Abstract
The KCNE2 single transmembrane–spanning voltage-gated potassium (Kv) channel β subunit is ubiquitously expressed and essential for normal function of a variety of cell types, often via regulation of the KCNQ1 Kv channel. A polymorphism upstream of KCNE2 is associated with reduced lung function in human populations, but the pulmonary consequences of KCNE2 gene disruption are unknown. Here, germline deletion of mouse Kcne2 reduced pulmonary expression of potassium channel α subunits Kcnq1 and Kcnb1 but did not alter expression of other Kcne genes. Kcne2 colocalized and coimmunoprecipitated with Kcnq1 in mouse lungs, suggesting the formation of pulmonary Kcnq1-Kcne2 potassium channel complexes. Kcne2 deletion reduced blood O2, increased CO2, increased pulmonary apoptosis, and increased inflammatory mediators TNF-α, IL-6, and leukocytes in bronchoalveolar lavage (BAL) fluids. Consistent with increased pulmonary vascular leakage, Kcne2 deletion increased plasma, BAL albumin, and the BAL:plasma albumin concentration ratio. Kcne2−/− mouse lungs exhibited baseline induction of the reperfusion injury salvage kinase pathway but were less able to respond via this pathway to imposed pulmonary ischemia/reperfusion injury (IRI). We conclude that KCNE2 regulates KCNQ1 in the lungs and is required for normal lung function and resistance to pulmonary IRI. Our data support a causal relationship between KCNE2 gene disruption and lung dysfunction.—Zhou, L., Köhncke, C., Hu, Z., Roepke, T. K., Abbott, G. W. The KCNE2 potassium channel β subunit is required for normal lung function and resilience to ischemia and reperfusion injury.
Keywords: KCNQ1, MiRP1, pulmonary, Kv channel, epithelial
Genes in the 5-member human KCNE family each encode single–transmembrane domain ion channel regulatory subunits termed minimal potassium channel subunit–related peptides (MiRPs) or, more commonly now, KCNE proteins. The KCNE proteins are primarily recognized for their regulation of voltage-gated potassium (Kv) channels (1, 2). KCNE2 is widely expressed in mammalian tissues, where it regulates many different Kv α subunits. Originally cloned from an expressed sequence tag from retinoblastoma tissue (2), KCNE2 was then detected in cardiac tissue from several different mammalian species, including Homo sapiens (3–8), and human KCNE2 gene variants were associated with increased predisposition to acquired Long QT syndrome, a cardiac arrhythmia syndrome involving delayed ventricular repolarization most commonly linked to drug block of the human ether-a-go-go-related gene potassium channel, which forms complexes with KCNE2 (2, 9). In mouse hearts, Kcne2 regulates the Kv4.2 and Kv1.5 channels, and Kcne2 deletion also causes delayed ventricular repolarization compared with wild-type mice when combined with the QT-prolonging agent sevoflurane (10).
KCNE2 is also expressed in many epithelia, where instead of regulating action potentials to control muscular contraction, it modulates the activity of channels controlling secretory processes and ion recycling. Thus, in parietal cells of the stomach, KCNE2 regulates KCNQ1, providing a K+ recycling pathway to permit K+ that enters the cell through the gastric H+/K+-ATPase to return to the stomach lumen, avoiding stagnation of this circuit (11–13); genetic deletion of either subunit impairs gastric acid secretion (14–17).
Similarly, in thyroid and choroid plexus epithelial cells, KCNE2 regulates KCNQ1 by direct physical interaction; these complexes themselves regulate the activity of sodium-coupled transporters: the myo-inositol transporter sodium-coupled myo-inositol transporter 1 in the choroid plexus and the sodim iodide symporter in the thyroid. Hence, germline deletion of Kcne2 reduces cerebrospinal fluid myo-inositol levels and also causes hypothyroidism (18–20). KCNQ1-KCNE2 channels probably also regulate insulin secretion from pancreatic β-cells, and germline deletion of either subunit dysregulates insulin secretion (21, 22).
KCNQ1 is also expressed in the lungs and airway epithelial cells in the trachea of mice. In addition, KCNQ1 is expressed in the human lung cancer cell line Calu-3, in which it is thought to regulate basal cAMP-stimulated Cl− secretion through the cystic fibrosis transmembrane conductance regulator (CFTR) (23–26). The Calu-3 cell line is a model for human submucosal serous cells of the lung, which ensure a sterile environment within the lungs, and CFTR is especially enriched in this cell type. Hence, CFTR dysfunction in serous cells is thought to be a major factor in the pathobiology of lung disease in cystic fibrosis (23, 25).
