Abstract
Effect of Runx3 gene on the cell proliferation and invasion of rectal cancer was investigated to explore potential new targets for targeted treatment of rectal cancer. The Runx3 overexpression group (OE group), blank plasmid control group, negative control and blank group of the rectal cancer HRC-9698 cell strain were set. The overexpressed Runx3 plasmid was transfected in OE group; the empty plasmid was transfected in blank plasmid control group; only liposome Lipofectamine was added to negative control group; only 1640 medium was used in blank group. RT-qPCR was used for detection of the mRNA expression of Runx3 in different groups; CCK-8 kit for detection of cell proliferation in different groups; Transwell chamber test for detection of cell strain invasion in different groups. The mRNA expression of Runx3 gene in OE group was the highest, significantly higher than that in blank plasmid control group, negative control and blank group (P<0.01). The OD values of overexpressed Runx3 at 96 h after transfection in OE group was significantly lower than each control group (P<0.01). At the same time-point, pairwise comparison in each group found that OE group was significantly lower than blank plasmid control, negative control and blank groups (all P<0.01). In the invasion experiment, the number of invasion cells in OE, blank plasmid control, negative control and blank groups were 38.63±9.33, 107.87±5.66, 110.93±4.33 and 112.86±6.66, respectively. OE group was significantly lower than each control group (P<0.01). Overexpression of Runx3 gene in vitro inhibits the cell proliferation of rectal cancer and blocks the cell invasion and metastasis. This study provides a new idea and a new molecular therapeutic target for molecular targeted therapy of rectal cancer.
Keywords: rectal cancer, Runx3, HRC-9698 cell strain, proliferation, invasion
Introduction
Rectal cancer occurs between the dentate line and the recto-sigmoid junction and is one of the most common malignancies in the digestive system (1). With the rapid development of the economic level, people's mental pressure on life is also increasing, the life pace is getting increasingly faster, the incidence of colorectal cancer (CRC) has increased year by year, and the incidence in youth has gradually increased (2). According to statistics, the top three new cancers in women in the United States in 2018 are breast cancer, lung cancer and CRCs, of which CRC accounts for 7% of new cancer cases (3). Globally CRC is the third most common cancer, accounting for 10% of all cancer cases (4). In many cancers, metastasis that is the main cause of death in patients begins with the spread of tumor cells at the primary site and leads to secondary tumors at distant anatomic site (5). Although significant progress has been made in understanding the molecular nature of metastasis, there are still many problems in the intracellular mechanism of controlling metastasis process (6).
Runx3, a member of the Runx family, functions as a transcription factor, and its encoded protein is an α subunit that can bind to CBFβ/PEBP2β to form a heterodimer in the enhancer or promoter of a target gene, so as to regulate the gene expression (7). It is the first member of the Runx family and plays a role as tumor suppressor gene in various cancers such as gastric, lung and liver cancers (8,9). Runx3, a downstream effector of the TGFβ signaling pathway, can participate in the inhibition of Wnt signal transduction, thereby exerting a carcinogenic effect (10). Studies have found that Runx3 expression is also downregulated in CRC, and Runx3 is associated with the prognosis, clinical stage, differentiation degree and lymph node metastasis of CRC (11,12). However, the Runx3-mediated mechanism that inhibits tumor metastasis is still unclear, and the effect of this gene on cell proliferation, invasion and migration of CRC has not been studied. This study explores potential new targets for targeted treatment of rectal cancer by investigating the effect of Runx3 gene on the cell proliferation and invasion of rectal cancer.
Materials and methods
Materials and reagents
HRC-9698 cells (Shanghai Zishi Biotechnology Co., Ltd.); Runx3 overexpression plasmid (Shandong Weizhen Biotechnology Co., Ltd.); fetal bovine serum, RPMI-1640 medium, 25% trypsin and 0.02% EDTA (Procell Life Science & Technology Co., Ltd.); TransScript Green Two-Step qRT-PCR SuperMix kit (TransGen BiotechBiotechnology Co., Ltd.); Lipofectamine® 3000 transfection reagent and TRIzol reagent (Thermo Fisher Scientific, Inc.); qPCR primer sequences (Shanghai Shenggong Biology Engineering Technology Service, Ltd.); CCK-8 kit (Shanghai Yisheng Biotechnology Co. Ltd.); Transwell chamber (Beijing Yiming Fuxing Biotechnology.).
