Abstract
This study’s objectives were to test correlations among groups of biomarkers that are associated with condylar morphology and to apply artificial intelligence to test shape analysis features in a neural network (NN) to stage condylar morphology in temporomandibular joint osteoarthritis (TMJOA). Seventeen TMJOA patients (39.9 ± 11.7 y) experiencing signs and symptoms of the disease for less than 10 y and 17 age- and sex-matched control subjects (39.4 ± 15.2 y) completed a questionnaire, had a temporomandibular joint clinical exam, had blood and saliva samples drawn, and had high-resolution cone beam computed tomography scans taken. Serum and salivary levels of 17 inflammatory biomarkers were quantified using protein microarrays. A NN was trained with 259 other condyles to detect and classify the stage of TMJOA and then compared to repeated clinical experts’ classifications. Levels of the salivary biomarkers MMP-3, VE-cadherin, 6Ckine, and PAI-1 were correlated to each other in TMJOA patients and were significantly correlated with condylar morphological variability on the posterior surface of the condyle. In serum, VE-cadherin and VEGF were correlated with one another and with significant morphological variability on the anterior surface of the condyle, while MMP-3 and CXCL16 presented statistically significant associations with variability on the anterior surface, lateral pole, and superior-posterior surface of the condyle. The range of mouth opening variables were the clinical markers with the most significant associations with morphological variability at the medial and lateral condylar poles. The repeated clinician consensus classification had 97.8% agreement on degree of degeneration within 1 group difference. Predictive analytics of the NN’s staging of TMJOA compared to the repeated clinicians’ consensus revealed 73.5% and 91.2% accuracy. This study demonstrated significant correlations among variations in protein expression levels, clinical symptoms, and condylar surface morphology. The results suggest that 3-dimensional variability in TMJOA condylar morphology can be comprehensively phenotyped by the NN.
Keywords: biomarkers, digital imaging/radiology, joint disease, bioinformatics, artificial intelligence, temporomandibular disorders (TMDs)
Introduction
Temporomandibular joint disorders (TMDs) are a major cause of orofacial pain of nondental origin (Gauer and Semidey 2015). TMD is the second most commonly occurring musculoskeletal condition resulting in pain and disability and affecting approximately 5% to 12% of the population (Schiffman et al. 2014). TMD involves a wide spectrum of syndromes: myofascial pain disorder, disk derangement disorders, and osteoarthritis (OA) (Gauer and Semidey 2015). Joint degeneration occurs from the loss in equilibrium of anabolic and catabolic processes involving chondrocyte initiation, proliferation, differentiation, and matrix synthesis and degradation (Wadhwa and Kapila 2008). Temporomandibular joint osteoarthritis (TMJOA) was once thought to be a “wear-and-tear” condition and of noninflammatory origin. It is now classified as a “low-inflammatory arthritic condition” (de Souza et al. 2012) and associated with inflammatory mediators that lead to harmful effects on the temporomandibular joint’s (TMJ’s) cartilage, bone, and synovium (Berenbaum 2013). TMJOA typically progresses very slowly (Rousseau and Delmas 2007), and the initial stages may be subclinical until the disease process has progressed (Su et al. 2014).
Advances in the health care field have led the drive to use biological markers as diagnostic markers of OA. Collecting saliva has become a popular trend to gain real-time levels of biomarkers due to its noninvasiveness, constant availability, and cost-effectiveness (Yan et al. 2009). Changes in protein levels have shown to be detectable in serum years before OA becomes radiographically evident (Ling et al. 2009). The disease process of OA is characterized by deterioration of the articular cartilage and its disc surfaces along with thickening and remodeling of the subchondral bone (Tanaka et al. 2008). Subchondral bone angiogenesis during early OA progression may facilitate increased crosstalk between cartilage and subchondral bone, leading to cartilage degradation (Sharma et al. 2013; Lepage et al. 2019).
Cevidanes et al. (2014) were the first to report an association between specific OA biomarkers and 3-dimensional (3D) morphological variations at specific anatomic regions on the TMJ condylar surface. Synovial fluid and serum samples were collected to measure levels of 50 biomarkers of arthritic inflammation (Cevidanes et al. 2014). Areas along the articular surface indicative of bone resorption were found particularly at the lateral pole of the condyle while bone apposition/reparative proliferation was found to occur on the condyle’s anterior surface (Cevidanes et al. 2014).
Computerized methods are a great help to clinicians to discover hidden patterns in data. These methods often employ data-mining and machine-learning algorithms, lending themselves as the computer-aided diagnosis tool that assists clinicians in making diagnostic decisions (Li 2018). Neural network (NN) applications in computer-aided diagnosis represent the main stream of computational intelligence in medical imaging (Qian et al. 2007). This study aims to combine a state-of-the-art machine-learning technique with a biological and clinical identification scheme to provide novel insights into the molecular basis of TMJOA. We hypothesized that variations in protein levels and clinical symptoms would correlate to the patterns of bone morphology on the condylar articular surfaces of TMJOA subjects, and there is a high degree of conformity between the NN and expert clinicians in classifying the condylar degree of OA.
Materials and Methods
Seventeen TMJOA subjects (aged 39.9 ± 11.7 y) who experienced signs and symptoms of the disease for less than 10 y and 17 age- and sex-matched asymptomatic control subjects (aged 39.4 ± 15.2 y) were consented and enrolled in this study (Appendix). Clinical and radiographic diagnosis of TMJOA followed the diagnostic criteria for temporomandibular disorders (DC/TMD) (Schiffman et al. 2014). The STROBE (Strengthening the Reporting of Observational Studies in Epidemiology) statement was reviewed, and this study was compliant with the checklist. The data acquisition and analysis in this study was approved by the University Institutional Review Board.
All TMJOA subjects had a clinical examination of their TMJs by an orofacial pain specialist while all control subjects had a dental specialist perform the clinical examination to rule out signs of TMD. Clinical information describing the signs and symptoms comprised and followed standardized DC/TMD clinical exam and questionnaire forms (Schiffman et al. 2014). Pain-related questions following a 0 to 10 visual analog scale and the amount of assisted and unassisted mouth opening measurements were included in the integrative analysis for the clinical markers. Age, a demographic variable, was also evaluated.
