Abstract
Identification of new molecular targets is needed for the treatment of colorectal cancer (CRC). Methylenetetrahydrofolate dehydrogenase 1 like (MTHFD1L), an enzyme in the folate cycle, is involved in formate generation and therefore in one-carbon metabolism. Here, we examined the expression and the role of MTHFD1L in CRC progression. Bioinformatics analysis of several public databases showed overexpression of MTHFD1L in CRC tissues as compared to normal tissues. Quantitative real-time PCR and Western blotting revealed that expressions of MTHFD1L RNA and protein were higher in CRC tissues compared to their corresponding normal tissues of CRC patients. Immunohistochemical staining demonstrated higher cytoplasmic MTHFD1L reactivity in tumor tissues compared to paired normal tissues. Further, to determine the functional relevance of MTHFD1L, it was knocked down by an siRNA in CRC cells. Silencing of MTHFD1L inhibited CRC cell proliferation, colony formation, invasion, and migration. Thus, to our knowledge for the first time in the literature, we show that MTHFD1L is involved in CRC progression and that blocking of MTHFD1L decreases the growth of colon cancer cells, thus providing an avenue to target this enzyme with small molecule inhibitors.
Introduction
Current chemotherapeutic drugs affect both tumor cells and normal proliferating cells, resulting in off-target side effects. Methotrexate and other anti-folate chemotherapeutics, which inhibit dihydrofolate reductase, are used for cancer treatment, but they have adverse side effects [1]. Thus, identification and targeting of cancer-specific metabolic enzymes overexpressed exclusively in tumors is an effective approach for cancer therapy.
Metabolic reprogramming is a hallmark of cancer growth and invasion [2]. Cancer cells rely on nucleotide metabolism, and the folate cycle is a necessary pathway for cancer growth [3], [4]. A cytoplasmic trifunctional enzyme, methylenetetrahydrofolate dehydrogenase, cyclohydrolase, and formyltetrahydrofolate synthetase 1 (MTHFD1) produces formate from 5,10-methylene-tetrahydrofolate in three reactions. The mitochondrial folate enzyme, MTHFD2/2 L, a bifunctional enzyme, and methylenetetrahydrofolate dehydrogenase 1 like (MTHFD1L), a monofunctional enzyme, perform these three reactions, generating formate in mitochondria; in cytoplasm, MTHFD1 alone performs this reaction [5], [6]. Intermediate compounds of these reactions are required for purine and pyrimidine synthesis [7]. MTHFD1L performs the last step of the folate cycle, generating formate in mitochondria and thus is involved in folate cycle maintenance [7].
The mitochondrial biosynthetic glycine pathway enzymes, MTHFD2 and MTHFD1L, which are involved in the synthesis of glycine, are associated with poor survival of breast cancer patients [2]. A study involving microarray gene profiling shows elevated expression of MTHFD2 across 1981 tumors in 19 cancer types [3]. MTHFD1L is involved in growth of hepatocellular carcinomas, and blocking MTHFD1L suppresses cancer cell growth [7]. Microarray gene-expression analysis of publicly available data shows a 2.38-fold elevation of the MTHFD1L gene in colon adenocarcinoma samples (n = 77) as compared to normal colon tissue (n = 117) [8].
In the present study, we investigated the functional role of MTHFD1L in colorectal cancer (CRC) progression. Our work demonstrated upregulation of MTHFD1L in CRC; its expression was independent of pathologic stage and patient's race, age, and gender. Silencing of MTHFD1L attenuated malignant phenotypes of colon cancer cells, such as proliferation, colony formation, invasion, and migration. In sum, we investigated the role of overexpressed MTHFD1L in the progression of colon cancer cells and found, for the first time, that it has the potential to serve as a target enzyme for therapeutic agents.
Materials and Methods
CRC Tissue Specimens
All specimens for this study, which was approved by the University of Alabama at Birmingham (UAB) Institutional Review Board, were provided by the Translational Anatomic Pathology Sections of the UAB Pathology Department. For assessing MTHFD1L gene expression by quantitative real-time PCR (qRT-PCR), 110 CRC tissues and their corresponding matched adjacent non-cancerous tissues (Stage I, 18; Stage II, 30; Stage III, 42; Stage IV, 20) were procured from patients who had undergone surgical resection. Immunoblot analysis was performed for some of the frozen CRC and their corresponding normal tissues. Immunohistochemical (IHC) analysis was accomplished with formalin-fixed, paraffin-embedded archival tissue blocks of those frozen CRC samples.