KCNE2 and KCNE3 are also expressed in Calu-3 cells, and it has been suggested that one or both of them regulates KCNQ1 there (24). Similar to KCNE3, KCNE2 converts KCNQ1 to a constitutively active noninactivating K+ channel, enabling KCNQ1-KCNE2 channels to be active at rest (27); KCNE2 also imparts pH sensitivity such that low extracellular pH augments KCNQ1-KCNE2 currents (28). The sum of these properties makes KCNQ1-KCNE2 channels ideal for their role in parietal cells (11–13).
A sequence polymorphism upstream of the human KCNE2 gene has been found to be associated with altered lung function (29, 30), which, together with the expression data from Calu-3 cells, suggested that KCNE2 may be important in human lung physiology, possibly via coassembly with KCNQ1. Here, we assessed the pulmonary functional effects of germline Kcne2 deletion in mice and found Kcnq1 remodeling, evidence of impaired lung function, pulmonary inflammation, and disrupted pulmonary injury response. We also found evidence that KCNE2 forms complexes with KCNQ1 in mouse lung tissue, suggesting that the pulmonary consequences of Kcne2 deletion may arise at least in part from disruption of KCNQ1-KCNE2 channels in lung tissue.
MATERIALS AND METHODS
Animals
We generated global-knockout Kcne2+/+ and Kcne2−/− C57BL/6 female mice from Kcne2+/+ × Kcne2−/− crosses using conventional methods as we previously described (15). The mean age was 13 mo. The Kcne2-mRuby (Kcne2RR) C57BL/6 mouse line, in which the Kcne2 gene is edited by clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) to generate Kcne2 tagged with mRuby, a bright monomeric red fluorescent protein variant optimized for imaging (31), was custom made to order by Cyagen Biosciences (Santa Clara, CA, USA). The gRNA-to mouse Kcne2 gene, the donor DNA containing 3xGGGGS-mRuby, and Cas9 mRNA were coinjected into fertilized mouse eggs to generate targeted knockin offspring. Founding and first–filial generation mice were identified by PCR followed by sequence analysis and then bred to wild-type mice to test germline transmission and for first–filial generation/second–filial generation animal generation. All mice were housed and used according to recommendations in the Guide for the Care and Use of Laboratory Animals [National Institutes of Health (NIH), Bethesda, MD, USA; 8th edition, 2011]. The Institutional Animal Care and Use Committees of Sichuan University (Permit 2015033A), Charité (Berlin, Germany), and University of California–Irvine approved the study.
Real-time quantitative PCR
We performed RNA extraction from lung tissue using the Qiagen RNeasy Kit (Qiagen, Hilden, Germany). Extracted RNA concentration was quantified by UV-spectrometric analysis using the NanoDrop system (Thermo Fisher Sceintific, Waltham, MA, USA). PCR was carried out by routine Taqman Real Time PCR (Thermo Fisher Scientific) using a housekeeping reference gene (18s). The primer sequences were as follows. KCNE2: forward 5′-CATGGTGATGATCGGCATGT-3′, reverse 5′-TCGCCGCTTCGACTTCA-3′; KCNQ1: forward 5′-CCTGGGCTCTGTAGTCTTCATTC-3′, reverse 5′-GGCCCAGAAAGCCAATGTAC-3′; KCND2: forward 5′-CAAACGAAGGGCACAGAAGAA-3′, reverse 5′-GCATTTGCACTCCCGCTTT-3′; KCND3: forward 5′-GGCAAGACCACCTCACTCATC-3′, reverse 5′-TCATGGTTAGTGGTCTTTTCTAAGCA-3′; KCNE1: forward 5′-TGAGCCTGCCCAATTCCA-3′, reverse 5′-CCGCCCTGTTCAGCTGTCT-3′; KCNE3: forward 5′-ACGGGACTGAGACCTGGTACA-3′, reverse 5′-AGCAAGTGACTGTGAAGGGTTGT-3′; KCNE4: forward 5′-GGTGCCCATGATGCTGAATAT-3′, reverse 5′-TCCCCTTCCATCGAGCAA-3′; KCNA4: forward 5′-GCGGGCACAGCAGATTG-3′, reverse 5′-TGAATATTGTGTGCCCTGAGTTCT-3′; KCNB1: forward 5′-CTTTGGGACCTGCTGGAGAA-3′, reverse 5′-GGAGATGATGGCCAGGATCTT-3′; KCNJ2: forward 5′-GAGCAAAGCATGCGTGTCA-3′, reverse 5′-TGTCTGGGTCTCGATGGAGAA-3′; KCNH2: forward 5′-CACCAATGGCATCGACATGA-3′, reverse 5′-GCGAGCGGTTCAGGTGTAG-3′; 18s: forward 5′-ACATCCAAGGAAGGCAGCAG-3′, reverse 5′-TTTTCGTCACTACCTCCCCG-3′.
