Abstract
The mannose receptor (CD206) functions in endocytosis and phagocytosis, and plays an important role in immune homeostasis. Tumor-associated macrophages express high level of CD206 and are thought to contribute to cancer progression through tumor immunosuppression, metastasis and angiogenesis. However, the significance of CD206 in the pathology of liver cancer has not been investigated. The present study evaluated the clinical significance of CD206 in the progression and prognosis of liver cancer in pathological tissues from 327 patients. Increased CD206 expression was observed in liver cancer samples compared with healthy adjacent liver tissue (42.8 vs. 62.4%; P<0.05). CD206 expression was significantly associated with tumor size (P=0.009) and metastasis (P=0.041). The recurrence free survival rate of patients with CD206-positive liver cancer was significantly decreased compared with patients with CD206-negative liver cancer (P=0.003). A Cox regression model revealed that liver cancer survival was independently associated with tumor size, metastasis and α-fetoprotein value. The results further revealed that CD206 expression in cancer stem cell (CSC)-like cells was comparable to other internationally recognized biomarkers. Additionally, when CD206 expression was silenced in the liver cancer cell lines HepG2 and PLC/PRF/5 using a short hairpin RNA approach, migration and invasion of the cells significantly decreased compared with controls (P<0.01). CD206 expression in liver cancer significantly influences distant metastasis and spread, resulting in poor patient prognosis. Furthermore, CD206 may be a potential biomarker in CSC-like cells to predict the occurrence of liver cancer.
Keywords: mannose receptor, liver cancer, cancer stem cells, biomarker, prognosis
Introduction
Liver cancer had the seventh highest age-adjusted incidence rate of all types of cancer in the United States of America in 2014, often with high mortality (1–3). Liver cancer requires the detection of small tumors that are often present in asymptomatic individuals (4). Liver cancer is diagnosed by imaging modalities, including computerized tomography and magnetic resonance imaging scans, followed by confirmation via liver biopsy, an invasive procedure (5–7). Although the diagnosis of liver cancer has improved rapidly in recent years, <5% of patients with liver cancer survive >5 years following diagnosis (8,9). Previous studies suggest that high invasiveness and metastasis are the main causes of poor prognosis (10,11). Early liver cancer diagnosis is of paramount importance to therapy and more effective biomarkers to predict the clinical outcome of liver cancer are required.
The mannose receptor (CD206), also known as C-type lectin, is expressed on the surface of macrophages and some subsets of immature dendritic cells (12). CD206 participates in antigen presentation, macrophage endocytosis and is considered a hallmark of tumor-associated macrophages (13). CD206 increased the growth and migration of microglia by promoting their activation (14). Additionally, CD206 expressed on lymphatic endothelium participates in the attachment of various cancer cells to lymphatic endothelium to promote lymphatic metastasis (15–17). Serum CD206 is elevated in patients with multiple myeloma and is a prognostic marker for overall survival (18,19). Furthermore, CD206 has been reported as a novel biomarker for the diagnosis of patients with colorectal and gastric cancer (20,21). Therefore, the investigation of the potential role of CD206 in liver cancer may be beneficial to patients.
The present study evaluated the clinical significance of CD206 in the progression and prognosis of liver cancer. A suspension culture was used to enrich liver cancer stem cell (CSC)-like cells, which acquire the properties of liver CSCs in term of self-renewal, differentiation, quiescence, chemo-resistance and tumorigenicity (22,23). The results obtained indicated that CD206 may act as a biomarker in CSC-like cells to predict liver cancer occurrence. CD206 promoted the motility and invasiveness of liver cancer cell lines in vitro. Furthermore, it was revealed that the upregulation of CD206 in liver cancer contributes to poor patient prognosis. Thus, novel therapeutic agents targeting CD206 may be beneficial for patients with liver cancer.
Materials and methods
Liver cancer samples and cell lines
The tissue microarrays (TMAs) used in the current study were purchased from Shanghai Biochip Co. Ltd. Written informed consent was obtained from all the patients and the protocol was approved by the Medical Ethics Review Committee of the Zhejiang Provincial People's Hospital. The experiment began in August 2013 and ended in August 2018. The inclusion criteria were as follows: i) diagnosis of hepatocellular carcinoma, ii) patients who signed informed consent and iii) patients with adequate hepatocellular carcinoma tissue to make tissue microarrays (cylindrical liver cancer tissue at least 1 mm in diameter and 1 mm in length). In the tissue microarrays, all the patients had complete clinical data. A total of 327 patients with liver cancer were divided into the following groups according to their respective characteristics: i) age (<55 or ≥55 years) (24), ii) gender (male or female), iii) tumor size (diameter <5 or ≥5 cm) (25), iv) tumor number (single or multiple) (26), v) Edmondson Grade (I+II or III) (27), vi) metastasis (M0 or M1) (28), vii) micro-vascular invasion (absent or present) (29,30), viii) Hepatitis B virus antigen (negative or positive) (31), ix) cirrhosis (negative or positive) (32) and x) α-fetoprotein (AFP; <20 or ≥20 µg/l) (33). The patients included 266 males and 61 females (age range, 31–83 years; median, 57.5 years). All the patients had follow-up records for 60 months. Survival time was calculated from the date of surgery to the deadline of 60 months. The liver cancer cell lines HepG2 and PLC/PRF/5 (Chinese Academy of Sciences) were cultured in medium DMEM (HyClone; GE Healthcare Life Sciences), containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 1% penicillin-streptomycin and 0.5% glutamine (2 mM) at 37°C in a humidified atmosphere (95% air and 5% CO2).
Immunohistochemical staining
The hepatocellular carcinoma tissue samples were fixed in 10% formalin for 24 h at 26°C and paraffin embedded. Histological sections were cut into 4 µm and stored at 4°C. The sections were dried for 2 h in an oven preheated to 65°C and dewaxed using xylene and standard procedures using graded alcohol at 37°C for 10 min each time for three times. Antigen retrieval was performed by heating the sections in citrate buffer (pH=6.0; 0.01 M) at 180°C for 3 min. Endogenous peroxidase activity was inhibited by 3% hydrogen peroxide solution at 37°C for 15 min. Sections were blocked using goat serum (cat. no. 31430; 1:500; Thermo Fisher Scientifc, Inc.) to prevent non-specific binding for 15 min at 37°C. Rabbit anti-CD206 primary monoclonal antibody (cat. no. ab64693; 1:400; Abcam) was used to incubate the sections at 4°C overnight. Following three washes with PBS, the biotin-labeled rabbit antibody (cat. no. 31402; 1:500; Thermo Fisher Scientific, Inc.) was used as a secondary antibody and incubated with sections. Horseradish enzyme labeled streptomycin albumen (Thermo Fisher Scientific, Inc.) was subsequently incubated for 0.5 h at 37°C. TMAs were stained with 3,3′-diaminobenzidine (Fuzhou Maixin Biotech Co., Ltd.) for 15 min at 37°C and counterstained with hematoxylin for 15 sec at 37°C. The sections were dehydrated at 37°C as follows: 75% alcohol immersion for 3 min, 85% alcohol immersion for 3 min, 95% alcohol immersion for 3 min, absolute alcohol immersion for 5 min and two rounds of xylene immersion for 5 min. The sections were dried, covered with neutral gum and mounted. The sections were observed using a fluorescence microscope (magnification, ×400).
