Abstract
Acute myeloid leukemia (AML) has a high mortality rate and its clinical management remains challenging. The aim of the present study was to identify the hub genes involved in AML. In order to do so, the gene expression data of the GSE9476 database, including 26 AML and 10 normal samples, were downloaded from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) were then identified via bioinformatics analysis. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses were performed on DEGs. Furthermore, the most upregulated genes were selected for further investigation in the Oncomine, gene expression profiling interactive analysis and UALCAN datasets. In total, 1,744 upregulated and 1,956 downregulated genes were detected. The GO and KEGG results revealed that upregulated genes were enriched in metabolic processes, while downregulated genes were associated with the immune response. Serine protease inhibitor Kazal-type 2 (SPINK2) ranked first among all the upregulated genes and was regarded as a hub gene in the development of AML. The overexpression of SPINK2 was validated in 12 patients with AML from the Linyi Central Hospital and in data from the Oncomine and Gene Expression Profiling Interactive Analysis (GEPIA) databases. Furthermore, the UALCAN and GEPIA datasets demonstrated that patients with high SPINK2 levels had shorter survival times. In conclusion, the results from the present study revealed that the SPINK2 gene was upregulated in patients with AML and that elevated SPINK2 expression was associated with poor outcomes in these patients.
Keywords: serine peptidase inhibitor Kazal-type 2, acute myeloid leukemia, prognosis, Oncomine dataset
Introduction
Acute myeloid leukemia (AML), a type of aggressive hematopoietic stem cell tumor, remains a considerable challenge in the clinical setting due to its high relapse rate (1,2). Lethal infections, bleeding and organ invasion are the main complications of AML (3). In the past decades, various studies have suggested that both environmental and genetic factors are important in the occurrence of AML (1,4–6). However, the current knowledge of the molecular mechanisms involved in the development of AML is limited, and early diagnosis remains difficult, which may result in delayed therapy. Thus, the identification of the key mechanisms regulating AML management and patient survival may aid in the development of AML-specific targeted therapies (3).
Microarray technology is a high-throughput and powerful tool to generate large quantities of data, including mRNA expression and DNA methylation (7). The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases are two common public platforms obtaining such data. These microarray results provide an opportunity to use bioinformatics to identify novel targets (8,9). Numerous public databases, including Oncomine (10), Gene Expression Profiling Interactive Analysis (GEPIA) (11) and UALCAN (12), provide several bioinformatic analysis tools, including differential expression, co-expression and comparative analyses, to identify novel tumor biomarkers and potential therapeutic targets through the use of the stored microarray data. Therefore, bioinformatics aid in the investigation of the underlying regulatory networks involved in different types of cancer, and constitute a powerful method of cancer research, including the early diagnosis of cancer, grading and prognostic prediction (8).
In the present study, microarray data were downloaded to investigate the hub genes affecting the development of AML from GEO in order to identify AML-associated genes via bioinformatics analysis. The present study further investigated those dysregulated genes at the molecular level and explored the potential candidate genes for prognosis in AML.
Materials and methods
Microarray data preprocessing and differentially expressed genes (DEGs) analysis
The original dataset GSE9476, provided by Stirewalt et al (13), was downloaded, which was based on the GPL96 Affymetrix Human Genome U133A Array platform (Affymetrix; Thermo Fisher Scientific, Inc.). The profiling dataset comprising 26 patients with AML and 10 normal peripheral blood samples was selected for further analysis (13). The raw data files were processed using the Affy package in R version 3.3.2 (https://www.r-project.org/). The limma package version 3.40.2.2 (http://www.bioconductor.org/packages/release/bioc/html/limma.html) was applied to identify the DEGs between AML samples and normal peripheral blood mononuclear cells (12). |Fold-change (FC)| ≥2 and false discovery rate (FDR) <0.05 were considered to indicate a statistically significant difference.
GO and KEGG pathway enrichment analyses
To obtain an insight into the function of the DEGs identified, these DEGs were divided into an upregulated (FC≥2 and FDR<0.05) and a downregulated group (FC≤-2 and FDR<0.05). Then, Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID; david.ncifcrf.gov/) online tool. FDR<0.05 was considered to indicate a statistically significant difference.