Immunohistochemistry
For immunohistochemistry of Kcnq1 in Kcne2+/+ vs. Kcne2−/− tissue, we used paraformaldehyde fixation of lung tissue and paraffin embedding (Shandon Pathcentre; Thermo Fisher Scientific) with microtome cutting of 5-µm sections. Antibodies (1/200 dilution) were rabbit anti-KCNQ1 (MilliporeSigma, Burlington, MA, USA) raised against epitope sequence TYEQLTVPRRGPDEGS (primary) and goat anti-rabbit IgG (Vector Laboratories, Burlingame, CA, USA) biotinylated antibody (secondary). Slides were washed with a PBS-Triton 0.5% mix, and the cell nuclei were stained with Mayer’s Hematoxylin (SigmaAldrich, St. Louis, MO, USA).
Fluorescence microscopy
For immunofluorescence of Kcne2 and Kcnq1 in mouse lungs, we used frozen lung tissue sections prepared from adult male and female Kcne2+/+ and Kcne2−/− mice. No sex-dependent differences in pulmonary Kcnq1 and Kcne2 localization were observed. Lungs were inflated ex vivo before freezing using 1:1 optimal cutting temperature:PBS, and then plastic molds containing lungs and optimal cutting temperature were placed into a dry ice-isopentane slurry and stored at −80°C before sectioning (10 µm) at the University of California–Irvine Pathology Core Facility. Frozen sections were fixed in ice-cold acetone (10 min), air-dried, then washed for 2 × 5 min in PBS containing 0.025% Tween-20 (PBS-T). Sections were then blocked for 2 h at room temperature in blocking buffer (PBS-T, 1% bovine serum albumin, 10% donkey serum), washed again, and then incubated overnight at 4°C with 1/1000 primary antibodies: goat anti-KCNQ1 (Santa Cruz Biotechnology, Dallas, TX, USA) in-house rabbit polyclonal anti-KCNE2 antibody, or both (15). The following day, sections were washed (2 × 5 min in PBS-T) and then incubated for 2 h in the dark with fluorescence-conjugated Alexa Fluor 488 or 594 donkey anti-goat IgG secondary antibody, anti-rabbit IgG secondary antibody, or both (1/500–1/1000 dilution; Thermo Fisher Scientific). Following a 2 × 5–min wash in PBS-T, 1 drop of Fluoromount G with DAPI (Thermo Fisher Scientific) was added to each section, and then slides were coverslipped, sealed with nail polish, and after drying were visualized using a BX-51 upright microscope equipped with a DP-72 camera and cellSens software (Olympus, Tokyo, Japan).
Lung sections from Kcne2RR mice were prepared from lungs fixed in paraformaldehyde (5%) and visualized as previously described, with similarly prepared wild-type mouse lung sections used as a negative control for the mRuby fluorescent signal.
Serum preparation and blood gas measurements
Blood was taken from the hearts immediately after euthanizing the mice; 0.3-ml blood samples were measured with a blood gas analyzer (ABL800 Flex; Danaher, Washington, DC, USA). The remainder of each blood sample was centrifuged at 4000 g for 10 min at 4°C, and then the serum samples were frozen at −20°C until assay.
Mouse model of lung ischemia/reperfusion injury
Sodium pentobarbital (50 mg/kg i.p.) was used for anesthesia. Mice were intubated with an endotracheal tube (PE90) attached to a mouse ventilator (Harvard Apparatus, Holliston, MA, USA) and ventilated with a tidal volume of 230 μl at a rate of 120 strokes/min. Once anesthetized, mice were placed on their right side, and a left anterolateral thoracotomy in the fifth intercostal space was performed. The chest was opened, and the left hilum was clamped, occluding the pulmonary artery, vein, and main stem bronchus. After 1 h of ligation, the clamp was released, and the left lung was reperfused for 2 h. Sham-treated mice underwent the same procedure but without clamping of the left pulmonary hilum. Anesthesia adequacy was controlled by monitoring the loss of the corneal reflex, the lack of response to toe pinching, and by evaluating heart rate. Mouse body temperature was maintained with a heating blanket. At the end of reperfusion, mice were euthanized with an overdose of sodium pentobarbital (200 mg/kg, i.p.), and death was monitored via cardiac activity and respiration. Left lungs were then surgically dissected and immediately frozen for further assessment.
