lobe; PL, parietal lobe; TL, temporal lobe.
Immunohistochemistry
For single and double labeling immunohistochemical experiments, 2‐ to 10‐μm‐thick serial sections were cut in frontal, sagittal or horizontal planes, and placed on silanized glass slides.
For bright‐field light microscopy, sections were deparaffinized and rehydrated in xylene and ethanol following standard protocols. Some primary antibodies required heat‐induced antigen retrieval (for list of antibodies and retrieval systems, see Table 1). Endogenous peroxidase was quenched using a 0.5% solution of hydrogen peroxide in TRIS‐buffered saline (TBS; 5 mm Tris‐HCl, 146 mm NaCl, pH 7.6) for 15 min. Following rinses with TBS, non‐specific binding was inhibited by incubation for 30 min with blocking buffer (ChemMate antibody diluent S2022, DakoCytomation, Glostrup, Denmark) or 10% goat serum (BI‐04‐009‐1A, In Vitro) at room temperature prior to overnight incubation with primary antibodies (Table 1) at 4 °C diluted in blocking buffer and then rinsed with TBS. The REAL EnVision Detection System (peroxidase/DAB+ rabbit/mouse, code K5007, DakoCytomation, Glostrup, Denmark) was used to detect mouse and rabbit primary antibodies. The sections were washed with TBS, followed by incubation for 10 min with 3,3’‐diamino‐benzidine chromogen solution. Positive staining was recognized as a brown color. The sections were counterstained with Mayer's hematoxylin (Ampliqon Laboratory Reagents, AMPQ00253.5000), dehydrated in graded alcohols and coverslipped with Pertex mounting medium (HistoLab, 00801).
Table 1.
List of primary and secondary antibodies
| Primary antibodies | Host IgG | Dilution | HIER | Producer | Code number |
|---|---|---|---|---|---|
| AQP4 | Rabbit IgG | 1:3000–1:6000 | M6 | AVIVA | OABB01958 |
| S100 | Mouse IgG2a | 1:2000 | M6 | Abcam | ab14849 |
| S100 | Rabbit IgG | 1:250 (F), 1:3000–1:6000 | – | Dako | Z0311 |
| GFAP | Mouse IgG1k | 1:50–1:200 | TEG/M6 | Dako | M0761 |
| GFAP | Rabbit IgG | 1:10000 | TEG | Dako | Z0334 |
| YKL‐40 | Mouse IgG2kb | 1:50/1:100 | – | * | 201.F9 |
| C3d | Rabbit IgG | 1:200–1:600 | M6 | Dako | A0063 |
| Secondary antibodies | Host IgG | Dilution | Target | Producer | Code number |
| Alexa Fluor 488 | Goat IgG | 1:100 | Anti‐mouse | Jackson | 115‐545‐166 |
| Alexa Fluor 594 | Goat IgG | 1:100 | Anti‐rabbit | Fisher | T20925 |
| Dylight 594 | Goat IgG | 1:100 | Anti‐mouse | Jackson | 115‐515‐146 |
| Dylight 594 Strept. | – | 1:200 | – | Vector | SA5594 |
| Biotin‐SP | Donkey IgG | 1:200 | Anti‐rabbit | Jackson | 711‐066‐152 |
F, fluorescence; HIER, heat‐induced epitope retrieval.
TEG: 1,211 g Trizma® Base (Sigma‐Aldrich T1503) + 0.190 g ethylene glycol tetraacetic acid (EGTA) buffer (Sigma‐Aldrich E‐4378) + 1000 mL distilled water (1dw) adjusted to pH 9. M6: 21.014 g citric acid monohydrate (Merck 100244) in 1dw + 29,410 g tri‐sodium citrate dihydrate (Merck 106448) in 1dw adjusted to pH 6.
Producers: Abcam, Cambridge, UK. Aviva Systems Biology, San Diego, CA, USA. Dako, Agilent, Glostrup, Denmark. Fisher Scientific, Toronto, CA. Jackson ImmunoResearch, Ely, UK. Vector Laboratories, Peterborough, UK.
Kind gift of Prof. Julia S. Johansen, Herlev Hospital, Copenhagen, Denmark, and Paul A. Price, UCSD, USA.
For immunofluorescence, sections were prepared as for bright‐field light microscopy. For double labeling, overnight incubation at 4 °C was performed either as a mix of primary antibodies or sequentially. Secondary fluorophore‐conjugated antibody or antibodies were then added for 30 min at room temperature with labeled polymer–HRP anti‐mouse or anti‐rabbit (DakoCytomation, EnVisionTM+ System/HRP K4007 or K4003) followed by Tyramide Signal Amplification with either Alexa Fluor 488 (Invitrogen, Molecular Probes, T20912) or Alexa Fluor 594 (Invitrogen, Molecular Probes, T20925) for 7 min at room temperature. Subsequently, the sections were incubated for 30 min at room temperature with either biotin‐SP‐conjugated F(ab’)2 fragment donkey anti‐rabbit antibodies (Jackson ImmunoResearch, 711‐066‐152) followed by streptavidin‐conjugated DyLight 594 (Vector Laboratories, SA5594), or with Alexa Fluor 488 goat anti‐mouse (Jackson ImmunoResearch, 115‐545‐166) or Dylight 594 goat anti‐mouse (Jackson ImmunoResearch, 115‐515‐145). Finally, a nuclear counterstain with DAPI (4’,6‐diamidino‐2‐phenylindole; Invitrogen, Molecular Probes, D1306) was added for 3 min before sections were coverslipped.
Fluorescent full slide scans of selected double immunolabeled sections were acquired using a Zeiss Axio Scan.Z1 slide scanner with a 20×/0.8 Plan‐Apochromat objective. For laser scanning confocal microscopy, a Zeiss LSM 710 or Zeiss LSM 780 Confocal Microscope was used with 20×/0.8 Plan‐Apochromat, 20×/0.8 Plan‐Neofluar or 40×/1.3 Oil Plan Neofluar objectives. Slide scan images and confocal images were analyzed and individual optical sections exported as TIFF files using Zeiss ZEN Blue Edition. Representative images chosen for figure editing were processed in Adobe photoshop CS6.
Results
GFAP, S100, AQP4 and YKL‐40 as astroglial markers
To evaluate astrocyte‐specific expression of the four selected astroglial markers GFAP, S100, AQP4 and YKL‐40, we explored RNA‐seq data of four corresponding candidate genes [GFAP, S100B, AQP4 and CHI3L1 (YKL‐40)] on immunopanned cell type‐specific cell suspensions (www.brainrnaseq.org), generously shared by the Barres Lab (Dr. Steven Sloan). Of the S100‐specific genes, we chose S100B because the S100B protein is abundant in the brain and well characterized as a glial marker, and the S100 antibody (Z0311, Dako, Agilent, Glostrup, Denmark) used for microscopy preferably reacts with S100B protein (for antibody reactivity details see product webpage or antibody datasheet). We found that GFAP and AQP4 were both expressed primarily in mature astrocytes, S100B was seen in both astrocytes and oligodendrocytes, and CHI3L1 was almost exclusively seen in astrocytes from glioblastoma core tumor tissue. Of the four genes evaluated, AQP4 seemed to be the most highly expressed and most selective gene for mature astrocytes [differences in RNA expression between GFAP, S100B, AQP4 and CHI3L1 are all significant (all P < 0.01, pairwise Wilcoxon signed rank tests with Bonferroni correction); only differences for mature astrocytes were analyzed] (Fig. 1A). Single‐cell RNA‐seq analysis on glioblastoma tumors, available at www.gbmseq.org (Darmanis et al. 2017), validated the specific expression of AQP4 in astrocytes, and expression of S100B in both astrocytes and oligodendrocytes, but also revealed expression of GFAP and S100B in oligodendrocyte progenitor cells (OPC). All four genes were expressed to some degree in neoplastic cells (data not shown, but available at www.gbmseq.org).
Figure 1.
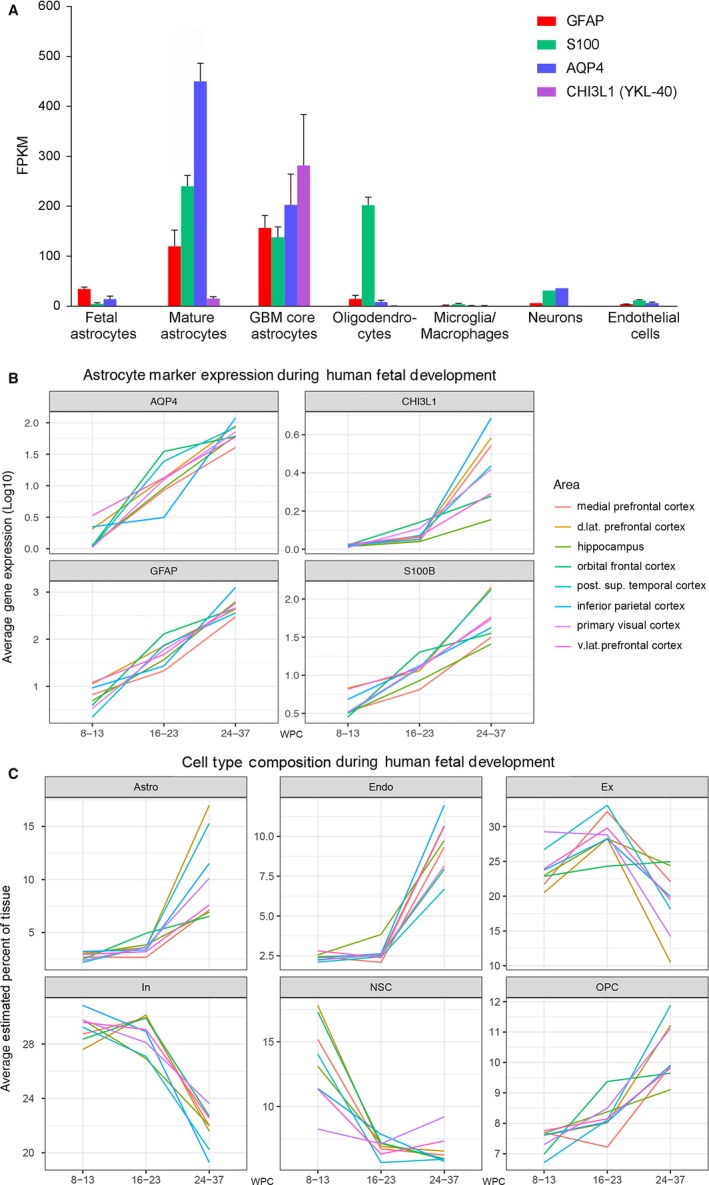
RNA expression of GFAP, S100B,AQP4 and CHI3L1‐40 in human developing and adult brain and estimation of cell‐type proportions in developing human brain. (A) RNA expression of GFAP, S100B,AQP4 and CHI3L1 (YKL‐40) in specific cell types in samples from human healthy temporal cortex (8–63 years, n = 12 for astrocytes, n = 5 for oligodendrocytes, n = 3 for microglia/macrophages, n = 1 for neurons, n = 2 for endothelial cells), astrocytes from fetal brain (16–16.5 wpc, n = 6) and astrocytes from glioblastoma tumor core (59–65 years, n = 3). Expression is listed as mean ± SEM (standard error of the mean) in FPKM. The RNA‐seq data in (A) was generously shared by the Barres Lab and is based on specific cell‐type suspensions derived by immunopanning (www.brainrnaseq.org) (Zhang et al. 2016). (B) is based a different dataset, consisting of RNA‐seq quantification of different brain regions in developing human brain and shows average gene expression (log10) (y‐axis) of GFAP, S100B, AQP4 and CHI3L1 (YKL‐40) (subplots) across all samples within three age intervals (early, mid‐ and late prenatal) (x‐axis) for selected brain regions (colors). Within the same dataset (C) shows average cell‐type proportions (y‐axis) for major cell types (subplots) within the same age groups (x‐axis) and regions (colors) as listed in (B). Specification of cell types (Box 2) is based on single‐cell RNA‐seq performed by Fan et al. (2018). RNA‐seq data for (B) and (C) originates from www.brainspan.org. For clarity, only cell types identified as making up more than 10% of cells, in at least one of the regions analyzed, are shown. Astro, astrocytes; d. lat, dorsolateral; Endo, endothelial cells; Ex, excitatory neurons; In, inhibitory neurons; NSC, neural stem cells; OPC, oligodendrocyte progenitor cells; post. sup, posterior superior; v.lat, ventrolateral.