Oncomine analysis
The differences in mRNA levels of serine protease inhibitor Kazal-type 2 (SPINK2) between the AML and control samples were evaluated using gene expression array datasets from Oncomine, a public cancer microarray database that is accessible online (10,14,15). The threshold was established at P<10−4 and FC>2. In addition, the data type was restricted to mRNA levels only.
GEPIA analysis
GEPIA possesses key customizable and interactive functions, including profiling plotting, differential expression analysis, patient survival analysis, similar gene detection and dimensionality reduction analysis (11). Comprehensive expression analyses using GEPIA greatly facilitate data mining in numerous areas of research, thus contributing to scientific discussions and the identification of novel therapies. The present study employed SPINK2 data into the GEPIA dataset to explore its differential expression and effect on the prognosis of patients with AML.
UALCAN dataset analysis
UALCAN provides easy access to publicly available cancer transcriptome data (namely, TCGA and MET500 transcriptome sequencing). It allows users to identify biomarkers and to perform in silico validations of potential genes of interest. In addition, it can depict gene expression and patient survival information based on gene expression (12). In the present study, SPINK2 was included into the UALCAN dataset to explore its effect on the prognosis of patients with AML.
RNA isolation, reverse transcription and quantitative (RT-q) PCR
Blood samples of 6 healthy donors and 12 patients with AML were collected from the left arm at the Linyi Central Hospital between January 2017 and October 2017. The age range of patients was 16–65 years and the male: Female ratio was 4:8. Blood samples were centrifuged at 1,500 g for 5 min and at 4°C, and the fraction containing blood cells was stored at −80°C for future analyses. All blood cells were lysed using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.), and total RNAs were extracted. First strand cDNA synthesis was conducted using 2 µg RNA using the SYBR Premix Ex Taq™ II kit (Takara Biotechnology Co.) complete with SYBR Green reagents (Bio-Rad Laboratories, Inc.). The reverse transcriptase reaction was performed for 60 min at 37°C, followed by 60 min at 42°C. qPCR was performed using a 7900HT real-time PCR system (Thermo Fisher Scientific, Inc.), with the Real-Time SYBR Green PCR Master Mix kit (Promega Corporation). The primers used were as follows: SPINK2, forward 5′-GAGTGGCGCAGGTAACAGAC-3′, reverse, 5′-ACCAAATTGAGGGATCAGAGAG-3′; and GAPDH, forward 5′-TGGTGAAGACGCCAGTGGA-3′ and reverse, 5′-GCACCGTAAGGCTGAGAAC-3′. The qPCR thermocycling conditions were as follows: 95°C for 30 sec, 35 cycles at 95°C for 5 sec, then 60°C for 30 sec. The relative mRNA expression was calculated using the 2−∆∆Cq method (16).
Statistical analysis
A paired t-test was used to determine the statistical significance of the differences between healthy donors and patients with AML. The Kaplan-Meier analysis and log rank were used to assess patient survival rates. In the present study, the cut-off of survival analysis was based on the median of expression level in the GEPIA and UALCAN datasets. Data were presented as the means ± standard deviation. Data were analyzed using SPSS 17.0 (SPSS, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
DEGs in AML
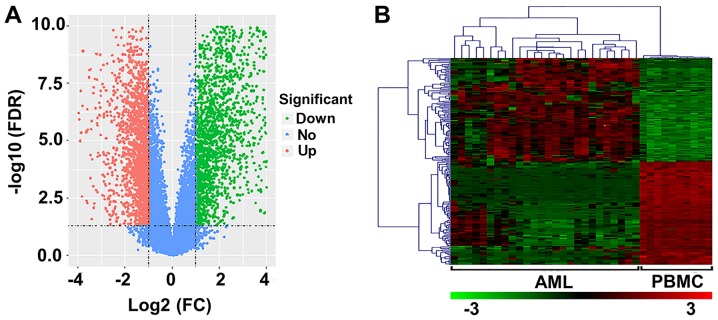
In the present study, 26 AML and 10 normal samples were analyzed. Upon preprocessing, 3,700 DEGs (FDR<0.05; log2 FC≥1) were identified, including 1,744 upregulated and 1,956 downregulated genes (Fig. 1A). The heat map of the top 100 upregulated and downregulated genes is presented in Fig. 1B.
Figure 1.
