Abstract
MicroRNAs (miRNAs) have been demonstrated to be involved in tumor progression of various human malignancies. The purpose of this study was to investigate the expression patterns and prognostic value of microRNA-106b (miR-106b) in osteosarcoma (OS) and to examine its functional role in OS progression. Reverse transcription-quantitative PCR (RT-qPCR) was used to estimate the expression of miR-106b in OS tissues and cells. The prognostic value of miR-106b in OS was evaluated by plotting Kaplan-Meier survival curves and performing Cox analyses. Cell experiments were carried out to examine the effects of miR-106b on OS cell proliferation, migration, and invasion. The expression of miR-106b was elevated in both OS tissues and cells compared with the expression in normal control tissues and cells (P<0.001). miR-106b expression was associated with metastasis (P=0.028) and Tumor-Node-Metastasis stage (P=0.017). Patients with high miR-106b expression levels had a poorer overall survival rate compared with those with low miR-106b expression levels (log-rank P=0.001). Multivariate Cox analyses indicated that miR-106b expression was an independent prognostic factor for patients with OS (hazard ratio=2.769; 95% confidence interval=1.369–5.599; P=0.005). The results of cell experiments implied that the upregulation of miR-106b could promote OS cell proliferation, migration and invasion, whereas the downregulation of miR-106b could suppress these functions (P<0.05). Taken together, this study's results indicated that the overexpression of miR-106b is associated with a poor prognosis for patients with OS and that overexpression promotes OS cell proliferation, migration, and invasion. This study may provide a novel prognostic biomarker and a candidate therapeutic target for OS treatment.
Keywords: microRNA-106b, prognosis, proliferation, migration, invasion, osteosarcoma
Introduction
Osteosarcoma (OS) is a type of bone cancer that can spread easily to other tissues or organs in the body (1). OS is considered the most frequent primary malignant bone tumor among children, adolescents and young adults and is responsible for approximately 5% of cancer-associated mortality worldwide (2). OS is an aggressive type of cancer and accounts for approximately 50% of bone sarcomas with metastasis (3). Statistical data have indicated that OS is more prevalent in males compared with females, and has an increasing incidence rate of 3.8 per 1,000,000 in males and 2.8 per 1,000,000 in females around the world (4). Although there is considerable progress in the improvement of therapeutic strategies for OS, such as surgical resection, chemotherapy, and radiotherapy, the outcomes and prognosis of patients with cancer remain poor (5). The 5-year survival rate for patients suffering from OS is <53.9% (6). Therefore, studies on novel functional molecules, which have pivotal roles during tumor progression, are important for OS treatment, as they may uncover potential therapeutic targets to improve cancer prognosis in patients with OS.
Accumulated evidence suggested that various genes are involved in tumor initiation and development and that genetic alterations are critical events during tumor progression in various types of human cancer, including colorectal carcinoma and gastric cancer (7,8). MicroRNAs (miRNAs), a class of small and noncoding RNAs, have a crucial gene regulation function that acts by binding to the 3′-untranslated regions (3′UTRs) of target genes (9). In addition, miRNAs have been reported to be involved in biological processes in both normal and tumor cells, such as cell proliferation, differentiation, invasion, migration, cell cycle and cell apoptosis (10–13). Emerging studies have shown that aberrant miRNA expression has been observed in tumor samples and that this expression exerts a functional role in the progression of various types of cancer, including retinoblastoma, gastric cancer and non-small cell lung cancer (14–16). Therefore, this study considered that the identification of functional miRNAs could provide novel prognostic biomarkers and effective therapeutic targets for the treatment of OS. MicroRNA-106b (miR-106b) is a member of the miR-106b-25 cluster, which has been found to be highly expressed in OS, according to Arabi et al (17). However, to the best of our knowledge, the clinical significance and functional role of miR-106b in OS have rarely been reported in previous studies.