It was surprising that none of the four candidate genes appeared to be prominently expressed in fetal astrocytes at 16–16.5 wpc in the brainrnaseq dataset (Fig. 1A), taking into account that all four genes have been described in fetal astrocytes in developing human brain (Table 2). To investigate this, we analyzed publicly available RNA‐seq gene expression in the BrainSpan Atlas of the Developing Human Brain, where we found consistent increasing expression of GFAP, S100B, AQP4 and CHI3L1 (YKL‐40) during the gestational period in seven areas of neocortex and in the hippocampal formation (Fig. 1B), with some spatiotemporal variation. Apart from in the inferior parietal cortex, AQP4 expression increased prominently from early‐ to midgestation (Fig. 1B). The same tendency was seen for GFAP and S100B.
Table 2.
First appearance of astroglial markers in radial glial cells and astrocytes in developing human brain
| Marker | Developmental stage* | Location | Developmental stages evaluated* | Location evaluated | References |
|---|---|---|---|---|---|
| GFAP | |||||
| Radial glial cells | 5.5–6 wpc (5.5–6 gw)?† | Lateral telencephalic wall | 5.5–23 wpc? (5.5–23 gw) | Lateral telencephalic wall | Zecevic (2004) |
| 6–8 wpc (8–10 gw)? | Cerebellum | 6 wpc–12 years (8 gw to 12 years)? | Cerebellum | Yachnis et al. (1993) | |
| 9 wpc (9 ow) | Fimbria, dentate gyrus, hippocampus | 5.5–20 wpc (5.5–20 ow) | Fimbria, dentate gyrus and hippocampus | Stagaard Janas et al. (1991b) | |
| 10 wpc (10 ow) | Frontoparietal cerebrum | 10–19 wpc (10–19 ow) | Frontoparietal cerebrum | Antanitus et al. (1976); Choi & Lapham (1978) | |
| 11 wpc (13 gw) | 7–38 wpc (9–40 gw) | Cortex | Middeldorp et al. (2010) | ||
| 13 wpc (15 gw) | VZ and SVZ around the third ventricle (radial glial cells?) | 5–23 wpc (7–25 gw) | Cerebrum | Sasaki et al. (1988) | |
| Astrocytes | 14 wpc (14 ow) | Fimbrio‐dentate border, hippocampus IZ | 4.5–20 wpc (4.5–20 ow) | Fimbria, dentate gyrus and hippocampus | Stagaard Janas et al. (1991a,1991b) |
| 14 wpc (16 gw) | Mainly SVZ in motor cortex | 14–22 wpc† (16–24 gw) | Motor cortex | Aquino et al. (1996) | |
| 15/17 wpc (17 wpc/gw)? | 16–40 wpc/gw? (16–40 wpc/gw) | Cortex, WM and GM | El‐Khoury et al. (2006) | ||
| 17 wpc (17 gw)? | SVZ (and few in CP) | 5.5–23 wpc? (5.5–23 gw) | Frontal and/or occipital cortex | Zecevic (2004) | |
| 18 wpc (20 gw) | IZ | 5–23 wpc (7–25 gw) | Cerebrum | Sasaki et al. (1988) | |
| Midgestation | CP | Human forebrain | Howard et al. (2008) | ||
| S100B | |||||
| Radial glial cells | 12 wpc (12 ow) | Fimbria (at 13 ow also in fornix) | 5.5–20 wpc (5.5–20 ow) | Fimbria, dentate gyrus and hippocampus | Stagaard Janas et al. (1991b) |
| 13 wpc (15 gw) | VZ and SVZ around the third ventricle (radial glial cells?) | 5–23 wpc (7–25 gw) | Cerebrum | Sasaki et al. (1988) | |
| Astrocytes | 6.5 wpc (6.5 ow) | Brainstem | 4.5–20 wpc (4.5–20 ow) | Brainstem | Stagaard Janas et al. (1991a) |
| 9.5 wpc (9.5 ow) | Fimbria, hippocampus and neocortex | 4.5–20 wpc (4.5–20 ow) | Fimbria, dentate gyrus, hippocampus and neocortex | Stagaard Janas et al. (1991a) | |
| 10 wpc (12 gw)? | All layers of hippocampus | 10–38 wpc (12–40 gw)? | Hippocampus, entorhinal cortex and occipital cortex | Tiu et al. (2000) | |
| 11 wpc (11 ow) | Fimbria, dentate gyrus and hippocampus | 5.5–20 wpc (5.5–20 ow) | Fimbria, dentate gyrus and hippocampus | Stagaard Janas et al. (1991b) | |
| 15/17 wpc (17 wpc/gw)? | 16–40 wpc/gw? (16–40 wpc/gw) | Cortex, WM and GM | El‐Khoury et al. (2006) | ||
| AQP4 | |||||
| Radial glial cells | Not found | Not found | Not found | Not found | Not found |
| Astrocytes | 12 wpc (14 gw) | End‐feet of astrocytes restricted to dorsal and ventral hippocampus | 12–38 wpc (14–40 gw) | Whole brain? | Gömöri et al. (2006) |
| 15/17 wpc (17 wpc/gw)? | GM > Cortex, WM and | 16–40 wpc/gw? (16–40 wpc/gw) | Cortex, WM and GM | El‐Khoury et al. (2006) | |
| 19–20 wpc (21–22 gw) | Neocortex | 12–38 wpc (14–40 gw) | Whole brain? | Gömöri et al. (2006) | |
| CHI3L1 (YKL‐40) | |||||
| Radial glial cells | 5–6 wpc | Few neuroepithelial cells/radial glial cells | 5–20 wpc | Forebrain | Bjørnbak et al. (2014) |
| 12 wpc | VZ (At 11 wpc only VZ in fimbria is immunoreactive for YKL‐40) | 8–20 wpc | Forebrain | Brøchner & Møllgård (2016) | |
| Astrocytes | 20 wpc | Fimbria | 5–20 wpc | Forebrain | Bjørnbak et al. (2014) |
| 15 wpc | SVZ in antihem (PSB) | 8–20 wpc | Forebrain | Brøchner & Møllgård (2016) | |
CP, cortical plate; GM, germinal matrix; gw, gestational weeks; IZ, intermediate zone; ow, ovulation weeks; wpc, weeks post conception; PSP, pallial–subpallial boundary; SVZ, subventricular zone; WM, white matter.
Approximated wpc (weeks post conception), original described developmental stage in brackets.
Two monoclonal GFAP antibodies yielded different results: monoclonal GFAP (Dako) was not expressed prior to 9–10 gw (gw in Zecevic, 2004; may be equivalent to wpc, therefore this is 9–10 wpc), whereas monoclonal GFAP (Chemicon) labeled radial glial cells at 6 gw (most probably 6 wpc).
GFAP was not detectable in prefrontal cortex before 14 wpc (16 gw) as evaluated by quantitative immunoblotting.
One problem with bulk RNA‐seq data is that observed expression changes can be due to either changes in usage of genes or to changes in cell proportions (or both). To further investigate this, we used midgestation cerebral cortex single‐cell expression and cell‐type annotation from Fan et al. (2018) to define gene expression profiles for the most common cell types. The expression profiles were then used to estimate the proportion of each of these cell types in the BrainSpan data (see methods and Box 2). Astrocytes (Astro) were estimated to constitute less than 5% of the tissue in all eight selected regions at both early‐ and midgestation, rising to more than 10% for some regions in the late prenatal period. Further, the proportion of neural stem cells (NSC) seemed to decrease rapidly from early‐ to midgestation for all regions except primary visual cortex, although it should be noted that the model was not good at predicting NSCs (Box 2). Comparing Fig. 1B and Fig. 1C it is evident that expression changes of GFAP, S100B and AQP4 are due not only to changes in cell‐type proportions (more intensive gliogenesis) but also reflect changes in usage of genes (e.g. maturation of early astrocytes). On the other hand, the observed increase in CHI3L1 expression (Fig. 1B) could be driven by an increase in the astrocytic cell population (Fig. 1C). Overall, it seems that even though the general expression of the four markers is low in the early to mid‐prenatal period, AQP4 is still able to distinguish astrocytic cells from other cell types (AQP4 has the 4th highest ratio of expression versus other cell types – see Box 2). Considering the low proposed proportion of astrocytes in the early and mid‐prenatal period and possible changes in usage of GFAP, S100 and AQP4, immunohistochemical analysis may contribute to a wider understanding of the distribution of GFAP, S100, AQP4 and CHI3L1.
Spatial pattern of GFAP and S100 immunoreactivity in the early prenatal period
To explain the described non‐conclusive evidence regarding GFAP, S100, AQP4 and YKL‐40 as astroglial markers during human brain development, we initially investigated immunoreactivity patterns of classical astrocyte markers GFAP and S100 in early prenatal period revealing diverging patterns of expression between and within regions (Fig. 2 and Fig. 3A,C).
Figure 2.
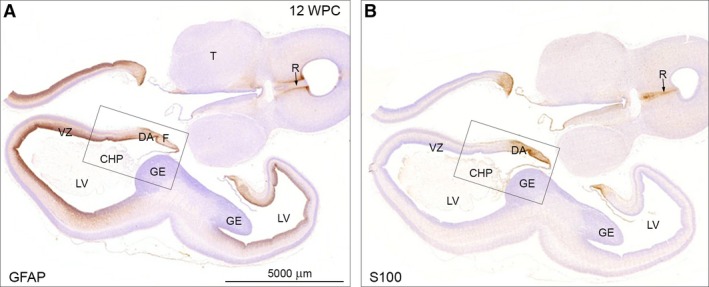
Adjacent horizontal sections of forebrain including thalamus (T) and brainstem of a 12 wpc (CRL: 74 mm) human fetal brain stained for GFAP (A) and S100 (B) shown at low magnification. (A) Note the GFAP‐positive staining of the entire VZ except the GE. Also that CHP and midline raphe (R, arrow) are negative. The framed area is shown in higher magnification in Fig. 3A. (B) The neighboring section immunostained for S100 depicts absence of S100‐reactivity in VZ outside the immunopositive DA and fimbria. The midline raphe (R) of the brainstem shows strong immunoreactivity for S100. The boxed area is shown in higher magnification in Fig. 3C together with other astroglial markers. CHP, choroid plexus of the lateral ventricle; DA, dentate anlage; F, fimbria; GE, ganglionic eminence; LV, lateral ventricle; R, raphe; T, thalamus; VZ, ventricular zone. A and B – same magnification. Scale bar: (A) 5000 μm.
Figure 3.