DEGs in patients with AML. (A) Volcano plot of DEGs. The abscissa and ordinates represent log2 FC and -log10 FDR value, respectively. The red and green dots correspond to upregulated and downregulated genes, respectively; blue dots correspond to non-DEGs. (B) Heat-map of the top 200 DEGs (100 upregulated and 100 downregulated genes). Red indicates upregulated genes and green indicates downregulated genes. DEGs, differentially expressed genes; AML, acute myeloid leukemia; FDR, false discovery rate; FC, fold change; PBMC, peripheral blood mononuclear cell.
GO term enrichment analysis for DEGs
The upregulated and downregulated genes were uploaded into the DAVID dataset to perform GO analysis. As presented in Table I, the most important biological processes (BPs) of overexpressed genes were ‘nitrogen compound metabolic process’, ‘cellular nitrogen compound metabolic process’ and ‘cellular metabolic process’. For the downregulated genes, BPs were significantly enriched in ‘immune response’, ‘immune system process’ and ‘defense response’ (Table II). For molecular function (MF) terms, upregulated DEGs were enriched in ‘RNA binding’, ‘poly (A) RNA binding’ and ‘nucleic acid binding’ (Table I), while the downregulated DEGs were enriched in ‘protein binding’, ‘binding’ and ‘anion binding’ (Table II). The most significant component functions (CFs) for upregulated DEGs were ‘intracellular organelle part’, ‘membrane-enclosed lumen’ and ‘organelle part’ (Table I), while the most significant CF terms for downregulated DEGs were ‘cytosol’, ‘cytoplasm’ and ‘cell periphery’ (Table II).
Table I.
GO Enrichment analysis of upregulated DEGs.
| A, Biological processes | |||
|---|---|---|---|
| GO ID | Pathway description | Gene count | FDR |
| GO.0006807 | Nitrogen compound metabolic process | 562 | 5.32×10−57 |
| GO.0034641 | Cellular nitrogen compound metabolic process | 537 | 4.68×10−56 |
| GO.0044237 | Cellular metabolic process | 712 | 4.33×10−50 |
| GO.0044238 | Primary metabolic process | 719 | 4.33×10−50 |
| GO.0071704 | Organic substance metabolic process | 722 | 7.52×10−47 |
| B, Molecular function | |||
| GO ID | Pathway description | Gene count | FDR |
| GO.000723 | RNA binding | 292 | 3.36×10−75 |
| GO.0044822 | Poly(A) RNA binding | 249 | 1.00×10−73 |
| GO.0003676 | Nucleic acid binding | 410 | 2.01×10−40 |
| GO.1901363 | Heterocyclic compound binding | 520 | 1.27×10−36 |
| GO.0097159 | Organic cyclic compound binding | 523 | 4.32×10−36 |
| C, Component function | |||
| GO ID | Pathway description | Gene count | FDR |
| GO.0044446 | Intracellular organelle part | 759 | 5.24×10−84 |
| GO.0031974 | Membrane-enclosed lumen | 527 | 1.44×10−81 |
| GO.0044422 | Organelle part | 764 | 1.44×10−81 |
| GO.0070013 | Intracellular organelle lumen | 516 | 9.85×10−81 |
| GO.0043233 | Organelle lumen | 519 | 7.00×10−80 |
GO, Gene Ontology; DEGs, differentially expressed genes; FDR, false discovery rate.
Table II.
GO Enrichment analysis of downregulated DEGs.
| A, Biological processes | |||
|---|---|---|---|
| GO ID | Pathway description | Gene count | FDR |
| GO.0006955 | Immune response | 264 | 1.30×10−72 |
| GO.0002376 | Immune system process | 316 | 1.43×10−63 |
| GO.0006952 | Defense response | 254 | 4.60×10−60 |
| GO.0007166 | Cell surface receptor signaling pathway | 308 | 1.34×10−55 |
| GO.0045087 | Innate immune response | 188 | 1.28×10−49 |
| B, Molecular function | |||
| GO ID | Pathway description | Gene count | FDR |
| GO.0005515 | Protein binding | 469 | 3.99×10−35 |
| GO.0005488 | Binding | 789 | 8.01×10−23 |
| GO.0043168 | Anion binding | 262 | 7.17×10−14 |
| GO.0019899 | Enzyme binding | 156 | 1.78×10−13 |
| GO.0060089 | Molecular transducer activity | 184 | 5.74×10−13 |
| C, Component function | |||
| GO ID | Pathway description | Gene count | FDR |
| GO.0005829 | Cytosol | 344 | 2.76×10−27 |
| GO.0005737 | Cytoplasm | 761 | 2.68×10−22 |
| GO.0071944 | Cell periphery | 413 | 7.57×10−19 |
| GO.0005886 | Plasma membrane | 403 | 1.46×10−17 |
| GO.0005622 | Intracellular | 884 | 6.42×10−14 |
GO, Gene Ontology; DEGs, differentially expressed genes; FDR, false discovery rate.
