Abstract
Oral mucosa provides the first line of defense against a diverse array of environmental and microbial irritants by forming the barrier of epithelial cells interconnected by multiprotein tight junctions (TJ), adherens junctions, desmosomes, and gap junction complexes. Grainyhead-like 2 (GRHL2), an epithelial-specific transcription factor, may play a role in the formation of the mucosal epithelial barrier, as it regulates the expression of the junction proteins. The current study investigated the role of GRHL2 in the Porphyromonas gingivalis (Pg)–induced impairment of epithelial barrier functions. Exposure of human oral keratinocytes (HOK-16B and OKF6 cells) to Pg or Pg-derived lipopolysaccharides (Pg LPSs) led to rapid loss of endogenous GRHL2 and the junction proteins (e.g., zonula occludens, E-cadherin, claudins, and occludin). GRHL2 directly regulated the expression levels of the junction proteins and the epithelial permeability for small molecules (e.g., dextrans and Pg bacteria). To explore the functional role of GRHL2 in oral mucosal barrier, we used a Grhl2 conditional knockout (KO) mouse model, which allows for epithelial tissue-specific Grhl2 KO in an inducible manner. Grhl2 KO impaired the expression of the junction proteins at the junctional epithelium and increased the alveolar bone loss in the ligature-induced periodontitis model. Fluorescence in situ hybridization revealed increased epithelial penetration of oral bacteria in Grhl2 KO mice compared with the wild-type mice. Also, blood loadings of oral bacteria (e.g., Bacteroides, Bacillus, Firmicutes, β-proteobacteria, and Spirochetes) were significantly elevated in Grhl2 KO mice compared to the wild-type littermates. These data indicate that Pg bacteria may enhance paracellular penetration through oral mucosa in part by targeting the expression of GRHL2 in the oral epithelial cells, which then impairs the epithelial barrier by inhibition of junction protein expression, resulting in increased alveolar tissue destruction and systemic bacteremia.
Keywords: oral mucosa, periodontal diseases, tight junctions, lipopolysaccharides, cell adhesion, gene knockout techniques
Introduction
Oral epithelium provides physical, chemical, and immunological barriers against microbial challenges in the oral cavity (Fujita et al. 2018). Oral epithelial cells are generally interconnected by tight junctions (TJs), adherens junctions, desmosomes, and gap junctions, providing the epithelial barrier (Meyle et al. 1999; Boivin and Schmidt-Ott 2017). Defects in the oral epithelial barrier may increase susceptibility to microbial infection, leading to local tissue destruction and systemic diseases (Nagpal et al. 2015). Hence, understanding the mechanisms that establish the epithelial barrier is important in elucidating the pathogenesis of oral inflammatory diseases linked to bacterial infection, such as periodontal diseases. Literatures indicate that Grainyhead-like (Grhl) family genes (e.g., Grhl1, Grhl2, and Grhl3), which determine epithelial phenotype, including cell polarity, motility, and differentiation, also determine epithelial barrier (Wang and Samakovlis 2012; Mlacki et al. 2014; Aue et al. 2015; Sumigray and Lechler 2015). In particular, our laboratory has shown that GRHL2 plays a unique role in the control of epithelial cell proliferation and differentia-tion, as well as epithelial plasticity, through transcriptional regulation of a diverse array of target genes, including p63, E-cadherin (E-cad), and microRNA-200 family (Mehrazarin et al. 2015; Chen et al. 2016). Thus, GRHL2 may be a determinant of the oral mucosal barrier and be a target of periodontal pathogens to initiate bacterial invasion.
Periodontal disease is a bacterial infectious disease associated with a wide diversity of bacterial pathogens. It initiates with accumulation of dental plaque in the gingival sulcus, which leads to release of microbial virulence factors and toxins (e.g., lipopolysaccharide [LPS]), through oral epithelium to stimulate the inflammatory process and tissue destruction (Ji et al. 2015). Loss of epithelial integrity is linked with microbial penetration into oral epithelial and underlying connective tissues (Ji et al. 2015). Prior studies showed that periodontal pathogens, like Porphyromonas gingivalis (Pg), abrogate the epithelial barrier in part through proteolytic degradation of the junction proteins (Katz et al. 2000, 2002; Groeger et al. 2010; Groeger and Meyle 2015) or through downregulation of TJ protein expression (Choi et al. 2014). The detailed molecular mechanism by which Pg or other invasive oral bacteria abrogates the mucosal barrier still remains unclear.
The current study investigated the mechanisms by which periodontal pathogens evade the oral mucosal barrier to cause tissue destruction. To that end, we used cultured Pg bacteria to determine its effects on GRHL2 and the junction protein expression in oral epithelial cells. Our data indicated that Pg inoculation induced precipitous loss of GRHL2 and the junction protein expression, which would impair the mucosal barrier effects. To delineate the functional role of GRHL2 in oral mucosal barrier, we used the Grhl2 conditional knockout (cKO) mouse model (Chen et al. 2018). Grhl2 knockout (KO) led to reduced expression of the junction proteins in the junctional epithelium and increased alveolar bone loss in the ligature-induced periodontitis model. Interestingly, Grhl2 KO allowed for increased penetration of oral bacteria and bacterial load in the systemic blood circulation compared to the wild-type (WT) mice. These data suggest that Pg bacteria may gain access through the mucosal barrier in part by targeting GRHL2, which is required for the oral mucosal barrier by maintaining the expression of the junction proteins.
