Abstract
Very little is known about long non-coding RNAs (lncRNAs) in the mammalian olfactory sensory epithelia. Deciphering the non-coding transcriptome in olfaction is relevant because these RNAs have been shown to play a role in chromatin modification and nuclear architecture reorganization, processes that accompany olfactory differentiation and olfactory receptor gene choice, one of the most poorly understood gene regulatory processes in mammals. In this study, we used a combination of in silico and ex vivo approaches to uncover a comprehensive catalogue of olfactory lncRNAs and to investigate their expression in the mouse olfactory organs. Initially, we used a novel machine-learning lncRNA classifier to discover hundreds of annotated and unannotated lncRNAs, some of which were predicted to be preferentially expressed in the main olfactory epithelium and the vomeronasal organ, the most important olfactory structures in the mouse. Moreover, we used whole-tissue and single-cell RNA sequencing data to discover lncRNAs expressed in mature sensory neurons of the main epithelium. Candidate lncRNAs were further validated by in situ hybridization and RT-PCR, leading to the identification of lncRNAs found throughout the olfactory epithelia, as well as others exquisitely expressed in subsets of mature olfactory neurons or progenitor cells.
Keywords: long non-coding RNAs, machine learning, olfaction, transcriptome
1. Introduction
In the last decades, several types of long non-coding RNAs (lncRNAs)—transcripts longer than 200 nt characterized by the absence of long open reading frames—have been shown to regulate a variety of biological processes,1 but the function of most of them remains enigmatic. Most lncRNAs are transcribed by RNA polymerase II2 and exhibit prominent tissue-specific expression.3 Even though they share some defining features, their modes of action involve a plethora of distinct molecular mechanisms, including modulation of nuclear architecture, chromatin modification, DNA methylation, transcription regulation, post-transcriptional processing, translation regulation, RNA stability control, biogenesis of miRNAs, and control of protein activity.4–6 An example of mammalian lncRNA with known molecular mechanism is Xist, a long non-coding transcript that recruits a series of molecular players to initiate large-scale silencing of one X chromosome in female cells.7 Most described lncRNAs recruit histone complex modifiers, such as polycomb-repressive complex 2, to initiate remodelling of various portions of the genome.8 Other lncRNAs seem to act by modulating DNA methylation, such as a recently described lncRNA that, once transcribed, controls DNA methylation at a nearby promoter in the protocadherin-α gene, leading to increased transcription of the corresponding protein-coding RNA through stochastic alternative promoter usage in sensory neurons of the mouse olfactory system.9
Olfaction is probably the most important sense for terrestrial animals,10 being crucial for the recognition of a range of environmental stimuli and for appropriate behavioural, physiological, and endocrine responses related to finding food, avoiding danger, and interacting with individuals of the same species. The two most important olfactory sensory organs in mammals are the main olfactory epithelium (MOE) and the vomeronasal organ (VNO), located in the nasal cavity. The MOE is related to the general perception of odours,10 while the VNO is typically associated with instinctive behaviours triggered by intra- and inter-specific communication cues, such as sex, aggression, territoriality, parental care, and defensive responses towards predators.11
During olfactory sensory neuron differentiation, several molecular mechanisms involving chromatin remodelling take place to shape the detection properties of these cells.12–15 For example, each MOE neuron singularly expresses only one type of odorant receptor gene allele chosen among 2,000 possible loci in the genome, a process shown to involve specific epigenetic changes in the genome.13,15–20 Given the role of lncRNAs in chromatin remodelling, we hypothesize they may play a role in controlling how olfactory neurons attain their unique molecular characteristics.
In this article, we used a combination of transcriptome analyses and a machine-learning model to provide for the first time a comprehensive list of lncRNAs in olfaction, including annotated as well as a large set of novel non-coding transcripts. We also identified lncRNAs preferentially expressed in the mouse olfactory organs and determined the temporal patterns of olfactory lncRNA expression during organismic development and sensory neuron differentiation. As an example of how the list of lncRNAs could be useful, we uncovered lncRNAs expressed in mature sensory neurons of the MOE as well as an lncRNA expressed in progenitor cells of the VNO.
2. Materials and methods
2.1. Transcriptome assembly
We used data from 5 different studies,21–25 totalling 49 samples, of which 43 are from adult mice (brain, cerebellum, cerebral cortex, heart, kidney, liver, MOE, and VNO) and 6 are from newborn animals (MOE and VNO). Splicing-aware read mapping was performed using STAR (version 2.5.1b)26 with a 2-pass mapping step to gather splice junctions detected during the first mapping step. The transcriptomes were assembled with Cufflinks (version 2.2.1)27 to the GRCm38 primary assembly masked version, downloaded from Ensembl, and Cuffmerge was used to merge together the resulting assemblies, using the ‘ref-gtf’ parameter to include GENCODE’s comprehensive gene annotation (release M9).
2.2. Candidate lncRNA identification in the assembled transcriptome
To identify possible lncRNAs in the transcriptome reconstructed by Cuffmerge, we adopted a two-step pipeline: firstly, we devised a model of classification of non-coding RNAs using a machine-learning approach with XGBoost (version 0.6 of the xgboost Python package),28 and this model was used to identify possible lncRNAs in the transcriptome; next, a genomic overlap filtering step was performed to eliminate non-coding RNAs that do not match the defining criteria for identifying lncRNAs, such as tRNAs, rRNAs, miRNAs, and untranslated region (UTR) fragments. For the latter, we first obtained the sequences of all annotated UTRs in the Ensembl database (release 87) using the biomaRt software (version 2.30).29 Next, the length distributions of the sequences obtained were inspected, and we selected their upper quartile [160 bp (5′-UTR) and 792 bp (3′-UTR)]. A GTF file containing intervals corresponding to the annotated coding regions plus the representative length values for the UTRs (artificial coding transcripts) was generated and we excluded one-exon transcripts that showed any overlap with the artificial coding transcripts. For transcripts with multiple exons, we excluded only transcripts whose overlap with the artificial coding transcripts comprised >25% of its length. More details on the machine learning and lncRNA identification via the bioinformatics pipeline can be found in the Supplementary Methods section and in the Supplementary Computational Notebook.
2.3. Quantification of transcript expression
Kallisto index was generated from the Cuffmerge assembled transcriptome. Transcript abundances were estimated using the ‘bias’ parameter to correct the quantification for sequence bias,30 and the parameter ‘bootstrap-samples’ was used to generate 100 bootstrap samples during the expectation-maximization step. For samples with unpaired reads (single-end), the parameters ‘fragment-length’ and ‘sd’ were set to 200 and 80 bp, respectively.31 Between-sample abundance normalization was applied to the raw abundance data (TPM values) using library size factors as computed by sleuth (version 0.30.0).32 Gene-level abundance was obtained summing up the normalized abundances of all isoforms. Several subsequent steps in our work used transcript level expression values because of the possibility of different isoforms of the same gene exhibiting different expression patterns among tissues and distinct molecular functions.
2.4. Identification of transcripts preferentially or differentially expressed in the olfactory organs
To quantify a transcript’s specificity of expression in a given tissue, we used the tspex Python package to calculate the SPM (specificity measure) metric (see Supplementary Methods for details), using log2(abundance + 1) values at both transcript and gene levels. Differential expression tests were performed to identify differentially expressed transcripts between two biological conditions (male vs. female and adult vs. newborn). For this, kallisto expression quantification incorporating bootstrap data was analysed with sleuth (version 0.30.0),32 using models with covariates indicating the tissue (MOE or VNO), sex (male or female), and age (adult or newborn). Likelihood ratio tests were performed at the transcript level and differentially expressed transcripts were selected at a 5% false-discovery rate threshold.
2.5. Analysis of MOE single-cell RNA-Seq libraries
The analysis of MOE scRNA-Seq data was performed with Monocle (version 2.10.0).33 Initially, raw abundance data (TPM) of the 93 scRNA-seq samples were converted to absolute abundance (estimated number of RNA molecules per cell) using the built-in Census algorithm (‘relative2abs’ function).34 Using DDRTree, the absolute abundance matrix of transcripts with variable expression was reduced to a two-dimensional space (‘reduceDimension’ function), in which the path of the pseudotime underlying the expression data was laid.35 Then, the algorithm assigned the position of each cell in that path, that is, the pseudotime associated with that sample (‘orderCells’ function). As the reconstruction of the route was done in an unsupervised way, we defined which end of the pseudotime corresponds to the beginning (precursor cells) and the end (mature neurons) of neurogenesis, based on the expression of Ascl1 (precursor cell marker) and Cnga2 (MOE mature sensory neuron marker). Finally, we performed likelihood ratio tests with the ‘differentialGeneTest’ function to identify transcripts that are differentially expressed between OMP-positive and OMP-negative cells. OMP-positive cells were those in which the absolute abundance of the Omp gene was >100. Transcripts were selected at a 5% false-discovery rate threshold and the ones with higher average expression in the OMP-positive cells were chosen for further analysis. Smooth spline curves representing transcript expression dynamics along pseudotime were obtained for the selected transcripts using the ‘genSmoothCurves’ function.