Bronchoalveolar lavage fluid collection and cell counts
The thorax was opened at the end of the experiment. Bronchoalveolar lavage (BAL) fluid was collected by cannulating the trachea with repeated instilling of 0.5 ml ice-cold sterile PBS containing heparin up to a total volume of 1.5 ml. The recovered BAL fluid was centrifuged at 1500 g for 10 min at 4°C. Supernatant fluids were stored at −80°C for ELISA analysis. Cell pellets were resuspended in PBS solution, and the total leukocytes were counted with the aid of a hemocytometer. The slides were visualized using Wright-Giemsa staining (MilliporeSigma), and cells were counted in a double-blind manner.
Determination of albumin and cytokine content in BAL fluid
Mouse albumin levels in BAL fluid or in plasma were measured in duplicate using a commercial ELISA kit (DLdevelop, Wuxi, China). The levels of inflammatory cytokines, including TNF-α and IL-6 in BAL, were quantified using ELISA according to the manufacturer’s instructions.
Wet:dry lung weight ratio
After the experiments, the left lower lobes of the lungs were taken and measured immediately after excision (wet weight). The lung tissue was then dried in an oven at 60°C for 5 d and reweighed to obtain dry weight. The wet:dry lung weight ratio (W/D) was calculated as an indicator of edema.
Lung tissue collection
After euthanizing the mice, the left lungs were dissected and embedded in 10% phosphate-buffered formalin overnight before cutting into 5-μm sections. Serial sections of transverse lung slices were deparaffinized in xylene and isopropanol and then mounted on glass slides for hematoxylin and eosin (H&E) staining or TUNEL.
Histologic evaluation
A numerical scoring system was used in this analysis for histologic evaluation based on an established lung injury scoring system (Supplemental Table S1) (32).
TUNEL assay
Cell death was assessed with the TUNEL kit according to the manufacturer’s instructions (Roche, Basel, Switzerland). A total of 10 visual fields from each slice were chosen randomly and analyzed in a blinded manner. TUNEL-positive apoptotic nuclei appeared fluorescent green and the apoptotic index was calculated as a ratio of the number of TUNEL-positive nuclei to the total nuclei population. Images were obtained and assessed using a fluorescence microscope (Carl Zeiss, Oberkochen, Germany).
Western blot analysis and immunoprecipitation
Frozen left lung tissue samples were homogenized with a precooled pestle grinder system (Thermo Fisher Scientific). RIPA buffer contained 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 1 mM EDTA, and 0.25% sodium deoxycholate mixed with phosphatase inhibitor cocktail (MilliporeSigma) and a protease inhibitor cocktail (MilliporeSigma). The homogenate was centrifuged at 10,000 g for 10 min at 4°C. The supernatant was collected, and protein concentration was determined by the bicinchoninic acid method (Thermo Fisher Scientific). An equal amount of protein (15 μg) was loaded onto 12% sodium dodecyl sulfate PAGE gels for electrophoresis. Proteins were then transferred onto nitrocellulose membranes (VWR International, Radnor, PA, USA) and blotted with primary antibodies. Primary rabbit antibodies included goat anti-KCNQ1 antibody (Santa Cruz Biotechnology), phosphorylated (p)ERK1/2 (Thr202/Tyr204), total (t)ERK1/2, (p)p38 MAPK (Thr180/Tyr182), (t)p38 MAPK, (p)stress-activated protein kinase/c-Jun N-terminal kinase (JNK) (Thr183/Tyr185), (t)JNK, (p)protein kinase B (AKT) (ser473), (t)AKT; (p)glycogen synthase kinase-3β (GSK-3β) (Ser9), (t)GSK-3β, (p)signal transducer and activator of transcription 3 (STAT-3) (Tyr705), and (t)STAT-3 (all 1:1000; Cell Signaling Technology, Danvers, MA, USA). After overnight incubation of primary antibodies, the membranes were incubated with a 1:5000 dilution of horseradish peroxidase–conjugated goat anti-rabbit IgG secondary antibody (Bio-Rad, Hercules, CA, USA). The membranes were developed by chemiluminescence Western blotting ECL (MilliporeSigma). Signals were visualized by an AmershamImager 600 system (GE Healthcare, Waukesha, WI, USA), and band densities were determined using ImageJ Data Acquisition Software (NIH). Band densities of phosphorylated proteins were normalized to the respective total protein signal densities.
