Abstract
Eluforsen (previously known as QR-010) is a 33-mer 2′-O-methyl modified phosphorothioate antisense oligonucleotide targeting the F508del mutation in the gene encoding CFTR protein of cystic fibrosis patients. In this study, eluforsen was incubated with endo- and exonucleases and mouse liver homogenates to elucidate its in vitro metabolism. Mice and monkeys were used to determine in vivo liver and lung metabolism of eluforsen following inhalation. We developed a liquid chromatography-mass spectrometry method for the identification and semi-quantitation of the metabolites of eluforsen and then applied the method for in vitro and in vivo metabolism studies. Solid-phase extraction was used following proteinase K digestion for sample preparation. Chain-shortened metabolites of eluforsen by 3′ exonuclease were observed in mouse liver in an in vitro incubation system and by either 3′ exonuclease or 5′ exonuclease in liver and lung samples from an in vivo mouse and monkey study. This study provides approaches for further metabolite characterization of 2′-ribose-modified phosphorothioate oligonucleotides in in vitro and in vivo studies to support the development of oligonucleotide therapeutics.
Keywords: eluforsen, antisense oligonucleotide, metabolism, cystic fibrosis, ion-pair, liquid chromatography, mass spectrometry
Introduction
Nonclinical evaluation of drug safety usually consists of standard animal toxicology studies, which includes assessment of drug exposure and primarily parent drug plasma concentrations. This information is used to predict and assess potential risks in clinical trials. The US Food and Drug Administration (FDA) Guidance for Industry about Safety Testing of Drug Metabolites has reported, “We encourage the identification of differences in drug metabolism between animals used in nonclinical safety assessments and humans as early as possible during the drug development process.”1 Therefore, metabolite profiling of the investigational drug in different species, which are used in nonclinical toxicity studies, is essential, since this information provides comprehensive insight into the potential risk during clinical trials. In human studies, measurement of only the parent compound is usually sufficient when the metabolite profiles in humans are similar to those in at least one of the animal species used in the nonclinical safety assessment.
Eluforsen is a 33-mer antisense oligonucleotide (ASO) product that is being developed for the chronic inhalation treatment of patients with cystic fibrosis (CF) that have the F508del mutation. It has been granted orphan drug designation in the United States (09/23/2013)2 and the European Union (07/10/2013)3 and received FDA fast-track designation (07/18/2016). Two clinical studies, an exploratory proof-of-concept trial and a phase 1b safety study (NCT02564354), have been completed. More than 85% of all CF patients are affected by ΔF508 (p.Phe508del) mutations in the gene responsible for the expression of the CF transmembrane conductance regulator (CFTR). The lack of CFTR function leads to a reduction of chloride transport, resulting in thick and sticky mucus in the lungs, pancreas, and other organs.4 Specifically, the mucus clogs the airways in the lungs resulting in extensive damage. All of the eluforsen nucleotides are modified with phosphorothioate (PS) and 2′-O-methyl ribose modifications. These modifications result in resistance to nuclease-mediated degradation, thereby greatly reducing its metabolic rate and increasing plasma and tissue half-life times for eluforsen.5, 6, 7, 8, 9, 10
Unlike the metabolism of small molecules, which is largely accomplished through CYP-mediated oxidative metabolic and/or conjugation pathways, oligonucleotides undergo different routes of metabolism.11, 12 Oligonucleotide metabolites are mainly formed by endo- and exonucleases that degrade nucleic acids by hydrolyzing phosphodiester bonds,13 which are ubiquitously present in many different cell types.7 Studies on the various FDA-approved oligonucleotides (e.g., Kynamro, Macugen, and Spinraza) have shown that oligonucleotides are primarily metabolized by intracellular endo- and exonucleases to produce chain-shortened oligonucleotide metabolites.14 The modifications present on the backbone of oligonucleotides determine the enzymatic activities leading to the generation of metabolites. Kynamro is a 20-nucleotide gapmer with 2′-O-methoxyethyl (MOE)-modified bases at the 5′ and 3′ ends with 10 internal nucleotides placed between the modified ends. All 20 nucleotides are modified by a PS backbone. Kynamro chain-shortened metabolites within tissues occur initially by endonucleases that cleave the molecule in the gap, resulting in the production of short oligonucleotides, which are further digested by exonucleases to produce a cascade of shorter oligonucleotides.14, 15, 16 However, Spinraza, which has a fully 2′-O-MOE ribose and PS backbone modifications, is metabolized more slowly and predominantly via exonuclease (3′ and 5′)-mediated hydrolysis producing chain-shortened metabolites in different tissues.17
Different techniques (e.g., complement factor H ELISA [hELISA], radiolabeling, or liquid chromatography-mass spectrometry) have been used for identification of ASO and its metabolites. Each technique has its own limitations. For preclinical development, bioanalytical methods are required to perform bioanalysis of eluforsen in various biological fluids (e.g., plasma, urine, tissue, etc.) in order to generate toxicokinetic (TK), pharmacokinetic (PK), bioavailability, and metabolic stability data of the parent molecule. Because of its unmatched sensitivity, hELISA methods are in place to determine the concentration of eluforsen in preclinical species and humans. However, the major limitation of hELISA is that this method generally does not distinguish between full-length oligonucleotides and its truncated metabolites.18, 19 Currently, mass spectrometry (MS) plays a key role in studies for the determination of metabolic stability of oligonucleotides in biological matrices, which is an important part of the drug development process.7, 18, 19, 20 liquid chromatography-mass spectrometry (LC-MS) technology, on one hand, has the capability to perform metabolite identification and on the other hand has the inherent ability to monitor individual target oligonucleotides, as well as their metabolites. Our group has developed LC-MS/MS methods for the detection of oligonucleotides.5, 21 In this study, we have developed an LC-MS approach for the metabolite identification of eluforsen and applied this method to the characterization and semi-quantitation of metabolites in in vitro incubations and in tissues obtained from in vivo animal studies.
Results
LC-MS Method Development for Identification of Metabolites
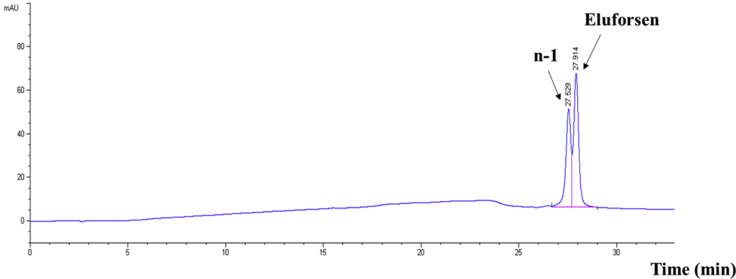
Based on the results of direct infusion experiments, the MS tuning parameters were optimized as described in the LC-MS conditions of the Materials and Methods section. The characterization of the effect of various alkylamine ion-pairing agents and fluorinated alcohols on the MS signal intensity of eluforsen were performed for the development of a sensitive LC-MS method. Initially, a concentration of 20 μg/mL of eluforsen in 50:50 methanol/water was directly infused into a Waters Synapt G2 HDMS quadrupole time-of-flight hybrid mass spectrometer. Subsequently, precise amounts of ion-pairing (IP) agents including N, N-diisopropylethylamine (DIEA) and N, N-dimethylcyclohexylamine (DMCHA), octylamine, tripropylamine, tributylamine, and N, N-dimethylbutylamine were added to the aliquots of the same solution and infused to the mass spectrometer. DIEA and DMCHA generated the highest electrospray ionization-mass spectrometry (ESI-MS) signal intensities for eluforsen compared to other alkylamine IP agents.22 In order to further optimize the mobile phase composition for higher MS sensitivity, the effect of various perfluorinated alcohols including hexafluoroisopropanol (HFIP) have been investigated as counter ions.22 For these experiments, the concentration of 25 mM perfluorinated alcohols along with 15 mM IP agents were utilized. DIEA and DMCHA still generated the strongest MS signal intensities even after the addition of HFIP. As a result, DMCHA performed well and slightly better than DIEA while maintaining equivalent LC separations. Figure 1 shows the HPLC chromatogram of eluforsen and its n-1 truncated oligonucleotide using the buffer system with DMCHA. Finally, the LC-MS method with 15 mM DMCHA and 25 mM HFIP was applied to analyze eluforsen and its metabolites for in vitro and in vivo metabolism studies.
Figure 1.
HPLC Separation of Eluforsen from Its 3′ n-1 Truncated Oligonucleotide with Ion-Pair DMCHA
DMCHA was selected as an ion-pair agent to provide good chromatographic separation after investigation of various alkylamines. 40 μg/mL of oligonucleotides were detected at 260 nm.