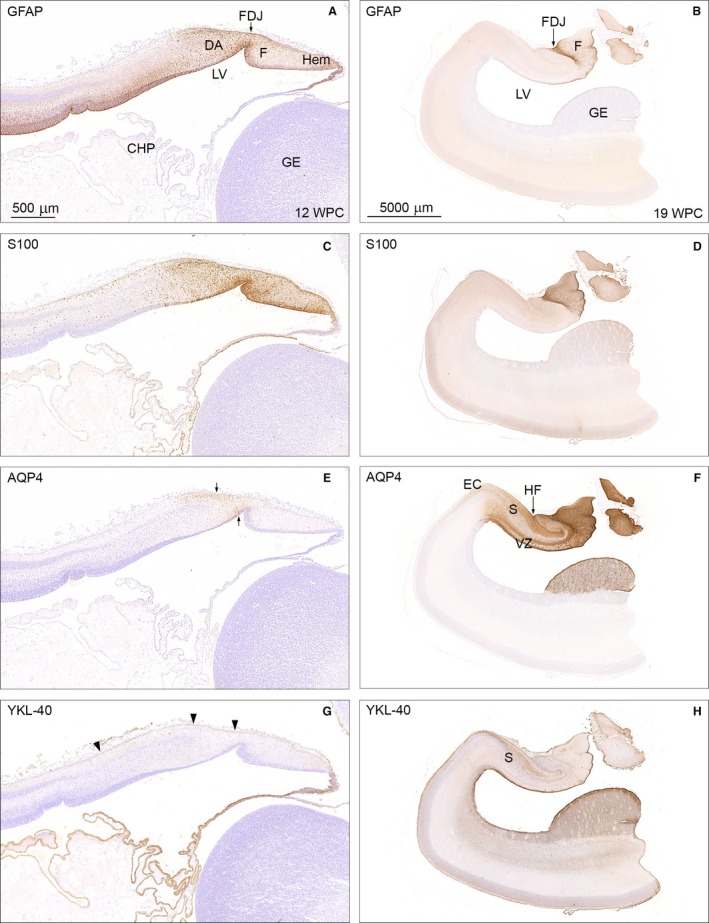
Temporal patterns of GFAP, S100, AQP4 and YKL‐40 immunoreactivity in 12 wpc (CRL: 74 mm) and 19 wpc (CRL: 165 mm) human fetal brain. Panel 1 (A,C,E,G) shows higher magnification of the framed areas in Figs 2A and B and of neighboring sections from the same 12 wpc fetus stained for AQP4 and YKL‐40. Panel 2 (B,D,F,H) illustrates the same region from a 19 wpc coronally sectioned fetus stained for the same astroglial markers: GFAP, S100, AQP4 and YKL‐40. At 12 wpc (panel 1) GFAP‐ and S100 immunoreactivity in (A) and (C) of the fimbria (F) and the dentate anlage (DA) separated by the fimbriodentate junction (FDJ) is strong but not entirely overlapping. In the marginal zone, S100‐reactivity continues into the hippocampus proper, which is devoid of GFAP staining. AQP4 and YKL‐40 immunoreactivity in (E) and (G) depicts the upcoming AQP4‐positive radial glial fibers in the DA between small arrows in (E) and the YKL‐40‐positive radial glial end feet (arrowheads) in (G). At 19 wpc the FDJ is strongly immunoreactive for GFAP (B), S100 (D) and AQP4 (F) but not for YKL‐40 (H). CHP, choroid plexus; DA, dentate anlage; EC, entorhinal cortex; F, fimbria; FDJ, fimbriodentate junction; GE, ganglionic eminence; Hem, hem; HF, hippocampal fissure; LV, lateral ventricle; S, subiculum; VZ, ventricular zone. Panel 1 (A, C, E and G) – same magnification, scale bar in (A): 500 μm. Panel 2 (B,D,F,H) – same magnification. Scale bar: (B) 5000 μm.
GFAP occupies the ventricular zone of the telencephalic wall (excluding the ganglionic eminence) and outlines the midline raphe in the brainstem (cf. Fig. 9B in Stagaard & Møllgård, 1989) with immunopositive radial glial cells (Fig. 2A), whereas S100 is not represented in these regions but instead reveals immunoreactivity in the choroid plexus and midline raphe itself (Fig. 2B). Both astroglial markers are present but not entirely overlapping in the hippocampal formation, a region of pronounced gliogenesis. GFAP and S100 show strong staining of fimbria and the dentate anlage (DA), but S100 immunostaining is most prominent in fimbria (Fig. 3C), and whereas S100 alone extends into the hippocampus proper in the marginal zone (Fig. 3C), GFAP continues into the hippocampus proper in the ventricular zone (Fig. 3A).
Temporal pattern of GFAP, S100, AQP4 and YKL‐40 from early prenatal period to midgestation
Establishing the presence of GFAP and S100 in the hippocampal formation in developing human brain, we decided to compare patterns of GFAP, S100, AQP4 and YKL‐40 from early prenatal period to midgestation in this region and found very little immunoreactivity of AQP4 and YKL‐40 (Fig. 3E and G) at 12 wpc, which is in accordance with the BrainSpan dataset (Fig. 1B). AQP4 is seen in radial glial fibers in DA (Fig. 3E) and YKL‐40 vaguely stains radial glial end feet (Fig. 3G) by 12 wpc.
At 19 wpc GFAP, S100 and AQP4 all intensely stain the fimbriodentate junction, and are present in the fimbria in varying degree (Fig. 3B,D,F). YKL‐40 has a very different pattern of immunoreactivity, staining the marginal zone in subiculum, entorhinal cortex and ordinary neocortex, the entire ventricular zone and ganglionic eminence (GE) including the subventricular zone (Fig. 3H). The most pronounced immunoreactivity at 19 wpc is observed following AQP4 staining (Fig. 3F), which apart from fimbria and FDJ also seen with GFAP and S100, further covers the region around the hippocampal fissure, the marginal zone of subiculum terminating at the entorhinal cortex and the GE. Although AQP4 and YKL‐40 are both present in the GE, AQP4 immunoreactivity is more prevalent, creating a sponge‐like appearance that is different from the crackled look of YKL‐40.
Regional and cellular AQP4 immunoreactivity at midgestation
As RNA expression data indicates, AQP4 is highly specific to the astroglial lineage (Fig. 1) and it seems to be the most widespread of the four markers in the hippocampus at midgestation (Fig. 3B,D,F,H). We therefor decided to investigate the spatial staining pattern of AQP4 in different regions of the cortical wall by analyzing hundreds of immunostained sections from frontal, parietal, temporal and visual cortical regions (see Box 1).
At midgestation, AQP4 immunoreactivity depicts a complicated system of water channels, creating a continuous brain‐wide water transport system from the hippocampal formation (Fig. 4I) following the rostral hippocampal extension via fimbria and fornix (Fig. 4H) along indusium griseum (Fig. 4H,G,F,E) to taenia tecta in the medial frontal cortex (Fig. 4D). The ventricular zone of the middle part of the medial wall displays uninterrupted AQP4 immunostaining (Fig. 4A‐P) from frontal cortical wall to the occipital lobe, where staining is barely visible (Fig. 4P), in contrast to that of the more rostral part of the visual cortical wall opposite the calcarine sulcus (Fig. 4M).
Figure 4.
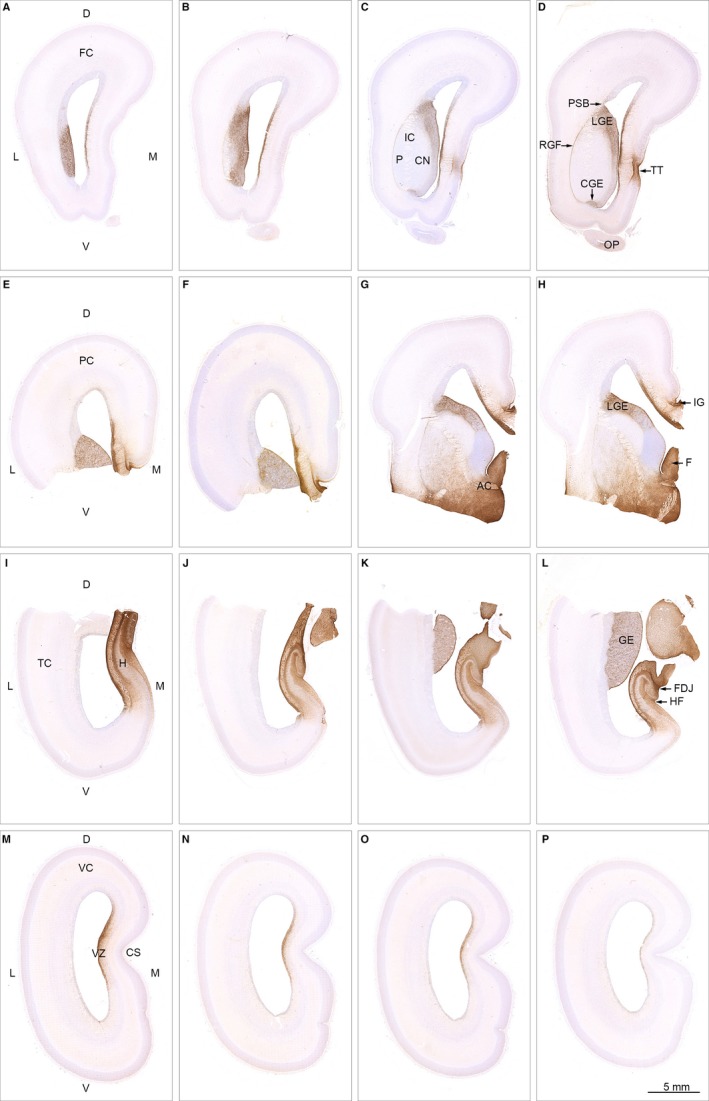
Spatial distribution of AQP4 in 19 wpc (CRL: 165 mm) human fetal brain. Sixteen sections immunostained for AQP4 from a series of more than 4000 coronal sections through the entire brain of a 19 wpc human fetus and taken from the material shown in Box 1 C (A–D from frontal lobe; E‐H from parietal lobe; I‐L from temporal lobe; M‐P from occipital lobe). At 19 wpc, APQ4 immunoreactivity delineates a continuous water transport system from the hippocampal formation (H) in (I) to taenia tecta (TT) in the medial frontal cortex (D) and via the basal forebrain (G,H) to the ganglionic eminence. The ventricular zone (VZ in M) of the medial wall also shows a continuous labeling from frontal to occipital lobe. The olfactory peduncle (OP in D) is weakly stained at this stage. Orientation: D, dorsal; L, lateral; M, medial; V, ventral. The entire cortical wall: FC, frontal cortex; PC, parietal cortex; TC, temporal cortex; VC, visual cortex. Subregions: AC, anterior commissure; CGE, caudal ganglionic eminence; CN, caudate nucleus; CS, calcarine sulcus; FDJ, fibriodentate junction; GE, ganglionic eminence; RGF, radial glia fiber fascicle; H, hippocampus; HF, hippocampal fissure; IC, internal capsule; IG, indusium griseum; LGE, lateral ganglionic eminence; OP, olfactory peduncle; PSB, pallial–subpallial boundary; TT, taenia tecta; VZ, ventricular zone. A‐P: same magnification. Scale bar: (P) 5 mm.
A continuous AQP4 immunoreactivity also labels the entire ganglionic eminence along the lateral cortical wall (Fig. 4A‐H,K,L) and AQP4‐positive reactivity is seen in the radial glia fiber fascicle (described by González‐Arnay et al. 2017), which extends from the pallial–subpallial boundary to the caudal ganglionic eminence along the deep surface of putamen, thus engaging putamen, internal capsule and head of the caudate nucleus (Fig. 4D).
To characterize the sponge‐like appearance of AQP4 in the ganglionic eminence, we used confocal laser scanning microscopy to evaluate AQP4 at the cellular level. GE is characterized by an uneven distribution of AQP4‐positive radial glial fibers resulting in a compartmentalized structure characterized by an intricate system resembling tunnels with thin and thick walls. Regularly spaced thin radial glial fibers with evenly spaced varicosities (Fig. 5B1) characterize the tortuous tunnels, whereas the matrix of the sponge‐like structure possesses a more dense network of thick, rather coarse radial glial fibers with larger and more intensely stained varicosities (Fig. 5B2). In both fiber‐sparse and fiber‐dense areas, radial glial fibers ensheathe blood vessels with astroglial end feet.
Figure 5.