The aim of the present study was to assess the expression patterns of miR-106b in OS tissues and cells, as well as its prognostic value for patients with OS. In addition, the effects of miR-106 on OS cell proliferation, migration, and invasion were also analyzed to further investigate its functional role during OS progression. The results of this study may provide novel insight on the prognosis and target therapy of OS regarding the role of miR-106b.
Materials and methods
Patients and tissues collection
The experimental protocols of this study were approved by the Ethics Committee of Ningbo No.2 Hospital (Ningbo, China). All patients provided written informed consent prior to surgery. A total of 134 patients, who were pathologically diagnosed with OS and received surgical resection at Ningbo No. 2 Hospital (Zhejiang, China) between January 2008 and July 2012, were recruited for this study. The patients with OS included 95 males and 39 females with a mean age of 19.95±13.17 years (range, 4–48 years). The patients met the following inclusion criteria: (1) None of the patients had received any anti-tumor therapy before the surgery; (2) the tumor tissues were pathologically diagnosed as OS by two experienced pathologists and (3) patients had a complete record in terms of demography and clinicopathological data. OS tissues and adjacent normal tissues were collected from the patients during the resection surgery and were immediately frozen in liquid nitrogen for subsequent RNA extraction. The tumors were classified into TNM stage I–II (n=61) and TNM stage III–IV (n=73) based on the guidelines of the American Joint Committee on Cancer (18). All patients had complete electronic medical records, and their demographic data and clinicopathological characteristics are summarized in Table I. A 5-year follow-up survey was conducted for the patients after the surgery, and the survival information was recorded for the survival analysis.
Table I.
Association between miR-106b expression and clinicopathological features of patients with OS.
| miR-106b expression | ||||
|---|---|---|---|---|
| Features | Total no. n=134 | Low (n=64) | High (n=70) | P-value |
| Age (years) | 0.982 | |||
| ≤40 | 92 | 44 | 48 | |
| >40 | 42 | 20 | 22 | |
| Sex | 0.811 | |||
| Female | 39 | 18 | 21 | |
| Male | 95 | 46 | 49 | |
| Tumor size (cm) | 0.941 | |||
| ≤5 | 80 | 38 | 42 | |
| >5 | 54 | 26 | 28 | |
| Differentiation | 0.151 | |||
| Well/moderate | 73 | 39 | 34 | |
| Poor | 61 | 25 | 36 | |
| Metastasis | 0.028a | |||
| Negative | 58 | 34 | 24 | |
| Positive | 76 | 30 | 46 | |
| TNM stage | 0.017a | |||
| I–II | 61 | 36 | 25 | |
| III–IV | 73 | 28 | 45 | |
TNM stage was determined according to the guidelines of the American Joint Committee on Cancer (18).
P<0.05. TNM, Tumor-Node-Metastasis; miR, microRNA; OS, osteosarcoma.
Cell culture and transfection
Four OS cell lines, including MG63, U2OS, HOS, and SaOS2, and one osteoblastic cell line, hFOB1.19, were purchased from the Cell Bank of the Chinese Academy of Sciences. All the cells were cultured in RPMI-1640 medium (HyClone; GE Healthcare Life Sciences), supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2. The expression of miR-106b was regulated in MG63 and HOS cells by cell transfection using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), since these cell lines exhibited higher expression levels of miR-106b. The vectors (50 nM) used in the transfection were as follows: miR-106b mimic, mimic negative control (NC), miR-106b inhibitor and inhibitor NC. All vectors were synthesized by Shanghai GenePharma Co., Ltd. with the sequences as follows: miR-106b mimic 5′-UAAAGUGCUGACAGUGCAGAU-3′, miR-106b inhibitor 5′-AUCUGCACUGUCAGCACUUUA-3′, mimic NC: 5′-UUCUCCGAACGUGUCACGUTT-3′, inhibitor NC: 5′-CAGUACUUUUGUGUAGUACAA-3′. After 48 h of cell transfection, the cells were used for subsequent cell experiments.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
The total RNA, including the miRNAs, was extracted from the collected tissues and all cell lines used in the present study with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. The concentration and quality of the RNA were evaluated using a NanoDrop 2000 (Thermo Fisher Scientific, Inc.). Reverse transcription was performed to synthesize cDNA using a PrimeScript RT Reagent kit (Takara Bio, Inc.), according to the manufacturer's protocols. The reaction condition was as follow: 42°C for 30 min, 85°C for 5 sec, storage at 4°C.