KEGG pathway analysis for DEGs
To gain further insight into the function of the genes in the interaction network, the DAVID database was used to identify the significant pathways associated with DEGs. According to the results of KEGG pathway analysis, upregulated DEGs were significantly enriched in ‘metabolic pathways’, ‘Huntington's disease’ and ‘ribosome’ (Table III). For the downregulated DEGs, the most significant pathways were ‘osteoclast differentiation’, ‘tuberculosis’ and ‘chemokine signaling pathway’ (Table III).
Table III.
KEGG pathway analysis of DEGs.
| A, Upregulated | ||
|---|---|---|
| Pathway description | Observed gene count | FDR |
| Metabolic pathways | 197 | 1.71×10−38 |
| Huntington's disease | 47 | 1.58×10−15 |
| Ribosome | 37 | 7.29×10−14 |
| Oxidative phosphorylation | 36 | 5.91×10−13 |
| Parkinson's disease | 37 | 7.94×10−13 |
| B, Downregulated | ||
| Pathway description | Observed gene count | FDR |
| Osteoclast differentiation | 46 | 1.66×10−21 |
| Tuberculosis | 53 | 6.51×10−21 |
| Chemokine signaling pathway | 53 | 4.78×10−20 |
| Natural killer cell-mediated cytotoxicity | 43 | 2.71×10−19 |
| Influenza A | 47 | 6.17×10−17 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes; FDR, false discovery rate.
SPINK2 expression is elevated in patients with AML
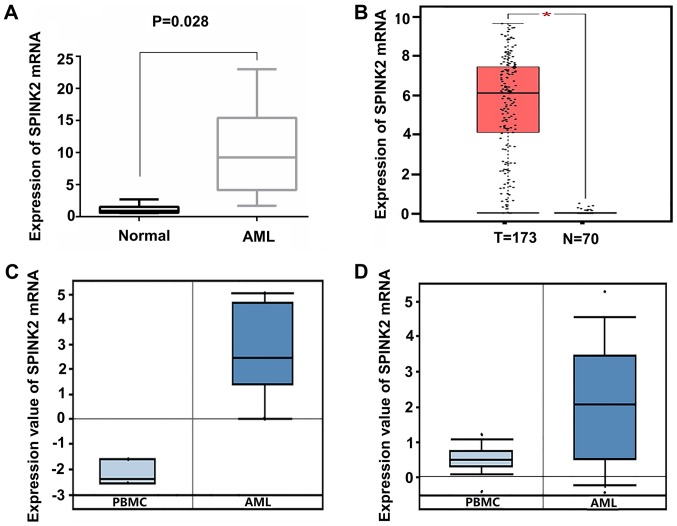
Among all the upregulated genes, SPINK2 ranked first (log2 FC, 6.59; FDR, 9.86×10−8). To validate the expression level of SPINK2 in AML, 6 healthy individuals and 12 patients with AML were recruited in the present study. qPCR results indicated that the levels of SPINK2 mRNA were significantly increased by 10.11-fold (P=0.028) in patients with AML compared with the healthy controls (Fig. 2A). The expression level of SPINK2 in the AML samples was validated in the Oncomine and GEPIA datasets. In the GEPIA database, 70 normal individuals and 173 patients with AML were included. The results suggested that the mRNA level of SPINK2 was elevated in patients with AML (P<0.01; Fig. 2B). From the Oncomine leukemia dataset of Stegmaier et al (14), SPINK2 expression was determined to be elevated by 20.21-fold in 9 AML samples compared with the PMBCs of 3 normal samples (P=2.12×10−5; Fig. 2C). The study on leukemia conducted by Haferlach et al (15) revealed that SPINK2 levels were elevated by 2.86-fold (P=8.15×10−42) in 542 AML samples compared with 74 normal samples (Fig. 2D). Therefore, the high expression of SPINK2 in AML was validated in larger cohorts.