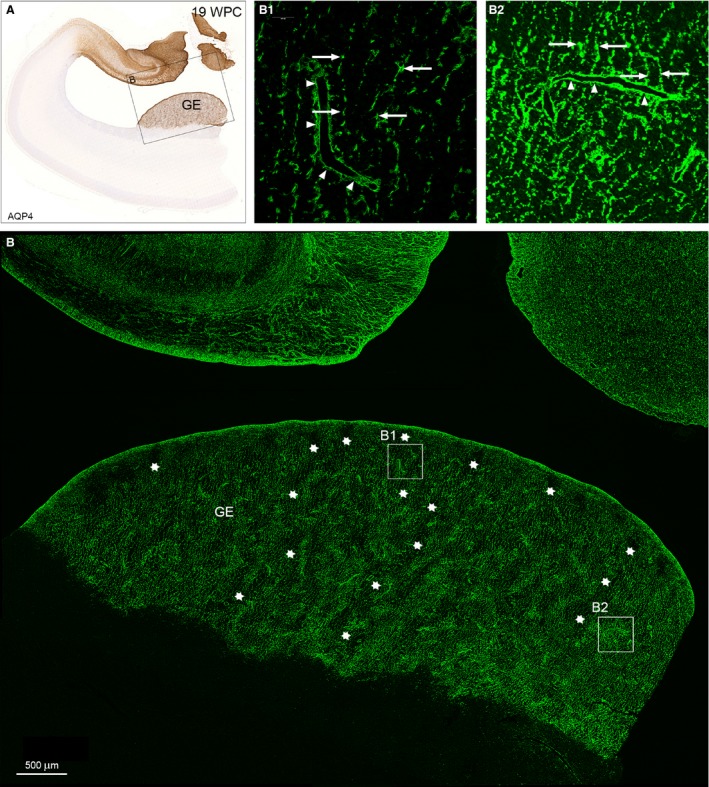
Distribution of AQP4 in the ganglionic eminence from a 19 wpc human fetal brain. A neighboring section to that of hippocampus and adjacent temporal cortex in Fig. 4K immunostained for AQP4 for bright‐field light microscopy (A) and confocal laser scanning microscopy (B). The framed area in (A) includes the ganglionic eminence (GE) in the lateral temporal wall and part of the strongly reacting hippocampal formation facing the lateral ventricle in the medial wall, which will be dealt with in Fig. 6. The GE in (B) is characterized by an uneven distribution of AQP4‐positive radial glial fibers resulting in a sponge‐like compartmentalized structure characterized by an intricate system resembling tunnels with thin and thick walls. The interior of some of the worm‐like tortuous tunnels is indicated by rows of white asterisks and shown in higher magnification in (B1). The more dense structure of the walls is depicted in (B2). (B1) Regularly spaced thin radial glial fibers with evenly spaced varicosities (arrows) characterize the tortuous tunnels of the AQP4‐positive ganglionic eminence. Side branches of the radial glial fibers ensheathe blood vessels (arrowheads) as astroglial end feet, although they are still part of the radial glial fiber system. (B2) The matrix of the sponge‐like structure possesses a more dense network of thick, rather coarse radial glial fibers with larger and more intensively stained varicosities (arrows), many of which seem to terminate on blood vessels as astroglial end feet (arrowheads). GE, ganglionic eminence. Scale bar: (B) 500 μm.
GFAP, S100 and AQP4 immunostaining in hippocampus and ganglionic eminence at midgestation
To further explore the distribution of AQP4 in the hippocampus at midgestation vs. GFAP and S100 we performed whole‐slide fluorescent scanning (Fig. 6), which does not provide sufficient resolution to investigate markers at the cellular level, but does reveal differences in patterning of the three markers. Combining bright‐field light microscopy (Fig. 3), laser scanning confocal microscopy (Fig. 5) and whole‐slide fluorescent scanning (Fig. 6) leads to the following observations.
Figure 6.
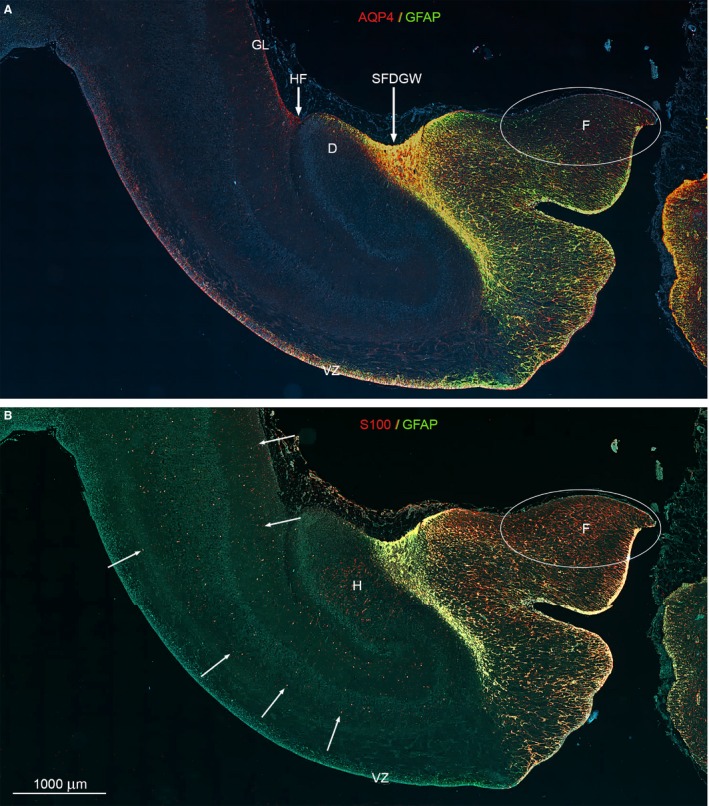
Whole‐slide fluorescent scanning of hippocampal formation from 19 wpc human fetal brain. Two immunofluorescent consecutive sections double‐labeled with antibodies against AQP4 and GFAP (A) and S100 and GFAP (B). Both sections are from the hippocampal formation adjacent to the bright‐field AQP4‐immunostained section shown in Fig. 5. (A, B) Include the entire wall of the hippocampal formation with the subpial fimbriodentate gliogenic wedge (SFDGW) region at the fimbriodentate junction and subiculum in the upper left corner – compare with Fig. 3F. The double‐labeled sections with DAPI‐stained nuclei shown in blue were subjected to whole‐slide fluorescent scanning. Radial glial cells in the ventricular zone (VZ) of the entire HF including the most distal part of fimbria is strongly positive for AQP4 as shown by the red and yellow color in (A). Within the cerebral wall, a diffuse fine dotted reactivity for AQP4 (red) can be distinguished in (A) (see also Fig. 3F) as opposed to the evenly distributed marked cellular S100 reactivity (red) indicated with white arrows in (B). The ventricular zone and the underlying future alveus are not labeled but there is an accumulation of S100‐positive cells in the hilus (H) of the dentate (D). (A, B) The subpial fimbriodentate gliogenic wedge (SFDGW) exhibits very strong immunoreactivity for the astroglial markers AQP4, GFAP and S100 and is mainly yellow with overlap, but also shows specific red (AQP4) patches in (A) and green (GFAP) patches in (B). D, dentate; F, fimbria; GL, glia limitans; H, hilus; HF, hippocampal fissure; SFDGW, subpial fimbriodentate gliogenic wedge; VZ, ventricular zone. (A–B) – same magnification. Scale bar: (B) 1000 μm.
The inner surface
Radial glial cells in the ventricular zone (VZ) of the entire hippocampal formation including the most distal part of fimbria is strongly positive for AQP4 (Fig. 6A), whereas radial glial cells in VZ stained for GFAP are confined to the hippocampus per se, i.e. do not react corresponding to subiculum (Figs 3B and 6A,B). In the fimbrial ventricular zone, all four markers are immunopositive, and S100 VZ immunoreactivity is confined to this region (Figs 3B,D,F,H and 6).
Cerebral wall and fimbria
Within the cerebral wall, a diffuse fine dotted reactivity characterizes AQP4 (Fig. 6A), whereas S100 reveals evenly distributed marked cellular reactivity (Fig. 6B). There is an accumulation of S100‐positive cells in the hilus of the dentate (Fig. 6B), but no reaction in VZ and the future alveus. Within the oval region outlined by a white ellipse in the upper part of fimbria in Fig. 6A,B there is weak reactivity for AQP4 (Fig. 6A) and virtually no reactivity for GFAP (Fig. 6A,B), in marked contrast to the strong S100 immunoreactivity of radial glia and of more differentiated cells (described in the literature as astrocytes) (Fig. 6B). S100‐positive cells span the cortical wall from the lateral ventricle to the outer surface (Fig. 6B), indicating the presence of an S100‐positive radial glial cell population as shown previously (Stagaard Janas et al. 1991a,1991b).
The outer surface
AQP4‐positive astrocytes extending from a cellular aggregation in the hippocampal fissure lay the foundation for the developing glia limitans (Fig. 6A). This glial cell accumulation is not immunoreactive for GFAP or S100 (Fig. 6). The subpial fimbriodentate gliogenic wedge (SFDGW) exhibits very strong immunoreactivity for the astroglial markers AQP4, GFAP and S100 with partial overlap (Fig. 6).
As revealed above, although partly overlapping, each of the four astroglial markers has its own characteristic distribution within the hippocampal formation.
Astrocytes in the subpial fimbriodentate gliogenic wedge immunostained with the functional marker C3d
GFAP, S100, AQP4 and YKL‐40 have all been associated with gliomas (see Fig. 1A; Kimura et al. 1986; Sawada et al. 2007; Qin et al. 2017; Gao et al. 2018) and reactive astrocytes (Mrak & Griffinbc, 2001; Sawada et al. 2007; Bonneh‐Barkay et al. 2010; Liddelow & Barres, 2017) and recent evidence suggests molecular similarities between fetal‐ and glioblastoma‐associated astrocytes (Zhang et al. 2016). We therefore decided to explore hippocampal distribution of complement factor C3 (C3d), a new player in brain development (Coulthard et al. 2018) and a key marker of A1‐reactive astrocytes (Liddelow et al. 2017; Clarke et al. 2018). We found accumulation of C3d‐immunoreactive irregular plump‐looking astrocytes with abundant cytoplasm and short processes in the subpial fimbriodentate gliogenic wedge bordering the outer surface of the fimbriodentate region of hippocampus (Fig. 7B). Double staining for GFAP and C3d (Fig. 7E) and S100 and C3d (Fig. 7H) shows strong overlap of immunoreactive astrocytes. In the middle of the neocortical wall, more mature S100‐positive astrocytes do not display C3d immunoreactivity (Fig. 7H’).
Figure 7.