Cell Lines
McCoy's media (Corning™ cellgro™, Fisher Scientific Co., Pittsburgh, PA) was used for growing colon cancer cell lines, HT29p53-mut (p53- R273H mutant), and SW480p53-mut (p53- R273H and P309S mutant) with different p53 gene status to assess whether different p53 mutation status impacts the expression of MTHFD1L. Media were supplemented with 10% FBS (Invitrogen, ThermoFisher Scientific, Carlsbad, CA) and penicillin–streptomycin. Cells were grown in a 5% CO2 cell culture incubator.
Quantitative Real-Time PCR Analysis
Total RNA from CRC tissues and cell lines was isolated using RNeasy mini kit (Zymo Research, Irvine, CA). Complementary DNA (cDNA) was prepared from RNA using Superscript III Reverse Transcriptase (Invitrogen, Carlsbad, CA). To determine the mRNA expression level of MTHFD1L, SYBR green qRT-PCR was performed using primers synthesized by Integrated DNA Technologies (Coralville, IA). A list of primers is in Table S1.
IHC Analysis
To evaluate the protein expression of MTHFD1L, IHC was accomplished with CRC tissue specimens as described previously [9], [10]. Deparaffinization, rehydration, antigen retrieval, reduction of endogenous activity, and blocking steps were performed sequentially before probing with an antibody against MTHFD1L (Cat# 16113–1-AP, Proteintech, Irvine, CA). ImmPRESS HRP anti-rabbit IgG (Cat# MP-7401, Vector laboratories, Burlingame, CA) as secondary antibody was used as a probe followed by detection of immunoreactivity with diaminobenzidine (Cat#SK-4100, Vector laboratories, Burlingame, CA). Counterstaining was accomplished with Vector Hematoxylin QS (Cat#H-3404, Vector laboratories, Burlingame, CA).
Immunoblot Analysis
Protein samples were separated on NuPAGE™ 4–12% Bis-Tris Midi Protein Gels, 20-well (Invitrogen, ThermoFisher Scientific, Carlsbad, CA) followed by transfer onto Immobilon-P PVDF membranes (EMD Millipore, Billerica, MA). Membranes were then incubated for 1 h in blocking buffer (Tris-buffered saline, 0.1% Tween [TBS-T], 5% nonfat dry milk) followed by overnight at 4 °C with MTHFD1L primary antibody. Horseradish peroxidase-conjugated secondary antibody was used to incubate the blots. Chemiluminescence signals were visualized using Luminata™ Crescendo chemiluminescence Western blotting substrate according to the manufacturer's protocol (EMD Millipore, Billerica, MA), and signals were acquired on Amersham Imager 600RGB (GE Healthcare Life Sciences, Chicago, IL). Antibody information is provided in Table S2.
RNA Interference
Colon cancer cells were transfected with an siRNA duplex (Table S3) purchased from GE healthcare Dharmacon, Inc. (Lafayette, CO), as described previously [9]. Reverse transfection was performed using Lipofectamine RNAiMAX (Life Technologies) by preparing the transfection mixture in 6-well plates and then seeding cells at a density of 1x106 per well.
Cell Proliferation Assay
Cells (5 × 103) transfected with MTHFD1L siRNA or control siRNA were seeded in triplicate wells in 12-well plates for three time points (2, 4, and 6 days). After trypsinization, cells were counted with a Z2 Coulter particle counter (Beckman Coulter, Brea, CA).
Colonogenic Assay
Colony formation was evaluated as described previously [11], [12]. MTHFD1L knockdown and control cells (2× 103) were seeded into 6-well plates in triplicate and were allowed to grow for 10 days followed by fixation with 5% glutaraldehyde and staining with crystal violet (Sigma-Aldrich, St. Louis, MO). An Amersham Imager 600RGB (GE Healthcare Life Sciences, Pittsburgh, PA) was used for taking photographs of the colonies.
Invasion Assay
The role of MTHFD1L in the invasion capacity of colon cancer cells was investigated as described previously [13], [14]. After MTHFD1L knockdown in colon cancer cells, Corning BioCoat™ Matrigel invasion chambers (Cat#354480, Corning, New York) were used to assess cell invasion. MTHFD1L knockdown and control cells (5× 104) in serum-free media were seeded onto 8-μm pore inserts with serum-containing media as a chemoattractant in the lower chamber. After 48 h, the cells that invaded through the Matrigel and migrated to the lower side of the membrane were fixed with 5% glutaraldehyde and stained with crystal violet, followed by imaging with a phase-contrast microscope.
