Abstract
Successful therapies for patients with breast cancer often lose their initial effectiveness. Thus, identifying new molecular targets is a constant goal in the fight against breast cancer. Gpn3 is a protein required for RNA polymerase II nuclear targeting in both yeast and human cells. We investigated here the effect of suppressing Gpn3 expression on cell proliferation in a progression series of isogenic cell lines derived from the nontumorigenic MCF-10A breast cells that recapitulate different stages of breast carcinogenesis. Gpn3 protein levels were comparable in all malignant derivatives of the nontumorigenic MCF-10A cells. shRNA-mediated inhibition of Gpn3 expression markedly decreased cell proliferation in all MCF-10A sublines. A fraction of the largest RNA polymerase II subunit Rpb1 was retained in the cytoplasm, but most Rpb1 remained nuclear after suppressing Gpn3 in all cell lines studied. Long-term proliferation experiments in cells with suppressed Gpn3 expression resulted in the eventual loss of all isogenic cell lines but MCF-10CA1d.cl1. In MCF-10CA1d.cl1 cells, Gpn3 knockdown reduced the proliferation of breast cancer stem cells as evaluated by mammosphere assays. After the identification that Gpn3 plays a key role in cell proliferation in mammary epithelial cells independent of the degree of transformation, we also analyzed the importance of Gpn3 in other human breast cancer cell lines from different subtypes. Gpn3 was also required for cell proliferation and nuclear translocation of RNA polymerase II in such cellular models. Altogether, our results show that Gpn3 is essential for breast cancer cell proliferation regardless of the transformation level, indicating that Gpn3 could be considered a molecular target for the development of new antiproliferative therapies. Importantly, our analysis of public data revealed that Gpn3 overexpression was associated with a significant decrease in overall survival in patients with estrogen receptor-positive and Human epidermal growth factor receptor 2 (HER2+) breast cancer, supporting our proposal that targeting Gpn3 could potentially benefit patients with breast cancer.
Keywords: Gpn3, cell proliferation, breast cancer, MCF-10A cells, Gpn3 mRNA levels, shRNA, RNA polymerase II, overall survival, breast cancer patients
Introduction
Breast cancer is the major cause of death due to cancer in women. Prevention is the best alternative, but once cancer has developed, the treatment options available include surgery and radiochemotherapy.1,2 Unfortunately, breast cancer heterogeneity represents an obstacle in cancer therapy, and in general, treatments may cause systemic toxicity and drug resistance.3 Tumors are formed by 2 types of cancer cells: (1) differentiated cells, which constitute the bulk of the tumor, and (2) breast cancer stem cells (BCSCs), which are thought to be the tumor-initiating cells. The BCSCs are also multidrug resistant, an important property for cancer relapse.4,5 Several BCSC features, such as the presence of distinct membrane markers, altered signaling pathways, and the activation of different genes has allowed to specifically target BCSCs and to demonstrate that this is an effective approach in advanced breast cancer therapy.6–9 Recent progress in nano medicine has provided new opportunities to create different smart delivery systems to tackle this disease.10–13
Gpn3, along with Gpn1 and Gpn2, constitutes the GTPase GPN-loop protein family, with all 3 paralogue genes being conserved in eukaryotic cells.14–16 Gpn3 physically associates with Gpn1 and Gpn2,and with several subunits of RNA polymerase II (RNAPII), including Rpb1, Rpb2, and Rpb3.17–19 RNA polymerase II is the enzyme that synthesizes all messenger RNAs (mRNAs) in eukaryotic cells and is composed of 12 subunits, which are assembled in the cytoplasm into a single protein complex that is subsequently targeted to the cell nucleus.20–23 Suppression experiments of Gpn1, Gpn2, or Gpn3 expression have demonstrated that all these 3 proteins are necessary for RNAPII nuclear localization.15,24–27 It has been proposed that Gpn3, along with Gpn1 and Gpn2, helps to assemble the RNAPII protein complex in the cytoplasm, indirectly affecting the nuclear transport of this enzyme, putatively mediated by Iwr1.15,28,29
We have previously demonstrated that Gpn3 is essential for proliferation in the nontumorigenic breast cell line MCF-12A.25 In contrast, MDA-MB-231 breast cancer cells can still proliferate in the absence of Gpn3,25 suggesting that tumorigenic cells can develop alternate mechanisms that support proliferation in the absence of Gpn3. Herein, we aimed to analyze the relationship between the degree of cell transformation and the importance of Gpn3 for cell proliferation and RNAPII nuclear targeting. As models, we employed isogenic, progressively malignant cell derivatives of the originally nontumorigenic MCF-10A mammary epithelial cells.30,31 We also evaluated the functional role of Gpn3 in BCSC proliferation using mammosphere assays. For comparison, we studied the effect of Gpn3 knockdown in the proliferation and RNAPII localization of 3 more human breast cancer cell lines from different subtypes: BT-20 (basal), SK-BR3 (HER2+), and BT-474 (luminal).32,33 We also tested the importance of Gpn3 in the proliferation of the luminal MCF7 breast cancer cell line. Finally, analysis of public data showed that GPN3 overexpression is a factor associated with a reduction in overall survival in patients with ER+ and HER2+ breast cancer.
Our investigation shows that although suppression of Gpn3 decreased RNAPII nuclear targeting only marginally, Gpn3 remained an essential protein for the proliferation of mammary cells, and this was true regardless of their transformation stage. These results, together with the clear negative effect that GPN3 overexpression has in ER+ and HER2+ breast cancer patients survival, make this protein a candidate to be considered as a novel therapeutic target in the fight against breast cancer.
Materials and Methods
Cell Culture Conditions
The series of MCF-10A cell lines were obtained from the Barbara Ann Karmanos Cancer Institute (Detroit, Michigan), and cells were cultured in Dulbecco modified Eagle medium (DMEM)-F12 supplemented with 5% horse serum, 20 ng/mL epidermal growth factor (EGF), 100 ng/mL cholera toxin, 10 μg/mL insulin, and 0.5 μg/mL hydrocortisone. BT-20, BT-474, SK-BR3, and MCF7A cell lines were obtained from American Type Culture Collection (Manassas, Virginia) and were grown in high-glucose DMEM containing 10% fetal bovine serum. All culture media contained 100 U/mL penicillin and 100 μg/mL streptomycin and were kept in a humidified air–CO2 atmosphere at 37°C.
