Abstract
Histone Lys-specific demethylases (KDMs) play a key role in many biological processes through epigenetic mechanisms. However, the role of KDMs in inflammatory responses to oral bacterial infection is poorly understood. Here, we show a novel regulatory role of KDM3C in inflammatory responses to oral bacterial infection. KDM3C expression is transiently suppressed in human and mouse macrophages exposed to LPS from Porphyromonas gingivalis (Pg LPS). Loss of KDM3C in both human and mouse macrophages led to notable induction of proinflammatory cytokines in response to Pg LPS stimulation. Also, KDM3C depletion led to strong induction of p65 phosphorylation and accelerated nuclear translocation in cells exposed to Pg LPS. Kdm3C knockout (KO) in mice led to increased alveolar bone destruction upon induction of experimental periodontitis or pulp exposure compared with those of the wild-type (WT) littermates. The Kdm3C KO mice also revealed an increased number of osteoclasts juxtaposed to the bony lesions. We also confirmed enhanced osteoclastogenesis by bone marrow–derived macrophages isolated from the Kdm3C KO compared with the WT controls. These findings suggest an anti-inflammatory function of KDM3C in regulating the inflammatory responses against oral bacterial infection through suppression of NF-κB signaling and osteoclastogenesis.—Lee, J. Y., Mehrazarin, S., Alshaikh, A., Kim, S., Chen, W., Lux, R., Gwack, Y., Kim, R. H., Kang, M. K. Histone Lys demethylase KDM3C demonstrates anti-inflammatory effects by suppressing NF-κB signaling and osteoclastogenesis.
Keywords: epigenetics, histone methylation, oral inflammation, JMJD1C, periodontitis
Oral inflammatory lesions resulting in periodontal bone loss at the alveolar crest or periapex are induced by inflammatory tissue responses to polymicrobial infection by pathogenic bacteria, which primarily include gram-negative obligate anaerobes [e.g., Porphyromonas gingivalis (Pg), Tannerella forsythia, and Treponema denticola] (1). These gram-negative anaerobes frequently trigger host inflammatory responses in part through the release of LPS, known as endotoxin, a component of the outer cell membrane recognized by TLR4 (2). The recognition of the endotoxin by the host cells then activates the inflammatory responses via NF-κB signaling through the myeloid differentiation primary response 88/IL-1 receptor (IL-1R)–associated kinase/TNF receptor associated factor 6 (TRAF6) signaling complex, leading to expression of proinflammatory cytokines (3, 4). These cytokines (e.g., IL-1β, IL-6, and TNF-α) play the key roles in many different pathophysiologic processes of inflammatory lesions, including the invasion of microbes or foreign substrates, and cause alveolar bone destruction in oral inflammatory lesions.
Growing evidence supports the importance of histone Lys-specific demethylases (KDMs) in epigenetic regulation of inflammatory responses. An earlier study showed a dynamic regulatory role of histone 3 Lys 9 (H3K9) methylation enrichment on the expression of inflammatory cytokines [e.g., Epstein-Barr virus (EBV)-induced molecular 1 ligand chemokine, macrophage-derived chemokine, and the p40 subunit of IL-12] (5). Subsequently, Lys-specific methyltransferase, G9a, was shown to bind at the promoter region of TNF in human monocytes to suppress the gene expression and elicit the endotoxin-tolerant phenotype through H3K9 methylation (6). The tolerant phenotype occurred with elevation of RelB, a repressive NF-κB subunit, in blood mononuclear cells demonstrating suppression of inflammatory cytokines (7). Interestingly, G9a is recruited to the cytokine gene promoters by RelB to trigger the target cytokine gene silencing through H3K9 methylation (8). These studies illustrate the dynamicity of the epigenetic gene regulation by histone Lys-modifying enzymes during inflammation.
In search of KDMs that directly regulate inflammatory signaling, De Santa et al. (9) reported the discovery of KDM6B, specific for Histone 3 Lys 27 trimethylation (H3K27me3), as an epigenetic factor that regulates proinflammatory responses to endotoxin challenge. This prior study showed rapid induction of KDM6B in macrophages exposed to LPS in a manner that depends on NF-κB activation, resulting in the elevated release of inflammatory mediators. In the current study, we identified KDM3C [also named Jumonj Domain-Containing (JMJD) 1C, JMJD1C or thyroid receptor-interacting protein 8 (TRIP8)] as an anti-inflammatory epigenetic regulator against oral bacterial infection in the pulp and periodontium. Our data demonstrate the role of KDM3C in regulation of the inflammatory responses to oral bacterial infection in cells and in animal models. We first identified the suppression of KDM3C expression upon cellular exposure to LPS among other Jumonji C (JmjC) domain–containing KDMs through PCR screening. Transient knockdown and complete knockout (KO) of KDM3C/Kdm3C in both human and mouse macrophages led to notable induction of IL-1β, IL-6, and TNF-α and robust induction of NF-κB subunit p65 activation when exposed to LPS from Pg (Pg LPS). Using a Kdm3C KO mouse model, we showed a significant increase in the alveolar and periapical bone destruction by oral inflammatory lesions with KDM3C depletion. Furthermore, the loss of KDM3C led to enhanced osteoclastogenesis by receptor activator of NF-κB ligand (RANKL) treatment in bone marrow–derived macrophages (BMDMs). These data suggest, for the first time, an anti-inflammatory function of KDM3C against oral bacterial infection in periapical and periodontal tissues, possibly through the mechanism that suppresses NF-κB signaling.
MATERIALS AND METHODS
Cell culture and reagents
Human monocytic Tohoku Hospital Pediatrics-1 (THP-1) cells were obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (Thermo Fisher Scientific) and 1% antibiotic-antimycotic solution. THP-1 cells were differentiated into macrophages with 100 nM phorbol 12-myristate 13-acetate (PMA; MilliporeSigma, Burlington, MA, USA). Human oral keratinocyte-16B (HOK-16B) cells were cultured in EpiLife Medium (Thermo Fisher Scientific) with Human Keratinocyte Growth Supplement (Thermo Fisher Scientific) and 60 µM calcium. Bone marrow cells were obtained by flushing the femurs and tibia of 6–8-wk-old Kdm3c wild-type (WT) or KO C57BL/6J mice. BMDMs were cultured in α–minimum essential medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum and 1% antibiotic-antimycotic solution and differentiated with 30 ng/ml M-CSF (R&D Systems, Minneapolis, MN, USA). To induce osteoclastogenesis, BMDMs were exposed to 100 ng/ml RANKL (R&D Systems). After 0–5 d, cells were fixed and stained for tartrate-resistant acid phosphatase (TRAP) activity or harvested for quantitative PCR (qPCR). To induce acute inflammatory signaling, PMA-differentiated THP-1 cells and M-CSF–differentiated BMDMs were challenged with Pg LPS from InvivoGen (San Diego, CA, USA) and LPS from Escherichia coli (Ec LPS) (MilliporeSigma) for various time points from 0 to 24 h, and the cells were harvested for biochemical assays.
For transient knockdown of human KDM3C pooled and validated small interfering RNA (siRNA) targeting human KDM3C mRNA (si-KDM3C) were obtained from Dharmacon (Lafayette, CO, USA). PMA-stimulated THP-1 cells were seeded in 6-well plates and transfected with 40 nM si-Kdm3C (Dharmacon) or the siRNA negative control (si-control; Dharmacon) using the Lipofectamine RNAiMax reagent (Thermo Fisher Scientific) according to the manufacturer’s guidelines. Sequences of si-control and si-KDM3C are described in Supplemental Table S3.
Mice
Heterozygous Kdm3C+/− mice were obtained from Riken (Tsukuba, Japan), courtesy of Dr. Makoto Tachibana (Tokushima University, Tokushima, Japan), and crossed to generate homozygous Kdm3C KO mice. Kdm3C WT and mutant alleles were identified by PCR according to the method published by Kuroki et al. (10). We used the WT primers 5′-CTCTGTAGTCCCCGCACTCA-3′ (forward) and 5′-CTAGCATTGAATCACAGGGGCTG-3′ (reverse) and mutant primers 5′-CCTTCTTGACGAGTTCTTCTGAGG-3′ (forward) and 5′-CTAGCATTGAATCACAGGGGCTG-3′ (reverse), yielding products of 670 bp (WT) and 500 bp (KO). All animal studies were performed according to the guidelines of the University of California–Los Angeles (UCLA) Institutional Animal Care and Use Committee.