Figure 2.
SPINK2 expression is elevated in patients with AML. (A) The results of the quantitative PCR suggested that the mRNA levels of SPINK2 were increased by 10.11-fold in 12 patients with AML (P=0.028). (B) In the GEPIA dataset, SPINK2 mRNA expression was elevated in patients with AML (*P<0.01). (C) In the study on leukemia conducted by Stegmaier et al (14), SPINK2 was elevated by 20.21-fold (P=2.12×10−5). (D) In the study by Haferlach et al (15) on leukemia, SPINK2 was increased by 2.86-fold (P=8.15×10−42). AML, acute myeloid leukemia; PBMC, peripheral blood mononuclear cells; SPINK2, serine protease inhibitor Kazal-type 2; AML, acute myeloid leukemia; GEPIA, gene expression profiling interactive analysis.
Upregulated SPINK2 is associated with poor outcomes in patients with AML
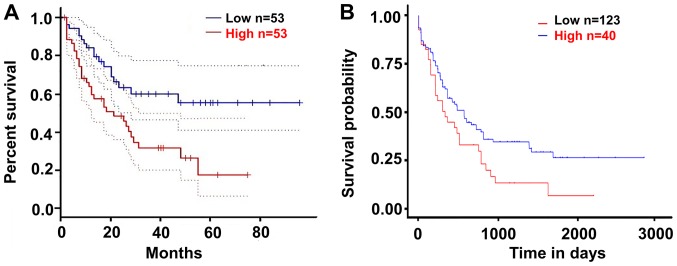
The present study validated the effect of SPINK2 on the prognosis of patients with AML using the GEPIA and UALCAN datasets. According to the median expression level of SPINK2, the patients were divided into a high-expression and a low-expression group. The results suggested that patients with high SPINK2 expression had shorter survival time than those with low SPINK2 levels in the GEPIA dataset (hazard ratio, 2.3; P=0.0039; Fig. 3A). In the UALCAN dataset, 163 patients with AML were analyzed for their expression levels of SPINK2. The Kaplan-Meier analysis demonstrated that patients with high SPINK2 expression had shorter survival time than patients with lower SPINK2 levels (P=0.024; Fig. 3B).
Figure 3.
Upregulated SPINK2 is associated with the poor prognosis of patients with acute myeloid leukemia. (A) In the GEPIA dataset, patients with high SPINK2 expression had poor outcomes (hazard ratio=2.3, P=0.0039). (B) In the UALCAN dataset, patients with high SPINK2 expression would have shorter survival time than those with lower SPINK2 levels (P=0.024). SPINK2, serine protease inhibitor Kazal-type 2; GEPIA, gene expression profiling interactive analysis.
Discussion
The pathogenesis of AML is heterogeneric and complex (17). To improve the outcome of AML treatment, it is important to understand the mechanism of AML (5,18). To detect DEGs in patients with AML, high-throughput platforms for the analysis of gene expression, such as microarrays, are increasingly used to identify novel molecular biomarkers and drug targets for clinical applications. Currently, microarray technology that combines bioinformatics analysis allows the wide exploration of the molecular mechanisms involved in the development and progression of AML. Using this method, Gal et al (19) compared DEGs of cluster of differentiation (CD) 34+ CD38− cells and CD34+ CD38+ cells using microarrays, and observed that 409 genes were 2-fold overexpressed or downregulated between the two cell populations. Previous focus on the Notch signaling pathway revealed that regulated leukemic stem cell self-renewal could be investigated to identify novel targets for therapy (19). Based on microarray data from 163 patients, Metzeler et al (20) developed a gene signature including an 86-probe set to predict the overall survival rate of patients with AML. When the authors applied the prognostic score in an independent cohort, this continuous score remained a significant predictor for the outcomes of patients with AML (20). Therefore, bioinformatics analysis could accelerate our understanding of AML (21,22).