Suppression of Gpn3 Expression
Cells were transduced with retroviral particles expressing either a Gpn3-targeting shRNA (g193) or a control shRNA (g239), as previously described.25 Briefly, retroviral particles were produced in HEK-293T/17 cells, diluted 1:1 with fresh culture medium containing polybrene (8 μg/mL), and added to the cell monolayer. After 6 hours, this cocktail with the viral particles was removed and fresh medium was added. Twenty-four hours after the beginning of infection, puromycin was added in fresh medium for 48 hours to eliminate noninfected cells.
Cell Proliferation
Cells transduced with the indicated shRNAs-coding retroviral particles were trypsinized, counted, and seeded in 12-well plates. The proliferation rate was determined by counting the number of cells under a light microscope on the indicated day postinfection. Briefly, media was first removed, and after rinsing the monolayer with phosphate-buffered saline (PBS), a solution of 0.05% trypsin was added and the plates incubated for 10 to 15 minutes at 37°C to detach the cells. Then serum-containing medium was added to stop trypsin action, and the cells were pipetted up and down to form a single-cell suspension. Cells were spun down at 900g, and the resulting pellets were resuspended in a small volume of media. An aliquot of each type of cells was added to a hemocytometer to determine the cell number under a light microscope. Proliferation of MCF7A and MCF-10A cells was also monitored using the Cell Titer-Glow Luminescence Assay kit (Promega, Madison, Wisconsin), by assessing relative adenosine triphosphate (ATP) levels. MCF7A or MCF-10A cells were infected with retroviral particles expressing the shRNAs g239 or g193 and 24 hours later selected with 2 μg/mL puromycin for 48 hours. MCF7 and MCF-10A cells were trypsinized and 1 × 103 cells were plated per well into 96-well, white, clear bottom plates. Relative ATP levels were determined at different time points after plating, according to the manufacturer’s instructions.
Western Blot Analysis
Immunoblotting was performed as reported previously.25 Briefly, total protein cell extracts were obtained in lysis buffer and mixed with an equal volume of 2× sodium dodecyl sulfate (SDS) loading buffer. Proteins were separated in SDS 10% polyacrylamide gels and electrotransferred to a polyvinyldifluoride membrane. Membranes were blocked with 5% nonfat dry milk in Tris-buffered saline with Tween 20 (TBST) and incubated with primary antibodies against Gpn3 (1:12 000), a polyclonal antibody generated in rabbits using a full-length, histidine-tagged Gpn3 recombinant protein as antigen, and α-tubulin (1:20, 00; Sigma-Aldrich, St Louis, Missouri). Then membranes were washed and incubated with anti-mouse or anti-rabbit secondary antibody conjugated with horseradish peroxidase (1:3000; Sigma-Aldrich), and the immunoreactivity was detected using the enhanced chemiluminescence system (Amersham, Little Chalfont, Buckinghamshire, United Kingdom).
Immunofluorescence Analysis
Cells were fixed with 3.7% formaldehyde in PBS for 15 minutes, rinsed with PBS, permeabilized with 0.2% Triton X-100, and blocked with 10% fetal bovine serum in PBS. Cells were subsequently incubated overnight with a mouse monoclonal anti-Rpb1 antibody (1:1000; COVANCE, Emeryville, California) at 4°C. Next day, cells were rinsed with PBS and incubated at room temperature for 1 hour with a goat anti-mouse Alexa Fluor 568-conjugated secondary antibody (1:3000; Life Technologies, Carlsbad, California). Nuclei were counterstained with DAPI (4,6-Diamidino-2-phenylindole, dihydrochloride). Slides were analyzed in an inverted fluorescence Olympus IX51 microscope (Olympus). Data were analyzed using the ImageJ software (NIH, Bethesda, Maryland), and the nucleus/cytoplasm fluorescence ratio (F n/c) was calculated with the formula: F n/c = (nucleus fluorescence-background)/(cytoplasm fluorescence-background).
Mammosphere Formation Assays
Sphere formation assay was performed as reported.34 Briefly, monolayer cells were trypsinized and counted to prepare single-cell suspensions. Subsequently, 100 viable cells/well were plated in 96-well ultra-low attachment plates (Corning, Lowell, Massachusetts) with MammoCult medium plus growth factors (StemCell Technologies, Vancouver, British Columbia, Canada) and incubated at 37°C in a 5% CO2 atmosphere. Micrographs were taken on day 7 (Eclipse Ti-U microscopy; Nikon), and the spheres with diameter >80 µm were quantified using ImageJ software. Three independent experiments were performed, each with 8 technical replicates. The results are presented as mammosphere-forming efficiency (MFE%), which was calculated with the following equation: MFE% = (number of mammospheres per well)/(number of cells seeded per well) × 100.
Survival Analysis
The role of GPN3 expression in survival was analyzed using the breast cancer cohort from The Cancer Genome Atlas data from Genomic Data Commons and the UCSC Xena platform.35 Primary tumor samples were classified into 2 different histological types: infiltrating ductal carcinoma (IDC; 793 samples) and infiltrating lobular carcinoma (ILC; 207 samples). Each type was then analyzed independently by creating subgroups based on GPN3 expression; the group with high GPN3 expression comprised samples with fragments per kilobase of transcript per million mapped reads (FPKM) within the upper quartile, whereas the group with low GPN3 expression included samples with FPKM within the lower quartile. Further comparisons were performed in the subsets of IDC estrogen receptor (ER)-positive tumors (479 samples) and IDC HER2-positive tumors (65 samples with highest HER2 immunoreactivity). All survival curves were compared by log-rank test.