Materials and Methods
Cells and Cell Culture
Primary NHKs were established from the oral epithelium of normal mucosa obtained from healthy patients using methods published elsewhere (Kang et al. 2000). NHK, HOK-16B, OKF6, and HOK-SI cells were cultured in keratinocyte growth medium (KGM) (Lonza) with 60 µM Ca++ (Kang et al. 2000). To overexpress GRHL2, we used a retroviral vector (LXSN-GRHL2) expressing GRHL2 according to methods described elsewhere (Chen et al. 2016). Endogenous GRHL2 was knocked down in HOK-16B using commercial GRHL2 shRNA vectors (ShGRHL2-1, -2) targeting different Grhl2 sequences (Origene).
Fn strain (ATCC 23726) was maintained on Columbia agar supplemented with 5% sheep blood or in Columbia broth (CB; Difco) under anaerobic conditions (10% H2, 10% CO2, 80% N2) at 37 °C. Pg (ATCC 33277) was maintained in the Columbia broth supplemented with hemin at 5 µg/mL and menadione at 1 µg/mL. Thiamphenicol at 5 µg/mL and clindamycin at 1 µg/mL (MP Biomedicals) were used for the selection and maintenance of strains possessing the catP and ermB determinants, respectively.
Generation of Mouse Periodontitis Model Using GRHL2 cKO Mice
Animal experiments were performed according to the protocol approved by the UCLA Institutional Animal Care and Use Committee. Grhl2 cKO was achieved with intraperitoneal (IP) administration of tamoxifen (Tmx) (75 mg/kg) in 8-wk-old K14/Grhl2 cKO mice for 7 d to induce homozygous deletion of Grhl2 in epithelial tissues, as described in our previous study (Chen et al. 2018). To induce experimental periodontitis, ligatures were placed around the left maxillary second molars using a 5-0 silk suture (Teleflex) with a surgical knot (Abe and Hajishengallis 2013). After 21 d, the mice were sacrificed, and maxillae were collected for µCT and immunohistochemical analyses, as previously described (Williams et al. 2014; Kim et al. 2018). To assess Pg bacterial penetration through oral mucosa, Grhl2 WT, heterozygous, and KO mice were orally inoculated with 1 × 109 Pg bacteria once every other day for 6 wk. Whole blood was collected through cardiac puncture, and genomic DNA was extracted to assess Pg bacterial loading in blood by PCR.
Detection of Bacteria by FISH
In situ hybridization was performed on formalin-fixed and paraffin-embedded tissue sections using AlexaFluor 488– labeled oligonucleotide probes (Alex488-POGI) specific for Pg 5′- CAATA CTCGTATCGCCCGTTATTC-3′ (Sunde et al. 2003). FISH was then performed according to the published methods (Rivera et al. 2013) using deparaffinized tissue sections of oral mucosal tissues with and without Pg inoculation. Stained slides were dried overnight and viewed using the Olympus FV101 confocal microscope.
Epithelial Permeability Assay
HOK-16B cells with or without GRHL2 knockdown or HOK-SI cells with or without GRHL2 overexpression were seeded on transwell culture inserts (Merck Millipore) and grown in KGM until confluence. The confluent monolayers were incubated for another 24 h in KGM in the absence or presence of different concentrations of Pg LPS (1 µg/mL and 5 µg/mL). Then, 5 µL of 10 mg/mL FITC-dextran (Sigma) was added to the upper compartment of the transwell chamber, and the fluorescence signal in the bottom compartment was determined at various time points using an Omega plate reader (BMG Labtech) at 485/520 nm. We also determined the epithelial permeability of Pg through the HOK-16B monolayer with or without GRHL2 knockdown in the transwell chamber system. After Pg (multiplicity of infection [MOI] = 100) was inoculated in the upper chamber, 20 µL medium from the bottom chamber was collected for quantitative PCR using the specific primers for Pg to investigate the level of Pg (Oppong et al. 2013; Abe-Yutori et al. 2017).
Reagents
The following antibodies were used in this study: glyceraldehyde 3-phosphate dehydrogenase (GAPDH), E-cad, β-cat, and occludin (Santa Cruz Biotech); p-p65 and TNF-α (Cell Signaling Technology); and CLDN-1, -3, and -4 and ZO-1 antibodies (Invitrogen). Alex488-POGI was purchased from Integrated DNA Technologies; anti-GRHL2 antibody and FITC-dextran were from Sigma-Aldrich. Pg LPS was purchased from InvivoGen.
Statistical Analysis
Statistical analysis was performed using the Student’s t test (2-tailed) for the quantitative reverse transcription PCR gene expression, Western blotting, and immunostaining experiments. P values <0.05 were considered significant. All data are expressed as mean ± SD. The number of mice in each group was determined by phenotypic changes observed with Grhl2 deficiency in the cell lines (Kang et al. 2009; Chen et al. 2016).
