Abstract
Breast cancer (BC) is one of the most aggressive malignancies worldwide among females. Matrix metalloproteinases (MMPs), as the most abundant class of non-serine proteases present in invasive and metastatic tumors, can regulate a variety of alterations in the microenvironment during tumor progression. However, the differential expression of MMPs and its prognostic values in BC is yet to be elucidated. In this research, using the ONCOMINE dataset, The Cancer Genome Atlas, Breast Cancer Gene-Expression Miner v4.1 (Bc-GenExMiner), Kaplan-Meier Plotter and cBioPortal, the transcriptional MMPs and survival outcome data of patients with BC was compared. It was indicated that mRNA levels of MMP1/3/9/10/11/12/13 were increased compared with non-tumor tissues, whereas mRNA expression of MMP2/16/19/23B/28 was lower in BC tissues. Kaplan-Meier plots showed that high mRNA levels of MMP2/10/16/19/20/23B/27 in patients with BC were associated with better recurrence-free survival. In contrast, high MMP1/8/9/11/12 conferred worse RFS rate. Meanwhile, high transcription levels of MMP1/3/11/12/13 predicted shorter distant metastasis-free survival, while high levels of MMP1/12 demonstrated worse overall survival in patients with BC. From Bc-GenExMiner, it was indicated that high expression of MMP16/20 was correlated with better prognosis, while MMP1/9/11/12/13/14/15 exerted a negative effect on patient prognosis. The integrative bioinformatics analysis performed in the present study suggests that MMP1/9/12/16, compared with other MMPs, are potentially appropriate targets for targeted therapy in patients with BC.
Keywords: matrix metalloproteinases, breast cancer, prognosis, bioinformatics analysis
Introduction
Breast cancer (BC) is one of the most common malignancies among women globally, with ~2.1 million newly diagnosed cases of BC in 2018 and 626,679 mortalities worldwide (1). On account of improvements in treatment and early detection by mammography, the 5-year overall survival (OS) rate has notably improved by 12% and the mortality rate has declined by 40% over the past three decades (2). However, due to the multi-aspect dysregulation of genes and the complicated mechanisms that involve several molecular and pathological subtypes, the efficiency of accurate target therapies for patients with BC remains unclear (3). The examination of BC pathogenesis and identifying novel targets for the treatment of BC is required for the development of medical science. Therefore, in order to reach the goal of individualized treatment, it is crucial and urgent to identify novel biological markers in the microenvironment in BC.
In invasive and metastatic tumors, the zinc-dependent matrix metalloproteases (MMPs) are the most plentiful class of non-serine proteases present, first identified in 1962 by Gross and Lapiere (4). They have the ability to degrade the components of the extracellular matrix (5). At present, >20 MMPs have been identified in humans (6), including collagenases, gelatinases, stromelysins, and the transmembrane type MMPs (7). MMPs can regulate a number of changes in the microenvironment during tumor progression (such as in signaling pathways that control cell growth, inflammation or angiogenesis, and may also function in a non-proteolytic manner) (8). Their enzymatic activity modulates the activities of a wide range of extracellular and intracellular proteins, including proliferation, migration, and adhesion (9). A previous study (10) showed that enhanced expression of MMPs is usually associated with BC invasion and metastasis, and acts as an important prognostic indicator. Therefore, an in-depth bioinformatics analysis associated with MMPs and patients with BC is required. In order to clarify the prognostic and potential therapeutic value of MMPs in BC treatment, this study analyzed the clinicopathological and survival data associated with MMPs in patients with BC, based on a number of large public databases.
Materials and methods
ONCOMINE data-mining and processing analysis
ONCOMINE version 4.5 (www.oncomine.org) is a database, which is used to promote data-mining the transcriptional expression levels of MMPs in 20 types of cancer (11). Using a Student's t-test, transcriptional expression levels of MMPs in cancer samples were compared with those in normal individuals. P<0.05 and fold change of >2 were considered to indicate a statistically significant difference.
Kaplan-Meier Plotter
The Kaplan-Meier Plotter (www.kmplot.com) is capable to assess the effect of any gene or gene combination on survival in breast, ovarian, lung, gastric and many other types of tumor (12). It was used to measure the prognostic value of MMPs mRNA transcription levels. In order to evaluate the recurrence-free survival (RFS), OS and distant metastasis-free survival (DMFS) rates of patients with BC, individuals were divided into two groups according to the median gene expression. The median value of MMPs expression calculated by the Kaplan-Meier Plotter was used to divide individuals into high and the low-expression groups. A log-rank test was used to analyze the Kaplan-Meier survival curves. The hazard ratio (HR) with 95% confidence intervals (CI) were calculated; P<0.05 was considered to indicate a statistically significant difference. The number of risks was presented below the curve.
Bc-GenExMiner
Breast Cancer Gene-Expression Miner version 4.1 (http://bcgenex.centregauducheau.fr) is a statistical mining tool for 36 well-known genome datasets and three classical mining functions: Expression, prognosis and correlation (13). The expression data was updated in December 2017 and the production of specific genes was compared with clinical parameters, including age, estrogen (ER), progesterone (PR) and HER2 status. The predictive module calculates the predictive values of target genes in BC and provides potential new predictive markers. To better assess the relationship between MMPs and breast cancer prognosis, Scarff, Bloom and Richardson grade (SBR) was used to evaluate the management of breast carcinoma, and the Nottingham prognostic index (NPI) was used for breast cancer prognostication. Welch's test was performed to compare the abnormal expression of MMPs between groups of patients according to different clinicopathological parameters. The HR with 95% CI were calculated; P<0.05 was considered statistically significant.
The cancer genome atlas (TCGA) data and cBioPortal
TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) contains many cancer-associated data, including gene expression and clinicopathological data. cBioPortal version 3.0 (www.cbioportal.org) is an open access database for exploring multiple cancer genes (14). In order to visualize the potential genes co-expressed with MMPs, the gene network was constructed using cBioPortal.
