Abstract
A current challenge in materials science and biotechnology is to express a specific and controlled functionality on the large interfacial area of a nanostructured material to create smart biohybrid systems for targeted applications. Here, we report on a biohybrid system featuring poly(vinyl alcohol) as the supporting synthetic polymer and bovine serum albumin as the biofunctional element. The optimal processing conditions to produce these self-standing composite membranes are determined, and the composition and distribution of the bioactive agent within the polymeric matrices are analyzed. A post-processing cross-linking using glutaraldehyde enables this functional membrane to be used as a chemical filter in aqueous environments. By demonstrating that our mats can remove large amounts of ketoprofen from water, we show that the combination of a BSA-induced biofunctionality with a nanostructured fibrous material allows for the development of an efficient biohybrid filtering device for the large and widely used family of nonsteroidal anti-inflammatory drugs (NSAIDs). The crystal structure of the complex between BSA and ketoprofen is determined for the first time and confirms the interaction between the two species.
Introduction
Filtration can be applied to capture airborne and waterborne contaminants such as chemical pollutants, allergens, and biological agents, and therefore, it plays an essential role in a wide range of human activities (e.g., water sanitation, food-chain control, health care, pharmaceutical and biotechnology industries, and others). Although extremely efficient under normal operation conditions, traditional filters based on sieving may prove ineffective in the separation of specific chemical contaminants, for which affinity-based filtration may be more efficient.
Among all of the different available techniques to produce yarns, electrospinning has gained great interest within the nanotechnology community for its ability to overcome challenges that are associated with the production of nanostructured filters.1 Electrospun nanofibrous mats are good candidates as filtering media due to their lightness, high permeability, and small and interconnected pores.2 The reduced porosity, together with the small fiber size, generates a slip flow regime, thus increasing the efficiency and providing a higher-resolution separation with respect to micromembranes.3,4
Although nanofibrous mats do not possess enough mechanical strength to withstand impingement or high flux during filtration, they can be processed as an active nanostructured filtering layer on top of a traditional filter.5 Their large specific surface area enables remarkable surface modification or the incorporation of active chemicals and functionalities at the nanoscale. Post-processing allows the addition of specific functionalities to a raw electrospun membrane or the removal/modification of the chemical structure of an electrospun polymer. Various post-electrospinning processes have been reported, depending on the composition of the mat and on the desired specific functionalities. They range from thermal or plasma surface treatments to physical vapor deposition, cross-linking, hydrolysis, oxidation, grafting, and acid treatments.6,7 All of these treatments act almost exclusively on the fiber surface, without affecting the bulk. However, this general approach entails considerable challenges, related to uniformity. In fact, post-modification can be inhomogeneous through the entire thickness of the electrospun mat. Moreover, fiber coating has to be uniform and ultrathin so as to not affect the pore size.
To overcome these issues, selected functionalities can be introduced directly during the electrospinning process by adding small molecules, oligomers, or even a second polymer to the feed solution. For this approach to work, the intrinsic affinity between the different components must be taken into consideration, for example, the solubility of the additive in the solvent of choice. In the case of biomolecules, particularly mild processing conditions are usually necessary to ensure the retention of both their biological activity and their native conformation. This direct functionalization usually leads to a uniform distribution of the additive within the fiber. Nevertheless, the preferential localization of the additive into specific fiber sites such as beads can also occur.8,9
To date, existing microporous membranes have been surface-modified to expose affinity ligands that interact with specific biomolecules, and they are widely used for affinity purification technologies.10 Amino acids, antigens, antibodies, dyes, and metal-affinity ligands have been immobilized onto sheets and fiber matrices, which have been successfully exploited for targeted separations. However, post-functionalization is often characterized by low yield and slow kinetics. For instance, the immobilization of a polysaccharide such as heparin onto an epoxy membrane to capture its complementary proteins requires a slow (8 days) membrane functionalization reaction.11
In biofunctionalized electrospun systems, the nanostructured scaffold with tridimensional porosity has been loaded with the biological component, leading to a decoration with a selected functionality onto the high surface area. Several biological components have been considered, ranging from proteins to enzymes,12−14 where an increased enzymatic activity was often reported once enzymes are immobilized in electrospun fibers,1 DNA strands,15 yeast,16 and cells.17
Following this approach, we report on a biohybrid electrospun filtering medium composed of a biocompatible polymer (poly(vinyl alcohol) (PVA)) and bovine serum albumin (BSA) as the biofunctional element. We show that the electrospinning conditions and the following cross-linking of the mat with glutaraldehyde (GA) impart water stability to the membrane without compromising the fibrous morphology of the tissue. Indeed, the physiological ability of BSA to bind and sequester many drugs, including most nonsteroidal anti-inflammatory drugs (NSAIDs), in the blood plasma is retained also when the protein is embedded into polymer nanofibers, and we demonstrate that this biohybrid membrane is an efficient system to filter an NSAID such as ketoprofen (KPF) from aqueous solutions. Owing to their analgesic and antipyretic properties, NSAIDs are widely prescribed to relieve pain and to reduce inflammation. As a result of their huge worldwide consumption and their metabolic profile, NSAIDs represent an increasingly important source of pollution of surface and underground waters.18,19 KPF is one of the most representative and widely used NSAIDs and it has been found in waste waters at concentrations as high as 2–10 μg/L20 up to 2 mg/L in the case of effluents of pharmaceutical production facilities.21 Therefore, it is considered as an important source of contamination of soil and ground water22 as it is particularly hazardous for water organisms.23 Unfortunately, the conventional wastewater treatment plants are not sufficiently effective to remove NSAID residues and additional water treatments are necessary.24 Therefore, efforts in developing tools and techniques to achieve a better removal of these pollutants from water can have an important environmental and socioeconomic impact.
To provide structural evidence of the direct interaction between BSA and ketoprofen and validate our BSA-loaded fibers as reliable chemical filtration tools for ketoprofen-contaminated waste waters, we set out to solve the X-ray crystal structure of the complex.
Materials and Methods
Materials
A lyophilized powder of bovine serum albumin ≥96% (BSA), poly(vinyl alcohol) >99% hydrolyzed (PVA), all buffer components, glutaraldehyde aqueous solution 50% (GA), hydrochloric acid min. 37% (HCl), and acetone ≥99.5% were purchased from Sigma Aldrich. Ketoprofen was purchased from Fluorochem and Coomassie brilliant blue G-250 from BioRad. The Thermo Scientific Pierce Coomassie Plus (Bradford) Protein Assay Kit was used during the Bradford protein assay. All chemicals and solvents were used as received. Crystallization plates were bought from Hampton Research (Aliso Vejo, CA).
Preparation of the Fully Hydrolyzed PVA Solution
A weighted amount of fully hydrolyzed poly(vinyl alcohol) PVA (Mw = 89 000–98 000 Da) was slowly added into distilled water, and the polymer was allowed to dissolve under moderate stirring. The solution was stirred at 95 °C for 1 h and then slowly cooled down to room temperature. This led to a clear PVA solution without precipitates.