Results
Gpn3 Is Necessary for Cell Proliferation and Survival in MCF-10A Derivatives of Increasing Malignancy
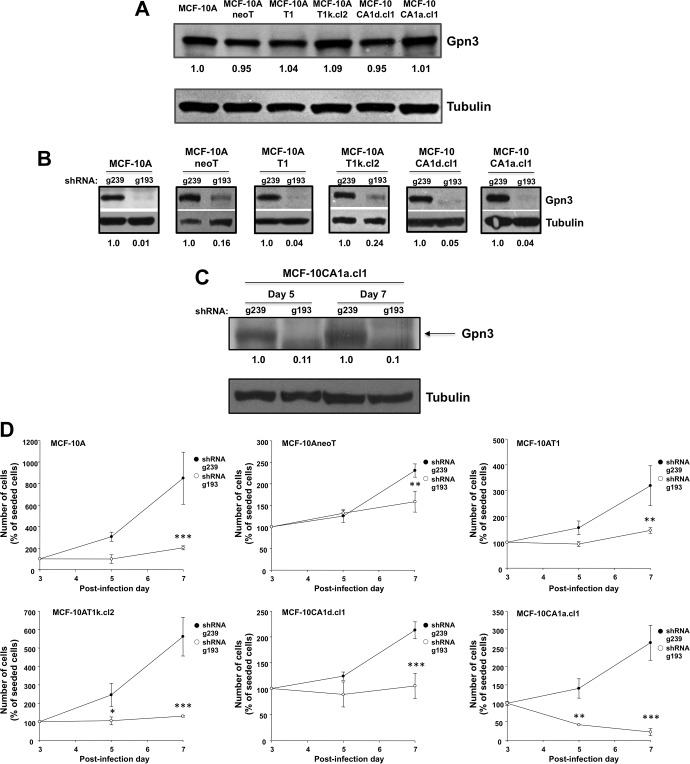
In order to analyze the relationship between the degree of cell transformation and the importance of Gpn3 for cell proliferation, we used isogenic cells of varying degrees of malignancy derived from the originally nontumorigenic MCF-10A cells: MCF-10AneoT, MCF-10AT1, MCF-10AT1k.cl2, MCF-10CA1d.cl1, and MCF-10CA1a.cl1 cells.30,31 First, we compared Gpn3 expression among these cell lines, finding that Gpn3 protein levels were similar in all 6 isogenic cell lines (Figure 1A). The role of Gpn3 on the cell proliferation rate was analyzed after suppressing endogenous Gpn3 expression by interference RNA. Expression of Gpn3 was successfully suppressed by the shRNA g193 but was unaffected by the shRNA g239 in all cell lines (Figures 1B and C). The proliferation rate was significantly reduced in cells after knocking down Gpn3 when compared to that of control (shRNA g293) cells, as indicated by the total cell number quantified at day 7 (Figure 1D). An equal number of cells were seeded 3 days after transducing the cells with either the shRNA control g239 or the shRNA g193 to suppress Gpn3 expression. After 4 days in culture (day 7 after transduction), MCF-10A, MCF-10AneoT, and MCF-10AT1 cells transduced with the shRNA g239 displayed a 7.49-, 1.3-, and 2.19-fold increase in cell number, but these cells showed only a 1.03-, 0.58-, and 0.46-fold increase when expressing the shRNA g193 (Figure 1D). Between days 3 and 7 post-transduction, MCF-10AT1k.cl2 and MCF-10CA1d.cl1 increased their cell number 4.6- and 1.1-fold when transduced with the shRNA g239 but only 0.3- and 0.05-fold when transduced with the shRNA g193 (Figure 1D). Finally, MCF-10CA1a.cl1 cells displayed a 1.64-fold increase with the shRNA g239, but the actual cell number decreased to 22% between day 3 and 7 when expressing the shRNA g193 (Figure 1D).
Figure 1.
Gpn3 suppression decreases cell proliferation in progressively malignant isogenic derivatives of the nontumorigenic MCF-10A breast cells. A, Gpn3 protein levels in MCF-10A cells, MCF-10AneoT, MCF-10AT1, MCF-10AT1k-cl2, MCF-10 CA1a.cl1, and MCF-10CA1d.cl1 cells. Total protein extracts of control, noninfected cells were analyzed by Western blot using a specific rabbit antibody raised against Gpn3. Tubulin was assessed with a monoclonal antibody and was used as a loading control. B, Gpn3 suppression efficiency in the increasingly malignant derivatives of the nontumorigenic MCF-10A cells, MCF-10AneoT, MCF-10AT1, MCF-10AT1k-cl2, MCF-10CA1a.cl1, and MCF-10CA1d.cl1. Total cell extracts from each cell type were prepared 5 days after infecting cells with retroviral particles expressing the shRNAs g193 or g239. Gpn3 protein levels were determined with a rabbit polyclonal antibody. Tubulin levels were assessed to control for protein loading. C, Comparative Gpn3 protein levels in MCF-10CA1d.cl1 cells at 5 and 7 days after infecting cells with retroviral particles expressing the shRNA control g239 or the shRNA g193, which is highly effective to suppress Gpn3 expression. D, Effect of suppressing Gpn3 expression on the proliferation of MCF-10AneoT, MCF-10AT1, MCF-10AT1k-cl2, MCF-10CA1a.cl1, and MCF-10CA1d.cl1. On the third day postinfection, 10 × 104 MCF-10A cells were seeded in 6-well plates, and on the fifth and seventh day postinfection, the cells were trypsinized and directly counted under the microscope in a hemocytometer as described in Material and Methods section. *P < .05; **P < .01; ***P < .001 versus g239-transduced cells at matching time points (2-way analysis of variance).
To establish whether Gpn3 was necessary for cell survival, we kept a long-term culture of all cell lines analyzed replacing the culture medium every other day. After 2 weeks, we lost 5 of the 6 cell lines analyzed, with only MCF-10CA1d.cl1 being able to survive (data not shown). These results demonstrated that although the impact of Gpn3 downregulation in cell proliferation varies in mammary epithelial cells with different transformation degree, Gpn3 is essential for long-term survival in 5 of 6 isogenic cell lines.