Saliva and blood samples were collected and measured (Appendix). Custom human Quantibody protein microarrays obtained from RayBiotech (Appendix Fig. 1) quantitatively assessed the saliva and serum samples for 17 specific biomarkers (Appendix Table 1) previously found to be expressed in synovial fluid and serum of TMJOA subjects: 6Ckine, ANG, BDNF, CXCL16, ENA-78, GM-CSF, IFN-γ, IL-1α, IL-6, MMP-3, MMP-7, PAI-1, TGF-β1, TIMP-1, TNF-α, VE-cadherin, and VEGF (Cevidanes et al. 2014). Each participant had duplicates run for the saliva and serum samples.
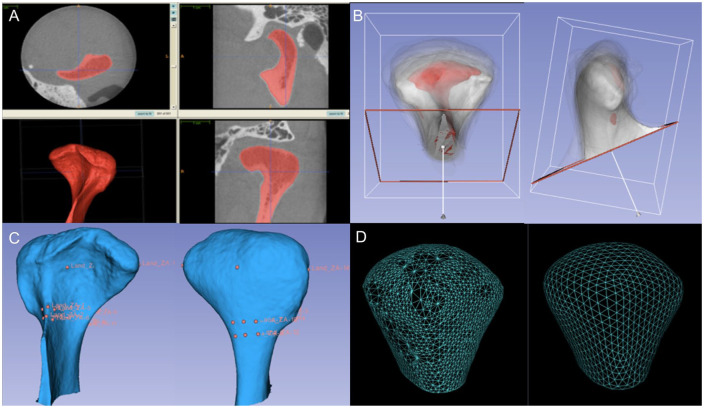
Subjects were scanned using a hr-CBCT scanner (Morita 3D Accuitomo; J. Morita MFG. CORP; see Appendix). The region of interest included the inferior border of the squamous portion of the temporal bone to the condylar neck, the narrowest portion of the condyle process. Reconstruction of the surface models of the right and left condyles from the cone beam computed tomography (CBCT) images of each subject was performed as shown in Figure 1A using ITK-SNAP v.2.4 (Yushkevich et al. 2006). Each subject had both TMJs scanned, but the biological and clinical data that refer to each subject would be analyzed without being side specific. The joint side of choice used for analysis on the 3D surface meshes data was the side with the most severe symptoms in the TMJOA group and the matching control condyle. The surface mesh reconstructions of all left condyles were mirrored in the sagittal plane using a 3D slicer (Fedorov et al. 2012) to be in the same orientation as the right condyles to facilitate bilateral comparisons. A validated regional superimposition technique (Schilling et al. 2014) was used for across-subject comparisons in a common coordinate system when all of the registered condylar surface models were cropped, as shown in Figure 1B, C. SPHARM-PDM (Styner et al. 2006; Paniagua et al. 2017) software was used to generate a mesh with 1,002 correspondent points (Fig. 1D) through spherical mapping and spherical parameterization of the surface to analyze the areas of most significant morphological variability (Brechbühler et al. 1995; Styner et al. 2006; Paniagua et al. 2011).
Figure 1.
(A) Segmentation (labeling) of a participant’s left condyle from the CBCT image using ITK-SNAP software. The ITK-SNAP user interface shows 3 orthogonal views (top 2 views and lower right view) of a volumetric image, linked by a common cursor (light blue crosshairs). A fourth panel (lower left view) was used to view the segmented structures in 3D. (B) The anterior and lateral views of the registered condyles. An arbitrary condyle of 1 OA subject was used as a template or reference for a common spatial orientation of all condyles. The reference condyle is shown in red, and the other TMJOA and control condyles are shown in white with 5% transparency. (C) Anterior and posterior surface model displaying the landmarks for surface registration. Sixteen landmarks were placed on the reference condyle. (D) The surface model on the left contains nonevenly distributed triangles, and the surface model on the right established correspondence between each of the 1,002 points on the condylar surface model after spherical mapping and spherical parameterization of the input volumes run in SPHARM-PDM. CBCT, cone beam computed tomography; OA, osteoarthritis; TMJOA, temporomandibular joint osteoarthritis; 3D, three-dimensional.
Deidentified patient data were stored in a flexible secure web-based system: Data Storage, Computation, and Integration (DSCI) (Appendix Fig. 2). The training of the NN was performed in the DSCI system, where the classifier learns from features extracted from the 3D meshes of the condyles. In total, 259 condyles (105 control and 154 from patients with a diagnosis of TMJOA), collected from previous studies (Gomes et al. 2015; Paniagua et al. 2017), were used to train the NN (Appendix). The 34 condyles from the subjects enrolled in the current study were then used to test the NN. The total study data set consisted of 3D surface meshes of 293 condyles. The NN module computed the average shape of each group of condylar dysmorphology as well as geometric features at each vertex of the mesh.
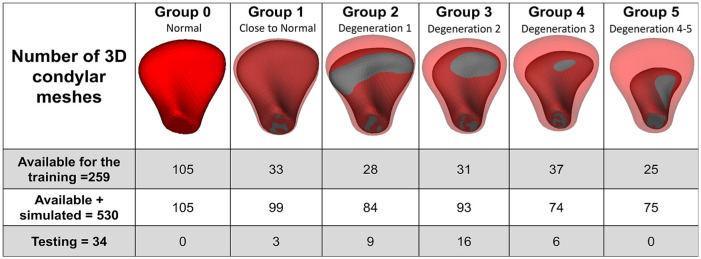
The NN in this study was trained to distinguish different degrees of shape deformation of TMJOA condyles. Two expert clinicians (A.R. and M.Y.) performed a consensus visualization and interpretation of 3D condylar surface morphology and classified the condylar morphology into 6 groups as shown in Figure 2, which included 5 subgroups with different degrees of condylar degeneration (groups 1–5). The clinicians scored each condyle based off of the shape, size, and morphology in a 3D overlay compared to the average control group condylar morphology. Group 0 included healthy control–shaped condyles, and group 5 included a condyle exhibiting the most degeneration and lacking any resemblance to a normal shape of a condyle. To test reproducibility, the clinicians’ classification was repeated for a subset of 46 condyles of the 259 condyles in the training data sets.
Figure 2.
Distribution of the training and testing data sets in 1 group representing the control/normal condyles and 5 subgroups characterizing different degrees of condylar degeneration following the clinicians’ visual classifications. Group 0, control; group 1, close to normal; group 2, degeneration 1; group 3, degeneration 2; group 4, degeneration 3; group 5, degeneration 4. 3D, 3-dimensional.