Preparation of BSA Solutions and PVA/BSA Blends
The aqueous BSA/PVA feed solution could not be directly prepared since bovine serum albumin undergoes irreversible denaturation at temperatures higher than 50 °C. Therefore, the two components were separately dissolved in distilled water (see the previous section for PVA solution preparation). The two freshly prepared samples were then mixed in well-defined proportions to obtain the final BSA/PVA blended solution, which was stirred for 1 h prior to electrospinning.
Preparation of Biohybrid Membranes
Electrospinning was performed by loading the freshly prepared feed solution in a 2.5 mL syringe with a 22 gauge needle (Hamilton Gastight model 1002 TLL), which was then placed on an infusion pump (KDS Scientific, model series 200) that provided a solution rate in the range of 0.05–0.4 mL/h. Electrospinning was carried out using a bottom–up vertical setup, with a tip-collector distance in the range of 12–30 cm. Voltage was applied to the needle by a high-voltage power supply (Spellman SL30P300) in the range of 12–24 kV. Electrospinning was performed onto either glass slides for optical characterization or silicon substrates for scanning electron microscopy characterization. Self-standing membranes were obtained with approximately 15 min of deposition onto glass slides and by subsequent peeling off. Samples were stored at 4 °C until use.
Chemical Cross-Linking of Electrospun Mats
Electrospun mats of PVA and the PVA/BSA blend were removed from the collector and immersed in a 0.15 M acetone glutaraldehyde aqueous solution bath with 0.05 M HCl.25 The bath comprised 1.5 mL of a 50% GA aqueous solution, 2.5 mL of a 1 M HCl stock solution, and 46 mL of acetone. The solution was kept under stirring for 1 h. Mats were gently taken out from the bath and rinsed in a clean bath to remove the excess of cross-linking solution.
Characterization
Optical images were obtained with an Olympus microscope with the use of an INFINITY microscope camera. Scanning electron microscopy (SEM) was performed with JEOL scanning electron microscope model JSM-6010LV, applying a potential of 5–8 kV. Fibers were deposited directly onto a silicon substrate, and no further metallization was needed. Image analysis was performed with ImageJ software (Rasband, W.S., ImageJ, US National Institutes of Health, Bethesda, Maryland, http://imagej.nih.gov/ij/, 1997–2016).
Transmission electron microscopy (TEM) was performed using a Philips CM 200 field-emission gun microscope. Fibers were deposited directly onto lacey carbon-coated copper grids and examined at an accelerating voltage of 80 kV. During the acquisition of TEM images, the samples did not undergo structural transformation. Low-beam current densities and short acquisition times were adopted.
Infrared spectra were recorded in the transmission mode using a Nicolet 6700 FTIR interferometer on freestanding membranes. The resolution was 2 cm–1, and 256 sample scans were acquired. The Omnic 7.1 software (Thermo Nicolet Instrumentation) was used to analyze the spectra. Spectra of pure BSA were obtained by depositing a few drops of a water–protein solution onto a zinc selenide substrate and analyzing after gentle solvent evaporation.
Bradford protein assays were performed using a Beckman Coulter DU730 Life Science UV–vis spectrophotometer, in semi-micro-cuvettes in PMMA. UV–vis absorption spectra were recorded on a Varian Cary 5000 spectrophotometer.
Bradford Assay for Protein Quantification
The amount of protein in the mat, both as-spun and after water immersion, was measured by the Bradford protein assay using Coomassie brilliant blue (CBB) G-250 and BSA as the reference protein. For a direct test, the absorbance at 595 nm of the dye solution, which is proportional to the amount of protein in solution, was measured. A standard curve of a known concentration of BSA in water was prepared and used as calibration for the measure of the samples. Specifically, BSA solutions of known concentrations were prepared in water; 250 μL of such solutions were mixed with 1.25 mL of CBB, left for 5 min, and subsequently probed at 595 nm in a spectrophotometer using CBB as the baseline.
The colorimetric assay was used (i) to quantify the BSA content present in the mat before cross-linking (loading test) and (ii) to evaluate the leaking of BSA from a cross-linked mat by immersion in water (leaking test). Regarding the loading test, a membrane of known mass was dissolved in water, and then the solution was probed for its BSA content using the standard curve as reference. For the leaking test, the cross-linked and water-insoluble mats were immersed in water for 1 h. After extracting the mat from the bath, the amount of released protein was determined by measuring the residual protein that was present in water. Measurements were carried out in triplicate, and mats of pure PVA were used as controls for both experiments.
Ketoprofen–Water Solution Filtration
Electrospun mats of cross-linked PVA or the PVA/BSA blend were used for water filtration tests. UV–vis absorption spectra of a ketoprofen–water solution at a known concentration (26 μM) were recorded before and after flowing the solution under gravity through the electrospun membrane. A weighted membrane was mounted on a filtering module attached with a luer lock connector to a glass syringe of 10 mL. The filtering module consisted of a metallic holder, where the self-standing electrospun mat was mounted, and a Teflon o-ring for the watertight seal. The UV–vis spectra of the filtered solution were recorded to calculate the residual concentration of ketoprofen. The amount of material filtered was retrieved and normalized based on the mat weight.
Preparation and Crystallization of the Bovine Serum Albumin Complex with Ketoprofen
Prior to crystallization, the protein was prepared as previously described.26 BSA was dissolved in a buffer composed of 10 mM Tris HCl, pH 7.5, and 150 mM NaCl (buffer A) and then purified with a single gel-filtration step on a Superdex 200 Increase 10/300GL size-exclusion chromatography column pre-equilibrated in the same buffer. The eluted protein was concentrated to 10 mg/mL using an Amicon-15 filter with a 10 000 MWCO. The BSA–ketoprofen complex was co-crystallized according to the procedure described for the bovine serum albumin complex with naproxen.27 A 10-fold molar excess of ketoprofen (1.5 mM solution in ethanol) was added to a 10 mg/mL solution of purified BSA in buffer A, and the mixture was incubated at 18 °C for 2 h. Crystals of the complex were obtained using the vapor-diffusion method at room temperature and using crystallization conditions similar to those of the BSA apo structure.26 Prior to X-ray data collection, crystals were frozen in a chemically identical solution supplemented with 1.5 mM ketoprofen and 25% (v/v) PEG 400 for cryoprotection. Data up to 2.80 Å resolution were collected at the X06DA-PXIII beamline at the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland). Diffraction images were processed using XDS.28 The structure was determined by molecular replacement using PHASER29 from the PHENIX suite30 and the apo structure of BSA (PDB ID: 4OR0) as the search probe. Refinement was carried out using PHENIX,30 whereas Coot31 was used for manual rebuilding. The program PROCHECK32 was used to assess the stereochemistry of the structure. Data collection and final refinement statistics are given in Table S1. All of the figures in the paper were generated using PyMOL.33
The final crystallographic coordinates of the crystal structure shown here are available in the RCSB (PDB accession code: 6QS9).