3. Results
3.1. A large set of unannotated lncRNAs identified by a novel computational pipeline involving machine learning
Our objective was to identify lncRNAs preferentially expressed in the olfactory organs, including novel non-coding RNAs. Therefore, we decided not to restrict our analyses to the mouse genome reference annotation. Instead, we used Cufflinks27 to assemble a new transcriptome using RNA sequencing (RNA-Seq) data from a variety of mouse tissues and organs (Supplementary Table S1), including adult whole brain, cerebellum, cerebral cortex, liver, kidney, heart, and the olfactory organs MOE and VNO.24 The stability of this dataset was checked by comparing with an independently assembled transcriptome using another transcript assembler, StringTie,36 leading up to largely overlapping results (Supplementary Fig. S1). The assembled transcriptome contained 226,171 transcripts, distributed in 58,980 loci, of which 15% were not present in the GENCODE reference annotation (release M9).
For this study, we decided to focus on intergenic lncRNAs, the loci of which are not shared with coding genes.37 Since many transcripts were not present in the GENCODE annotation, we used a two-step in silico strategy to classify RNAs into potentially coding or non-coding species and identify intergenic lncRNAs. This pipeline included (i) a novel machine-learning-based model to determine potentially non-coding transcripts, and (ii) a step to exclude transcripts that overlap with coding genes and genes for other kinds of non-coding RNAs.
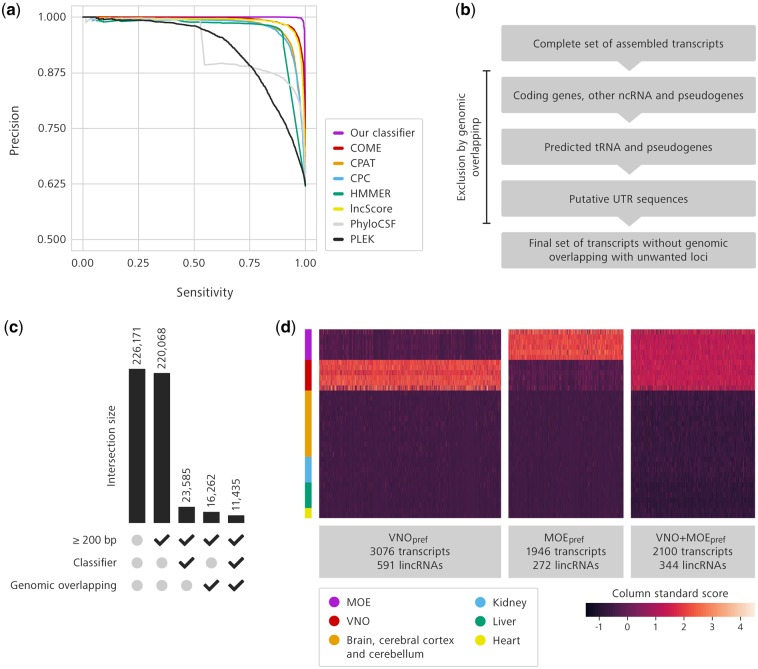
We created a new non-coding RNA classification model, which uses a more efficient machine-learning algorithm and a larger set of informative features than currently available lncRNA predictors.38–42 It consists of an ensemble of 500 decision trees with a total of 21 features to classify transcripts into coding or non-coding (Supplementary Table S2). We selected transcripts annotated as lncRNA or protein-coding in the GENCODE (release M11) and then randomly assigned them to test or training sets, containing 20 and 80% of the total transcripts, respectively, while maintaining the proportion of coding to non-coding RNAs in each set. The model generated with the training set was used to classify lncRNAs in the test dataset (see Supplementary Methods and Supplementary Computational Notebook for details), and the results were compared with those obtained with other lncRNA prediction software (COME, CPAT, CPC, HMMER,43 lncScore, PhyloCSF,44 and PLEK) on the same test dataset (Fig. 1a). Our model exhibits a better trade-off between sensitivity and precision, and better overall performance, as summarized by a range of metrics, including accuracy, precision, sensitivity, specificity, and ROC curves (Fig. 1a, Table 1, and Supplementary Fig. S2a).
Figure 1.
An improved pipeline identifies hundreds of long non-coding RNAs preferentially expressed in the olfactory organs. (a) Precision-recall curve of our lncRNA classifier (purple) in comparison with other currently available classification models. (b) Schematic representation of the sequential removal of transcripts displaying genomic overlap with unwanted genomic loci. (c) Bar plot representing the filtering steps for lncRNA identification. Each bar represents the number of transcripts kept by the combination of sieves represented by the check marks below (selection of long transcripts, selection of transcripts that pass our classifier, and selection of transcripts that do not overlap with unwanted genomic loci). The numbers of filtered transcripts are shown above each bar. (d) Heatmap of expression values for lncRNAs preferentially expressed in the MOE (MOEpref), in the VNO (VNOpref), or in both olfactory organs (VNO+MOEpref). Abundance values were normalized by standardized score calculation, which represents deviation of expression from the average. Columns are transcripts and lines are distinct RNA-Seq libraries from tissues represented by the colour code on the left.
Table 1.
Comparison between performance of lncRNA classifiers
| Classifier | Accuracy | Sensitivity | Specificity | Precision | Area under the precision-recall curve | Area under the ROC curve |
|---|---|---|---|---|---|---|
| Our classifier | 0.9972 | 0.9898 | 0.9989 | 0.9952 | 0.9996 | 0.9999 |
| COME | 0.9285 | 0.9639 | 0.9202 | 0.7391 | 0.9965 | 0.9859 |
| CPAT | 0.8661 | 0.9624 | 0.8436 | 0.5905 | 0.9906 | 0.9620 |
| CPC | 0.7487 | 0.9865 | 0.6930 | 0.4296 | 0.9895 | 0.9585 |
| HMMER | — | — | — | — | 0.9842 | 0.9326 |
| lncScore | 0.9374 | 0.9463 | 0.9353 | 0.7742 | 0.9949 | 0.9808 |
| PhyloCSF | 0.6079 | 0.8675 | 0.5471 | 0.4566 | 0.9689 | 0.8891 |
| PLEK | 0.7120 | 0.9069 | 0.6664 | 0.5446 | 0.9640 | 0.8586 |
For the computation of each performance metric, we considered lncRNAs as the positive class and coding transcripts as the negative class.
Next, we filtered the assembled transcriptome to remove transcripts overlapping with protein-coding genes and pseudogenes, and excluded long non-coding transcripts that represent or share the same genomic locus with other classes of non-coding RNAs, such as rRNA, tRNA, snoRNA, ribozymes, and miRNA (Fig. 1b).
Transcript assembly software typically return fragments of UTRs of mRNAs as independent transcripts,45 which can be misclassified as non-coding RNA. As not all coding transcripts in GENCODE have annotated UTR regions, it is possible that fragments of UTRs may have escaped our first step of filtering. Therefore, an additional step was used to remove candidate lncRNAs lying within 160 and 792 bp from the 5′- and 3′-ends of coding sequences (CDS), respectively. These values were determined from the upper quartile value of UTR lengths calculated from annotated RNAs in GENCODE (Supplementary Fig. S2b).
In total, our classification model identified 23,585 non-coding RNAs that are longer than 200 bp (Fig. 1c), mapping onto 19,448 loci. Of these, 11,435 transcripts in 10,158 loci do not overlap with protein-coding genes, pseudogenes, or other non-coding RNA categories (Fig. 1c). Only 12.8% of all loci (1,301/10,158) have been previously annotated as lncRNAs in GENCODE’s M9 release. Additionally, when we mapped the predicted lncRNA loci onto a reference annotation focussed on non-coding transcripts, NONCODE v5, >45% (4,624/10,158) of our classified lncRNAs fall onto previously mapped non-coding loci.
The resulting comprehensive list of candidate intergenic lncRNAs (Supplementary Spreadsheet), which includes transcripts from both unannotated and annotated loci, was chosen for subsequent investigation.
3.2. A multitude of lncRNAs is preferentially expressed in the olfactory organs
Usually, lncRNAs exhibit unique patterns of expression, characterized by preferential or specific expression in tissues or cell types.3 Because we were interested in identifying lncRNAs potentially involved in the molecular mechanisms of olfaction, we determined the tissue-specificity of expression for the lncRNAs identified in the previous section. We calculated each transcript’s SPM (specificity measure), a parameter that measures the specificity of a gene’s expression in a given tissue or organ.46 As a means of comparison, we calculated the SPM for genes known to be preferentially expressed in the olfactory organs, such as Trpc247 for the VNO, and Cnga248 for the MOE, as well as Omp,49 which is expressed in both olfactory organs (Supplementary Table S3).
We chose to select genes that are either preferentially expressed in the VNO (VNOpref transcripts), or preferentially expressed in the MOE (MOEpref transcripts), or preferentially expressed in both tissues as compared with other organs/tissues (VNO+MOEpref transcripts). Moreover, we selected SPM cut-off values of 0.9, 0.9, and 0.55 for VNOpref, MOEpref, and VNO+MOEpref transcripts, respectively, based on SPM values for the marker genes mentioned earlier.