To estimate the expression levels of miR-106b, RT-qPCR was carried out using a 7300 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) with the SYBR green I Master Mix kit (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. U6 was selected as an internal control in the reactions. The thermocycling conditions included initial denaturation at 95°C for 10 min, denaturation at 95°C for 30 sec, 56°C for 30 sec, 72°C for 15 sec for a total of 40 cycles. The sequences of primer were as follows: miR-106b, forward: 5′-GGGGCTAAAGTGCTGACAGT-3′, reverse: 5′-GGAGCAGCAAGTACCCACAG-3′; U6, forward: 5′-GCTTCGGCAGCACATATACTAAAAT-3′, reverse: 5′-CGCTTCACGAATTTGCGTGTCAT-3′. The relative expression value of miR-106b was computed using the 2−ΔΔCq method (19) and was normalized to U6.
Cell proliferation analysis
To analyze the effect of miR-106b on cell proliferation of MG63 and HOS cells, an MTT assay was performed. The stably transfected OS cells were seeded into 96-well plates, and the cell density was adjusted to 4×105 cells/well. The cell culture plates were stored at 37°C with 5% CO2 for 72 h. During the incubation, the 10 µl MTT (5 mg/ml) was added to the wells every 24 h, and the samples were cultured for another 4 h. After removing of media, the cells were treated with 150 µl DMSO for 10 min. The absorbance was measured at a wavelength of 490 nm by a microplate reader (Thermo Fisher Scientific, Inc.) to evaluate the ability of OS cell proliferation.
Cell migration and invasion analysis
The abilities of cell migration and invasion of MG63 and HOS cells were examined by Transwell analysis using Transwell chambers with 8 µm pore size (Corning, Inc.). The stably transfected OS cells were seeded into the upper chambers (4×105 cells/well), which were filled with serum-free RPMI-1640 medium. The medium was supplemented with 10% FBS and was added to the lower chambers. The chambers were all cultured at 37°C in a humidified incubator with 5% CO2 for 48 h. After incubation, the migratory cells in the lower chambers were fixed with precooled 3.7% formaldehyde for 5 min at room temperature and stained using 0.1% crystal violet for 15 min at room temperature. The cell number was counted under a microscope (magnification, ×200) in 7 randomly-selected fields of view. For the invasion analysis, the Transwell chambers used were coated with Matrigel (Corning, Inc.).
Statistical analysis
The data are expressed as the mean ± standard deviation. All the statistical analyses were performed using SPSS 18.0 software (SPSS Inc.) and GraphPad Prism 5.0 software (GraphPad Software, Inc.). The expression values of miR-106b in the clinical samples were checked by Kolmogorov-Smirnov test. The comparisons of the groups were assessed using Student's t-test and one-way analysis of variance followed by Tukey's multiple comparison test. The association of miR-106b expression with clinicopathological features was analyzed using the χ2 test. The survival analysis was carried out using the Kaplan-Meier method and the log-rank test. Cox regression analysis was used to confirm the prognostic value of miR-106b in patients with OS. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-106b in OS tissues and cells
The expression values of miR-106b corresponded to a Gaussian distribution, and were significantly upregulated in OS tissues compared with that in the paired normal tissues (P<0.001; Fig. 1A). Similar results were observed in the OS cell lines, in which miR-106b expression was higher in the four OS cell lines than the expression in the normal cells (P<0.01; Fig. 1B).
Figure 1.
Expression of miR-106b in OS tissues and cell lines. (A) Expression of miR-106 was significantly increased in OS tissues compared with that in the matched normal controls. (B) Expression of miR-106b in the four OS cell lines (MG63, U2OS, HOS and SaOS2) was upregulated compared with that in the normal cell line hFOB1.19. **P<0.01 and ***P<0.001 vs. hFOB1.19. miR, microRNA; OS, osteosarcoma.