The study was approved by the Ethics Committee of China-Japan Union Hospital of Jilin University (Changchun, China).
Culture method for HRC-9698 cell strain
HRC-9698 cells are suspension cells. The cells were aspirated with a pipette into a RPMI-1640 medium with 9 ml of 10% fetal bovine serum; centrifuged at 800 × g for 5 min at 4°C and resuspended; the resuspended cells were aspirated with a pipette into a new medium and incubated in an incubator at 37°C and 5% CO2. After the cells grew to ~70% confluence, they were digested with 0.25% trypsin and 0.02% EDTA mixture, inoculated into a six-well plate, and divided into 4 groups (OE, blank plasmid control, negative control and blank groups), 3 complex wells per group.
Liposome transfection
Transfection was performed when the cell confluence reached 80%. The Runx3 overexpression vector and liposome Lipofectamine® 3000 were added to OE group, the empty plasmid and liposome Lipofectamine 3000 to blank plasmid control group, only liposome Lipofectamine 3000 to negative control group, and equal amounts of complete medium to blank group but it was not transfected. Operation was performed strictly in accordance with Lipofectamine 3000 transfection reagent instruction and the cells were incubated at room temperature for 5 min. The prepared plasmid DNA and liposome complexes were added to the cells that were incubated at 37°C and 5% CO2 for 2 days, and then the transfected cells were detected and analyzed.
RT-qPCR detection of Runx3 gene expression
RNA extraction and cDNA synthesis: TRIzol and chloroform reagent were used for extraction of total RNA from each group of cells, and the UV spectrophotometer for detection of RNA concentration and purity, A260/A280.
The ratio is considered as qualified between 1.8–2.0. The reverse transcription reaction system was prepared according to Table I, mixed gently, incubated at 42°C for 15 min, heated at 85°C for 5 sec to inactivate TransScript® RT/RI (TransGen BiotechBiotechnology Co., Ltd.) and gDNA remover.
Table I.
Preparation of reaction system.
| Reverse transcription | PCR |
|---|---|
| Total RNA 1 µg | Upstream and downstream sequences (10 µM) 0.4 µl |
| 5X TransScript® All-in-One | Passive reference Dve |
| SuperMix for qPCR 4 µl | (50X) (optional) 0.4 µl |
| gDNA remover 1 µl | 2X TransStart® Tin Green |
| aPCR SuperMix 0.4 µl | |
| RNase-free water to 20 µl | cDNA 2 µl |
| RNase-free water to 20 µl |
qPCR reaction with TransScript Green Two-Step qRT-PCR SuperMix kit (TransGen BiotechBiotechnology Co., Ltd.): the qPCR reaction system was prepared according to Table I, and two-step method was used for amplification, pre-denaturation at 94°C for 30 sec, at 94°C for 5 sec, and at 60°C for 30 sec for 40 cycles. GAPDH was used as an internal reference, and the upstream and downstream sequences were 5′-GGTGGTGCTAAGCGTGTTA-3′ and 5′-CCCTCCACAATGCCAA-3′, respectively; the upstream and downstream sequences of the target gene Runx3 were 5′-GGCGGTACCATGGCATCGAACAGCATCTTC-3′ and 5′-GCGCTCGAGTCAGTAGGGCCGCCACACGG-3′, respectively. The experimental results were analyzed using the2−ΔΔCq method (13).
CCK-8 test of proliferation
Cell suspensions (1×105) were prepared and placed in a 96-well plate, 100 µl per well; 3 complex wells per group; pre-incubated in an incubator at 37°C and 5% CO2 for 1 day; incubated continually in incubator and 10 µl of CCK-8 solution was added at 12 h. After the incubation for 3 h, a microplate reader (Bio-Rad Laboratories, Inc.) was used for determination of OD450 nm as the initial value. Then, 10 µl of CCK-8 solution was added to each well every 24 h (24, 48, 72 and 96 h). After incubated at 37°C and 5% CO2 for 3 h, a microplate reader was used for determination of the absorbance at 450 nm.