Results
Exposure of Human Oral Keratinocytes to Pg Inhibits GRHL2 and Intercellular Adhesion Molecule Expression
To investigate the role of GRHL2 in oral mucosal barrier, we first tested the effects of Pg inoculation on the expression of GRHL2, adherens complex (E-cad and β-catenin [β-cat]), and TJ proteins (e.g., claudins [CLDNs] and zonula occludens [ZO]). Within hours after bacterial inoculation, the levels of GRHL2, the adherens, and TJ proteins were strongly suppressed in cultured human oral keratinocytes (HOKs) (Fig. 1A). We further tested the effect of Pg inoculation on the junction gene expression in the presence or absence of another periodontal pathogen, Fusobacterium nucleatum (Fn) (Fig. 1B). GRHL2, E-cad, and β-cat levels as well as the TJ gene expression levels were significantly suppressed by Pg alone or in combination with Fn, but Fn inoculation alone elicited mild effects. To determine the kinetics of these protein expression changes exerted by Pg, we performed Western blotting within minutes after bacterial inoculation. The suppression of GRHL2 and ZO-1 by Pg was detectable rapidly within 15 min of bacterial inoculation, and the suppressive effect progressively increased over the 60-min period (Fig. 1C). This effect was specific to Pg because Fn inoculation did not provoke any effect on GRHL2 or junction protein levels.
Figure 1.
Cellular exposure to Porphyromonas gingivalis (Pg) suppressed the expression of Grainyhead-like 2 (GRHL2) and junction proteins in cultured oral keratinocytes. (A) Western blotting was performed with HOK-16B and OKF6 cells after exposure to Pg bacteria to assess the levels of GRHL2 and junction proteins, zonula occludens (ZO)–1, β-catenin (β-cat), and E-cadherin (E-cad). (B) Quantitative reverse transcription polymerase chain reaction was performed with HOK-16B exposed to Pg (multiplicity of infection [MOI] = 100 or 200) alone or in combination with Fusobacterium nucleatum (Fn) (MOI = 100) for 24 h to assess the messenger RNA levels of GRHL2 and various junction molecules (e.g.,E-cad; β-cat; claudin (CLDN)–1, -3, -4; and ZO-1, -2, -3). Error bars indicate mean/SD. *Statistically significant (P < 0.05) compared with control cells. (C) HOK-16B cells were inoculated with Pg, Fn, or combined Pg/Fn at the indicated MOI and harvested at 15, 30, 45, or 60 min after inoculation. Western blotting was performed for GRHL2, ZO-1, β-cat, E-cad, and claudins. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was probed for loading control.
To assess whether the diminution of GRHL2 expression and the junction protein upon Pg inoculation was linked to inflammatory signaling, we exposed cells to purified LPS from Pg and performed Western blotting. Pg LPS exposure readily induced the inflammatory signaling, as evinced by increasing tumor necrosis factor–α (TNF-α) and phosphor-p65 levels, and suppressing the levels of GRHL2 and the junction proteins (Fig. 2A, B). Interestingly, overexpression of GRHL2 ameliorated the inhibitory effects of Pg LPS on the expression of GRHL2 and the junction proteins in HOK-SI cells (Fig. 2C). To explore the role of GRHL2 in epithelial barrier function, we assessed the permeability of a small-molecule, fluorescein isothiocyanate (FITC)–conjugated dextran (4 kDa) across the confluent monolayer of HOK-SI cells with or without GRHL2 overexpression. With ectopic expression of GRHL2 in HOK-SI cells, the level of FITC-dextran penetration was markedly decreased, reflecting reduced epithelial permeability (Fig. 2D). Increased FITC-dextran penetration was observed through the control HOK-SI cells exposed to Pg LPS, while GRHL2 overexpression inhibited Pg LPS-induced epithelial permeability of FITC-dextran. These data indicate that GRHL2 is required for the maintenance of the epithelial barrier and that Pg impairs epithelial barrier by targeting GRHL2.
Figure 2.
Grainyhead-like 2 (GRHL2) and junction protein levels were lost in oral keratinocytes exposed to Porphyromonas gingivalis (Pg) lipopolysaccharide (LPS). (A) HOK-16B cells were exposed to Pg LPS (5 µg/mL) for up to 24 h, and Western blotting was performed for GRHL2, E-cadherin (E-cad), claudin (CLDN)–1, CLDN-3, CLDN-4, and occludin (OCLN) protein levels. We also probed for p-p65 and tumor necrosis factor–α (TNF-α) in the cells to assess the level of inflammatory signaling after Pg LPS treatment. HOK-16B cells with Grhl2 knockdown by ShRNA (HOK-16B/ShGRHL2) were included as the positive control in which endogenous GRHL2 was abrogated. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used for loading control. (B) Quantitative reverse transcription polymerase chain reaction was performed for gene expression levels of Grhl2 and junction molecule genes (e.g., CLDN-1, -3, -4 and OCLN) in HOK-16B exposed to Pg LPS (5 µg/mL) for various time points. (C) HOK-SI with GRHL2 overexpression (LXSN-GRHL2) and control cells transfected with empty viral vector (LXSN) were exposed to Pg LPS (5 µg/mL) for up to 24 h, and Western blotting was performed for GRHL2, E-cad, CLDN-3, CLDN-4, and OCLN protein levels. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used for loading control. (D) Permeability of FITC-dextran across a confluent HOK-SI monolayer was determined using the cells with GRHL2 overexpression (LXSN-GRHL2 vs. LXSN) or exposed to Pg LPS (5 µg/mL) for 24 h. Error bars indicate mean/SD. *Statistically significant (P < 0.05) compared with the control cells.