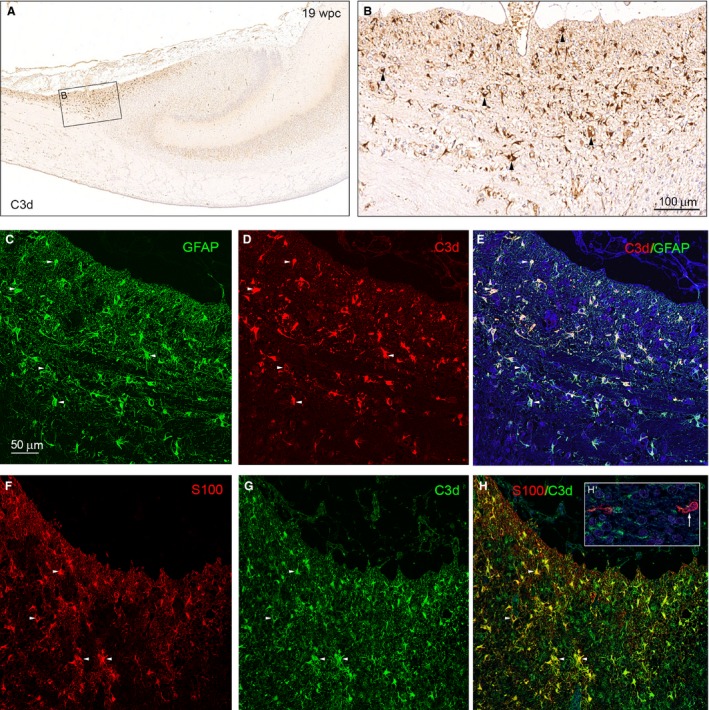
C3d‐immunoreactive astrocytes of the subpial fimbriodentate gliogenic wedge. Bright‐field (A, B) and double‐immunofluorescence microscopy (C–H) of adjacent coronal sections of hippocampus from the subpial fimbriodentate gliogenic wedge described in Fig. 6 immunostained with antibodies against complement C3d (A, B), C3d and GFAP(C–E) and C3d and S100 (F–H). (A) Shows an accumulation of C3d‐immunoreactive cells in the subpial fimbriodentate gliogenic wedge. The boxed area shown in higher magnification in (B), demonstrates numerous C3d‐positive astrocytes (arrowheads). Double staining for GFAP (green) and C3d (red) in (C) and (D) shows a strong overlap of immunoreactive astrocytes in (E). Individual cells are labeled with 5 small white arrowheads. Double staining for S100 (red, F) and C3d (green, G) also shows a strong overlap of immunoreactive astrocytes (yellow) in (H). Four individual cells are labeled with small arrowheads demonstrating that early developing S100‐positive astrocytes possess C3d. The insert in (H) labeled (H’) depicts two more mature S100‐positive but C3d‐negative astrocytes (one indicated by small white arrow) from the middle of the neocortical wall. Scale bar in (B): 100 μm. (C‐H) Same magnification. Scale bar: (C) 50 μm.
Immunoreactivity patterns of GFAP, S100, AQP4 and YKL‐40 in the brainstem and cerebellum at midgestation
To explore staining patterns for all four astroglial markers in other gliogenic areas and among different groups of radial glial cells, we compared immunohistochemical expression patterns of GFAP, S100, AQP4 and YKL‐40 in cerebellum and the brainstem.
As expected, there were marked differences in staining patterns. GFAP, S100, AQP4 and YKL‐40 show an overlapping pattern of immunoreactivity in the dorsal subpial gliogenic region extending from the tectum mesencephali towards the cerebello‐mesencephalic junction (Fig. 8A‐D), but whole‐slide fluorescent scanning microscopy reveals separate although partly overlapping cell populations (Fig. 8A‐D), with a stream of YKL‐40 and AQP4‐positive cells migrating from the gliogenic hook between the dorsal brainstem and rostral cerebellum towards the center of cerebellum (Fig. 8 AB,CD).
Figure 8.
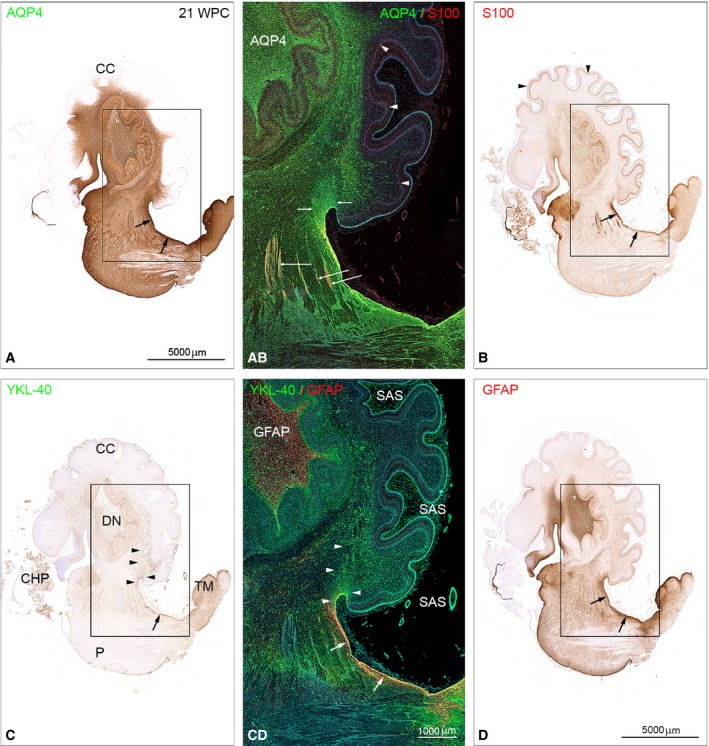
Distribution of AQP4, S100 and YKL‐40, GFAP in brainstem and cerebellum from a 21 wpc (CRL: 200 mm) human fetal brain. Adjacent sagittal sections immunostained with antibodies against AQP4, S100 (A,B and AB) and YKL‐40, GFAP (C,D and CD). Note the marked differences in staining patterns with the entire brainstem and the core of cerebellum showing strong AQP4 immunoreactivity in contrast to a lack of staining of the cerebellar cortex (CC) in (A), whereas Bergman glia in cortex (arrowheads) in (B) exhibits a pronounced staining for S100. (C) A stream of YKL‐40‐positive cells migrates from the gliogenic hook between the dorsal brainstem and rostral cerebellum towards the center of cerebellum (arrowheads). All four astroglial markers show an overlapping pattern of immunoreactivity in the dorsal subpial gliogenic region (small black arrows) extending from the tectum mesencephali (TM in C) towards the cerebello‐mesencephalic junction. The boxed areas in (A‐D) are shown in (AB) and (CD) as double‐immunofluorescent sections stained with the same four antibodies and with nuclei labeled with DAPI shown in blue. The double‐labeled sections were subjected to whole‐slide fluorescent scanning and representative panels are displayed. (AB) and (CD) clearly demonstrate that GFAP‐, S100‐, AQP4‐ and YKL40‐positive cells represent separate although partly overlapping cell populations in the dorsal subpial gliogenic region (white arrows in CD). The radial glial cells of cerebellum (Bergmann glia) are strongly positively stained for S100 (arrowheads in AB and B) but not for the other astroglial markers. Cell aggregations seen along fiber tracts connecting cerebellum and upper brainstem seem to be distinctly immunoreactive for S100 (long white arrows in AB). The fiber tracts are surrounded by AQP4‐positive small cells (green). The migrating cell population from the dorsal subpial gliogenic region that is positively stained for YKL‐40 in (CD) (marked with white arrowheads) seems to be immunoreactive also for AQP4 (small white arrows in AB). Note the strongly GFAP‐immunoreactive core of the dentate nucleus (DN) in (CD) and a similar reactivity for AQP4 in AB. In the subarachnoid space (SAS) the numerous small leptomeningeal cells (green) and the smooth muscle cells (green) in small arteries are also YKL‐40‐positive. CC, cerebellar cortex; CHP, choroid plexus of the 4th ventricle; DN, dentate nucleus; P, pons; SAS, subarachnoid space; TM, tectum mesencephali. (A‐D) Same magnification. Scale bars: (A) 5000 μm; (AB,CD) same magnification; (CD) 1000 μm.
The entire brainstem and the core of cerebellum show strong AQP4 immunoreactivity (Fig. 8A,AB), but lack of staining of the cerebellar cortex. Radial glial cells of cerebellum (Bergmann glia) which have their nuclei between the prospective white matter and the Purkinje cell layer at midgestation (corresponding to bRG in developing neocortex) are strongly positively stained for S100 (Fig. 8B) but not for the other astroglial markers (Fig. 8). Cell aggregations along fiber tracts connecting cerebellum and upper brainstem are distinctly immunoreactive for S100 (Fig. 8AB,B). These cell clusters are also positively stained for OSP (oligocyte‐specific protein) (not shown) indicating that some S100 cell populations might belong to an oligodendroglial rather than an astroglial cell lineage. The fiber tracts are surrounded by AQP4‐positive small cells.
Choroid plexus (CHP) epithelial cells are immunoreactive for both S100 and YKL‐40 (Fig. 8B,C) although double‐immunofluorescence microscopy at the cellular level shows staining of different epithelial cell segments (not shown).
Discussion
With increased awareness of the importance and complexity of the astroglial lineage, it is necessary to reevaluate existing definitions and descriptions of this heterogeneous population of cells in the developing human brain. Although use of GFAP and S100 as astrocytic markers has many limitations, including inter‐ and intraregional variability of expression in healthy CNS astrocytes, dynamic regulation by other signaling molecules and expression in radial glial cells and other cell types (Sofroniew & Vinters, 2010), they are still frequently used to define astrocytes. AQP4 is now gaining impact as an astrocyte marker, but has not been thoroughly investigated in developing human brain.
Use of markers in description of astrogliogenesis
Transcriptomic analyses of purified cell populations and single cells
Single cell technology and creation of purified cell populations are major steps towards defining cell types and their function in the CNS. It has been very difficult to purify human resting astrocytes from adult human brain and little is known about the maturation process of human astrocytes. Zhang et al. (2016) used an immunopanning‐based technique with an antibody against HepaCAM to generate purified (>95%) cultures of primary adult, juvenile and fetal (15‐18 wpc) human astrocytes followed by transcriptome profiling. Fetal astrocytes were described as highly proliferative, whereas adult/juvenile astrocytes were post‐mitotic, similar to OPCs and mature oligodendrocytes. This study is groundbreaking and seminal for the understanding of human fetal and mature astrocyte function.
Zhang and co‐workers found CHI3L1 (YKL‐40) to be the 10th most differentially expressed gene between fetal (immature astrocytes) and mature astrocytes, with a 172‐fold increase in mature astrocytes and very low expression in fetal astrocytes. We have previously found YKL‐40 protein expression in subgroups of possible astroglial progenitors and fetal astrocytes (Bjørnbak et al. 2014; Brøchner & Møllgård, 2016). Based on the transcriptome profiling (http://www.brainrnaseq.org) performed by Zhang et al. (2016) we did not find noteworthy expression of any of the four markers in fetal astrocytes, which is in contradiction to our immunohistochemical findings of GFAP, S100, AQP4 and YKL‐40 in astrocytes in hippocampus at midgestation. These discrepancies could be a result of RNA to protein differences, restriction of marker sensitivity (Hepa‐CAM, GFAP, S100, AQP4, YKL‐40), interregional and temporal heterogeneity and/or different definitions of astrocyte precursors/astroglial progenitors/fetal astrocytes. It is also very likely that there are several stages of astrocyte maturation. Zhang et al. reported that a substantial number of genes are differentially expressed in fetal and mature astrocytes; among others, fetal astrocytes express high levels of proliferative genes and gap junction genes are only expressed by mature astrocytes. When the results of Zhang et al. are compared with the finding of local highly proliferative astrocytic cells with gap junctions morphologically resembling mature astrocytes in postnatal transgenic mouse cortex (Ge et al. 2012), it seems there may be unknown maturation steps or subgroups on the path between the described fetal and mature astrocytes.
Recently, several single‐cell RNA analyses have described different cell types in the developing and adult human brain (Darmanis et al. 2017; Nowakowski et al. 2017; Fan et al. 2018), providing detailed descriptions of specific cell types. Nowakowski and colleagues pursued spatiotemporal single‐cell RNA expression in developing ventral (medial ganglionic eminence) and dorsal telencephalon (cerebral cortex) to identify restricted trajectories of neuronal differentiation; they found several different expression patterns in radial glial cells displaying topographical, typological and temporal hierarchies. The hypothesis that expression profiles of the astroglial lineage differ between regions, within single regions and with age, corresponds well with our findings and emphasizes that these factors should be evaluated when using cell‐type descriptions based on single‐cell transcriptomics.