Induction of oral inflammatory lesions
Mice were anesthetized with 100 mg/kg of ketamine hydrochloride and 5 mg/kg of xylazine. To induce pulpal exposure, maxillary first molars were exposed using 1/4 round bur on a dental drill and size-15 endodontic file under dissecting microscope (BM-LED Stereo Microscope; Meiji Techno, Saitama, Japan) as previously described by He et al. (11). Following pulpal exposure, the cavity was left open to the oral cavity for 21 d, and the maxillae were harvested for micro–computed tomography (μ-CT) and histologic analyses. To induce experimental periodontitis, a ligature was placed around the left maxillary second molar using 6-0 silk suture (Teleflex, Wayne, PA, USA) with a surgical knot (12). After 21 d, the mice were euthanized, and maxillae were collected for further analysis. The mice tissues were harvested and analyzed for extent of alveolar bone loss by μ-CT and histology by hematoxylin and eosin (H&E) staining.
μ-CT analysis
The harvested maxillae were fixed in 4% paraformaldehyde (Thermo Fisher Scientific) for 24 h and scanned for µ-CT using Scanco μ-CT 40 (Scanco Medical, Brüttisellen, Switzerland) using 1-mm aluminum filter and a voxel size of 10 μm3 in high-resolution settings at 60 kVp and 166 µA. The reconstruction was performed by NRecon Reconstruction software (Bruker, Billerica, MA, USA), and 3-dimensional images were reconstructed using DataViewer CTVOx and CTvol software (Bruker). Morphologic parameters of apical periodontitis in maxilla were assessed using the CTAn software (Bruker) as previously described by Bouxsein et al. (13). The extent of periapical radiolucent area was measured by calculating the difference between the bone volume and tissue volume from the first molar midroot to the apex of the radiolucent area. To measure the alveolar bone height changes in the periodontitis model, we calculated the distance from the cemento-enamel junction to the alveolar crest from the infected site and normalized with the control site. After the scanning, the tissues were decalcified in 5% EDTA and 4% sucrose in PBS (pH 7.4) for 3 wk at 4°C and sectioned for histologic analyses.
Histology analysis and TRAP histochemical staining
Paraffin-embedded histologic sections (5 µm) were mounted and stained with H&E for histologic examination. To detect osteoclasts in Kdm3c WT and KO mice in situ, the histologic sections were stained with the TRAP solution (MilliporeSigma). Quantitative measurement of the TRAP staining was performed on digital pictures taken through Olympus microscope (Olympus, Tokyo, Japan) at ×100 magnification by counting the TRAP+ multinucleated cells with more than 5 nuclei (14).
RNA extraction and real-time qPCR
Total RNA was isolated from cells using the Trizol reagent (Thermo Fisher Scientific) following the manufacturer’s instructions. DNA-free total RNA (5 µg) with Random Primer (Thermo Fisher Scientific) was used for reverse transcription by SuperScript II Reverse Transcriptase (Thermo Fisher Scientific). Real-time qPCR was performed using a QuantStudio 3 (Thermo Fisher Scientific) and PowerUp Sybr Green Master Mix (Thermo Fisher Scientific) to measure the expression of genes under the following conditions: 45 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 60 s. The comparative Ct (ΔΔCt) method was used to determine the relative gene expression levels. For normalization of the cycling threshold values obtained with the experimental samples, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was amplified in the same condition. Primer sequences of the genes are detailed in the Supplemental Table S1. All experiments were independently repeated at least 3 times to ensure reproducibility.
Western blotting
Cells were harvested in cold PBS by scraping and lysed in the lysis buffer [20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1 time Roche Complete Protease Inhibitor cocktail (Roche, Basel, Switzerland), and 1×PhosStop phosphatase inhibitor]. After elution using the 4× sample buffer, protein extracts were boiled, separated by SDS-PAGE, transferred to a PVDF membrane filter with 0.45-µm pore size (MilliporeSigma), and subjected to Western blot analysis. Chemiluminescence signal was detected by using the Clarity or Clarity Max Western ECL Blotting Substrate (Bio-Rad, Hercules, CA, USA). Each experiment was performed 3 times. The following antibodies were used for Western blot analyses: mouse anti-KDM3C (D356-3) from Medical & Biologic Laboratories (Nagoya, Japan); human anti-KDM3C (sc-101073), anti–IL-1β (sc-1250), anti-p65 (sc-8008), anti–IκB-α (sc-56710), and anti-GAPDH (sc-25778) from Santa Cruz Biotechnology (Dallas, TX, USA); mouse anti–TNF-α (11948S), human anti–TNF-α (6945S), mouse anti–IL-6 (12912S), and anti–phosphorylated-p65 (3033S) from Cell Signaling Technology (Danvers, MA, USA).
ELISA for cytokines
After stimulation of cells with Pg LPS, the cytokine levels in the cell culture supernatants from KDM3C–knocked-down THP-1 and control cells or Kdm3c WT and KO BMDMs were measured using TNF-α, IL-6, and Il-1β ELISA kits according to the manufacturer’s instructions (Thermo Fisher Scientific). All reactions were independently repeated at least 3 times to ensure reproducibility, and the values were calculated on the basis of a standard curve constructed for each assay.
In situ immunofluorescence staining
BMDMs from Kdm3C WT and KO mice were seeded on a 4-well chamber slide (Thermo Fisher Scientific) and stimulated with Pg LPS. The cells were then washed with PBS twice and fixed in 4% formaldehyde for 20 min, followed by blocking for 1 h with PBS containing 5% bovine serum albumin and incubation with primary antibodies at 4°C for overnight. Anti-p65 antibody (sc-8008, 1:100; Santa Cruz Biotechnology) was used to detect nuclear translocation of the NF-κB subunit p65. After 3 washes with PBS, slides were incubated for 1 h with 488-conjugated goat anti-mouse IgG (1:500; Thermo Fisher Scientific) at room temperature in the dark. Cells were counterstained with DAPI (MilliporeSigma), and slides were mounted with ImmunoHistoMount (MilliporeSigma). An FV10i confocal laser scanning microscope (Olympus) was used to image the fluorescent signals. Stained cells with intranuclear signal were counted via ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed by using the Magnify ChIP system according to the manufacturer’s instructions (Thermo Fisher Scientific). Briefly, cells were cross-linked with 1% formaldehyde, and the reaction was stopped with 125 mM glycine. After protein-DNA crosslinking, cells were lysed, and the chromatin was sonicated to yield DNA fragments of 300–500 bp in length. Cellular lysates were diluted in ChIP dilution buffer, and chromatin solutions were complexed with 4 μg anti-KDM3C antibody (Santa Cruz Biotechnology), anti–H3K9 monomethylation (H3K9me1) antibody (ab9045; Abcam, Cambridge, United Kingdom), anti–H3K9 dimethylation (H3K9me2) antibody (ab1220; Abcam), or 1 μg mouse IgG–conjugated Magnetic Protein A/G beads overnight. The immune complexes were precipitated and eluted, and the precipitated DNA was used for PCR amplification to assess the enrichment of KDM3C on the target gene promoters. qPCR was performed, and samples pulled down with IgG were used as a negative control. qPCR readout was normalized relative to the amount of amplification from the input. The primer sequences for DNA amplification are detailed in Supplemental Table S2.
Statistical analysis
Statistical analyses were performed with an unpaired Student’s t test for 2 groups using Prism 6 software (GraphPad Software, La Jolla, CA, USA). A value of P < 0.05 was considered as a statistically significant difference. The graphs represent mean values and include sd unless stated otherwise. All experiments were repeated 2 or 3 times. The sample size of the animals used in our experiments and of the public datasets are indicated in the respective figure legends.