This approach led us to identify 3,076 VNOpref, 1,946 MOEpref, and 2,100 VNO+MOEpref transcripts, of which 591, 272, and 344, respectively, were classified as lncRNAs (Fig. 1d; Supplementary Spreadsheet). Our improved long intergenic non-coding RNA identification pipeline was specifically devised to identify unannotated non-coding transcripts, and, thus, we expected that few of the lncRNAs listed above would have been annotated in the genome. In fact, only 6.6% (39/591) of the VNOpref, 4.0% (11/272) of the MOEpref, and 7.6% (26/344) of the VNO+MOEpref lncRNAs are annotated in GENCODE’s M9 release (Supplementary Spreadsheet). When the lncRNAs identified by our classifier were compared with the non-coding specific annotation NONCODE (v5), 14.21% (84/591) of the VNOpref, 11.76% (32/272) of the MOEpref, and 30.81% (106/344) of the VNO+MOEpref lncRNAs correspond to annotated loci (Supplementary Spreadsheet). Even though these numbers are higher than those found with GENCODE, they are still much lower than the percentage of total lncRNAs in NONCODE, probably due to the fact that the olfactory organs possess peculiar molecular characteristics, such that most of the lncRNAs we detected were probably not previously identified in other studies.
Among the lncRNAs identified in our study, most of the annotated genes are either not expressed or expressed at low levels in the olfactory organs (Supplementary Fig. S3). This probably reflects the fact that most of the lowly expressed lncRNAs are expressed at higher levels in other tissues and organs, in the context of which they have been described. Importantly, most of the lncRNAs that are highly expressed in the olfactory organs have not been previously annotated (Supplementary Fig. S3) and represent novel non-coding RNA species.
3.3. Spatial expression patterns for olfactory lncRNAs
Next, we analysed the expression of selected candidate lncRNAs by RT-PCR and in situ hybridization to confirm the in silico expression quantification and to gain further relevant information about their patterns of expression in the olfactory organs. We chose to evaluate several lncRNAs in each of the three preferential expression groups (VNOpref, MOEpref, or VNO+MOEpref). For each group, we selected (i) lncRNAs whose expression falls above the third quartile of TPM (high expression group), (ii) lncRNAs whose expression falls in the interquartile range (medium expression group), and (iii) lncRNAs of low expression that falls below the first quartile (Supplementary Fig. S4), totalling seven genes analysed per tissue preferential group.
Of these, the only selected transcripts that map onto annotated lncRNA loci in GENCODE are one VNOpref lncRNA (VNO-D), five VNO+MOEpref lncRNAs (VNO/MOE-A, VNO/MOE-B, VNO/MOE-D, VNO/MOE-F, VNO/MOE-G), and one MOEpref lncRNA (MOE-A), all of which have no known biological function (Supplementary Spreadsheet). Moreover, all selected transcripts were chosen among lncRNAs with multiple exons, since these tend to be bona fide transcripts and not the result of purposeless run-off transcription or transcript misassembly.50
Next, we performed reverse transcription-PCR (RT-PCR) experiments, with the dual purpose of confirming the complete expressed sequences for each candidate lncRNA and the preferential nature of their expression in olfactory organs. We designed specific PCR primers for each lncRNA (Supplementary Table S4), positioning them on exons near the ends of each transcript.
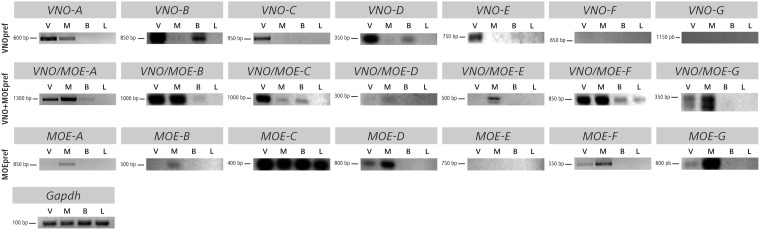
Out of the 16 lncRNA genes in the high and medium expression groups, only one gene, MOE-E, could not be amplified by RT-PCR (Fig. 2). The majority of the remaining genes in these expression level categories was found to be expressed in the olfactory organs according to the in silico quantifications. For example, VNO-A was more highly expressed in the VNO than in the MOE libraries (Table 2), and the RT-PCR data are in agreement with these expression values (Fig. 2). MOE-C, which was quantified in silico to be more highly expressed in the MOE than in the VNO, was amplified by RT-from both olfactory organs and from other tissues (Fig. 2). Moreover, VNO/MOE-E, which exhibits similar abundance levels in the MOE and VNO RNA-Seq libraries, was amplified from MOE tissue. Of the five genes in the low expression category (below the first quartile), two could not be amplified (VNO-F and VNO-G; Fig. 2) and these have the lowest TPM values (<2.4) across all 21 genes analysed (Table 2).
Figure 2.
RT-PCR amplification of selected lncRNAs from olfactory organs, brain, and liver tissues. Agarose electrophoresis gels showing amplified bands in RT-PCR experiments for selected lncRNA candidates in the VNOpref, VNO+MOEpref, and MOEpref preferential expression categories from cDNA prepared from the vomeronasal organ (V), main olfactory epithelium (M), whole brain (B), and liver (L). The first line of images represents investigation of VNOpref lncRNAs, including transcripts in three expression level groups (see Table 2 for details). The middle and bottom lines show transcripts in the VNO+MOEpref and MOEpref categories, respectively. The numbers on the left of each band are the approximate sizes for the amplified bands, which largely overlaps with the in silico transcript sequence assemblies (Supplementary FASTA file). Note that VNO/MOE-G displays two amplified bands from VNO and MOE, but both were purified and used as probes for in situ investigations in Fig. 3. The last image is the endogenous control gene GAPDH, which exhibits unchanged expression levels as judged by band intensity across all samples.
Table 2.
Selected lncRNAs preferentially expressed in one or both the olfactory organs
| Transcript | Strand | GENCODE M9 annotation | Expression group | VNO |
MOE |
||
|---|---|---|---|---|---|---|---|
| Abundance | SPM | Abundance | SPM | ||||
| VNO-A | − | High | 453.23 | 0.97 | 4.87 | 0.25 | |
| VNO-B | + | High | 26.22 | 0.99 | 0.88 | 0.14 | |
| VNO-C | + | High | 7.40 | 1.00 | 0.00 | 0.00 | |
| VNO-D | − | Gm33206 | Medium | 5.00 | 0.99 | 0.00 | 0.00 |
| VNO-E | − | Medium | 4.57 | 1.00 | 0.06 | 0.02 | |
| VNO-F | − | Low | 2.38 | 1.00 | 0.00 | 0.00 | |
| VNO-G | + | Low | 1.71 | 1.00 | 0.00 | 0.00 | |
| MOE-A | n.d. | Gm31557 | High | 3.67 | 0.17 | 17.62 | 0.98 |
| MOE-B | − | High | 1.80 | 0.11 | 11.93 | 0.99 | |
| MOE-C | + | Medium | 0.29 | 0.11 | 7.48 | 0.94 | |
| MOE-D | + | Medium | 0.12 | 0.09 | 6.46 | 1.00 | |
| MOE-E | + | Medium | 0.30 | 0.18 | 6.29 | 0.98 | |
| MOE-F | − | Low | 0.09 | 0.11 | 3.86 | 0.99 | |
| MOE-G | − | Low | 0.00 | 0.00 | 2.56 | 1.00 | |
| VNO/MOE-A | − | Tmem74bos | High | 33.00 | 0.66 | 92.17 | 0.72 |
| VNO/MOE-B | − | BC051077 | High | 14.55 | 0.65 | 35.40 | 0.68 |
| VNO/MOE-C | + | Medium | 8.52 | 0.76 | 9.73 | 0.59 | |
| VNO/MOE-D | − | Gm20675 | Medium | 6.51 | 0.73 | 9.00 | 0.68 |
| VNO/MOE-E | + | Medium | 5.04 | 0.70 | 5.29 | 0.56 | |
| VNO/MOE-F | + | Gm12996 | Medium | 4.70 | 0.70 | 5.72 | 0.59 |
| VNO/MOE-G | + | Platr3 | Low | 2.55 | 0.79 | 3.42 | 0.60 |
‘Expression group’ refers to the assignments based on expression quartiles for each set of preferentially expressed lncRNAs, according to Supplementary Fig. S4. ‘Abundance data’ means transcripts per million (TPM). n.d., unable to determine expression strand.
All amplified bands were sequenced, revealing experimentally determined transcript sequences that match the in silico data (Supplementary FASTA file). Sizes of amplified fragments for these loci were concordant with each amplicon’s expected length, as determined by in silico assembly of the corresponding lncRNA transcripts. Exon-intron boundaries were shown to be mostly correct (Supplementary Fig. S5), except in a few cases where the introns were a few nucleotides longer or shorter. The exceptions are: MOE-D, which, though amplified as one band, revealed two slightly distinct splicing variants after sequencing; VNO/MOE-G, which amplified as two bands; and MOE-F, for which the experimentally determined transcript was slightly shorter than the expected size of the lncRNA (Fig. 2), because two exons predicted in silico were missing in the PCR amplified band (Supplementary FASTA file).