Results
Discrepancies in MMP expression in patients with BC
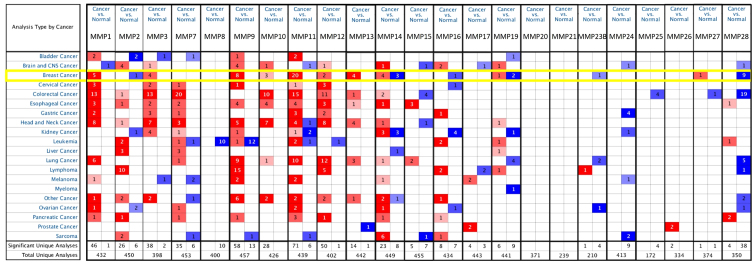
A total of 23 MMPs were identified using the ONCOMINE database. The expression level of MMPs was analyzed in 20 types of cancer, and compared with that in normal individuals. Fig. 1 shows the transcription levels of MMPs in various types of cancers. High expression was defined when MMPs expression was higher than that of adjacent non-tumors, and vice versa. The mRNA expression of MMP1/3/9/10/11/12/13 was upregulated in BC specimens in different studies (Fig. 1). According to the TCGA breast dataset, MMP1 was indicated to be highly expressed in several types of BC compared with normal tissues and the fold-change was 11.611 in invasive BC (Table SI). Richardson et al (15) reported that the fold-change of MMP1 in ductal BC was 21.13, whereas studies by Sorlie et al (16,17) demonstrated fold changes of 2.963 and 2.482 for MMP1 in ductal BC. Compared with normal tissues, MMP3 was overexpressed in some types of BC. The fold-change of invasive lobular BC in TCGA data was 5.945, while Curtis et al (18) reported that the fold-change of MMP3 in invasive lobular BC was 2.008. According to the TCGA dataset, MMP9 was overexpressed in almost all types of BC and normal breast tissues, including intraductal cribriform BC with fold-change=5.463, mucinous BC with fold-change=4.783 and invasive BC with fold-change=3.245. Curtis et al (18) reported that the fold-changes of MMP9 in medullary BC and ductal BC in situ were 9.238 and 6.629, respectively. In different datasets for MMP9, a fold-change of 4.378 in invasive ductal BC compared with normal breast was observed (18). Radvanyi et al (19) (fold-change=4.043) and Turashvili et al (20) (fold-change=3.640) found similar trends.
Figure 1.
Transcriptional levels of MMPs in different types of tumors (the threshold including fold-change ≥2; gene rank ≥top 10% and P-value ≤1.0×10−4. Red indicates high expression of MMPs mRNA level and blue indicates low expression with statistically significant P<0.05 cancer vs. normal tissue. MMPs, matrix metalloproteinases.
According to TCGA dataset, the transcriptional levels of MMP10 showed fold-changes of 6.767, 4.265, and 4.130, in invasive BC, invasive ductal BC and invasive lobular BC, respectively. On the other hand, Turashvili et al (20) reported invasive ductal BC with an MMP10 fold-change of 2.596. Radvanyi et al (19) and Karnoub et al (21) reported that MMP11 expression increased significantly in ductal BC in situ and invasive ductal BC stroma, with a fold-change of 2.639 and 188.233 respectively. Compared with normal breast tissue, upregulated expression of MMP12 and MMP13 were also found in most types of BC (Table SI).
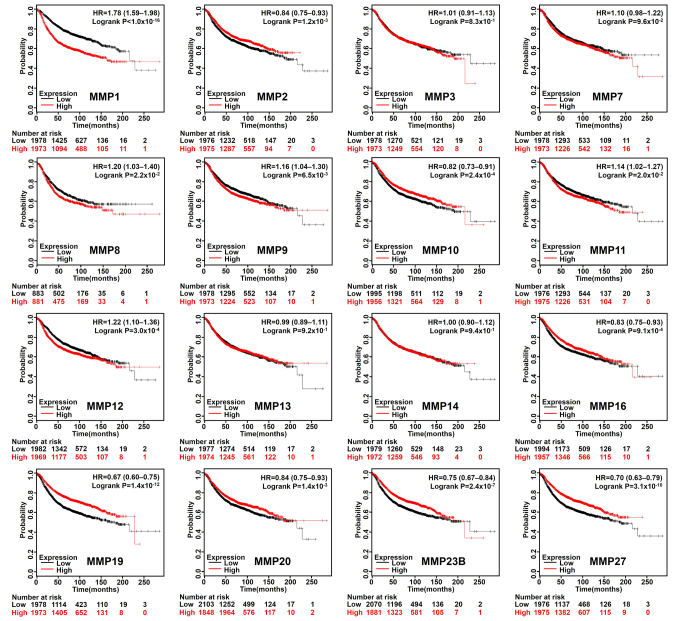
Increased expression of MMP2/10/16/19/20/23B/27 and reduced expression of MMP1/3/8/9/11/12/13 are associated with better RFS, DMFS or OS rate in patients with BC
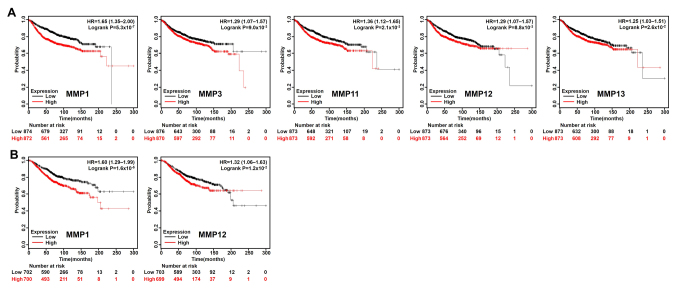
Based on the median expression value of each MMP in all samples, all types of patients with BC were divided into two groups to detect MMP expression (high expression vs. low expression). The Kaplan-Meier curves of survival data showed that increased expression of MMP 2/10/16/19/20/23B/27 and reduced expression of MMP 1/8/9/11/12 were associated with better RFS rate (P<0.05) in all BC types (Fig. 2). In addition, patients with BC with lower MMP1/3/11/12/13 transcriptional expression levels indicated better DMFS rate (Fig. 3A), and lower mRNA levels of MMP1/12 exhibited better OS rate than those with high expression (Fig. 3B). To further examine the role of MMPs in BC prognosis, the Bc-GenExMiner version 4.1 was used to validate this research. MMP1/9/11/12/13/14/15 has a significant negative impact on the prognosis of patients. High expression of MMP16/20 was associated with better prognosis (Table I).