RESULTS
KDM3C regulates proinflammatory cytokine expression in human macrophages challenged with Pg LPS
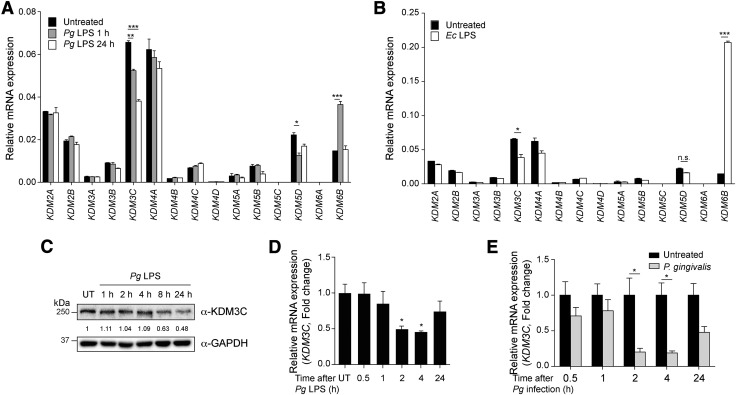
To investigate the role of KDMs in inflammatory responses triggered with Pg LPS, we screened for differential mRNA expression levels of the human KDMs in THP-1 cells exposed to Pg LPS. Because macrophages play a crucial role in the host defense against bacterial infection (15), fully differentiated THP-1 macrophages were exposed to Pg LPS, and the KDM levels were determined by real-time qPCR. Among the 15 KDMs screened, Pg LPS stimulation led to significant down-regulation of KDM3C and KDM5D as well as transient up-regulation of KDM6B known as JMJD3 (Fig. 1A). Exposure to Ec LPS also resulted in similar down-regulation of KDM3C (Fig. 1B), indicating that KDM3C expression is altered by both Pg and Ec LPS. It has been reported that KDM6B is up-regulated by LPS treatment and mediates the induction of multiple downstream inflammatory genes (16, 17). To confirm the altered expression of KDM3C in response to Pg LPS, Western blotting was performed for KDM3C in THP-1 cells stimulated with Pg LPS. We found a progressive decrease in the KDM3C protein level during the 24-h period after exposure to Pg LPS (Fig. 1C). The KDM3C mRNA level also decreased after Pg LPS treatment in a time-dependent manner (Fig. 1D). Furthermore, the exposure of THP-1 to live Pg bacteria led to a similar decrease in KDM3C mRNA expression (Fig. 1E). Also, we inoculated the cultured HOK-16B cells with live Pg or Fusobacterium nucleatum (Fn) to examine their effect on the expression of KDM3C. Western blotting showed a precipitous loss of the KDM3C protein level in the cells exposed to Pg, whereas the protein level remained unchanged in the cells exposed to Fn (Supplemental Fig. S1). Thus, down-regulation of KDM3C by bacterial infection may in part depend on the bacterial species.
Figure 1.
KDM3C is negatively regulated by Pg and its LPS treatment in THP-1 cells. A, B) The mRNA expression profiling of KDMs in PMA-differentiated THP-1 cells treated with Pg LPS (1 µg/ml, 1 and 24 h) (A) or Ec LPS (1 µg/ml, 1 h) (B). The mRNA expression of KDMs was assessed by using real-time qPCR. Expressions values were normalized by GAPDH. C) Changes in the levels of KDM3C protein expression was determined by Western blotting with time-dependent Pg LPS (1 µg/ml) exposure in THP-1 cells. Molecular mass markers are indicated. GAPDH was used as a loading control. KDM3C and GAPDH fold change values, which were normalized to untreated (UT) control, were obtained by using densitometry. D) KDM3C mRNA expression in Pg LPS–treated THP-1 cells was determined by real-time qPCR. E) THP-1 cells were infected with Pg for the indicated time points, and the expression of KDM3C mRNA was determined by real-time qPCR. Data are representative of at least 2 or more independent experiments and were statistically analyzed by Student’s t test; n.s., not significant. *P < 0.05, **P < 0.01, ***P < 0.001 compared with the untreated controls.
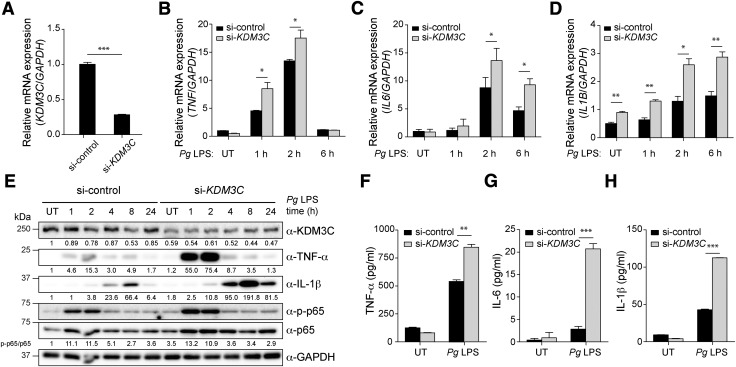
To assess the functional role of KDM3C in inflammation, we knocked down the KDM3C level in THP-1 cells using a pool of different siRNAs specific for human KDM3C (si-KDM3C), whereas control cells were similarly transfected with nonspecific siRNA (si-control) (Fig. 2A). Compared with the si-control THP-1 cells, Pg LPS stimulation in the THP-1 cells with KDM3C knockdown led to an elevated mRNA level of TNF-α, IL-1β, and IL-6 (Fig. 2B–D). This was confirmed by Western blotting for KDM3C that showed increased activation of NF-κB signaling (i.e., p65 phosphorylation) with KDM3C knockdown (Fig. 2E). The ELISA data further corroborated the result, showing increased expression of TNF-α, IL-1β, and IL-6 in cells with KDM3C knockdown compared with the control cells (Fig. 2F–H). Collectively, these data suggest a potential suppressive role of KDM3C in inflammatory signaling in THP-1 cells challenged with Pg LPS.
Figure 2.
KDM3C acts as a negative regulator of Pg LPS–induced proinflammatory signaling pathway in human THP-1 cells. A) si-KDM3C or si-control was transiently transfected into THP-1 cells. Real-time qPCR was used to measure endogenous KDM3C mRNA levels. B–D) Pg LPS–induced mRNA expressions of proinflammatory cytokines were assessed in THP-1 cells transfected with si-control or si-KDM3C by using real-time qPCR. Cells were harvested at the indicated times of Pg LPS (1 µg/ml) treatment. mRNA expressions of proinflammatory cytokines TNF (B), IL-6 (C), and IL-1B (D) were determined. E) Western blotting analysis of the levels of KDM3C, TNF-α, IL-1β, total p65 (NF-κB subunit), and phosphorylated p65 (p-p65) in control and KDM3C–knocked-down THP-1 cell treated with Pg LPS at indicated times. GAPDH was used as a loading control. The normalized intensities of the KDM3C, TNF-α, and IL-1β to GAPDH were calculated and shown as fold change below individual lanes. p-p65:p65 ratio was calculated and shown as fold change. These data are representative of at least 3 independent experiments. F–H) Cytokine levels in culture supernatants were measured by ELISA. THP-1 cells transfected with si-control or si-KDM3C were stimulated with Pg LPS (1 µg/ml) for 24 h. Secretions of cytokines, TNF-α (F), IL-6 (G), and IL-1β (H) were analyzed. UT, untreated. Data are representative of at least 3 independent experiments and statistically analyzed by Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001 compared with control cells (si-control).
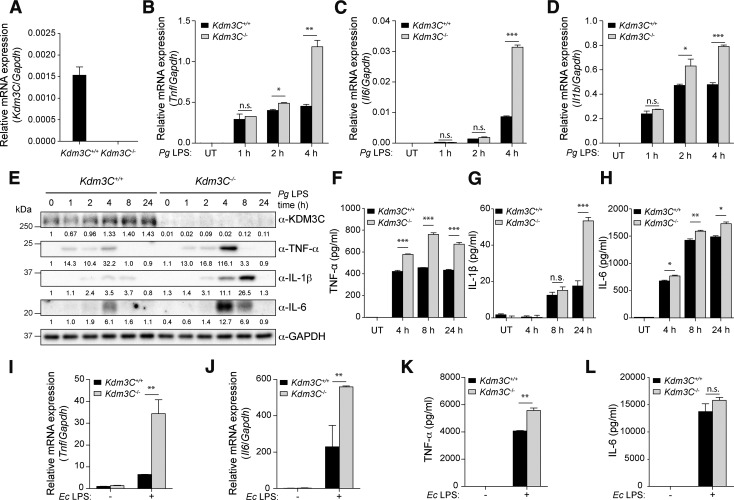
Kdm3C KO enhances proinflammatory responses to Pg LPS in BMDMs
To further validate the suppressive role of KDM3C in inflammation, we harvested bone marrow cells from Kdm3C WT and KO mice and induced their differentiation to BMDMs using M-CSF. We then confirmed Kdm3C KO in these cells by real-time qPCR (Fig. 3A). Fully differentiated BMDMs were then exposed to Pg LPS, and the levels of TNF-α, IL-6, IL-1β were determined. Real-time qPCR and Western blotting revealed that the expression of these cytokines was strongly induced by Pg LPS in BMDMs of both Kdm3C WT and KO mice, but the level of induction was increased in BMDMs from the Kdm3C KO mice compared with those of the WT (Fig. 3B–E). This finding was further confirmed by ELISA, which showed enhanced secretion of TNF-α, IL-1β, and IL-6 from BMDMs with Kdm3C KO (Fig. 3F–H). With Ec LPS stimulation, the BMDMs from Kdm3C KO mice expressed higher levels of these cytokines than those of the WT mice (Fig. 3I–L). These findings demonstrate the anti-inflammatory effects of KDM3C through suppression of proinflammatory cytokine production in response to bacterial challenge.