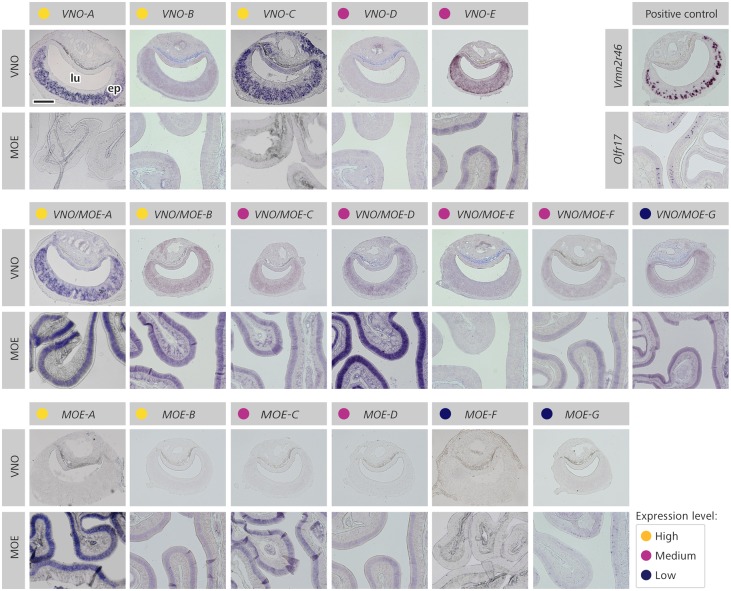
Next, we conducted in situ hybridization for the lncRNA candidates with specific cRNA probes to investigate their spatial patterns of expression in the olfactory organs. For all genes in the high expression category (yellow labels in Fig. 3), we detected clear in situ hybridization signal in the sensory epithelium and verified preferential expression data concordant with the in silico quantification. For example, VNO-A and MOE-A exhibit strong expression in the VNO and MOE neuroepithelia, respectively, while VNO/MOE-A shows expression in both olfactory organs (Fig. 3; higher magnification images are shown in Supplementary Fig. S6). For lncRNAs in the medium expression group (interquartile TPM range), strong or intermediate staining levels were seen (magenta labels in Fig. 3), except for VNO-D in the VNO and for VNO/MOE-E in the MOE.
Figure 3.
Spatial patterns of expression for selected lncRNAs in the olfactory epithelia of MOE and VNO. Representative microscopy images of olfactory tissue sections subjected to chromogenic in situ hybridization staining (purple) with riboprobes for selected preferentially expressed lncRNAs. lncRNA names are as listed on Table 2. The top, middle, and bottom lines of images represent transcripts in the VNOpref, VNO+MOEpref, and MOEpref groups, respectively. At the end of the first line, controls of hybridization are shown for comparison, including expression of an olfactory receptor gene (Olfr17) for the MOE, and staining for vomeronasal organ Vmn2r46 probe for the VNO. Expression level groups ‘high’, ‘medium’, and ‘low’ were determined according to expression quartiles (Supplementary Fig. S4) and are indicated by yellow, magenta, and blue circles, respectively. lu, VNO lumen; ep, MOE or VNO sensory epithelia. Scale bar = 100 μm.
Most lncRNAs with discernible in situ hybridization staining in either olfactory organ exhibit a homogeneous spatial pattern of expression in the neuroepithelium (Fig. 3 and Supplementary Fig. S6). Moreover, our data show that most lncRNAs expressed in the MOE are found throughout the neuroepithelium turbinates, without detectable preferential expression in one or more particular dorsal-ventral zones51,52 (Supplementary Fig. S7c and d).
On the other hand, two exceptional lncRNAs exhibit a remarkable punctate staining pattern in the olfactory organs. VNO-E seems to be expressed throughout the VNO epithelium, but more highly so in groups of cells lining the base of this olfactory organ and near the epithelial margins (Fig. 3 and Supplementary Fig. S6), the same location of the VNO’s progenitor cells.53 Additionally, MOE-G is not expressed in the whole MOE, but in a very defined subpopulation of MOE cells (Fig. 3 and Supplementary Figs S6 and S7a and b), which seems to correspond to one of the MOE’s odorant receptor dorsal-ventral expression zones.51,52
For some lncRNA candidates, we performed in situ hybridization with probes in the opposite strand, as a control. In the absence of protein-coding information, we synthesized these probes for each lncRNA after confirming the expressed strand with an independent set of MOE RNA-Seq libraries produced via strand-specific SOLiD technology (Table 2). This approach was particularly relevant for intronless transcripts, for which we could not use conserved intron 5′ and 3′ boundary sequences as a proxy to locate the expressed strand. Most of these lncRNA loci showed no significant staining with control probes in the opposite strand, except VNO/MOE-A, whose expression could be detected with probes in both orientations (Supplementary Fig. S8), suggesting that transcription from this locus is entailed from both strands.
In sum, the novel machine-learning-based pipeline described here allowed us to identify, for the first time, a comprehensive set of long non-coding transcripts in the olfactory organs of mammals. Importantly, we identified hundreds of putative lncRNAs preferentially expressed in the VNO or MOE. We further tested a subset of these transcripts with TPMs encompassing the dynamic range of expression, via a combination of RT-PCR and in situ hybridization experiments, which largely confirmed the preferential expression of high- and medium-TPM lncRNAs in either one or both the olfactory organs. Together, these data strongly suggest that the transcripts identified by our bioinformatics pipeline constitute the long non-coding olfactory transcriptome of mice.
3.4. Putative olfactory lncRNA loci are not clustered with olfactory receptor genes or enhancers
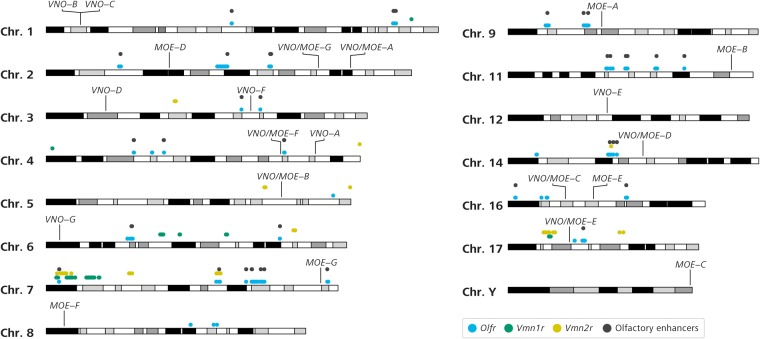
Loci for all lncRNA transcripts analysed in the previous section were mapped onto the chromosomes in comparison with the location of genes known to be preferentially expressed in the olfactory organs, such as receptor genes in the Or (odorant receptor) family expressed in the MOE or in the vomeronasal receptor V1r and V2r families expressed in the VNO, as well as known olfactory receptor enhancers, such as the H, P, J, and Greek Islands elements, shown to influence the expression of certain groups of odorant receptors in the MOE.14,16,54–59
The selected lncRNAs appear not to cluster with those genes and are not in close proximity to the enhancers (Fig. 4). Moreover, when we analysed the distance between all loci for lncRNAs with preferential expression in the olfactory organs and olfactory regulatory sequences, no enhancers were located within 5 kb from the nearest lncRNA gene (not shown).
Figure 4.
lncRNA loci are not near olfactory enhancers or receptor genes. Chromosomal location of lncRNA loci (named according to Table 2) on the mouse chromosomes, relative to odorant receptor (Olfr) gene loci (blue dots), vomeronasal V1R receptor gene (Vmn1r) loci (green dots) and vomeronasal V2R receptor gene (Vmn2r) loci (yellow dots). Regulatory elements known to be involved in olfactory receptor gene control (P, H, J, and Greek Islands elements) are depicted as grey dots. Only chromosomes with lncRNA loci are shown. Black and greyscale bars represent cytogenetic bands, as retrieved from UCSC’s Genome Browser.
3.5. lncRNA expression in the olfactory organs is not influenced by gender
The VNO is responsible for initiating a series of gender-specific responses, such as aggression between males,60,61 choice of sexual partners,60,62,63 courtship behaviour, and lordosis behaviour.64 Although the MOE is not typically associated with the generation of instinctive behaviours, signalling through this organ is fundamental to the display of dimorphic sexual behaviours in males and females.65,66 Therefore, it is plausible to suspect that the animal’s gender is, to some extent, encoded at the sensory interface of the olfactory organs, in the form of variations in the repertoire of expressed genes, including lncRNAs.