Staining patterns and evaluation
The degree of CD206 immunostaining was evaluated blindly by three pathologists. CD206 expression evaluation was based on the intensity of stained tumor cells. A total of five fields of view were randomly selected in each microarray. Scores were used to represent the intensity of the staining in the cytoplasm or membrane in the microarrays. ‘0’ was non-stained, ‘1’ was weakly stained as light yellow, ‘2’ was moderately stained as brown and ‘3’ was heavily stained as dark brown. In total, five fields of view were observed and scored according to the aforementioned rules. CD206 scores were calculated with scores of 0–5 and 6–12 representing the low and high expression groups, respectively.
Sphere culture and sphere passage
PLC/PRF/5 cells were resuspended to a cell density of 5000 cells/ml, washed to remove serum and suspended in serum-free DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 20 ng/ml human recombinant basic fibroblast growth factor (Merck KGaA), 20 ng/ml human recombinant epidermal growth factor (Merck KGaA), 2% B27 supplement without vitamin A (Gibco; Thermo Fisher Scientific, Inc.), 100 IU/ml penicillin and 100 µg/ml of streptomycin in ultra-low attachment 6-well plates (Corning Inc.). Fresh DMEM/F12 with 20 ng/ml human recombinant basic fibroblast growth factor, 20 ng/ml human recombinant epidermal growth factor, 2% B27 was added to the ultra-low attachment plates every two days. Four days later, the tumor spheres were collected by gentle centrifugation with 100 × g for 4 min at 37°C and digested by Accutase (Sigma-Aldrich; Merck KGaA) for 5 min at 37°C to form a single cell suspension for subsequent experiments. Tumor spheres were centrifuged with 100 × g for 3 min at 37°C to remove the enzyme and resuspended with 20 ng/ml human recombinant basic fibroblast growth factor, 20 ng/ml human recombinant epidermal growth factor and 2% B27 in ultra-low attachment 6-well plates (Corning Inc.) and allowed to reform spheres. The spheres were passaged every 4 days.
Reverse-transcription quantitative polymerase chain reaction (RT-qPCR)
HepG2 and PLC/PRF/5 cells transfected with shCD206 and shNC were washed three times with PBS, and the total RNA was extracted using Trizol (Invitrogen; Thermo Fisher Scientific, Inc.). The extacted RNA was reverse-transcribed into cDNA using PrimeScript™ 1st Strand cDNA Synthesis kit (Takara Biotechnology Co., Ltd.). qPCR was performed using a KAPA SYBR Green qPCR kit (Roche Diagnostics). The following primer pairs were used: CD206 forward, 5′-GCAGAAGGAGTAACCCACCC-3′ and reverse, 5′-TGGCAAATGAAGGCGTTTGG-3′; Nanog homeobox (Nanog) forward, 5′-AAGGCCTCAGCACCTACCTA-3′ and reverse, 5′-ACATTAAGGCCTTCCCCAGC-3′; POU class 5 homeobox 1 (Oct4) forward, 5′-GCCCGAAAGAGAAAGCGAAC-3′ and reverse, 5′-AACCACACTCGGACCACTCG-3′; SRY-box 2 (Sox2) forward, 5′-TTTGTCGGAGACGGAGAAGC-3′ and reverse, 5′-TAACTGTCCATGCGCTGGTT-3′; MYC-binding protein (c-Myc) forward, 5′-GCATACATCCTGTCCGTCCA-3′ and reverse, 5′-CGTCGTTTCCGCAACAAGTC-3′; CD44 molecule (CD44) forward, 5′-AGCAACTGAGACAGCAACCA-3′ and reverse, 5′-CGTACCAGCCATTTGTGTTGT-3′ and GAPDH forward, 5′-GCTCCCTCTTTCTTTGCAGC-3′ and reverse, 5′-GTTGTCATGGATGACCTTGGC-3′. The following thermocycling conditions were used: Pre-denaturing at 95°C for 5 min, 35 cycles of 95°C for 10 sec, 60°C for 30 sec and, 72°C for 30 sec and extension at 72°C for 10 min. mRNA levels were quantified using the 2−ΔΔCq method (20) and normalized to GAPDH. RT-qPCR was performed in triplicate.
Transfection
The short hairpin (sh) CD206 (5′-GCAGAAGGAGTAACCCACCC-3′) and sh negative control (NC) (5′-GATCCGACTTCATAAGGCTTC-3′) were purchased from GeneCopoeia, Inc. The QIAGEN Plasmid Mini kit (Qiagen GmbH), EndoFectin-Lenti™ (GeneCopoeia, Inc.) and TiterBoost™ reagents (GeneCopoeia, Inc.) were used to generate plasmids delivering shNC and shCD206. The plasmids were co-transfected into 293Ta cells (Chinese Academy of Sciences) with Lenti-Pac™ HIV packaging mix (cat. no. HPK-LvTR-20, GeneCopoeia, Inc.). The lentiviral particles were purified by centrifugation at 3,500 × g and 4°C for 25 min. Lentiviral particles were stored at −80°C until use. HepG2 and PLC/PRF/5 cells seeded in 24-well plate (1×105 cells/well). The lenti-shCD206 and lenti-shNC were used to transfect HepG2 and PLC/PRF/5 cell at a multiplicity of infection of 30. DMEM was replaced 24 h following transfection. Puromycin at 1–10 µg/ml was used to screen the HepG2 and PLC/PRF/5 cells stably expressing shNC and shCD206.