Our training database contained fewer samples for some of the disease stage groups. In order not to overtrain the NN for one of the groups, the training procedure required the same number of meshes in each training group (LeCun et al. 2015). To increase the number of data sets in each training group, we simulated data by adding perlin noise (VTK.org 2017) of small magnitude to each coordinate in the shape, and then the features were recomputed, which provided us with 530 total 3D condylar meshes, including the 259 condyles from the training database (de Dumast et al. 2018). Data were simulated to ensure that 74 meshes were available per group (Fig. 2). For groups that had more than 74 meshes, the preprocessing step randomly selected 74 condyles. Thereafter, the NN was used to classify the stage of TMJOA of the testing data sets (34 condylar surface meshes from the subjects enrolled in the current study) and then compared to the clinical experts’ classifications twice.
Statistical Analysis
Data analysis was performed using the SPSS software (version 24.0; SPSS, Inc.). We applied multivariate functional shape data analysis (MFSDA) to test the integrating information of 3D mesh coordinates, clinical markers, and levels of biological markers. The multivariate varying coefficient model (Zhu et al. 2012; Huang et al. 2017) in MFSDA was introduced to build an integrative statistical model of clinical, biological, and imaging markers (Appendix).
Results
Eleven of the 17 proteins (ANG, MMP-3, MMP-7, PAI-1, TIMP-1, VE-cadherin, 6Ckine, CXCL16, ENA-78, IL-1α, VEGF) were found to have protein concentrations of quantifiable levels and found to have a coefficient of determination value greater than 0.96 (Appendix Fig. 3). These biomarkers were quantified and used for further analyses in conjunction with the clinical and imaging markers (Tables 1 and 2, Appendix Table 2). When comparing the serum and saliva levels for each biomarker between the control and TMJOA groups, there was no significant difference found (Appendix Table 3).
Table 1.
Preprocessing Statistics prior to Running MFSDA: Clinical Variables.
| Pearson Correlation Coefficients and P Values |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinical Variables | Status | BPY | CP | WP | AP | PL | WPMO | UMO | AMO | H | MS |
| Begin pain years | TMJOA | 0.203 | 0.462 | 0.400 | 0.003 | 0.197 | –0.432 | –0.484 | 0.441 | 0.457 | |
| Current facial pain rate | TMJOA | 0.435 | 0.568 | 0.608 | 0.113 | 0.099 | –0.373 | –0.362 | 0.173 | 0.370 | |
| Facial worst pain rate | TMJOA | 0.062 | 0.017a | 0.900 | –0.057 | –0.108 | –0.149 | –0.145 | 0.390 | 0.193 | |
| Average rate 6 mo | TMJOA | 0.111 | 0.010a | 0.000b | –0.049 | –0.201 | –0.201 | –0.202 | 0.235 | 0.293 | |
| Pain location | TMJOA | 0.989 | 0.665 | 0.827 | 0.851 | 0.303 | –0.051 | –0.065 | –0.287 | 0.129 | |
| Range without pain mouth openingc | TMJOA | 0.449 | 0.704 | 0.679 | 0.440 | 0.237 | 0.086 | –0.017 | –0.247 | –0.099 | |
| Range unassisted mouth openingc | TMJOA | 0.083 | 0.140 | 0.568 | 0.439 | 0.846 | 0.742 | 0.981 | –0.375 | –0.795 | |
| Range assisted mouth openingc | TMJOA | 0.049 | 0.154 | 0.578 | 0.437 | 0.804 | 0.948 | 0.000b | –0.385 | –0.789 | |
| Headaches | TMJOA | 0.077 | 0.508 | 0.121 | 0.365 | 0.264 | 0.338 | 0.138 | 0.127 | 0.308 | |
| Muscle soreness | TMJOA | 0.065 | 0.144 | 0.459 | 0.254 | 0.622 | 0.707 | 0.000b | 0.000b | 0.229 | |
Pearson correlation coefficients are shown. The values above the main diagonal describe the Pearson correlation coefficients and below the main diagonal represent the P values. Gray-shaded values describe only the variables after multivariate functional shape data analysis statistics that presented statistically significant correlations with specific regions of morphological variability (as shown in Fig. 3).
AMO, range assisted mouth opening; AP, average rate 6 mo; BPY, begin pain years; CP, current facial pain rate; H, headache; MFSDA, multivariate functional shape data analysis; MS, muscle soreness; PL, pain location; TMJOA, temporomandibular joint osteoarthritis; UMO, range unassisted mouth opening; WP, facial worst pain rate; WPMO, range without pain mouth opening.
P < .05.
P < .001.
Measured in millimeters.
Table 2.
Preprocessing Statistics prior to Running MFSDA: Biological Variables.
| Pearson Correlation Coefficients and P Values |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biological Variables | Fluid | Status | ANG | MMP-3 | MMP-7 | PAI-1 | TIMP-1 | VE-Cadherin | 6Ckine | CXCL16 | ENA-78 | IL-1α | VEGF |
| ANG | Serum | TMJOA | 0.387 | 0.194 | –0.295 | 0.042 | –0.365 | 0.293 | 0.557 | 0.413 | 0.034 | –0.058 | |
| MMP-3 | Serum | TMJOA | 0.125 | 0.595 | –0.184 | –0.098 | –0.029 | 0.050 | 0.712 | 0.017 | –0.079 | 0.058 | |
| MMP-7 | Serum | TMJOA | 0.456 | 0.012a | 0.218 | 0.277 | 0.413 | 0.120 | 0.551 | 0.384 | –0.265 | 0.093 | |