The present study analyzed the gene expression patterns of patients with AML and healthy individuals by computational methods, and identified 1,744 upregulated and 1,956 downregulated genes in patients with AML. GO and KEGG functional enrichment analyses demonstrated that the upregulated genes were mainly associated with metabolic processes, which was in agreement with the results of a previous study (23). Furthermore, the genes involved in the immune response and cell surface signaling were downregulated according to our results. Cancer cells are able to avoid the immune response via a number of mechanisms (24). Tumor cells escape the immune response either by employing mechanisms to suppress the immune response, or by downregulating the expression of immunogenic molecules (25). Controlling the immune system in AML may facilitate the treatment of AML. Notably, the expression level of SPINK2 was the most altered among all the upregulated genes. Further analysis validated the upregulation of SPINK2 in patients with AML from the Oncomine and GEPIA datasets. Analysis further suggested that patients with AML and high expression levels of SPINK2 would have poor outcomes. Therefore, the findings from the present study indicated that SPINK2 may serve an important role in the development of AML.
SPINK2, also known as human seminal plasma inhibitor II, belongs to the SPINK family, which consists of members containing ≥1 conserved Kazal domain composed of 6 cysteine residues (26–28). The expression of SPINK2 is closely associated with cancer development, and high transcript levels of SPINK2 can be detected in patients with primary cutaneous follicle center cell lymphoma (29). SPINK2 serves as a classification marker for lymphoma, as well as a predictive marker of response to cancer therapy. Chen et al (30) reported that SPINK2 was significantly elevated in the majority of the leukemia cell lines investigated, and served an important role in tumor progression and response to treatment. Hoefnagel et al (29) proposed a possible interaction between SPINK2 and its currently unknown cancer-associated target proteinase, which appeared to be essential for tumor progression and metastasis. In the present study, patients with AML and high levels of SPINK2 had reduced survival times. Thus, we hypothesize that SPINK2 may serve an important role in the process of AML. However, the molecular mechanism of SPINK2 affecting the tumorigenic process remains unclear. Thus, further functional studies are required to deeply investigate how SPINK2 affects the development of AML.
There are some limitations to the present study. First, the GSE9476 dataset provides the complete data of AML and healthy people, but a larger dataset, including more patients, would be more robust. Of note, the GSE9476 dataset was only used to investigate the DEGs, and so the results from the present study require the analysis of additional datasets in order to be validated. The different expression levels of SPINK2 were validated in the present study by analyzing 70 healthy individuals and 173 patients with AML from the GEPIA database, and by analyzing 542 patients with AML and 74 healthy individuals in the report of leukemia by Haferlach et al (15). Furthermore, the prognosis of patients with AML were validated by analyzing 106 patients with AML from the GEPIA dataset (11) and 163 patients with AML from the UALCAN dataset (12). Therefore, despite the limited sample size, the results are credible. Secondly, AML has several subgroups, and the DEGs in patients with different subtypes were not analyzed in the present study. Thirdly, although the upregulated levels of SPINK2 mRNA in the Oncomine dataset were investigated in the present study, the protein levels of SPINK2 were not. Additional AML samples are required to validate these results. Finally, the present study only determined that SPINK2 may contribute to the development of AML; further studies are required that investigate the underlying molecular mechanisms of SPINK2 in the progression of AML.
To conclude, the present study has provided a comprehensive investigation of dysregulated genes involved in the progression of AML. SPINK2 could be regarded as a hub gene in patients with AML, and elevated levels of SPINK2 were associated with the poor prognosis in these patients. However, further molecular biology investigations are needed to confirm the mechanisms of SPINK2 in AML.
Acknowledgements
The authors would like to thank Dr Jun Zhang (Hematology Department, Linyi Central Hospital) for providing critical review and elaborated revision of this manuscript. In addition, the authors would like to thank Dr Guoming Deng and Dr Ebun Omoyinmi (Hematology Department, Linyi Central Hospital) for their helpful discussion regarding this report.
Glossary
Abbreviations
- AML
acute myeloid leukemia
- DEGs
differentially expressed genes
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- SPINK2
serine protease inhibitor Kazal-type 2
- GEO
Gene Expression Omnibus
- TCGA
The Cancer Genome Atlas
- FC
fold change
- FDR
false discovery rate
- BP
biological process
- MF
molecular function
- CF
component function
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
CX, JZ, YX and GZ curated the data. CX and GZ performed the investigations. CX and XW designed the study. JZ performed the statistical analysis. CX and XW wrote the manuscript.