Association between miR-106b expression and the clinicopathological characteristics of patients with OS
To investigate the potential role of miR-106b in OS development, the association between miR-106b expression and the clinicopathological data of patients with OS was assessed. The mean miR-106b expression value (2.071) was used as a cut-off value to classify miR-106b into low and high expression groups. The analysis results presented in Table I revealed that the expression of miR-106b was associated with metastasis (P=0.028) and Tumor-Node-Metastasis (TNM) stage (P=0.017). However, no association between miR-106b and other clinicopathological parameters, including age, sex, tumor size, and differentiation, was found in this analysis (P>0.05).
Prognostic value of miR-106b in patients with OS
The focus of the present study was on the clinical significance of miR-106b in OS prognosis. First, the survival information collected from the 5-year follow-up survey was used to perform a Kaplan-Meier survival analysis, and it was indicated that patients with high miR-106b expression had shorter survival times than those with low miR-106b expression (log-rank P=0.001; Fig. 2). Second, the expression of miR-106b and other clinical parameters were included in the Cox regression analysis. The multivariate Cox analysis indicated that miR-106b expression [hazard ratio (HR)=2.769; 95% confidence interval (CI)=1.369–5.599; P=0.005)] and metastasis (HR=2.235; 95% CI=1.166–4.284; P=0.015) were two independent prognostic factors in patients with OS (Table II).
Figure 2.

Kaplan-Meier survival curve based on the miR-106b expression for patients with OS. The patients with high miR-106b expression had a poorer overall survival time than those with low miR-106b expression (log-rank P=0.001). miR, microRNA; OS, osteosarcoma.
Table II.
Multivariate Cox regression analysis for miR-106b in patients with OS.
| Multivariate analysis | |||
|---|---|---|---|
| Variables | HR | 95% CI | P-value |
| miR-106b | 2.769 | 1.369–5.599 | 0.005b |
| Age | 1.070 | 0.564–2.030 | 0.835 |
| Sex | 1.080 | 0.575–2.030 | 0.810 |
| Tumor size | 1.304 | 0.727–2.341 | 0.373 |
| Differentiation | 1.062 | 0.590–1.913 | 0.840 |
| Metastasis | 2.235 | 1.166–4.284 | 0.015a |
| TNM stage | 1.261 | 0.692–2.299 | 0.448 |
TNM stage was determined according to the guidelines of the American Joint Committee on Cancer (18).
P<0.05
P<0.01. TNM, Tumor-Node-Metastasis; miR, microRNA; OS, osteosarcoma; CI, confidence interval; HR, hazard ratio.
Effects of miR-106b on cell proliferation of OS cells
Given the dysregulation of miR-106b in OS samples, it was hypothesized that miR-106b may be involved in the tumor progression of OS. Therefore, the effects of miR-106b expression on OS cell biological behaviors were examined by regulating the expression of miR-106b. This was done using an miR-106 mimic and an miR-106 inhibitor. The OS cell lines MG63 and HOS were used for the cell experiments, as they had a higher expression of miR-106b. According to the RT-qPCR results, the expression of miR-106b was elevated in the cells transfected with the miR-106b mimic (P<0.001), and was decreased in the cells transfected with the miR-106b inhibitor (P<0.001; Fig. 3A and B). The results of the MTT assay demonstrated that cell proliferation was promoted by the overexpression of miR-106b, however it was inhibited by miR-106b reduction in both MG63 and HOS cells (P<0.05; Fig. 3C and D).
Figure 3.
Effects of miR-106b expression on cell proliferation in OS cells. Expression of miR-106b was upregulated by transfection with an miR-106b mimic, while it was downregulated by transfection with an miR-106b inhibitor in both (A) MG63 and (B) HOS cells. OS cell proliferation was measured by MTT assay, and was promoted by the overexpression of miR-106b, however was suppressed by the knockdown of miR-106b in both (C) MG63 and (D) HOS cells. *P<0.05, **P<0.01, ***P<0.001 vs. untreated. miR, microRNA; OS, osteosarcoma; OD, optical density.