| PAI-1 | Serum | TMJOA | 0.251 | 0.480 | 0.401 | 0.668 | 0.321 | –0.170 | –0.266 | 0.162 | –0.311 | 0.205 | |
| TIMP-1 | Serum | TMJOA | 0.873 | 0.707 | 0.281 | 0.003b | 0.213 | –0.189 | –0.133 | 0.176 | –0.053 | –0.118 | |
| VE-cadherin | Serum | TMJOA | 0.150 | 0.911 | 0.099 | 0.208 | 0.411 | 0.405 | –0.140 | 0.206 | 0.276 | 0.516 | |
| 6Ckine | Serum | TMJOA | 0.254 | 0.847 | 0.645 | 0.514 | 0.468 | 0.106 | 0.274 | 0.651 | 0.569 | 0.751 | |
| CXCL16 | Serum | TMJOA | 0.020a | 0.001b | 0.022a | 0.302 | 0.610 | 0.591 | 0.287 | 0.469 | –0.018 | 0.074 | |
| ENA-78 | Serum | TMJOA | 0.100 | 0.949 | 0.128 | 0.534 | 0.500 | 0.427 | 0.005b | 0.057 | 0.114 | 0.347 | |
| IL-1α | Serum | TMJOA | 0.896 | 0.763 | 0.304 | 0.225 | 0.841 | 0.284 | 0.017a | 0.944 | 0.662 | 0.505 | |
| VEGF | Serum | TMJOA | 0.824 | 0.826 | 0.723 | 0.431 | 0.652 | 0.034a | 0.001b | 0.779 | 0.173 | 0.039a | |
| ANG | Saliva | TMJOA | 0.221 | 0.601 | 0.187 | 0.453 | 0.158 | 0.094 | 0.163 | 0.048 | 0.305 | 0.487 | |
| MMP-3 | Saliva | TMJOA | 0.393 | 0.600 | 0.946 | 0.219 | 0.988 | 0.962 | 0.297 | 0.077 | 0.404 | 0.175 | |
| MMP-7 | Saliva | TMJOA | 0.011a | 0.011a | 0.560 | 0.155 | 0.601 | 0.563 | 0.384 | 0.286 | 0.346 | 0.407 | |
| PAII | Saliva | TMJOA | 0.471 | 0.000c | 0.019a | 0.181 | 0.947 | 0.951 | 0.233 | -0.050 | 0.338 | 0.032 | |
| TIMP-1 | Saliva | TMJOA | 0.068 | 0.399 | 0.553 | 0.488 | 0.185 | 0.099 | 0.391 | 0.087 | 0.379 | 0.699 | |
| VE-cadherin | Saliva | TMJOA | 0.545 | 0.000c | 0.011a | 0.000c | 0.478 | 0.979 | 0.301 | 0.075 | 0.393 | 0.133 | |
| 6Ckine | Saliva | TMJOA | 0.720 | 0.000c | 0.019a | 0.000c | 0.705 | 0.000c | 0.243 | –0.032 | 0.360 | 0.017 | |
| CXCL16 | Saliva | TMJOA | 0.533 | 0.247 | 0.128 | 0.368 | 0.121 | 0.241 | 0.347 | 0.557 | 0.186 | 0.688 | |
| ENA-78 | Saliva | TMJOA | 0.855 | 0.768 | 0.266 | 0.850 | 0.741 | 0.774 | 0.902 | 0.020a | 0.353 | 0.434 | |
| IL-1α | Saliva | TMJOA | 0.234 | 0.107 | 0.174 | 0.185 | 0.134 | 0.119 | 0.156 | 0.475 | 0.165 | 0.321 | |
| VEGF | Saliva | TMJOA | 0.047 | 0.501 | 0.105 | 0.903 | 0.002b | 0.610 | 0.948 | 0.002b | 0.082 | 0.208 | |
| ANG | Serum | Control | 0.647 | 0.097 | –0.172 | –0.124 | 0.268 | 0.134 | 0.749 | –0.036 | 0.528 | –0.110 | |
| MMP-3 | Serum | Control | 0.005b | 0.12 | –0.01 | –0.001 | 0.485 | 0.063 | 0.850 | –0.209 | 0.778 | 0.203 | |
| MMP-7 | Serum | Control | 0.712 | 0.640 | 0.324 | –0.206 | 0.048 | –0.189 | 0.355 | –0.039 | 0.077 | 0.043 | |
| PAII | Serum | Control | 0.508 | 0.978 | 0.205 | 0.059 | 0.232 | –0.584 | 0.090 | 0.496 | –0.192 | 0.572 | |
| TIMP-1 | Serum | Control | 0.637 | 0.997 | 0.428 | 0.822 | –0.134 | –0.404 | –0.187 | –0.360 | –0.326 | 0.434 | |
| VE-cadherin | Serum | Control | 0.297 | 0.048a | 0.855 | 0.371 | 0.609 | 0.132 | 0.435 | –0.022 | 0.596 | 0.541 | |
| 6Ckine | Serum | Control | 0.608 | 0.810 | 0.467 | 0.014a | 0.107 | 0.614 | 0.085 | –0.081 | 0.449 | –0.364 | |
| CXCL16 | Serum | Control | 0.001b | 0.000c | 0.163 | 0.732 | 0.472 | 0.081 | 0.746 | 0.149 | 0.760 | 0.044 | |
| ENA-78 | Serum | Control | 0.890 | 0.421 | 0.881 | 0.043a | 0.156 | 0.933 | 0.756 | 0.568 | –0.058 | –0.117 | |
| IL-1α | Serum | Control | 0.029a | 0.000c | 0.769 | 0.461 | 0.201 | 0.012a | 0.071 | 0.000c | 0.825 | 0.064 | |
| VEGF | Serum | Control | 0.674 | 0.435 | 0.870 | 0.016a | 0.082 | 0.025 | 0.151 | 0.868 | 0.654 | 0.807 | |
| ANG | Saliva | Control | 0.072 | 0.738 | 0.323 | 0.506 | 0.418 | 0.559 | 0.671 | 0.199 | 0.363 | 0.411 | |
| MMP-3 | Saliva | Control | 0.783 | 0.325 | –0.093 | 0.459 | –0.065 | –0.030 | 0.408 | –0.212 | 0.862 | 0.158 | |
| MMP-7 | Saliva | Control | 0.001b | 0.203 | 0.494 | 0.527 | 0.349 | 0.444 | 0.710 | –0.066 | 0.397 | 0.333 | |
| PAII | Saliva | Control | 0.206 | 0.724 | 0.044a | 0.178 | –0.115 | –0.146 | 0.431 | –0.242 | 0.106 | 0.303 | |
| TIMP-1 | Saliva | Control | 0.038a | 0.064 | 0.030a | 0.495 | 0.264 | 0.340 | 0.686 | 0.169 | 0.634 | 0.692 | |
| VE-cadherin | Saliva | Control | 0.095 | 0.804 | 0.169 | 0.659 | 0.305 | 0.702 | 0.194 | 0.565 | –0.138 | 0.159 | |
| 6Ckine | Saliva | Control | 0.020a | 0.910 | 0.074 | 0.577 | 0.182 | 0.002b | 0.147 | 0.591 | –0.064 | 0.186 | |
| CXCL16 | Saliva | Control | 0.003b | 0.104 | 0.001b | 0.085 | 0.002b | 0.456 | 0.574 | 0.059 | 0.621 | 0.687 | |
| ENA-78 | Saliva | Control | 0.445 | 0.414 | 0.801 | 0.350 | 0.517 | 0.018a | 0.013a | 0.823 | –0.188 | 0.233 | |
| IL-1α | Saliva | Control | 0.153 | 0.000c | 0.115 | 0.685 | 0.006b | 0.596 | 0.807 | 0.008b | 0.471 | 0.370 | |
| VEGF | Saliva | Control | 0.101 | 0.545 | 0.192 | 0.237 | 0.002 | 0.543 | 0.475 | 0.002b | 0.367 | 0.143 | |
Pearson correlation coefficients are shown. The values above the main diagonal describe the Pearson correlation coefficients and below the main diagonal represent the P values. Gray-shaded values describe only the variables after MFSDA statistics that presented statistically significant correlations with specific regions of morphological variability (as shown in Fig. 3).