Thus, we conducted in silico differential expression tests to investigate whether lncRNA expression is gender-biased or gender-specific. Differential expression analysis between males and females revealed few transcripts differentially expressed between the sexes in the olfactory organs (Supplementary Fig. S9 and Supplementary Table S5). Importantly, none of the differentially expressed genes were lncRNAs. Even though these results are counterintuitive, they are in agreement with another report that did not detect gender-biased expression of olfactory receptors in the MOE or VNO.24
3.6. A catalogue of lncRNAs expressed in mature olfactory sensory neurons
The differentiation of olfactory neurons is a complex process that involves morphological changes and the acquisition of peculiar molecular characteristics. For example, the differentiating cell undergoes a series of molecular events that culminate with the stochastic choice of one olfactory receptor type to express out of thousands of receptor genes in the genome (singular receptor choice), a phenomenon not completely understood.67 It is thought to involve epigenetic modifications known to take place during olfactory differentiation,68 as well as nuclear architecture reorganization.12,69 Because lncRNAs have been shown to act through epigenetic mechanisms,3 it is conceivable to hypothesize that they play a functional role in one or more of the many cell types along the differentiation lineage, including mature olfactory neurons.
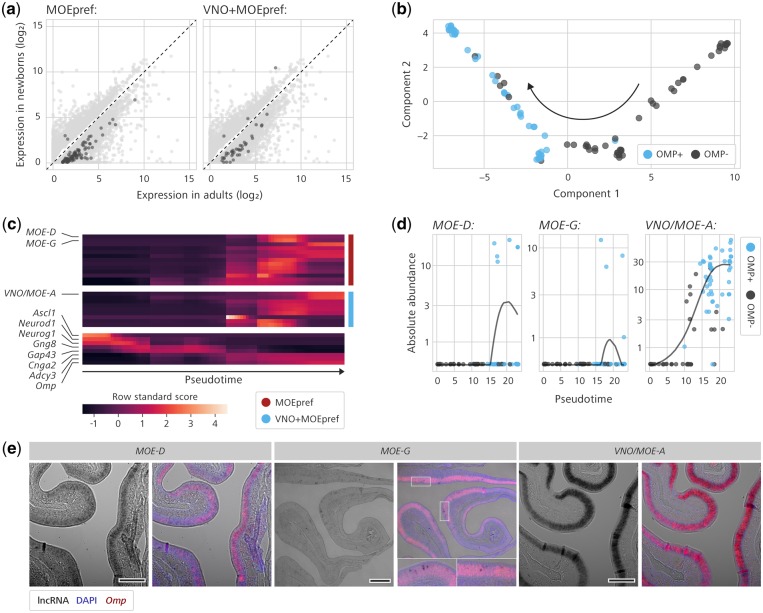
In order to obtain a comprehensive catalogue of olfactory lncRNAs expressed in mature MOE sensory neurons, we took a two-step approach. First, we reasoned that lncRNAs more highly expressed in adults would be enriched for non-coding transcripts expressed in mature neurons, because the adult epithelium contains a comparatively much larger population of mature neurons than newborns.70 The list of transcripts differentially expressed between adults and newborns is extensive (6,784 of 226,171 transcripts) and 67.6% of these are more highly expressed in adults than in newborns (dots below the diagonal 45° dashed line in Fig. 5a). Importantly, we counted 268 lncRNAs (out of 11,435 in total) differentially expressed between adults and newborns, 233 of which (86.9%) are more abundant in the adult MOE.
Figure 5.
Discovery of lncRNAs differentially expressed in mature olfactory sensory neurons. (a) Mean expression levels (TPM) of all transcripts differentially expressed between adult (x-axis) and newborn (y-axis) in the mouse MOE. Each dot represents a differentially expressed transcript. Dots highlighted in dark grey colour are MOEpref (left) and VNO+MOEpref (right) lncRNA transcripts differentially expressed between adults and newborns. (b) Reconstructed trajectory of cell differentiation in the MOE, represented in a two-dimensional space (constructed using the DDRTree algorithm), based on gene expression analysis of 93 single-cell RNA-Seq transcriptomes.71 The black arrow indicates the presumptive progressive temporal sequence of events that transpires from the different single cells along a differentiation pseudotime. Single cells are colour-coded according to presence or absence of OMP expression (cut-off was absolute abundance ≥ 100). (c) Horizontal lines are abundance profiles across the 93 single cells ordered according to the pseudotime determined in (b), for 22 lncRNAs more highly expressed in adults and differentially expressed in OMP-positive single cells (see Supplementary Spreadsheet for complete list). These lncRNAs were taken as putative non-coding species expressed in mature olfactory sensory neurons. Expression of marker genes for several stages along the olfactory differentiation is shown at the bottom. Abundance values were normalized by standardized score calculation. The preferential expression classes (MOEpref or VNO+MOEpref) are shown on the right. Three lncRNA candidates are highlighted (MOE-D, MOE-G, and VNO/MOE-A), because they are multi-exon and were chosen for subsequent investigation. (d) Expression abundance data (y-axis) for lncRNAs MOE-D, MOE-G, and VNO/MOE-A along the olfactory differentiation path. The x-axis represents the pseudotime as measured in the bidimensional space. Black and blue dots represent OMP-negative and OMP-positive single cells, respectively. Black lines are smooth spline curves representing transcript expression dynamics along pseudotime, as determined by Monocle 2 software. (e) For each gene (MOE-D, MOE-G, and VNO/MOE-A), the left panel is a lower magnification microscopy black-and-white image of in situ hybridization on MOE sections, showing expression of the lncRNA (dark staining) throughout the sensory epithelium (ep) for MOE-D and VNO/MOE-A, and a striking punctate spatial pattern for MOE-G. The right panels are co-labelling for lncRNA (purple) and OMP (fluorescent staining in red). DAPI-stained nuclei are shown as blue overlaid fluorescence. For MOE-G, higher magnification images of insets in top, right panel are displayed at the bottom, showing details of co-localization of OMP and lncRNA. Size bar is 100 μm.
Considering only the lncRNAs preferentially expressed in the MOE (MOEpref), we found 93 lncRNAs (out of 272 in total) that are differentially expressed between adults and newborns (dark dots in Fig. 5a, left), 91 of which are more highly expressed in adults. When we focussed on the lncRNAs preferentially expressed in both olfactory organs relative to other tissues (VNO+MOEpref), we found 39 lncRNAs (out of 344 in total) that are differentially expressed between the ages in MOE tissue (dark dots in Fig. 5a, right), 36 of which are prevalently found in adults.
Because we were interested in identifying lncRNAs expressed in mature olfactory neurons, which are positive for Olfactory Marker Protein (OMP) transcripts,49 we further filtered the combined list of 91 MOEpref and 36 VNO+MOEpref lncRNA transcripts more highly expressed in adults by selecting those expressed in OMP-positive single MOE cells. These cells are part of a panel of 93 dissociated main olfactory epithelial cells in distinct stages of differentiation.71
The dynamics of lncRNA expression in the MOE was analysed using single-cell RNA-Seq (scRNA-Seq) data from the dissociated cells. This approach reconstructed the olfactory lineage differentiation path, as revealed by the position of individual scRNA-Seq samples on a bidimensional space constructed using expression data (Fig. 5b) and comparison with marker genes for several stages along olfactory differentiation, including OMP (Fig. 5c). Of the initial list of 127 (91 + 36) lncRNAs more highly expressed in adults, 9 VNO+MOEpref and 13 MOEpref non-coding transcripts (Supplementary Spreadsheet) were found to be differentially expressed in the OMP-positive subpopulation in comparison with OMP-negative single cells (Fig. 5c), with varying temporal patterns of expression along the differentiation pseudotime.
We chose to validate the expression of multi-exon lncRNAs from this short list of olfactory transcripts expressed in mature sensory neurons. Three transcripts matched these criteria—VNO/MOE-A, MOE-D, and MOE-G (Fig. 5d)—and we performed double in situ hybridization experiments with probes for OMP and each of these lncRNAs. Chromogenic detection of hybridized MOE-G probe revealed a striking punctate spatial expression pattern in the MOE epithelium (Fig. 3); when combined with fluorescent detection of OMP probe, cells staining positive for the lncRNA clearly co-express the OMP marker (Fig. 5e), indicating that these are mature olfactory neurons and corroborating the in silico prediction. Additionally, we investigated lncRNAs VNO/MOE-A and MOE-D using the same approach: these transcripts are expressed throughout the MOE epithelium (Fig. 3) and we confirmed them to be expressed in mature olfactory neurons by double in situ hybridization with OMP (Fig. 5e).
Together, our analyses combined whole tissue and single-cell transcriptome data, differential expression, and in situ validation experiments to produce a list of putative lncRNAs expressed in mature olfactory sensory neurons.
3.7. One lncRNA is expressed in neural progenitor cells of the VNO
Both the MOE and VNO continuously generate new neurons throughout adult life via differentiation from a stock of progenitor cells located deep within the neuroepithelium.72 Olfactory neurogenesis is unique, but few genes have been shown to be specifically expressed in progenitor cells of the olfactory organs. In particular, little is known about the molecular characteristics of the progenitor subpopulation in the VNO.