Results and Discussion
As the surface-affinity ligand for drug removal from waste water, bovine serum albumin (BSA) was chosen. BSA is one of the most abundant plasma proteins contributing to the circulatory system, accounting for over 60% of the total protein content in the blood.34 Although several specialized transport proteins are known to be present in the plasma, serum albumin is highly promiscuous in binding and transporting several different species, such as fatty acids, which are driven to the heart and skeletal muscles and converted into energy. BSA is also known to bind to a wide selection of drugs and other bioactive molecules and promote their transport within the blood stream.34
Biomolecules alone are rather challenging to electrospin as they lack the suitable viscoelasticity, a fundamental property that is required for the stabilization of the spun jet.35,36 Previous studies have shown that BSA aqueous solutions cannot generate fibers at any input voltage for electrospinning.37 Only denatured protein solutions have afforded nanofibers made entirely of BSA.35,38
To overcome this limitation, BSA can be blended with hydrophilic synthetic polymers such as polyethylene oxide37 and poly(vinyl alcohol) (PVA).39,40 Alternatively, an emulsion of a biocompatible synthetic polymer such as polycaprolactone supplemented with surfactants can also undergo electrospinning.41 Following this approach, it has been shown that electrospinning does not cause any significant changes, damages, or unfolding to the conformation of BSA.37
As the wettability of filtering media plays a significant role, poly(vinyl alcohol) (PVA) was chosen as the structural polymer for the biohybrid membrane, since it combines good mechanical properties with excellent hydrophilicity.42 PVA fiber formation is strongly influenced by a number of electrospinning parameters, ranging from the solution properties to the presence of additives or the setup and environmental conditions.43,44 Although the addition of strong bases such as NaOH is known to help fiber formation of fully hydrolyzed PVA,45 electrospinning of the polymer in distilled water without any additive may help reduce the risk of structural or chemical BSA modifications. By electrospinning aqueous PVA solutions at different polymer concentrations, we collected fibers at <18 wt % PVA, whereas we observed the formation of an unprocessable gel when 18 wt % PVA is reached. The characteristic features of the collected mats under different processing conditions are reported in Table 1.
Table 1. Electrospinning Conditions and Fiber Morphology for PVA and the Blend PVA/BSA.
| feed solution | voltage (kV) | feed rate (mL/h) | tip-collector distance (cm) | note on sample morphology |
|---|---|---|---|---|
| 10 wt % PVA | 12–24 | 0.1–0.4 | 18–25 | highly beaded and defective fibers |
| 12 wt % PVA | 15–21 | 0.05–0.1 | 20–30 | defect-free fibers and very few beaded fibers |
| 14 wt % PVA | 15–18 | 0.05–0.2 | 25–28 | homogeneous and defect-free fibers |
| 15 wt % PVA | 15–19 | 0.1–0.3 | 12–18 | homogeneous and defect-free fibers |
| 16 wt % PVA | 15–17 | 0.2 | 13–18 | few defect-free fibers; low production yield |
| 12 wt % PVA (10–100)% BSA | 17–19 | 0.3 | 18 | homogeneous and defect-free fibers |
| 13 wt % PVA (10–100)% BSA | 17–19 | 0.3 | 18 | homogeneous and defect-free fibers |
| 14 wt % PVA (10–100)% BSA | 17–19 | 0.3 | 18 | homogeneous and defect-free fibers |
We found that the critical PVA concentration to obtain defect-free and highly homogeneous fibers is ≥14 wt % (Figure 1a) and that the fiber diameter increases with increasing polymer concentration in the electrospinning solution. We did not measure the diameter of the beaded fibers that resulted from electrospinning 10 wt % PVA solutions since these extremely heterogeneous samples did not allow the calculation of a reliable average diameter (see Figure S1). The addition of BSA to the feed solution did not have any negative effect on the electrospinning process, as smooth and flawless fibers were obtained under all of the tested conditions (Figure 1b). It is worth noting that a remarkable reduction of the fiber diameter was observed when even a low amount of BSA (10%) was added (Figure 1c). This can be explained by considering that the isoelectric point of BSA is about 4.7 at 25 °C, meaning that BSA carries a net negative charge when added to a neutral solution.38 Therefore, the effect of BSA on the electrospinning solution is comparable to the addition of salts, which causes an increase in the conductivity and a decrease in the fiber diameter due to the stronger elongational forces acting on the polymer jet.40,46
Figure 1.
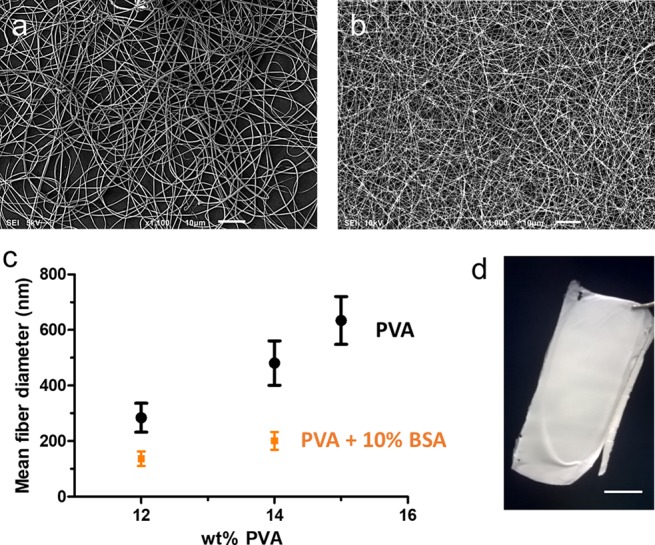
Scanning electron microscope (SEM) images of electrospun samples: (a) 14 wt % PVA (scale bar 10 μm), (b) PVA/BSA (14 wt % PVA–10% BSA) (scale bar 10 μm), and (c) average fiber diameter (mean obtained over n = 100 measurements) measured for the PVA fiber (black dots) and the PVA loaded with 10 wt % BSA (orange squares). (d) PVA/BSA self-standing electrospun membrane (12 wt % PVA–10% BSA) as removed from the collector after electrospinning (scale bar 1 cm).
The biohybrid sample composition of 12 wt % PVA and 10% BSA was chosen as the reference for all of the following tests due to the uniformity and the small diameter of the fibers thus obtained (137 ± 26 nm), which provides a large functional surface area to the environment. Self-standing mats were obtained by depositing the fibers onto a static collector, located at ca. 18 cm from the needle tip, for about 15 min (Figure 1d).