Figure 3.
Deletion of KDM3C shows the increased inflammatory response to Pg LPS in BMDMs. A) BMDMs from Kdm3C WT (Kdm3C+/+) and KO (Kdm3C−/−) mice were cultured, and real-time qPCR was used to measure endogenous Kdm3C mRNA levels. B-D) Real-time qPCR analysis of mRNA from Kdm3C+/+ and Kdm3C−/− BMDMs treated with Pg LPS (1 µg/ml) as indicated. mRNA expression levels of cytokines, Tnf (B), Il6 (C), and Il1b (D), were determined. E) Western blotting analysis of the levels of indicated proteins in Kdm3C+/+ and Kdm3C−/− BMDMs treated with Pg LPS at indicated times. GAPDH was used as a loading control. Protein expression was quantified, and the ratio of normalized protein is shown as fold change. F–H) Secretions of cytokines TNF-α (F), IL-1β (G), and IL-6 (H) into the culture supernatant from Kdm3C+/+ and Kdm3C−/− BMDMs after stimulation of Pg LPS (4, 8, 24 h) were measured by ELISA. I–L) mRNA expression levels of Tnf (I) and Il6 (J) and secretion levels of TNF-α (K) and IL-6 (L) were determined upon Ec LPS (1 µg/ml) exposure in Kdm3C+/+ and Kdm3C−/− BMDMs. UT, untreated. Data are representative of at least 3 independent experiment and statistically analyzed by Student’s t test; n.s., not significant. *P < 0.05, **P < 0.01, ***P < 0.001 compared with WT cells.
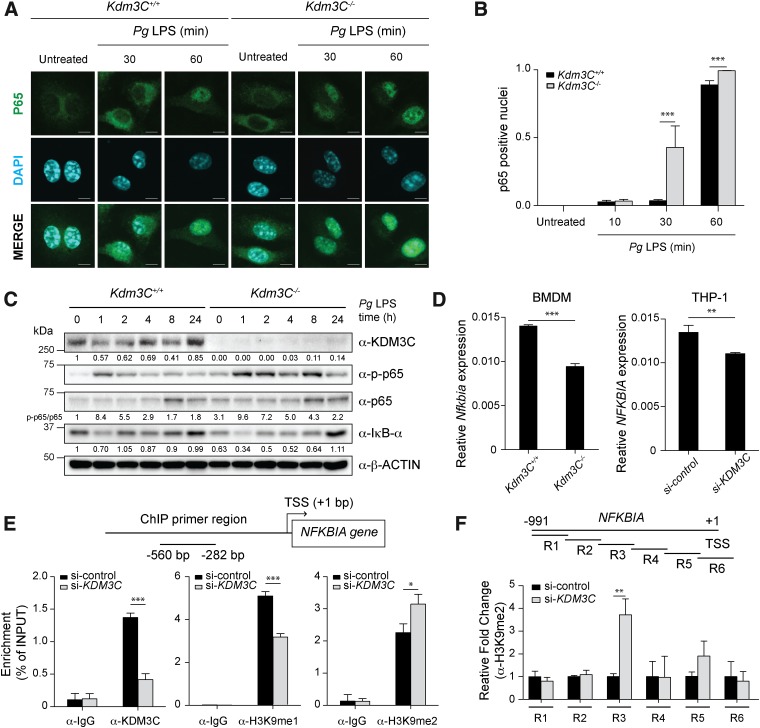
KDM3C attenuates NF-κB activation in response to inflammatory signaling
Because LPS-induced proinflammatory cytokine expression is primarily regulated by NF-κB signaling (18), we next examined whether KDM3C modulates NF-κB activation. BMDMs derived from Kdm3C WT and KO mice were exposed to Pg LPS, and p65 nuclear translocation was determined by confocal microscopy. Cytoplasmic distribution of p65 protein was noted in unstimulated BMDMs from both Kdm3C WT and KO mice (Fig. 4A). After 30-min exposure to Pg LPS, p65 nuclear translocation was apparent in the BMDMs with Kdm3C KO, whereas those of the WT mice did not show nuclear translocation until 60 min after Pg LPS stimulation (Fig. 4A, B). Consistent with the accelerated nuclear translocation, p65 phosphorylation was also increased in the BMDMs with Kdm3C KO mice, whereas the level of IκB-α was reduced in these cells compared with those of Kdm3C WT mice (Fig. 4C). These differences in the kinetics of NF-κB activation were observed over a 24-h time period. To further delineate the differences in NF-κB activation in the 2 cell groups, we compared the p65 phosphorylation levels in BMDMs derived from Kdm3C WT and KO mice immediately after LPS stimulation during 0–120 min. The KDM3C level was suppressed in the 10 min after Pg LPS stimulation in BMDMs from the WT mice and then recovered gradually during the subsequent 120-min time period (Supplemental Fig. S2A). Induced p65 phosphorylation was observed in 10 min after Pg LPS stimulation in the BMDMs of Kdm3C KO mice compared with those of the WT mice (Supplemental Fig. S2B). Interestingly, the IκB-α protein and mRNA [NF-κB inhibitor α (Nfkbia)] expression level decreased in the BMDMs with Kdm3C KO, suggesting that KDM3C may be required for the expression of IκB-α (Fig. 4C, D). This relationship was also observed in the THP-1 cells with KDM3C knockdown (Fig. 4D). Because the IκB-α protein has a critical role in suppression of NF-κB signaling, the anti-inflammatory effect of KDM3C may involve the transcriptional regulation of NFKBIA/Nfkbia.
Figure 4.
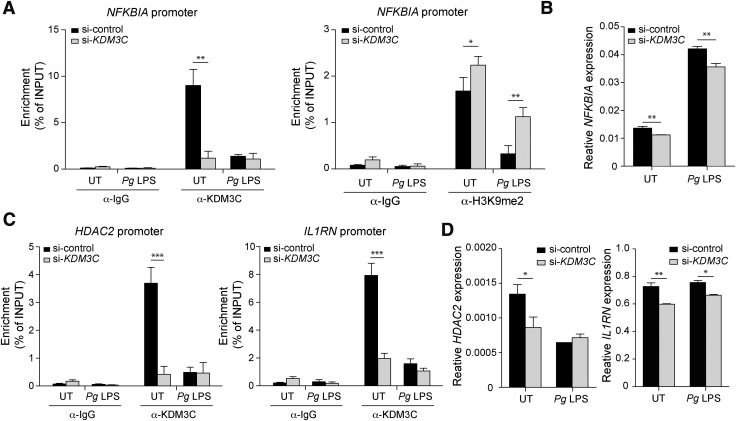
KDM3C deletion enhances and accelerates activation and translocation of NF-κB in mouse BMDMs. A) NF-κB subunit p65 localization was determined in BMDMs from Kdm3C WT (Kdm3C+/+) and Kdm3C KO (Kdm3C−/−) mice after Pg LPS (1 µg/ml) treatment for 0, 30, and 60 min. Representative images from the immunofluorescence assay. Scale bars, 5 µm. Green, p65; cyan, DAPI staining (nuclei). B) Nuclear-located p65 cells were scored after Pg LPS treatments for 0, 10, 30, and 60 min. Cells were counted in 7 random areas of each image in duplicate experiments and described as a ratio of cells containing nuclear-located p65 for total cells. In each region, at least 150 cells were blindly counted. Statistics were performed using 1-way ANOVA and the Bonferroni method. ***P < 0.001 compared with Kdm3C+/+ BMDMs. C) Western blotting analysis of the levels of phosphorylated p65 (p-p65), total p65, IκB-α, and KDM3C in Kdm3C+/+ and Kdm3C−/− BMDMs upon time-dependent Pg LPS exposure. β-actin was used as a loading control. The normalized intensities of the proteins were calculated and shown as fold change. The p-p65:p65 ratio was calculated and shown as fold change. These data are representative of at least 3 independent experiments. D) Expression levels of NFKBIA/Nfkbia, encoding IκB-α, were determined by real-time qPCR in the Kdm3C+/+ and Kdm3C−/− BMDMs (left) or in the KDM3C–knocked-down (si-KDM3C) and control (si-control) THP-1 cells (right). **P < 0.01, ***P < 0.001 compared with Kdm3C+/+ BMDMs (left) or control THP-1 cells (right). E) KDM3C binding and H3K9me1, H3K9me2 levels to the NFKBIA promoter in control and KDM3C–knocked-down THP-1 cells. Cells were chromatin immunoprecipitated with an antibody against KDM3C, H3K9me1, H3K9me2, or IgG controls. The precipitated DNA was then amplified by qPCR using primers illustrated at the top. *P < 0.05, ***P < 0.001 compared with control cells. TSS, Transcription start site. F) The positions of primer pairs are indicated as R1 to R6. ChIP assays using primers (R1 ∼ R6) to the NFKBIA promoter to determine the location of H3K9me2 binding, which is regulated by KDM3C, on the NFKBIA promoter in THP-1 cells.