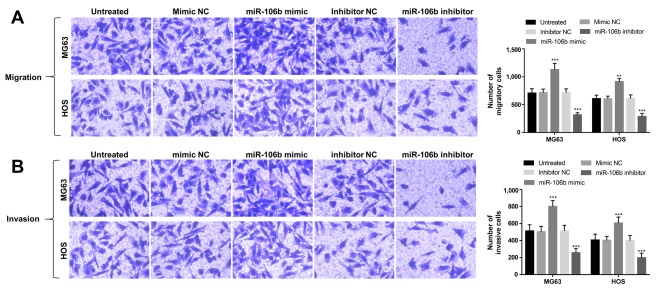
Effects of miR-106b on cell migration and invasion of OS cells
The abilities of cell migration and invasion in the transfected OS cells were also analyzed using Transwell analysis. As shown in Fig. 4A, this study found that the upregulation of miR-106b could enhance cell migration, whereas the downregulation of miR-106b could suppress cell migration in MG63 and HOS cells (P<0.01). In addition, OS cell invasion was also promoted by the overexpression of miR-106b, but was inhibited by the knockdown of miR-106b (P<0.01; Fig. 4B).
Figure 4.
Effects of miR-106b expression on cell migration and invasion in MG63 and HOS cells. (A) Cell migration was enhanced by miR-106b upregulation, however was inhibited with miR-106b downregulation. Magnification, ×200. (B) The overexpression of miR-106b may promote cell invasion, whereas the reduction of miR-106b may suppress cell invasion in the two OS cell lines. Magnification, ×200; **P<0.01, ***P<0.001 vs. untreated. miR, microRNA; OS, osteosarcoma.
Discussion
Emerging evidence has shown that miRNAs play pivotal roles in the initiation and development of various types of human cancer, and therefore, are applied in targeted therapy for these malignant diseases (20,21). In addition, the clinical significance of miRNAs has also attracted increasing attention for their dramatic diagnostic and prognostic value in human cancers (22,23). OS, as the most frequent primary malignancy among adolescents and children, has become a serious health burden worldwide (24). Regardless of the advances in diverse therapeutic strategies, including aggressive surgical resection and adjuvant chemotherapy, the outcomes for patients diagnosed with OS remain dismal. Therefore, the identification of the functional miRNAs that are involved in OS progression, is urgently needed for the improvement of OS treatment. Currently, some members of miRNAs have been observed in OS samples with deregulated expression levels and critical roles. A study scheduled by Zhang et al (25) showed that miR-33a-5p was downregulated in OS tissues and could suppress the cell growth of OS cells. The increased expression of miR-148a detected in OS tissues was shown to be involved in OS growth in vitro and in vivo (26). Mao et al (27) demonstrated that OS cell migration and invasion could be suppressed by miR-195, indicating that miR-195 served as a potential therapeutic target for OS treatment. In addition, the clinical value of miRNAs has also been assessed in the previous studies for patients with OS. Li et al (28) have indicated that the increased serum miR-17 was associated with the poor overall survival of patients with OS and could play as an efficient prognostic biomarker of OS. In the present study, the aberrant expression of miR-106b was indicated in both OS tissues and cells. Therefore, it was hypothesized that miR-106 may be involved in OS progression with an important role.