According to the literature,47 we used FTIR spectroscopy to analyze the BSA conformation in the electrospun tissue. When we compared the FTIR spectra of the electrospun samples of pure PVA and of the PVA–BSA blend, the characteristic vibrational modes of BSA were clearly visible in the biohybrid system (Figure S5). BSA has two characteristic vibrational modes at 1541–1550 and 1651–1660 cm–1, which can be assigned to amide I (ν C=O) and amide II (δ N-H) bands, respectively, and do not overlap with the PVA spectra. When BSA is blended with PVA, no shift of either the PVA or the BSA fingerprint bands is observed, which indicates that no protein conformational change has been induced by either an interaction between the two components of the blend or by the electrospinning process itself. This indicates that PVA and the reported electrospinning conditions are suitable for the fabrication of the biohybrid composite membrane. However, for the membrane to be used for water filtration, cross-linking is required to prevent the mat from dissolving in water during operation. Cross-linking was achieved by immersion of the as-spun membrane in a bath of glutaraldehyde (GA) in acetone. GA was chosen as the cross-linking agent for PVA since it is known to be more effective than other chemicals such as formaldehyde or glycidyl acrylate.48 Moreover, it limits swelling of the matrix, which is a significant drawback of physical post-electrospinning processing methods such as heating-induced increased crystallinity.42 Acetone is a poor solvent for both PVA49 and BSA,50 whereas a small HCl content, which catalyzes the cross-linking reaction, is not hazardous for the protein. SEM micrographs of the as-spun sample, the sample obtained after cross-linking treatment, and after water immersion of the cross-linked membranes are reported in Figure 2a–c. The cross-linking treatment turned out to be fully effective; indeed, after the process, the fibrous morphology was completely maintained and no change in the fiber diameter was observed.
Figure 2.
Cross-linking of the PVA/BSA membrane with glutaraldehyde. (a) SEM micrograph of PVA 12 wt %–BSA 10% as-spun fibers (bar = 5 μm). (b) SEM micrograph of the sample after cross-linking with GA. The sample was immersed into an acetone bath containing 0.15 M glutaraldehyde and 0.05 M HCl for 1 h and gently dried (bar = 5 μm). (c) SEM micrograph of the cross-linked mat after overnight immersion in water and gentle drying (bar = 5 μm). (d) FTIR of a mat of PVA before (red line) and after (blue line) cross-linking with GA. (e) FTIR of a mat of PVA 12 wt %–BSA 10% before (purple line) and after (red line) cross-linking with GA. Relevant vibrational modes are highlighted for PVA and PVA/BSA, before and after cross-linking.
The effect of PVA cross-linking with GA is clearly evidenced by FTIR spectroscopy, which shows a decrease in the ν(-OH) band at 3315–3387 cm–1 and the appearance of a new, intense band in the 1150–1085 cm–1 region, corresponding to the C–O–C stretching mode of the bonds of the acetal family formed by GA with the -OH groups of PVA (Figure 2d). New bands are also present in the 1720–1740 and the 2830–2695 cm–1 regions, which are related to the C=O and C–H stretching, respectively. These signals are consistent with the reaction of only one aldehyde of the cross-linker with an -OH group of PVA, which leads to a residual aldehyde signal, as described previously in the literature.51,52
Similar bands are also visible after cross-linking of the biohybrid PVA–BSA system (Figure 2e). It is worth noting that the vibrational bands of amide I and amide II of BSA are not modified after the cross-linking process, thus indicating that the cross-linking does not affect the BSA structure. However, GA forms covalent bonds with both the hydroxyl groups of the PVA chains and with the BSA amine groups exposed on the fiber surface. Although the former reaction is predominant, cross-linking with glutaraldehyde can involve both biohybrid system components and stabilizes the protein within the polymeric matrix. Analogous situations have been previously reported for other proteins and enzymes.40,53,54
The Bradford assay55 was used for BSA quantification in the fibrous mat (loading test) and as a direct quantification of the possible BSA release when the membrane is used in aqueous media (leaking test).54,56,57 It is known that, although they are often added to the feed solution to successfully electrospin PVA, the presence of surfactants interferes with the readout of the Bradford test, leading to misrepresented data. Hence, no additives were used in our membranes.
Calibration curves (Figure S7) for the direct Bradford assay were used for the quantification of the amount of BSA that was present in solution or in the mat. Bradford tests allowed the measurement of (i) the amount of BSA in the as-spun biohybrid matrix dissolved in water (loading test) and (ii) the amount of protein released by a cross-linked membrane (leaking test). Results are reported in Table 2.
Table 2. Quantification of BSA in Water-Soluble Mats (Loading Test) and of the Protein Released from Cross-Linked Mats (Leaking Tests) by the Bradford Assay.
| sample | nominal BSA content in the electrospinning solution (%) | Bradford assay results on BSA content (%) |
|---|---|---|
| loading test | ||
| control PVA mat | 0 | 0.0 ± 0.1 |
| PVA–BSA mat | 10 | 9.5 ± 0.2 |
| leaking test | ||
| control PVA mat | 0 | 0.0 ± 0.1 |
| PVA–BSA mat | 10 | 0.2 ± 0.2 |
For the loading test, PVA/BSA membranes of a known mass were dissolved in a known volume of water, which was then probed with the Bradford assay. A BSA concentration of 9.5 ± 0.2% was determined, which is in good agreement with the nominal concentration of BSA in the electrospinning solution (10%). Therefore, the nominal concentration of the protein in the electrospinning solution is maintained in the final membrane sample. This protein content is preserved also after the cross-linking process, as no significant amount of protein released by a cross-linked sample was measured by the leaking test. For a cross-linked mat with a nominal BSA concentration of 10%, the protein released after 1 h of dipping in water is as low as 0.21 ± 0.2% and increases to just 1.17 ± 0.6% for a 30% BSA mat. The absence of BSA release was also confirmed by FTIR analysis of the cross-linking bath; indeed, no BSA fingerprint could be observed, also suggesting that no significant leaking of the protein occurs during the cross-linking process (Figure S6). Therefore, we can conclude that the concentration of BSA that is found in the final cross-linked mat is the same as the nominal concentration of BSA in the electrospinning solution.
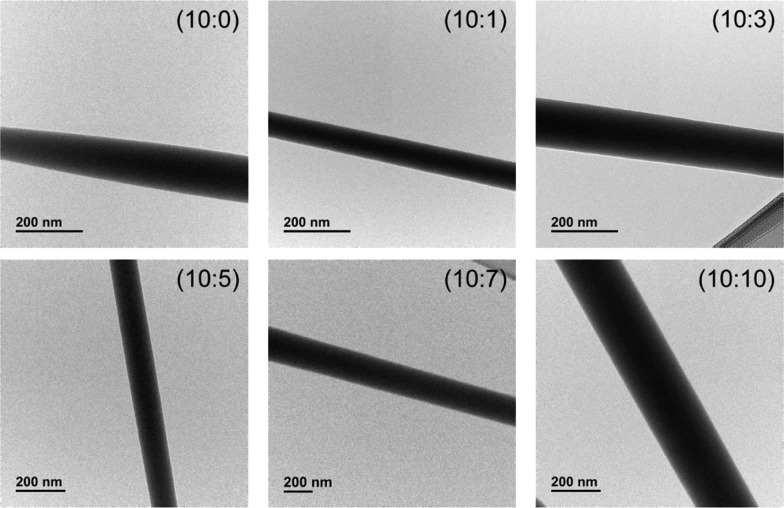
Although the PVA fiber surface is decorated with BSA, as clearly shown by the blue coloration assumed by the biohybrid cross-linked membrane when it is immersed in a CBB solution (Figure S8), the protein does not segregate at the fiber surface during the electrospinning process, but it is uniformly dispersed in the whole polymer material. Indeed, transmission electron microscopy (TEM) images of the electrospun fibers with different BSA contents show constant contrast along the fibers, without any core–sheath structure58,59 with BSA segregated in an outer layer (Figure 3).