Wound Healing Assay
A wound-healing assay, performed as described previously [15], was used to investigate the migratory capacity of colon cancer cells. MTHFD1L knockdown and control colon cancer cells (1 × 106) in triplicate were seeded onto 35-mm Petri dishes. A wound was mechanically created on confluent monolayers by use of sterile 200-μl pipette tips, and photomicrographs were taken at 0 h and at 24 h with an inverted phase-contrast microscope with a 4X objective.
3D Spheroid Model
The Cultrex® 3D spheroid BME cell invasion assay (Cat# 3500–096-K, Trevigen, Gaithersburg, MD) was used to evaluate the spheroid forming capacity of colon cancer cells. MTHFD1L knockdown and control colon cancer cells (5× 103) were seeded with spheroid formation ECM solution in 96-well plates in triplicate according to the manufacturer's instructions, followed by centrifugation at 200 g for 3 min to position the cells in the center and then incubated at 37 °C in a 5% CO2 incubator for 72 h. After 72 h of incubation, invasion matrix (50 μl) was added to the wells followed by centrifugation at 200 g for 3 min at 4 °C. After 1 h, 100 μl of warm media supplemented with FBS was added. After 4 days of incubation, images were taken using a 4X objective.
Statistical Analysis
Student's two-tailed t-test was applied for results of all experiments. Data were expressed as means ± standard deviation, and a P-value of <.05 was considered significant.
Results
Overexpression of MTHFD1L in Colorectal Cancer Tissues
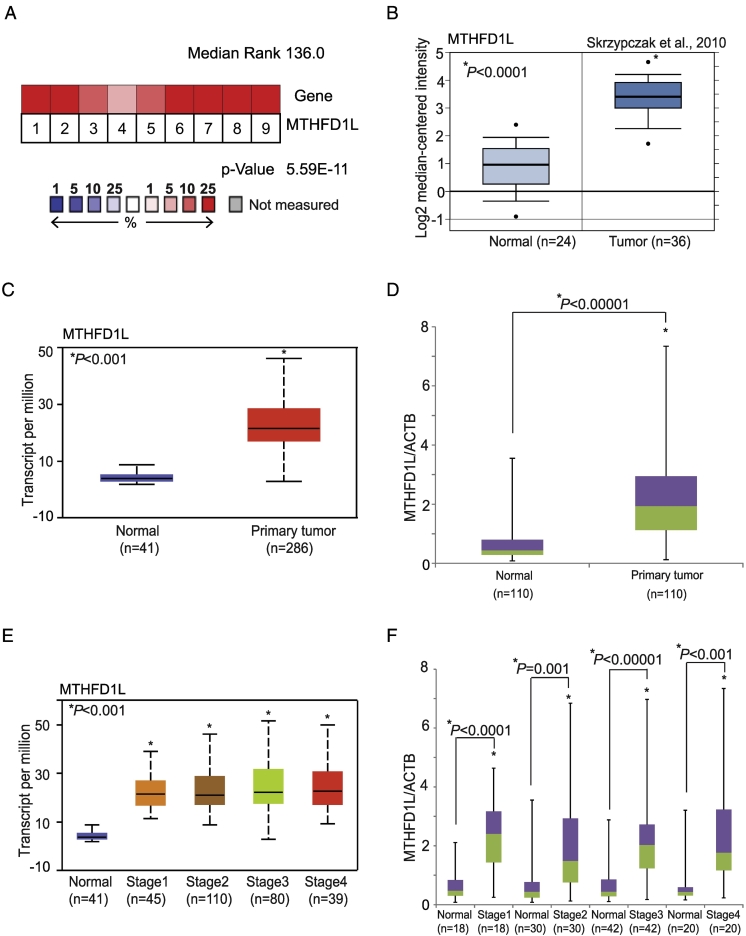
MTHFD1L gene expression was characterized using Oncomine [Oncomine™ Platform (Life Technologies, Ann Arbor, MI)] [16]. As shown in multiple, independent profiling studies, there was elevated MTHFD1L expression in CRC tissues (Figure 1A). Microarray datasets from a previous study have showed MTHFD1L overexpression in CRC (n = 36) as compared to normal colon (n = 24) (Figure 1B) [17]. Datasets from Kaiser et al., 2007 [19] (Supplementary Figure S1A) and Hong et al., 2010 [20] (Supplementary Figure S1B) also showed upregulated MTHFD1L in CRC. The bioinformatics portal UALCAN (http://ualcan.path.uab.edu) [18], which allows analysis of data generated by TCGA, showed over-expression of MTHFD1L in colon adenocarcinomas compared to normal colon (Figure 1C). We implemented a validation strategy by using qRT-PCR analysis of paired tumor and adjacent non-cancerous tissue and found overexpression of MTHFD1L in CRC tissues (n = 110) relative to normal colon tissue (n = 110) (Figure 1D). Further data mining using UALCAN for MTHFD1L expression in different pathologic stages showed MTHFD1L overexpression irrespective of the stage of the cancer (P < .01) (Figure 1E). The analysis of cancer tissues using qRT-PCR validated the TCGA data from UALCAN showing high expression of MTHFD1L (P < .01) in