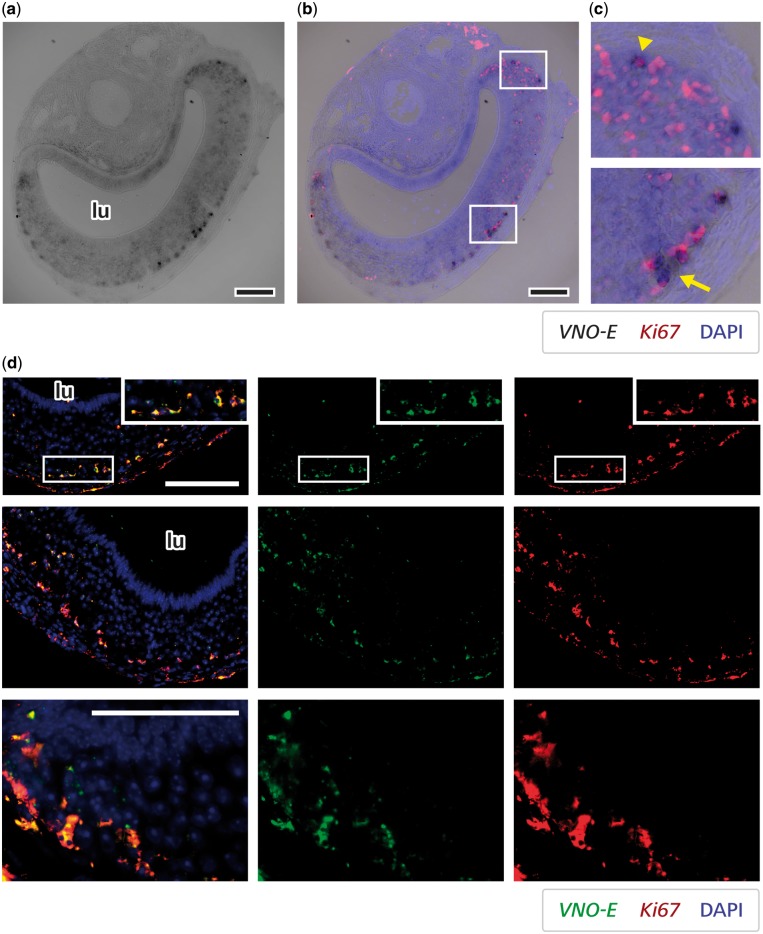
We used in situ hybridization to analyse the spatial pattern of expression for one of the lncRNAs discovered in our study, VNO-E, which is preferentially expressed in the VNO. Our initial analyses determined that such non-coding transcript is expressed in a small subpopulation of cells near the base and corners of the epithelium (Fig. 3), where progenitor cells are localized. We performed double in situ hybridization to investigate whether the VNO-E lncRNA is in fact expressed in progenitor cells, using a probe for marker Ki67.73Ki67-positive cells are sparsely distributed along the base and corners of the VNO (Fig. 6), and a fraction of these co-express VNO-E overall, suggesting that this lncRNA is expressed in vomeronasal progenitors. Interestingly, it seems that the co-expression levels are more pronounced in Ki67 cells near the base as compared with the progenitor population at the VNO epithelium corners (Fig. 6), but the functional significance of this apparent difference remains to be determined.
Figure 6.
A lncRNA is expressed in vomeronasal progenitor cells. (a) Lower magnification microscopy black-and-white images of VNO sections stained by in situ hybridization for lncRNA VNO-E probe (dark staining). (b) Co-labelling for VNO-E probe (chromogenic detection) and Ki67 (fluorescent red staining). DAPI-stained nuclei are shown as blue overlaid fluorescence. (c) Higher magnification images of insets in centre panel showing striking localization near the base of the vomeronasal neuroepithelium (arrow) and around the corners (arrowhead). (d) Microscopy images of VNO sections subjected to double fluorescent in situ hybridizations for lncRNA VNO-E (green) and Ki67 (red). The first two rows of images depict co-localization between signals for the two genes in cells near the base of the epithelium, confirming the data presented in (a-c). The third row of pictures are high magnification images showing the co-localization of VNO-E and Ki67 in the vast majority of VNO-E-positive cells at the corners of the VNO neuroepitelium, where Ki67 staining is concentrated (neural progenitors). lu, VNO lumen. DAPI is pseudo-coloured in blue. Size bar is 100 μm.
4. Discussion
In this study, we used a bioinformatics pipeline containing a novel machine-learning classifier tool to identify long intergenic non-coding RNAs (lncRNAs) in whole-olfactory organ de novo transcriptome assemblies. We discovered a range of unannotated lncRNAs preferentially expressed in the olfactory organs, providing the first comprehensive report of lncRNAs in olfaction. We also performed differential expression and single-cell RNA-Seq analyses to gather spatial and temporal expression information. Selected lncRNA candidates were identified in mature olfactory sensory neurons, including one exquisite transcript that exhibits a striking punctate pattern in a subpopulation of these cells. Moreover, we identified transcripts expressed in olfactory progenitor cells. Our work will lay the foundation for future studies on the function of non-coding RNAs in olfaction.
4.1. An improved bioinformatics pipeline uncovers olfactory lncRNAs
The lncRNA classification model adopted in our study yields improved classification quality for the identification of lncRNAs over previously published tools (Fig. 1a and Table 1), because it uses a modern machine-learning algorithm and a novel set of features (Supplementary Fig. S10) not used in previously reported prediction algorithms (see Section 2 for details).
Olfactory organs are unique from the molecular and cellular standpoints, and only few studies analysed high-throughput olfactory tissue transcriptomes. Only 1,301 out of the 10,158 lncRNA loci in this study have been annotated in the GENCODE’s M9 release reference mouse genome, and 4,624 have been annotated in the NONCODE v5 version (Supplementary Spreadsheet). Seven lncRNA candidates chosen for further investigation were previously categorized as non-coding transcripts in the GENCODE M9 mouse genome annotation, none of which has assigned biological function.74
Our first-hand discovery of lncRNAs in the olfactory organs is relevant, because non-coding transcribed species have been shown to regulate chromatin state, leading to large-scale changes in transcriptional landscape,75 processes that take place during olfactory differentiation, for example as part of the regulation of olfactory receptor genes,12,17 one of the molecular hallmarks of sensory neuron differentiation. Therefore, the olfactory lncRNAs we describe here may help the identification of new molecular mechanisms in olfaction.
4.2. Preferential expression of lncRNAs in the olfactory organs
The olfactory system displays striking molecular and functional characteristics, including the singular expression of receptor genes, which gives rise to a highly heterogeneous sensory epithelium, in contrast to the much more uniform nature of gene expression in other tissues. Therefore, it makes sense to look for molecular players preferentially or specifically expressed in these sensory structures.
We discovered hundreds of lncRNA candidates preferentially expressed in the olfactory organs (VNOpref or MOEpref groups), or expressed in both organs relative to other mouse tissues (VNO+MOEpref). For several selected lncRNA candidates shown in silico to be expressed in one or both of the olfactory organs, confirmatory experiments were performed in situ (Figs 3, 5e, and 6). Some transcripts exhibited widely distributed expression across the sensory epithelium, whereas another exhibited expression in a restricted subpopulation of mature sensory cells (MOE-G), which is reminiscent of the punctate expression pattern of olfactory receptor genes, each of which is expressed in a limited subpopulation of sensory cells widely distributed across the olfactory organ.76,77 Finally, we found one exquisite transcript in the VNO, VNO-E, which is expressed in the little understood progenitor cells of that sensory organ.
4.3. lncRNA expression in the olfactory organs is influenced by age, but not by gender
The VNO has long been recognized as a sensory organ that chiefly detects pheromones,11 substances released by an individual and received by other individuals of the same species, ultimately producing changes in behaviour. Sex pheromones are the most obvious examples, and numerous cases of gender-specific behaviours controlled by pheromones have been described, including the male’s gender discrimination of sexual partners60,62 and female lordosis sexual receptivity behaviour,64 to name just a few. Therefore, it seems conceivable that some olfactory genes expressed in the VNO would be gender-specific. However, over the years, it became apparent that gender-biased genes are not the norm.24 In agreement with these results, we could not find any lncRNAs differentially expressed between the sexes (Supplementary Fig. S9 and Supplementary Table S5).
On the other hand, we found dozens of lncRNA candidates differentially expressed between adults and newborns (Fig. 5). The presence of age-biased transcripts could be due to several possible reasons, including differences in the cellular composition of the olfactory organs according to age, differences resulting from developmental mechanisms specific to either stage, or functional differences between the olfactory organs in adults and newborns. For example, some behaviours are exhibited differentially according to age, including suckling in juveniles, and nursing, infanticide, pup grooming, intermale aggression, and male and female sexual behaviours.60–62,64,78–82 It remains to be determined whether the age-biased lncRNAs identified here are involved simply with the control of olfactory organ development or with the regulation of genes related to olfactory stimulus detection and function.