Figure 3.
TEM bright-field micrographs of electrospun biohybrid fibers with 12 wt % PVA and different amounts of BSA (PVA:BSA ratios of 10:0, 10:1, 10:3, 10:5, 10:7, and 10:10).
Biohybrid PVA–BSA membranes were used for the chemical filtration of an aqueous ketoprofen solution. Electrospun membranes were produced with 12 wt % PVA–10% BSA loading, cross-linked, dried, and weighted for the quantification of the total protein content. Membranes were placed in a filtering syringe module, which was mounted onto the syringe tip and kept fixed via a Teflon ring (Figure 4).
Figure 4.
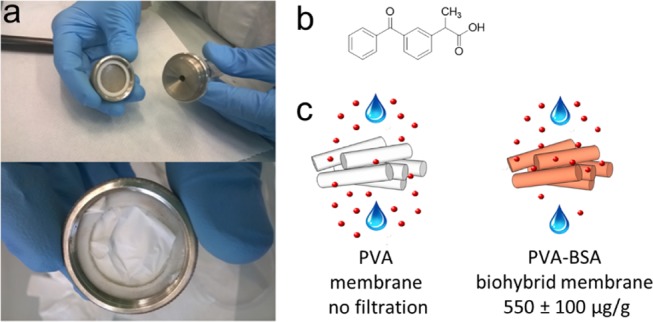
Chemical filtration of ketoprofen from water with a biohybrid PVA–BSA membrane. (a) Electrospun membrane is placed in a filter module and then mounted on the syringe tip with a Luer Lock connection. (b) Chemical structure of ketoprofen used as the target in the filtration experiments. (c) Normalized quantities of ketoprofen filtered from a solution of 26 μM concentration with respect to the weight of the membrane are zero for the pure PVA membrane and 550 ± 100 μg/g for the 12 wt % PVA–10% BSA biohybrid membrane.
The amount of ketoprofen removed from the water by the membrane was measured by comparing the UV–vis absorption spectra of the solution before and after filtration (Figure S10). The molar extinction coefficient of ketoprofen in water was previously calculated to determine the concentration of the drug in solution from the absorption spectra (Figure S9).
The ratio of the mass of drug to the mass of membrane is calculated as an indication of the ability of the biohybrid filter to bind ketoprofen in solution. The mass of the drug filtered was divided by the mat weight, and it was found that the biohybrid membrane was able to filter a mass of 550 ± 100 μg of ketoprofen per gram of membrane. To demonstrate that the filtration of ketoprofen occurs via the interaction of the drug with the protein and is not due to physical filtration by a sieving mechanism, a PVA cross-linked membrane without BSA was also tested. No significant change in the concentration of ketoprofen before and after filtration was detected, indicating that no sieving occurs.
In the literature, there are a number of studies involving BSA as a model biomolecule to study drug–protein interactions. To this end, particularly relevant are the availability of structural data26,27,60,61 and the fact that BSA shows 80% homology with human serum albumin34 (HSA). Ketoprofen has been shown to interact with BSA and to bind to the protein without affecting its overall architecture.62,63 However, the crystal structure of the ketoprofen–BSA complex has so far been elusive, whereas BSA has been successfully co-crystallized with naproxen,27 which also belongs to the class of NSAIDs.
In this work, the crystal structure of the BSA–ketoprofen complex was solved, and the structure shown herein provides clear evidence of the direct interaction between BSA and ketoprofen and the formation of a stable complex.
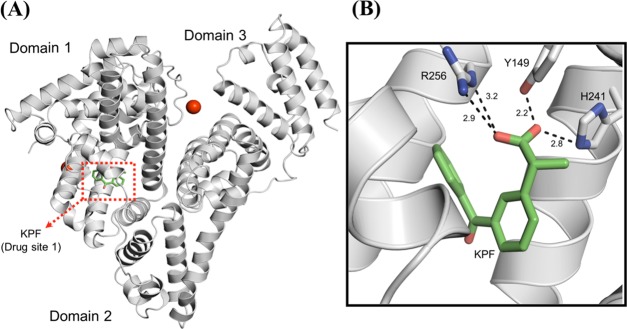
The BSA–ketoprofen complex crystallized in the monoclinic C2 space group with the unit cell parameters a = 212.49 Å, b = 44.37 Å, c = 142.71 Å, and β = 113.24°. As with all of the other BSA structures described in the literature,26,60,64 the crystal contains two molecules in the asymmetric unit. No significant variations are observed when the structure is compared to other BSA crystal structures (RMSD = 0.531 Å to 4OR0 chain A, Figure 5A), with the overall fold characterized by a heart-shaped molecule composed of three structurally similar helical domains.26 In this structure, one ketoprofen molecule occupies the drug site 1, which is located in the subdomain IIA of BSA, similar to naproxen27 or 3,5-diiodosalicylic acid.60 The carboxyl group of ketoprofen forms hydrogen bonds with the hydroxyl group of Tyr149, the imidazole nitrogen atoms of His241, and the guanidine moiety of Arg256 (Figure 5B). Overall, the ligand is embedded in a mostly hydrophobic pocket lined by one tryptophan (Trp213), four leucines (Leu218, Leu233, Leu237, and Leu259), two isoleucines (Ile263 and Ile289), two alanines (Ala260 and Ala290), and one phenylalanine (Phe222).
Figure 5.
Overall crystal structure of the BSA–ketoprofen complex and details of the KPF binding mode. (A) Cartoon representation of the BSA monomer showing the three-domain architecture of the protein and the drug site 1. Calcium ions and the ketoprofen molecule are represented by red spheres and green sticks, respectively. (B) Drug site 1 of BSA in complex with KPF. The hydrogen-bonding interactions between the ligand and the polar residues in the binding site are shown as dashed black lines. Distances are expressed in Ångstrom.
Conclusions
We demonstrated that a biohybrid electrospun membrane consisting of BSA-loaded PVA fibers effectively removes ketoprofen from an aqueous solution by chemical filtration. This biohybrid filter was obtained by electrospinning, in a one-step procedure, a feed solution containing both the polymer and the protein. Control over the spinning parameters led to homogeneous and defect-free fibers as thin as 137 ± 26 nm with a uniform distribution of the protein in the fiber section. Cross-linking with glutaraldehyde made fibers insoluble, thus enabling the use of these membranes for filtering aqueous solutions. Neither the electrospinning process nor the cross-linking reaction caused any modification to the protein structure, as shown by the unvaried characteristic amide I and amide II FTIR bands of the protein during all of the fabrication steps. The colorimetric Bradford protein assay allowed the quantification of BSA loaded in the membrane, which matched the nominal protein concentration of the electrospinning solution, thus demonstrating that no BSA leakage occurs during either the cross-linking reaction or the filtration step.