Figure 2.
Kaplan-Meier curve shows the differences in recurrence-free survival according to the mRNA level of MMPs in patients with breast cancer (n=3,951). High, higher than the median gene expression; Low, lower than the median gene expression MMP, matrix metalloproteinase; HR, hazard ratio.
Figure 3.
Kaplan-Meier Plotter reveals the DMFS and OS rate differences according to the mRNA level of MMPs in patients with BC. (A) Patients with BC with decreased expression of MMP1/3/11/12/13 have better DMFS rate (n=1,746). (B) Patients with BC with decreased expression of MMP1/12 have better OS rate (n=1,402). High, higher than the median gene expression; low, lower than the median gene expression; DMFS, distant metastasis-free survival; OS, overall survival; MMP, matrix metalloproteinase; BC, breast cancer.
Table I.
Prognostic association of MMPs expression in breast cancer according to bc-GenExMiner version 4.1.
| MMPs | Event | P-value | HR | 95% CI | Good prognosis' RNA level |
|---|---|---|---|---|---|
| MMP1 | ROD | <0.0001 | 1.20 | 1.15–1.26 | Low |
| MMP2 | ROD | 0.5267 | 0.98 | 0.94–1.03 | NA |
| MMP3 | ROD | 0.4391 | 1.02 | 0.97–1.07 | NA |
| MMP7 | ROD | 0.2956 | 1.03 | 0.98–1.07 | NA |
| MMP8 | ROD | 0.5543 | 1.02 | 0.96–1.07 | NA |
| MMP9 | ROD | 0.0004 | 1.09 | 1.04–1.14 | Low |
| MMP10 | ROD | 0.5019 | 0.98 | 0.94–1.03 | NA |
| MMP11 | ROD | <0.0001 | 1.13 | 1.08–1.19 | Low |
| MMP12 | ROD | <0.0001 | 1.11 | 1.06–1.16 | Low |
| MMP13 | ROD | 0.0020 | 1.08 | 1.03–1.13 | Low |
| MMP14 | ROD | 0.0001 | 1.11 | 1.05–1.16 | Low |
| MMP15 | ROD | 0.0006 | 1.09 | 1.04–1.15 | Low |
| MMP16 | ROD | 0.0198 | 0.93 | 0.88–0.99 | High |
| MMP17 | ROD | 0.6791 | 0.99 | 0.94–1.04 | NA |
| MMP19 | ROD | 0.8597 | 1.01 | 0.94–1.07 | NA |
| MMP20 | ROD | 0.0313 | 0.95 | 0.90–1.00 | High |
| MMP21 | ROD | 0.4320 | 0.97 | 0.90–1.05 | NA |
| MMP23B | ROD | 0.1280 | 0.93 | 0.84–1.02 | NA |
| MMP24 | ROD | 0.2675 | 0.97 | 0.91–1.03 | NA |
| MMP25 | ROD | 0.1522 | 0.96 | 0.90–1.02 | NA |
| MMP26 | ROD | 0.8713 | 1.00 | 0.95–1.06 | NA |
| MMP27 | ROD | 0.0691 | 0.95 | 0.91–1.00 | NA |
| MMP28 | ROD | 0.7088 | 0.99 | 0.93–1.05 | NA |
CI, confidence interval; ROD, relapse or death; high, Higher than the median gene expression; HR, hazard ratio; low, lower than the median gene expression; NA, not applicable as there was no significant correlation between the expression of MMPs and the progression of the tumor; MMP, matrix metalloproteinases; BC, breast cancer.
Association between abnormal expression of MMPs and the clinicopathological parameters of patients with BC
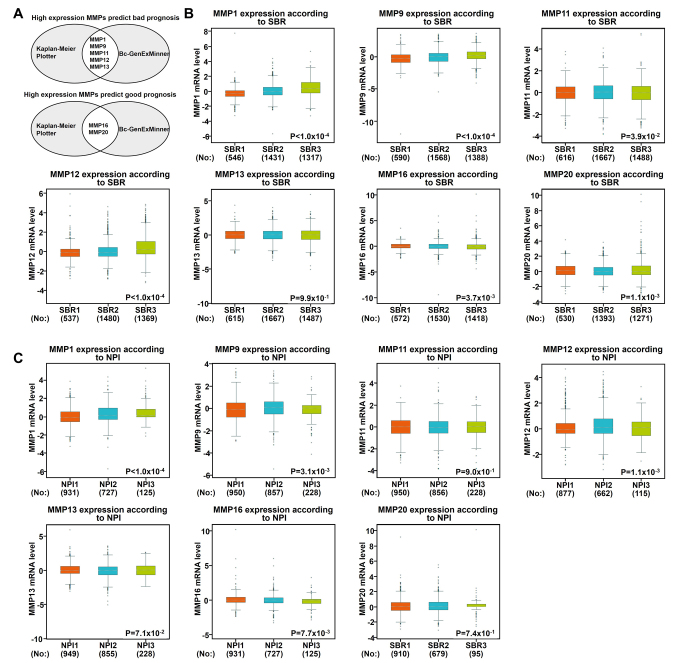
Using the aforementioned databases, this study subsequently focused on whether MMP1/9/11/12/13 and MMP16/20 play a key role in the progression of BC (Fig. 4A). Scarff, Bloom and Richardson grade (SBR) is one of the important pathological parameters to be evaluated for the management of breast carcinoma (22,23). The survival rate of patients with poorly differentiated cancer (grades II and III) was lower than that of patients with well-differentiated cancer (grade I) (23). Using the Bc-GenExMiner software, it was observed that patients with high-grade tumors (grades II and III) appeared to exhibit high levels of MMP 1/9/11/12 and low levels of MMP 16/20 (Fig. 4B). In addition to molecular subtypes, the Nottingham prognostic index (NPI) is also a helpful prognostic model for patients with BC (24). In Fig. 4C, it was indicated that MMP1/9/12/16 were associated with NPI: Patients with high-grade tumors expressed high levels of MMP1/9/12 and lower levels of MMP16, which was consistent with our previous data mentioned above.