Optimization and Comparison of Sample Preparation Methods for Metabolism Studies
Designing proper sample preparation is critical in the MS-based analysis due to the significant impact on the quality and reproducibility of sample preparation.7, 19 Appropriate sample extraction and preparation methods need to be chosen depending on the analyte of interest, analytical method used, and experimental goals. Biological samples for therapeutic oligonucleotides contain significant amounts of unwanted cellular materials including proteins and lipids that cause signal suppression and nonspecific binding of therapeutic oligonucleotides.23, 24 Many approaches, including liquid-liquid extraction (LLE),18, 21, 25 trizol extraction,26 proteinase K digestion,27 solid-phase extraction (SPE),5, 18, 25, 28, 29 and magnetic bead extraction methods,30 can be applied successfully to isolate oligonucleotides from biological samples prior to LC-MS analysis.
Proteinase K is a general enzyme used to hydrolyze all proteins with a quick digestion time. It can be easily modified to improve the quality of the approach by adding dithiothreitol to reduce protein disulfide bonds and adding guanidinium chloride to increase the rate of protein degradation.31 EDTA can be used to inhibit nuclease activity during the proteinase K digestion by acting through the sequestration of metal atoms that are essential cofactors for several families of nucleases.32, 33 The approach can minimize the loss of oligonucleotides caused by nonspecific adsorption during sample preparation by greatly reducing the number of sample transfer steps.27 Additionally, the recovery for oligonucleotides has been determined to be greater than 90%.27 It can be readily used with other methods, including SPE. Hence, proteinase K digestion was chosen in this study to remove contaminating proteins prior to the extraction of oligonucleotides from biological samples.
To assess the effectiveness of sample preparation methods for metabolic studies of eluforsen, samples were incubated with mouse liver homogenates for 7 days. After proteinase K digestion, the samples were extracted using either anion-exchange SPE or the immunoaffinity capture approach using streptavidin-coated magnetic beads with a biotinylated capture strand. A biotinylated capture strand that was complementary with eluforsen and bound to streptavidin magnetic beads was used to selectively capture eluforsen and its metabolites. As a result, only n-1, n-2, and n-3 metabolites (3′ end shortmers) of eluforsen were detected using the streptavidin magnetic-bead method, while more 3′ end shortmers (from n-1 to n-8) were observed using the SPE method. These results indicate that the highly selective capture strand loses affinity from 3′ n-4 shortmers onward when using the magnetic-bead approach. It should be noted that, in this stage, 5′ end shortmers were not observed using either approach. Hence, to recover a wide variety of different types of oligonucleotides, anion-exchange SPE method rather than a magnetic-bead-based approach was used following proteinase K digestion in order to maximize the coverage of metabolites.
Determination of In Vitro-Generated Metabolites by Endo- and Exonucleases
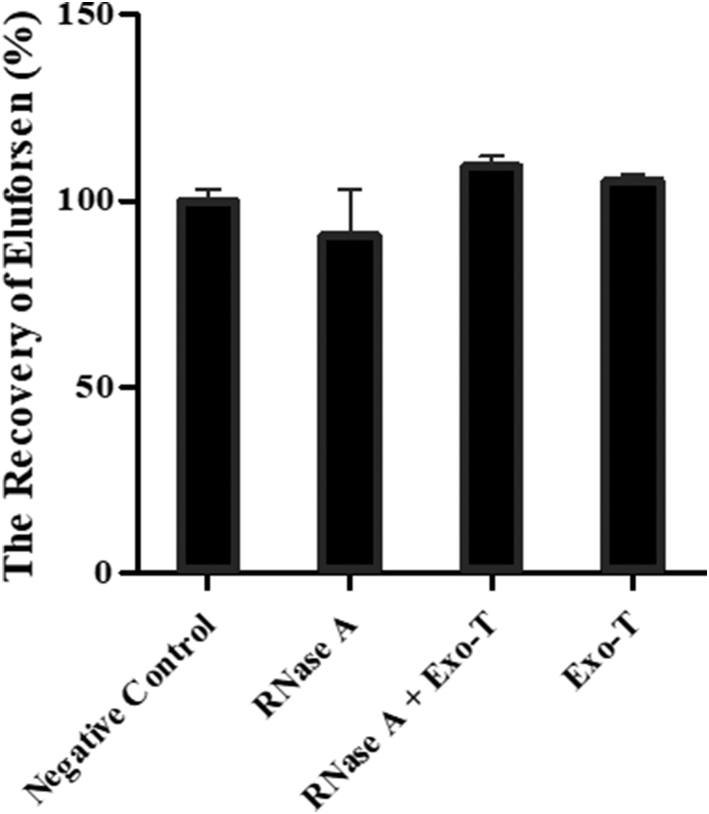
Eluforsen was incubated with purified endo- (RNase A) and exonuclease (Exo-T) enzymes to identify the possible metabolites that could be formed in vitro. An RNA strand was used as a positive control, which was totally degraded after 1 h of incubation with RNase A. The samples were analyzed by LC-MS. As shown in Figure 2, the recoveries of eluforsen were 100.00% ± 3.35%, 90.84% ± 12.23%, 109.17% ± 2.90%, and 105.46% ± 1.73% (mean ± SEM) in the buffer only, RNase A, both enzymes, and Exo-T, respectively. Eluforsen did not show any metabolism after 2 days of incubation with RNase A and Exo-T. Since eluforsen is fully modified with PSs and 2′-O-methylribonucleosides, it is highly resistant against digestion by endo- and exonuclease enzymes. This result is consistent with previous studies involving enzymatic incubations of PS-containing oligonucleotides when using purified enzymes34 and nucleases derived from cells.35 Because the purified enzymes were not able to digest eluforsen, we proceeded to test mouse liver homogenate to identify metabolites.36
Figure 2.
Comparison of the Recovery (%) of Eluforsen after 2-Day Incubations with Purified Nucleases
1 μg of eluforsen was incubated with either endonuclease (RNase A), exonuclease (Exo-T), or both enzymes (double-digestion) at 37°C for 2 days. A negative control was prepared by incubation of eluforsen in buffer but without any endo- and exonuclease. The recoveries of eluforsen were 100.00% ± 3.35%, 90.84% ± 12.23%, 109.17% ± 2.90%, and 105.46% ± 1.73% (mean ± standard error mean) in the buffer only, RNase A, both enzymes, and Exo-T, respectively.
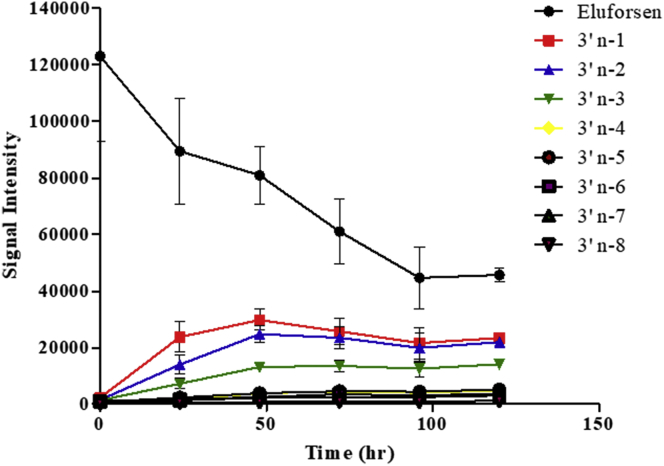
Determination of the Rate of Formation of Shortmers of Eluforsen with Mouse Liver Homogenates
In vitro metabolic studies of eluforsen were performed by incubation with mouse liver homogenates for 0, 24, 48, 72, 96, and 120 h to determine the rate of formation of shortmers of eluforsen. Subsequently, the samples were analyzed by LC-MS after sample preparation using a combination of proteinase K digestion and SPE extraction. Table 1 and Figure 3 show the results obtained in a series of short-time incubations with mouse liver homogenates. Table 1 shows 3′ end shortmers generated after 120 h of incubation with mouse liver homogenates; 5′ end shortmers were not observed during incubation with mouse liver homogenates. The results suggest that 3′ exonucleases are the major enzyme responsible for metabolizing eluforsen in mouse liver homogenates as supported by the high level of formation of 3′ end shortmers. As shown in Figure 3, 3′ end shortmers were mainly produced in the order of n-1, n-2, and n-3. The result suggests that the 3′-poly(U) tail appears to be the most metabolically susceptible portion of eluforsen in mouse liver homogenates. We could not observe the presence of any oxidation of the PS backbone, conversion of a 2′-O-Me to a hydroxyl group, or simple loss of any of the bases (e.g., depurination or depyrimidination).
Table 1.