Subcellular Distribution of Rpb1 in MCF-10A Transformed Cells After Suppression of Gpn3 Expression
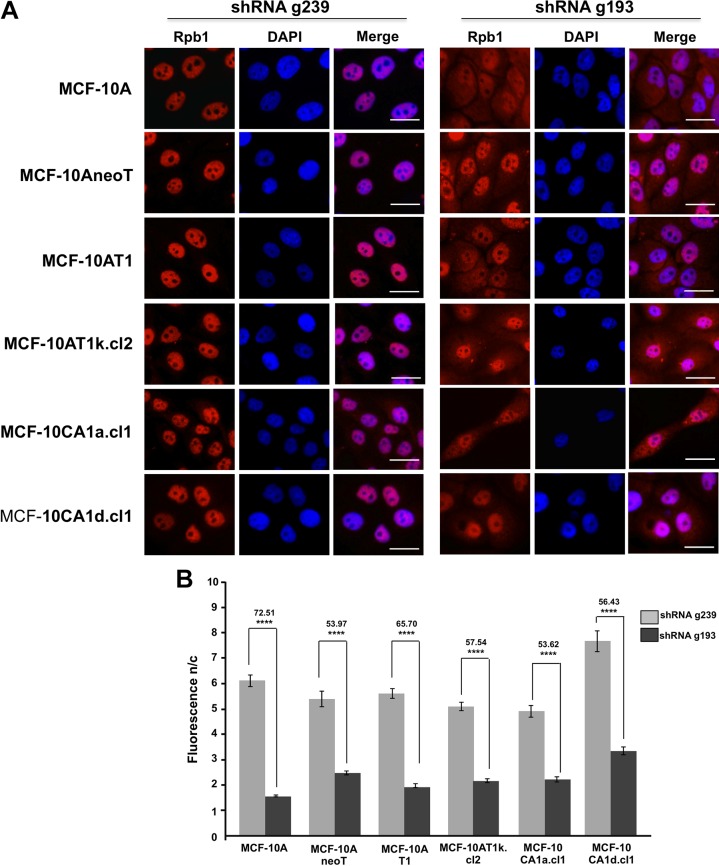
RNA polymerase II is the cellular enzyme that synthesizes, among others, all mRNAs.20,22 Reduced expression of Gpn3 causes the cytoplasmic retention of RNAPII in MDA-MB-468 and MCF-12A cells.25 Thus, we study the relationship between the degree of MCF-10A cell transformation and the importance of Gpn3 for RNAPII nuclear localization of the RNAPII subunit Rpb1. Immunofluorescence experiments revealed that in control, shRNA 239-expressing cells, Rpb1 is localized almost exclusively in the cell nucleus (Figure 2A). However, in the shRNA g193-expressing cells, we observed a partial cytoplasmic retention of Rpb1 (Figure 2A). To quantitate the importance of Gpn3 in the subcellular distribution of Rpb1, we calculated the nucleus–cytoplasm Rpb1 fluorescence ratio (Fn/c) in both control cells and in cells with suppressed Gpn3 expression. These results confirmed that suppression of Gpn3 expression resulted in a partial retention of Rpb1 in the cytoplasm of all cell types examined (Figure 2B). Importantly, we found that the role played by Gpn3 in RNAPII nuclear targeting is maintained regardless of the degree of cell transformation. It is noteworthy that although Gpn3 does not play an essential role in RNAPII nuclear accumulation in our conditions, as substantial Rpb1 signal is still detected in the cell nucleus after suppressing Gpn3 expression, Gpn3 is indeed an essential protein for cell proliferation, as shown in Figure 1D.
Figure 2.
RNA polymerase II (RNAPII) nuclear accumulation in isogenic increasingly malignant derivatives of MCF-10A breast cells after suppressing Gpn3 expression. A, Subcellular distribution of RNAPII in MCF-10A cells, MCF-10AneoT, MCF-10AT1, MCF-10AT1k-cl2, MCF-10CA1a.cl1, and MCF-10CA1d.cl1 cells in the presence (shRNA g239) or absence (shRNA g193) of Gpn3. The subcellular distribution of Rpb1, the largest subunit of the RNA polymerase II, was analyzed by immunofluorescence. Rpb1 was stained in red, and nuclei were counterstained with DAPI (4,6-Diamidino-2-phenylindole, dihydrochloride; blue). B, Results shown in (A) were quantified using ImageJ software. The nuclear/cytoplasm (n/c) Rpb1 fluorescence ratio was calculated for at least 300 cells and are means ± standard deviation. Values comparing bars of the same cell type represent the percentage of inhibition in Rpb1 n/c fluorescence ratio after suppressing Gpn3 expression. ****P < .0001 (unpaired t test with Welch correction for heterogeneous variances).
Gpn3 Is Partially Required for Cell Proliferation in BCSCs
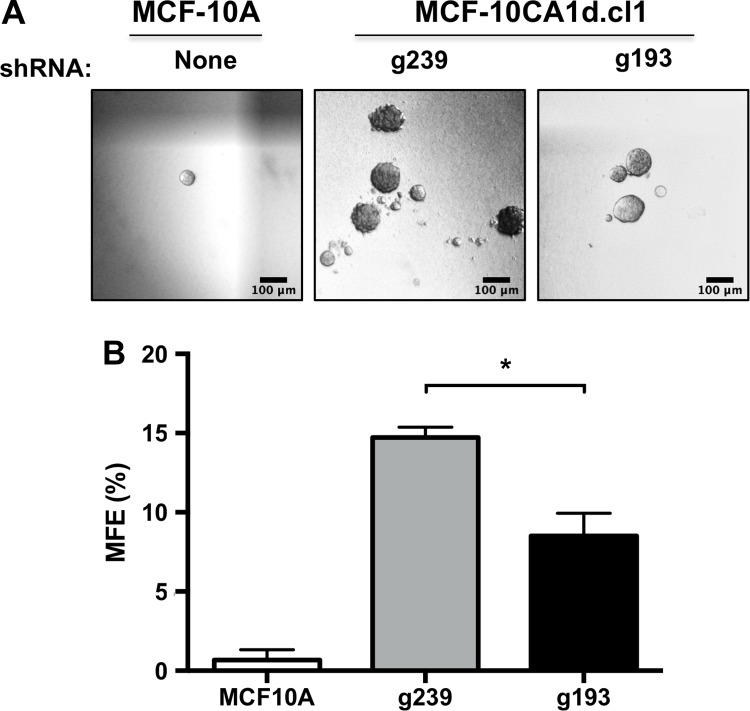
Using the only cell line capable of surviving without Gpn3 expression, MCF-10CA1d.cl1, we evaluated the role of Gpn3 in clonogenicity using mammosphere assays.36,37 As control for the assay, we used the nontransformed original MCF-10A cells. The average MFE% was 0.66% for MCF-10A, 14.72% for MCF-10CA1d.cl1-g239, and 8.50% for MCF-10CA1d.cl1-g193 (Figure 3). These results indicate that Gpn3 reduces the MFE, suggesting that, besides its demonstrated role in bulk cell proliferation (Figure 1D), Gpn3 participates in the expansion of MCF-10CA1d.cl1 BCSCs. Thus, Gpn3 may be a good target for the development of new therapies aimed to reduce both stem and non-stem cancer cells. However, we did not perform serial passages of mammospheres to determine mammosphere self-renewing, which constitutes a limitation of our study.