GRHL2 Is Required for Oral Epithelial Barrier Function
The functional role of GRHL2 in the oral epithelial barrier was further explored through GRHL2 knockdown in HOK-16B cells, in which E-cad, CLDN-1, CLDN-3, CLDN-4, OCLN, and ZO-1 were suppressed (Fig. 3A, B). On the contrary, when GRHL2 was overexpressed in normal human keratinocytes (NHKs), gene expression of junction molecules was strongly induced (Fig. 3C, Appendix Fig. 1). Immunofluorescent staining (IFS) revealed that the loss of GRHL2 in HOK-16B cells ameliorated the expression of E-cad, ZO-1, and CLDN-4 (Fig. 3D). Hence, GRHL2 appears to directly regulate the expression of the junction molecules in HOKs. Then the permeability of FITC-dextran through the monolayer of HOK-16B cells with or without GRHL2 knockdown was evaluated. The level of fluorescent signal was markedly increased through the confluent monolayer of HOK-16B cells with GRHL2 knockdown, reflecting increased epithelial permeability. Similarly, increased FITC-dextran penetration was observed through the epithelial cells after exposure to Pg LPS (Fig. 3E). We also compared the penetration of Pg across the monolayer of oral epithelial cells with or without GRHL2 knockdown by detecting the presence of Pg in the bottom compartment of the transwell after inoculation of Pg in the upper chamber. With GRHL2 knockdown in HOK-16B cells, Pg penetration began to increase in 6 h after Pg inoculation and significantly increased by 24 h (Fig. 3F). These data indicate the functional significance of GRHL2 in maintaining the epithelial barrier of oral keratinocytes against small molecules and Pg in vitro.
Figure 3.
Grainyhead-like 2 (GRHL2) is required for the junction protein expression and the maintenance of epithelial barrier function in oral epithelial cells. (A) GRHL2 was knocked down in HOK-16B cells by 2 independent Grhl2 ShRNAs (Sh1 and Sh2). Western blotting was performed to assess the level of GRHL2, claudin (CLDN)–1, and CLDN-4. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was probed for loading control. (B) In HOK-16B cells with GRHL2 knockdown, quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed to assess the level of GRHL2, β-catenin (β-cat), E-cadherin (E-cad), CLDN-1, CLDN-3, CLDN-4, occludin (OCLN), and zonula occludens (ZO)–1 messenger RNA (mRNA). (C) qRT-PCR was performed with total RNAs isolated from normal human keratinocyte (NHK)/GRHL2 or NHK/LXSN to assess the mRNA levels of junction molecules (β-cat; E-cad; CLDN-1, -3, -4; OCLN; and ZO-1). (D) Immunofluorescent staining was performed to examine GRHL2 and junction protein (E-cad, ZO-1, and CLDN-4) expression in HOK-16B cells with or without GRHL2 knockdown. (E) Permeability of FITC-dextran across confluent HOK-16B monolayer was determined using the cells exposed to Porphyromonas gingivalis (Pg) LPS (1 or 5 µg/mL) for 24 h or the cells with GRHL2 knockdown (ShGRHL2 vs. ShContr). Error bars indicate mean/SD. *Statistically significant (P < 0.05) compared with the control cells. (F) Permeability of Pg across HOK-16B cell monolayer with or without GRHL2 knockdown was determined at different time intervals after Pg bacterial inoculation using the transwell culture system. Pg bacterial content in the bottom chamber was determined by quantitative polymerase chain reaction using Pg-specific primer sets. Error bars indicate mean/SD. *Statistically significant (P < 0.05) compared with the control cells.
GRHL2 Is Required for the Maintenance of Oral Mucosal Barrier In Vivo
To evaluate the role of GRHL2 in oral mucosal barrier function in vivo, we generated Grhl2 cKO mice by crossing Grhl2 floxed (fl/fl) mice with K14-CreERT mice, which allowed inducible Grhl2 KO in epithelial tissues through intraperitoneal (IP) administration of tamoxifen (Tmx). The resulting Grhl2 KO mice revealed absence of GRHL2 expression and downregulation of the adhesion molecules (e.g., β-cat; CLDN-1, -3, -4; and ZO-1) in the epidermis (Fig. 4A–C). Next, we analyzed if there is difference in the oral microbial loads on the surface of oral mucosa or in the blood between Grhl2 KO and WT mice. The relative microbial loads on the buccal mucosa or tongue were randomly distributed between the Grhl2 WT and KO mice (Appendix Fig. 2). However, the microbial loads in the blood were consistently increased in the Grhl2 KO mice compared to the WT controls (Fig. 4D). These data suggested the possibility that the oral mucosa in Grhl2 KO mice was more permeable to oral microbiota compared to the WT controls.