MFSDA, multivariate functional shape data analysis; TMJOA, temporomandibular joint osteoarthritis.
P < .05.
P < .01.
P < .001.
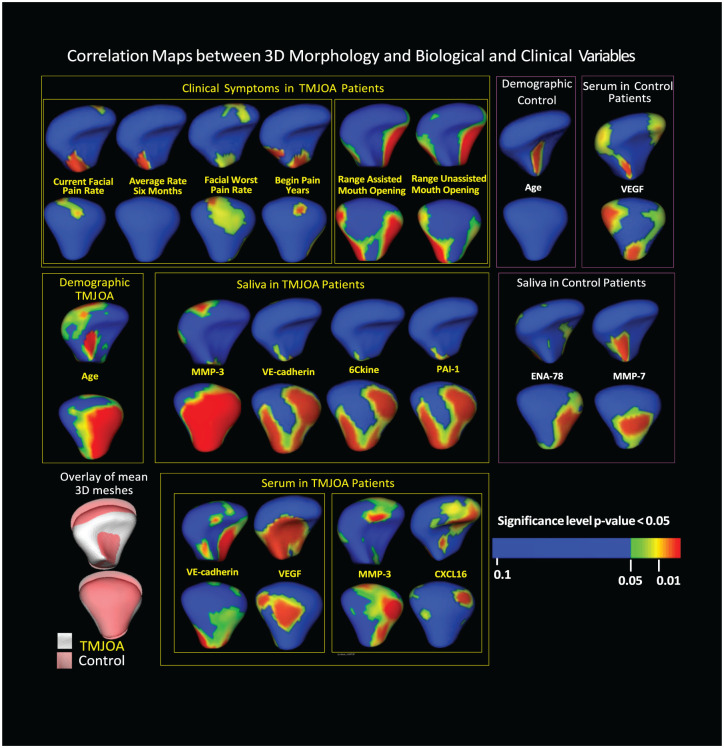
The MFSDA model tested correlations with morphology of subjects’ age, pain-related clinical variables, ranges of mouth opening, and the 11 biological markers that were expressed at the best confidence levels in saliva and serum samples (Fig. 3). Age showed significant Pearson correlations with morphological variability on the anterior surface of control condyles and the lateral pole and posterior surface of the TMJOA condyles, which are areas of resorptive changes in TMJOA. The clinical markers “current facial pain rate,” “average rate 6 mo,” and “facial worst pain rate,” were correlated among themselves and together with “begin pain years,” which showed statistically significant associations with the superior-posterior articular surfaces. “Range assisted mouth opening” and “range unassisted mouth opening” demonstrated statistically significant associations with variability in medial and lateral poles of the condyles.
Figure 3.
MFSDA statistics graphically displays statistically significant Pearson correlations between biological (from serum and saliva) and clinical marker levels, as well as specific locations on the 3D morphology of the mandibular condyle. MFSDA, multivariate functional shape data analysis; TMJOA, temporomandibular joint osteoarthritis; 3D, 3-dimensional.
Expression levels of MMP-3, VE-cadherin, 6Ckine, and PAI-1 were correlated among themselves in saliva in the TMJOA group and showed significant Pearson correlations with condylar morphological variability on the posterior surface of the condyle. In serum, VE-cadherin and VEGF levels were correlated with one another and with significant morphological variability on the anterior surface of the condyle, a region that is typically associated with bone proliferation, while MMP-3 and CXCL16 levels were found to have statistically significant associations with variability on the anterior surface, lateral pole, and superior-posterior surface of the condyle. In the control group, it was found that expression levels of MMP-7 and ENA-78 in saliva and VEGF in serum showed significant Pearson correlations with condylar morphological variability. The level of MMP-7 in saliva was correlated with morphological variability on the anterior surface of the condyle near the condylar neck and posterior surface. The level of ENA-78 in saliva was associated with morphological variability on the lateroposterior surface of the condyle. In serum, the level of VEGF was correlated with significant variability in small regions of the medial pole of the articular surface, as well as anterior and posterior surfaces of the condylar neck.
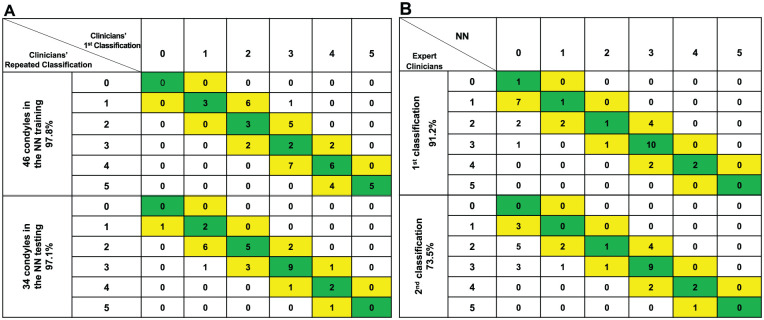
The performance of the 2 clinical experts’ assessments is considered the “control” for the NN to be compared against. From the different combinations of features that were used to train the network, the features that led to higher accuracy of the morphological classification compared to the clinical experts’ assessments were normal vectors, mean curvature, and the distances to the average meshes at each mesh vertex. The tests of repeatability of the clinicians’ classification for both the training and testing data sets and the NN performance are shown in the confusion matrices (Stehman 1997) in Figure 4. Each column represents the instances in the NN classification group, and each row represents the group instances as assessed by the consensus between 2 clinical experts. Agreement between the clinician consensus classifications and the NN classification is located in the main diagonal of the table. Cells adjacent to the main diagonal (1 diagonal to the right and 1 diagonal to the left) indicate the classification of the degree of degeneration was within 1 group difference. The repeated clinician consensus classification had 97.8% agreement on degree of degeneration within 1 group difference for the training data sets and 97.1% for the testing data sets. Predictive analytics of the NN’s staging of TMJOA compared to the repeated clinicians’ consensus revealed 73.5% and 91.2% degree of conformity.