miR-106 is a member of the miR-106b-25 cluster, which has been described to play oncogenic roles in some types of cancer, such as hepatocellular carcinoma (29) and myeloid leukemia (30). The dysregulation of miR-106b has been observed in some cancers, and its expression variation trends are diverse in different types of tumor. For example, the decreased expression of miR-106b was detected in breast cancer, and the miR-106b reduction could enhance breast cancer cell migration and invasion (31). Furthermore, the increased miR-106b expression was found in hepatocellular carcinoma tissues compared with that in normal controls and may be involved in tumor progression of this malignant disease (32). Xu et al (33) reported that the expression of miR-106b was significantly increased in tumor tissues collected from paediatric patients with OS, and was associated with cell proliferation, migration and invasion of OS cell line U2OS. In this study, the expression of miR-106b in a larger research cohort with 134 patients with OS was measured by RT-qPCR, and was found to be upregulated compared with the adjacent normal tissues. Similarly, the increased expression of miR-106b was also observed in four OS cell lines compared to that in the normal cells. Therefore, this study considered that miR-106b may be an oncogenic miRNA in OS. In addition, the overexpression of miR-106b was associated with positive metastasis and the advanced TNM stage of patients with OS, indicating that miR-106b was involved in the tumor development of OS.
Given the deregulated expression of miR-106b in OS, the clinical significance of miR-106b in OS prognosis was further analyzed. According to the Kaplan-Meier survival analysis, the patients with OS with high miR-106b expression had a shorter survival time compared with those with low miR-106b expression, suggesting that the overexpression of miR-106b was associated with poor overall survival time of patients with OS. Furthermore, the results of multivariate Cox analysis implied that miR-106b was an independent prognostic factor in patients with OS. Therefore, this study suggests that the overexpression of miR-106b served as a novel and non-invasive prognostic biomarker of OS.
To further explore the functional role of miR-106b in OS progression, the effects of miR-106b expression on cell proliferation, migration and invasion were examined in OS cells. The results of the MTT assay revealed that OS cell proliferation was promoted by the overexpression of miR-106b, however was suppressed by miR-106b-knockdown. According to Transwell analysis, it was found that the upregulation of miR-106b could enhance OS cell migration and invasion, whereas the downregulation of miR-106b could inhibit these functions. All the data above indicated that the dysregulation of miR-106b may serve as a therapeutic target for the treatment of OS. miR-106b has been demonstrated to promote the cell proliferation, migration, and invasion of medulloblastoma cells by directly targeting PTEN (34). Tumor cell proliferation, migration, and invasion were also found to be enhanced by the overexpression of miR-106b in esophageal squamous cell carcinoma, as these tumor-promoting effects were exerted via the downregulation of Smad7 (35). Although this data also indicated that miR-106b may promote tumor progression in OS, the molecular mechanisms underlying the role of miR-106b in OS remain elusive and need to be examined in further studies.
In conclusion, the data in this study revealed that the upregulated expression of miR-106b serves as a candidate prognostic biomarker in patients with OS, and this overexpression of miR-106b promotes OS cell proliferation, migration, and invasion, suggesting that miR-106b may be a potential therapeutic target in OS treatment. This study provides a novel prognostic biomarker for patients with OS, and the strategies to downregulate miR-106b may be effective therapeutic methods in OS treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
KX and BW were responsible for the overall design of the study, clinical research, clinical data analyses and manuscript writing. WX and SZ performed the cell experiments and analyzed the associated data.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Ningbo No. 2 Hospital (Ningbo, China). Written informed consent was obtained from each patient.