Figure 4.
Grainyhead-like 2 (Grhl2) knockout (KO) suppresses the gene expression of junction proteins in vivo. (A) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed with total RNAs isolated from Grhl2 wild-type (WT), Grhl2 heterozygote (+/–), and Grhl2 KO mice epidermis for junction molecules (e.g., β-catenin; claudin [CLDN]–1, -3, -4; and zonula occludens [ZO]–1). Data were derived from 3 independent experiments and qRT-PCR assays were performed in triplicates. (B) Western blotting was performed with epidermal tissues isolated from Grhl2 WT and Grhl2 KO mice for GRHL2 and the junction proteins, CLDN-1, -3, and -4. (C) Western blotting signals were quantitated by densitometric analysis and plotted with the mean values for Grhl2 WT and KO mice groups. (D) Quantitative polymerase chain reaction was performed to assess the level of oral bacterial species, Bacteroidetes, Firmicutes, Spirochetes, β-proteobacteria, Bacillus, and Lactobacillus, in the whole blood harvested from Grhl2 WT and KO mice. Bars indicate mean/SD. *Statistically significant (P < 0.05) compared with WT mice.
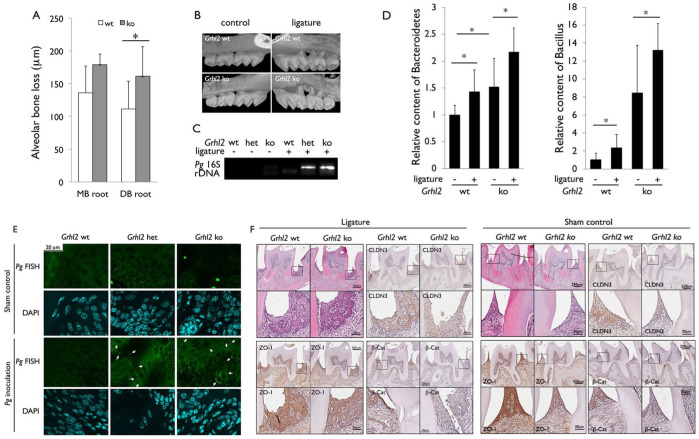
We then determined the effect of Grhl2 KO on alveolar bone loss in the experimental periodontitis model by placement of ligature around maxillary second molars. After 21 d of ligature placement, we compared the level of alveolar bone loss by micro–computed tomography (µCT) of the dentoalveolar complex and found that Grhl2 KO led to increased alveolar bone loss compared to the WT littermates (Fig. 5A, B). We also compared the blood level of Pg in Grhl2 WT, hetero (+/–), and KO mice after intraoral inoculation of the bacteria by polymerase chain reaction (PCR) using primers against Pg-specific 16S ribosomal DNA (rDNA). Compared to Grhl2 WT mice, those with Grhl2 hetero or KO genotype revealed increased Pg content in the blood after the ligature placement (Fig. 5C). Also, the blood levels of Bacteroides and Bacillus were significantly elevated in the Grhl2 KO mice compared with the WT mice with or without the ligature placement (Fig. 5D). Fluorescence in situ hybridization (FISH) using the Pg-specific probe revealed increased bacterial presence in the tongue epithelium from Grhl2 KO or hetero (+/–) mice inoculated with Pg compared to WT mice (Fig. 5E). These results indicate that Grhl2 KO damaged the oral mucosal barrier and led to increased penetration of Pg into the oral mucosa. Immunohistochemical staining revealed reduced protein levels of CLDN-3, ZO-1, and β-cat at the junctional epithelium in the Grhl2 KO mice compared with the WT littermates (Fig. 5F). Collectively, these data suggest that the loss of the junction protein expression in Grhl2 KO mice impaired the oral epithelial barrier function, leading to increased bacterial penetration in local tissues and in systemic circulation.
Figure 5.
Grainyhead-like 2 (Grhl2) knockout (KO) in oral epithelium led to increased alveolar bone loss and bacterial penetration to systemic circulation in the ligature-induced periodontitis mouse model. (A, B) Mice with ligatures placed around the maxillary second molars for 3 wk showed increased alveolar bone loss in Grhl2 KO mice compared to Grhl2 wild-type (WT). After 3 wk, micro–computed tomography images of maxillary molars from the mice were obtained and reconstructed. DB, distobuccal; MB, mesiobuccal. n = 5. (C) Whole blood was collected by cardiac puncture to assess the level of Porphyromonas gingivalis (Pg) bacteria in Grhl2 WT and KO mice, which were orally inoculated with 1 × 109 live Pg once every 2 d for 6 wk; n = 3. (D) Genomic DNA was extracted from the whole blood collected from Grhl2 WT and KO mice at 3 wk after the placement of the ligature, and quantitative polymerase chain reaction was performed to assess the levels of bacterial species of Bacteroidetes and Bacillus. (E) Pg-specific fluorescence in situ hybridization (FISH) was performed using tongue epithelia from Grhl2 WT, hetero (+/–), and KO mice after Pg inoculation in the oral cavity. White arrows indicate the FISH signals representing interepithelial Pg penetration. (F) Representative hematoxylin and eosin staining of the maxillary second molars with the junctional epithelium was shown, alongside with the immunohistochemical staining for claudin (CLDN)–3, zonula occludens (ZO)–1, and β-catenin (β-cat) in the junctional epithelium from the Grhl2 WT and KO mice with ligatures (or sham control) placed around the maxillary second molars for 3 wk.