In Vitro-Generated Eluforsen Metabolites after Incubation with Purified Nucleases and Mouse Liver Homogenates and In Vivo-Generated Metabolites in Liver and Lung Samples of Mice and Monkeys
| Metabolites |
In Vitro-Generated Metabolites |
In Vivo-Generated Metabolites |
||||
|---|---|---|---|---|---|---|
| Incubation with Purified Nucleases (after 7 Days) | Incubation with Mouse Liver Homogenates (after 5 Days) | Mouse |
Monkey |
|||
| Liver | Lung | Liver | Lung | |||
| 3′ n-1 | – | 18.97% | 13.03% | 12.09% | 9.97% | 6.72% |
| 3′ n-2 | – | 17.80% | 7.14% | 9.44% | 8.07% | 5.41% |
| 3′ n-3 | – | 11.48% | 5.72% | 8.91% | 1.75% | 3.38% |
| 3′ n-4 | – | 3.94% | 4.25% | 6.60% | 1.01% | 2.84% |
| 3′ n-5 | – | 4.21% | 3.71% | 5.55% | 1.12% | 3.47% |
| 3′ n-6 | – | 3.03% | 4.58% | 4.79% | 1.97% | 4.19% |
| 3′ n-7 | – | 2.53% | 3.25% | 4.20% | 0.97% | 3.22% |
| 3′ n-8 | – | 0.98% | 1.40% | – | – | 2.78% |
| 5′ n-1 | – | – | 15.41% | 19.88% | 7.48% | 7.62% |
| 5′ n-2 | – | – | 7.31% | 6.94% | 1.24% | 3.27% |
| 5′ n-3 | – | – | 8.25% | 8.98% | 3.18% | 4.24% |
| 5′ n-4 | – | – | 4.90% | 9.05% | 1.68% | 4.50% |
| 5′ n-5 | – | – | 10.29% | 15.03% | 6.77% | 7.01% |
| 5′ n-6 | – | – | – | – | – | 2.37% |
| 5′ n-7 | – | – | – | – | – | 2.25% |
| 5′ n-8 | – | – | – | – | – | – |
Data reported as the mean percentage.
Figure 3.
Determination of the Rate of Formation of 3′ End Shortmers of Eluforsen with Mouse Liver Homogenates
50 μg/mL of eluforsen was incubated for 0, 24, 48, 72, 96, and 120 h with mouse liver homogenates. The intensities values represent the mean ± SD (n = 3).
Determination of In Vivo-Generated Metabolites of Eluforsen in Liver and Lung Samples of Mice and Monkeys Using LC-MS/MS
Drug metabolism studies usually involve identification of circulating metabolites in plasma or serum, as this enables cross-species comparison between animals and humans. However, following inhalation administration of eluforsen, systemic exposure in serum was too low to serve as a matrix for metabolite profiling investigations (data not shown). Metabolite profiles were therefore characterized in lung and liver obtained from mice and monkeys following 13 weeks of inhalation delivery of eluforsen. Lung is the intended pharmacological target organ of eluforsen and the main organ for disposition following the inhalation delivery of eluforsen, and the liver is a key organ for accumulation of oligonucleotides following systemic exposure,36, 37 as well as the main organ for metabolism of xenobiotics including therapeutics.2
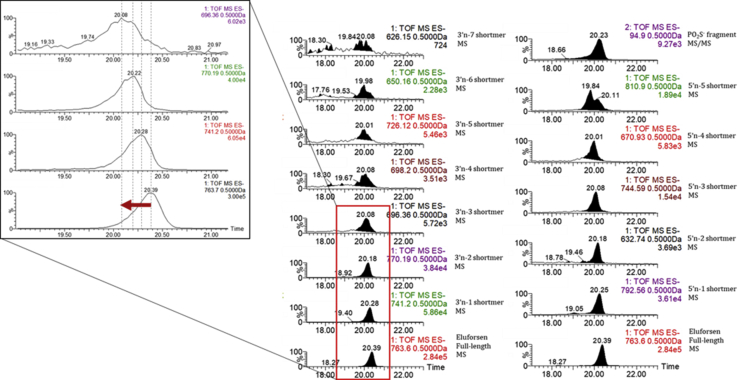
From the 13-week inhalation studies, 18 liver samples (from eight monkeys, 10 mice) and 13 lung samples (from eight monkeys, five mice), collected at the end of the 13-week treatment period, were prepared to identify in vivo-generated eluforsen metabolites. The liver and lung samples were homogenized by following the protocol as detailed in the Materials and Methods section. Mouse lung and liver homogenates were preincubated with proteinase K followed by SPE for making matrix-matched standard calibration curves to mimic the loss of recovery from the biological matrix. As shown in Figure S1, eluforsen and its metabolites were identified from the first mass analyzer precursor ion chromatogram and MSE spectra. The MS/MS fragmentation patterns of eluforsen and its metabolites were not specific enough to fully sequence each metabolite (Figure S2). However, for each metabolite, there were several specific fragment ions present that reinforced the initial assignment made using the molecular weight. The limitations of using MS/MS in qualitative analysis have been previously addressed because major MS/MS fragment ions to fully characterize each metabolite were generally not observed.38 To calculate signal intensities, an ion peak for each analyte was extracted using the m/z value of the first mass analyzer precursor ion at a specific charge state and used to determine the area of the peak. As shown in Table 2, different charge states were selected as precursor ions, as seen in the first mass analyzer to avoid overlapped m/z series for each analyte. Six calibration curves (Figure S3) were made separately using signal intensities of eluforsen in different charge states (−12, −13, −14, −15, −16, and −17). The calibration curves of eluforsen were used to estimate the concentrations of its metabolites, which, although semiquantitative, could still be used to approximate the metabolic stability of eluforsen in in vitro and in vivo systems. The calibration curve with the same charge state as each analyte was used to estimate the concentration of each analyte. PO2S− fragment ions (m/z 94.9 seen in MS/MS data) were observed to confirm the presence of therapeutic oligonucleotides in in vivo samples.37
Table 2.
Sequences, Selected Charge States, and m/z of Eluforsen and Its Potential Metabolites
| Name | Sequence (from 5′ to 3′) | Selected Charge State | The Number of Phosphorothioate Oxidations (m/z) | ||||||
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | |||
| Eluforsen | AUC AUA GGA AAC ACC AAA GAU GAU AUU UUC UUU | −15 | 763.60 | 762.54 | 761.47 | 760.40 | 759.32 | 758.25 | 757.18 |
| 3′ n-1 | AUC AUA GGA AAC ACC AAA GAU GAU AUU UUC UU | −15 | 741.20 | 740.12 | 739.05 | 737.98 | 736.91 | 735.84 | 734.76 |
| 3′ n-2 | AUC AUA GGA AAC ACC AAA GAU GAU AUU UUC U | −14 | 770.19 | 769.04 | 767.89 | 766.74 | 765.60 | 764.45 | 763.30 |
| 3′ n-3 | AUC AUA GGA AAC ACC AAA GAU GAU AUU UUC | −15 | 696.36 | 695.29 | 694.21 | 693.14 | 692.07 | 691.00 | 689.93 |
| 3′ n-4 | AUC AUA GGA AAC ACC AAA GAU GAU AUU UU | −16 | 631.82 | 630.81 | 629.81 | 628.80 | 627.80 | 626.80 | 625.79 |
| 3′ n-5 | AUC AUA GGA AAC ACC AAA GAU GAU AUU U | −14 | 698.20 | 697.05 | 695.91 | 694.76 | 693.61 | 692.46 | 691.32 |
| 3′ n-6 | AUC AUA GGA AAC ACC AAA GAU GAU AUU | −13 | 726.12 | 724.88 | 723.65 | 722.41 | 721.18 | 719.94 | 718.71 |
| 3′ n-7 | AUC AUA GGA AAC ACC AAA GAU GAU AU | −14 | 650.16 | 649.02 | 647.87 | 646.72 | 645.57 | 644.43 | 643.28 |
| 3′ n-8 | AUC AUA GGA AAC ACC AAA GAU GAU A | −14 | 626.15 | 625.00 | 623.85 | 622.70 | 621.56 | 620.41 | 619.26 |
| 5′ n-1 | UC AUA GGA AAC ACC AAA GAU GAU AUU UUC UUU | −14 | 792.56 | 791.41 | 790.26 | 789.12 | 787.97 | 786.82 | 785.67 |
| 5′ n-2 | C AUA GGA AAC ACC AAA GAU GAU AUU UUC UUU | −17 | 632.74 | 631.79 | 630.85 | 629.90 | 628.96 | 628.01 | 627.07 |
| 5′ n-3 | AUA GGA AAC ACC AAA GAU GAU AUU UUC UUU | −14 | 744.59 | 743.44 | 742.30 | 741.15 | 740.00 | 738.85 | 737.71 |
| 5′ n-4 | UA GGA AAC ACC AAA GAU GAU AUU UUC UUU | −15 | 670.93 | 669.86 | 668.79 | 667.72 | 666.65 | 665.58 | 664.51 |
| 5′ n-5 | A GGA AAC ACC AAA GAU GAU AUU UUC UUU | −12 | 810.90 | 809.56 | 808.22 | 806.88 | 805.54 | 804.20 | 802.86 |