The fibrous membrane decorated with properly folded BSA, uniformly distributed in the fiber, was designed and exploited to interact with ketoprofen, which is a relevant and widespread pollutant in waste water. The filtration test was successful due to the specific chemical interaction between the protein and the target drug. In fact, the crystalline structure of the BSA–ketoprofen complex, which was solved for the first time, clearly indicates that the drug forms a strong interaction in a well-described binging site for the protein, named drug site number 1.
The affordability of the raw materials, the control over the processing conditions, and the facile industrial scalability of the process allows for the integration of this biohybrid system in macroscale devices, which is a crucial step for any chemical filtration application. Moreover, these experiments provide a conceptual framework and an experimental platform for follow-up experiments in different technological settings, ranging from biosensing to probes, and target immobilization for biotechnology, nanomaterial self-assembly, and environmental applications.
Acknowledgments
R.C thanks the CERCA Programme/Generalitat de Catalunya and the BEST Postdoctoral Fellowship, funded by the European Commission under Horizon 2020s Marie Skłodowska-Curie Actions COFUND scheme (Grant Agreement no. 712754) and by the Severo Ochoa programme of the Spanish Ministry of Science and Competitiveness (Grant SEV-2014-0425 (2015–2019)).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.9b01442.
SEM and FTIR characterization of electrospun membranes of PVA and PVA/BSA before and after cross-linking with GA; the Bradford assay calibration curve and filtration tests; crystal structure data of the complex BSA–ketoprofen (PDF)
Author Present Address
∥ Institute for Bioengineering of Catalonia (IBEC), The Barcelona Institute of Science and Technology, c/ Baldiri Reixac 10–12, 08028 Barcelona, Spain (R.C.).
Author Contributions
This manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Greiner A.; Wendorff J. H. Electrospinning: A Fascinating Method for the Preparation of Ultrathin Fibers. Angew. Chem., Int. Ed. 2007, 46, 5670–5703. 10.1002/anie.200604646. [DOI] [PubMed] [Google Scholar]
- Ma H.; Burger C.; Hsiao B. S.; Chu B. Ultra-Fine Cellulose Nanofibers: New Nano-Scale Materials for Water Purification. J. Mater. Chem. 2011, 21, 7507–7510. 10.1039/c0jm04308g. [DOI] [Google Scholar]
- Bao L.; Seki K.; Niinuma H.; Otani Y.; Balgis R.; Ogi T.; Gradon L.; Okuyama K. Verification of Slip Flow in Nanofiber Filter Media through Pressure Drop Measurement at Low-Pressure Conditions. Sep. Purif. Technol. 2016, 159, 100–107. 10.1016/j.seppur.2015.12.045. [DOI] [Google Scholar]
- Choi J.; Yang B. J.; Bae G. N.; Jung J. H. Herbal Extract Incorporated Nanofiber Fabricated by an Electrospinning Technique and Its Application to Antimicrobial Air Filtration. ACS Appl. Mater. Interfaces 2015, 7, 25313–25320. 10.1021/acsami.5b07441. [DOI] [PubMed] [Google Scholar]
- Ahmed F. E.; Lalia B. S.; Hashaikeh R. A Review on Electrospinning for Membrane Fabrication: Challenges and Applications. Desalination 2015, 356, 15–30. 10.1016/j.desal.2014.09.033. [DOI] [Google Scholar]
- Kalaoglu-Altan O. I.; Sanyal R.; Sanyal A. Orthogonally “Clickable” Biodegradable Nanofibers: Tailoring Biomaterials for Specific Protein Immobilization. ACS Omega 2019, 4, 121–129. 10.1021/acsomega.8b03041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagitha P.; Reshmi C. R.; Sundaran S. P.; Sujith A. Recent Advances in Post-Modification Strategies of Polymeric Electrospun Membranes. Eur. Polym. J. 2018, 105, 227–249. 10.1016/j.eurpolymj.2018.05.033. [DOI] [Google Scholar]
- Li T. X.; Ding X.; Sui X.; Tian L. L.; Zhang Y.; Hu J. Y.; Yang X. D. Sustained Release of Protein Particle Encapsulated in Bead-on-String Electrospun Nanofibers. J. Macromol. Sci., Part B: Phys. 2015, 54, 887–896. 10.1080/00222348.2015.1051210. [DOI] [Google Scholar]
- Ren G.; Xu X.; Liu Q.; Cheng J.; Yuan X.; Wu L.; Wan Y. Electrospun Poly(Vinyl Alcohol)/Glucose Oxidase Biocomposite Membranes for Biosensor Applications. React. Funct. Polym. 2006, 66, 1559–1564. 10.1016/j.reactfunctpolym.2006.05.005. [DOI] [Google Scholar]
- Klein E. Affinity Membranes: A 10-Year Review. J. Membr. Sci. 2000, 179, 1–27. 10.1016/S0376-7388(00)00514-7. [DOI] [Google Scholar]
- Lütkemeyer D.; Bretschneider M.; Büntemeyer H.; Lehmann J. Membrane Chromatography for Rapid Purification of Recombinant Antithrombin III and Monoclonal Antibodies from Cell Culture Supernatant. J. Chromatogr. A 1993, 639, 57–66. 10.1016/0021-9673(93)83088-A. [DOI] [PubMed] [Google Scholar]
- Zong M.-H.; Wu H.; Wen Y.; Wen P.; Linhardt R. J. Encapsulation of Bioactive Compound in Electrospun Fibers and Its Potential Application. J. Agric. Food Chem. 2017, 65, 9161–9179. 10.1021/acs.jafc.7b02956. [DOI] [PubMed] [Google Scholar]
- Zeng J.; Aigner A.; Czubayko F.; Kissel T.; Wendorff J. H.; Greiner A. Poly(Vinyl Alcohol) Nanofibers by Electrospinning as a Protein Delivery System and the Retardation of Enzyme Release by Additional Polymer Coatings. Biomacromolecules 2005, 6, 1484–1488. 10.1021/bm0492576. [DOI] [PubMed] [Google Scholar]
- Han D.; Filocamo S.; Kirby R.; Steckl A. J. Deactivating Chemical Agents Using Enzyme-Coated Nanofibers Formed by Electrospinning. ACS Appl. Mater. Interfaces 2011, 3, 4633–4639. 10.1021/am201064b. [DOI] [PubMed] [Google Scholar]
- Ner Y.; Grote J. G.; Stuart J. A.; Sotzing G. A. Enhanced Fluorescence in Electrospun Dye-Doped DNA Nanofibers. Soft Matter 2008, 4, 1448–1453. 10.1039/b717581g. [DOI] [PubMed] [Google Scholar]