Discussion
The current study elucidated a novel mechanism by which microbial pathogens alter the oral mucosal barrier and enhance inflammatory bone destruction at the alveolar crest. Our data indicated that pathogenic bacteria, such as Pg, effectively suppressed the expression of junction proteins in part through downregulation of GRHL2. This was demonstrated in our in vitro experiments in which inoculation of cells with live Pg led to precipitous loss of the junction proteins along with loss of GRHL2 level. Interestingly, this suppressive effect was specific to Pg, while similar inoculation with Fn yielded mild or no effects. Loss of GRHL2 and junction protein levels was also induced by Pg LPS. Thus, the suppressive effect of Pg on GRHL2 and junction protein may in part be linked to proinflammatory effects as well as other unknown mechanisms elicited by Pg.
The functional role of GRHL2 in the regulation of junction proteins was demonstrated in HOK-16B cells with GRHL2 knockdown, which revealed concomitant loss of gene expression of the junction molecules, as well as increased penetration of Pg and small molecules. This was also confirmed in our in vivo model, in which we conditionally knocked out Grhl2 in oral epithelium (Chen et al. 2018). Grhl2 KO led to decreased expression of CLDN-1, CLDN-3, CLDN-4, and ZO-1 and increased blood loading of oral bacteria. Consequently, the Grhl2 KO mice showed increased alveolar bone loss compared with the WT littermates in the ligature-induced periodontitis model. Immunohistochemical assessment of the periodontal lesions confirmed decreased protein levels of ZO-1, β-cat, and CLDN-3 in the junctional epithelium of Grhl2 KO mice compared with the WT, suggesting an impaired mucosal barrier that led to increased bacterial penetration with Grhl2 KO. These findings are in keeping with earlier studies that showed increased bacterial penetration in gingival tissues with periodontal disease. Saglie et al. (1986) reported abundance of Bacteroides and Capnocytophaga species in advanced periodontitis, as well as Actinobacillus actinomycetemcomitans in juvenile periodontitis, while healthy gingiva lacked bacterial presence in situ. A subsequent study used immunohistochemical staining of Pg-specific thiol proteinase protein as a marker of the bacteria to show increased intracellular Pg penetration in the gingival epithelial cells of patients with chronic periodontitis (Rautemaa et al. 2004). Our current study primarily deals with the role of GRHL2 in regulation of junction protein expression in light of establishing the oral mucosal barrier.
Although it is unknown whether GRHL2 alters intracellular invasion of pathogenic bacteria into epithelial cells, maintenance of intact junction protein expression by GRHL2 appears to be critical to impede paracellular bacterial invasion. Prior studies have shown that pathogenic oral bacteria have also devised other unique mechanisms to abrogate the cell adhesion complexes to promote invasion through oral mucosa. Some Pg strains release gingipains to trigger proteolytic degradation of epithelial cell junction complexes (e.g., occludin and E-cad), thereby aiding in paracellular bacterial penetration through the oral mucosa (Katz et al. 2000, 2002). The functional role of the gingipains in altering the mucosal barrier was demonstrated by use of the mutant Pg strain defective in gingipain expression (KDP136), which led to a reduced inhibitory effect on the epithelial barrier (Groeger et al. 2010). Our current study used Pg strain ATCC33277, which is gingipain positive. Thus, it remains to be determined whether Pg abolishes GRHL2 expression in a gingipain-dependent manner or whether a Pg inhibitory effect on GRHL2 is additive to gingipain’s proteolysis of TJ proteins to impair the mucosal barrier. Likewise, Bacteroides fragilis, a common colonic microflora, is known to abrogate the intestinal barrier function by proteolytic degradation of TJ proteins, like ZO-1, through release of its toxin fragilysin (Obiso et al. 1997). As such, alteration of TJ complex stability has a significant impact on the epithelial barrier functions against a microbial challenge.
Increased microbial penetration would have profound health effects that may result in local destruction of alveolar tissues and systemic pathologies. Increased periodontal bone loss was evinced in the mouse model of Grhl2 KO, which led to significant downregulation of junction proteins (e.g., CLDN-1, CLDN-3, CLDN-4, and ZO-1), suggesting that local microbial infiltration may facilitate tissue damage. Also, our current bacteriological studies revealed increased abundance of oral microflora in systemic blood circulation in the Grhl2 KO mice compared to the WT controls. These data indicated that the oral mucosa in Grhl2 KO mice became more permeable to oral microbiota compared to the WT mice, although there remains the possibility that the microbial permeability in other mucosal epithelia (e.g., intestinal mucosa) may similarly be affected by Grhl2 KO. Further experiments will test these possibilities. Beyond the local alveolar tissue destruction, increased bacteremia through epithelial barriers has broad pathological consequences in systemic diseases. The current study highlights the crucial role of GRHL2 in establishing the oral mucosal barrier in part through regulation of junction protein expression. Interestingly, our study revealed for the first time that some periodontal pathogens like Pg inhibit the junction protein expression by targeting GRHL2 and abrogate the epithelial barrier. Further research will elucidate the mechanism by which microbial challenges regulate GRHL2 expression and oral mucosal barrier functions, in light of its importance in preserving the local alveolar tissues and systemic health.