Figure 3.
Effect of Gpn3 knockdown in mammosphere formation. Viable MCF10A and MCF-10CA1d.cl1 cells expressing a control shRNA (g239) or MCF-10CA1d.cl1 cells expressing the Gpn3-targeting shRNA (g193) were seeded to test their capacity to grow mammospheres. Graph shows the mean number of mammospheres ± standard error of mean (SEM) from 3 independent experiments. Representative pictures are shown. Bar = 100 μm. *P < .05 versus g239 (Student t test).
Gpn3 Is Required for Cell Proliferation and Long-Term Survival in Breast Cancer Cells From Different Subtypes
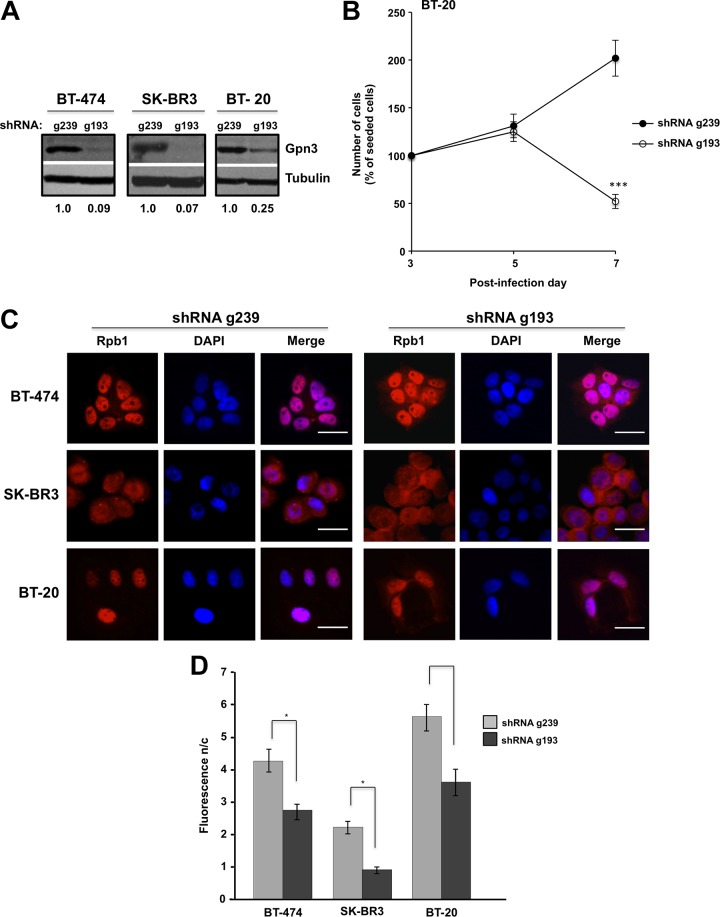
We then studied the role of Gpn3 in breast cancer cells with different phenotypic characteristics: BT-474, SK-BR3, and BT-20.32,33 Gpn3 depletion by the shRNA 193 was corroborated by Western blot (Figure 4A). We observed that knocking down Gpn3 expression blocked the proliferation of BT-20 cells and eventually led to a reduction in cell number (Figure 4B). Similarly, we previously showed that suppression of Gpn3 expression in BT-474 or SK-BR3 cells blocked the cell proliferation.25 We also examined here the importance of Gpn3 in the proliferation of luminal MCF7A breast cancer cells (Figure 5). Gpn3 expression was suppressed by the shRNA g193 in MCF7A cells as effectively as in MCF-10A cells (Figure 5A). To evaluate the effect of suppressing Gpn3 expression in the proliferation of MCF7 cells, we assessed relative cellular ATP levels employing the Cell Titer-Glow Luminescence Assay kit (Promega) as indicated in Materials and Methods section. By measuring cellular ATP levels, it was clear that cell proliferation was completely blocked in MCF7A cells expressing the shRNA g193 compared to MCF7A cells expressing the control shRNA g239 (Figure 5B). As a positive control, we also evaluated here the proliferation of MCF-10A cells after suppressing Gpn3 expression. The results obtained in MCF10A cells by assessing ATP levels (Figure 5B) were very similar as those obtained by directly counting the cell number under the microscope (Figure 1D), supporting the employment of this method to evaluate the importance of Gpn3 in cell proliferation. We also determined the effect of long-term suppression of Gpn3 expression on cell proliferation and survival by keeping a continuous culture of SK-BR3, BT-20, and BT-474 cells expressing the shRNAs g239 or g193. All cells expressing the control shRNA displayed normal cell proliferation. In contrast, the 3 cell lines with reduced Gpn3 expression were lost after 2 weeks in culture beyond the 7-day typical period of our experiments (data not shown). Altogether, these results corroborate that Gpn3 is necessary for breast cancer cell proliferation and suggest a role for Gpn3 in long-term breast cancer cell survival.
Figure 4.
Gpn3 is essential for BT-20 breast cancer cell proliferation. A, Gpn3 suppression efficiency in BT-474, SK-BR3, and BT-20 cells was determined by Western blot with a Gpn3 rabbit polyclonal antibody. Cells were infected with retroviruses expressing the shRNAs g193 to suppress Gpn3 expression or the ineffective shRNA g239 as a control. B, Cell proliferation of BT-20 cells after suppressing Gpn3 expression. Cell number was assessed on the fifth and seventh day postinfection. ***P < .001 g193 versus g239-transduced cells at matching time point (2-way analysis of variance [ANOVA]). C, Rpb1 subcellular distribution in BT-474, SK-BR3, and BT-20 cells after suppressing Gpn3 expression. Rpb1 was visualized by immunofluorescence on the seventh day postinfection and stained red; nuclei were counterstained with DAPI (4,6-Diamidino-2-phenylindole, dihydrochloride; blue). D, The nucleus/cytoplasm fluorescence ratio (F n/c) for Rpb1 distribution was calculated in at least 300 cells from (C) with ImageJ. *P < .05 (unpaired t test with Welch correction for heterogeneous variances).
Figure 5.