For Kcnq1 and Kcne2 coimmunoprecipitation, mouse lung tissue was homogenized using a BeadRuptor Elite bead mill with porcelain beads (Omni International, Kennesaw, GA, USA), precleared with protein A agarose beads (Thermo Fisher Scientific), and then incubated overnight at 4°C with in-house anti-KCNE2 antibodies raised in rabbit (15) or goat anti-KCNQ1 pAb (Santa Cruz Biotechnology). The following day, antibody-protein complexes were precipitated using protein A beads and separated on 4–12% Bis-Tris gels (Thermo Fisher Scientific), transferred onto PVDF membranes (Bio-Rad), and then Western blotted using 1/1000 dilution of in-house anti-KCNE2 antibodies (15) or rabbit anti-KCNQ1 pAbs (MilliporeSigma) in 5% milk (overnight at 4°C) as we previously described (10). Following washes in Tris-buffered saline–Tween (3 × 5 min), primary antibodies were probed using goat anti-rabbit IgG secondary antibody (Thermo Fisher Scientific) at 1 in 5000 in 1% milk and Tris-buffered saline–Tween. Bands were visualized using Immobilon Forte Western HRP Substrate (MilliporeSigma) and a Gbox system (Syngene, Cambridge, United Kingdom).
Statistical analysis
All values are expressed as means ± sem. Unpaired 2-tailed Student’s t tests were used for 2-value comparisons. For multiple comparisons over 3 groups, 1-way ANOVA was used followed by the Newman-Keuls test or Dunnett’s T3 test depending on the equality of homogeneity of variance. Values were considered to be significant at P < 0.05.
RESULTS
Germline Kcne2 deletion reduces pulmonary expression of KCNQ1 and KCNB1
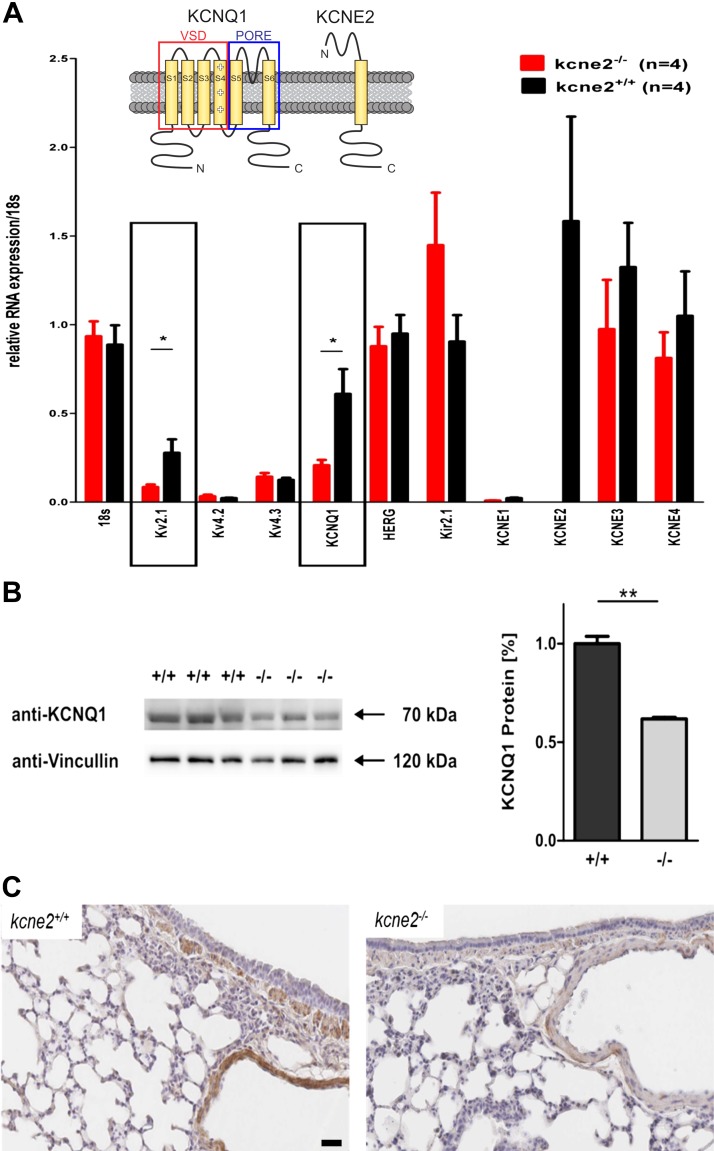
Using real-time quantitative PCR (qPCR), we quantified transcripts of Kv channel α and β subunits in wild-type mouse lung tissue and examined the effects of Kcne2 deletion (Fig. 1A). Kcne2 was relatively highly expressed in normal lung tissue, confirming our previous result (33). Kcne1 was absent but Kcne3 and Kcne4 were readily detectable, and their expression was unchanged by Kcne2 deletion. Among several known Kcne2-partner Kv α subunits that were quantified, only transcripts for Kcnb1 and Kcnq1 were altered by Kcne2 deletion, with each showing down-regulation. We confirmed this down-regulation at the protein level for Kcnq1 (Fig. 1B). Similarly, Kcnq1 protein was detectable in several locations in the lung, including epithelial cells surrounding bronchioles, and was down-regulated in Kcne2−/− mice (Fig. 1C).