different stages [Stage 1 (n = 18), Stage 2 (n = 30), Stage 3 (n = 42), and Stage 4 (n = 20)]; expression was independent of the stage of the tumor (Figure 1F). Further TCGA data procured through UALCAN also showed overexpression of MTHFD1L (P < .01) independent of patient's race (Supplementary Figure S1C), tumor histologic type (Supplementary Figure S1D), gender (Supplementary Figure S1E), and age (Supplementary Figure S1F). We validated this data of MTHFD1L expression using qRT-PCR analysis and found upregulation of MTHFD1L expression (P < .01) independent of patient's race [Caucasian (n = 79) vs African-American (n = 23)] (Supplementary Figure S2A), tumor histotypes [Adenocarcinoma (n = 92) vs Mucinous adenocarcinoma (n = 18)] (Supplementary Figure S2B), gender [Male (n = 57) vs Female (n = 53)] (Supplementary Figure S2C) and age [21–40 years (n = 4), 41–60 years (n = 33), 61–80 years (n = 55) and 81–100 years (n = 18)] (Supplementary Figure S2D). This data showed overexpression of MTHFD1L in CRC as compared to normal colon irrespective of the tumor stage and histologic type, and patient's race, gender and age.
Figure 1.
Elevated expression of MTHFD1L in CRC tissues. (A) Gene expression analysis of CRC datasets showed upregulated expression of MTHFD1L in CRC tissues [16]. (B) Dataset from Skrzypczak et al., 2010 [17] showing overexpression of MTHFD1L in CRCs (n = 36) as compared to normal colon tissues (n = 24). (C) TCGA data acquired from UALCAN[18] with MTHFD1L expression in normal colon (n = 41) and CRCs tissues (n = 286). (D) qRT-PCR analysis showing mRNA levels of MTHFD1L in CRCs (n = 110) and matched adjacent non-cancerous tissue (n = 110). (E) Stage-wise expression of MTHFD1L transcripts from UALCAN in CRC tissues Stage I (n = 45), Stage II (n = 110), Stage (n = 80), and Stage IV (n = 39) and normal (n = 41). (F) MTHFD1L mRNA expression in paired samples from different stages of CRC [(Stage 1 (n = 18), Stage 2 (n = 30), Stage 3 (n = 42), and Stage 4 (n = 20)].
MTHFD1L Protein is Up-Regulated in Tissues of CRC Patients
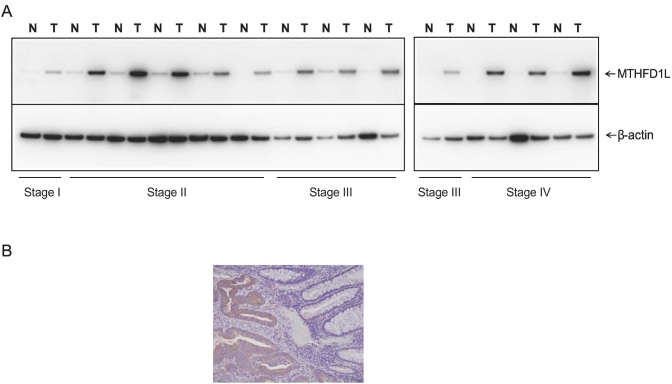
To investigate whether MTHFD1L upregulation was also apparent at the protein level, we performed immunoblot analyses and analyzed stage-wise expression of MTHFD1L protein in matched normal and cancer tissues. MTHFD1L protein expression was upregulated in various pathological stages of CRC tissues compared with matched normal colon tissues (Figure 2A). To locate MTHFD1L protein expression in human tissues, IHC analysis was performed on colon tissue sections containing both normal and CRC tissue. The expression of MTHFD1L was localized predominantly in the cytoplasm of the tumor cells as compared to normal colon tissue sections (Figure 2B). These results indicated overexpression of MTHFD1L protein in CRCs irrespective of disease stage.