Western blot analysis
HepG2 and PLC/PRF/5 cells stably expressing shCD206 or shNC were lysed using lysis buffer (Beyotime Institute of Biotechnology), 1% complete mini-protease inhibitor cocktail (Roche Diagnostics) and 5 mM sodium fluoride. The total protein was quantified using a bicinchoninic acid assay (Thermo Fisher Scientific, Inc.) and heated for 10 min at 100°C. A total of 30 µg protein/lane was separated via SDS-PAGE on a 12% gel and transferred to a nitrocellulose membrane (Merck KGaA). The membrane was blocked for 1 h at 37°C and immunoblotted with primary antibodies against CD206 (cat. no. ab8918; 1:1,000; Abcam), Nanog (cat. no. ab109250; 1:1,000; Abcam), Oct4 (cat. no. ab18976; 1:1,000; Abcam), Sox2 (cat. no. ab79351; 1:1,000; Abcam), c-Myc (cat. no. ab39688; 1:1,000; Abcam), CD44 (cat. no. ab189524; 1:1,000; Abcam) and GAPDH (cat. no. ab9485; 1:5,000; Abcam) overnight at 4°C. Membranes were subsequently washed with TBST and incubated with horseradish peroxidase-conjugated goat anti-rabbit (cat no. HA1001; 1:50,000; HuaBio) or anti-mouse antibodies (cat. no. HA1006; 1:5,000; HuaBio) for 1 h at room temperature. Protein bands were detected using a ChemiDoc™ MP Imaging system (Bio-Rad Laboratories, Inc.) with a super enhanced chemiluminescence detection kit (Applygen Technologies, Inc.).
Wound healing assay
HepG2 and PLC/PRF/5 cells transfected with lenti-shNC and lenti-shCD206 were seeded on separate 6-well plates at a concentration of 1×106 cells/ml. When the liver cancer cells reached ~90% confluence, a marker pen was used to draw a horizontal line on the bottom of the 6-well plates. A cross was drawn every 0.5 cm and 5 lines were drawn across each well. A 20 µl pipette tip was used to scratch the cell layer perpendicular to the horizontal lines. PBS was used to wash the cells three times and serum-free DMEM was added into the plates. The cells were cultured at 37°C with 5% CO2 for 24 h. Images at 5 fields of view were taken and migration as calculated as follows: (Width of the scratch at 0 h-width of the scratch at 24 h)/width of the scratch at 0 h ×100%.
Cell invasion assay
HepG2 and PLC/PRF/5 cells transfected with shCD206 and shNC were serum starved for 24 h prior to the invasion assay. Transwell inserts were placed into 24-well plates, 24-well transwell plate (8.0 µm pore size; Corning Inc.) coated with 20 µg Matrigel and incubated for 120 min at 37°C to promote uniform gel formation. A total of 100 µl cell suspension at a concentration of 3×104 cells/ml was added to the upper chamber, and 500 µl 10% FBS-DMEM medium containing cells transfected with shCD206 or shNC were added to the lower chamber. In the control group, 500 µl 10% FCS-DMEM medium was added to lower chamber. Following incubation for 48 h at 37°C with 5% CO2, the cells were fixed with 4% formaldehyde at 37°C for 15 min. Cells were stained with 0.5% crystal violet dye at 37°C for 10 min. Cells on the Matrigel and microporous membrane layer were removed using a cotton swab and cells which invaded the lower microporous membrane were retained. The number of cells in five randomly selected fields of view was counted using an inverted light microscope (magnification, ×100).
Statistical analysis
The Statistical Package for the Social Sciences (version 13.0; SPSS Inc.) was used for statistical analysis. Data are presented as the mean ± standard deviation. For comparison between 2 groups, significant differences were determined using the Student's t-test or Wilcoxon rank test. The χ2 or Fisher exact tests were used to assess CD206 expression in patients with HCC. The Kaplan-Meier method and the log-rank test were used to analyze survival curves. The multivariate Cox proportional hazards regression model was used to analyze the univariate factors with prognostic significance in HCC. P<0.05 was considered to indicate a statistically significant difference.
Results
CD206 expression in liver cancer samples and healthy tissue
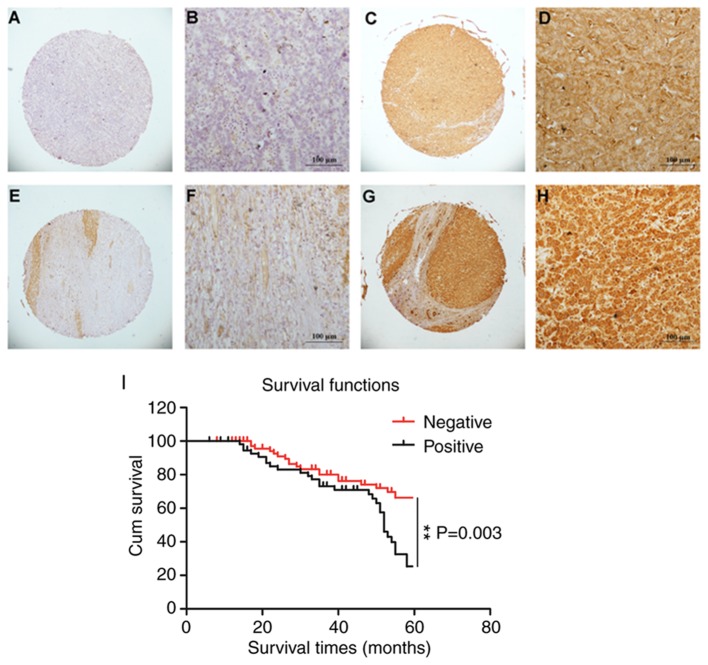
Immunohistochemical methods were used to assess the expression of CD206 in liver cancer tissue. The staining for CD206 was observed predominantly in the membrane and cytoplasm of liver cancer cells. Three pathologists independently evaluated the liver cancer tissue microarrays under ×40 (Fig. 1A, C, E and G) and ×200 magnification (Fig. 1B, D, F and H). CD206 showed positive expression in the cell membrane and cytoplasm of liver cancer cells. High levels of CD206 expression were detected in 204/327 (62.4%) of the patients with liver cancer. Positive expression was observed in adjacent healthy liver tissue (5 mm from the liver tumor and histopathologically confirmed) in 140/327 (42.8%) patients. Upon dividing samples into high expression (>6 points) and low expression groups (0–5 points), the expression value of CD206 in liver cancer tissues was 7.69±3.11 points, and the expression value of CD206 in adjacent healthy liver tissue was 3.60±2.17 points (P<0.05). Taken together, these data demonstrated elevated CD206 expression in liver cancer tumors.
Figure 1.