Transwell invasion experiment
Preparation of cell suspension: serum-free 1640 medium was used to culture cells for 12 h to remove serum influence and prepare cell suspension. The cells were digested with 0.25% trypsin and 0.02% EDTA mixture, the culture solution was discarded by centrifugation at 3,000 × g for 8 min at 4°C after digestion, and the cells were washed 1–2 times with PBS, resuspend with serum-free medium, and counted to adjust cell density to 5×105/ml; 100 µl of cell suspensions were added into the Transwell upper chamber of a 24-well plate; 1640 medium with 600 µl of 20% fetal bovine serum was added to the lower chamber; incubated at 37°C and 5% CO2 for 12 h; the upper non-migrated cells were gently wiped off with a cotton swab and washed 3 times with PBS; fixed with 4% paraformaldehyde for 10 min; stained with 0.1% crystal violet for 20 min. The counts with five fields under a microscope (Olympus Corporation) at 400 times were observed, and five field counts were randomly selected in each well to obtain the average value. The invasion experiment was performed on Matrigel-coated cabs purchased from BD Company and diluted at the ratio of 1:8.
Statistical analysis
SPSS20.0 (IBM Corp.) software statistical package was used for test and analysis of the data. The measurement data are expressed as mean ± standard deviation, and whether there was a difference in results of the four groups was analyzed using ANOVA single factor, then LSD was used to test whether there was a difference between the two. The counting data was expressed as rate and tested using Chi-square. The comparison among different time-points in the same group was analyzed using repeated measurement variance. The 2−ΔΔCq method was used to statistically analyze the RT-qPCR results. The significance level is α=0.05.
Results
RT-qPCR detection of Runx3 gene expression
RT-qPCR was used for detection of the Runx3 gene expression in the four groups of cells. The results of OE group, blank plasmid control group, negative control group and blank group were 242.55±13.97, 1.31±0.10, 1.45±0.06 and 1.55±0.12, respectively. The difference was not statistically significant among blank plasmid control, negative control and blank groups (P>0.05). They were significantly higher in OE group than those in blank plasmid control, negative control and blank groups (all P<0.01) (Fig. 1).
Figure 1.
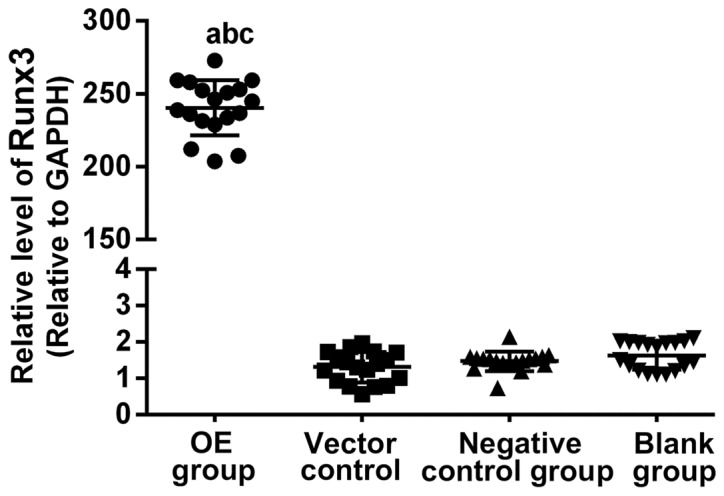
RT-qPCR detection of Runx3 gene expression. RT-qPCR was used for detection of the Runx3 gene expression in the four groups of cells, and it was found that the difference was not statistically significant among blank plasmid control, negative control and blank groups (P>0.05). They were significantly higher in OE group than those in blank plasmid control, negative control and blank groups (all P<0.01). aP<0.01, compared to blank plasmid control group; bP<0.01, compared to negative control group; cP<0.01, compared to blank group. OE group, overexpression group.
CCK-8 test
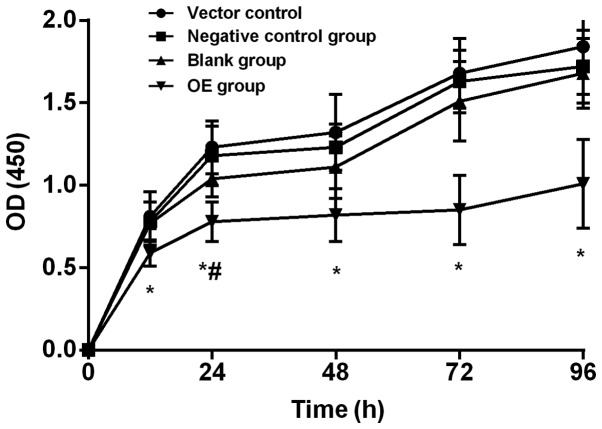
The cell proliferation in each group was determined at 12, 24, 48, 72 and 96 h using the CCK-8 method. The OD values measured at the five time-points in each group by the CCK-8 test are shown in Table II. At the same time-point, pairwise comparison in each group found that OE group was significantly lower than blank plasmid control, negative control and blank groups (all P<0.01), and the difference was not statistically significant among the latter three groups (P>0.05). CCK-8 proliferation curve is shown in Fig. 2. Comparison among different time-points in the same group found that the proliferation curves of the four groups all showed an upward trend, and the OD values at 24 and 72 h were significantly higher than those at previous time-point in blank plasmid control, negative control and blank groups, and the OD values at 24 h were significantly higher than those at previous time-point in OE group (all P<0.01).