4.4. Single-cell RNA-Seq and differential expression analysis reveal lncRNAs expressed in mature olfactory sensory neurons
The olfactory sensory epithelia are exposed to the environment and subject to chemical and biotic injury. To cope with the constant loss of neurons, the olfactory organs harbour permanent pools of stem and progenitor cells, which continuously replenish each organ with newly generated mature olfactory neurons.83 In the MOE, progenitor cells give rise to mature olfactory sensory neurons,72 and recent single olfactory neuron transcriptome analyses have shed light on the complex dynamics of gene expression along such differentiation process.71,84,85
We used a combination of bulk RNA-Seq from whole MOE in adults and newborns and single-cell transcriptome analysis to discover 22 olfactory lncRNAs that are at the same time more highly expressed in adults (where the proportion of mature neurons is higher) and in OMP-expressing single cells (which marks mature olfactory neurons) (Fig. 5c). The expression of multi-exon lncRNA candidates fitting these criteria was confirmed in mature olfactory neurons by double in situ hybridization with probes for the lncRNA and OMP (Fig. 5e).
Due to the role of lncRNAs in chromatin modification in other cell types,4 we hypothesize that these non-coding RNAs play a similar role in olfaction. One possibility is that they may participate in the regulation of olfactory neuron differentiation. Another exciting function for these non-transcribed species is that they may regulate olfactory receptor expression (singular gene choice) or olfactory detection in mature sensory neurons. We anticipate that the comprehensive list of lncRNA candidates and the validated transcripts we provide here will pave the road to better understand the molecular processes of olfaction.
Ethics approval and consent to participate
Animals used in this study were obtained directly from our vivarium facility, and procedures were carried out in accordance with Animal Protocol no. 1883-1, approved on June 2009 by the Institute of Biology’s Institutional Animal Care and Use Committee (Committee for Ethics in Animal Use in Research), at the University of Campinas, or by animal protocol numbers 19/2013 and 60/2017 approved by the University of São Paulo Chemistry Institute’s IACUC. The protocols follow the guidelines established by the National Council for Animal Experimentation Control (CONCEA-Brazil).
Supplementary Material
Acknowledgements
We thank Gonçalo A. G. Pereira, José A. Yunes, and Alexandre B. Cardoso for providing resources, Ana P. F. Ferreira and Erica M. R. Bandeira for technical support and Gustavo G. L. Costa for insightful suggestions to the classification model. We thank the staff of the Life Sciences Core Facility (LaCTAD) from University of Campinas (UNICAMP), for help with confocal microscopy. We also thank Anamaria A. Camargo and Fernanda Koyama for help with RNA sequencing using SOLiD.
Accession numbers
The SOLiD RNA-seq libraries generated in this study are available in the SRA repository under the PRJNA497158 accession.
Funding
This work was supported by Sao Paulo Research Foundation (FAPESP) grants to F.P. (#2015/50371-0), B.M. (#2016/24471-0) and M.C. (#2013/08293-7), by FAPESP fellowships to A.C. (#2016/05379-6), T.N. (#2018/11860-4) and J.N. (#2011/22611-6), and fellowships from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) to T.N., P.N. and L.F.
References
- 1. Wapinski O., Chang H.Y.. 2011, Long noncoding RNAs and human disease, Trends Cell Biol., 21, 354–61. [DOI] [PubMed] [Google Scholar]
- 2. Khalil A.M., Guttman M., Huarte M., et al. 2009, Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression, Proc. Natl. Acad. Sci. USA, 106, 11667–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ransohoff J.D., Wei Y., Khavari P.A.. 2018, The functions and unique features of long intergenic non-coding RNA, Nat. Rev. Mol. Cell Biol., 19, 143–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang K.C., Chang H.Y.. 2011, Molecular mechanisms of long noncoding RNAs, Mol. Cell, 43, 904–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhat S.A., Ahmad S.M., Mumtaz P.T., et al. 2016, Long non-coding RNAs: mechanism of action and functional utility, Noncoding RNA Res., 1, 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Quinodoz S., Guttman M.. 2014, Long noncoding RNAs: an emerging link between gene regulation and nuclear organization, Trends Cell Biol., 24, 651–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao J., Sun B.K., Erwin J.A., Song J.J., Lee J.T.. 2008, Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome, Science, 322, 750–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Signal B., Gloss B.S., Dinger M.E.. 2016, Computational approaches for functional prediction and characterisation of long noncoding RNAs, Trends Genet., 32, 620–37. [DOI] [PubMed] [Google Scholar]
- 9. Canzio D., Nwakeze C.L., Horta A., et al. 2019, Antisense lncRNA transcription drives stochastic protocadherin α promoter choice, Cell, 177, 639–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Munger S.D., Leinders-Zufall T., Zufall F.. 2009, Subsystem organization of the mammalian sense of smell, Annu. Rev. Physiol., 71, 115–40. [DOI] [PubMed] [Google Scholar]
- 11. Tirindelli R., Dibattista M., Pifferi S., Menini A.. 2009, From pheromones to behavior, Physiol. Rev., 89, 921–56. [DOI] [PubMed] [Google Scholar]
- 12. Armelin-Correa L.M., Gutiyama L.M., Brandt D.Y.C., Malnic B.. 2014, Nuclear compartmentalization of odorant receptor genes. Proc. Natl. Acad. Sci. USA, 111, 2782–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nagai M.H., Armelin-Correa L.M., Malnic B.. 2016, Monogenic and monoallelic expression of odorant receptors, Mol. Pharmacol., 90, 633–9. [DOI] [PubMed] [Google Scholar]
- 14. Monahan K., Schieren I., Cheung J., Mumbey-Wafula A., Monuki E.S., Lomvardas S.. 2017, Cooperative interactions enable singular olfactory receptor expression in mouse olfactory neurons, Elife, 6, e28620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Magklara A., Lomvardas S.. 2013, Stochastic gene expression in mammals: lessons from olfaction, Trends Cell Biol., 23, 449–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lomvardas S., Barnea G., Pisapia D.J., Mendelsohn M., Kirkland J., Axel R.. 2006, Interchromosomal interactions and olfactory receptor choice, Cell, 126, 403–13. [DOI] [PubMed] [Google Scholar]
- 17. Magklara A., Yen A., Colquitt B.M., et al. 2011, An epigenetic signature for monoallelic olfactory receptor expression, Cell, 145, 555–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khan M., Vaes E., Mombaerts P.. 2011, Regulation of the probability of mouse odorant receptor gene choice, Cell, 147, 907–21. [DOI] [PubMed] [Google Scholar]
- 19. Fuss S.H., Omura M., Mombaerts P.. 2007, Local and cis effects of the H element on expression of odorant receptor genes in mouse, Cell, 130, 373–84. [DOI] [PubMed] [Google Scholar]
- 20. Degl’Innocenti A., D’Errico A.. 2017, Regulatory features for odorant receptor genes in the mouse genome, Front. Genet., 8, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brawand D., Soumillon M., Necsulea A., et al. 2011, The evolution of gene expression levels in mammalian organs, Nature, 478, 343–8. [DOI] [PubMed] [Google Scholar]
- 22. Dillman A.A., Hauser D.N., Gibbs J.R., et al. 2013, MRNA expression, splicing and editing in the embryonic and adult mouse cerebral cortex, Nat. Neurosci., 16, 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fushan A.A., Turanov A.A., Lee S.G., et al. 2015, Gene expression defines natural changes in mammalian lifespan, Aging Cell, 14, 352–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ibarra-Soria X., Levitin M.O., Saraiva L.R., Logan D.W., 2014, The olfactory transcriptomes of mice, PLoS Genet., 10, e1004593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ibarra-Soria X., Nakahara T.S., Lilue J., et al. 2017, Variation in olfactory neuron repertoires is genetically controlled and environmentally modulated, Elife, 6, 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dobin A., Davis C.A., Schlesinger F., et al. 2013, STAR: ultrafast universal RNA-seq aligner, Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trapnell C., Williams B.A., Pertea G., et al. 2010, Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation, Nat. Biotechnol., 28, 511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen T., Guestrin C.. 2016, Xgboost: A scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, p.785–94. Association for Computing Machinery, New York, NY, USA.
- 29. Durinck S., Spellman P.T., Birney E., Huber W., 2009, Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt, Nat. Protoc., 4, 1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roberts A., Trapnell C., Donaghey J., Rinn J.L., Pachter L., 2011, Improving RNA-Seq expression estimates by correcting for fragment bias, Genome Biol., 12, R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Trapnell C., Roberts A., Goff L., et al. 2012, Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks, Nat. Protoc., 7, 562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pimentel H., Bray N.L., Puente S., Melsted P., Pachter L., 2017, Differential analysis of RNA-seq incorporating quantification uncertainty, Nat. Methods, 14, 687–90. [DOI] [PubMed] [Google Scholar]
- 33. Qiu X., Mao Q., Tang Y., et al. 2017, Reversed graph embedding resolves complex single-cell trajectories, Nat. Methods, 14, 979–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qiu X., Hill A., Packer J., Lin D., Ma Y.A., Trapnell C., 2017, Single-cell mRNA quantification and differential analysis with census, Nat. Methods, 14, 309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mao Q., Wang L., Goodison S., Sun Y.. 2015, Dimensionality reduction via graph structure learning. In: Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. Association for Computing Machinery, New York, NY, USA.