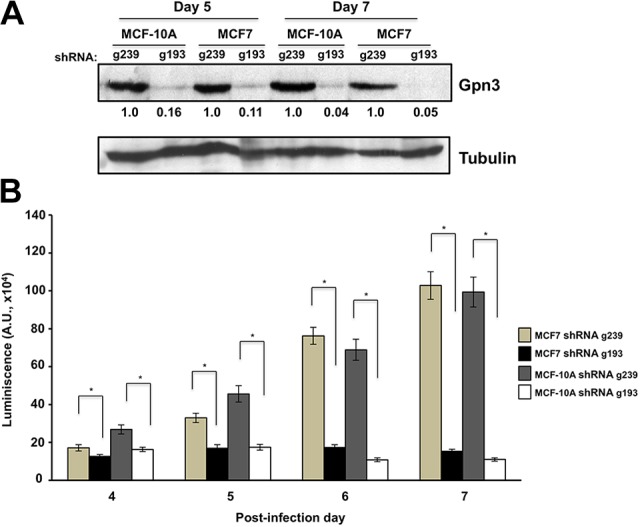
Gpn3 is essential for the proliferation of luminal MCF7 breast cancer cells. A, Gpn3 expression in MCF7 and MCF-10A human breast cells was effectively suppressed by the shRNA g193 but not by the shRNA control g239. MCF7 and MCF-10A cells were infected with retroviruses expressing either the shRNA g239 or the shRNA g193, followed by a 48-hour exposure to puromycin to eliminate noninfected cells as described in Materials and Methods section. Gpn3 cellular levels were determined at the indicated time points after infection in whole cell extracts by Western blotting using a rabbit polyclonal antibody. B, Cell proliferation of MCF7 cells depends on Gpn3. Puromycin-resistant MCF7 and MCF-10A cells from above were seeded in 96-well plates, and cell proliferation was assessed at the indicated time points with the Promega CellTiter-Glo Luminescent Cell Viability Assay, where the amount of ATP is proportional to the number of cells, as described in Materials and Methods section. MCF-10A cells were examined in parallel, as we had previously shown that the proliferation of these cells is completely dependent on the presence of Gpn3 (Sánchez-Olea et al 38). Results shown are means ± standard deviation (n = 8). A Student t test was performed to determine whether the difference between connected bars (shRNA g239- vs shRNA g193-expressing cells) was statistically significant. *P < .05.
Suppression of Gpn3 Expression Causes the Cytoplasmic Retention of a Fraction of Rpb1
By immunofluorescence, we found that Rpb1 localizes almost exclusively in the cell nuclei of BT-474 and BT-20 cells transduced with the control shRNA g239 (Figure 4C), showing a Fn/c)of 4.16 and 7.09, respectively (Figure 4D). In contrast, a fraction of this protein was also found in the cytoplasm of SK-BR3 cells (Fn/c = 2.08; Figures 4C and D). In all 3 cell lines, GPN3 knockdown induced the retention of a fraction of Rpb1 in the cytoplasm (Figure 4C), decreasing the Fn/c (Figure 4D). As most RNAPII remains nuclear in BT-474, BT-20, and SK-BR3 mammary cells after suppressing Gpn3 expression, these results suggest that the essentiality of Gpn3 for proliferation in these cells is unlikely due to the well-described role of this protein in RNAPII nuclear targeting.
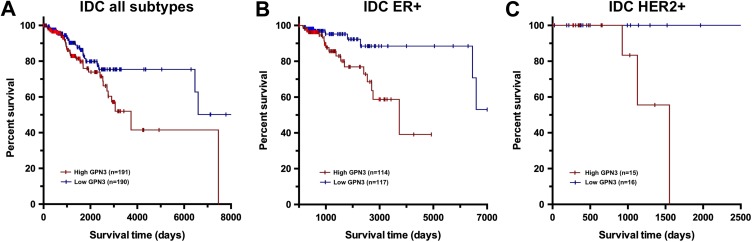
GPN3 Overexpression Shortens Life Expectancy in Patients With Breast Cancer
In order to validate the proposal that Gpn3 targeting could be beneficial for patients with breast cancer, we analyzed the relationship between Gpn3 expression and clinical outcome. Using publicly available data from patients with breast cancer, we found that overexpression of the GPN3 mRNA is associated with a decrease in overall survival in patients with IDC (Figure 6A). On the other hand, we found no association for patients with ILC (data not shown). When patients with IDC were further grouped by subtype, GPN3 expression was significantly associated with bad prognosis in ER+ (Figure 6B) and HER2+ tumors (Figure 6C), suggesting that GPN3 expression may be more relevant for these subtypes. These results show that the level of GPN3 expression is indeed relevant for the survival time of patients with breast cancer and support the idea that GPN3 can be considered a molecular target for extending the lives of patients with breast cancer.
Figure 6.
GPN3 overexpression is an unfavorable prognosis marker in patients with breast cancer. The role of GPN3 expression on patient survival was assessed in RNAseq data from The Cancer Genome Atlas (TCGA) using as cutoffs the 75 and 25 percentiles for the high- (red lines) and low-expressing (blue lines) groups, respectively. A, Comparison in infiltrating ductal carcinoma (IDC) primary tumors; P = .039 (log-rank test). B, Analysis in IDC samples positive for estrogen receptor (ER); P = .004 (log-rank test). C, Analysis in IDC samples with HER2 overexpression; P = .014 (log-rank test).