- Canbolat M. F.; Gera N.; Tang C.; Monian B.; Rao B. M.; Pourdeyhimi B.; Khan S. A. Preservation of Cell Viability and Protein Conformation on Immobilization within Nanofibers via Electrospinning Functionalized Yeast. ACS Appl. Mater. Interfaces 2013, 5, 9349–9354. 10.1021/am4022768. [DOI] [PubMed] [Google Scholar]
- Jayasinghe S. N. Cell Electrospinning: A Novel Tool for Functionalising Fibres, Scaffolds and Membranes with Living Cells and Other Advanced Materials for Regenerative Biology and Medicine. Analyst 2013, 138, 2215–2223. 10.1039/c3an36599a. [DOI] [PubMed] [Google Scholar]
- Luo Y.; Guo W.; Ngo H. H.; Nghiem L. D.; Hai F. I.; Zhang J.; Liang S.; Wang X. C. A Review on the Occurrence of Micropollutants in the Aquatic Environment and Their Fate and Removal during Wastewater Treatment. Sci. Total Environ. 2014, 473–474, 619–641. 10.1016/j.scitotenv.2013.12.065. [DOI] [PubMed] [Google Scholar]
- Heberer T. Occurrence, Fate, and Removal of Pharmaceutical Residues in the Acquatic Environment: A Review of Recent Research Data. Toxicol. Lett. 2002, 131, 5–17. 10.1111/j.1439-0388.1936.tb00094.x. [DOI] [PubMed] [Google Scholar]
- Jelic A.; Gros M.; Ginebreda A.; Cespedes-Sánchez R.; Ventura F.; Petrovic M.; Barcelo D. Occurrence, Partition and Removal of Pharmaceuticals in Sewage Water and Sludge during Wastewater Treatment. Water Res. 2011, 45, 1165–1176. 10.1016/j.watres.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Amasha M.; Baalbaki A.; Ghauch A. A Comparative Study of the Common Persulfate Activation Techniques for the Complete Degradation of an NSAID: The Case of Ketoprofen. Chem. Eng. J. 2018, 350, 395–410. 10.1016/j.cej.2018.05.118. [DOI] [Google Scholar]
- WHO, W. H. organization . Pharmaceuticals in Drinking-Water. Geneva World Heal. Organ, 2012. https://apps.who.int/iris/handle/10665/44630
- Burkina V.; Zlabek V.; Zamaratskaia G. Effects of Pharmaceuticals Present in Aquatic Environment on Phase I Metabolism in Fish. Environ. Toxicol. Pharmacol. 2015, 40, 430–444. 10.1016/j.etap.2015.07.016. [DOI] [PubMed] [Google Scholar]
- Rivera-Utrilla J.; Sánchez-Polo M.; Ferro-García M. Á; Prados-Joya G.; Ocampo-Pérez R. Pharmaceuticals as Emerging Contaminants and Their Removal from Water. A Review. Chemosphere 2013, 93, 1268–1287. 10.1016/j.chemosphere.2013.07.059. [DOI] [PubMed] [Google Scholar]
- Tang Z.; Wei J.; Yung L.; Ji B.; Ma H.; Qiu C.; Yoon K.; Wan F.; Fang D.; Hsiao B. S.; et al. UV-Cured Poly(Vinyl Alcohol) Ultrafiltration Nanofibrous Membrane Based on Electrospun Nanofiber Scaffolds. J. Membr. Sci. 2009, 328, 1–5. 10.1016/j.memsci.2008.11.054. [DOI] [Google Scholar]
- Majorek K. A.; Porebski P. J.; Dayal A.; Zimmerman M. D.; Jablonska K.; Stewart A. J.; Chruszcz M.; Minor W. Structural and Immunologic Characterization of Bovine, Horse, and Rabbit Serum Albumins. Mol. Immunol. 2012, 52, 174–182. 10.1016/j.molimm.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujacz A.; Zielinski K.; Sekula B. Structural Studies of Bovine, Equine, and Leporine Serum Albumin Complexes with Naproxen. Proteins: Struct., Funct., Bioinf. 2014, 82, 2199–2208. 10.1002/prot.24583. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. Phaser Crystallographic Software. J. Appl. Crystallogr. 2007, 40, 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkóczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L. W.; Kapral G. J.; Grosse-Kunstleve R. W.; et al. PHENIX: A Comprehensive Python-Based System for Macromolecular Structure Solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Features and Development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R. A.; MacArthur M. W.; Moss D. S.; Thornton J. M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. 10.1107/S0021889892009944. [DOI] [Google Scholar]
- Schrödinger L.The PyMOL Molecular Graphics System, version 1.8; Schrodinger LLC: New York, NY, 2015.
- Peters T. Jr. Serum Albumin. Adv. Protein Chem. 1985, 37, 161–245. 10.1016/S0065-3233(08)60065-0. [DOI] [PubMed] [Google Scholar]
- Dror Y.; Ziv T.; Makarov V.; Wolf H.; Admon A.; Zussman E. Nanofibers Made of Globular Proteins. Biomacromolecules 2008, 9, 2749–2754. 10.1021/bm8005243. [DOI] [PubMed] [Google Scholar]
- Formenti S.; Castagna R.; Momentè R.; Bertarelli C.; Briatico-Vangosa F. The Relevance of Extensional Rheology on Electrospinning: The Polyamide/Iron Chloride Case. Eur. Polym. J. 2016, 75, 46–55. 10.1016/j.eurpolymj.2015.12.003. [DOI] [Google Scholar]
- Kowalczyk T.; Nowicka A.; Elbaum D.; Kowalewski T. A. Electrospinning of Bovine Serum Albumin. Optimization and the Use for Production of Biosensors. Biomacromolecules 2008, 9, 2087–2090. 10.1021/bm800421s. [DOI] [PubMed] [Google Scholar]
- Regev O.; Khalfin R.; Zussman E.; Cohen Y. About the Albumin Structure in Solution and Related Electro-Spinnability Issues. Int. J. Biol. Macromol. 2010, 47, 261–265. 10.1016/j.ijbiomac.2010.04.012. [DOI] [PubMed] [Google Scholar]
- Risdian C.; Nasir M.; Rahma A.; Rachmawati H. The Influence of Formula and Process on Physical Properties and the Release Profile of PVA/BSA Nanofibers Formed by Electrospinning Technique. J. Nano Res. 2015, 31, 103–116. 10.4028/www.scientific.net/JNanoR.31.103. [DOI] [Google Scholar]