Patient consent for publication
Patients provided written informed consent for the publication of any associated data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 3.McTiernan A, Jinks RC, Sydes MR, Uscinska B, Hook JM, van Glabbeke M, Bramwell V, Lewis IJ, Taminiau AH, Nooij MA, et al. Presence of chemotherapy-induced toxicity predicts improved survival in patients with localised extremity osteosarcoma treated with doxorubicin and cisplatin: A report from the European Osteosarcoma Intergroup. Eur J Cancer. 2012;48:703–712. doi: 10.1016/j.ejca.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 5.Li XP, Cao GW, Sun Q, Yang C, Yan B, Zhang MY, Fu YF, Yang LM. Cancer incidence and patient survival rates among the residents in the Pudong New Area of Shanghai between 2002 and 2006. Chin J Cancer. 2013;32:512–519. doi: 10.5732/cjc.012.10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment-where do we stand? A state of the art review. Cancer Treat Rev. 2014;40:523–532. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Feng Y, Dong YW, Song YN, Xiao JH, Guo XY, Jiang WL, Lu LG. MicroRNA-449a is a potential predictor of colitis-associated colorectal cancer progression. Oncol Rep. 2018;40:1684–1694. doi: 10.3892/or.2018.6566. [DOI] [PubMed] [Google Scholar]
- 8.Peng X, Zha L, Chen A, Wang Z. HOXA5 is a tumor suppressor gene that is decreased in gastric cancer. Oncol Rep. 2018;40:1317–1329. doi: 10.3892/or.2018.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou B, Li Z, Yang H, He N. Extracellular miRNAs: Origin, function and biomarkers in hepatic diseases. J Biomed Nanotechnol. 2014;10:2865–2890. doi: 10.1166/jbn.2014.1993. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z, Tang ZY, He Y, Liu LF, Li DJ, Chen X. miRNA-205 is a candidate tumor suppressor that targets ZEB2 in renal cell carcinoma. Oncol Res Treat. 2014;37:658–664. doi: 10.1159/000368792. [DOI] [PubMed] [Google Scholar]
- 11.Zhao J, Xu J, Zhang R. MicroRNA-539 inhibits colorectal cancer progression by directly targeting SOX4. Oncol Lett. 2018;16:2693–2700. doi: 10.3892/ol.2018.8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen K, Chen Y, Chen Z, Shi Y, He Z, Ding B, Wang C, Yu L. miR-134 increases the antitumor effects of cytarabine by targeting Mnks in acute myeloid leukemia cells. Onco Targets Ther. 2018;11:3141–3147. doi: 10.2147/OTT.S143465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Li L, Liu Z, Yuan Q, Lu X. Downregulation of MiR-431 expression associated with lymph node metastasis and promotes cell invasion in papillary thyroid carcinoma. Cancer Biomark. 2018;22:727–732. doi: 10.3233/CBM-181253. [DOI] [PubMed] [Google Scholar]
- 14.Liu H, Cao B, Zhao Y, Liang H, Hao F. Upregulated miR-221/222 promotes cell proliferation and invasion and is associated with invasive features in retinoblastoma. Cancer Biomark. 2018;22:621–629. doi: 10.3233/CBM-170721. [DOI] [PubMed] [Google Scholar]
- 15.Liu N, Yang H, Wang H. miR-598 acts as a tumor suppressor in human gastric cancer by targeting IGF-1R. Onco Targets Ther. 2018;11:2911–2923. doi: 10.2147/OTT.S166597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W, Wan X, Mu Z, Li F, Wang L, Zhao J, Huang X. MiR-1256 suppresses proliferation and migration of non-small cell lung cancer via regulating TCTN1. Oncol Lett. 2018;16:1708–1714. doi: 10.3892/ol.2018.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arabi L, Gsponer JR, Smida J, Nathrath M, Perrina V, Jundt G, Ruiz C, Quagliata L, Baumhoer D. Upregulation of the miR-17-92 cluster and its two paraloga in osteosarcoma-reasons and consequences. Genes Cancer. 2014;5:56–63. doi: 10.18632/genesandcancer.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. 7th. New York: Springer; 2010. AJCC cancer staging manual. [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Xiang J, Wu Y, Li DS, Wang ZY, Shen Q, Sun TQ, Guan Q, Wang YJ. miR-584 suppresses invasion and cell migration of thyroid carcinoma by regulating the target oncogene ROCK1. Oncol Res Treat. 