Table II.
CCK-8 test results of each group at different time-points.
| Items | Blank plasmid control group | Negative control group | Blank group | OE group | F-value | P-value |
|---|---|---|---|---|---|---|
| 12 h | 0.88±0.15 | 0.78±0.12 | 0.77±0.13 | 0.59±0.08a | 5.96 | 0.02 |
| 24 h | 1.23±0.16b | 1.18±0.18b | 1.04±0.11b | 0.78±0.12a,b | 17.34 | <0.001 |
| 48 h | 1.32±0.23 | 1.23±0.14 | 1.11±0.19 | 0.82±0.16a | 12.72 | <0.001 |
| 72 h | 1.68±0.21b | 1.63±0.19b | 1.51±0.24b | 0.85±0.21a | 29.34 | <0.001 |
| 96 h | 1.84±0.29 | 1.72±0.22 | 1.68±0.21 | 1.01±0.27a | 20.24 | <0.001 |
| F-value | 32.03 | 42.84 | 36.6 | 6.26 | ||
| P-value | <0.001 | <0.001 | <0.001 | <0.001 |
P<0.05, at the same time-point, compared to blank plasmid control, negative control and blank groups
P<0.05, compared to previous time-point in the same group. OE group, overexpression group.
Figure 2.
CCK-8 test. The cell proliferation in each group was determined at 12, 24, 48, 72 and 96 h. The OD values measured at the five time-points in each group by the CCK-8 test are shown in Table I. At the same time-point, pairwise comparison in each group found that OE group was significantly lower than blank plasmid control, negative control and blank groups (all P<0.01), and the difference was not statistically significant among the latter three groups (P>0.05). Comparison among different time-points in the same group found that the proliferation curves of the four groups all showed an upward trend, and the OD values at 24 and 72 h were significantly higher than those at previous time-point in blank plasmid control, negative control and blank groups, and the OD values at 24 h were significantly higher than those at previous time-point in OE group (all P<0.01). *P<0.05, at the same time-point, compared to blank plasmid control group, negative control group and blank group. #P<0.05, compared to previous time-point in the same group. OE group, overexpression group.
Transwell invasion experiment
The number of invasion cells is shown in Table III. In OE group, blank plasmid control, negative control and blank groups were 38.63±9.33, 107.87±5.66, 110.93±4.33 and 112.86±6.66, respectively. The percentage of invasion was calculated as the mean number of invasion in blank group of 100%. As shown in Fig. 3, the number of invasion cells in OE group was significantly lower than that in blank plasmid control, negative control and blank groups, and there was no significant difference among the latter three groups. The experimental results were statistically significant (P<0.01).
Table III.
Comparison of number of invasion cells.
| Groups | Number of invasion cells | (%) |
|---|---|---|
| OE | 38.63±9.33a–c | 34.23a–c |
| Blank plasmid control | 107.87±5.66 | 95.58 |
| Negative control | 110.93±4.33 | 98.23 |
| Blank | 112.86±6.66 | 100.00 |
P<0.01, compared to blank plasmid control group
P<0.01, compared to negative control group
P<0.01, compared to blank group. OE group, overexpression group.
Figure 3.