Figure 1.
Kcne2 deletion stimulates pulmonary Kv channel remodeling. A) Real-time qPCR quantification of Kv α and β subunit transcript expression in Kcne2+/+ and Kcne2−/− mouse lung tissue (n = 4 mice/genotype). *P < 0.05. Upper inset: schematic of KCNQ1 and KCNE2 topology. B) Western blot quantification of KCNQ1 protein in Kcne2+/+ and Kcne2−/− mouse lung tissue (n = 3 mice/genotype). Left, blot; right, band density normalized to mean wild-type band density. **P < 0.01. C) Exemplar Kcne2+/+ and Kcne2−/− mouse lung tissue histology with KCNQ1 immunostaining (brown). HERG, human ether-a-go-go-related gene; Kir2.1, inward rectifier potassium channel 2; VSD, voltage-sensing domain. Scale bar, 20 µm.
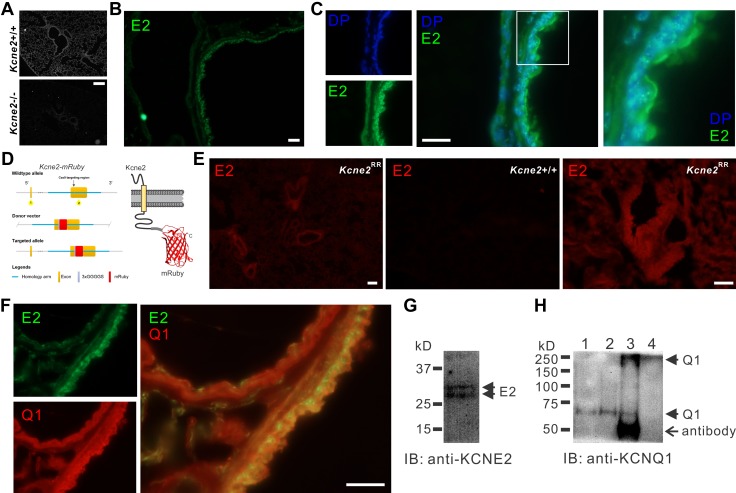
To verify Kcne2 protein expression in mouse lungs and test the hypothesis that Kcne2 regulates Kcnq1 in mouse lungs, we adopted 3 complementary approaches. First, we probed Kcne2 expression by immunofluorescence in Kcne2+/+ mouse lung sections, using Kcne2−/− lung sections as a negative control. We observed the specific Kcne2 protein signal surrounding the bronchioles and to a lesser extent in alveoli (Fig. 2A, B). In cells lining the bronchi and bronchioles, Kcne2 was primarily localized at or close to the cell membrane (Fig. 2C). Second, using CRISPR/Cas9, we generated a mutant mouse line in which the native Kcne2 gene was edited to add an mRuby fluorescent tag (Kcne2RR mice) (Fig. 2D). Fluorescence microscopy of Kcne2RR mice lungs revealed Kcne2-mRuby protein distribution around the bronchi and bronchioles and, to a lesser extent, in alveoli; wild-type mice imaged as a negative control using equivalent exposure showed minimal background red fluorescence (Fig. 2E). Kcne2 colocalized with Kcnq1 in cells lining the bronchi and bronchioles (Fig. 2F). Third, Kcnq1 protein was immunoprecipitated from mouse lung tissue using anti-Kcne2 antibody, which itself detected Kcne2 using Western blotting (Fig. 2G, H). These data suggest that Kcne2 forms channel complexes with Kcnq1 in mouse lungs, especially in cells lining the bronchioles. We next tested the effects of Kcne2 deletion on lung function and resilience to injury.