CD206 staining in liver cancer and healthy adjacent tissues. Low expression of CD206 in normal tissue (score=0–5), magnification (A) ×40 and (B) ×200. High expression of CD206 in normal tissue (score=6–12), magnification (C) ×40 and (D) ×200. Low expression of CD206 in liver cancer tissue (score=4), magnification (E) ×40 and (F) ×200. High expression of CD206 in liver cancer tissue (score=16), magnification (G) ×40 and (H) ×200. (I) Association between CD206 expression and survival time. CD206, mannose receptor. P<0.01, as indicated.
Patient characteristics and the association between CD206 expression, liver cancer clinicopathological features and prognosis
The present study investigated whether CD206 expression is associated with the progression of liver cancer. CD206 immunopositivity was not associated with gender, age, tumor number, Edmondson grade, microvascular invasion, hepatitis B virus antigen, cirrhosis and AFP (Table I). The survival time for patients with CD206-negative liver cancer was 51.517±1.781 months and was significantly increased compared with patients with CD206-postitive liver cancer (46.067±2.183 months). The Kaplan-Meier survival curves indicated that CD206 expression was significantly associated with overall survival in patients with liver cancer (P=0.003; Fig. 1I). Additionally, prognosis factors in liver cancer were analyzed by Cox-regression analysis. CD206 positivity was significantly associated with tumor size (P=0.039), metastasis (P=0.022) and AFP value (P=0.002). There was no statistically significant association between CD206 expression and gender (P>0.05), age (P>0.05), cirrhosis (P>0.05), metastasis (P>0.05), AFP (P>0.05) and tumor number (P>0.05) as demonstrated by multivariate analysis (Table II). However, CD206 was significantly associated with the Edmondson grade (P=0.009).
Table I.
Expression of CD206 in liver cancer tissue.
| CD206 expression | |||||
|---|---|---|---|---|---|
| Clinical parameter | Number | Low | Ηigh | χ2 | P-value |
| Age (years) | 1.601 | 0.206 | |||
| <55 | 126 | 42 | 84 | ||
| ≥55 | 201 | 81 | 120 | ||
| Gender | 0.745 | 0.388 | |||
| Male | 266 | 103 | 163 | ||
| Female | 61 | 20 | 41 | ||
| Sizea | 6.913 | 0.009 | |||
| <5 | 191 | 83 | 108 | ||
| ≥5 | 128 | 37 | 81 | ||
| Tumour number | 0.003 | 0.956 | |||
| Single | 269 | 101 | 168 | ||
| Multiple | 58 | 22 | 36 | ||
| Edmondson gradea | 3.771 | 0.052 | |||
| I+II | 202 | 68 | 134 | ||
| III | 119 | 53 | 66 | ||
| Metastasisa | 4.159 | 0.041 | |||
| M0 | 294 | 105 | 189 | ||
| M1 | 27 | 15 | 12 | ||
| Microvascular invasiona | 0.118 | 0.732 | |||
| Absence | 122 | 49 | 73 | ||
| Presence | 121 | 46 | 75 | ||
| Hepatitis B virus antigena | 3.101 | 0.078 | |||
| Negtive | 62 | 29 | 3 | ||
| Positive | 259 | 90 | 169 | ||
| Cirrhosis | 0.008 | 0.928 | |||
| Negtive | 110 | 41 | 69 | ||
| Positive | 217 | 82 | 135 | ||
| α-fetoproteina | 0.090 | 0.764 | |||
| <20 | 143 | 49 | 94 | ||
| ≥20 | 123 | 40 | 83 | ||
Total number was <327 due to incomplete pathological data.
Table II.
Univariate and multivariate Cox regression analysis of the clinicopathological parameters in patients with liver cancer.
| Univariate analysis | Multivariate analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameter | Number | Regression coefficient | HR | 95% CI | P-value | Regression coefficient | HR | 95.0% CI | P-value |
| Sex (male/female) | 266/61 | −0.124 | 0.883 | 0.556–1.403 | 0.598 | −0.186 | 0.830 | 0.371–1.857 | 0.650 |
| Age (<55/≥55 years) | 126/201 | −0.027 | 0.973 | 0.571–1.658 | 0.920 | −0.493 | 0.611 | 0.289–1.293 | 0.198 |
| Tumor size (<50/≥50 mm)a | 191/128 | 0.489 | 1.630 | 1.025–2.592 | 0.039 | 0.254 | 1.289 | 0.557–2.982 | 0.553 |
| Tumor number (single/multiple)a | 268/58 | 0.059 | 1.060 | 0.582–1.933 | 0.848 | 1.008 | 2.740 | 1.051–7.143 | 0.039 |
| Edmondson grade (I+II/III)a | 202/119 | 0.455 | 1.577 | 0.989–2.515 | 0.056 | 1.021 | 2.775 | 1.291–5.965 | 0.009 |
| Metastasis (M0/M1)a | 294/27 | 0.160 | 1.173 | 0.629–2.188 | 0.022 | 1.293 | 3.644 | 1.314–10.104 | 0.013 |
| Microvascular invasion (−/+)a | 122/121 | 0.606 | 1.834 | 1.089–3.087 | 0.615 | 0.310 | 1.364 | 0.578–3.219 | 0.479 |
| Hepatitis B virus (−/+)a | 62/259 | 0.002 | 1.002 | 0.558–1.800 | 0.994 | −0.526 | 0.591 | 0.180–1.937 | 0.385 |
| Cirrhosis (−/+) | 110/217 | −0.283 | 0.754 | 0.456–1.247 | 0.271 | 0.901 | 2.463 | 0.912–6.655 | 0.076 |
| α-fetoprotein (<20/≥20 µg/l)a | 143/123 | 0.920 | 2.510 | 1.395–4.517 | 0.002 | 0.647 | 1.910 | 0.855–4.264 | 0.114 |
Total number was <327 due to incomplete pathological data. HR, hazard ratio; CI, confidence interval.
CD206 as a biomarker in cancer stem cells may be used to predict liver cancer
The suspension culture in growth factor-defined serum-free medium may be used to enrich cells associated with the traits of CSCs (34,35). To validate whether liver cancer cell lines acquire these traits when passed through the suspension culture, the liver cancer cell line PLC/PRF/5 was subjected to specific serum-free medium. In ultra-low attachment plates, PLC/PRF/5 gradually formed non-adherent spheroid bodies, termed sphere cells, following culture for 4–6 days (Fig. 2A). RT-qPCR was used to assess the expression of recognized biomarker genes (Nanog, Oct4, Sox2, c-Myc and CD44) and CD206 in the CSC spheres and the parent cells from which they were derived (36–38). The results revealed that, although the mRNA levels of CD206 did not increase to the same levels as Nanog, CD44 and Sox2, CD206 expression was comparable to Oct4, and was significantly increased compared with c-Myc (Fig. 2B). These results were validated at the protein level through western blot analysis, where increased expression of CD206 was observed in the sphere cells compared with the parent cells (Fig. 2C). Taken together, these data suggest that CD206, similar to the biomarkers Nanog, CD44, Sox2, Oct4 and c-Myc, has the potential to predict cancer occurrence and progression in CSC-like cells.