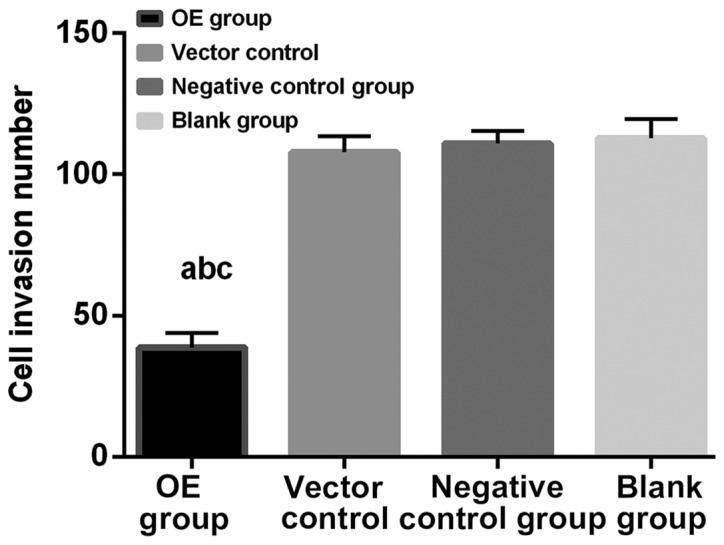
Transwell invasion experiment. The number of invasion cells in OE group was significantly lower than that in blank plasmid control, negative control and blank groups, and there was no significant difference among the latter three groups. The experimental results were statistically significant (P<0.01). aP<0.01, compared to blank plasmid control group; bP<0.01, compared to negative control group; cP<0.01, compared to blank group. OE group, overexpression group.
Discussion
CRC is one of the ‘four killers’ that threaten human health (14). After the age of 50, the incidence of CRC rapidly increases. It generally undergoes precancerous lesions for several decades, so precancerous screening before the age of 50 can early detect and treat it, so as to reduce its mortality (15). Early radical surgery can greatly reduce the mortality of rectal cancer, but due to the particularity of the anatomical position of it, the operation is difficult to complete, and it is difficult to retain the anus and function after operation, bringing great inconvenience to the patient's postoperative life (16). Runx3 that is downregulated in a variety of tumor tissues has been found to be closely related to the differentiation degree and clinical stage of rectal cancer (17,18). The study of the effect of Runx3 on the cell proliferation, invasion and migration of CRC can help us to understand the antitumor effect and the manner of action of Runx3, and provide more theoretical basis for the clinical study and drug development of CRC cells.
In this study, the Runx3 gene expression in each group was detected by RT-qPCR and was significantly higher in OE group than that in blank plasmid control, negative control and blank groups, indicating that Runx3 overexpression vector successfully transfected into OE group of cells in this experiment. CCK-8 proliferation experiment found that at the same time-point, pairwise comparison in each group found that OE group was significantly lower than blank plasmid control, negative control and blank groups. Comparison among different time-points in the same group found that the proliferation curves of the four groups all showed an upward trend, suggesting that the Runx3 overexpression inhibited cell proliferation. Transwell invasion experiment revealed that the number of invasion cells in OE group was significantly lower than that in blank plasmid control, negative control and blank groups, indicating that the Runx3 overexpression inhibited the invasion of OE group. In a study of the effect of Runx3 on CRC metastasis and angiogenesis, Kim et al (19) found that compared to normal paracancerous colonic tissues, Runx3 expression decreased in CRC tissues and was significantly associated with TNM stage. The Runx3 overexpression inhibits the migration and invasion of CRC cells due to high expression of matrix metalloproteinase-2 and −9, similar to the results of this study. Whether Runx3 also inhibits cell invasion by matrix metalloproteinase-2 and −9 in rectal cancer requires further validation. Wang et al (20) also found that miR-301a promoted the cell growth, migration and invasion of gastric cancer by downregulating the Runx3 expression. However, only one HRC-9698 rectal cancer cell strain was used in this study, and it does not truly represent the most natural occurrence and progression of rectal cancer. What is the Runx3 expression in normal rectal cancer cell strains? How does it function? Both need to be further validated with several cell lines.