Figure 2.
Kcne2 colocalizes with Kcnq1 in mouse lungs. A) Immunofluorescence detection of Kcne2 protein in Kcne2+/+ but not Kcne2−/− mouse lung tissue. Scale bar, 40 µm. B) Immunofluorescence detection of Kcne2 (E2) protein (green) in Kcne2+/+ mouse lung tissue. Scale bar, 10 µm. C) Immunofluorescence detection of Kcne2 protein (green) in Kcne2+/+ mouse lung tissue compared with location of the nucleus stained with DAPI (DP; blue). Left, single channel images; center, merged; right, close-up images of boxed region from center panel. Scale bar, 10 µm. D) Schematic of CRISPR/Cas9 targeting approach to generate Kcne2RR mice. E) Fluorescence detection of Kcne2-mRuby (red) in Kcne2RR but not Kcne2+/+ mouse lung tissue. Scale bars, 20 µm. F) Immunofluorescence detection of Kcne2 protein (green) and Kcnq1 protein (Q1) (red) in Kcne2+/+ mouse lung tissue. Left, single channel images; right, merged. Scale bar, 10 µm. G) Western immunoblot (IB) of Kcne2 in mouse lung lysate. H) Anti-Kcnq1 Western IB showing detection of Kcnq1 from mouse lung lysates immunoprecipitated (IP) with anti-Kcne2 antibody. Lanes: 1, lysate; 2, anti-Kcnq1 IP; 3, anti-Kcne2 IP; 4, supernatant after initial bead precipitation from anti-Kcne2 IP. Kcnq1 bands at MWs corresponding to both the monomeric (60-kD) and the tetrameric (240-kD) forms are visible in the anti-Kcne2 lane (lane 3). No Kcnq1 was detectable in the supernatant after the Kcne2 IP (lane 4).
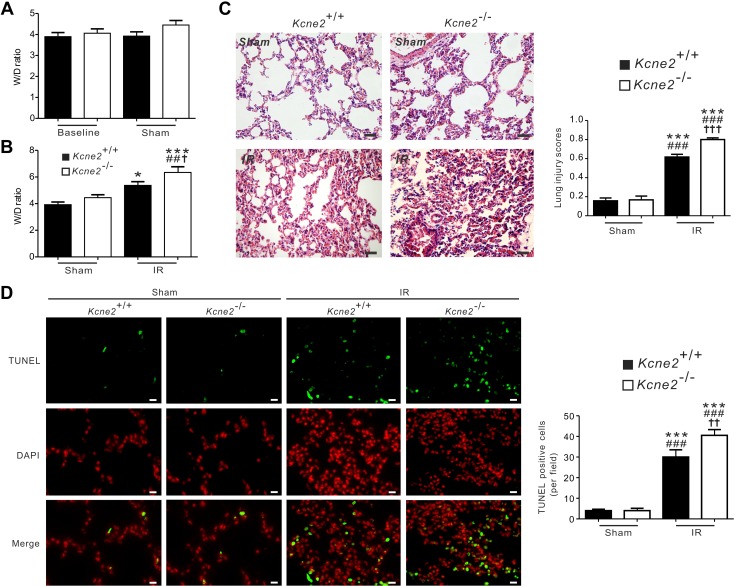
Kcne2 deletion increases pulmonary damage and inflammatory cytokine production at baseline
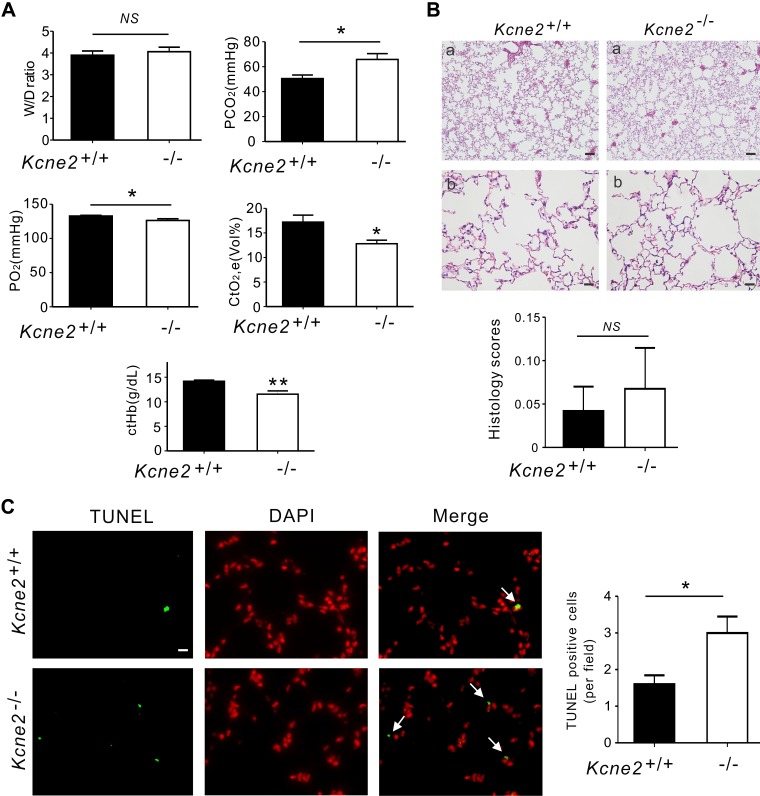
Kcne2 deletion did not cause lung edema because equal W/D was found in baseline Kcne2+/+ and Kcne2−/− mice (P = 0.5763). We also analyzed arterial blood gases from Kcne2−/− mice and their wild-type littermates. Kcne2−/− mice had high partial pressure of carbon dioxide (65.8 ± 4.7 mmHg), low partial pressure of oxygen (126.3 ± 2.5 mmHg), and low concentration of total oxygen (12.8 ± 0.7 mmHg) and total hemoglobin, with elevated concentration of Na+, Ca2+, and Cl− concentrations, demonstrating a mixed respiratory and metabolic imbalance (Fig. 3A and Supplemental Fig. S1). Although