| 5′ n-6 | GGA AAC ACC AAA GAU GAU AUU UUC UUU | −15 | 624.56 | 623.49 | 622.42 | 621.35 | 620.28 | 619.21 | 618.14 |
| 5′ n-7 | GA AAC ACC AAA GAU GAU AUU UUC UUU | −15 | 599.54 | 598.47 | 597.40 | 596.33 | 595.26 | 594.19 | 593.12 |
| 5′ n-8 | A AAC ACC AAA GAU GAU AUU UUC UUU | −15 | 574.52 | 573.45 | 572.38 | 571.31 | 570.24 | 569.17 | 568.10 |
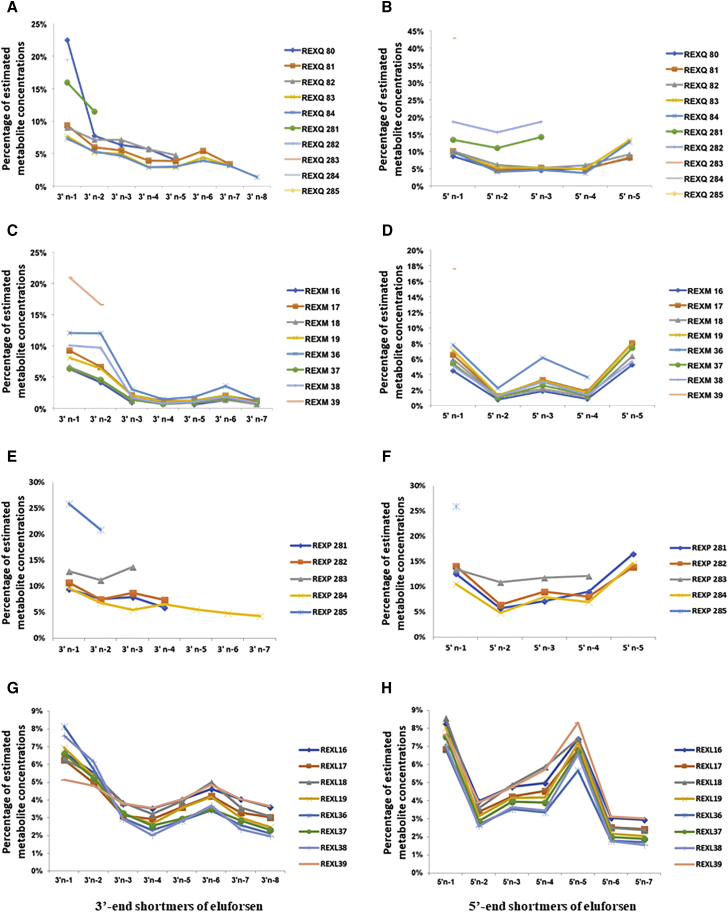
Figure 4 shows the representative chromatograms of one in vivo sample. Eluforsen and its metabolites were observed in both in vivo mouse and monkey liver samples. The estimated mean percentages for in vivo-generated metabolites in liver and lung samples of mice and monkeys are presented in Table 1. 3′ end shortmers (from n-1 to n-8) and 5′ end shortmers (from n-1 to n-5) were observed in both mouse and monkey samples. 3′ end shortmers generated in mouse and monkey liver samples ranged from 1.40% to 13.03% and from 0.97% to 9.97%, respectively. 5′ end shortmers generated in mouse and monkey liver samples ranged from 4.90% to 15.41% and from 1.24% to 7.48%, respectively. The results suggest that both 3′ exonuclease and 5′ exonuclease were responsible for metabolizing eluforsen in in vivo mouse and monkey liver. As shown in Figures 5A and 5B for mouse samples and Figures 5C and 5D for monkey samples, trends of metabolite generation were very similar between samples. Of note, the 5′ n-3 and 5′ n-5 shortmers were present at higher amounts than expected based on the presence of other shortmers. As these metabolites had an adenine at the end of their sequence, this observation may suggest that adenine provides increased nuclease stability compared to the other nucleotides in the sequence.
Figure 4.
Representative Extracted Ion Chromatograms of Eluforsen and Its Metabolites
In vivo samples were analyzed by LC-MS after sample preparation using a combination of proteinase K digestion and SPE extraction. Mobile phase A consisted of 15 mM DMCHA and 25 mM HFIP in 5% methanol, and mobile phase B containing 95% methanol was used as an optimized buffer system.
Figure 5.
Trends of Generation of Eluforsen Metabolites in In Vivo Liver and Lung Samples of Mice and Monkeys
(A) 3′ end shortmers in mouse liver samples. (B) 5′ end shortmers in mouse liver samples. (C) 3′ end shortmers in monkey liver samples. (D) 5′ end shortmers in monkey liver samples. (E) 3′ end shortmers in mouse lung samples. (F) 5′ end shortmers in mouse lung samples. (G) 3′ end shortmers in monkey lung samples. (H) 5′ end shortmers in monkey lung samples.
Higher levels of eluforsen and its metabolites were observed in mouse and monkey lung samples than in the liver samples. Interestingly, the identifiable presence of some oxidations (1–6) of the PS backbone were identified in mouse lung samples (Figure S4). Therefore, different precursor ion series were used to identify metabolites based on the number of oxidations of the PS backbone as shown in Table 2. As before, different charge states were selected as precursor ions to avoid overlapped m/z series for each analyte. 3′ end shortmers (from n-1 to n-7) and 5′ end shortmers (from n-1 to n-5) were observed in mouse lung samples, and 3′ end shortmers (from n-1 to n-8) and 5′ end shortmers (from n-1 to n-7) were observed in the monkey lung samples (Table 1). 3′ end shortmers generated in mouse and monkey lung samples ranged from 4.20% to 12.09% and from 2.78% to 6.72%, respectively. 5′ end shortmers generated in mouse and monkey lung samples ranged from 6.94% to 19.88% and from 2.25% to 7.62%, respectively. Like the result of liver samples, these results suggest that both 3′ exonuclease and 5′ exonuclease were essential for metabolizing eluforsen in mouse and monkey lung. Figures 5E and 5F for mouse samples, and Figures 5G and 5H for monkey samples suggest similar tendencies of metabolite generation between samples. While both 3′ and 5′ shortmers were observed, conversion of a 2′-O-Me to a hydroxyl group and simple loss of any of the bases (depurination or depyrimidation) were not identified in the in vivo samples. Consistent with the results from the liver samples, an increased prevalence of 5′ n-3 and 5′ n-5 shortmers was also observed in lung samples.
Discussion
Ion-pair reversed-phase LC-MS (IP-RP-LC-MS) has been routinely used in the development of oligonucleotide-based therapeutics. To improve mobile phase systems with good chromatographic separation and enhanced MS signal intensity for the metabolite identification of eluforsen, various alkylamines and fluorinated modifiers have been investigated in this study, as previously reported.21, 22, 39 Ultimately, 15 mM DMCHA containing 25 mM HFIP was chosen as our buffer after comprehensive investigation to assess the effect of alkylamines and fluorinated alcohols on both chromatographic separation and MS signal intensity.
Employing effective sample preparation methods that combine one or two techniques between proteinase K digestion, LLE, SPE, and magnetic bead approaches, have been successfully used for the isolation of oligonucleotides prior to LC-MS analysis.5, 18, 27, 30 Proteinase K digestion has been widely used to remove proteins during the oligonucleotide analysis of biological samples using LC-MS,5, 27 and this approach was associated with more than 90% recovery.27 Due to difficulties associated with the loss of affinity of a variety of metabolites from a capture strand, the magnetic-bead approach was not applied in the metabolite identification process despite its high specificity and good recovery (80%–95%).30 To maximize coverage of metabolites of eluforsen, SPE, with a recovery range of 60%–80%,5 was therefore used for relatively nonspecific extraction of metabolites generated in biological samples after proteinase K digestion. Using this approach, we found more metabolites than using a magnetic-bead approach.
Owing to the mass spectral complexity of larger macromolecules, analysis of therapeutic oligonucleotides faces many technical challenges.7 While small-molecule analysis often has a single m/z species for identification that can be chosen, larger macromolecules provide complex primary mass spectra. Specifically, the monoisotopic mass of eluforsen is 11,461.18, which is approximately 1.5 times higher than other therapeutic oligonucleotides (e.g., Spinraza, 7,501 Da;17 Kynamro, 7,594.9 Da40), resulting in the production of higher charge states than that of smaller oligonucleotides. In terms of technical aspects, mass spectral complexity of the 33-mer eluforsen was shown to have overlapping signals at the same m/z values from isotopic distributions as well as metabolites commonly generated by sequential removal of nucleotides from the full-length oligo. Hence, careful selection of charge states and m/z values were necessary to unambiguously identify eluforsen and its metabolites.