Figure 4.
Association between mRNA expression levels of MMPs, and NPI or SBR grade status according to bc-GenExMiner version 4.1. (A) Charts exhibit the number of MMPs identified as potential good or bad prognostic targets of breast cancer between Bc-GenExMiner and Kaplan-Meier Plotter. (B) The association between mRNA expression levels of MMPs and SBR grade status according to bc-GenExMiner. (C) The association between mRNA expression levels of MMPs and NPI status according to bc-GenExMiner. A Welch's test was used to evaluate global significant differences between groups; P<0.05 was considered as statistically significant. MMP, matrix metalloproteinase; NPI, Nottingham Prognosis Index; SBR, Scarff Bloom & Richardson grade.
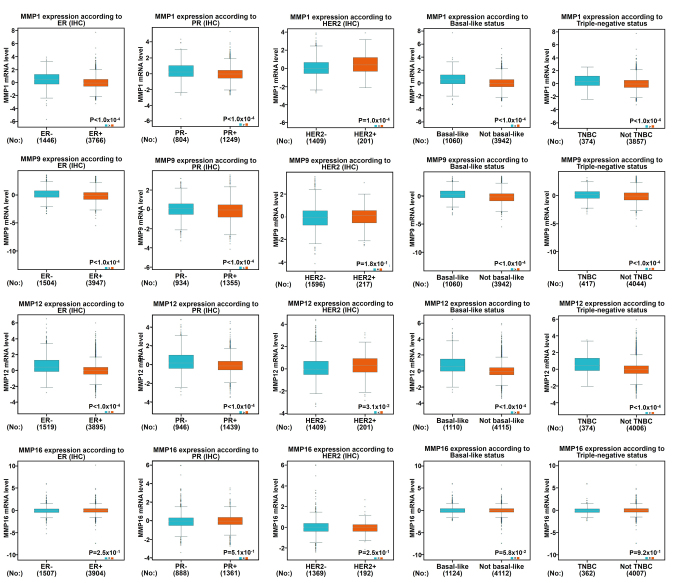
Regarding the age standard (Fig. S1), there was no significant difference between the ≤51-year-old group and the >51-year-old group, except MMP12/16, indicating that the MMP12/16 level was decreased in the elderly group. The expression of MMP9/12/16 in patients with lymph node-positive BC was lower than that in negative lymph node patients, while the expression of MMP1 indicated opposite results. As shown in Fig. 5, when compared with patients who were ER-negative or PR-negative, it was indicated that patients who were ER-positive or PR-positive were more likely to express low levels of MMP1/9/12. The growth of cancer cells in ER-positive and PR-positive patients is dependent on estrogen and can be treated by blocking estrogens with drugs, such as tamoxifen, as they have previously been reported to exhibit a better prognostic trend (25). Therefore, from these results, it was indicated that low expression of MMP1/9/12 could predict a better prognosis. Furthermore, the expression of MMP1/12 was positively associated with HER2 status. Triple-negative breast cancer (TNBC) is highly sensitive to chemotherapy; however it has a strong invasive ability and is characterized by rapid progression. In addition, there is a lack of endocrine therapy and targeted therapy, and its therapeutic effect is worse than other subtypes (26). The molecular characteristics of TNBC include ER (−), PR (−) and HER2 (−) (26). This study indicated a significant increase of MMP1/9/12 expression in patients with TNBC, and the same pattern was found in base-like BC (Fig. 5). However, no relevance between the mRNA expression of MMP16 and the ER/PR/HER status in BC could be identified.
Figure 5.
Association between mRNA expression levels of MMP1/9/12/16, and the ER, PR or HER2 status and pathological parameters of breast carcinoma. The data of IHC samples from bc-GenExMiner datasets were analyzed. P<0.05 was considered as statistically significant. ER, estrogen; PR, progesterone; MMP, matrix metalloproteinase.
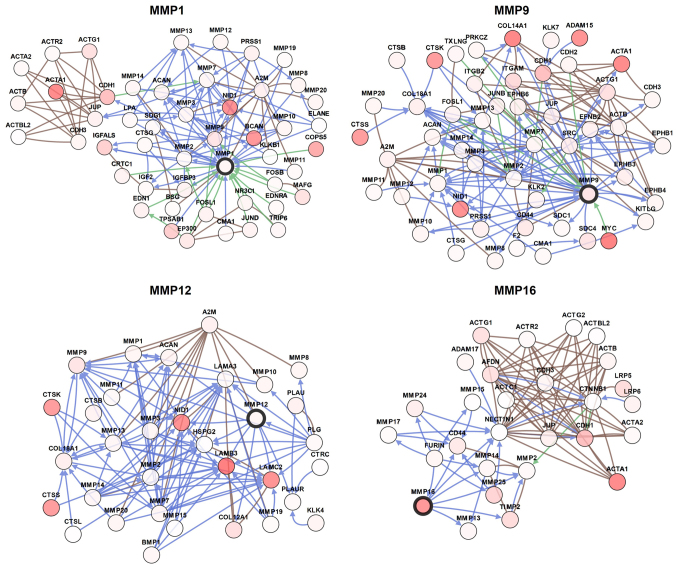
Combined with this information, multiple bioinformatics analyses identified MMP1/9/12/16 as potential therapeutic targets for patients with BC. In order to visualize the potential genes co-expressed with MMP1/9/12/16, the gene network was constructed as shown in Fig. 6. The network was designed to recognize potential interactions between MMP1/9/12/16 and a number of other key proteins. The network analysis can provide a series of gene candidates to help clarify the molecular mechanism of MMPs involvement in BC.