- Fazel R.; Torabi S.-F.; Naseri-Nosar P.; Ghasempur S.; Ranaei-Siadat S.-O.; Khajeh K. Electrospun polyvinyl alcohol/bovine serum albumin biocomposite membranes for horseradish peroxidase immobilization. Enzyme Microb. Technol. 2016, 93–94, 1–10. 10.1016/j.enzmictec.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Hu J.; Prabhakaran M. P.; Ding X.; Ramakrishna S. Emulsion Electrospinning of Polycaprolactone: Influence of Surfactant Type towards the Scaffold Properties. J. Biomater. Sci., Polym. Ed. 2015, 26, 57–75. 10.1080/09205063.2014.982241. [DOI] [PubMed] [Google Scholar]
- Bolto B.; Tran T.; Hoang M.; Xie Z. Crosslinked Poly(Vinyl Alcohol) Membranes. Prog. Polym. Sci. 2009, 34, 969–981. 10.1016/j.progpolymsci.2009.05.003. [DOI] [Google Scholar]
- Theron S. A.; Zussman E.; Yarin A. L. Experimental Investigation of the Governing Parameters in the Electrospinning of Polymer Solutions. Polymer 2004, 45, 2017–2030. 10.1016/j.polymer.2004.01.024. [DOI] [Google Scholar]
- Koski A.; Yim K.; Shivkumar S. Effect of Molecular Weight on Fibrous PVA Produced by Electrospinning. Mater. Lett. 2004, 58, 493–497. 10.1016/S0167-577X(03)00532-9. [DOI] [Google Scholar]
- Son W. K.; Ho Youk J.; Seung Lee T.; Park W. H. Effect of PH on Electrospinning of Poly(Vinyl Alcohol). Mater. Lett. 2005, 59, 1571–1575. 10.1016/j.matlet.2005.01.025. [DOI] [Google Scholar]
- Zong X.; Kim K.; Fang D.; Ran S.; Hsiao B. S.; Chu B. Structure and Process Relationship of Electrospun Bioabsorbable Nanofiber Membranes. Polymer 2002, 43, 4403–4412. 10.1016/S0032-3861(02)00275-6. [DOI] [Google Scholar]
- Xu Y.; Hanna M. A. Morphological and Structural Properties of Two-Phase Coaxial Jet Electrosprayed BSA-PLA Capsules. J. Microencapsulation 2008, 25, 469–477. 10.1080/02652040802049513. [DOI] [PubMed] [Google Scholar]
- Migneault I.; Dartiguenave C.; Bertrand M. J.; Waldron K. C. Glutaraldehyde: Behavior in Aqueous Solution, Reaction with Proteins, and Application to Enzyme Crosslinking. Biotechniques 2004, 37, 790–802. 10.2144/04375RV01. [DOI] [PubMed] [Google Scholar]
- Tang C.; Saquing C. D.; Sarin P. K.; Kelly R. M.; Khan S. A. Nanofibrous Membranes for Single-Step Immobilization of Hyperthermophilic Enzymes. J. Membr. Sci. 2014, 472, 251–260. 10.1016/j.memsci.2014.08.037. [DOI] [Google Scholar]
- Lin J. J.; Meyer J. D.; Carpenter J. F.; Manning M. C. Stability of Human Serum Albumin during Bioprocessing: Denaturation and Aggregation during Processing of Albumin Paste. Pharm. Res. 2000, 17, 391–396. 10.1023/A:1007564601210. [DOI] [PubMed] [Google Scholar]
- Mansur H. S.; Sadahira C. M.; Souza A. N.; Mansur A. A. P. FTIR Spectroscopy Characterization of Poly (Vinyl Alcohol) Hydrogel with Different Hydrolysis Degree and Chemically Crosslinked with Glutaraldehyde. Mater. Sci. Eng. C 2008, 28, 539–548. 10.1016/j.msec.2007.10.088. [DOI] [Google Scholar]
- Farid O.; Mansour F.; Habib M.; Robinson J.; Tarleton S. Investigating the Sorption Influence of Poly(Vinyl Alcohol) (PVA) at Different Crosslinking Content. J. Environ. Chem. Eng. 2016, 4, 293–298. 10.1016/j.jece.2015.08.007. [DOI] [Google Scholar]
- Barbosa O.; Ortiz C.; Berenguer-Murcia Á.; Torres R.; Rodrigues R. C.; Fernandez-Lafuente R. Glutaraldehyde in Bio-Catalysts Design: A Useful Crosslinker and a Versatile Tool in Enzyme Immobilization. RSC Adv. 2014, 4, 1583–1600. 10.1039/C3RA45991H. [DOI] [Google Scholar]
- Moradzadegan A.; Ranaei-Siadat S.-O.; Ebrahim-Habibi A.; Barshan-Tashnizi M.; Jalili R.; Torabi S.-F.; Khajeh K. Immobilization of Acetylcholinesterase in Nanofibrous PVA/BSA Membranes by Electrospinning. Eng. Life Sci. 2010, 10, 57–64. 10.1002/elsc.200900001. [DOI] [Google Scholar]
- Bradford M. M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Valmikinathan C. M.; Defroda S.; Yu X. Polycaprolactone and Bovine Serum Albumin Based Nanofibers for Controlled Release of Nerve Growth Factor. Biomacromolecules 2009, 10, 1084–1089. 10.1021/bm8012499. [DOI] [PubMed] [Google Scholar]
- Homaeigohar S.; Dai T.; Elbahri M. Biofunctionalized Nanofibrous Membranes as Super Separators of Protein and Enzyme from Water. J. Colloid Interface Sci. 2013, 406, 86–93. 10.1016/j.jcis.2013.05.076. [DOI] [PubMed] [Google Scholar]
- Niu Q.; Mu X.; Nie J.; Ma G. Potential Fabrication of Core-Shell Electrospun Nanofibers from a Two-Step Method: Electrospinning and Photopolymerization. J. Ind. Eng. Chem. 2016, 38, 193–199. 10.1016/j.jiec.2016.05.009. [DOI] [Google Scholar]
- Qian W.; Yu D. G.; Li Y.; Liao Y. Z.; Wang X.; Wang L. Dual Drug Release Electrospun Core-Shell Nanofibers with Tunable Dose in the Second Phase. Int. J. Mol. Sci. 2014, 15, 774–786. 10.3390/ijms15010774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekula B.; Zielinski K.; Bujacz A. Crystallographic Studies of the Complexes of Bovine and Equine Serum Albumin with 3,5-Diiodosalicylic Acid. Int. J. Biol. Macromol. 2013, 60, 316–324. 10.1016/j.ijbiomac.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Bujacz A. Structures of Bovine, Equine and Leporine Serum Albumin. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2012, 68, 1278–1289. 10.1107/S0907444912027047. [DOI] [PubMed] [Google Scholar]
- Maruthamuthu M.; Kishore S. Binding of Ketoprofen with Bovine Serum Albumin. Proc. Indian Acad. Sci. 1987, 99, 187–193. 10.1007/BF02880901. [DOI] [Google Scholar]
- Misra P. P.; Kishore N. Differential Modulation in Binding of Ketoprofen to Bovine Serum Albumin in the Presence and Absence of Surfactants: Spectroscopic and Calorimetric Insights. Chem. Biol. Drug Des. 2013, 82, 81–98. 10.1111/cbdd.12136. [DOI] [PubMed] [Google Scholar]
- Bujacz A.; Zielinski K.; Sekula B. Structural Studies of Bovine, Equine, and Leporine Serum Albumin Complexes with Naproxen. Proteins: Struct., Funct., Bioinf. 2014, 82, 2199–2208. 10.1002/prot.24583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.