Examination of the metabolic profiles of eluforsen indicated that it was not degraded by endo- and exonucleases after 48 hr (Figure 2) with these nucleases in vitro. PS modification that substitutes a sulfur atom for a nonbridging oxygen in the phosphodiester backbone of oligonucleotides is known to provide nuclease resistance.35, 41, 42, 43, 44, 45, 46, 47 Hence, the PS molecule was stable against in vitro nucleolytic degradation. It has been reported that the PS groups of both the Rp- and Sp-diastereomers were resistant against the staphylococcal nucleases, DNase I and II,48 and Iwamoto et al.49 have shown that Sp linkages increase the metabolic stability relative to Rp linkages in rat liver homogenates. 2′-ribose modification also results in resistance to nuclease-mediated degradation.8, 9, 10 Therefore, a fully 2′-O-methyl ribose modification provides increased stability of eluforsen against degradation by endonucleases, whereas MOE gapmers were found to be initially metabolized by endonucleases and then exonucleases to produce chain-shortened metabolites.15, 16, 36
Eluforsen was metabolized by cellular nucleases in mouse liver homogenates, which was consistent with the metabolism of other PS oligonucleotides in rat liver homogenates.47 The different results obtained between purified enzymes and liver homogenate might be due to the presence of other molecules and enzymes in liver homogenate that may enhance the interaction between the oligonucleotides and endo- and exonucleases, but this needs further investigation. The result that only 3′ end shortmers were observed suggests that the metabolism of eluforsen occurred mainly through 3′ exonuclease activity in mouse liver homogenates. Specifically, the fact that pyrimidine-rich oligonucleotides can be metabolized faster47 may explain why the 3′-poly(U) tail was observed as the most metabolically active portion in mouse liver homogenates.
It was demonstrated that both 3′ exonuclease and 5′ exonuclease were responsible for metabolism of eluforsen in in vivo liver and lung tissues from mice and monkeys (Figure 5). In the in vivo data, increased concentrations of 5′ n-3 and 5′ n-5 shortmers appeared to result from increased stability from the presence of adenine. The result can be explained by the fact that pyrimidine nucleotides (U, C) were metabolized faster than purines (A, G).47 It is still unknown whether metabolites can be generated by the simultaneous activities of both nucleases. However, no metabolites were observed resulting from losses of nucleotides from both ends of the molecule. Overall, the typical pattern of chain-shortened metabolites observed in other systems12, 14, 15, 36, 47, 50, 51 was shown in this study. Additionally, some identifiable oxidations of PSs, substitutions of sulfur for oxygen, were detected in in vivo mouse lung samples. It has been well known that nucleolytic degradation is a central metabolic pathway for oligonucleotides.47, 52 Our result shows that eluforsen is metabolized through oxidative metabolic pathways in in vivo mouse lungs. While oligonucleotides are not primarily metabolized via flavin-containing monooxygenases (FMOs) or cytochromes P450-driven metabolic oxidation processes,53, 54, 55 there have been some observations that PSs undergo metabolic oxidation in in vivo samples.55, 56
Comparison of the metabolites generated in mice and monkeys showed the similar tendencies of metabolite generation between the two species. The results are consistent with a previous study of the PS oligonucleotide (ISIS 2503), where similar patterns of metabolites were observed in the plasma of mice and monkeys.57 As shown in Table 1, the sum of mean percentages of 3′ end shortmers in mouse liver, monkey liver, mouse lung, and monkey lung samples were 43.08%, 24.86%, 51.58%, and 32.01% equivalents, respectively. The sum of mean percentages of 5′ end shortmers in mouse liver, monkey liver, mouse lung, and monkey lung samples were 46.16%, 20.35%, 59.88%, and 31.26% equivalents, respectively. Overall, metabolites in mice were approximately 20%–30% higher than those of monkeys. The result of a more active metabolism of eluforsen in mice than in monkeys is consistent with that of a previously reported study.57 Metabolites in lung samples were approximately 10% higher than those in liver samples. Eluforsen and its metabolites showed greater accumulation in the lung relative to liver after inhalation administration. Increased drug concentrations within target organs is a pivotal factor for the development of delivery strategies.58
Since 2′-O-ribose-modified PS oligonucleotides like eluforsen have a predictive metabolism via exonuclease resulting in chain shortening of the sequence from the 3′ and/or 5′ end, the metabolites do not differ qualitatively between animal species and humans. The identified metabolites in tissues of the animal species are therefore considered to reflect metabolites that may be present in humans following inhalation exposure to eluforsen. Since systemic exposure following inhalation administration of eluforsen in humans is negligible (data not shown), metabolites in human plasma will also be negligible. The metabolite profile of eluforsen is sufficiently characterized in lung and liver of animal species, and additional studies in human material are not considered to provide additional relevant information.
The impact of the modifications on the metabolism patterns of oligonucleotides has been reported in several studies. Kynamro, a 2′-O-MOE gapmer, is initially metabolized by endonucleases and then exonucleases to produce chain-shortened metabolites.15, 16, 36 Spinraza, a PS-modified oligonucleotide with 2′-O-MOE ribose modifications throughout the PS backbone, is primarily metabolized by 3′ and 5′ exonucleases.17 In our study, eluforsen was predominantly metabolized by 3′ and 5′ exonuclease-mediated hydrolysis producing chain-shortened metabolites, which is consistent with the results from Spinraza.17
Conclusions
We have developed an LC-MS method for analysis of fully phosphorothioated 2′-O-Me-modified oligonucleotides and its metabolites generated in in vitro and in vivo studies. The metabolism of eluforsen was evaluated in in vitro test systems with nucleases and mouse liver homogenates and in vivo liver and lung samples of mice and monkeys following inhalation administration. We have demonstrated that the RNA-based oligonucleotide with full PS modifications and fully modified ribose with 2′-O-methyl offers increased stability against base hydrolysis by nucleases. Purified enzymes were not able to digest eluforsen. Incubation with liver homogenate lead to generation of n-1 to n-8 from the 3′ end, while no metabolites were formed from the 5′ end. Chain-shortened metabolites from both 3′ and 5′ ends were formed in in vivo liver and lung of both species (mice and monkeys), and the results indicated that similar metabolites were generated in the two species. Ultimately, we believe that these metabolism studies will contribute toward improving the identification of metabolites from other modified therapeutic oligonucleotides and thereby advance nonclinical drug safety evaluations for this important class of therapies.
Materials and Methods
Chemicals and Reagents
Eluforsen, a 33-mer PS oligonucleotides with 2′-O-methyl-modified 2′-ribose sites, was provided by ProQR Therapeutics (Leiden, the Netherlands). Its sequence was 5′-AUC AUA GGA AAC ACC AAA GAU GAU AUU UUC UUU-3′. All alkylamines, including DMCHA and DIEA, as well as 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) and LC-MS grade methanol, acetonitrile, and water were purchased from Sigma Aldrich (St. Louis, MO, USA). 1,1,1,3,3,3-hexafluoro-2-methyl-2-propanol (HFMIP) was supplied from Fisher Scientific (Toronto, ON, Canada). RNase A was acquired from Thermo Fisher Scientific (Waltham, MA, USA), and Exo T, the reaction buffer (10× NE buffer 4), and streptavidin magnetic beads were purchased from New England Biolabs (Ipswich, MA, USA). Magnesium acetate, EDTA, DTT, guanidine hydrochloride, Triton X-100, tetrahydrofuran (THF), ammonium bicarbonate (NH4HCO3), tris (2-carboxyethyl) phosphine (TCEP), and sodium chloride (NaCl) were obtained from Sigma Aldrich (St. Louis, MO, USA) as well. Proteinase K was purchased from Gold Biotechnology (St. Louis, MO, USA). Mouse liver homogenates and mouse lung tissue used as blanks to generate calibration curves were acquired from BioIVT (Westbury, NY, USA).
In Vitro Incubation with Purified Enzymes
For endo (RNase A)- and exonuclease (Exo-T), 1 μg of eluforsen was incubated with 20 units of either endonuclease (20/0), exonuclease (0/20), or endonuclease and exonuclease (20/20) at 37°C for 1, 2, and 7 days. Heat inactivation at 65°C for 20 min was used to stop the reaction after the incubation period. Eluforsen was incubated in buffer only without endo- and exonuclease for 1, 2, and 7 days as a negative control. An RNA strand with sequence 5′-CGUACUAGUGGUCCUAAUCGUAC-3′ was used as a positive control.