Figure 2.
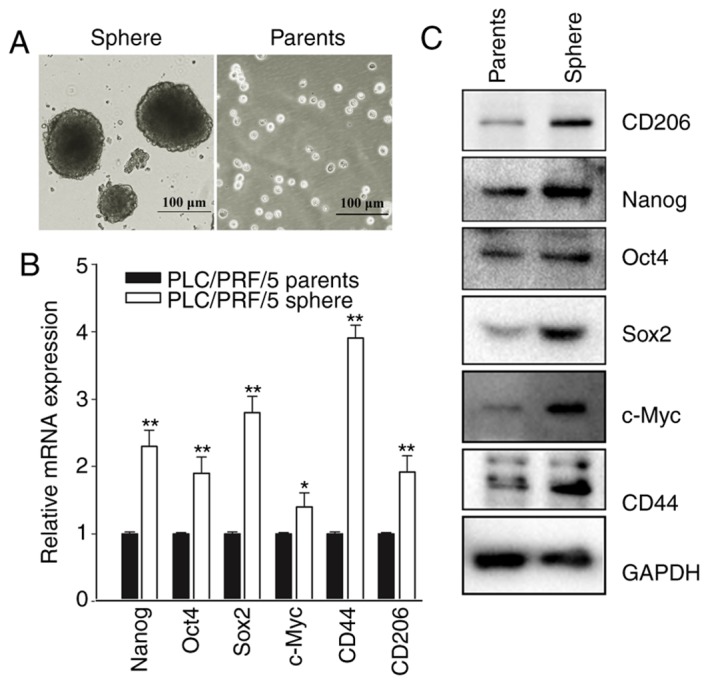
CD206 is a biomarker in cancer stem cells to predict the progression of liver cancer. (A) The liver cancer cell line PLC/PRF/5 formed sphere bodies in suspension culture/ (B) mRNA levels of Nanog, Oct4, Sox2, c-Myc, CD44 and CD206. *P<0.05, **P<0.01 vs. PLC/PRF/5 parents. (C) The expression level of CD206 increased in a similar manner to the expression of the internationally recognized biomarker proteins Nanog, Oct4, Sox2, c-Myc and CD44. CD206, mannose receptor; Nanog, Nanog homeobox; Oct4, POU class 5 homeobox 1; Sox2, SRY-box 2; c-Myc, MYC binding protein; CD44, CD44 molecule.
CD206 silencing decreases liver cancer cell motility and invasion
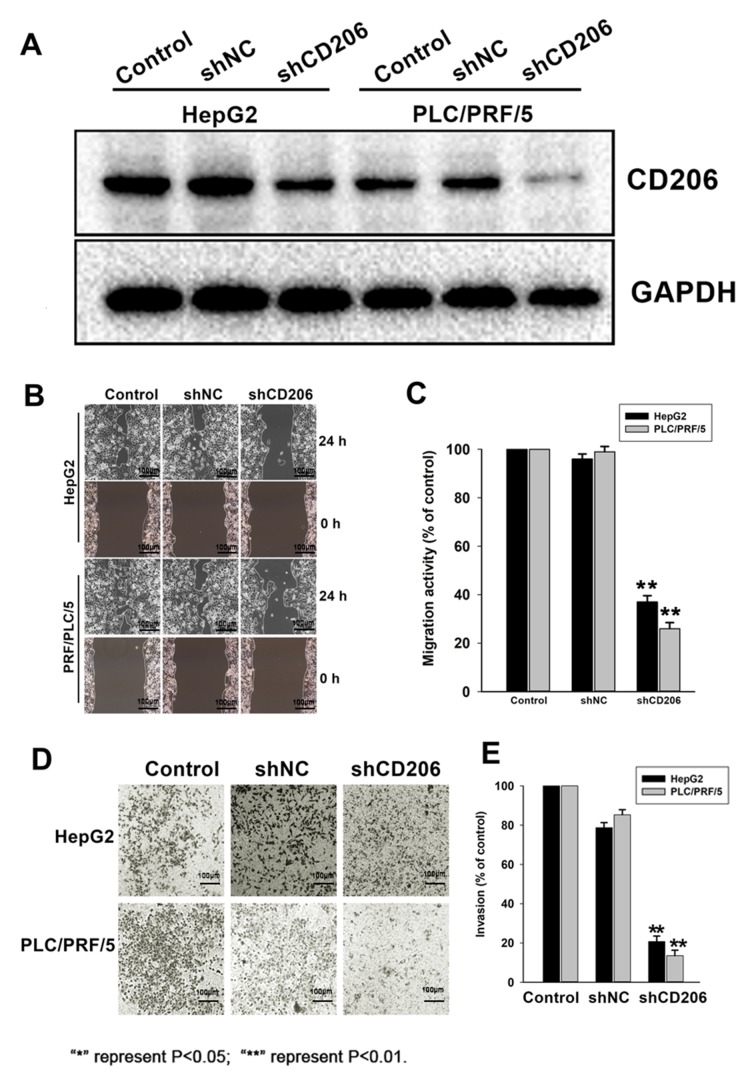
To assess the role of CD206 in liver cancer cell migration and invasion, HepG2 and PLC/PRF/5 cells were infected with lentiviruses expressing CD206 shRNA (shCD206) or a scrambled sequence (shNC) to obtain stable expression cell lines. CD206 silencing following shCD206 lentiviral transfection was confirmed by Western Blot analysis (Fig. 3A). Migration (Fig. 3B and C) and invasion assays (Fig. 3D and E) revealed that CD206 silencing significantly decreased HepG2 and PLC/PRF/5 cell migration and invasion compared with the shNC group (P<0.05). These observations indicated that CD206 promotes the motility and invasiveness of liver cancer cell lines.
Figure 3.