Figure 6.
Prospective gene networks of the main MMPs (screened out by cBioPortal). Indications of colored lines: Blue, controls state change of; brown, controls transport of; green, controls expression of; darker red: A higher degree of alteration frequency. MMP, matrix metalloproteinase.
Discussion
MMPs have been identified as an important family of proteinases associated with tumorigenesis and as key mediators of tumor progression (27). MMPs can mediate various events in the microenvironment during the progression of tumors, such as inflammation, angiogenesis, proliferation, migration, and adhesion (27). Due to their role in cancer, MMPs have been speculated to serve as an effective therapeutic target (28). However, in clinical trials, marimastat, a broad-spectrum MMPs inhibitor, failed to prolong progression-free survival of patients with metastatic BC (28). Despite the recent research illustrating the complex role of MMPs, the role of different MMPs in the progression and metastasis of BC remains elusive (29). To the best of our knowledge, this research is the first comprehensive bioinformatics study to elucidate specific MMPs, and their prognostic value and biological function in breast cancer.
A previous study demonstrated that MMP1 mRNA expression in BC tissues is higher than in normal breast tissue (30). In addition, previously published data reported that patients with BC and high MMP1 expression are associated with poor prognoses, such as shorter relapse-free-survival (31), which is consistent with our findings. In contrast, Kulic et al (32) reported that patients with lower expression levels of MMP1 had significantly shorter 5-year survival rates than those with higher levels of MMP1, suggesting a poor prognostic role of low levels of serum MMP1. An animal experiment verified that MMP2 plays a role in the development of BC metastases, regulated by astrocyte factors and the ERK1/2 signaling pathway (33). Results on whether MMP2 is associated with worse prognosis in patients with BC have been contradictory. High MMP2 expression was reported to be indicative of relatively poor patient prognosis in BC (34), while a meta-analysis indicated that worse OS rate was not linked to MMP2-positive patients (35).
As a direct downstream target of microRNA-519d, MMP3 acted as an executive molecule for the carcinogenic effect of microRNA-519d in BC (36). A mouse model revealed that peroxidases induced the transcription of the MMP3 gene, in turn, MMP3 promoted tumor growth, invasion and metastasis (37). MMP3 has been reported to contribute to BC by participating in cancer invasion and metastasis, especially in ER-/PR-tumors (38). Vizoso et al (39) found that the high expression of MMP7 was associated with shorter recurrence-free survival in patients with BC. Kim et al (40) reported that compared with luminal A and HER2-overexpressing subtype, MMP7 expression was significantly higher in the basal-like subtype. Furthermore, findings have identified MMP7 as a new mechanism through which forkhead box C1 can regulate the aggressive phenotype of basal-like breast cancer and the tendency of metastasis of these cancers (41). However, circulating MMP7 expression levels are poor predictors of BC development (42); therefore, an in-depth study analysis is required. MMP8 was initially thought to be produced only by neutrophils, however it was subsequently found to be expressed by a large variety of other cell types (43), including breast cancer cells (44). It has been reported that MMP8 has a suppressive effect on tumor progression and metastasis, particularly in protecting against lymph node metastasis, while the molecular mechanisms underlying these effects are unclear (45).
MMP9 has been reported to be mainly expressed in the positive region of proliferating cell nuclear antigen, and mediates different signaling pathways to promote or inhibit BC progression or metastasis (46). Wang et al (47) reported that activation of MMP9 by Kruppel like factor 8 serves as a new signal transduction mechanism for invasion and metastasis of human BC. In another study, recombinant WNT-5A was demonstrated to activate CD42 in BC cells, whereas WNT-5A signaling and constitutively active Cdc42 both decreased MMP9 activity, thereby inhibiting the migration and metastasis of BC cells (48). MMP10 was first cloned from the human adenocarcinoma cDNA library (49). Previously, senescent cells were determined to secrete increased levels of MMPs, whereas MMP10 is uniformly upregulated in fibroblasts undergoing senescence (50). MMP11 is expressed in the stromal fibroblasts associated with epithelial cancer cells, and the high level of MMP11 has been linked to the progression of cancer and the adverse prognosis of BC (51). MMP11, a proliferation-related gene, is reported to be significantly associated with DMFS rate in HR−/ HER2+ BC (52). A number of previous studies have shown that MMP11 functioned in BC via insulin-like growth factor-1 signaling or as a downstream target of oncogene or tumor suppressor microRNA (53,54).
Several studies have reported that MMP12 has a dual role in cancer, including anti-angiogenesis (55), or tumor invasion and metastasis (56). An animal experiment showed that overexpressed MMP12 converts plasminogen to angiostatin by cleaving the plasminogen precursor molecule, which then inhibits angiogenesis (55). Another study has identified MMP12 as a potential mediator for CXCR4 to increase invasion (57). A number of studies have reported that MMP13 is highly overexpressed in BC tissue, which is a potential tumor marker for BC diagnosis and serves as a therapeutic target for bone metastasis of BC (58,59). Membrane type I-MMP14 is known to be involved in the initiation and progression of angiogenesis through multiple mechanisms (60,61). The MMP14 inhibitor has been reported to impede angiogenesis, and delay tumor progression and the formation of metastatic lesions; therefore, a selective MMP14 inhibitor may be a potential therapeutic strategy for the treatment of BC (62). To the best of our knowledge, there are a limited number of studies reporting on the relevance between MMP16 and BC.