To explore the regulatory role of KDM3C on I-κBα expression, we performed ChIP in THP-1 cells with KDM3C knockdown and assessed the enrichment of KDM3C, H3K9me2, and H3K9me1 on the NFKBIA promoter region. We detected and confirmed binding of KDM3C at the promoter region (−580 to −282 bp) of NFKBIA (Fig. 4E). In cells with KDM3C knockdown, the NFKBIA promoter was enriched with an elevated level of H3K9me2 and a reduced level of H3K9me1 compared with the control cells (Fig. 4E). To determine the specific region of the NFKBIA promoter enriched with H3K9me2 upon KDM3C knockdown, ChIP assay was performed with several oligonucleotide pairs spanning the different regions of the NFKBIA promoter (Fig. 4F). KDM3C knockdown led to increased H3K9me2 enrichment at a region located from −632 to −451 bp, which is juxtaposed to the promoter region occupied by KDM3C (Fig. 4F).
We next explored the dynamic role of KDM3C in NFKBIA promoter regulation during inflammatory signaling. We confirmed binding of KDM3C at the NFKBIA promoter region by ChIP and found that the KDM3C binding level was reduced upon Pg LPS stimulation in the control THP-1 cells, whereas KDM3C binding at the promoter was negligible in the cells with KDM3C knockdown (Fig. 5A). KDM3C knockdown also led to increased enrichment of H3K9me2 in the cells with and without Pg LPS exposure, resulting in reduced NFKBIA mRNA expression (Fig. 5A, B). This finding supports the possibility that KDM3C regulates the NFKBIA gene expression through an epigenetic mechanism. To explore the regulatory role of KDM3C on broader NF-κB signaling network genes, we compared the expression of histone deacetylase (HDAC) 1, HDAC2, IL-33, NF-κB inhibitor β, NF-κB inhibitor δ, NF-κB inhibitor ζ, IL-1 receptor-associated kinase M (IRAKM), RELA, and IL-1RN in THP-1 cells with and without KDM3C knockdown (Supplemental Fig. S3). Among them, we further examined the expression of HDAC2 and IL-1RN, both of which have inhibitory roles in NF-κB signaling (19, 20). ChIP assay revealed KDM3C binding at the promoter regions of HDAC2 and IL-1RN in unstimulated THP-1 cells, and the binding was lost upon exposure to Pg LPS (Fig. 5C). When KDM3C was knocked down, the levels of HDAC2 and IL-1RN mRNA were decreased in cells, reflecting the requirement of KDM3C enrichment at the promoters for the gene expression (Fig. 5D). Taken together, these data suggest that the anti-inflammatory effect of KDM3C occurs in part through epigenetic regulation of the NF-κB signaling network genes.
Figure 5.
KDM3C regulates gene expression of NF-κB inhibitors and DNA binding. A) KDM3C binding and H3K9me2 levels to the NFKBIA promoter were analyzed with ChIP assays in control and KDM3C–knocked-down THP-1 cells with or without Pg LPS challenge for 1 h. B) Expression levels of NFKBIA gene were determined by real-time qPCR. C) The enrichment of KDM3C at HDAC2 and IL-1RN promoter regions was validated by ChIP assays. D) Expression levels of HDAC2 and IL-1RN mRNA were determined by real-time qPCR. UT, untreated. Data were statistically analyzed by Student’s t test. *P < 0.05, **P < 0.01 compared with control cells.
Kdm3C deficiency leads to increased alveolar bone loss in oral inflammatory lesions
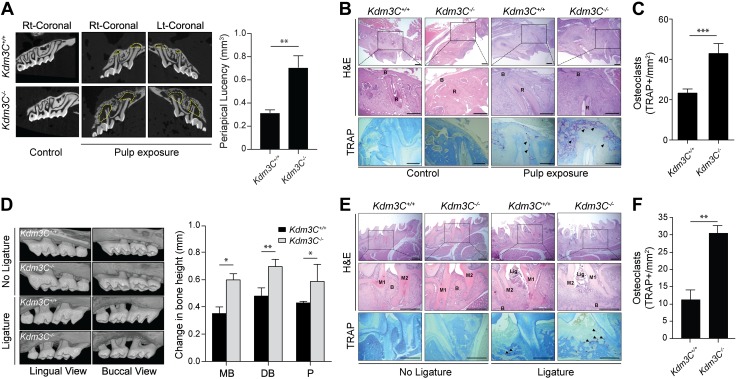
We next determined the role of KDM3C in modulating the inflammatory responses to oral bacterial infections. This was accomplished by pulpal exposure to induce apical periodontitis in maxillary first molars or by ligature placement on maxillary second molars to induce periodontitis in Kdm3C WT and KO mice. After 21 d of postpulpal exposure, µ-CT scans revealed enlarged periapical lesions in the Kdm3C KO mice compared with those of the WT littermates (Fig. 6A and Supplemental Fig. S4). H&E and TRAP staining confirmed increased infiltration of inflammatory cells and an abundance of osteoclasts at the periapex of Kdm3C KO mice compared with the WT mice (Fig. 6B, C). Likewise, ligature-induced periodontitis led to increased alveolar bone loss and an abundance of osteoclasts in Kdm3C KO mice (Fig. 6D–F).
Figure 6.
Loss of KDM3C exacerbates inflammatory response to oral infection. A) Pulp exposure was done on both sides of the maxillary first molar from Kdm3C WT (Kdm3C+/+) and Kdm3C KO (Kdm3C−/−) mice. After 21 d, the maxilla was harvested for µ-CT analysis. Size of periapical lesions was measured by calculating the total loss of bone volume using µ-CT images. Data were expressed as means ± sem with 10 mice per group. B) Representative H&E- and TRAP-stained images. Arrows depict TRAP-stained areas. C) Quantification of TRAP+ multinucleated osteoclasts number per square millimeter. D) Experimental periodontitis was induced by the ligature placement around the maxillary second molar for 21 d. µ-CT images of maxillary molars from Kdm3C+/+ and Kdm3C−/− mice were reconstructed. E) Representative H&E- and TRAP-stained photographs obtained. F) Quantification of TRAP+ osteoclasts number per square millimeter (n = 3). Arrows depict TRAP-stained areas. B, bone; DB, distobuccal; Lig.; ligature; M1, first molar; M2, second molar; MB, mesiobuccal; P, palatal roots; R, root. Scale bars, 500 µm. *P < 0.05, **P < 0.01, ***P < 0.001 compared with Kdm3C+/+.
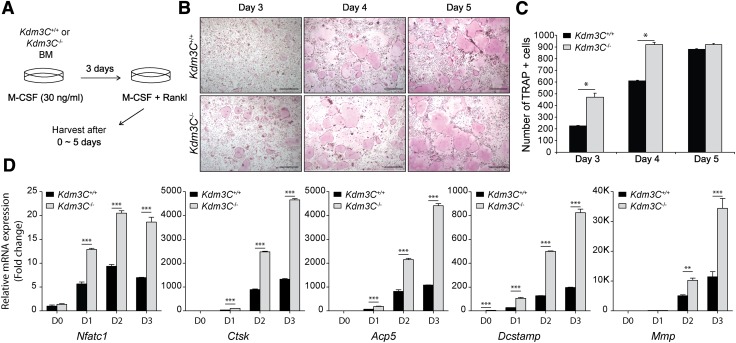
To gain further insights into the impact of Kdm3C KO on osteoclastogenesis, we isolated bone marrow cells from the Kdm3C WT and KO mice and differentiated them ex vivo. This was achieved by culturing the cells in the presence of M-CSF for 3 d, followed by exposure to RANKL and M-CSF for 0–5 d (Fig. 7A). Fully differentiated osteoclasts were detected as TRAP+ multinucleated cells. The kinetics of RANKL-induced osteoclast differentiation was significantly increased in the BMDMs isolated from the Kdm3C KO mice compared with those of WT (Fig. 7B, C). Consistently, mRNA expressions of the osteoclast markers [e.g., nuclear factor of activated T cells 1 (Nfatc1); cathepsin K; acid phosphatase 5, tartrate resistant; dendrocyte expressed seven transmembrane protein; and matrix metallopeptidase 9] were elevated in the BMDMs of the Kdm3C KO mice upon stimulation with RANKL compared with those of WT mice (Fig. 7D). These data support our in vivo findings that Kdm3C KO led to an increased alveolar bone loss in oral inflammatory lesions and illustrate a possible mechanistic connection between KDM3C and osteoclastogenesis.