Decreased cell migration and invasion following CD206 silencing in HepG2 and PLC/PRF/5 cells. (A) Western blot analysis demonstrating that CD206 expression was reduced following shCD206 transfection. (B) Significant impairment of cell migration in HepG2 and PLC/PRF/5 cells transfected with shCD206 compared with controls was observed. (C) Quantification of the migration of shCD206 transfected cells demonstrating that cell migration was significantly decreased compared with controls. (D) Significant reduction in invasion was observed in HepG2 and PLC/PRF/5 cells transfected with shCD206 compared with shNC controls. (E) Quantification of the invasion of shCD206 transected cells, indicating reduced invasiveness compared with controls. **P<0.01 vs. shNC treatment. CD206, mannose receptor; sh, short hair; NC, negative control.
Discussion
Liver cancer is a multigene disease characterized by a high degree of malignancy, rapid development, low survival rates and late detection (9,39). Surgical interventions are often ineffective due to late diagnosis (40). Novel liver cancer diagnostic and therapeutic targets are therefore required.
CD206 is a pattern recognition receptor that identifies the extracellular domains of specific carbohydrate molecules and is highly expressed on the surface of macrophages and immature dendritic cells (41). In the present study, CD206 was expressed in the cytoplasm and on the plasma membrane of liver cancer cells. Additionally, CD206 upregulation was observed in liver cancer tissue compared with healthy adjacent tissue obtained from patients with liver cancer. CD206 expression was not significantly associated with gender, age, tumor number, Edmondson grade, microvascular invasion, hepatitis B virus antigen and cirrhosis, but had a positive association with tumor size, metastasis and the AFP value. The present study therefore demonstrated a preliminary association between CD206 and the occurrence and development of liver cancer. Previous studies investigating liver cancer-associated proteins have demonstrated an association between marker expression and cancer staging. The present study revealed that CD206 expression was associated with tumor size, metastasis and the AFP value, indicating that its expression is associated with poor liver cancer prognosis. Furthermore, the association between CD206 expression and survival time revealed that the levels of CD206 were positively associated with poor prognosis. Taken together, these data suggested that high CD206 expression promotes the rapid growth and metastasis of liver cancer.
Uncontrolled self-renewal directly contributes to the progression of liver cancer and other types of carcinomas. The same molecular pathways that regulate self-renewal in normal stem cells also control CSCs (22,38,42). The present study revealed that CD206 is an important biomarker for liver cancer progression in CSCs, similar to other known markers including Nanog, Oct4, Sox2, c-Myc and CD44 (15,43–46). The expression of these markers in CSCs are recognized as evidence of stem cell carcinogenesis (37,47,48). The present study detected the expression levels of CD206 in CSCs and demonstrated high expression at both the mRNA and protein levels, which were comparable with the levels of Oct4 and c-Myc. The results obtained in the current study suggested that CD206 may be used as a biomarker in CSCs to predict liver cancer, highlighting its diagnostic value.
The present study investigated the direct effects of CD206 knockdown on liver cancer cells lines. The migration and invasion abilities of liver cancer cells decreased when CD206 was silenced, suggesting that high CD206 expression levels promote tumor metastasis and poor prognosis in patients.
Hepatitis C virus (HCV) infection is a main cause of liver cancer (49,50). However, the present study did not investigate the important role of HCV infection in liver cancer and this is a limitation of the study. Future studies investigating the association between HCV infection and CD206 expression are required.
In summary, the present study revealed that CD206 may be used as a diagnostic tool to screen for liver cancer, and its upregulation in liver tumors represents a therapeutic target to reduce liver cancer cell metastasis. Future investigation of the efficacy of CD206 inhibitors on liver cancer cells as well as an evaluation of the potential toxic effects on normal hepatocytes is required. Such studies may lay the theoretical foundations for future clinical liver cancer diagnosis and treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Science Foundation of China (grant nos. 81602706 and 81570198), Zhejiang Medical Technology Plan Project (grant no. WKJ-ZJ-1709) and the State Administration of Traditional Chinese Medicine of Zhejiang (grant nos. 2016ZZ007 and 2017ZB006).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
SW, LZ and XT conceived and designed the study. WF, XY and FH performed the experiments. SW wrote the paper. SW, LZ and XM reviewed and edited the manuscript. All authors read and approved the manuscript.
Ethical approval and consent to participate
The study was approved by the Ethics Committee of Zhejiang Provincial People's Hospital and all patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J Carcinog. 2017;16:1. doi: 10.4103/jcar.JCar_9_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altekruse SF, Henley SJ, Cucinelli JE, McGlynn KA. Changing hepatocellular carcinoma incidence and liver cancer mortality rates in the United States. Am J Gastroenterol. 2014;109:542–553. doi: 10.1038/ajg.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 4.Tsai WC, Kung PT, Wang YH, Kuo WY, Li YH. Influence of the time interval from diagnosis to treatment on survival for early-stage liver cancer. PLoS One. 2018;13:e0199532. doi: 10.1371/journal.pone.0199532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ronot M, Clift AK, Vilgrain V, Frilling A. Functional imaging in liver tumours. J Hepatol. 2016;65:1017–1030. doi: 10.1016/j.jhep.2016.06.024. [DOI] [PubMed] [Google Scholar]
- 6.Marrero JA, Ahn J, Rajender Reddy K, Americal College of Gastroenterology ACG clinical guideline: The diagnosis and management of focal liver lesions. Am J Gastroenterol. 2014;109:1328–1348. doi: 10.1038/ajg.2014.213. [DOI] [PubMed] [Google Scholar]