In Vitro Incubation with Mouse Liver Homogenates
The homogenization buffer consisted of 100 mM Tris-HCl (pH 8.0), 1 mM magnesium acetate, and 1× penicillin/streptomycin mixture. 5 g of untreated mouse liver (BioIVT, Westbury, NY, USA) was placed in a glass vessel, and 10 mL of the homogenization buffer was added. These were homogenized mechanically with a Polytron (Kinematica, Lucerne, Switzerland). The resulting homogenate was further diluted with the homogenization buffer to make several 400-μL aliquots of liver homogenate. Based on the optimized ratio, 10 μL of liver homogenate was added to 390 μL of buffer solution. 50 μg/mL of eluforsen was added to each tube, and then the samples were incubated for 0, 24, 48, 72, 96, and 120 h for the determination of the rate of formation of shortmers of eluforsen.
In Vivo Metabolism Studies of Eluforsen
The tissue samples were collected from inhalation toxicity studies of eluforsen in mice and in cynomolgus monkeys. The studies complied with the OECD Principles of Good Laboratory Practice, ENV/MC/CHEM (98)17 (issued January 1998), and FDA Title 21 Code of Federal Regulations Part 58, Good Laboratory Practice for Non-clinical Studies, issued 22 December 1978 Federal Register plus subsequent amendments. The studies were conducted in an AAALAC (Association for Assessment and Accreditation of Laboratory Animal Care) facility and all animal studies were conducted using protocols and methods approved by the Institutional Animal Care Committee and carried out in accordance with the Guide to the Care and Use of Experimental Animals adopted and promulgated by the Canadian Council on Animal Care and by the NIH (Bethesda, MD, USA). After 13-week inhalation administration of eluforsen (10 mg/kg), FVB/NCrl mice (n = 10 [5 male, 5 female]) exposed 1 or 3 times/week and cynomolgus monkeys (n = 8 [4 male, 4 female]) exposed 3 times/week, were euthanized upon completion of the treatment period of the study. Upon sacrifice, lung and liver tissue samples were collected, snap frozen in liquid nitrogen, and stored frozen (≤60°C) until shipment and analysis (Table 3).
Table 3.
In Vivo Samples from the Eluforsen 13-Week Inhalation Study in Cynomolgus Monkey (REXM and REXL) and FVB/NCrl Mice (REXQ and REXP)
| Sample Number | Tissue | Gender |
| REXM 16-19 | liver | M |
| REXM 36-39 | liver | F |
| REXL 16-19 | lung | M |
| REXL 36-39 | lung | F |
| REXQ 80-84 | liver | F |
| REXQ 281-285 | liver | M |
| REXP 281-285 | lung | F |
Tissue Homogenization
Liver and lung samples from mice and monkeys were prepared into appropriately sized amounts for processing (10 mg to 300 mg per tube). Beads with the same mass as the tissue were added to the tube. Two volumes of buffer were added to each sample prior to homogenization using a Bullet blender (Next Advance, Averill Park, NY, USA). After centrifugation, homogenates were separated from the debris and beads. The tissue homogenates were stored at −80°C ± 10°C.
Proteinase K Digestion
The proteinase K digestion buffer (60 mM Tris, 100 mM EDTA, 400 mM guanidine hydrochloride, and 0.1% Triton X-100 at pH 9) was stored at 4°C. 20 mM DTT was added to aliquots of the buffer before use. 200 mg/mL of proteinase K solution was prepared in water and was stored at −20°C. 20 μL of digestion buffer and 15 μL of proteinase K for in vivo samples were added into 100 μL of each homogenate sample. All samples were vortexed for at least 1 min, then heated at 55°C while shaking (250 g) for 3 h. Samples were then processed with either SPE or using streptavidin magnetic beads.
SPE
SPE was performed using Phenomenex Clarity OTX cartridges (Phenomenex, Torrance, CA, USA) after the proteinase K digestion. As previously described,5 the equilibration buffer (50 mM NH4OAc) was prepared in water and the pH adjusted with acetic acid to 5.5. The wash buffer was made by mixing the equilibration buffer with acetonitrile in a 1:1 ratio. The elution buffer was made by mixing 50 mL of solution (100 mM NH4HCO3 and 1 mM TCEP in water at pH 9.5) with 40 mL of acetonitrile and 10 mL of THF. The SPE cartridges were conditioned using 1 mL of methanol and then 1 mL of equilibration buffer. Tissue samples were then mixed with 900 μL of Clarity OTX buffer and loaded onto the column. The cartridges were washed using 4 mL of washing buffer, and the analytes were eluted with 0.5 mL × 2 of elution buffer. The collected solutions were evaporated to near dryness under vacuum and reconstituted with deionized water.
Magnetic Bead Approaches
A proteinase K digestion step was added at the beginning of the extraction protocol. Streptavidin magnetic beads with biotinylated capture strand (5′-biotin-AAAGAAAATATCATCT-3′) were used to selectively capture eluforsen and its metabolites. Elution buffer containing 10 mM Tris-HCl (pH 7.5) and 1 mM EDTA was prewarmed in a 65°C bath and a low-salt buffer consisting of 0.15 M NaCl, 20 mM Tris-HCl (pH 7.5), and 1 mM EDTA was chilled on ice. 50 μg of biotinylated capture strand was dissolved in 500 μL of wash/binding buffer containing 0.5 M NaCl, 20 mM Tris-HCl (pH 7.5), and 1 mM EDTA. 200 μL (1,500 μg) of hydrophilic streptavidin-coated magnetic beads were added into a new RNase-free microcentrifuge tube. 200 μL of wash/binding buffer was added to the beads, and they were vortexed to create a suspension. The magnet was applied for approximately 30 s, and the supernatant was discarded. 50 μL of biotinylated capture solution was added to the magnetic beads, and the mixture was vortexed. This sample was incubated at room temperature for 10 min with occasional agitation. The magnet was then applied, and the supernatant was discarded. The beads were washed by adding 200 μL of wash/binding buffer and then vortexing. The magnet was applied, and the supernatant was discarded. The wash step was repeated two more times. 150 μL of 2× wash/binding buffer consisting of 1 M NaCl, 40 mM Tris-HCl (pH 7.5), and 2 mM EDTA was added to the samples. They were heated at 65°C for 5 min and then quickly chilled on ice for 3 min. The samples from the previous step were added into the previously prepared biotinylated capture strand that was bound to streptavidin-coated magnetic beads, vortexed to suspend the particles, then incubated at room temperature for 20 min with occasional agitation. The magnet was applied, and the supernatant was removed. 200 μL of wash/binding buffer was added, and the tubes were vortexed to suspend the beads. The magnet was applied, and the supernatant was discarded. Washing with fresh wash buffer was repeated two more times. 200 μL of cold low-salt buffer was added to the beads, and the tubes were vortexed to suspend them. The magnet was applied, and the supernatant was discarded. 20 μL of prewarmed elution buffer was added followed by vortexing to suspend the beads and incubated at 65°C in a bath for 2 min. The magnet was applied, and the supernatant was transferred to a new auto-sampler vial. Elution was repeated with 20 μL of fresh elution buffer. The magnet was applied, and the supernatant was added to the first oligonucleotide elution.
LC-MS/MS Conditions
The samples were analyzed using a Waters Acquity UPLC connected to a Waters Synapt G2 Q-TOF mass spectrometer (Waters, Milford, MA, USA). The LC separation was performed on a Phenomenex Clarity 2.6-μm Oligo-XT 100 Å, 2.1 × 100-mm column (Phenomenex, Torrance, CA, USA) at 60°C. The flow rate and injection volume were 0.5 mL/min and 15 μL, respectively. Mobile phase A consisted of 15 mM DMCHA and 25 mM HFIP in 5% methanol, and mobile phase B was 95% methanol. The gradient conditions were as follows: (time [in min], % mobile phase B), (0.00, 0), (5.00, 0), (25.00, 35), (25.01, 100), (30.00, 100), (30.01, 0), (35.00, 0). The capillary voltage was −2.0 kV, the cone voltage was 25 V, and the extraction cone voltage was 2 V. The source temperature was 125°C, and the desolvation temperature was 450°C. The flow rate of the desolvation gas (nitrogen) was 1,000 L/h. The MS data acquisition was performed using the MSE sensitivity mode with a 1 s scan time. The low collision energy was set to zero, and high collision energy ramping was from 20 to 30 eV. Additionally, the UPLC flow-through was diverted from the mass spectrometer during the initial 1 min and the final 10 min.
Selections of Charge States and m/z of Eluforsen and Its Potential Metabolites
To optimize the metabolite identification, different charge states were selected as precursor ions seen in the first mass analyzer to avoid overlapped m/z series for each analyte. For this reason, several calibration curves with different charge states corresponding to each analyte were needed to estimate concentrations. Table 2 shows m/z values for eluforsen and its potential metabolites used for identification.