In this study, MMP1/9/12 mRNA levels were indicated to be significantly overexpressed in BC and increased in higher SBR grades, which suggested a rapid growth of metastatic tumors. This result is in line with the aforementioned studies. In addition, high levels of MMP 1/9/12 indicated that RFS rate in all patients with BC is relatively short; thus, these MMPs may be considered as potential therapeutic targets. In the present study, the transcriptional expression and prognostic value of all MMPs in BC was systematically analyzed, providing a deeper understanding of the complex mechanisms in the molecular biology of BC. Integrative bioinformatics analysis showed that MMP1/9/12/16 could be potential targets for the precise treatment of BC compared with other MMPs; however, the lack of clinical samples is a limitation of the present study. Additional clinical trials are required to validate the diagnostic potentials of these MMPs. In addition, more in-depth experiments, such as single-cell sequencing, are essential to examine the latent interaction mechanism between cancer cells and stromal cells in BC.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YZ carried out the design of this study. YZ and HX performed the statistical analysis and drafted the manuscript. WY and ML made substantial contributions to acquisition of data and helped to revise the manuscript. HL, WP and LW participated in its design and coordination and also helped to draft the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 3.Chen E, Qin X, Peng K, Xu X, Li W, Cheng X, Tang C, Cui Y, Wang Z, Liu T. Identification of potential therapeutic targets among CXC chemokines in breast tumor microenvironment using integrative bioinformatics analysis. Cell Physiol Biochem. 2018;45:1731–1746. doi: 10.1159/000487782. [DOI] [PubMed] [Google Scholar]
- 4.Gross J, Lapiere CM. Collagenolytic activity in amphibian tissues: A tissue culture assay. Proc Natl Acad Sci USA. 1962;48:1014–1022. doi: 10.1073/pnas.48.6.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi E, Tateyama H, Akatsu H, Yamakawa Y, Fujii Y, Eimoto T. Expression of matrix metalloproteinases 2 and 7 in tumor cells correlates with the World Health Organization classification subtype and clinical stage of thymic epithelial tumors. Hum Pathol. 2003;34:1253–1258. doi: 10.1016/j.humpath.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 6.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui N, Hu M, Khalil RA. Biochemical and biological attributes of matrix metalloproteinases. Prog Mol Biol Transl Sci. 2017;147:1–73. doi: 10.1016/bs.pmbts.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alaseem A, Alhazzani K, Dondapati P, Alobid S, Bishayee A, Rathinavelu A. Matrix metalloproteinases: A challenging paradigm of cancer management. Semin Cancer Biol. 2019;56:100–115. doi: 10.1016/j.semcancer.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez-Garcia B, Eiro N, Marin L, González-Reyes S, González LO, Lamelas ML, Vizoso FJ. Expression and prognostic significance of fibronectin and matrix metalloproteases in breast cancer metastasis. Histopathology. 2014;64:512–522. doi: 10.1111/his.12300. [DOI] [PubMed] [Google Scholar]
- 11.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 13.Jezequel P, Campone M, Gouraud W, Guérin-Charbonnel C, Leux C, Ricolleau G, Campion L. bc-GenExMiner: An easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012;131:765–775. doi: 10.1007/s10549-011-1457-7. [DOI] [PubMed] [Google Scholar]
- 14.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 16.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radvanyi L, Singh-Sandhu D, Gallichan S, Lovitt C, Pedyczak A, Mallo G, Gish K, Kwok K, Hanna W, Zubovits J, et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci USA. 2005;102:11005–11010. doi: 10.1073/pnas.0500904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turashvili G, Bouchal J, Baumforth K, Wei W, Dziechciarkova M, Ehrmann J, Klein J, Fridman E, Skarda J, Srovnal J, et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer. 2007;7:55. doi: 10.1186/1471-2407-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 22.Le Doussal V, Tubiana-Hulin V, Friedman S, Hacene K, Spyratos F, Brunet M. Prognostic value of histologic grade nuclear components of Scarff-Bloom-Richardson (SBR). An improved score modification based on a multivariate analysis of 1262 invasive ductal breast carcinomas. Cancer. 1989;64:1914–1921. doi: 10.1002/1097-0142(19891101)64:9<1914::AID-CNCR2820640926>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 23.Bansal C, Singh US, Misra S, Sharma KL, Tiwari V, Srivastava AN. Comparative evaluation of the modified Scarff-Bloom-Richardson grading system on breast carcinoma aspirates and histopathology. Cytojournal. 2012;9:4. doi: 10.4103/1742-6413.92550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu K, Lee CH, Tan PH, Hong GS, Wee SB, Wong CY, Tan P. A molecular signature of the Nottingham prognostic index in breast cancer. Cancer Res. 2004;64:2962–2968. doi: 10.1158/0008-5472.CAN-03-2430. [DOI] [PubMed] [Google Scholar]