Figure 2.
MTHFD1L protein overexpression in CRC tissues. (A) Immunoblot analysis to showing MTHFD1L protein expression in different stages of CRCs by probing of lysates of paired CRCs and matched adjacent non-cancerous tissues with MTHFD1L antibody. β-Actin was used as a loading control. (B) Photographs of MTHFD1L immunostaining using MTHFD1L antibody in tissue containing CRC cells and normal cells.
Role of MTHFD1L in Colon Cancer Cell Growth
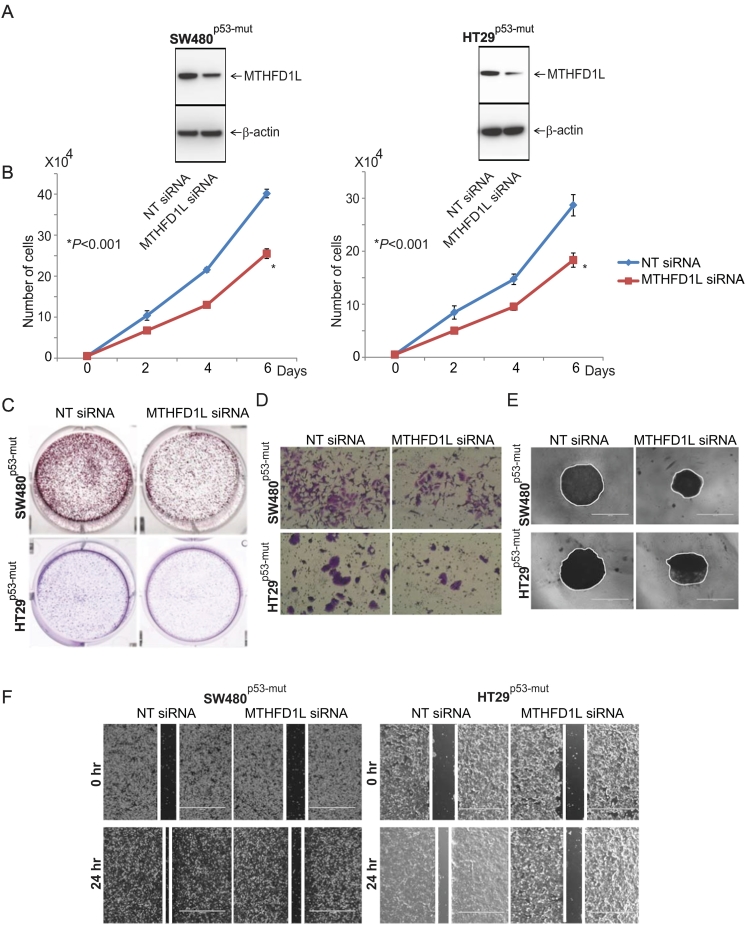
We performed cell growth assays to understand the functional relevance of MTHFD1L in colon cancer cells. We used lentivirus-mediated gene silencing to knock down MTHFD1L in colon cancer cells, HT29p53-mut and SW480p53-mut (p53 mutated). Immunoblot analysis confirmed target knockdown using MTHFD1L siRNA (Figure 3A). Further, we studied the effect of MTHFD1L on proliferation and colony formation of colon cancer cells. For SW480 and HT29 cells, MTHFD1L knockdown reduced proliferation by 36% (P < .01) at 6 days as compared to cells exposed to control siRNA (Figure 3B). Colony formation of cancer cells was also reduced after MTHFD1L knockdown as compared to control cells. (Figure 3C).
Figure 3.
Silencing of MTHFD1L reduced malignant phenotypes of colon cancer. (A) Colon cancer cells, SW480 and HT29, were transiently transfected with MTHFD1L siRNA, and immunoblot analyses were performed to show knockdown of MTHFD1L. (B) Proliferation assay of cells transfected with MTHFD1L siRNA or control siRNA at 2, 4, and 6 days (∗P < .001). (C) Representative images of colony formation after transfection of cells with control or MTHFD1L siRNA. (D) Cells transfected with MTHFD1L siRNA or control siRNA and plated in 8-μm pore invasion chambers with Matrigel. After 48 h, cells that invaded through the pores were fixed and stained. Photographs of invaded cells are shown. (E) Spheroid formation assay after transfection of MTHFD1L siRNA or control siRNA. (F) Wound healing assay after transfection with MTHFD1L siRNA or control siRNA.