Figure 4.
Confusion matrices: rows and columns show the classification of 5 stages of condylar degeneration, where 0 is the control group. (A) Agreement of the expert clinicians’ first classification and expert clinicians’ repeated consensus classification for the training and testing data sets. (B) Agreement of the expert clinicians’ repeated consensus and the NN classification. The main diagonal cells (highlighted in green) show when the group was classified by the NN exactly the same as the clinicians. The cells in 1 diagonal to the right and 1 diagonal to the left show number of condyles where the prediction differed from clinicians by only 1 group and still indicates an acceptable estimate of condylar characterization in TMJOA patients and disease staging. NN, neural network; TMJOA, temporomandibular joint osteoarthritis.
Discussion
This is the first study to test biomarkers in pairs or groups to evaluate their correlation with condylar morphology and to apply artificial intelligence to test the shape analysis features in a NN for the staging of condylar morphology in TMJOA. Although significant progress has been made in TMJOA research in recent years, very little is known about the molecular mechanisms of OA initiation and progression. The lateral surface of condyles usually demonstrates resorption in TMJOA patients, with resultant flattening on the lateroposterior condylar (Nah 2012). There has been a need for the development of a comprehensive diagnostic model that integrates clinical, morphological, and biomolecular assessments.
The first part of this investigation aimed to detect levels of known inflammatory biomarkers in systemic (serum and saliva) samples, to identify clinical markers, and then to correlate these markers to 3D models of TMJOA. The previous pilot quantitative assays of localized joint synovial fluid and serum samples from TMJOA patients (Cevidanes et al. 2014) were limited by the inability to test biomarkers in pairs or groups to evaluate whether or not there is cross-reactivity between them that is associated with condylar morphology. The present investigation tested biomarkers in groups to likely be a more accurate representation of the in vivo state.
No significant difference being found when comparing biomarkers in serum and saliva between the (asymptomatic) controls and TMJOA groups may be explained by the normal presence of these proinflammatory proteins in systemic fluids of both groups. The biomarkers that were measured and evaluated in this study serve various physiologic and pathophysiologic processes and may simply play different roles in the progressive degeneration in OA compared to control groups. The application of MFSDA statistics determined a comprehensive model of the integrative correlations between biological and clinical marker levels and morphological condylar surface changes at the 1,002 vertices of 3D meshes. The regional correlations between biological markers and morphology for the control group may indicate the roles of these biomarkers in the physiological remodeling with maintenance of homeostasis that occurs in healthy TMJs. In the TMJOA group, the pain-related variables tested were correlated among themselves and showed statistically significant associations only with the superior-posterior articular surfaces, while range of mouth opening variables were correlated among themselves and with the morphological variability in the medial and lateral poles of the condyles.
Interestingly, levels of VE-cadherin, VEGF, MMP-3, and CXCL16 in serum were highly correlated to areas of bone apposition/reparative proliferation that occurs on the anterior surface of condyles and leads to characteristic changes in condylar torque and morphology. Bone resorption with flattening and reshaping of the lateral pole of the condyle involves molecular pathways with interaction of 5 proteins measured in this study: VE-cadherin and MMP-3 both in saliva and serum, CXCL16 in serum, and 6Ckine and PAI-1 in saliva. In a previous study (Cevidanes et al. 2014), these same serum biomarkers, as well as ENA-78, 6Ckine, TIMP-1, ANG, PAI-1, GM-CSF, IFN-γ, IL-1α, IL-6, TNF-α, TGF-β1, BDNF, and other biomarkers not evaluated in this study, were found to be correlated with morphological variability on different regions of the condylar surface. It should be noted that the statistical shape analysis, MFSDA, used in the present study was more rigorous and robust than the multivariate analysis of covariance (MANCOVA) used in the previous study (Cevidanes et al. 2014).
The second part of this investigation aimed to classify different degrees of 3D joint degeneration through novel phenotyping using a NN. The deep learning architecture chosen for this study was able to capture complex morphology patterns. Interestingly, the confusion matrix found adequate agreement between the clinical experts and the NN when classifying the testing data set condyles. The differences in classification between the clinicians’ assessment and the NN may be due to the need to increase the training database to improve NN classification, limitations in the clinicians’ visual perception, or the fact the registration and correspondence/homology of vertices in the surface meshes affect the computation of shape features in the NN (de Dumast et al. 2018).
Variability in patient symptoms and imaging findings can create challenges in diagnosing, which leads to frequent disagreement among clinicians and sometimes misdiagnosis. The difficulties of TMJOA diagnosis include the subjectivity of radiographic interpretation and the pain threshold of an individual. The diagnosis itself may be the biggest barrier in creating biomarker disease profiles for diseased and healthy groups (Ebrahim et al. 2017). This study sample of biological and standardized DC/TMD clinical survey data consisted of only 17 TMJOA and 17 asymptomatic controls, while the NN TMJOA morphology disease staging used a larger training sample for which only imaging data and clinical diagnosis are available. A more accurate NN may be trained when larger standardized omics data become available. This study’s DSCI data management system has the capability to integrate, securely store, compute, and analyze all omics, beyond the clinical, biological, and 3D meshes morphology included in this study. The developments in this study and advances in data science in the TMJ health and disease field may aid researchers to gain further insight into biomarkers for diagnosis purposes to help guide treatment choices for TMJOA.
Previous studies have collected and evaluated specific synovial fluid biomarkers in the TMJ of subjects with internal joint derangement (Kubota et al. 1997; Srinivas et al. 2001; Tominaga et al. 2004; Yoshida et al. 2006). In this study, venipuncture, minimally invasive in comparison to arthrocentesis, was performed to evaluate biomarkers. While venipuncture involves insertion of a needle to collect blood from a vein, arthrocentesis involves intravenous or general anesthesia to be performed along with monitoring of respiratory drive throughout the procedure (Mehra and Arya 2015). Both methods may share similar complications in swelling, hematoma, and possible nerve damage (Galena 1992; Vaira et al. 2018); however, arthrocentesis is impractical in everyday practice.