Author Contributions
Conceptualization, J.K., B.B., M.G.B., C.H.; Methodology, J.K., B.B., M.G.B., E.v.d.H., C.H.; Validation, E.v.d.H.; Investigation, J.K., B.B., C.P., C.d.B.; Resources, E.v.d.H., C.P., C.d.B.; Writing – Original Draft, J.K., M.G.B.; Writing – Review & Editing, B.B., M.G.B., C.H., C.P., C.d.B.; Visualization, J.K.; Supervision, M.G.B.; Project Administration, C.H.; Funding Acquisition, M.G.B., E.v.d.H.
Acknowledgments
This work was supported by ProQR Therapeutics.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.07.006.
Supplemental Information
References
- 1.US Food Drug Administration (2008). Guidance for Industry–Safety Testing of Drug Metabolites. February 2008. https://www.fda.gov/media/72279/download.
- 2.US Food and Drug Administration (2013). List of orphan designations and approvals. https://www.accessdata.fda.gov/scripts/opdlisting/oopd/listResult.cfm.
- 3.European Medicines Agency (2013). European public assessment reports: Orphan medicines. https://www.ema.europa.eu/en/medicines/field_ema_web_categories%253Aname_field/Human/ema_group_types/ema_orphan.
- 4.Van Goor F., Hadida S., Grootenhuis P.D.J., Burton B., Stack J.H., Straley K.S., Decker C.J., Miller M., McCartney J., Olson E.R. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc. Natl. Acad. Sci. USA. 2011;108:18843–18848. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen B., Bartlett M. A one-step solid phase extraction method for bioanalysis of a phosphorothioate oligonucleotide and its 3′ n-1 metabolite from rat plasma by uHPLC-MS/MS. AAPS J. 2012;14:772–780. doi: 10.1208/s12248-012-9394-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patil S.D., Rhodes D.G., Burgess D.J. DNA-based therapeutics and DNA delivery systems: a comprehensive review. AAPS J. 2005;7:E61–E77. doi: 10.1208/aapsj070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGinnis A.C., Chen B., Bartlett M.G. Chromatographic methods for the determination of therapeutic oligonucleotides. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2012;883-884:76–94. doi: 10.1016/j.jchromb.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Pallan P.S., Allerson C.R., Berdeja A., Seth P.P., Swayze E.E., Prakash T.P., Egli M. Structure and nuclease resistance of 2′,4′-constrained 2′-O-methoxyethyl (cMOE) and 2′-O-ethyl (cEt) modified DNAs. Chem. Commun. (Camb.) 2012;48:8195–8197. doi: 10.1039/c2cc32286b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seth P.P., Siwkowski A., Allerson C.R., Vasquez G., Lee S., Prakash T.P., Wancewicz E.V., Witchell D., Swayze E.E. Short antisense oligonucleotides with novel 2′-4′ conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals. J. Med. Chem. 2009;52:10–13. doi: 10.1021/jm801294h. [DOI] [PubMed] [Google Scholar]
- 10.Seth P.P., Vasquez G., Allerson C.A., Berdeja A., Gaus H., Kinberger G.A., Prakash T.P., Migawa M.T., Bhat B., Swayze E.E. Synthesis and biophysical evaluation of 2′,4′-constrained 2‘O-methoxyethyl and 2’,4′-constrained 2'O-ethyl nucleic acid analogues. J. Org. Chem. 2010;75:1569–1581. doi: 10.1021/jo902560f. [DOI] [PubMed] [Google Scholar]
- 11.Geary R.S., Norris D., Yu R., Bennett C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Geary R.S. Antisense oligonucleotide pharmacokinetics and metabolism. Expert Opin. Drug Metab. Toxicol. 2009;5:381–391. doi: 10.1517/17425250902877680. [DOI] [PubMed] [Google Scholar]
- 13.Schildkraut I. Nuclease. In: Brenner S., Miller J.H., editors. Encyclopedia of Genetics. Academic Press; 2001. pp. 1357–1358. [Google Scholar]
- 14.Geary R.S., Baker B.F., Crooke S.T. Clinical and preclinical pharmacokinetics and pharmacodynamics of mipomersen (kynamro(®)): a second-generation antisense oligonucleotide inhibitor of apolipoprotein B. Clin. Pharmacokinet. 2015;54:133–146. doi: 10.1007/s40262-014-0224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu R.Z., Kim T.-W., Hong A., Watanabe T.A., Gaus H.J., Geary R.S. Cross-species pharmacokinetic comparison from mouse to man of a second generation antisense oligonucleotide, ISIS 301012, targeting human apolipoprotein B-100. Drug Metab. Dispos. 2006;35:460–468. doi: 10.1124/dmd.106.012401. [DOI] [PubMed] [Google Scholar]
- 16.Crooke S.T., Geary R.S. Clinical pharmacological properties of mipomersen (Kynamro), a second generation antisense inhibitor of apolipoprotein B. Br. J. Clin. Pharmacol. 2013;76:269–276. doi: 10.1111/j.1365-2125.2012.04469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.US Food and Drug Administration (2016). FDA Product label. SPINRAZA (nusinersen) injection, for intrathecal use. December 2016. https://www.accessdata.fda.gov/drugsatfda_docs/label/2016/209531lbl.pdf.
- 18.Zhang G., Lin J., Srinivasan K., Kavetskaia O., Duncan J.N. Strategies for bioanalysis of an oligonucleotide class macromolecule from rat plasma using liquid chromatography-tandem mass spectrometry. Anal. Chem. 2007;79:3416–3424. doi: 10.1021/ac0618674. [DOI] [PubMed] [Google Scholar]
- 19.Basiri B., Bartlett M.G. LC-MS of oligonucleotides: applications in biomedical research. Bioanalysis. 2014;6:1525–1542. doi: 10.4155/bio.14.94. [DOI] [PubMed] [Google Scholar]
- 20.Lin Z.J., Li W., Dai G. Application of LC-MS for quantitative analysis and metabolite identification of therapeutic oligonucleotides. J. Pharm. Biomed. Anal. 2007;44:330–341. doi: 10.1016/j.jpba.2007.01.042. [DOI] [PubMed] [Google Scholar]
- 21.Chen B., Bartlett M.G. Evaluation of mobile phase composition for enhancing sensitivity of targeted quantification of oligonucleotides using ultra-high performance liquid chromatography and mass spectrometry: application to phosphorothioate deoxyribonucleic acid. J. Chromatogr. A. 2013;1288:73–81. doi: 10.1016/j.chroma.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Basiri B., van Hattum H., van Dongen W.D., Murph M.M., Bartlett M.G. The role of fluorinated alcohols as mobile phase modifiers for LC-MS analysis of oligonucleotides. J. Am. Soc. Mass Spectrom. 2017;28:190–199. doi: 10.1007/s13361-016-1500-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Annesley T.M. Ion suppression in mass spectrometry. Clin. Chem. 2003;49:1041–1044. doi: 10.1373/49.7.1041. [DOI] [PubMed] [Google Scholar]
- 24.Panuwet P., Hunter R.E., Jr., D’Souza P.E., Chen X., Radford S.A., Cohen J.R., Marder M.E., Kartavenka K., Ryan P.B., Barr D.B. Biological Matrix Effects in Quantitative Tandem Mass Spectrometry-Based Analytical Methods: Advancing Biomonitoring. Crit. Rev. Anal. Chem. 2016;46:93–105. doi: 10.1080/10408347.2014.980775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng P., Chen X., Zhang G., Zhong D. Bioanalysis of an oligonucleotide and its metabolites by liquid chromatography-tandem mass spectrometry. J. Pharm. Biomed. Anal. 2010;52:571–579. doi: 10.1016/j.jpba.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 26.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 27.McGinnis A.C., Cummings B.S., Bartlett M.G. Ion exchange liquid chromatography method for the direct determination of small ribonucleic acids. Anal. Chim. Acta. 2013;799:57–67. doi: 10.1016/j.aca.2013.08.040. [DOI] [PubMed] [Google Scholar]
- 28.Johnson J.L., Guo W., Zang J., Khan S., Bardin S., Ahmad A., Duggan J.X., Ahmad I. Quantification of raf antisense oligonucleotide (rafAON) in biological matrices by LC-MS/MS to support pharmacokinetics of a liposome-entrapped rafAON formulation. Biomed. Chromatogr. 2005;19:272–278. doi: 10.1002/bmc.450. [DOI] [PubMed] [Google Scholar]