To investigate the effect of MTHFD1L on the invasion capacity of colon cancer cells, we performed Transwell membrane assays with HT29p53-mut and SW480p53-mut cells. Transfection with MTHFD1L siRNA reduced the invasion capacity compared with cells transfected with control siRNA (Figure 3D). To understand the role of MTHFD1L in this process, we performed 3D spheroid formation assays. 3D spheroid cultures recapitulate in vivo growth conditions of cancer [21]. MTHFD1L knockdown reduced spheroid formation of HT29p53-mut and SW480p53-mut colon cancer cells (Figure 3E).
Further, we performed a wound-healing assay to elucidate the role of MTHFD1L in the spread of tumor cells to distant organs. Colon cancer cells (1 × 106) transfected with MTHFD1L siRNA were seeded in 6-well plates; artificial wounds were created in confluent monolayers using 200 μl pipet tips; and images were acquired at 0 and 24 h. The photographs showed a decrease in the motility of MTHFD1L knockdown cells compared to those transfected with control siRNA (Figure 3F). In sum, these data show that suppression of MTHFD1L decreases cell proliferation, colony formation, invasion, motility, and spheroid formation and support a role for MTHFD1L in the progression of colon cancer cells. Blocking of MTHFD1L expression attenuated malignant phenotypes of colon cancer cells, suggesting a role of this enzyme in the malignant properties of colon cancer. It is noteworthy that none of these cell biological features measured by altering the MTHFD1L status (silenced vs. not silenced) differed based on the p53 status of the cells.
Discussion
CRC occurrence and progression involves a series of genetic and epigenetic events affecting cell structure and expression of various oncogenes and tumor suppressors [22], [23]. The folate cycle has co-factors and co-substrates in one-carbon metabolism that influence CRC progression [24]. In the current study, we demonstrated that, in CRCs, the folate metabolic enzyme MTHFD1L was overexpressed at both mRNA and protein levels and then explored its functional relevance by performing gene knockdown.
Meta-analysis of TCGA data helps cancer researchers in identifying potential prognostic biomarkers. For instance, a study of mitochondrial folate metabolism shows that mRNA expression of enzymes SHMT2, MTHFD2, and ALDH1L2 associated with this metabolism are higher in CRC tissues as compared to normal tissue [25]. Using the Oncomine analysis, we found that, as compared with normal colon tissues, MTHFD1L expression is upregulated in CRC tissues [16]. Acquired through TCGA from UALCAN, RNAseq data of CRC and normal tissue confirmed overexpression of MTHFD1L in CRCs. In addition, we showed, by qRT-PCR analysis of paired CRC and normal tissues, that MTHFD1L mRNA was upregulated in CRCs as compared to normal tissue. Furthermore, there was overexpression of MTHFD1L mRNA in various tumor stages and histologic types. Overexpression was not affected by patient's race, age, or gender. In addition, IHC analysis showed MTHFD1L expression in CRCs, but it was minimally present in adjacent noncancerous colon tissues. Collectively, our data show that MTHFD1L is upregulated in CRC and indicate that it could be involved in the pathogenesis of the disease.
Previous studies showed the role of MTHFD1L in tumorigenesis and concluded that this enzyme is necessary for the survival of hepatocellular carcinoma and esophageal squamous cell carcinoma [7], [26]. In the present study, we demonstrated that MTHFD1L was overexpressed in CRCs and could be required for colon cancer cell proliferation and invasion. Gene silencing of MTHFD1L resulted in reduced cell migration and growth of colon cancer cells. Our results demonstrated that MTHFD1L expression status in various pathologic stages of CRCs and MTHF1L-regulated cellular functions in cultures are not associated with p53 mutations and MSI stable status. Thus, these findings suggest that MTHFD1L is oncogenic and is involved in the invasion and metastasis of colon cancer cells.
In summary, in CRCs, there is elevated expression of MTHFD1L mRNA and protein, and expression of this enzyme is independent of the tumor stage and histotype as well as the race, gender, and age of patients. Further, the elevated expression of MTHFD1L is involved in progression of these cancers; knocking down MTHFD1L reduces colon cancer cell growth. In conclusion, our findings suggest that increased expression of MTHFD1L is involved in the progression of CRC. Furthermore, in the treatment of CRC, MTHFD1L has potential for targeted inhibition by small molecules.
Acknowledgments
The authors thank Dr. Donald Hill, a faculty member in the UAB Comprehensive Cancer Center, University of Alabama at Birmingham, Birmingham, for critical reading and editing of this manuscript.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
Funding/support: UAB O'Neal Comprehensive Cancer Center and UAB School of Medicine Developmental funds
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2019.07.011.