- 7.Elsayes KM, Hooker JC, Agrons MM, Kielar AZ, Tang A, Fowler KJ, Chernyak V, Bashir MR, Kono Y, Do RK, et al. 2017 version of LI-RADS for CT and MR Imaging: An update. Radiographics. 2017;37:1994–2017. doi: 10.1148/rg.2017170098. [DOI] [PubMed] [Google Scholar]
- 8.Ryerson AB, Eheman CR, Altekruse SF, Ward JW, Jemal A, Sherman RL, Henley SJ, Holtzman D, Lake A, Noone AM, et al. Annual report to the nation on the status of cancer, 1975–2012, featuring the increasing incidence of liver cancer. Cancer. 2016;122:1312–1337. doi: 10.1002/cncr.29936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sia D, Villanueva A, Friedman SL, Llovet JM. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152:745–761. doi: 10.1053/j.gastro.2016.11.048. [DOI] [PubMed] [Google Scholar]
- 10.Bhoori S, Mazzaferro V. Current challenges in liver transplantation for hepatocellular carcinoma. Best Pract Res Clin Gastroenterol. 2014;28:867–879. doi: 10.1016/j.bpg.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Chen W, Fan W, Ru G, Huang F, Lu X, Zhang X, Mou X, Wang S. Gemcitabine combined with an engineered oncolytic vaccinia virus exhibits a synergistic suppressive effect on the tumor growth of pancreatic cancer. Oncol Rep. 2019;41:67–76. doi: 10.3892/or.2018.6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staines K, Hunt LG, Young JR, Butter C. Evolution of an expanded mannose receptor gene family. PLoS One. 2014;9:e110330. doi: 10.1371/journal.pone.0110330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SH, Charmoy M, Romano A, Paun A, Chaves MM, Cope FO, Ralph DA, Sacks DL. Mannose receptor high, M2 dermal macrophages mediate nonhealing Leishmania major infection in a Th1 immune environment. J Exp Med. 2018;215:357–375. doi: 10.1084/jem.20171389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng H, Geil Nickell CR, Chen KY, McClain JA, Nixon K. Increased expression of M1 and M2 phenotypic markers in isolated microglia after four-day binge alcohol exposure in male rats. Alcohol. 2017;62:29–40. doi: 10.1016/j.alcohol.2017.02.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salmi M, Karikoski M, Elima K, Rantakari P, Jalkanen S. CD44 binds to macrophage mannose receptor on lymphatic endothelium and supports lymphocyte migration via afferent lymphatics. Circ Res. 2013;112:1577–1582. doi: 10.1161/CIRCRESAHA.111.300476. [DOI] [PubMed] [Google Scholar]
- 16.van Lessen M, Shibata-Germanos S, van Impel A, Hawkins TA, Rihel J, Schulte-Merker S. Intracellular uptake of macromolecules by brain lymphatic endothelial cells during zebrafish embryonic development. Elife. 2017;6(pii):e25932. doi: 10.7554/eLife.25932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azad AK, Rajaram MV, Metz WL, Cope FO, Blue MS, Vera DR, Schlesinger LS. γ-Tilmanocept, a new radiopharmaceutical tracer for cancer sentinel lymph nodes, binds to the mannose receptor (CD206) J Immunol. 2015;195:2019–2029. doi: 10.4049/jimmunol.1402005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersen MN, Andersen NF, Rodgaard-Hansen S, Hokland M, Abildgaard N, Moller HJ. The novel biomarker of alternative macrophage activation, soluble mannose receptor (sMR/sCD206): Implications in multiple myeloma. Leuk Res. 2015;39:971–975. doi: 10.1016/j.leukres.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Bianchi G, Munshi NC. Pathogenesis beyond the cancer clone(s) in multiple myeloma. Blood. 2015;125:3049–3058. doi: 10.1182/blood-2014-11-568881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kume H, Muraoka S, Kuga T, Adachi J, Narumi R, Watanabe S, Kuwano M, Kodera Y, Matsushita K, Fukuoka J, et al. Discovery of colorectal cancer biomarker candidates by membrane proteomic analysis and subsequent verification using selected reaction monitoring (SRM) and tissue microarray (TMA) analysis. Mol Cell Proteomics. 2014;13:1471–1484. doi: 10.1074/mcp.M113.037093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodgaard-Hansen S, Rafique A, Christensen PA, Maniecki MB, Sandahl TD, Nexo E, Moller HJ. A soluble form of the macrophage-related mannose receptor (MR/CD206) is present in human serum and elevated in critical illness. Clin Chem Lab Med. 2014;52:453–461. doi: 10.1515/cclm-2013-0451. [DOI] [PubMed] [Google Scholar]
- 22.Sun C, Shui B, Zhao W, Liu H, Li W, Lee JC, Doran R, Lee FK, Sun T, Shen QS, et al. Central role of IP3R2-mediated Ca2+ oscillation in self-renewal of liver cancer stem cells elucidated by high-signal ER sensor. Cell Death Dis. 2019;10:396. doi: 10.1038/s41419-019-1613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao J, Zhao M, Liu J, Zhang X, Pei Y, Wang J, Yang X, Shen B, Zhang J. RACK1 promotes self-renewal and chemoresistance of cancer stem cells in human hepatocellular carcinoma through stabilizing nanog. Theranostics. 2019;9:811–828. doi: 10.7150/thno.29271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X, Li S, Wang H, Chen W, Mou X, Wang S. Expression of coxsackie and adenovirus receptor is correlated with inferior prognosis in liver cancer patients. Oncol Lett. 2019;17:2485–2490. doi: 10.3892/ol.2018.9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrizzo A, Buonaguro L. Application of the Immunoscore as prognostic tool for hepatocellular carcinoma. J Immunother Cancer. 2016;4:71. doi: 10.1186/s40425-016-0182-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo Y, Pandey A, Ghasabeh MA, Pandey P, Varzaneh FN, Zarghampour M, Khoshpouri P, Ameli S, Li Z, Hu D, Kamel IR. Prognostic value of baseline volumetric multiparametric MR imaging in neuroendocrine liver metastases treated with transarterial chemoembolization. Eur Radiol. 2019 Mar 15; doi: 10.1007/s00330-019-06100-3. doi: 10.1007/s00330-019-06100-3 (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 27.Colecchia A, Scaioli E, Montrone L, Vestito A, Di Biase AR, Pieri M, D'Errico-Grigioni A, Bacchi-Reggiani ML, Ravaioli M, Grazi GL, Festi D. Pre-operative liver biopsy in cirrhotic patients with early hepatocellular carcinoma represents a safe and accurate diagnostic tool for tumour grading assessment. J Hepatol. 2011;54:300–305. doi: 10.1016/j.jhep.2010.06.037. [DOI] [PubMed] [Google Scholar]