This study established associations between the biochemical/clinical indicators and TMJOA morphology, as well as used a preliminary NN to link TMJOA morphology to clinical diagnoses. The associations computed with the MFSDA statistical modeling, as shown in Figure 3, were based on the diagnosis of TMJOA or asymptomatic control using the DC/TMD diagnostic criteria. Even though the current NN disease staging is limited by subjective clinician classification of 3D morphological variability and sole dependence on morphologic assessments, this study is the first to train a NN for TMJOA staging of severity of bone degenerative disease. Future studies will train the NN to also include biochemical/clinical indicators, as well as objective quantitative radiomic features of the subchondral bone structure, when larger samples including such data are collected. The future mining of high-dimensional clinical, biological, and imaging patient data has the potential to allow clinicians to address the heterogeneity among the TMJOA patients and guide personalized management of the disease.
Conclusions
Levels of VE-cadherin, VEGF, MMP-3, and CXCL16 in serum, as well as MMP-3, VE-cadherin, 6Ckine, and PAI-1 in saliva, were significantly correlated with specific regions of condylar morphological variability. The ranges of mouth opening were the clinical variables with most significant associations with morphological variability at the medial and lateral condylar poles. The NN presented a high degree of conformity in classifying and categorizing condyles based on the stage and degree of OA the mandibular condyle has exhibited.
Author Contributions
B. Shoukri, L. Cevidanes, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; J.C. Prieto, M. Styner, H. Zhu, C. Huang, B. Paniagua, contributed to design, data analysis, and interpretation, drafted the manuscript; A. Ruellas, contributed to design, data analysis, and interpretation, drafted and critically revised the manuscript; M. Yatabe, contributed to data analysis and interpretation, critically revised the manuscript; J. Sugai, contributed to design, data acquisition, and analysis, drafted the manuscript; S. Aronovich, E. Benavides, contributed to data acquisition and interpretation, critically revised the manuscript; L. Ashman, contributed to data acquisition, critically revised the manuscript; P. de Dumast, N.T. Ribera, C. Mirabel, L. Michoud, L.R. Gomes, contributed to data analysis and interpretation, drafted the manuscript; Z. Allohaibi, contributed to data analysis, drafted the manuscript; M. Ioshida, L. Bittencourt, contributed to data acquisition and analysis, drafted the manuscript; L. Fattori, contributed to data acquisition, drafted the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034519865187 for Minimally Invasive Approach for Diagnosing TMJ Osteoarthritis by B. Shoukri, J.C. Prieto, A. Ruellas, M. Yatabe, J. Sugai, M. Styner, H. Zhu, C. Huang, B. Paniagua, S. Aronovich, L. Ashman, E. Benavides, P. de Dumast, N.T. Ribera, C. Mirabel, L. Michoud, Z. Allohaibi, M. Ioshida, L. Bittencourt, L. Fattori, L.R. Gomes and L. Cevidanes in Journal of Dental Research
Acknowledgments
We are indebted and grateful to Dr. William V. Giannobile for the helpful discussions along with his support and guidance throughout the study. We sincerely thank all the subjects for participating in this study.
Footnotes
A supplemental appendix to this article is available online.
The research in this publication was supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health under grants R01DE024450 and R21DE025306.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
Research materials generated during and/or analyzed during this study are available from the corresponding author upon request.
References
- Berenbaum F. 2013. Osteoarthritis as an inflammatory disease (osteoarthritis is not osteoarthrosis!). Osteoarthritis Cartilage. 21(1):16–21. [DOI] [PubMed] [Google Scholar]
- Brechbühler C, Gerig G, Kübler O. 1995. Parametrization of closed surfaces for 3-D shape description. Comput Vis Image Underst. 61(2):154–170. [Google Scholar]
- Cevidanes LH, Walker D, Schilling J, Sugai J, Giannobile W, Paniagua B, Benavides E, Zhu H, Marron JS, Jung BT, et al. 2014. 3D osteoarthritic changes in TMJ condylar morphology correlates with specific systemic and local biomarkers of disease. Osteoarthritis Cartilage. 22(10):1657–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Dumast P, Mirabel C, Cevidanes L, Ruellas A, Yatabe M, Ioshida M, Ribera NT, Michoud L, Gomes L, Huang C. 2018. A web-based system for neural network based classification in temporomandibular joint osteoarthritis. Comput Med Imaging Graph. 67:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza RF, Lovato da, Silva CH, Nasser M, Fedorowicz Z, Al-Muharraqi MA. 2012. Interventions for managing temporomandibular joint osteoarthritis. Cochrane Database Syst Rev. 4:CD007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim FH, Ruellas AC, Paniagua B, Benavides E, Jepsen K, Wolford L, Goncalves JR, Cevidanes LH. 2017. Accuracy of biomarkers obtained from cone beam computed tomography in assessing the internal trabecular structure of the mandibular condyle. Oral Surg Oral Med Oral Pathol Oral Radiol. 124(6):588–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin J-C, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M. 2012. 3D slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging. 30(9):1323–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galena HJ. 1992. Complications occurring from diagnostic venipuncture. J Fam Pract. 34(5):582–584. [PubMed] [Google Scholar]
- Gauer RL, Semidey MJ. 2015. Diagnosis and treatment of temporomandibular disorders. Am Fam Physician. 91(6):378–386. [PubMed] [Google Scholar]
- Gomes LR, Gomes M, Jung B, Paniagua B, Ruellas AC, Gonçalves JR, Styner MA, Wolford L, Cevidanes L. 2015. Diagnostic index of three-dimensional osteoarthritic changes in temporomandibular joint condylar morphology. J Med Imaging (Bellingham). 2(3):034501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Thompson P, Wang Y, Yu Y, Zhang J, Kong D, Colen RR, Knickmeyer RC, Zhu H; Alzheimer’s Disease Neuroimaging Initiative. 2017. FGWAS: functional genome wide association analysis. NeuroImage. 159:107–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota E, Imamura H, Kubota T, Shibata T, Murakami KI. 1997. Interleukin 1β and stromelysin (MMP3) activity of synovial fluid as possible markers of osteoarthritis in the temporomandibular joint. J Oral Maxillofac Surg. 55:20–27. [DOI] [PubMed] [Google Scholar]