Figure 7.
Loss of Kdm3C accelerates the in vitro osteoclast differentiation. A) Schematic drawing of the in vitro bone marrow osteoclast differentiation assay. BMDMs generated from Kdm3C WT (Kdm3C+/+) and Kdm3C KO (Kdm3C−/−) mice were cultured with M-CSF (30 ng/ml) and RANKL (100 ng/ml) for 0 ∼ 5 d to induce the osteoclast differentiation. B) Development of osteoclasts was monitored at different time points by TRAP staining. Representative images of the cultures. Scale bars, 500 µm. C) Quantification of TRAP+ multinucleated (>5 nuclei) osteoclasts per well. D) Effect of Kdm3C deficiency on the expression of the osteoclast marker genes Nfatc1 (encoding NFATc1), Ctsk (encoding Cathepsin K), acid phosphatase 5, tartrate resistant (Acp5, encoding TRAP), dendrocyte expressed seven transmembrane protein (Dcstamp), and matrix metallopeptidase 9 (Mmp9) were measured by real-time qPCR. Day (D) after incubation with RANKL. These data are representative of 3 independent experiments and statistically analyzed by Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, compared with WT.
Expression of KDM3C mRNA is reduced in inflamed gingiva
To test whether our findings from the cultured cells and the animal models have relevance to the human diseases, we analyzed the KDM3C mRNA expression levels in publicly available microarray datasets between healthy and diseased gingival tissues (21). The level of KDM3C mRNA was significantly decreased in the gingiva obtained from patients with chronic or aggressive periodontitis compared with healthy tissues (Supplemental Fig. S5A). The levels of proinflammatory cytokines IL-6 and IL-1B were also up-regulated in the inflamed gingival tissues compared with the healthy controls (Supplemental Fig. S5B, C). These data are in keeping with our current findings to support the role of KDM3C in modulating the inflammatory responses to microbial challenge.
DISCUSSION
The current study demonstrates a novel role for KDM3C in epigenetic regulation of inflammatory responses to bacterial endotoxin challenge, in part through modulation of NF-κB signaling and osteoclastogenesis. Our data showed suppression of KDM3C in THP-1 cells and BMDMs challenged by Pg LPS or Pg bacteria inoculation and a significant increase in inflammatory signaling with Kdm3C KO as evinced by an enhanced level of proinflammatory cytokines and activation of NF-κB signaling. Furthermore, KDM3C ablation by gene KO in vivo led to increased alveolar bone loss after pulpal infection or ligature placement, suggesting its anti-inflammatory effects in oral inflammatory lesions. Although we acknowledge the pathogenic differences between pulpal and periodontal infection, the focus of the current investigation lies on the anti-inflammatory role of KDM3C in tissue responses to oral bacterial infection. Increased alveolar bone loss occurred with an enhanced level of mature osteoclasts in the Kdm3C KO mice compared with the WT littermates, which suggests a possible role of KDM3C in suppressing osteoclastogenesis. Earlier studies demonstrated that inflammatory bone resorption is mediated by NF-κB–mediated activation of proinflammatory cytokines (22, 23). Also, NF-κB plays an essential role in RANKL-induced osteoclastogenesis, as evinced in regulation of NFATc1 by NF-κB (24). The role of the NF-κB signaling in bone resorption is further supported by impaired osteoclastogenesis and occurrence of osteopetrosis phenotype in mice with double KO in NF-κB subunits p50 and p52 (25). These findings further corroborate our current data that support the anti-inflammatory role of KDM3C through suppression of NF-κB signaling and osteoclastogenesis.
Although earlier studies showed functional roles of KDM3C in varied pathophysiologic processes, including stemness, steroidogenesis, and cancer (26–28), the current study is the first report demonstrating the regulatory role of KDM3C in inflammatory signaling. Although the association between KDM3C and inflammatory regulation has not been reported previously, a dynamic involvement of H3K9 methylation in inflammatory signaling has been reported (5). In this prior study, the LPS-induced inflammatory signaling in human cultured dendritic cells led to transient loss of H3K9 methylation at the promoter regions of inducible inflammatory genes (e.g., macrophage-derived chemokine, EBV-induced molecular 1 ligand chemokine, and the p40 subunit of IL-12), coincident with the recruitment of RNA polymerase II and the gene expression. Thus, H3K9 demethylation by KDM3C and methylation by histone methyltransferases (HMTs) may determine the dynamicity of epigenetic responses to inflammatory signaling.
There are several members of HMTs specific for H3K9, including suppressor of variegation 3-9 homolog (SUV39H) 1, SUV39H2, G9a-Like Protein 1 (euchromatic histone lysine methyltransferase (EHMT) 1), G9a (EHMT2), positive regulatory domain 2 (PRDM2), SET domain bifurcated (SETDB) 1, and SETDB2 (29–33), which in general lead to transcriptional repression. H3K9 methylation by SUV39H1 generates the binding sites for Heterochromatin Protein 1 (HP1), which results in heterochromatic chromatin and transcriptional repression (34, 35). Interestingly, some of these HMTs exhibit a negative role in NF-κB–mediated inflammatory responses through H3K9 methylation on the target gene promoters. G9a-Like Protein 1 suppresses a large fraction of NF-κB target genes through methylation of H3K9 on the gene promoters (36). Also, hypermethylation of H3K9 by G9a is linked with the silencing of TNF-α transcription during endotoxin tolerance phenotyping in THP-1 cells (37). The functional significance of G9a enrichment for cytokine gene silencing was shown by knockdown of G9a and resulting re-expression of TNF-α in the tolerant cells. Therefore, these prior studies indicate that H3K9 methylation by the specific HMTs may have unique relevance for the epigenetic regulation of inflammatory signaling, with the net effect determined by the balance between the HMTs and KDM3C on inflammatory gene targets.
The ChIP analysis with human THP-1 cells found KDM3C enrichment at the promoter regions of the target genes that elicit negative effects on NF-κB signaling (e.g., NFKBIA, HDAC2, and IL-1RN). In particular, NFKBIA encodes for IκB-α, which is the most crucial regulator of NF-κB signaling through physical association with NF-κB heterodimer and masking the nuclear localization signal of p65 (38). IκB-α also inhibits NF-κB activity through interfering with the DNA-binding ability of the heterodimers (39). Hence, NF-κB activation requires inactivation of IκB-α, which occurs with its phosphorylation by the IκB kinase and ubiquitin-mediated protein degradation (40). With ablation of KDM3C expression in THP-1 and BMDMs, our current data showed a significant decrease in IκB-α expression level. Moreover, we found a reduction in the H3K9me2 mark on the promoter regions of NFKBIA in THP-1 cells with KDM3C knockdown. Also, a recent study reported that the H3K9-specific HMT SUV39H1 is required for the activation of NF-κB signaling by addition of histone methyl-mark on the promoter region of NFKBIA, leading to gene silencing (41). Hence, NFKBIA promoter histone molecules may directly be modified by KDM3C during inflammatory responses as a mechanism to attenuate NF-κB activation. NF-κB activity is also modulated by HDACs, as reported in an earlier study that showed increased proinflammatory responses in microglial cells treated with HDAC inhibitor, trichostatin A, and suberoylanilide hydroxamic acid (vorinostat) (42). This potentiation of inflammatory responses by the HDAC inhibitors occurred in an NF-κB–dependent mechanism. Also, To et al. (43) reported a negative correlation between HDAC2 level and NF-κB activity in human skeletal muscle samples and showed enhanced NF-κB activity with HDAC2 knockdown in a cell culture model. Moreover, it was reported that LPS treatment reduced the HDAC2 expression and its inhibitory activity on NF-κB–mediated gene expression (44). These previous studies demonstrated the inhibitory effects of HDAC2 on the NF-κB signaling. Our data also showed that KDM3C binds to the promoter regions of HDAC2 to modulate the gene expression; this mechanism may mediate the inhibitory effects of KDM3C on NF-κB signaling. Furthermore, our data also indicate KDM3C binding at the promoter region of IL-1RN, which encodes for IL-1R antagonist. IL-1R agonist suppresses the IL-1α and IL-1β signals by binding to the IL-1Rs and therefore NF-κB activation induced by IL-1R signaling (45). A recent study demonstrated that experimental periodontitis induced by oral infection of Aggregatibacter actinomycetemcomitans was facilitated by IL-1 signals and was exacerbated in Il-1rn KO mice (46). Hence, it may be possible that the enhanced alveolar bone loss in Kdm3C KO mice exposed in oral inflammatory lesions may have resulted in part from reduced Il-1rn expression and thus increased NF-κB signaling.