- 28.Kong M, Hong SE. Optimal follow-up duration for evaluating objective response to radiotherapy in patients with hepatocellular carcinoma: A retrospective study. Chin J Cancer. 2015;34:79–85. doi: 10.5732/cjc.014.10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gan W, Zhang MX, Wang JX, Fu YP, Huang JL, Yi Y, Jing CY, Fan J, Zhou J, Qiu SJ. Prognostic impact of lactic dehydrogenase to albumin ratio in hepatocellular carcinoma patients with Child-Pugh I who underwent curative resection: A prognostic nomogram study. Cancer Manag Res. 2018;10:5383–5394. doi: 10.2147/CMAR.S194035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Couri T, Pillai A. Goals and targets for personalized therapy for HCC. Hepatol Int. 2019;13:125–137. doi: 10.1007/s12072-018-9919-1. [DOI] [PubMed] [Google Scholar]
- 31.Zhu R, Huang H, Zhang H, Wang Z, Hu X, Zhai W, Lin Y, Wang J, Zhu H. Prognostic analysis in chronic hepatitis B patients: A retrospective study of 216 cases about Scheuer scores, in situ expression of viral antigens and tissue hepatitis B virus DNA levels. Liver Int. 2006;26:82–89. doi: 10.1111/j.1478-3231.2005.01199.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhu AX, Kang YK, Yen CJ, Finn RS, Galle PR, Llovet JM, Assenat E, Brandi G, Pracht M, Lim HY, et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased alpha-fetoprotein concentrations (REACH-2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019;20:282–296. doi: 10.1016/S1470-2045(18)30937-9. [DOI] [PubMed] [Google Scholar]
- 33.Shim JH, Yoon DL, Han S, Lee YJ, Lee SG, Kim KM, Lim YS, Lee HC, Chung YH, Lee YS. Is serum alpha-fetoprotein useful for predicting recurrence and mortality specific to hepatocellular carcinoma after hepatectomy? A test based on propensity scores and competing risks analysis. Ann Surg Oncol. 2012;19:3687–3696. doi: 10.1245/s10434-012-2416-1. [DOI] [PubMed] [Google Scholar]
- 34.Zhong M, Zhong C, Cui W, Wang G, Zheng G, Li L, Zhang J, Ren R, Gao H, Wang T, et al. Induction of tolerogenic dendritic cells by activated TGF-beta/Akt/Smad2 signaling in RIG-I-deficient stemness-high human liver cancer cells. BMC Cancer. 2019;19:439. doi: 10.1186/s12885-019-5670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei X, You X, Zhang J, Zhou C. MicroRNA-1305 inhibits the stemness of LCSCs and Tumorigenesis by repressing the UBE2T-dependent Akt-signaling pathway. Mol Ther Nucleic Acids. 2019;16:721–732. doi: 10.1016/j.omtn.2019.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bamodu OA, Kuo KT, Yuan LP, Cheng WH, Lee WH, Ho YS, Chao TY, Yeh CT. HDAC inhibitor suppresses proliferation and tumorigenicity of drug-resistant chronic myeloid leukemia stem cells through regulation of hsa-miR-196a targeting BCR/ABL1. Exp Cell Res. 2018;370:519–530. doi: 10.1016/j.yexcr.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 37.Choi HS, Kim JH, Kim SL, Deng HY, Lee D, Kim CS, Yun BS, Lee DS. Catechol derived from aronia juice through lactic acid bacteria fermentation inhibits breast cancer stem cell formation via modulation Stat3/IL-6 signaling pathway. Mol Carcinog. 2018;57:1467–1479. doi: 10.1002/mc.22870. [DOI] [PubMed] [Google Scholar]
- 38.Mani SK, Zhang H, Diab A, Pascuzzi PE, Lefrancois L, Fares N, Bancel B, Merle P, Andrisani O. EpCAM-regulated intramembrane proteolysis induces a cancer stem cell-like gene signature in hepatitis B virus-infected hepatocytes. J Hepatol. 2016;65:888–898. doi: 10.1016/j.jhep.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu M, Jiang L, Guan XY. The genetic and epigenetic alterations in human hepatocellular carcinoma: A recent update. Protein Cell. 2014;5:673–691. doi: 10.1007/s13238-014-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orcutt ST, Anaya DA. Liver resection and surgical strategies for management of primary liver cancer. Cancer Control. 2018;25:1073274817744621. doi: 10.1177/1073274817744621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scodeller P, Simon-Gracia L, Kopanchuk S, Tobi A, Kilk K, Saalik P, Kurm K, Squadrito ML, Kotamraju VR, Rinken A, et al. Precision targeting of tumor macrophages with a CD206 binding peptide. Sci Rep. 2017;7:14655. doi: 10.1038/s41598-017-14709-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maehara O, Ohnishi S, Asano A, Suda G, Natsuizaka M, Nakagawa K, Kobayashi M, Sakamoto N, Takeda H. Metformin regulates the expression of CD133 through the AMPK-CEBPβ pathway in hepatocellular carcinoma cell lines. Neoplasia. 2019;21:545–556. doi: 10.1016/j.neo.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palla AR, Piazzolla D, Abad M, Li H, Dominguez O, Schonthaler HB, Wagner EF, Serrano M. Reprogramming activity of NANOGP8, a NANOG family member widely expressed in cancer. Oncogene. 2014;33:2513–2519. doi: 10.1038/onc.2013.196. [DOI] [PubMed] [Google Scholar]
- 44.Jerabek S, Merino F, Scholer HR, Cojocaru V. OCT4: Dynamic DNA binding pioneers stem cell pluripotency. Biochim Biophys Acta. 2014;1839:138–154. doi: 10.1016/j.bbagrm.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Guvench O. Revealing the mechanisms of protein disorder and N-glycosylation in CD44-hyaluronan binding using molecular simulation. Front Immunol. 2015;6:305. doi: 10.3389/fimmu.2015.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida GJ. Emerging roles of Myc in stem cell biology and novel tumor therapies. J Exp Clin Cancer Res. 2018;37:173. doi: 10.1186/s13046-018-0964-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki Y, Haraguchi N, Takahashi H, Uemura M, Nishimura J, Hata T, Takemasa I, Mizushima T, Ishii H, Doki Y, et al. SSEA-3 as a novel amplifying cancer cell surface marker in colorectal cancers. Int J Oncol. 2013;42:161–167. doi: 10.3892/ijo.2012.1713. [DOI] [PubMed] [Google Scholar]
- 48.Botchkina GI, Zuniga ES, Das M, Wang Y, Wang H, Zhu S, Savitt AG, Rowehl RA, Leyfman Y, Ju J, et al. New-generation taxoid SB-T-1214 inhibits stem cell-related gene expression in 3D cancer spheroids induced by purified colon tumor-initiating cells. Mol Cancer. 2010;9:192. doi: 10.1186/1476-4598-9-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanda T, Goto T, Hirotsu Y, Moriyama M, Omata M. Molecular mechanisms driving progression of liver cirrhosis towards hepatocellular carcinoma in chronic Hepatitis B and C infections: A review. Int J Mol Sci. 2019;20(pii):E1358. doi: 10.3390/ijms20061358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zahra M, Azzazy H, Moustafa A. Transcriptional regulatory networks in Hepatitis C virus-induced hepatocellular carcinoma. Sci Rep. 2018;8:14234. doi: 10.1038/s41598-018-32464-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.