Discussion
Cancer has been one of the principal causes of death in humans for decades. The identification of new therapeutic molecular targets that might limit tumor growth is a constant goal.5–7 Breast cancer is widely heterogeneous in its etiology and is influenced by genetic factors, and until now no single therapy has proved to be effective to treat all patients. Assays aimed at elucidating Gpn3 cellular function revealed that deletion of the GPN3 gene in Saccharomyces cerevisiae results in loss of cell viability,39 and silencing of Gpn3 in Caenorhabditis elegans caused embryonic lethality40 Suppression of Gpn3 expression markedly reduced cell proliferation in MCF-10A, but much less in MDA-MB-231 and not at all in HeLa cells.38 In contrast, Gpn3 was necessary for BT-474 and SK-BR3 cell proliferation25 as well as for that of BT-20 and MCF7A cells (this work). We have previously investigated the molecular consequences of suppressing Gpn3 expression in MCF-10A cells.38 Knocking down Gpn3 in MCF-10A cells resulted in a clear inability of the cells to proliferate, as the failure to increase cell number over time was associated with a marked decrease in the fraction of cells expressing the proliferation marker Ki67 (see figure 6B in the study by Sánchez-Olea et al 38). Flow cytometry experiments revealed that suppression of Gpn3 expression in MCF10A cells clearly increased the fraction of cells in the G1 phase of the cell cycle and caused a parallel decrease in the fraction of cells in S and G2/M phases (see figures 4A and 6A in the study by Sánchez-Olea et al 38), indicating that suppressing Gpn3 expression causes an early G1 cell cycle arrest. Consistently, Gpn3 suppression in MCF-10A cells caused a marked decrease in cyclin A1 and cyclin B1 protein levels (figure 6C in the study by Sánchez-Olea et al 38), both of which are expressed in the cell cycle after G1. Thus, in the short term, interfering with Gpn3 function seems to be more associated with causing a cytostatic effect than with inducing cell death. Consistent with these findings, we have not detected here any signs of apoptotic cell death, such as activation of caspases or PARP cleavage, after suppressing Gpn3 expression (results not shown). These results are consistent with Gpn3 being originally isolated as a protein important to induce or maintain Apaf-1 in a competent state to be activated by a proapoptotic stimulus.38 Although the cell lines were eventually lost in a long-term cell culture after suppressing Gpn3, the loss of cells was very slow, indicating a lack of synchrony in the process, a fact that will increase the difficulty of identifying the molecular mechanism involved. However, regardless of the precise mechanism, we also found here that Gpn3 is essential for cell proliferation in 5 isogenic breast cell lines with different transformation level. This is important from a therapeutic perspective because Gpn3 could putatively be targeted in breast cancer cells regardless of their transformation stage. We did not find supporting evidence that in breast cancer cells the need of Gpn3 for cell viability was related to the best known Gpn3 cellular function, that is, to mediate RNAPII nuclear targeting. Although we detected a clear and significant Rpb1 retention in the cytoplasm in all cell types examined after suppressing Gpn3 expression, most Rpb1 still localized to the cell nucleus even in the absence of Gpn3. This may be the reason for our inability to detect Rpb1 in the cytoplasm in cell fractionation experiments after suppressing Gpn3 expression (results not shown). Notably, in these conditions, cell proliferation was abolished, indicating that in addition to RNAPII nuclear targeting Gpn3 is involved in some other still to identify critical cellular function. Thus, tumorigenic cells seem to have developed a molecular mechanism for RNAPII nuclear localization that is independent of Gpn3. Interestingly, although Rpb1 nuclear targeting was barely affected in BT-20 cells, cell proliferation was completely abolished in these cells, supporting our proposal that targeting Gpn3 would be an effective mechanism to prevent cell proliferation in breast cancer cells. Our results showed that no compensatory mechanisms are activated in cancer cells after suppressing Gpn3 expression is an important finding, especially in the context of preventing the expansion of breast cancer cells by inhibiting Gpn3 function. Our results showing that Gpn3 plays a still unknown but critical role in cell proliferation is precisely what makes Gpn3 a potential molecular target for breast cancer therapy. Although there is much to investigate and learn about the molecular mechanisms controlled by Gpn3 in cells, the relationship between GPN3 overexpression and a reduced overall survival observed for patients with ER+ and HER2+ breast cancer make us to propose Gpn3 as an extremely promising molecular target in the fight against breast cancer. Although it is not an ideal situation, very often successful therapies that target essential proteins are commonly used in the clinic, including inhibition of topoisomerase I or II by camptothecin or doxorubicin, respectively, or of the mitotic microtubule system by taxanes. The anticipated result will be that, although global inhibition of Gpn3 would affect this protein in both normal and cancer cells, the overall balance will be beneficial for the patient due to the higher proliferative rate of cancer cells compared to normal cells.
Abbreviations
- ATP
adenosine triphosphate
- BCSC
breast cancer stem cells
- DMEM
Dulbecco modified Eagle medium
- ER
estrogen receptor
- IDC
infiltrating ductal carcinoma
- ILC
infiltrating lobular carcinoma
- FKPM
fragments per kilobase of transcript per million mapped reads
- F n/c
nucleus/cytoplasm fluorescence ratio
- MFE
mammosphere-forming efficiency
- mRNA
messenger RNA
- PBS
phosphate-buffered saline
- SDS
sodium dodecyl sulfate
- RNAPII
RNA polymerase II
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship and/or publication of this article: Funding was provided by the Consejo Nacional de Ciencia y Tecnología grant numbers 254075 (MRC), A1-S-21070 (RSO) and PAPPIIT UNAM IN219719 (MAVV).
ORCID iD: Marco Antonio Velasco Velazquez  https://orcid.org/0000-0001-9717-0265
https://orcid.org/0000-0001-9717-0265
Mónica Raquel Calera  https://orcid.org/0000-0002-9351-9180
https://orcid.org/0000-0002-9351-9180
References
- 1. Corradini S, Niyazi M, Niemoeller OM. et al. Adjuvant radiotherapy after breast conserving surgery—a comparative effectiveness research study. Radiother Oncol. 2015;114(1):28–34. [DOI] [PubMed] [Google Scholar]
- 2. Zhao B, Hemann MT, Lauffenburger DA. Modeling tumor clonal evolution for drug combinations design. Trends Cancer. 2016;2(3):144–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bao B, Ahmad A, Azmi AS, Ali S, Sarkar FH. Overview of cancer stem cells (CSCs) and mechanisms of their regulation: implications for cancer therapy. Curr Protoc Pharmacol. 2014;Chapter 14:Unit 14.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Velasco-Velazquez MA, Homsi N, De La Fuente M, Pestell RG. Breast cancer stem cells. Int J Biochem Cell Biol. 2012;44: 573–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang F, Xu J, Tang L, Guan X. Breast cancer stem cell: the roles and therapeutic implications. Cell Mol Life Sci. 2016;74(6):951–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prud’homme GJ. Cancer stem cells and novel targets for antitumor strategies. Curr Pharm Des. 2012;18(19):2838–2849. [DOI] [PubMed] [Google Scholar]