Ethics approval and consent to participate
The present study was approved by the Academic Committee of Linyi Central Hospital. Written informed consent was obtained from all patients included within the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zhu B, Zhang J, Wang X, Chen J, Li C. Correlation between acute myeloid leukemia and IL-17A, IL-17F, and IL-23R gene polymorphism. Int J Clin Exp Pathol. 2015;8:5739–5743. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Quotti Tubi L, Canovas Nunes S, Brancalion A, Doriguzzi Breatta E, Manni S, Mandato E, Zaffino F, Macaccaro P, Carrino M, Gianesin K, et al. Protein kinase CK2 regulates AKT, NF-κB and STAT3 activation, stem cell viability and proliferation in acute myeloid leukemia. Leukemia. 2017;31:292–300. doi: 10.1038/leu.2016.209. [DOI] [PubMed] [Google Scholar]
- 3.Godwin CD, Gale RP, Walter RB. Gemtuzumab ozogamicin in acute myeloid leukemia. Leukemia. 2017;31:1855–1868. doi: 10.1038/leu.2017.187. [DOI] [PubMed] [Google Scholar]
- 4.Belson M, Kingsley B, Holmes A. Risk factors for acute leukemia in children: A review. Environ Health Perspect. 2007;115:138–145. doi: 10.1289/ehp.9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 6.Schuz J, Erdmann F. Environmental exposure and risk of childhood leukemia: An overview. Arch Med Res. 2016;47:607–614. doi: 10.1016/j.arcmed.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 7.Yin F, Shu L, Liu X, Li T, Peng T, Nan Y, Li S, Zeng X, Qiu X. Microarray-based identification of genes associated with cancer progression and prognosis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2016;35:127. doi: 10.1186/s13046-016-0403-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin F, Yi S, Wei L, Zhao B, Li J, Cai X, Dong C, Liu X. Microarray-based identification of genes associated with prognosis and drug resistance in ovarian cancer. J Cell Biochem. 2019;120:6057–6070. doi: 10.1002/jcb.27892. [DOI] [PubMed] [Google Scholar]
- 9.Raich T, Powell S. Identification of bacterial and fungal pathogens from positive blood culture bottles: A microarray-based approach. Methods Mol Biol. 2015;1237:73–90. doi: 10.1007/978-1-4939-1776-1_8. [DOI] [PubMed] [Google Scholar]
- 10.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stirewalt DL, Meshinchi S, Kopecky KJ, Fan W, Pogosova-Agadjanyan EL, Engel JH, Cronk MR, Dorcy KS, McQuary AR, Hockenbery D, et al. Identification of genes with abnormal expression changes in acute myeloid leukemia. Genes Chromosomes Cancer. 2008;47:8–20. doi: 10.1002/gcc.20500. [DOI] [PubMed] [Google Scholar]
- 14.Stegmaier K, Ross KN, Colavito SA, O'Malley S, Stockwell BR, Golub TR. Gene expression-based high-throughput screening (GE-HTS) and application to leukemia differentiation. Nat Genet. 2004;36:257–263. doi: 10.1038/ng0404-427b. [DOI] [PubMed] [Google Scholar]
- 15.Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Béné MC, De Vos J, Hernández JM, Hofmann WK, Mills KI, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: Report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol. 2010;28:2529–2537. doi: 10.1200/JCO.2009.23.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Wang YX, Zhang TJ, Yang DQ, Yao DM, Yang L, Zhou JD, Deng ZQ, Ma JC, Guo H, Wen XM, et al. Reduced miR-215 expression predicts poor prognosis in patients with acute myeloid leukemia. Jpn J Clin Oncol. 2016;46:350–356. doi: 10.1093/jjco/hyv204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryotokuji T, Yamaguchi H, Ueki T, Usuki K, Kurosawa S, Kobayashi Y, Kawata E, Tajika K, Gomi S, Kanda J, et al. Clinical characteristics and prognosis of acute myeloid leukemia associated with DNA-methylation regulatory gene mutations. Haematologica. 2016;101:1074–1081. doi: 10.3324/haematol.2016.143073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gal H, Amariglio N, Trakhtenbrot L, Jacob-Hirsh J, Margalit O, Avigdor A, Nagler A, Tavor S, Ein-Dor L, Lapidot T, et al. Gene expression profiles of AML derived stem cells; similarity to hematopoietic stem cells. Leukemia. 2006;20:2147–2154. doi: 10.1038/sj.leu.2404401. [DOI] [PubMed] [Google Scholar]