At the same time, studies (8) have found that Runx3 not only has the ability to inhibit the invasion and metastasis of various cancer cells, but also has many other potential functions and clinical effects. Bauer et al (21) found that the Runx3 expression in leukocytes is the key to affect DMBA/TPA two-step method, thus inducing the occurrence of skin tumors. The loss of Runx3 in leukocytes significantly increased the content of antitumor cytokine thymic stromal lymphopoietin (TSLP), and significantly decreased the content of pro-tumor cytokine interleukin-17a and osteopontin. This unique combination of cytokines enabled the tumor microenvironment to be polarized to an effective antitumor state, which means that Runx3 expression facilitates inflammation-mediated tumor progression in skin tumors. It is contrary to this study and other literature reports that Runx3 has antitumor effect, which may be related to the biological function of TSLP. TSLP, a multifunctional IL-7-like cytokine, can act on lymphocytes, DC cells and tumor cells. On the one hand, TSLP plays an antitumor role in colon, skin early breast and pancreatic cancers; on the other hand, it exerts tumor-promoting effects in gastric cancer, lung cancer, cervical cancer and acute lymphoblastic leukemia (22). In addition, Runx3 has an inhibitory effect on various tumors such as gastric, colon and breast cancers, but has a carcinogenic effect on basal cell carcinoma, head and neck cancer and ovarian cancer (23)
In conclusion, overexpression of Runx3 gene in vitro inhibits the cell proliferation of rectal cancer and blocks the cell invasion and metastasis. This study provides a new idea and a new molecular therapeutic target for molecular targeted therapy of rectal cancer.
Acknowledgements
Not applicable.
Funding
This study was supported by Jilin Province High Technology Industrialization Program (2014Y083), Jilin Provincial Science and Technology Development Program (20140204028XY), Jilin Provincial Direct Health Special Program (2016, 2017), Jilin Natural Science Foundation Program (20160101115JC) and Jilin Provincial Youth Science Technology Innovation Leader and Team Program (20180519025JH).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YF drafted the manuscript. YF and SG were mainly devoted to cell culture and transfection. YG and DS performed PCR. XW and ZC were responsible for the CCK-8 assay. All authors read and approved the final study.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of China-Japan Union Hospital of Jilin University (Changchun, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Maingon P, Huguet F. Rectal Cancer: The Radiation Oncologist: The Great Watchmaker. Semin Radiat Oncol. 2016;26:173–175. doi: 10.1016/j.semradonc.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Cienfuegos JA, Rotellar F, Baixauli J, Beorlegui C, Sola JJ, Arbea L, Pastor C, Arredondo J, Hernández-Lizoáin JL. Impact of perineural and lymphovascular invasion on oncological outcomes in rectal cancer treated with neoadjuvant chemoradiotherapy and surgery. Ann Surg Oncol. 2015;22:916–923. doi: 10.1245/s10434-014-4051-5. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 4.Suárez-Villanueva S, Ayala-Madrigal ML, Peregrina-Sandoval J, Macías-Gómez N, Ramírez-Ramírez R, Muñiz-Mendoza R, Moreno-Ortiz JM, Centeno-Flores M, Maciel-Gutiérrez V, Cabrales E, et al. RUNX3 gene polymorphisms and haplotypes in Mexican patients with colorectal cancer. Genet Mol Res. 2015;14:15505–15510. doi: 10.4238/2015.November.30.28. [DOI] [PubMed] [Google Scholar]
- 5.Almaraz RT, Tian Y, Bhattarcharya R, Tan E, Chen SH, Dallas MR, Chen L, Zhang Z, Zhang H, Konstantopoulos K, et al. Metabolic flux increases glycoprotein sialylation: implications for cell adhesion and cancer metastasis. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M112.017558. M112.017558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su SC, Hsieh MJ, Yang WE, Chung WH, Reiter RJ, Yang SF. Cancer metastasis: Mechanisms of inhibition by melatonin. J Pineal Res. 2017;62:62. doi: 10.1111/jpi.12370. [DOI] [PubMed] [Google Scholar]