- 29.Dai G., Wei X., Liu Z., Liu S., Marcucci G., Chan K.K. Characterization and quantification of Bcl-2 antisense G3139 and metabolites in plasma and urine by ion-pair reversed phase HPLC coupled with electrospray ion-trap mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2005;825:201–213. doi: 10.1016/j.jchromb.2005.05.049. [DOI] [PubMed] [Google Scholar]
- 30.Ye G., Beverly M. The use of strong anion-exchange (SAX) magnetic particles for the extraction of therapeutic siRNA and their analysis by liquid chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 2011;25:3207–3215. doi: 10.1002/rcm.5221. [DOI] [PubMed] [Google Scholar]
- 31.Jeanpierre M. A rapid method for the purification of DNA from blood. Nucleic Acids Res. 1987;15:9611. doi: 10.1093/nar/15.22.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dattagupta J.K., Fujiwara T., Grishin E.V., Lindner K., Manor P.C., Pieniazek N.J., Saenger R., Suck D. Crystallization of the fungal enzyme proteinase K and amino acid composition. J. Mol. Biol. 1975;97:267–271. doi: 10.1016/s0022-2836(75)80039-8. [DOI] [PubMed] [Google Scholar]
- 33.Hilz H., Wiegers U., Adamietz P. Stimulation of proteinase K action by denaturing agents: application to the isolation of nucleic acids and the degradation of ‘masked’ proteins. Eur. J. Biochem. 1975;56:103–108. doi: 10.1111/j.1432-1033.1975.tb02211.x. [DOI] [PubMed] [Google Scholar]
- 34.Stein C.A., Subasinghe C., Shinozuka K., Cohen J.S. Physicochemical properties of phosphorothioate oligodeoxynucleotides. Nucleic Acids Res. 1988;16:3209–3221. doi: 10.1093/nar/16.8.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoke G.D., Draper K., Freier S.M., Gonzalez C., Driver V.B., Zounes M.C., Ecker D.J. Effects of phosphorothioate capping on antisense oligonucleotide stability, hybridization and antiviral efficacy versus herpes simplex virus infection. Nucleic Acids Res. 1991;19:5743–5748. doi: 10.1093/nar/19.20.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baek M.-S., Yu R.Z., Gaus H., Grundy J.S., Geary R.S. In vitro metabolic stabilities and metabolism of 2′-O-(methoxyethyl) partially modified phosphorothioate antisense oligonucleotides in preincubated rat or human whole liver homogenates. Oligonucleotides. 2010;20:309–316. doi: 10.1089/oli.2010.0252. [DOI] [PubMed] [Google Scholar]
- 37.Husser C., Brink A., Zell M., Müller M.B., Koller E., Schadt S. Identification of GalNAc-Conjugated Antisense Oligonucleotide Metabolites Using an Untargeted and Generic Approach Based on High Resolution Mass Spectrometry. Anal. Chem. 2017;89:6821–6826. doi: 10.1021/acs.analchem.7b01244. [DOI] [PubMed] [Google Scholar]
- 38.Studzińska S., Rola R., Buszewski B. Development of a method based on ultra high performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry for studying the in vitro metabolism of phosphorothioate oligonucleotides. Anal. Bioanal. Chem. 2016;408:1585–1595. doi: 10.1007/s00216-015-9266-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGinnis A.C., Grubb E.C., Bartlett M.G. Systematic optimization of ion-pairing agents and hexafluoroisopropanol for enhanced electrospray ionization mass spectrometry of oligonucleotides. Rapid Commun. Mass Spectrom. 2013;27:2655–2664. doi: 10.1002/rcm.6733. [DOI] [PubMed] [Google Scholar]
- 40.Kastle Therapeutics LLC (2016). KYNAMRO-mipomersen sodium injection, solution, https://rxdruglabels.com/lib/rx/rx-meds/kynamro-2/.
- 41.Campbell J.M., Bacon T.A., Wickstrom E. Oligodeoxynucleoside phosphorothioate stability in subcellular extracts, culture media, sera and cerebrospinal fluid. J. Biochem. Biophys. Methods. 1990;20:259–267. doi: 10.1016/0165-022x(90)90084-p. [DOI] [PubMed] [Google Scholar]
- 42.Crooke R. Cellular uptake, distribution and metabolism of phosphorothioate, phosphorodiester and methylphosphonate oligonucleotides. In: Crooke S.T., Lebleu B., editors. Antisense Research and Applications. CRC Press; 1993. p. 427. [Google Scholar]
- 43.Crooke R.M., Graham M.J., Cooke M.E., Crooke S.T. In vitro pharmacokinetics of phosphorothioate antisense oligonucleotides. J. Pharmacol. Exp. Ther. 1995;275:462–473. [PubMed] [Google Scholar]
- 44.Crooke S.T., Lebleu B. CRC Press; 1993. Antisense Research and Applications. [Google Scholar]
- 45.Gao W.Y., Han F.S., Storm C., Egan W., Cheng Y.-C. Phosphorothioate oligonucleotides are inhibitors of human DNA polymerases and RNase H: implications for antisense technology. Mol. Pharmacol. 1992;41:223–229. [PubMed] [Google Scholar]
- 46.Brown D.A., Kang S.-H., Gryaznov S.M., DeDionisio L., Heidenreich O., Sullivan S., Xu X., Nerenberg M.I. Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. J. Biol. Chem. 1994;269:26801–26805. [PubMed] [Google Scholar]
- 47.Crooke R.M., Graham M.J., Martin M.J., Lemonidis K.M., Wyrzykiewiecz T., Cummins L.L. Metabolism of antisense oligonucleotides in rat liver homogenates. J. Pharmacol. Exp. Ther. 2000;292:140–149. [PubMed] [Google Scholar]
- 48.Spitzer S., Eckstein F. Inhibition of deoxyribonucleases by phosphorothioate groups in oligodeoxyribonucleotides. Nucleic Acids Res. 1988;16:11691–11704. doi: 10.1093/nar/16.24.11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iwamoto N., Butler D.C.D., Svrzikapa N., Mohapatra S., Zlatev I., Sah D.W.Y., Meena, Standley S.M., Lu G., Apponi L.H. Control of phosphorothioate stereochemistry substantially increases the efficacy of antisense oligonucleotides. Nat. Biotechnol. 2017;35:845–851. doi: 10.1038/nbt.3948. [DOI] [PubMed] [Google Scholar]
- 50.Crooke S.T. Therapeutic applications of oligonucleotides. Annu. Rev. Pharmacol. Toxicol. 1992;32:329–376. doi: 10.1146/annurev.pa.32.040192.001553. [DOI] [PubMed] [Google Scholar]
- 51.Crooke S.T., Bennett C.F. Progress in antisense oligonucleotide therapeutics. Annu. Rev. Pharmacol. Toxicol. 1996;36:107–129. doi: 10.1146/annurev.pa.36.040196.000543. [DOI] [PubMed] [Google Scholar]
- 52.Crooke R. vitro cellular uptake, distribution, and metabolism of oligonucleotides. In: Crooke S.T., editor. Antisense Research and Application. Springer; 1998. pp. 103–140. [Google Scholar]
- 53.Geary R.S., Leeds J.M., Henry S.P., Monteith D.K., Levin A.A. Antisense oligonucleotide inhibitors for the treatment of cancer: 1. Pharmacokinetic properties of phosphorothioate oligodeoxynucleotides. Anticancer Drug Des. 1997;12:383–393. [PubMed] [Google Scholar]
- 54.Crooke S.T. Basic principles of antisense therapeutics. In: Crooke S.T., editor. Antisense Research and Application. Springer; 1998. pp. 1–50. [Google Scholar]
- 55.Nicklin P., Craig S., Phillips J. Pharmacokinetic properties of phosphorothioates in animals—absorption, distribution, metabolism and elimination. In: Crooke S.T., editor. Antisense Research and Application. Springer; 1998. pp. 141–168. [Google Scholar]
- 56.Cohen A.S., Bourque A.J., Wang B.H., Smisek D.L., Belenky A. A nonradioisotope approach to study the in vivo metabolism of phosphorothioate oligonucleotides. Antisense Nucleic Acid Drug Dev. 1997;7:13–22. doi: 10.1089/oli.1.1997.7.13. [DOI] [PubMed] [Google Scholar]
- 57.Yu R.Z., Geary R.S., Leeds J.M., Watanabe T., Moore M., Fitchett J., Matson J., Burckin T., Templin M.V., Levin A.A. Comparison of pharmacokinetics and tissue disposition of an antisense phosphorothioate oligonucleotide targeting human Ha-ras mRNA in mouse and monkey. J. Pharm. Sci. 2001;90:182–193. doi: 10.1002/1520-6017(200102)90:2<182::aid-jps9>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 58.Rani K., Paliwal S. A review on targeted drug delivery: Its entire focus on advanced therapeutics and diagnostics. Sch. J. App. Med. Sci. 2014;2:328–331. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.