- LeCun Y, Bengio Y, Hinton G. 2015. Deep learning. Nature. 521(7553):436–444. [DOI] [PubMed] [Google Scholar]
- Lepage SI, Robson N, Gilmore H, Davis O, Hooper A, St. John S, Kamesan V, Gelis P, Carvajal D, Hurtig M. 2019. Beyond cartilage repair: the role of the osteochondral unit in joint health and disease. Tissue Eng Part B Rev. 25(2):114–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. 2018. Data mining and machine learning methods for dementia research. In: Perneczky RG, editor. Biomarkers for Alzheimer’s disease drug development. New York (NY): Humana Press. p. 363–370. [DOI] [PubMed] [Google Scholar]
- Ling SM, Patel DD, Garnero P, Zhan M, Vaduganathan M, Muller D, Taub D, Bathon JM, Hochberg M, Abernethy DR, et al. 2009. Serum protein signatures detect early radiographic osteoarthritis. Osteoarthritis Cartilage. 17(1):43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra P, Arya V. 2015. Temporomandibular joint arthrocentesis: outcomes under intravenous sedation versus general anesthesia. J Oral Maxillofac Surg. 73(5):834–842. [DOI] [PubMed] [Google Scholar]
- Nah K-S. 2012. Condylar bony changes in patients with temporomandibular disorders: a CBCT study. Imaging Sci Dent. 42(4):249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniagua B, Cevidanes L, Walker D, Zhu H, Guo R, Styner M. 2011. Clinical application of SPHARM-PDM to quantify temporomandibular joint osteoarthritis. Comput Med Imaging Graph. 35(5):345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniagua B, Pascal L, Prieto J, Vimort JB, Gomes L, Yatabe M, Ruellas AC, Budin F, Pieper S, Styner M, et al. 2017. Diagnostic index: an open-source tool to classify TMJ OA condyles. Proc SPIE Int Soc Opt Eng. 10137. pii: 101372H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Zhukov T, Song D, Tockman MS. 2007. Computerized analysis of cellular features and biomarkers for cytologic diagnosis of early lung cancer. Anal Quant Cytol Histol. 29(2):103–111. [PubMed] [Google Scholar]
- Rousseau JC, Delmas PD. 2007. Biological markers in osteoarthritis. Nat Clin Pract Rheumatol. 3(6):346–356. [DOI] [PubMed] [Google Scholar]
- Schiffman E, Ohrbach R, Truelove E, Look J, Anderson G, Goulet JP, List T, Svensson P, Gonzalez Y, Lobbezoo F, et al. 2014. Diagnostic criteria for temporomandibular disorders (DC/TMD) for clinical and research applications: recommendations of the international RDC/TMD consortium network and orofacial pain special interest group. J Oral Facial Pain Headache. 28(1):6–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling J, Gomes LC, Benavides E, Nguyen T, Paniagua B, Styner M, Boen V, Goncalves JR, Cevidanes LH. 2014. Regional 3D superimposition to assess temporomandibular joint condylar morphology. Dentomaxillofac Radiol. 43(1):20130273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Jagga S, Lee S-S, Nam J-S. 2013. Interplay between cartilage and subchondral bone contributing to pathogenesis of osteoarthritis. Int J Mol Sci. 14(10):19805–19830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas R, Sorsa T, Tjäderhane L, Niemi E, Raustia A, Pernu H, Teronen O, Salo T. 2001. Matrix metalloproteinases in mild and severe temporomandibular joint internal derangement synovial fluid. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 91(5):517–525. [DOI] [PubMed] [Google Scholar]
- Stehman SV. 1997. Selecting and interpreting measures of thematic classification accuracy. Remote Sens Environ. 62(1):77–89. [Google Scholar]
- Styner M, Oguz I, Xu S, Brechbuhler C, Pantazis D, Levitt JJ, Shenton ME, Gerig G. 2006. Framework for the statistical shape analysis of brain structures using SPHARM-PDM. Insight J. 1071:242–250. [PMC free article] [PubMed] [Google Scholar]
- Su N, Liu Y, Yang X, Luo Z, Shi Z. 2014. Correlation between bony changes measured with cone beam computed tomography and clinical dysfunction index in patients with temporomandibular joint osteoarthritis. J CranioMaxillofac Surg. 42:1402–1407. [DOI] [PubMed] [Google Scholar]
- Tanaka E, Detamore MS, Mercuri LG. 2008. Degenerative disorders of the temporomandibular joint: etiology, diagnosis, and treatment. J Dent Res. 87(4):296–307. [DOI] [PubMed] [Google Scholar]
- Tominaga K, Habu M, Sukedai M, Hirota Y, Takahashi T, Fukuda J. 2004. IL-1β, IL-1 receptor antagonist and soluble type II IL-1 receptor in synovial fluid of patients with temporomandibular disorders. Arch Oral Biol. 49(6):493–499. [DOI] [PubMed] [Google Scholar]
- Vaira LA, Raho MT, Soma D, Salzano G, Dell’aversana Orabona G, Piombino P, De Riu G. 2018. Complications and post-operative sequelae of temporomandibular joint arthrocentesis. Cranio. 36(4):264–267. [DOI] [PubMed] [Google Scholar]
- VTK.org. Vtkperlinnoise class reference. 2017. [accessed 2017. July 24]. http://www.vtk.org/doc/nightly/html/classvtkPerlinNoise.html.
- Wadhwa S, Kapila S. 2008. TMJ disorders: future innovations in diagnostics and therapeutics. J Dent Educ. 72(8):930–947. [PMC free article] [PubMed] [Google Scholar]
- Yan W, Apweiler R, Balgley BM, Boontheung P, Bundy JL, Cargile BJ, Cole S, Fang X, Gonzalez-Begne M, Griffin TJ. 2009. Systematic comparison of the human saliva and plasma proteomes. Proteomics Clin Appl. 3(1):116–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Takatsuka S, Hatada E, Nakamura H, Tanaka A, Ueki K, Nakagawa K, Okada Y, Yamamoto E, Fukuda R. 2006. Expression of matrix metalloproteinases and aggrecanase in the synovial fluids of patients with symptomatic temporomandibular disorders. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 102(1):22–27. [DOI] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. 2006. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 31(3):1116–1128. [DOI] [PubMed] [Google Scholar]
- Zhu H, Li R, Kong L. 2012. Multivariate varying coefficient model for functional responses. Ann Stat. 40(5):2634–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034519865187 for Minimally Invasive Approach for Diagnosing TMJ Osteoarthritis by B. Shoukri, J.C. Prieto, A. Ruellas, M. Yatabe, J. Sugai, M. Styner, H. Zhu, C. Huang, B. Paniagua, S. Aronovich, L. Ashman, E. Benavides, P. de Dumast, N.T. Ribera, C. Mirabel, L. Michoud, Z. Allohaibi, M. Ioshida, L. Bittencourt, L. Fattori, L.R. Gomes and L. Cevidanes in Journal of Dental Research