2015;38:436–440. doi: 10.1159/000438967. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Li S. miR-205-5p inhibits cell migration and invasion in prostatic carcinoma by targeting ZEB1. Oncol Lett. 2018;16:1715–1721. doi: 10.3892/ol.2018.8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng Y, Chen Y. Increased expression of miR-29a and its prognostic significance in patients with cholangiocarcinoma. Oncol Res Treat. 2017;40:128–132. doi: 10.1159/000455869. [DOI] [PubMed] [Google Scholar]
- 23.Dan B, Luo J, Li K, Chen S. Prognostic value of miR-375 for survival outcomes in various cancers: A systematic review and meta-analysis. Oncol Res Treat. 2018;41:47–50. doi: 10.1159/000481708. [DOI] [PubMed] [Google Scholar]
- 24.Zhou W, Hao M, Du X, Chen K, Wang G, Yang J. Advances in targeted therapy for osteosarcoma. Discov Med. 2014;17:301–307. [PubMed] [Google Scholar]
- 25.Zhang J, Wang D, Xiong J, Chen L, Huang J. MicroRNA-33a-5p suppresses growth of osteosarcoma cells and is downregulated in human osteosarcoma. Oncol Lett. 2015;10:2135–2141. doi: 10.3892/ol.2015.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Wang Y, Xu T, Li C, Wu J, He Q, Wang G, Ding C, Liu K, Tang H, Ji F. Increased expression of microRNA-148a in osteosarcoma promotes cancer cell growth by targeting PTEN. Oncol Lett. 2016;12:3208–3214. doi: 10.3892/ol.2016.5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mao JH, Zhou RP, Peng AF, Liu ZL, Huang SH, Long XH, Shu Y. microRNA-195 suppresses osteosarcoma cell invasion and migration in vitro by targeting FASN. Oncol Lett. 2012;4:1125–1129. doi: 10.3892/ol.2012.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li S, Gao Y, Wang Y, Wang K, Dai ZP, Xu D, Liu W, Li ZL, Zhang ZD, Yang SH, Yang C. Serum microRNA-17 functions as a prognostic biomarker in osteosarcoma. Oncol Lett. 2016;12:4905–4910. doi: 10.3892/ol.2016.5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan W, Li Y, Lim SG, Tan TM. miR-106b-25/miR-17-92 clusters: Polycistrons with oncogenic roles in hepatocellular carcinoma. World J Gastroenterol. 2014;20:5962–5972. doi: 10.3748/wjg.v20.i20.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verboon LJ, Obulkasim A, de Rooij JD, Katsman-Kuipers JE, Sonneveld E, Baruchel A, Trka J, Reinhardt D, Pieters R, Cloos J, et al. MicroRNA-106b~25 cluster is upregulated in relapsed MLL-rearranged pediatric acute myeloid leukemia. Oncotarget. 2016;7:48412–48422. doi: 10.18632/oncotarget.10270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni X, Xia T, Zhao Y, Zhou W, Wu N, Liu X, Ding Q, Zha X, Sha J, Wang S. Downregulation of miR-106b induced breast cancer cell invasion and motility in association with overexpression of matrix metalloproteinase 2. Cancer Sci. 2014;105:18–25. doi: 10.1111/cas.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yen CS, Su ZR, Lee YP, Liu IT, Yen CJ. miR-106b promotes cancer progression in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol. 2016;22:5183–5192. doi: 10.3748/wjg.v22.i22.5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu M, Zhang YY, Wang HF, Yang GS. The expression and function of miRNA-106 in pediatric osteosarcoma. Eur Rev Med Pharmacol Sci. 2017;21:715–722. [PubMed] [Google Scholar]
- 34.Li KK, Xia T, Ma FM, Zhang R, Mao Y, Wang Y, Zhou L, Lau KM, Ng HK. miR-106b is overexpressed in medulloblastomas and interacts directly with PTEN. Neuropathol Appl Neurobiol. 2015;41:145–164. doi: 10.1111/nan.12169. [DOI] [PubMed] [Google Scholar]
- 35.Dai F, Liu T, Zheng S, Liu Q, Yang C, Zhou J, Chen Y, Sheyhidin I, Lu X. MiR-106b promotes migration and invasion through enhancing EMT via downregulation of Smad 7 in Kazakh's esophageal squamous cell carcinoma. Tumour Biol. 2016;37:14595–14604. doi: 10.1007/s13277-016-5338-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.