- 36. Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L., 2015, StringTie enables improved reconstruction of a transcriptome from RNA-seq reads, Nat. Biotechnol., 33, 290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ma L., Bajic V.B., Zhang Z., 2013, On the classification of long non-coding RNAs, RNA Biol., 10, 925–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu L., Xu Z., Hu B., Lu Z.J., 2017, COME: a robust coding potential calculation tool for lncRNA identification and characterization based on multiple features, Nucleic Acids Res., 45, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang L., Park H.J., Dasari S., Wang S., Kocher J.P., Li W., 2013, CPAT: coding-potential assessment tool using an alignment-free logistic regression model, Nucleic Acids Res., 41, e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao J., Song X., Wang K., 2016, LncScore: alignment-free identification of long noncoding RNA from assembled novel transcripts, Sci. Rep., 6, 34838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kong L., Zhang Y., Ye Z.Q., et al. 2007, CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine, Nucleic Acids Res., 35, W345–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li A., Zhang J., Zhou Z., 2014, PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme, BMC Bioinformatics, 15, 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eddy S.R., 1995, Multiple alignment using hidden Markov models, Proc. Int. Conf. Intell. Syst. Mol. Biol., 3, 114–20. [PubMed] [Google Scholar]
- 44. Lin M.F., Jungreis I., Kellis M., 2011, PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions, Bioinformatics, 27, i275–i282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen J., Shishkin A.A., Zhu X., et al. 2016, Evolutionary analysis across mammals reveals distinct classes of long non-coding RNAs, Genome Biol., 17, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pan J.B., Hu S.C., Wang H., Zou Q., Ji Z.L., 2012, PaGeFinder: quantitative identification of spatiotemporal pattern genes, Bioinformatics, 28, 1544–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liman E.R., Corey D.P., Dulac C., 1999, TRP2: a candidate transduction channel for mammalian pheromone sensory signaling, Proc. Natl. Acad. Sci. USA, 96, 5791–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Berghard A., Buck L.B., Liman E.R., 1996, Evidence for distinct signaling mechanisms in two mammalian olfactory sense organs, Proc. Natl. Acad. Sci. USA, 93, 2365–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Farbman A.I., Margolis F.L., 1980, Olfactory marker protein during ontogeny: immunohistochemical localization, Dev. Biol., 74, 205–15. [DOI] [PubMed] [Google Scholar]
- 50. Ulitsky I., 2016, Evolution to the rescue: using comparative genomics to understand long non-coding RNAs, Nat. Rev. Genet., 17, 601–14. [DOI] [PubMed] [Google Scholar]
- 51. Ressler K.J., Sullivan S.L., Buck L.B., 1993, A zonal organization of odorant receptor gene expression in the olfactory epithelium, Cell, 73, 597–609. [DOI] [PubMed] [Google Scholar]
- 52. Miyamichi K., Serizawa S., Kimura H.M., Sakano H., 2005, Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb, J. Neurosci., 25, 3586–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Martinez-Marcos A., Jia C., Quan W., Halpern M., 2005, Neurogenesis, migration, and apoptosis in the vomeronasal epithelium of adult mice, J. Neurobiol., 63, 173–87. [DOI] [PubMed] [Google Scholar]
- 54. Zhang X., Firestein S., 2002, The olfactory receptor gene superfamily of the mouse, Nat. Neurosci., 5, 124–33. [DOI] [PubMed] [Google Scholar]
- 55. Zhang X., Zhang X., Firestein S., 2007, Comparative genomics of odorant and pheromone receptor genes in rodents, Genomics, 89, 441–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shi P., Zhang J., 2007, Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land, Genome Res., 17, 166–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Godfrey P.A., Malnic B., Buck L.B., 2004, The mouse olfactory receptor gene family, Proc. Natl. Acad. Sci. USA, 101, 2156–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Markenscoff-Papadimitriou E., Allen W.E., Colquitt B.M., et al. 2014, Enhancer interaction networks as a means for singular olfactory receptor expression, Cell, 159, 543–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Iwata T., Niimura Y., Kobayashi C., et al. 2017, A long-range cis-regulatory element for class i odorant receptor genes, Nat. Commun., 8, 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stowers L., Holy T.E., Meister M., Dulac C., Koentges G., 2002, Loss of sex discrimination and male-male aggression in mice deficient for TRP2, Science, 295, 1493–500. [DOI] [PubMed] [Google Scholar]
- 61. Chamero P., Marton T.F., Logan D.W., et al. 2007, Identification of protein pheromones that promote aggressive behaviour, Nature, 450, 899–902. [DOI] [PubMed] [Google Scholar]
- 62. Leypold B.G., Yu C.R., Leinders-zufall T., Kim M.M., Zufall F., Axel R., 2002, Altered sexual and social behaviors in trp2 mutant mice, Proc. Natl. Acad. Sci. USA, 99, 6376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nyby J., Wysocki C.J., Whitney G., Dizinno G., 1977, Pheromonal regulation of male mouse ultrasonic courtship (Mus musculus), Anim. Behav., 25, 333–41. [DOI] [PubMed] [Google Scholar]
- 64. Haga S., Hattori T., Sato T., et al. 2010, The male mouse pheromone ESP1 enhances female sexual receptive behaviour through a specific vomeronasal receptor, Nature, 466, 118–22. [DOI] [PubMed] [Google Scholar]
- 65. Yoon H., Enquist L.W., Dulac C., 2005, Olfactory inputs to hypothalamic neurons controlling reproduction and fertility, Cell, 123, 669–82. [DOI] [PubMed] [Google Scholar]
- 66. Keller M., Douhard Q., Baum M.J., Bakker J., 2017, Destruction of the main olfactory epithelium reduces female sexual behavior and olfactory investigation in female mice, Chem. Senses, 31, 315–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rodriguez I., 2013, Singular expression of olfactory receptor genes, Cell, 155, 274–7. [DOI] [PubMed] [Google Scholar]
- 68. Clowney E.J., LeGros M. A., Mosley C.P., et al. 2012, Nuclear aggregation of olfactory receptor genes governs their monogenic expression, Cell, 151, 724–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Monahan K., Horta A., Lomvardas S., 2019, LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice, Nature, 565, 448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kondo K., Suzukawa K., Sakamoto T., et al. 2010, Age-related changes in cell dynamics of the postnatal mouse olfactory neuroepithelium: cell proliferation, neuronal differentiation, and cell death, J. Comp. Neurol., 518, 1962–75. [DOI] [PubMed] [Google Scholar]
- 71. Hanchate N.K., Kondoh K., Lu Z., et al. 2015, Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis, Science, 350, 1251–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Calof A.L., Bonnin A., Crocker C., et al. 2002, Progenitor cells of the olfactory receptor neuron lineage, Microsc. Res. Tech., 58, 176–88. [DOI] [PubMed] [Google Scholar]
- 73. Oboti L., Ibarra-Soria X., Pérez-Gómez A., et al. 2015, Pregnancy and estrogen enhance neural progenitor-cell proliferation in the vomeronasal sensory epithelium, BMC Biol., 13, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ramos A.D., Diaz A., Nellore A., et al. 2013, Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo, Cell Stem Cell, 12, 616–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rinn J.L., Kertesz M., Wang J.K., et al. 2007, Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs, Cell, 129, 1311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Malnic B., Hirono J., Sato T., Buck L.B., 1999, Combinatorial receptor codes for odors, Cell, 96, 713–23. [DOI] [PubMed] [Google Scholar]
- 77. Buck L., Axel R., 1991, A novel multigene family may encode odorant receptors: a molecular basis for odor recognition, Cell, 65, 175–87. [DOI] [PubMed] [Google Scholar]
- 78. Nakahara T.S., Cardozo L.M., Ibarra-Soria X., et al. 2016, Detection of pup odors by non-canonical adult vomeronasal neurons expressing an odorant receptor gene is influenced by sex and parenting status, BMC Biol., 14, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wu Z., Autry A.E., Bergan J.F., Watabe-Uchida M., Dulac C.G., 2014, Galanin neurons in the medial preoptic area govern parental behaviour, Nature, 509, 325–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kimchi T., Xu J., Dulac C., 2007, A functional circuit underlying male sexual behaviour in the female mouse brain, Nature, 448, 1009–14. [DOI] [PubMed] [Google Scholar]
- 81. Ferrero D.M., Moeller L.M., Osakada T., et al. 2013, A juvenile mouse pheromone inhibits sexual behaviour through the vomeronasal system, Nature, 502, 368–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Logan D.W., Brunet L.J., Webb W.R., Cutforth T., Ngai J., Stowers L., 2012, Learned recognition of maternal signature odors mediates the first suckling episode in mice, Curr. Biol., 22, 1998–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Brann J.H., Firestein S.J., 2014, A lifetime of neurogenesis in the olfactory system, Front. Neurosci., 8, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tan L., Li Q., Xie X.S., 2015, Olfactory sensory neurons transiently express multiple olfactory receptors during development, Mol. Syst. Biol., 11, 844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Fletcher R.B., Das D., Gadye L., et al. 2017, Deconstructing olfactory stem cell trajectories at single-cell resolution, Cell Stem Cell, 20, 817–30.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.