The ligature-induced experimental periodontitis model is one of the widely used preclinical models to initiate experimental periodontitis in different animals, ranging from mice to nonhuman primates (12, 47, 48). However, there are limitations with this model that should be considered with data interpretation. Although the ligature aids in facilitating bacterial colonization and induced periodontal bone loss, it does not precisely reflect the human microbiome of periodontal disease. In addition, the structure of the mice periodontal apparatus does not precisely reflect that of humans. Despite these limitations, the ligature model remains one of the versatile systems with which to study the pathogenic events of periodontitis, such as gingival inflammatory responses to bacterial infection. The current study utilized the ligature model to study the mechanisms of host-tissue responses to microbial challenges in the periodontium and to determine the role of KDM3C in the inflammatory response in vivo rather than focus on the microbiota associated with the disease progress.
In this study, we have shown an inhibitory role of KDM3C in inflammatory signaling by utilizing Pg LPS. However, the periodontal disease is associated with a consortium of oral bacterial species rather than a single dominant species (49). The role of KDM3C in the response against the bacterial infection may differ depending on the microbial profile of the infection. The differential response of KDM3C on bacterial species was evinced in HOK-16B cells inoculated with Pg or Fn. KDM3C was precipitously lost in HOK-16B exposed to Pg, whereas inoculation of cells with Fn did not elicit any changes in KDM3C expression (Supplemental Fig. S1). Hence, the anti-inflammatory effects of KDM3C may depend on the microbial content of the oral bacterial infection, and further studies on the dynamics between KDM3C and different bacteria species may clarify the species-specific mechanism of KDM3C in the inflammatory tissue responses.
In summary, we have shown a novel inhibitory effect of KDM3C in the inflammatory signaling mechanism that appears to involve NF-κB–dependent pathways. Functional experiments revealed elevated susceptibility of Kdm3C KO mice to oral bacterial infection with increased alveolar bone loss compared with the WT mice, further confirming the anti-inflammatory role of KDM3C. Consistent with the increased alveolar bone loss, BMDMs derived from the Kdm3C KO mice demonstrated an increased rate of RANKL-induced osteoclastogenesis, which may result from the elevated NF-κB signaling. By using ChIP analyses, we identified several KDM3C target loci that exhibit regulatory effects on NF-κB activity, which include NFKBIA, HDAC2, and IL-1RN. Based on these experiments, we propose a model that may explain the role of KDM3C in inflammatory responses. Upon oral bacterial infection, the KDM3C level is reduced, allowing for methylation of H3K9me2 moieties and reduced expression of KDM3C target genes, which may negatively regulate the NF-κB signaling to allow for proinflammatory responses. These findings provide insight into the epigenetic mechanisms by which KDM3C regulates inflammatory response and present a possibility of developing novel anti-inflammatory therapeutics in management and treatment of oral inflammatory conditions.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
This work was supported, in part, by grants from the U.S. National Institutes of Health, National Institute of Dental and Craniofacial Research (R21DE028269, R56DE024593, and R03DE024259) and a grant from the American Association of Endodontists Foundation. M.K.K. is also supported by the Jack A. Weichman Endowed Fund. The authors declare no conflicts of interest.
Glossary
- μ-CT
micro–computed tomography
- BMDM
bone marrow–derived macrophage
- ChIP
chromatin immunoprecipitation
- Ec LPS
LPS from Escherichia coli
- Fn
Fusobacterium nucleatum
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- H3K9
histone 3 Lys 9
- H3K9me1
H3K9 monomethylation
- H3K9me2
H3K9 dimethylation
- HDAC
histone deacetylase
- H&E
hematoxylin and eosin
- HMT
histone methyltransferase
- HOK-16B
human oral keratinocyte-16B
- IL-1R
IL-1 receptor
- KDM
Lys-specific demethylase
- KO
knockout
- NFATc1
nuclear factor of activated T cells 1
- NFKBIA
NF-κB inhibitor α
- Pg
Porphyromonas gingivalis
- Pg LPS
LPS from Pg
- PMA
phorbol 12-myristate 13-acetate
- qPCR
quantitative PCR
- RANKL
receptor activator of NF-κB ligand
- si-control
siRNA negative control
- si-Kdm3C
siRNA targeting human Kdm3C mRNA
- siRNA
small interfering RNA
- THP-1
Tohoku Hospital Pediatrics-1
- TRAP
tartrate-resistant acid phosphatase
- WT
wild type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
J. Y. Lee and S. Mehrazarin designed the research; J. Y. Lee, S. Mehrazarin, A. Alshaikh, and S. Kim performed the experimental work, analyzed data, and provided technical assistance; W. Chen, R. Lux, Y. Gwack, and R. H. Kim participated in and coordinated the study; M. K. Kang designed and supervised the experimental work; J. Y. Lee and M. K. Kang wrote the manuscript; and all authors interpreted the data and commented on and approved the final manuscript.
REFERENCES
- 1.Darveau R. P. (2010) Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8, 481–490 [DOI] [PubMed] [Google Scholar]
- 2.Poltorak A., He X., Smirnova I., Liu M. Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., Freudenberg M., Ricciardi-Castagnoli P., Layton B., Beutler B. (1998) Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282, 2085–2088 [DOI] [PubMed] [Google Scholar]
- 3.Lu Y. C., Yeh W. C., Ohashi P. S. (2008) LPS/TLR4 signal transduction pathway. Cytokine 42, 145–151 [DOI] [PubMed] [Google Scholar]
- 4.Zhou Q., Desta T., Fenton M., Graves D. T., Amar S. (2005) Cytokine profiling of macrophages exposed to Porphyromonas gingivalis, its lipopolysaccharide, or its FimA protein. Infect. Immun. 73, 935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saccani S., Natoli G. (2002) Dynamic changes in histone H3 Lys 9 methylation occurring at tightly regulated inducible inflammatory genes. Genes Dev. 16, 2219–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Gazzar M., Liu T., Yoza B. K., McCall C. E. (2010) Dynamic and selective nucleosome repositioning during endotoxin tolerance. J. Biol. Chem. 285, 1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoza B. K., McCall C. E. (2011) Facultative heterochromatin formation at the IL-1 beta promoter in LPS tolerance and sepsis. Cytokine 53, 145–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X., El Gazzar M., Yoza B. K., McCall C. E. (2009) The NF-kappaB factor RelB and histone H3 lysine methyltransferase G9a directly interact to generate epigenetic silencing in endotoxin tolerance. J. Biol. Chem. 284, 27857–27865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Santa F., Narang V., Yap Z. H., Tusi B. K., Burgold T., Austenaa L., Bucci G., Caganova M., Notarbartolo S., Casola S., Testa G., Sung W. K., Wei C. L., Natoli G. (2009) Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 28, 3341–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuroki S., Akiyoshi M., Tokura M., Miyachi H., Nakai Y., Kimura H., Shinkai Y., Tachibana M. (2013) JMJD1C, a JmjC domain-containing protein, is required for long-term maintenance of male germ cells in mice. Biol. Reprod. 89, 93 [DOI] [PubMed] [Google Scholar]
- 11.He Y., Gan Y., Lu J., Feng Q., Wang H., Guan H., Jiang Q. (2017) Pulpal tissue inflammatory reactions after experimental pulpal exposure in mice. J. Endod. 43, 90–95 [DOI] [PubMed] [Google Scholar]
- 12.Abe T., Hajishengallis G. (2013) Optimization of the ligature-induced periodontitis model in mice. J. Immunol. Methods 394, 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouxsein M. L., Boyd S. K., Christiansen B. A., Guldberg R. E., Jepsen K. J., Muller R. (2010) Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J. Bone Miner. Res. 25, 1468–1486 [DOI] [PubMed] [Google Scholar]
- 14.Van ’t Hof R. J., Rose L., Bassonga E., Daroszewska A. (2017) Open source software for semi-automated histomorphometry of bone resorption and formation parameters. Bone 99, 69–79 [DOI] [PubMed] [Google Scholar]
- 15.Lam R. S., O’Brien-Simpson N. M., Lenzo J. C., Holden J. A., Brammar G. C., Walsh K. A., McNaughtan J. E., Rowler D. K., Van Rooijen N., Reynolds E. C. (2014) Macrophage depletion abates Porphyromonas gingivalis-induced alveolar bone resorption in mice. J. Immunol. 193, 2349–2362 [DOI] [PubMed] [Google Scholar]