- 7.Slattery ML, Lundgreen A, Herrick JS, Caan BJ, Potter JD, Wolff RK. Associations between genetic variation in RUNX1, RUNX2, RUNX3, MAPK1 and eIF4E and riskof colon and rectal cancer: Additional support for a TGF-β-signaling pathway. Carcinogenesis. 2011;32:318–326. doi: 10.1093/carcin/bgq245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barghout SH, Zepeda N, Vincent K, Azad AK, Xu Z, Yang C, Steed H, Postovit LM, Fu Y. RUNX3 contributes to carboplatin resistance in epithelial ovarian cancer cells. Gynecol Oncol. 2015;138:647–655. doi: 10.1016/j.ygyno.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 9.Kang KA, Zhang R, Kim GY, Bae SC, Hyun JW. Epigenetic changes induced by oxidative stress in colorectal cancer cells: Methylation of tumor suppressor RUNX3. Tumour Biol. 2012;33:403–412. doi: 10.1007/s13277-012-0322-6. [DOI] [PubMed] [Google Scholar]
- 10.Chen F, Liu X, Bai J, Pei D, Zheng J. The emerging role of RUNX3 in cancer metastasis (Review) Oncol Rep. 2016;35:1227–1236. doi: 10.3892/or.2015.4515. [DOI] [PubMed] [Google Scholar]
- 11.Moon JW, Lee SK, Lee JO, Kim JH, Kim N, Kim J, Kim HS, Park SH. Demethylation of RUNX3 by vincristine in colorectal adenocarcinoma cells. Anticancer Res. 2014;34:133–140. [PubMed] [Google Scholar]
- 12.Nishina S, Shiraha H, Nakanishi Y, Tanaka S, Matsubara M, Takaoka N, Uemura M, Horiguchi S, Kataoka J, Iwamuro M, et al. Restored expression of the tumor suppressor gene RUNX3 reduces cancer stem cells in hepatocellular carcinoma by suppressing Jagged1-Notch signaling. Oncol Rep. 2011;26:523–531. doi: 10.3892/or.2011.1336. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Slattery ML, Lundgreen A, Herrick JS, Wolff RK, Caan BJ. Genetic variation in the transforming growth factor-β signaling pathway and survival after diagnosis with colon and rectal cancer. Cancer. 2011;117:4175–4183. doi: 10.1002/cncr.26018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nie K, Shi L, Chen Q, Hu X, Jabbour SK, Yue N, Niu T, Sun X. Rectal cancer: Assessment of neoadjuvant chemoradiation outcome based on radiomics of multiparametric MRI. Clin Cancer Res. 2016;22:5256–5264. doi: 10.1158/1078-0432.CCR-15-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Acuna SA, Chesney TR, Baxter NN. Pathologic outcomes of laparoscopic vs open mesorectal excision for rectal cancer. JAMA Surg. 2017;152:986–987. doi: 10.1001/jamasurg.2017.1724. [DOI] [PubMed] [Google Scholar]
- 17.Kim YS, Chae YK, Choi YS, Min JH, Ahn SW, Yoon JW, Lee SE, Lee YK. A comparative study of emergence agitation between sevoflurane and propofol anesthesia in adults after closed reduction of nasal bone fracture. Korean J Anesthesiol. 2012;63:48–53. doi: 10.4097/kjae.2012.63.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slattery ML, Lundgreen A. The influence of the CHIEF pathway on colorectal cancer-specific mortality. PLoS One. 2014;9:e116169. doi: 10.1371/journal.pone.0116169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim BR, Kang MH, Kim JL, Na YJ, Park SH, Lee SI, Kang S, Joung SY, Lee SY, Lee DH, et al. RUNX3 inhibits the metastasis and angiogenesis of colorectal cancer. Oncol Rep. 2016;36:2601–2608. doi: 10.3892/or.2016.5086. [DOI] [PubMed] [Google Scholar]
- 20.Wang M, Li C, Yu B, Su L, Li J, Ju J, Yu Y, Gu Q, Zhu Z, Liu B. Overexpressed miR-301a promotes cell proliferation and invasion by targeting RUNX3 in gastric cancer. J Gastroenterol. 2013;48:1023–1033. doi: 10.1007/s00535-012-0733-6. [DOI] [PubMed] [Google Scholar]
- 21.Bauer O, Hantisteanu S, Lotem J, Groner Y. Carcinogen-induced skin tumor development requires leukocytic expression of the transcription factor Runx3. Cancer Prev Res (Phila) 2014;7:913–926. doi: 10.1158/1940-6207.CAPR-14-0098-T. [DOI] [PubMed] [Google Scholar]
- 22.Milford TA, Su RJ, Francis OL, Baez I, Martinez SR, Coats JS, Weldon AJ, Calderon MN, Nwosu MC, Botimer AR, et al. TSLP or IL-7 provide an IL-7Rα signal that is critical for human B lymphopoiesis. Eur J Immunol. 2016;46:2155–2161. doi: 10.1002/eji.201646307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee CW, Chuang LS, Kimura S, Lai SK, Ong CW, Yan B, Salto-Tellez M, Choolani M, Ito Y. RUNX3 functions as an oncogene in ovarian cancer. Gynecol Oncol. 2011;122:410–417. doi: 10.1016/j.ygyno.2011.04.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.