- 20.Metzeler KH, Hummel M, Bloomfield CD, Spiekermann K, Braess J, Sauerland MC, Heinecke A, Radmacher M, Marcucci G, Whitman SP, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008;112:4193–4201. doi: 10.1182/blood-2008-02-134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delas MJ, Sabin LR, Dolzhenko E, Knott SR, Munera Maravilla E, Jackson BT, Wild SA, Kovacevic T, Stork EM, Zhou M, et al. lncRNA requirements for mouse acute myeloid leukemia and normal differentiation. ELife. 2017;6(pii):e25607. doi: 10.7554/eLife.25607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer SE, Qin T, Muench DE, Masuda K, Venkatasubramanian M, Orr E, Suarez L, Gore SD, Delwel R, Paietta E, et al. DNMT3A haploinsufficiency transforms FLT3ITD myeloproliferative disease into a rapid, spontaneous, and fully penetrant acute myeloid leukemia. Cancer Ddiscov. 2016;6:501–515. doi: 10.1158/2159-8290.CD-16-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salvestrini V, Zini R, Rossi L, Gulinelli S, Manfredini R, Bianchi E, Piacibello W, Caione L, Migliardi G, Ricciardi MR, et al. Purinergic signaling inhibits human acute myeloblastic leukemia cell proliferation, migration, and engraftment in immunodeficient mice. Blood. 2012;119:217–226. doi: 10.1182/blood-2011-07-370775. [DOI] [PubMed] [Google Scholar]
- 24.Austin R, Smyth MJ, Lane SW. Harnessing the immune system in acute myeloid leukaemia. Crit Rev Oncol Hematol. 2016;103:62–77. doi: 10.1016/j.critrevonc.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 25.Davidson-Moncada J, Viboch E, Church SE, Warren SE, Rutella S. Dissecting the immune landscape of acute myeloid leukemia. Biomedicines. 2018;6(pii):E110. doi: 10.3390/biomedicines6040110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dietrich MA, Slowinska M, Karol H, Adamek M, Steinhagen D, Hejmej A, Bilińska B, Ciereszko A. Serine protease inhibitor Kazal-type 2 is expressed in the male reproductive tract of carp with a possible role in antimicrobial protection. Fish Shellfish Immunol. 2017;60:150–163. doi: 10.1016/j.fsi.2016.11.041. [DOI] [PubMed] [Google Scholar]
- 27.Kherraf ZE, Christou-Kent M, Karaouzene T, Amiri-Yekta A, Martinez G, Vargas AS, Lambert E, Borel C, Dorphin B, Aknin-Seifer I, et al. SPINK2 deficiency causes infertility by inducing sperm defects in heterozygotes and azoospermia in homozygotes. EMBO Mol Med. 2017;9:1132–1149. doi: 10.15252/emmm.201607461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee B, Park I, Jin S, Choi H, Kwon JT, Kim J, Jeong J, Cho BN, Eddy EM, Cho C. Impaired spermatogenesis and fertility in mice carrying a mutation in the Spink2 gene expressed predominantly in testes. J Biol Chem. 2011;286:29108–29117. doi: 10.1074/jbc.M111.244905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoefnagel JJ, Dijkman R, Basso K, Jansen PM, Hallermann C, Willemze R, Tensen CP, Vermeer MH. Distinct types of primary cutaneous large B-cell lymphoma identified by gene expression profiling. Blood. 2005;105:3671–3678. doi: 10.1182/blood-2004-04-1594. [DOI] [PubMed] [Google Scholar]
- 30.Chen T, Lee TR, Liang WG, Chang WS, Lyu PC. Identification of trypsin-inhibitory site and structure determination of human SPINK2 serine proteinase inhibitor. Proteins. 2009;77:209–219. doi: 10.1002/prot.22432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.