- 7. de Souza VB, Schenka AA. Cancer stem and progenitor-like cells as pharmacological targets in breast cancer treatment. Breast Cancer (Auckl). 2015;9(suppl 2):45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi AR, Park JR, Kim RJ. et al. Inhibition of Wnt1 expression reduces the enrichment of cancer stem cells in a mouse model of breast cancer. Biochem Biophys Res Commun. 2012;425(2):436–442. [DOI] [PubMed] [Google Scholar]
- 9. Iyevleva AG, Imyanitov EN. Cytotoxic and targeted therapy for hereditary cancers. Hered Cancer Clin Pract. 2016;14(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sanchez-Moreno P, Ortega-Vinuesa JL, Peula-Garcia JM, Marchal JA, Boulaiz H. Smart drug-delivery systems for cancer nanotherapy. Curr Drug Targets. 2018;19(4):339–359. [DOI] [PubMed] [Google Scholar]
- 11. Ediriwickrema A, Saltzman WM. Nanotherapy for Cancer: targeting and multifunctionality in the future of cancer therapies. ACS Biomater Sci Eng. 2015;1(2):64–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee H, Dam DH, Ha JW, Yue J, Odom TW. Enhanced human epidermal growth factor receptor 2 degradation in breast cancer cells by lysosome-targeting gold nanoconstructs. ACS Nano. 2015;9(10):9859–9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mignani S, Bryszewska M, Klajnert-Maculewicz B, Zablocka M, Majoral JP. Advances in combination therapies based on nanoparticles for efficacious cancer treatment: an analytical report. Biomacromolecules. 2015;16(1):1–27. [DOI] [PubMed] [Google Scholar]
- 14. Leipe DD, Wolf YI., Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317(1):41–72. [DOI] [PubMed] [Google Scholar]
- 15. Minaker SW, Filiatrault MC, Ben-Aroya S, Hieter P, Stirling PC. Biogenesis of RNA polymerases II and III requires the conserved GPN small GTPases in Saccharomyces cerevisiae. Genetics. 2013;193(3):853–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gras S, Chaumont V, Fernandez B. et al. Structural insights into a new homodimeric self-activated GTPase family. EMBO Rep. 2007;8(6):569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mendez-Hernandez LE, Pérez-Mejía AE, Lara-Chacón B. et al. Gpn1 and Gpn3 associate tightly and their protein levels are mutually dependent in mammalian cells. FEBS Lett. 2014;588(21):3823–3829. [DOI] [PubMed] [Google Scholar]
- 18. Alonso B, Beraud C, Meguellati S. et al. Eukaryotic GPN-loop GTPases paralogs use a dimeric assembly reminiscent of archeal GPN. Cell Cycle. 2013;12(3):463–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alonso B, Chaussinand G, Armengaud J, Godon C. A role for GPN-loop GTPase yGPN1 in sister chromatid cohesion. Cell Cycle. 2011;10(11):1828–1837. [DOI] [PubMed] [Google Scholar]
- 20. Young RA. RNA polymerase II. Annu Rev Biochem.1991;60:689–715. [DOI] [PubMed] [Google Scholar]
- 21. Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292(5523):1863–1876. [DOI] [PubMed] [Google Scholar]
- 22. Bernecky C, Herzog F, Baumeister W, Plitzko JM, Cramer P. Structure of transcribing mammalian RNA polymerase II. Nature. 2016;529(7587):551–554. [DOI] [PubMed] [Google Scholar]
- 23. Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Mol Biol. 2004;11(5):394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carre C, Shiekhattar R. Human GTPases associate with RNA polymerase II to mediate its nuclear import. Mol Cell Biol. 2011;31(19):3953–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Calera MR, Zamora-Ramos C, Araiza-Villanueva MG, Parcs/Gpn3 is required for the nuclear accumulation of RNA polymerase II. Biochim Biophys Acta. 2011;813(10):1708–1716. [DOI] [PubMed] [Google Scholar]
- 26. Forget D, Lacombe AA, Cloutier P. et al. The protein interaction network of the human transcription machinery reveals a role for the conserved GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II. Mol Cell Proteomics. 2010;9(12):2827–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Staresincic L, Walker J, Dirac-Svejstrup AB, Mitter R, Svejstrup JQ. GTP-dependent binding and nuclear transport of RNA polymerase II by Npa3 protein. J Biol Chem. 2011;286(41):35553–35561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Niesser J, Wagner FR, Kostrewa D, Muhlbacher W, Cramer P. Structure of GPN-loop GTPase Npa3 and implications for RNA polymerase II assembly. Mol Cell Biol. 2015;36(5):820–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zeng F, Hua Y, Liu X. et al. Gpn2 and Rba50 directly participate in the assembly of the Rpb3 subcomplex in the biogenesis of RNA polymerase II. Mol Cell Biol. 2018;38(13) e00091–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Worsham MJ, Pals G, Schouten JP. et al. High-resolution mapping of molecular events associated with immortalization, transformation, and progression to breast cancer in the MCF10 model. Breast Cancer Res Treat. 2006;96(2):177–186. [DOI] [PubMed] [Google Scholar]
- 31. Qu Y, Han B, Yu Y. et al. Evaluation of MCF10A as a reliable model for normal human mammary epithelial cells. PLoS One. 2015;10(7):e0131285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Holliday DL, Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011;13(4):215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Subik K, Lee JF, Baxter L. et al. The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by immunohistochemical analysis in breast cancer cell lines. Breast Cancer (Auckl). 2010;4:35–41. [PMC free article] [PubMed] [Google Scholar]
- 34. Vasquez-Bochm LX, Velázquez-Paniagua M, Castro-Vázquez SS. et al. Transcriptome-based identification of lovastatin as a breast cancer stem cell-targeting drug. Pharmacol Rep. 2019;71(3):535–544. [DOI] [PubMed] [Google Scholar]
- 35. Goldman M, Craft B, Hastie M. et al. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. bioRxiv. 2019; doi:10.1101/326470. [Google Scholar]
- 36. Grimshaw MJ, Cooper L, Papazisis K. et al. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. 2008;10(3): R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shaw FL, Harrison H, Spence K. et al. A detailed mammosphere assay protocol for the quantification of breast stem cell activity. J Mammary Gland Biol Neoplasia. 2012;17(2):111–117. [DOI] [PubMed] [Google Scholar]
- 38. Sanchez-Olea R, Ortiz S, Barreto O. et al. Parcs is a dual regulator of cell proliferation and apaf-1 function. J Biol Chem. 2008;283(36):24400–24405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Giaever G, Chu AM, Ni L. et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418(6896):387–391. [DOI] [PubMed] [Google Scholar]
- 40. Gonczy P, Echeverri C, Oegema K. et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408(6810):31–336. [DOI] [PubMed] [Google Scholar]