Contributor Information
Upender Manne, Email: upendermanne@uabmc.edu.
Sooryanarayana Varambally, Email: svarambally@uabmc.edu.
Appendix A. Supplementary data
Supplementary Figure S1. MTHFD1L gene expression in colon adenocarcinoma- Gene expression profiling studies acquired from two different data sets, (A) Kaiser et al., 2007 and (B) Hong et al., 2010 showed elevated expression MTHFD1L in colorectal tumors as compared to normal colon. TCGA data from UALCAN showed MTHFD1L expression with respect to patient’s race (C), tumor histotypes (D), patient’s gender (E) and age (F). Related to figure 1. Supplementary Figure S2. MTHFD1L expression in colon cancer. qRT-PCR to show MTHFD1L expression in patient’s race (A), tumor histotypes (B), patient’s gender (C) and different age groups (D). Related to figure 1.
Supplementary tables
References
- 1.Miller DR. A tribute to Sidney Farber-- the father of modern chemotherapy. Br J Haematol. 2006;134:20–26. doi: 10.1111/j.1365-2141.2006.06119.x. [DOI] [PubMed] [Google Scholar]
- 2.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, Huang J, Asplund A, Mootha VK. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng Y, Lin TY, Lee G, Paddock MN, Momb J, Cheng Z, Li Q, Fei DL, Stein BD, Ramsamooj S. Mitochondrial One-Carbon Pathway Supports Cytosolic Folate Integrity in Cancer Cells. Cell. 2018;175:1546–60 e17. doi: 10.1016/j.cell.2018.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacFarlane AJ, Perry CA, McEntee MF, Lin DM, Stover PJ. Mthfd1 is a modifier of chemically induced intestinal carcinogenesis. Carcinogenesis. 2011;32:427–433. doi: 10.1093/carcin/bgq270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moruzzi S, Guarini P, Udali S, Ruzzenente A, Guglielmi A, Conci S, Pattini P, Martinelli N, Olivieri O, Tammen SA. One-carbon genetic variants and the role of MTHFD1 1958G>A in liver and colon cancer risk according to global DNA methylation. PLoS One. 2017;12 doi: 10.1371/journal.pone.0185792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee D, Xu IM, Chiu DK, Lai RK, Tse AP, Lan Li L, Law CT, Tsang FH, Wei LL, Chan CY. Folate cycle enzyme MTHFD1L confers metabolic advantages in hepatocellular carcinoma. J Clin Invest. 2017;127:1856–1872. doi: 10.1172/JCI90253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugiura T, Nagano Y, Inoue T, Hirotani K. A novel mitochondrial C1-tetrahydrofolate synthetase is upregulated in human colon adenocarcinoma. Biochem Biophys Res Commun. 2004;315:204–211. doi: 10.1016/j.bbrc.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 9.Chakravarthi BV, Goswami MT, Pathi SS, Dodson M, Chandrashekar DS, Agarwal S, Nepal S, Hodigere Balasubramanya SA, Siddiqui J, Lonigro RJ. Expression and Role of PAICS, a De Novo Purine Biosynthetic Gene in Prostate Cancer. Prostate. 2017;77:10–21. doi: 10.1002/pros.23243. [DOI] [PubMed] [Google Scholar]
- 10.Agarwal S, Parashar D, Gupta N, Jagadish N, Thakar A, Suri V, Kumar R, Gupta A, Ansari AS, Lohiya NK. Sperm associated antigen 9 (SPAG9) expression and humoral response in benign and malignant salivary gland tumors. Oncoimmunology. 2014;3 doi: 10.4161/2162402X.2014.974382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakravarthi B, Rodriguez Pena MDC, Agarwal S, Chandrashekar DS, Hodigere Balasubramanya SA, Jabboure FJ, Matoso A, Bivalacqua TJ, Rezaei K, Chaux A. A Role for De Novo Purine Metabolic Enzyme PAICS in Bladder Cancer Progression. Neoplasia. 2018;20:894–904. doi: 10.1016/j.neo.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saini S, Agarwal S, Sinha A, Verma A, Parashar D, Gupta N, Ansari AS, Lohiya NK, Jagadish N, Suri A. Gene silencing of A-kinase anchor protein 4 inhibits cervical cancer growth in vitro and in vivo. Cancer Gene Ther. 2013;20:413–420. doi: 10.1038/cgt.2013.32. [DOI] [PubMed] [Google Scholar]