GFAP, S100, AQP4 and YKL‐40 as astroglial markers
GFAP, S100, AQP4 and YKL‐40 have all previously been described in astrocytes in developing human brain in different settings (see Table 2). Our results established the presence of all four markers in partly overlapping subpopulations of radial glial cells and astrocytes, although each marker also had unique immunoreactivity profiles, such as S100 in Bergmann glia cell bodies in cerebellum and AQP4 in the ganglionic eminence. GFAP, S100, AQP4 and YKL‐40 also revealed more subtle intraregional cellular diversity. We suggest that AQP4 immunopositive radial glial cells are destined for the astroglial lineage, and that AQP4 is the most comprehensive and specific astrocyte marker in developing human midgestation brain compared to GFAP, S100 and YKL‐40, based on both RNA expression data and immunohistochemistry. Assuming that AQP4 is specific to the astroglial lineage, our results clearly show that GFAP should no longer be used as a general astroglial marker in brain development, since it overlooks several populations of AQP4‐positive astrocytes and radial glial cells. In addition, GFAP is not restricted to the astroglial lineage, since GFAP also characterizes radial glial cells and adult stem cells with neurogenic potential (Silver & Steindler, 2009). Nevertheless, GFAP could still be relevant under specific conditions accounting for topographical, typological and temporal differences in expression patterns. S100 is important in oligodendrocyte differentiation (Deloulme et al. 2004) and we found possible S100‐positive oligodendrocyte precursors along fiber tracts in the brainstem. Apart from combined SSEA‐4 and YKL‐40 staining in astroglial progenitors and astrocytes (Brøchner & Møllgård, 2016), YKL‐40 has been found in pericytes, leptomeningeal cells and choroid plexus epithelium in developing human brain (Bjørnbak et al. 2014) and in neurons in adult human brain (results not shown), and has a staining pattern very different from that of other astroglial markers. Since neither S100 nor YKL‐40 are specific to the astroglial lineage, but still seem to describe distinct subpopulations of astroglial cells, they should, as with GFAP, be used with caution when describing astroglial cells in a developmental setting.
Although AQP4 antibodies do not stain all fetal astrocytes and only subpopulations of radial glial cells, AQP4 may be the best single marker so far describing astrogliogenesis at midgestation. Our results, however, generally underscore that it is not possible to describe the astroglial lineage with one pan‐astrocytic marker in developing human brain.
Establishment of an AQP4 brain‐wide water transport system in cerebral cortex at midgestation
Prior to the first description of a glymphatic system, two studies from 2006 examined the distribution of AQP4 in the developing human brain. El‐Khoury and co‐workers (2006) compared astrocytic end feet coverage of blood vessels in the germinal matrix in the thalamostriate groove by GFAP, AQP4 and S100B immunoreactivity with that of blood vessels in the frontal cortex and frontal white matter in human fetuses of 16–40 wpc. They found no differences in AQP4 reactivity between the three regions. Gömöri et al. (2006) described AQP4 immunolabeling in cerebral cortex of fetuses aged 14–40 gestational weeks (gw). AQP4 was detected first in the dorsal hippocampus (14 gw), later in astrocytes in the marginal zone facing the pial surface (22 gw) and in the developing ependymal lining (24 gw). These authors also showed, but did not comment on, a strong reactivity in the fimbrio‐dentate junction and indusium griseum (Figs 2A and 4E in Gömöri et al. 2006). These findings are supported by the present study. After the proposal of a glymphatic system, Fallier‐Becker and co‐workers (2014) described the onset of AQP4 expression in the developing mouse brain. Their well‐documented immunohistochemical findings including the description of the initial staining of radial glial cells, are consistent with our results from developing human brain.
In the glymphatic clearance system (Iliff et al. 2012; Xie et al. 2013) AQP4 is a key constituent which controls water homeostasis and movement in the CNS. In the adult brain, the glymphatic system contributes to keeping extracellular ionic concentrations and fluid levels efficiently regulated and to the removal of various metabolites and neurotoxic molecules by facilitating the circulation of cerebrospinal fluid (CSF) through a brain‐wide network of paravascular pathways supplemented by subpial and subependymal routes (for recent reviews see Abbott et al. 2018; Plog & Nedergaard, 2018). So far there is no published evidence about the initial establishment and further development of a glymphatic system in the fetal brain characterized by moderate secretion and slow turnover of CSF with a remarkable high plasma protein concentration (Saunders et al. 2018) and changing human brain barrier mechanisms (Møllgård et al. 2017). Furthermore, the maintenance of a stable ionic composition in fetal brain interstitial fluid is only beginning to be studied (for recent review, see Saunders et al. 2018).
In the present study, we evaluated when the key constituent of the glymphatic system, AQP4, is first present at the regional level in the human developing cerebral cortex, whether associated with radial glia, astrocyte progenitor cells or early astrocytes. In the cerebral cortex at midgestation we discovered and described the appearance of a continuous complicated AQP4‐positive brain‐wide water transport system, which continued via the diencephalon down into the core of the brainstem as shown in Box 1. An AQP4‐positive ependyma and subependymal layer covered the inside of the corpus callosum and could be followed in all slides from parietal cortex. Although the final transition from a subpial radial glia end feet layer to a fully developed glia limitans is reported first to take place between 25–28 wpc (Kadhim et al. 1988), we demonstrated that already at 19 wpc a stream of AQP4‐positive but GFAP‐negative astrocytes migrated from the hippocampal fissure towards the entorhinal cortex to form the initial outer pial‐facing layer of an early developing glia limitans.
The exceptionally strong and early AQP4 immunolabeling of the hippocampal formation at an earlier time point than the rest of the brain indicates a need for a particularly well‐developed glymphatic function, e.g. supplying nutrients via CSF for cell proliferation or myelin formation, keeping extracellular ionic concentrations and fluid levels efficiently regulated to allow early synaptic function. Actually, an early myelination in the hippocampal formation already from midgestation, visualized by myelin basic protein immunoreactivity (Abraham et al. 2010), and a very early onset of synapse formation in hippocampal archicortex around 16 gw (Kostovic et al. 1989) might explain (at least some of) the needs for an early functional glymphatic system in this region.
Different classes and subtypes of radial glial cells at midgestation
At midgestation the radial glia scaffold in the developing human cerebral cortex has become discontinuous, causing a transformation of classical radial glial cells to (1) apical truncated (or just truncated) radial glial cells in the ventricular zone, with distal processes terminating on blood vessels in outer subventricular zone, and (2) basal radial glial cells (bRGs)/outer radial glial cells (oRGs) (Nowakowski et al. 2016; for further references, see Introduction).
We consider the AQP4‐positive sponge‐like radial glial cell complex in ganglionic eminences to consist of apical truncated radial glial cells. At this stage of cortical development it is conceivable that this radial glial cell subtype is committed to an astroglial cell lineage that will differentiate to ependymal cells or to protoplasmic or fibrillary astrocytes in the region from the ventricular surface to cortical plate. The numerous tortuous tunnels penetrating the ‘sponge’ are occupied by AQP4‐negative but vimentin‐positive radial glial subtypes (not shown) that could be involved in interneuron or oligodendrocyte production.
Subpial gliogenic, neurogenic and mixed glio‐neurogenic proliferative regions in the marginal zone exemplified in this study by the fimbriodentate junction and hippocampal fissure might well represent dense accumulations of basal radial glial cells derived from an early disintegration of an archicortical radial glia scaffold. Subpopulations of basal radial glial cells are probably not lineage‐specified from the beginning but give rise to local neurogenesis, particularly around the hippocampal fissure (see seminal papers by Meyer and coworkers: Abraham et al. 2004; Meyer, 2010) and gliogenesis in the subpial fimbriodentate gliogenic wedge. An early disintegration of the radial glia scaffold at the fimbriodentate junction and the creation of ‘proliferative matrices’ with development of astroglial cells from basal radial glia was already described in 1992 in the developing E15 hamster (Sievers et al. 1992). Even earlier, Choi & Lapham (1980) pointed out that some radial glial cells in the developing human cerebellum already have features of apical truncated radial glial cells in early stages (12 wpc), whereas a second set exhibited basal radial glia morphology at the same stage (see their Fig. 3A). At 20 wpc the basal radial glial cells now equivalent to Bergmann glial cells arise in the intermediate zone, with their cell bodies below the Purkinje cell layer, and reach the pial surface. In the present study, these cell bodies show a very strong immunoreactivity for S100.
‘Reactive astrocytes’ in brain development
Upon injury or disease, astrocytes respond by means of reactive astrogliosis, which can be both detrimental or beneficial, or both (Sofroniew, 2009; Sofroniew & Vinters, 2010; Liddelow & Barres, 2017). Astrogliosis involves a spectrum of molecular, cellular and functional changes (Sofroniew & Vinters, 2010; Liddelow & Barres, 2017). These changes are context‐dependent, vary with severity of the insult (Sofroniew, 2009; Sofroniew & Vinters, 2010; Anderson et al. 2014) and result in diverse changes in gene expression and cell function and varying degrees of hypertrophy, upregulation of GFAP, proliferation, loss of individual astrocyte domains and, in some cases, glial scar formation (Sofroniew, 2009; Sofroniew & Vinters, 2010; Anderson et al. 2014). Two different reactive states, have been described in mice treated either with a systemic injection of lipopolysaccharide (LPS), or received middle cerebral artery occlusion (MCAO), termed A1 (LPS) and A2 (MCAO), respectively (see Liddelow et al. 2017), but it is still uncertain whether reactive astrocytes exist as a continuum of these two states, or whether there are several yet undetected reactive profiles (Liddelow & Barres, 2017) and whether heterogeneity of reactive astrocytes is already established during development (Liddelow & Barres, 2017). Gene‐expression profiling has revealed a cohort of genes associated with both astrocyte precursor populations (GLAST‐dsRed+/CD15+) in mouse spinal cord and reactive astrocytes in human white matter injury (Chaboub et al. 2016), suggesting that there may be some overlap between developing and reactive astrocytes. GFAP, S100, AQP4 and YKL‐40 have all been described in reactive astrocytes (Mrak & Griffinbc, 2001; Sawada et al. 2007; Bonneh‐Barkay et al. 2010; Liddelow & Barres, 2017), but comparing immunopanned RNA‐seq data on GFAP, S100B, AQP4 and CHI3L1 (YKL‐40) in Fig. 1A, it seems that CHI3L1 is the only marker clearly upregulated in tumor‐associated astrocytes compared with mature resting astrocytes. Further, CHI3L1 (YKL‐40) is one of the 50 most induced genes in LPS (A1) astrocytes and present in both LPS and MCAO (A2) reactive astrocytes in mice (Zamanian et al. 2012). Hence, YKL‐40 positive radial glia and astrocytes in developing human brain may share common features with at least subpopulations of reactive astrocytes. The complement component C3 has recently been discovered to be specifically upregulated in A1 reactive astrocytes (Liddelow et al. 2017). We found a subpopulation of C3d‐positive plump‐looking astrocytes in the fimbriodentate junction colocalizing with GFAP and S100 (Fig. 7), but not with YKL‐40 (data not shown). We therefore speculate that there may be several subpopulations of ‘developmental’ astrocytes which share features and molecular traits with reactive astrocytes, but it is not yet clear when these developmental astrocytes lose their reactive signature, which functions they control and whether they are predestined to resume a reactive expression pattern upon insult in the adult CNS.
Concluding remarks
Established astroglial markers GFAP, S100 and AQP4 are neither sensitive nor specific enough to embrace the entire population of astrocytic/astroglial cells in the developing human brain at midgestation and cannot distinguish between different maturation steps.
Although well‐described and often used, GFAP is not a reliable pan‐astroglial marker in developing human brain.
AQP4 seems to be the most sensitive and specific marker for characterizing astrocytes in developing human brain at midgestation.
Developmental age, region and, if possible, astroglial subtype should be considered when choosing appropriate astroglial markers.
AQP4 characterizes a brain‐wide water transport system in cerebral cortex at midgestation.
AQP4 may characterize radial glial cells destined for the astroglial lineage.
YKL‐40 and C3d, previously found in reactive astrocytes, stain different subpopulations of astrocytes/astroglial progenitors in developing hippocampus at midgestation and may characterize specific groups of ‘developmental astrocytes’.
Conflict of interest
The authors have no conflict of interest to declare.