Author Contributions
W. Chen, M.K. Kang, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; A. Alshaikh, contributed to design, data acquisition, analysis, and interpretation, critically revised the manuscript; S. Kim, J. Kim, C. Chun, S. Mehrazarin, J. Lee, R. Lux, R.H. Kim, K.H. Shin, N.H. Park, K. Walentin, K.M. Schmidt-Ott, contributed to design, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034519865184 for Porphyromonas gingivalis Impairs Oral Epithelial Barrier through Targeting GRHL2 by W. Chen, A. Alshaikh, S. Kim, J. Kim, C. Chun, S. Mehrazarin, J. Lee, R. Lux, R.H. Kim, K.H. Shin, N.H. Park, K. Walentin, K.M. Schmidt-Ott and M.K. Kang in Journal of Dental Research
Acknowledgments
The authors also thank Dr. Paulo Camargo for critical review of the manuscript.
Footnotes
A supplemental appendix to this article is available online.
This work was supported in part by the grants from the National Institute of Dental and Craniofacial Research (NIDCR)/National Institutes of Health (NIH) (1R56DE024593, R03DE024259, R21DE028269). M.K. Kang is also supported by the Jack A. Weichman Endowed Fund. K.M. Schmidt-Ott is supported by the German Research Foundation (Deutsche Forschungsgemeinschaft) and the Urological Research Foundation (Stiftung Urologische Forschung).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
ORCID iD: K.M. Schmidt-Ott  https://orcid.org/0000-0002-7700-7142
https://orcid.org/0000-0002-7700-7142
References
- Abe T, Hajishengallis G. 2013. Optimization of the ligature-induced periodontitis model in mice. J Immunol Methods. 394(1–2):49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe-Yutori M, Chikazawa T, Shibasaki K, Murakami S. 2017. Decreased expression of E-cadherin by Porphyromonas gingivalis–lipopolysaccharide attenuates epithelial barrier function. J Periodontal Res. 52(1):42–50. [DOI] [PubMed] [Google Scholar]
- Aue A, Hinze C, Walentin K, Ruffert J, Yurtdas Y, Werth M, Chen W, Rabien A, Kilic E, Schulzke JD, et al. 2015. A grainyhead-like 2/ovo-like 2 pathway regulates renal epithelial barrier function and lumen expansion. J Am Soc Nephrol. 26(11):2704–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin FJ, Schmidt-Ott KM. 2017. Transcriptional mechanisms coordinating tight junction assembly during epithelial differentiation. Ann NY Acad Sci. 1397(1):80–99. [DOI] [PubMed] [Google Scholar]
- Chen W, Kang KL, Alshaikh A, Varma S, Lin YL, Shin KH, Kim R, Wang CY, Park NH, Walentin K, et al. 2018. Grainyhead-like 2 (GRHL2) knockout abolishes oral cancer development through reciprocal regulation of the map kinase and TGF-beta signaling pathways. Oncogenesis. 7(5):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Yi JK, Shimane T, Mehrazarin S, Lin YL, Shin KH, Kim RH, Park NH, Kang MK. 2016. Grainyhead-like 2 regulates epithelial plasticity and stemness in oral cancer cells. Carcinogenesis. 37(5):500–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Kim YC, Ji S, Choi Y. 2014. Increased bacterial invasion and differential expression of tight-junction proteins, growth factors, and growth factor receptors in periodontal lesions. J Periodontol. 85(8):e313–e322. [DOI] [PubMed] [Google Scholar]
- Fujita T, Yoshimoto T, Kajiya M, Ouhara K, Matsuda S, Takemura T, Akutagawa K, Takeda K, Mizuno N, Kurihara H. 2018. Regulation of defensive function on gingival epithelial cells can prevent periodontal disease. Jpn Dent Sci Rev. 54(2):66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeger S, Doman E, Chakraborty T, Meyle J. 2010. Effects of Porphyromonas gingivalis infection on human gingival epithelial barrier function in vitro. Eur J Oral Sci. 118(6):582–589. [DOI] [PubMed] [Google Scholar]
- Groeger SE, Meyle J. 2015. Epithelial barrier and oral bacterial infection. Periodontol 2000. 69(1):46–67. [DOI] [PubMed] [Google Scholar]
- Ji S, Choi YS, Choi Y. 2015. Bacterial invasion and persistence: critical events in the pathogenesis of periodontitis? J Periodontal Res. 50(5):570–585. [DOI] [PubMed] [Google Scholar]
- Kang MK, Bibb C, Baluda MA, Rey O, Park NH. 2000. In vitro replication and differentiation of normal human oral keratinocytes. Exp Cell Res. 258(2):288–297. [DOI] [PubMed] [Google Scholar]