- 13.Chakravarthi B, Chandrashekar DS, Agarwal S, Balasubramanya SAH, Pathi SS, Goswami MT, Jing X, Wang R, Mehra R, Asangani IA. miR-34a Regulates Expression of the Stathmin-1 Oncoprotein and Prostate Cancer Progression. Mol Cancer Res. 2018;16:1125–1137. doi: 10.1158/1541-7786.MCR-17-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jagadish N, Parashar D, Gupta N, Agarwal S, Suri V, Kumar R, Suri V, Sadasukhi TC, Gupta A, Ansari AS. Heat shock protein 70-2 (HSP70-2) is a novel therapeutic target for colorectal cancer and is associated with tumor growth. BMC Cancer. 2016;16:561. doi: 10.1186/s12885-016-2592-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakravarthi BV, Goswami MT, Pathi SS, Robinson AD, Cieslik M, Chandrashekar DS, Agarwal S, Siddiqui J, Daignault S, Carskadon SL. MicroRNA-101 regulated transcriptional modulator SUB1 plays a role in prostate cancer. Oncogene. 2016;35:6330–6340. doi: 10.1038/onc.2016.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D, Pachlewski J, Oledzki J, Ostrowski J. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser S, Park YK, Franklin JL, Halberg RB, Yu M, Jessen WJ, Freudenberg J, Chen X, Haigis K, Jegga AG. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007;8:R131. doi: 10.1186/gb-2007-8-7-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong Y, Downey T, Eu KW, Koh PK, Cheah PY. A 'metastasis-prone' signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin Exp Metastasis. 2010;27:83–90. doi: 10.1007/s10585-010-9305-4. [DOI] [PubMed] [Google Scholar]
- 21.Ishiguro T, Ohata H, Sato A, Yamawaki K, Enomoto T, Okamoto K. Tumor-derived spheroids: Relevance to cancer stem cells and clinical applications. Cancer Sci. 2017;108:283–289. doi: 10.1111/cas.13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L, Zhang Y, Wong CC, Zhang J, Dong Y, Li X, Kang W, Chan FKL, Sung JJY, Yu J. RNF6 Promotes Colorectal Cancer by Activating the Wnt/beta-Catenin Pathway via Ubiquitination of TLE3. Cancer Res. 2018;78:1958–1971. doi: 10.1158/0008-5472.CAN-17-2683. [DOI] [PubMed] [Google Scholar]
- 23.Tomkovich S, Yang Y, Winglee K, Gauthier J, Muhlbauer M, Sun X, Mohamadzadeh M, Liu X, Martin P, Wang GP. Locoregional effects of microbiota in a preclinical model of colon carcinogenesis. Cancer Res. 2017;77:2620–2632. doi: 10.1158/0008-5472.CAN-16-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gylling B, Van Guelpen B, Schneede J, Hultdin J, Ueland PM, Hallmans G, Johansson I, Palmqvist R. Low folate levels are associated with reduced risk of colorectal cancer in a population with low folate status. Cancer Epidemiol Biomarkers Prev. 2014;23:2136–2144. doi: 10.1158/1055-9965.EPI-13-1352. [DOI] [PubMed] [Google Scholar]
- 25.Miyo M, Konno M, Colvin H, Nishida N, Koseki J, Kawamoto K, Tsunekuni K, Nishimura J, Hata T, Takemasa I. The importance of mitochondrial folate enzymes in human colorectal cancer. Oncol Rep. 2017;37:417–425. doi: 10.3892/or.2016.5264. [DOI] [PubMed] [Google Scholar]
- 26.Yang YS, Yuan Y, Hu WP, Shang QX, Chen LQ. The role of mitochondrial folate enzyme MTHFD1L in esophageal squamous cell carcinoma. Scand J Gastroenterol. 2018;53:533–540. doi: 10.1080/00365521.2017.1407440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. MTHFD1L gene expression in colon adenocarcinoma- Gene expression profiling studies acquired from two different data sets, (A) Kaiser et al., 2007 and (B) Hong et al., 2010 showed elevated expression MTHFD1L in colorectal tumors as compared to normal colon. TCGA data from UALCAN showed MTHFD1L expression with respect to patient’s race (C), tumor histotypes (D), patient’s gender (E) and age (F). Related to figure 1. Supplementary Figure S2. MTHFD1L expression in colon cancer. qRT-PCR to show MTHFD1L expression in patient’s race (A), tumor histotypes (B), patient’s gender (C) and different age groups (D). Related to figure 1.
Supplementary tables