Author contributions
C.B.H., C.B.B. and K.M. conceived the study and performed immunohistochemistry and immunofluorescence. K.V.‐S. and C.B.H. analyzed and described the publicly available RNA expression data. Final description and interpretation of the results was carried out by all authors. C.B.H. produced the first version of the manuscript and all authors approved the final version.
Acknowledgements
We would like to thank Pernille S. Froh, Ha Nguyen and Keld B. Ottosen, Department of Cellular and Molecular Medicine, Faculty of Health and Medical Sciences, University of Copenhagen, for excellent technical assistance. We would also like to thank Dr. Steven Sloan, the Barres Lab, for advice regarding RNA expression data. Furthermore, we acknowledge the Core Facility for Integrated Microscopy (CFIM), Faculty of Health and Medical Sciences, University of Copenhagen, for expert assistance. This work was supported by a Graduate Scholarship from The Faculty of Health and Medical Sciences, University of Copenhagen, Denmark (C.B.H.), a grant from Læge Sofus Carl Emil Friis og hustru Olga Doris Friis’ Legat (C.B.B.) and Vera and Carl Johan Michaelsen Foundation (#34077 to K.M.).
References
- Abbott NJ, Ronnback L, Hansson E (2006) Astrocyte‐endothelial interactions at the blood‐brain barrier. Nat Rev Neurosci 7, 41–53. [DOI] [PubMed] [Google Scholar]
- Abbott NJ, Pizzo ME, Preston JE, et al. (2018) The role of brain barriers in fluid movement in the CNS: is there a ‘glymphatic’ system? Acta Neuropathol 135, 387–407. [DOI] [PubMed] [Google Scholar]
- Abraham H, Perez‐Garcia CG, Meyer G (2004) p73 and Reelin in Cajal‐Retzius cells of the developing human hippocampal formation. Cereb Cortex 14, 484–495. [DOI] [PubMed] [Google Scholar]
- Abraham H, Vincze A, Jewgenow I, et al. (2010) Myelination in the human hippocampal formation from midgestation to adulthood. Int J Dev Neurosci 28, 401–410. [DOI] [PubMed] [Google Scholar]
- Allen NJ (2013) Role of glia in developmental synapse formation. Curr Opin Neurobiol 23, 1027–1033. [DOI] [PubMed] [Google Scholar]
- Alvarez‐Buylla A, Garcia‐Verdugo JM, Tramontin AD (2001) A unified hypothesis on the lineage of neural stem cells. Nat Rev Neurosci 2, 287–293. [DOI] [PubMed] [Google Scholar]
- Anderson MA, Ao Y, Sofroniew MV (2014) Heterogeneity of reactive astrocytes. Neurosci Lett 565, 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriezen WL (1893) The neuroglia elements in the human brain. Br Med J 2, 227–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antanitus DS, Choi BH, Lapham LW (1976) The demonstration of glial fibrillary acidic protein in the cerebrum of the human fetus by indirect immunofluorescence. Brain Res 103, 613–616. [DOI] [PubMed] [Google Scholar]
- Aquino DA, Padin C, Perez JM, et al. (1996) Analysis of glial fibrillary acidic protein, neurofilament protein, actin and heat shock proteins in human fetal brain during the second trimester. Brain Res Dev Brain Res 91, 1–10. [DOI] [PubMed] [Google Scholar]
- Assentoft M, Larsen BR, MacAulay N (2015) Regulation and function of AQP4 in the central nervous system. Neurochem Res 40, 2615–2627. [DOI] [PubMed] [Google Scholar]
- Astrand R, Unden J, Romner B (2013) Clinical use of the calcium‐binding S100B protein. Methods Mol Biol 963, 373–384. [DOI] [PubMed] [Google Scholar]
- deAzevedo LC, Fallet C, Moura‐Neto V, et al. (2003) Cortical radial glial cells in human fetuses: depth‐correlated transformation into astrocytes. J Neurobiol 55, 288–298. [DOI] [PubMed] [Google Scholar]
- Barry DS, Pakan JM, McDermott KW (2014) Radial glial cells: key organisers in CNS development. Int J Biochem Cell Biol 46, 76–79. [DOI] [PubMed] [Google Scholar]
- Bayraktar OA, Fuentealba LC, Alvarez‐Buylla A, et al. (2014) Astrocyte development and heterogeneity. Cold Spring Harb Perspect Biol 7, a020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Haim L, Rowitch DH (2017) Functional diversity of astrocytes in neural circuit regulation. Nat Rev Neurosci 18, 31–41. [DOI] [PubMed] [Google Scholar]
- Berg DA, Bond AM, Ming GL, et al. (2018) Radial glial cells in the adult dentate gyrus: what are they and where do they come from? F1000Res, 7, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørnbak C, Brøchner CB, Larsen LA, et al. (2014) Brain barriers and a subpopulation of astroglial progenitors of developing human forebrain are immunostained for the glycoprotein YKL‐40. J Histochem Cytochem 62, 369–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrini M, Fulmore CA, Tartt AN, et al. (2018) Human hippocampal neurogenesis persists throughout aging. Cell Stem Cell 22, 589–599.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneh‐Barkay D, Wang G, Starkey A, et al. (2010) In vivo CHI3L1 (YKL‐40) expression in astrocytes in acute and chronic neurological diseases. J Neuroinflammation 7, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneh‐Barkay D, Bissel SJ, Kofler J, et al. (2012) Astrocyte and macrophage regulation of YKL‐40 expression and cellular response in neuroinflammation. Brain Pathol 22, 530–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brøchner CB, Møllgård K (2016) SSEA‐4 and YKL‐40 positive progenitor subtypes in the subventricular zone of developing human neocortex. Glia 64, 90–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystron I, Blakemore C, Rakic P (2008) Development of the human cerebral cortex: boulder Committee revisited. Nat Rev Neurosci 9, 110–122. [DOI] [PubMed] [Google Scholar]
- Chaboub LS, Manalo JM, Lee HK, et al. (2016) Temporal profiling of astrocyte precursors reveals parallel roles for Asef during development and after injury. J Neurosci 36, 11904–11917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BH (1986) Glial fibrillary acidic protein in radial glia of early human fetal cerebrum: a light and electron microscopic immunoperoxidase study. J Neuropathol Exp Neurol 45, 408–418. [DOI] [PubMed] [Google Scholar]
- Choi BH, Kim RC (1984) Expression of glial fibrillary acidic protein in immature oligodendroglia. Science 223, 407–409. [DOI] [PubMed] [Google Scholar]
- Choi BH, Kim RC (1985) Expression of glial fibrillary acidic protein by immature oligodendroglia and its implications. J Neuroimmunol 8, 215–235. [DOI] [PubMed] [Google Scholar]
- Choi BH, Lapham LW (1978) Radial glia in the human fetal cerebrum: a combined Golgi, immunofluorescent and electron microscopic study. Brain Res 148, 295–311. [DOI] [PubMed] [Google Scholar]
- Choi BH, Lapham LW (1980) Evolution of Bergmann glia in developing human fetal cerebellum: a Golgi, electron microscopic and immunofluorescent study. Brain Res 190, 369–383. [DOI] [PubMed] [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CC, et al. (2005) Thrombospondins are astrocyte‐secreted proteins that promote CNS synaptogenesis. Cell 120, 421–433. [DOI] [PubMed] [Google Scholar]
- Chung WS, Clarke LE, Wang GX, et al. (2013) Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature 504, 394–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke LE, Liddelow SA, Chakraborty C, et al. (2018) Normal aging induces A1‐like astrocyte reactivity. Proc Natl Acad Sci U S A 115, E1896–e1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthard LG, Hawksworth OA, Woodruff TM (2018) Complement: the emerging architect of the developing brain. Trends Neurosci 41, 373–384. [DOI] [PubMed] [Google Scholar]
- Dahl D, Rueger DC, Bignami A, et al. (1981) Vimentin, the 57 000 molecular weight protein of fibroblast filaments, is the major cytoskeletal component in immature glia. Eur J Cell Biol 24, 191–196. [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Croote D, et al. (2017) Single‐Cell RNA‐seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep 21, 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloulme JC, Raponi E, Gentil BJ, et al. (2004) Nuclear expression of S100B in oligodendrocyte progenitor cells correlates with differentiation toward the oligodendroglial lineage and modulates oligodendrocytes maturation. Mol Cell Neurosci 27, 453–465. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Garcia‐Verdugo JM, Alvarez‐Buylla A (1997) Cellular composition and three‐dimensional organization of the subventricular germinal zone in the adult mammalian brain. J Neurosci 17, 5046–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drukarch B, Schepens E, Stoof JC, et al. (1998) Astrocyte‐enhanced neuronal survival is mediated by scavenging of extracellular reactive oxygen species. Free Radic Biol Med 25, 217–220. [DOI] [PubMed] [Google Scholar]
- Edwards MA, Yamamoto M, Caviness VS Jr (1990) Organization of radial glia and related cells in the developing murine CNS. An analysis based upon a new monoclonal antibody marker. Neuroscience 36, 121–144. [DOI] [PubMed] [Google Scholar]
- El‐Khoury N, Braun A, Hu F, et al. (2006) Astrocyte end‐feet in germinal matrix, cerebral cortex, and white matter in developing infants. Pediatr Res 59, 673–679. [DOI] [PubMed] [Google Scholar]
- Falk S, Gotz M (2017) Glial control of neurogenesis. Curr Opin Neurobiol 47, 188–195. [DOI] [PubMed] [Google Scholar]
- Fallier‐Becker P, Vollmer JP, Bauer HC, et al. (2014) Onset of aquaporin‐4 expression in the developing mouse brain. Int J Dev Neurosci 36, 81–89. [DOI] [PubMed] [Google Scholar]
- Fan X, Dong J, Zhong S, et al. (2018) Spatial transcriptomic survey of human embryonic cerebral cortex by single‐cell RNA‐seq analysis. Cell Res 28, 730–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Hatten ME, Heintz N (1994) Brain lipid‐binding protein (BLBP): a novel signaling system in the developing mammalian CNS. Neuron 12, 895–908. [DOI] [PubMed] [Google Scholar]
- Fietz SA, Kelava I, Vogt J, et al. (2010) OSVZ progenitors of human and ferret neocortex are epithelial‐like and expand by integrin signaling. Nat Neurosci 13, 690–699. [DOI] [PubMed] [Google Scholar]
- Gao H, Zhang IY, Zhang L, et al. (2018) S100B suppression alters polarization of infiltrating myeloid‐derived cells in gliomas and inhibits tumor growth. Cancer Lett 439, 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge WP, Jia JM (2016) Local production of astrocytes in the cerebral cortex. Neuroscience 323, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge WP, Miyawaki A, Gage FH, et al. (2012) Local generation of glia is a major astrocyte source in postnatal cortex. Nature 484, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gömöri E, Pal J, Abraham H, et al. (2006) Fetal development of membrane water channel proteins aquaporin‐1 and aquaporin‐4 in the human brain. Int J Dev Neurosci 24, 295–305. [DOI] [PubMed] [Google Scholar]
- González‐Arnay E, González‐Gómez M, Meyer G (2017) A Radial Glia Fascicle Leads Principal Neurons from the Pallial‐Subpallial Boundary into the Developing Human Insula. Front Neuroanat 11, 111, 10.3389/fnana.2017.00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gos T, Schroeter ML, Lessel W, et al. (2013) S100B‐immunopositive astrocytes and oligodendrocytes in the hippocampus are differentially afflicted in unipolar and bipolar depression: a postmortem study. J Psychiatr Res 47, 1694–1699. [DOI] [PubMed] [Google Scholar]
- Gotz M, Sirko S, Beckers J, et al. (2015) Reactive astrocytes as neural stem or progenitor cells: in vivo lineage, In vitro potential, and Genome‐wide expression analysis. Glia 63, 1452–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachem S, Aguirre A, Vives V, et al. (2005) Spatial and temporal expression of S100B in cells of oligodendrocyte lineage. Glia 51, 81–97. [DOI] [PubMed] [Google Scholar]
- Hamilton LK, Truong MK, Bednarczyk MR, et al. (2009) Cellular organization of the central canal ependymal zone, a niche of latent neural stem cells in the adult mammalian spinal cord. Neuroscience 164, 1044–1056. [DOI] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PR, et al. (2010) Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561. [DOI] [PubMed] [Google Scholar]