- 16.De Santa F., Totaro M. G., Prosperini E., Notarbartolo S., Testa G., Natoli G. (2007) The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130, 1083–1094 [DOI] [PubMed] [Google Scholar]
- 17.Lee H. T., Kim S. K., Kim S. H., Kim K., Lim C. H., Park J., Roh T. Y., Kim N., Chai Y. G. (2014) Transcription-related element gene expression pattern differs between microglia and macrophages during inflammation. Inflamm. Res. 63, 389–397 [DOI] [PubMed] [Google Scholar]
- 18.Vanden Berghe W., Plaisance S., Boone E., De Bosscher K., Schmitz M. L., Fiers W., Haegeman G. (1998) p38 and extracellular signal-regulated kinase mitogen-activated protein kinase pathways are required for nuclear factor-kappaB p65 transactivation mediated by tumor necrosis factor. J. Biol. Chem. 273, 3285–3290 [DOI] [PubMed] [Google Scholar]
- 19.Ashburner B. P., Westerheide S. D., Baldwin A. S., Jr (2001) The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol. 21, 7065–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haddad J. J., Lauterbach R., Saade N. E., Safieh-Garabedian B., Land S. C. (2001) Alpha-melanocyte-related tripeptide, Lys-d-Pro-Val, ameliorates endotoxin-induced nuclear factor kappaB translocation and activation: evidence for involvement of an interleukin-1beta193-195 receptor antagonism in the alveolar epithelium. Biochem. J. 355, 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demmer R. T., Behle J. H., Wolf D. L., Handfield M., Kebschull M., Celenti R., Pavlidis P., Papapanou P. N. (2008) Transcriptomes in healthy and diseased gingival tissues. J. Periodontol. 79, 2112–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mundy G. R. (1993) Role of cytokines in bone resorption. J. Cell. Biochem. 53, 296–300 [DOI] [PubMed] [Google Scholar]
- 23.Pfeilschifter J., Chenu C., Bird A., Mundy G. R., Roodman G. D. (1989) Interleukin-1 and tumor necrosis factor stimulate the formation of human osteoclastlike cells in vitro. J. Bone Miner. Res. 4, 113–118 [DOI] [PubMed] [Google Scholar]
- 24.Takatsuna H., Asagiri M., Kubota T., Oka K., Osada T., Sugiyama C., Saito H., Aoki K., Ohya K., Takayanagi H., Umezawa K. (2005) Inhibition of RANKL-induced osteoclastogenesis by (-)-DHMEQ, a novel NF-kappaB inhibitor, through downregulation of NFATc1. J. Bone Miner. Res. 20, 653–662 [DOI] [PubMed] [Google Scholar]
- 25.Franzoso G., Carlson L., Xing L., Poljak L., Shores E. W., Brown K. D., Leonardi A., Tran T., Boyce B. F., Siebenlist U. (1997) Requirement for NF-kappaB in osteoclast and B-cell development. Genes Dev. 11, 3482–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakajima R., Okano H., Noce T. (2016) JMJD1C exhibits multiple functions in epigenetic regulation during spermatogenesis. PLoS One 11, e0163466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe S., Watanabe K., Akimov V., Bartkova J., Blagoev B., Lukas J., Bartek J. (2013) JMJD1C demethylates MDC1 to regulate the RNF8 and BRCA1-mediated chromatin response to DNA breaks. Nat. Struct. Mol. Biol. 20, 1425–1433 [DOI] [PubMed] [Google Scholar]
- 28.Xiao F., Liao B., Hu J., Li S., Zhao H., Sun M., Gu J., Jin Y. (2017) JMJD1C ensures mouse embryonic stem cell self-renewal and somatic cell reprogramming through controlling microRNA expression. Stem Cell Reports 9, 927–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters A. H., O’Carroll D., Scherthan H., Mechtler K., Sauer S., Schofer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., Opravil S., Doyle M., Sibilia M., Jenuwein T. (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 [DOI] [PubMed] [Google Scholar]
- 30.Tachibana M., Ueda J., Fukuda M., Takeda N., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y. (2005) Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 19, 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dodge J. E., Kang Y. K., Beppu H., Lei H., Li E. (2004) Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell. Biol. 24, 2478–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H., An W., Cao R., Xia L., Erdjument-Bromage H., Chatton B., Tempst P., Roeder R. G., Zhang Y. (2003) mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell 12, 475–487 [DOI] [PubMed] [Google Scholar]
- 33.Kim K. C., Geng L., Huang S. (2003) Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res. 63, 7619–7623 [PubMed] [Google Scholar]
- 34.Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 [DOI] [PubMed] [Google Scholar]
- 35.Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., Allshire R. C., Kouzarides T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 [DOI] [PubMed] [Google Scholar]
- 36.Ea C. K., Hao S., Yeo K. S., Baltimore D. (2012) EHMT1 protein binds to nuclear factor-kappaB p50 and represses gene expression. J. Biol. Chem. 287, 31207–31217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El Gazzar M., Yoza B. K., Chen X., Hu J., Hawkins G. A., McCall C. E. (2008) G9a and HP1 couple histone and DNA methylation to TNFalpha transcription silencing during endotoxin tolerance. J. Biol. Chem. 283, 32198–32208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacobs M. D., Harrison S. C. (1998) Structure of an IkappaBalpha/NF-kappaB complex. Cell 95, 749–758 [DOI] [PubMed] [Google Scholar]
- 39.Léveillard T., Verma I. M. (1993) Diverse molecular mechanisms of inhibition of NF-kappa B/DNA binding complexes by I kappa B proteins. Gene Expr. 3, 135–150 [PMC free article] [PubMed] [Google Scholar]
- 40.Karin M. (1999) How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene 18, 6867–6874 [DOI] [PubMed] [Google Scholar]
- 41.Santos-Barriopedro I., Bosch-Presegue L., Marazuela-Duque A., de la Torre C., Colomer C., Vazquez B. N., Fuhrmann T., Martinez-Pastor B., Lu W., Braun T., Bober E., Jenuwein T., Serrano L., Esteller M., Chen Z., Barcelo-Batllori S., Mostoslavsky R., Espinosa L., Vaquero A. (2018) SIRT6-dependent cysteine monoubiquitination in the PRE-SET domain of Suv39h1 regulates the NF-kappaB pathway. Nat. Commun. 9, 101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suuronen T., Huuskonen J., Pihlaja R., Kyrylenko S., Salminen A. (2003) Regulation of microglial inflammatory response by histone deacetylase inhibitors. J. Neurochem. 87, 407–416 [DOI] [PubMed] [Google Scholar]
- 43.To M., Swallow E. B., Akashi K., Haruki K., Natanek S. A., Polkey M. I., Ito K., Barnes P. J. (2017) Reduced HDAC2 in skeletal muscle of COPD patients. Respir. Res. 18, 99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ni W., Lin N., He H., Zhu J., Zhang Y. (2014) Lipopolysaccharide induces up-regulation of TGF-alpha through HDAC2 in a rat model of bronchopulmonary dysplasia. PLoS One 9, e91083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding G. J., Fischer P. A., Boltz R. C., Schmidt J. A., Colaianne J. J., Gough A., Rubin R. A., Miller D. K. (1998) Characterization and quantitation of NF-kappaB nuclear translocation induced by interleukin-1 and tumor necrosis factor-alpha. Development and use of a high capacity fluorescence cytometric system. J. Biol. Chem. 273, 28897–28905 [DOI] [PubMed] [Google Scholar]
- 46.Izawa A., Ishihara Y., Mizutani H., Kobayashi S., Goto H., Okabe E., Takeda H., Ozawa Y., Kamiya Y., Sugita Y., Kubo K., Kamei H., Kikuchi T., Mitani A., Hayashi J., Nishihara T., Maeda H., Noguchi T. (2014) Inflammatory bone loss in experimental periodontitis induced by Aggregatibacter actinomycetemcomitans in interleukin-1 receptor antagonist knockout mice. Infect. Immun. 82, 1904–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oz H. S., Puleo D. A. (2011) Animal models for periodontal disease. J. Biomed. Biotechnol. 2011, 754857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kajikawa T., Meshikhes F., Maekawa T., Hajishengallis E., Hosur K. B., Abe T., Moss K., Chavakis T., Hajishengallis G. (2017) Milk fat globule epidermal growth factor 8 inhibits periodontitis in non-human primates and its gingival crevicular fluid levels can differentiate periodontal health from disease in humans. J. Clin. Periodontol. 44, 472–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Socransky S. S., Haffajee A. D., Cugini M. A., Smith C., Kent R. L., Jr (1998) Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25, 134–144 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.