- 25.Gluck S. Extending the clinical benefit of endocrine therapy for women with hormone receptor-positive metastatic breast cancer: Differentiating mechanisms of action. Clin Breast Cancer. 2014;14:75–84. doi: 10.1016/j.clbc.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 26.Dawood S. Triple-negative breast cancer: Epidemiology and management options. Drugs. 2010;70:2247–2258. doi: 10.2165/11538150-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 27.Javadian M, Gharibi T, Shekari N, Abdollahpour-Alitappeh M, Mohammadi A, Hossieni A, Mohammadi H, Kazemi T. The role of microRNAs regulating the expression of matrix metalloproteinases MMPs) in breast cancer development, progression, and meta stasis. J Cell Physiol. 2019;234:5399–5412. doi: 10.1002/jcp.27445. [DOI] [PubMed] [Google Scholar]
- 28.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: Trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 29.Shay G, Lynch CC, Fingleton B. Moving targets: Emerging roles for MMPs in cancer progression and metastasis. Matrix Biol. 2005;44-46:200–206. doi: 10.1016/j.matbio.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohrmann A, Kammerer U, Kapp M, Dietl J, Anacker J. Expression of matrix metalloproteinases (MMPs) in primary human breast cancer and breast cancer cell lines: New findings and review of the literature. BMC Cancer. 2009;9:188. doi: 10.1186/1471-2407-9-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bostrom P, Soderstrom M, Vahlberg T, Söderström KO, Roberts PJ, Carpén O, Hirsimäki P. MMP-1 expression has an independent prognostic value in breast cancer. BMC Cancer. 2011;11:348. doi: 10.1186/1471-2407-11-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulic A, Dedic Plavetic N, Vrbanec J, Sirotković-Skerlev M. Low serum MMP-1 in breast cancer: A negative prognostic factor? Biomarkers. 2012;17:416–421. doi: 10.3109/1354750X.2012.678885. [DOI] [PubMed] [Google Scholar]
- 33.Mendes O, Kim HT, Lungu G, Stoica G. MMP2 role in breast cancer brain metastasis development and its regulation by TIMP2 and ERK1/2. Clin Exp Metastasis. 2007;24:341–351. doi: 10.1007/s10585-007-9071-0. [DOI] [PubMed] [Google Scholar]
- 34.Li H, Qiu Z, Li F, Wang C. The relationship between MMP-2 and MMP-9 expression levels with breast cancer incidence and prognosis. Oncol Lett. 2017;14:5865–5870. doi: 10.3892/ol.2017.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren F, Tang R, Zhang X, Madushi WM, Luo D, Dang Y, Li Z, Wei K, Chen G. Overexpression of MMP family members functions as prognostic biomarker for breast cancer patients: A Systematic review and meta-analysis. PLoS One. 2015;10:e0135544. doi: 10.1371/journal.pone.0135544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu C, Liu X, Bai X, Zhao T, Wang M, Xu R, Li M, Hu Y, Li W, Yang LU, et al. MiR-519d suppresses breast cancer tumorigenesis and metastasis via targeting MMP3. Int J Biol Sci. 2014;14:228–236. doi: 10.7150/ijbs.22849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panagopoulos V, Leach DA, Zinonos I, Ponomarev V, Licari G, Liapis V, Ingman WV, Anderson P, DeNichilo MO, Evdokiou A. Inflammatory peroxidases promote breast cancer progression in mice via regulation of the tumour microenvironment. Int J Oncol. 2017;50:1191–1200. doi: 10.3892/ijo.2017.3883. [DOI] [PubMed] [Google Scholar]
- 38.Slattery ML, John E, Torres-Mejia G, Stern M, Lundgreen A, Hines L, Giuliano A, Baumgartner K, Herrick J, Wolff RK. Matrix metalloproteinase genes are associated with breast cancer risk and survival: The Breast Cancer Health Disparities Study. PLoS One. 2013;8:e63165. doi: 10.1371/journal.pone.0063165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vizoso FJ, Gonzalez LO, Corte MD, Rodríguez JC, Vázquez J, Lamelas ML, Junquera S, Merino AM, García-Muñiz JL. Study of matrix metalloproteinases and their inhibitors in breast cancer. Br J Cancer. 2007;96:903–911. doi: 10.1038/sj.bjc.6603666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim GE, Lee JS, Choi YD, Lee KH, Lee JH, Nam JH, Choi C, Kim SS, Park MH, Yoon JH, Kweon SS. Expression of matrix metalloproteinases and their inhibitors in different immunohistochemical-based molecular subtypes of breast cancer. BMC Cancer. 2014;14:959. doi: 10.1186/1471-2407-14-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sizemore ST, Keri RA. The forkhead box transcription factor FOXC1 promotes breast cancer invasion by inducing matrix metalloprotease 7 (MMP7) expression. J Biol Chem. 2012;287:24631–24640. doi: 10.1074/jbc.M112.375865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aroner SA, Rosner BA, Tamimi RM, Tworoger SS, Baur N, Joos TO, Hankinson SE. Plasma matrix metalloproteinase 1, 3, and 7 levels and breast cancer risk in the Nurses' Health study. Cancer Causes Control. 2014;25:1717–1723. doi: 10.1007/s10552-014-0462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Lint P, Libert C. Matrix metalloproteinase-8: Cleavage can be decisive. Cytokine Growth Factor Rev. 2006;17:217–223. doi: 10.1016/j.cytogfr.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 44.Decock J, Long JR, Laxton RC, Shu XO, Hodgkinson C, Hendrickx W, Pearce EG, Gao YT, Pereira AC, Paridaens R, et al. Association of matrix metalloproteinase-8 gene variation with breast cancer prognosis. Cancer Res. 2007;67:10214–10221. doi: 10.1158/0008-5472.CAN-07-1683. [DOI] [PubMed] [Google Scholar]