- Howard BM, Zhicheng M, Filipovic R, et al. (2008) Radial glia cells in the developing human brain. Neuroscientist 14, 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadecola C, Nedergaard M (2007) Glial regulation of the cerebral microvasculature. Nat Neurosci 10, 1369–1376. [DOI] [PubMed] [Google Scholar]
- Iliff JJ, Wang M, Liao Y, et al. (2012) A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid beta. Sci Transl Med, 4, 147ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi T, Dakin KA, Stevens B, et al. (2006) Astrocytes promote myelination in response to electrical impulses. Neuron 49, 823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Nardelli J (2016) Cellular and molecular introduction to brain development. Neurobiol Dis 92, 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadhim HJ, Gadisseux JF, Evrard P (1988) Topographical and cytological evolution of the glial phase during prenatal development of the human brain: histochemical and electron microscopic study. J Neuropathol Exp Neurol 47, 166–188. [DOI] [PubMed] [Google Scholar]
- Kempermann G, Gage FH, Aigner L, et al. (2018) Human adult neurogenesis: evidence and remaining questions. Cell Stem Cell 23, 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Budka H, Soler‐Federsppiel S (1986) An immunocytochemical comparison of the glia‐associated proteins glial fibrillary acidic protein (GFAP) and S‐100 protein (S100P) in human brain tumors. Clin Neuropathol 5, 21–27. [PubMed] [Google Scholar]
- Kostovic I, Seress L, Mrzljak L, et al. (1989) Early onset of synapse formation in the human hippocampus: a correlation with Nissl‐Golgi architectonics in 15‐ and 16.5‐week‐old fetuses. Neuroscience 30, 105–116. [DOI] [PubMed] [Google Scholar]
- Levitt P, Rakic P (1980) Immunoperoxidase localization of glial fibrillary acidic protein in radial glial cells and astrocytes of the developing rhesus monkey brain. J Comp Neurol 193, 815–840. [DOI] [PubMed] [Google Scholar]
- Liddelow SA, Barres BA (2017) Reactive astrocytes: production, function, and therapeutic potential. Immunity 46, 957–967. [DOI] [PubMed] [Google Scholar]
- Liddelow SA, Guttenplan KA, Clarke LE, et al. (2017) Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Kwon HJ, Huang Z (2012) A functional requirement for astroglia in promoting blood vessel development in the early postnatal brain. PLoS ONE 7, e48001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malatesta P, Hartfuss E, Gotz M (2000) Isolation of radial glial cells by fluorescent‐activated cell sorting reveals a neuronal lineage. Development 127, 5253–5263. [DOI] [PubMed] [Google Scholar]
- Marín‐Padilla M (1995) Prenatal development of fibrous (white matter), protoplasmic (gray matter), and layer I astrocytes in the human cerebral cortex: a Golgi study. J Comp Neurol 357, 554–572. [DOI] [PubMed] [Google Scholar]
- Merkle FT, Mirzadeh Z, Alvarez‐Buylla A (2007) Mosaic organization of neural stem cells in the adult brain. Science 317, 381–384. [DOI] [PubMed] [Google Scholar]
- Meyer G (2010) Building a human cortex: the evolutionary differentiation of Cajal‐Retzius cells and the cortical hem. J Anat 217, 334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middeldorp J, Boer K, Sluijs JA, et al. (2010) GFAPδ in radial glia and subventricular zone progenitors in the developing human cortex. Development 137, 313–321. [DOI] [PubMed] [Google Scholar]
- Miller FD, Gauthier AS (2007) Timing is everything: making neurons versus glia in the developing cortex. Neuron 54, 357–369. [DOI] [PubMed] [Google Scholar]
- Møllgård K, Dziegielewska KM, Holst CB, et al. (2017) Brain barriers and functional interfaces with sequential appearance of ABC efflux transporters during human development. Sci Rep 7, 11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Deneen B (2015) Astrocyte development: a Guide for the Perplexed. Glia 63, 1320–1329. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Krencik R, Ullian EM, et al. (2012) Astrocytes and disease: a neurodevelopmental perspective. Genes Dev 26, 891–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Kelley KW, Tsai HH, et al. (2014) Astrocyte‐encoded positional cues maintain sensorimotor circuit integrity. Nature 509, 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow T, Song MR, Ghosh A (2001) Sequential specification of neurons and glia by developmentally regulated extracellular factors. Development 128, 3585–3594. [DOI] [PubMed] [Google Scholar]
- Mrak RE, Griffinbc WS (2001) The role of activated astrocytes and of the neurotrophic cytokine S100B in the pathogenesis of Alzheimer's disease. Neurobiol Aging 22, 915–922. [DOI] [PubMed] [Google Scholar]
- Nowakowski TJ, Pollen AA, Sandoval‐Espinosa C, et al. (2016) Transformation of the radial glia scaffold demarcates two stages of human cerebral cortex development. Neuron 91, 1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Bhaduri A, Pollen AA, et al. (2017) Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plog BA, Nedergaard M (2018) The glymphatic system in central nervous system health and disease: past, present, and future. Annu Rev Pathol 13, 379–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G, Li X, Chen Z, et al. (2017) Prognostic value of YKL‐40 in patients with glioblastoma: a systematic review and meta‐analysis. Mol Neurobiol 54, 3264–3270. [DOI] [PubMed] [Google Scholar]
- Rakic P (1972) Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol 145, 61–83. [DOI] [PubMed] [Google Scholar]
- Rakic P (1988) Specification of cerebral cortical areas. Science 241, 170–176. [DOI] [PubMed] [Google Scholar]
- Rakic P (2003) Elusive radial glial cells: historical and evolutionary perspective. Glia 43, 19–32. [DOI] [PubMed] [Google Scholar]
- Reemst K, Noctor SC, Lucassen PJ, et al. (2016) The indispensable roles of microglia and astrocytes during brain development. Front Hum Neurosci 10, 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reillo I, de Juan Romero C, Garcia‐Cabezas MA, et al. (2011) A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb Cortex 21, 1674–1694. [DOI] [PubMed] [Google Scholar]
- Rowitch DH, Kriegstein AR (2010) Developmental genetics of vertebrate glial‐cell specification. Nature 468, 214–222. [DOI] [PubMed] [Google Scholar]
- Sasaki A, Hirato J, Nakazato Y, et al. (1988) Immunohistochemical study of the early human fetal brain. Acta Neuropathol 76, 128–134. [DOI] [PubMed] [Google Scholar]
- Saunders NR, Dziegielewska KM, Møllgård K, et al. (2018) Physiology and molecular biology of barrier mechanisms in the fetal and neonatal brain. J Physiol, 596, 5723–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada T, Kato Y, Kobayashi M (2007) Expression of aquaporin‐4 in central nervous system tumors. Brain Tumor Pathol 24, 81–84. [DOI] [PubMed] [Google Scholar]
- Schmechel DE, Rakic P (1979) A Golgi study of radial glial cells in developing monkey telencephalon: morphogenesis and transformation into astrocytes. Anat Embryol (Berl) 156, 115–152. [DOI] [PubMed] [Google Scholar]
- Shibata T, Yamada K, Watanabe M, et al. (1997) Glutamate transporter GLAST is expressed in the radial glia‐astrocyte lineage of developing mouse spinal cord. J Neurosci 17, 9212–9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidman RL, Rakic P (1973) Neuronal migration, with special reference to developing human brain: a review. Brain Res 62, 1–35. [DOI] [PubMed] [Google Scholar]
- Sievers J, Hartmann D, Pehlemann FW, et al. (1992) Development of astroglial cells in the proliferative matrices, the granule cell layer, and the hippocampal fissure of the hamster dentate gyrus. J Comp Neurol 320, 1–32. [DOI] [PubMed] [Google Scholar]
- Silbereis J, Heintz T, Taylor MM, et al. (2010) Astroglial cells in the external granular layer are precursors of cerebellar granule neurons in neonates. Mol Cell Neurosci 44, 362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DJ, Steindler DA (2009) Common astrocytic programs during brain development, injury and cancer. Trends Neurosci 32, 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Bhardwaj R, Wilczynska KM, et al. (2011) A complex of nuclear factor I‐X3 and STAT3 regulates astrocyte and glioma migration through the secreted glycoprotein YKL‐40. J Biol Chem 286, 39893–39903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart IH, Dehay C, Giroud P, et al. (2002) Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb Cortex 12, 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Speed T (2003) Normalization of cDNA microarray data. Methods 31, 265–273. [DOI] [PubMed] [Google Scholar]
- Sofroniew MV (2009) Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci 32, 638–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew MV, Vinters HV (2010) Astrocytes: biology and pathology. Acta Neuropathol 119, 7–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrells SF, Paredes MF, Cebrian‐Silla A, et al. (2018) Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 555, 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagaard M, Møllgård K (1989) The developing neuroepithelium in human embryonic and fetal brain studied with vimentin‐immunocytochemistry. Anat Embryol (Berl) 180, 17–28. [DOI] [PubMed] [Google Scholar]
- Stagaard Janas M, Nowakowski RS, Møllgård K (1991a) Glial cell differentiation in neuron‐free and neuron‐rich regions. II. Early appearance of S‐100 protein positive astrocytes in human fetal hippocampus. Anat Embryol (Berl) 184, 559–569. [DOI] [PubMed] [Google Scholar]
- Stagaard Janas M, Nowakowski RS, Terkelsen OB, et al. (1991b) Glial cell differentiation in neuron‐free and neuron‐rich regions. I. Selective appearance of S‐100 protein in radial glial cells of the hippocampal fimbria in human fetuses. Anat Embryol (Berl) 184, 549–558. [DOI] [PubMed] [Google Scholar]
- Steiner J, Bernstein HG, Bielau H, et al. (2007) Evidence for a wide extra‐astrocytic distribution of S100B in human brain. BMC Neurosci 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata H (2015) Diverse subtypes of astrocytes and their development during corticogenesis. Front Neurosci 9, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiu SC, Chan WY, Heizmann CW, et al. (2000) Differential expression of S100B and S100A6(1) in the human fetal and aged cerebral cortex. Brain Res Dev Brain Res 119, 159–168. [DOI] [PubMed] [Google Scholar]
- Ullian EM, Sapperstein SK, Christopherson KS, et al. (2001) Control of synapse number by glia. Science 291, 657–661. [DOI] [PubMed] [Google Scholar]
- Ullian EM, Christopherson KS, Barres BA (2004) Role for glia in synaptogenesis. Glia 47, 209–216. [DOI] [PubMed] [Google Scholar]
- Watkins TA, Emery B, Mulinyawe S, et al. (2008) Distinct stages of myelination regulated by gamma‐secretase and astrocytes in a rapidly myelinating CNS coculture system. Neuron 60, 555–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Kang H, Xu Q, et al. (2013) Sleep drives metabolite clearance from the adult brain. Science 342, 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Xiao M, Li S, et al. (2017) Aquaporins in Nervous System. Adv Exp Med Biol 969, 81–103. [DOI] [PubMed] [Google Scholar]
- Yachnis AT, Rorke LB, Lee VM, et al. (1993) Expression of neuronal and glial polypeptides during histogenesis of the human cerebellar cortex including observations on the dentate nucleus. J Comp Neurol 334, 356–369. [DOI] [PubMed] [Google Scholar]
- Zamanian JL, Xu L, Foo LC, et al. (2012) Genomic analysis of reactive astrogliosis. J Neurosci 32, 6391–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecevic N (2004) Specific characteristic of radial glia in the human fetal telencephalon. Glia 48, 27–35. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sloan SA, Clarke LE, et al. (2016) Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 89, 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]