- Kang X, Chen W, Kim RH, Kang MK, Park NH. 2009. Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. Oncogene. 28(4):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz J, Sambandam V, Wu JH, Michalek SM, Balkovetz DF. 2000. Characterization of Porphyromonas gingivalis–induced degradation of epithelial cell junctional complexes. Infect Immun. 68(3):1441–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz J, Yang QB, Zhang P, Potempa J, Travis J, Michalek SM, Balkovetz DF. 2002. Hydrolysis of epithelial junctional proteins by Porphyromonas gingivalis gingipains. Infect Immun. 70(5):2512–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Kim S, Song M, Lee C, Yagita H, Williams DW, Sung EC, Hong C, Shin KH, Kang MK, et al. 2018. Removal of pre-existing periodontal inflammatory condition before tooth extraction ameliorates medication-related osteonecrosis of the jaw-like lesion in mice. Am J Pathol. 188(10):2318–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrazarin S, Chen W, Oh JE, Liu ZX, Kang KL, Yi JK, Kim RH, Shin KH, Park NH, Kang MK. 2015. The p63 gene is regulated by grainyhead-like 2 (GRHL2) through reciprocal feedback and determines the epithelial phenotype in human keratinocytes. J Biol Chem. 290(32):19999–20008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyle J, Gultig K, Rascher G, Wolburg H. 1999. Transepithelial electrical resistance and tight junctions of human gingival keratinocytes. J Periodontal Res. 34(4):214–222. [DOI] [PubMed] [Google Scholar]
- Mlacki M, Darido C, Jane SM, Wilanowski T. 2014. Loss of grainy head-like 1 is associated with disruption of the epidermal barrier and squamous cell carcinoma of the skin. PLoS One. 9(2):e89247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal R, Yamashiro Y, Izumi Y. 2015. The two-way association of periodontal infection with systemic disorders: an overview. Mediators Inflamm. 2015:793898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obiso RJ, Jr, Azghani AO, Wilkins TD. 1997. The Bacteroides fragilis toxin fragilysin disrupts the paracellular barrier of epithelial cells. Infect Immun. 65(4):1431–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppong GO, Rapsinski GJ, Newman TN, Nishimori JH, Biesecker SG, Tukel C. 2013. Epithelial cells augment barrier function via activation of the toll-like receptor 2/phosphatidylinositol 3-kinase pathway upon recognition of Salmonella enterica serovar Typhimurium curli fibrils in the gut. Infect Immun. 81(2):478–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautemaa R, Jarvensivu A, Kari K, Wahlgren J, DeCarlo A, Richardson M, Sorsa T. 2004. Intracellular localization of Porphyromonas gingivalis thiol proteinase in periodontal tissues of chronic periodontitis patients. Oral Dis. 10(5):298–305. [DOI] [PubMed] [Google Scholar]
- Rivera MF, Lee JY, Aneja M, Goswami V, Liu L, Velsko IM, Chukkapalli SS, Bhattacharyya I, Chen H, Lucas AR, et al. 2013. Polymicrobial infection with major periodontal pathogens induced periodontal disease and aortic atherosclerosis in hyperlipidemic ApoE(null) mice. PLoS One. 8(2):e57178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saglie FR, Smith CT, Newman MG, Carranza FA, Jr, Pertuiset JH, Cheng L, Auil E, Nisengard RJ. 1986. The presence of bacteria in the oral epithelium in periodontal disease: II. Immunohistochemical identification of bacteria. J Periodontol. 57(8):492–500. [DOI] [PubMed] [Google Scholar]
- Sumigray KD, Lechler T. 2015. Cell adhesion in epidermal development and barrier formation. Curr Top Dev Biol. 112:383–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunde PT, Olsen I, Gobel UB, Theegarten D, Winter S, Debelian GJ, Tronstad L, Moter A. 2003. Fluorescence in situ hybridization (FISH) for direct visualization of bacteria in periapical lesions of asymptomatic root-filled teeth. Microbiology. 149(Pt 5):1095–1102. [DOI] [PubMed] [Google Scholar]
- Wang S, Samakovlis C. 2012. Grainy head and its target genes in epithelial morphogenesis and wound healing. Curr Top Dev Biol. 98:35–63. [DOI] [PubMed] [Google Scholar]
- Williams DW, Lee C, Kim T, Yagita H, Wu H, Park S, Yang P, Liu H, Shi S, Shin KH, et al. 2014. Impaired bone resorption and woven bone formation are associated with development of osteonecrosis of the jaw-like lesions by bisphosphonate and anti-receptor activator of NF-κB ligand antibody in mice. Am J Pathol. 184(11):3084–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034519865184 for Porphyromonas gingivalis Impairs Oral Epithelial Barrier through Targeting GRHL2 by W. Chen, A. Alshaikh, S. Kim, J. Kim, C. Chun, S. Mehrazarin, J. Lee, R. Lux, R.H. Kim, K.H. Shin, N.H. Park, K. Walentin, K.M. Schmidt-Ott and M.K. Kang in Journal of Dental Research