- 45.Decock J, Hendrickx W, Vanleeuw U, Van Belle V, Van Huffel S, Christiaens MR, Ye S, Paridaens R. Plasma MMP1 and MMP8 expression in breast cancer: Protective role of MMP8 against lymph node metastasis. BMC Cancer. 2008;8:77. doi: 10.1186/1471-2407-8-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu B, Cui J, Sun J, Li J, Han X, Guo J, Yi M, Amizuka N, Xu X, Li M. Immunolocalization of MMP9 and MMP2 in osteolytic metastasis originating from MDA-MB-231 human breast cancer cells. Mol Med Rep. 2016;14:1099–1106. doi: 10.3892/mmr.2016.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Lu H, Urvalek AM, Li T, Yu L, Lamar J, DiPersio CM, Feustel PJ, Zhao J. KLF8 promotes human breast cancer cell invasion and metastasis by transcriptional activation of MMP9. Oncogene. 2011;30:1901–1911. doi: 10.1038/onc.2010.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prasad CP, Chaurasiya SK, Axelsson L, Andersson T. WNT-5A triggers Cdc42 activation leading to an ERK1/2 dependent decrease in MMP9 activity and invasive migration of breast cancer cells. Mol Oncol. 2013;7:870–883. doi: 10.1016/j.molonc.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muller D, Quantin B, Gesnel MC, Millon-Collard R, Abecassis J, Breathnach R. The collagenase gene family in humans consists of at least four members. Biochem J. 1988;253:187–192. doi: 10.1042/bj2530187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thakur S, Nabbi A, Klimowicz A, Riabowol K. Stromal ING1 expression induces a secretory phenotype and correlates with breast cancer patient survival. Mol Cancer. 2015;14:164. doi: 10.1186/s12943-015-0434-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Vega RC, Sanchez MLF, Eiro N, Vizoso FJ, Sperling M, Karst U, Medel AS. Multimodal laser ablation/desorption imaging analysis of Zn and MMP-11 in breast tissues. Anal Bioanal Chem. 2018;410:913–922. doi: 10.1007/s00216-017-0537-x. [DOI] [PubMed] [Google Scholar]
- 52.Han J, Choi YL, Kim H, Choi JY, Lee SK, Lee JE, Choi JS, Park S, Choi JS, Kim YD, et al. MMP11 and CD2 as novel prognostic factors in hormone receptor-negative, HER2-positive breast cancer. Breast Cancer Res Treat. 2017;164:41–56. doi: 10.1007/s10549-017-4234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kasper G, Reule M, Tschirschmann M, Dankert N, Stout-Weider K, Lauster R, Schrock E, Mennerich D, Duda GN, Lehmann KE. Stromelysin-3 over-expression enhances tumourigenesis in MCF-7 and MDA-MB-231 breast cancer cell lines: Involvement of the IGF-1 signalling pathway. BMC Cancer. 2007;7:12. doi: 10.1186/1471-2407-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kwon YJ, Hurst DR, Steg AD, Yuan K, Vaidya KS, Welch DR, Frost AR. Gli1 enhances migration and invasion via up-regulation of MMP-11 and promotes metastasis in ERalpha negative breast cancer cell lines. Clin Exp Metastasis. 2011;28:437–449. doi: 10.1007/s10585-011-9382-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Margheri F, Serrati S, Lapucci A, Anastasia C, Giusti B, Pucci M, Torre E, Bianchini F, Calorini L, Albini A, et al. Systemic sclerosis-endothelial cell antiangiogenic pentraxin 3 and matrix metalloprotease 12 control human breast cancer tumor vascularization and development in mice. Neoplasia. 2009;11:1106–1115. doi: 10.1593/neo.09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delassus GS, Cho H, Eliceiri GL. New signaling pathways from cancer progression modulators to mRNA expression of matrix metalloproteinases in breast cancer cells. J Cell Physiol. 2011;226:3378–3384. doi: 10.1002/jcp.22694. [DOI] [PubMed] [Google Scholar]
- 57.Hernandez L, Magalhaes MA, Coniglio SJ, Condeelis JS, Segall JE. Opposing roles of CXCR4 and CXCR7 in breast cancer metastasis. Breast Cancer Res. 2011;13:R128. doi: 10.1186/bcr3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang HJ, Yang MJ, Yang YH, Hou MF, Hsueh EJ, Lin SR. MMP13 is potentially a new tumor marker for breast cancer diagnosis. Oncol Rep. 2009;22:1119–1127. doi: 10.3892/or_00000544. [DOI] [PubMed] [Google Scholar]
- 59.Nannuru KC, Futakuchi M, Varney ML, Vincent TM, Marcusson EG, Singh RK. Matrix metalloproteinase (MMP)-13 regulates mammary tumor-induced osteolysis by activating MMP9 and transforming growth factor-beta signaling at the tumor-bone interface. Cancer Res. 2010;70:3494–3504. doi: 10.1158/0008-5472.CAN-09-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alfranca A, Lopez-Oliva JM, Genis L, López-Maderuelo D, Mirones I, Salvado D, Quesada AJ, Arroyo AG, Redondo JM. PGE2 induces angiogenesis via MT1-MMP-mediated activation of the TGFbeta/Alk5 signaling pathway. Blood. 2008;112:1120–1128. doi: 10.1182/blood-2007-09-112268. [DOI] [PubMed] [Google Scholar]
- 61.Turunen SP, Tatti-Bugaeva O, Lehti K. Membrane-type matrix metalloproteases as diverse effectors of cancer progression. Biochim Biophys Acta Mol Cell Res. 2017;1864:1974–1988. doi: 10.1016/j.bbamcr.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 62.Ager EI, Kozin SV, Kirkpatrick ND, Seano G, Kodack DP, Askoxylakis V, Huang Y, Goel S, Snuderl M, Muzikansky A, et al. Blockade of MMP14 activity in murine breast carcinomas: Implications for macrophages, vessels, and radiotherapy. J Natl Cancer Inst. 2015;107(pii):djv017. doi: 10.1093/jnci/djv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009;11:R7. doi: 10.1186/bcr2222. (Table S1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. (Table S1) [DOI] [PubMed] [Google Scholar]
- 65.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. (Table S1) [DOI] [PubMed] [Google Scholar]
- 66.Gluck S, Ross JS, Royce M, McKenna EF, Jr, Perou CM, Avisar E, Wu L. TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine ± trastuzumab. Breast Cancer Res Treat. 2012;132:781–791. doi: 10.1007/s10549-011-1412-7. (Table S1) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.