one Kcne2−/− lung showed slight histopathological alterations such as thickened alveolar walls and neutrophil infiltration, H&E-stained sections of Kcne2+/+ and Kcne2−/− lungs didn’t reveal statistically significant histologic structure and architecture differences between the 2 genotypes, perhaps because of variability within the Kcne2−/− mouse group (Fig. 3B). We quantified apoptosis using TUNEL staining of Kcne2+/+ and Kcne2−/− lung tissues and examined the sections by immunofluorescence microscopy. To identify apoptotic cells, double labeling was performed. Apoptotic cells were visualized by colocalized double positivity of TUNEL-green and DAPI-red. Also indicative of more pulmonary damage in Kcne2−/− mice compared with Kcne2+/+ mice, Kcne2 deletion significantly increased the number of TUNEL-positive cells in lung tissues compared with Kcne2+/+ mice (Fig. 3C).
Figure 3.
Kcne2 deletion increases pulmonary damage and inflammatory cytokine production at baseline. A) Lung and blood gas parameters of Kcne2+/+ and Kcne2−/− mice as indicated. W/D of left lung sections from Kcne2+/+ and Kcne2−/− mice; n = 8–10. Partial pressure of carbon dioxide (P co2) values of Kcne2+/+ and Kcne2−/− mice; n = 6–7. Partial pressure of oxygen (P o2) values from Kcne2+/+ and Kcne2−/− mice; n = 6–7. Concentration of total oxygen (Ct o2) and concentration of total hemoglobin (CtHb) values of Kcne2+/+ and Kcne2−/− mice; n = 6–7. B) Upper, effect of Kcne2 deletion on lung histology. Lung tissues were stained with H&E and examined under a light microscope; n = 4–5 mice/genotype. Scale bar, 5 μm (a); scale bar, 20 μm (b). Lower, quantification of morphologic evaluation of lung sections of both Kcne2+/+ and Kcne2−/− mice; n = 4–5 mice/genotype. C) Left: representative micrographs of TUNEL apoptosis assay performed on lung tissue collected from Kcne2+/+ and Kcne2−/− mice. TUNEL-positive cells were stained green (TUNEL) and nuclei were stained red (DAPI). Arrows indicate TUNEL-positive cells. Right: graph showing the quantification of TUNEL-positive cells; n = 5 mice each genotype. Scale bar, 20 μm. NS, no difference between both genotypes. *P < 0.05, **P < 0.01 between genotypes.
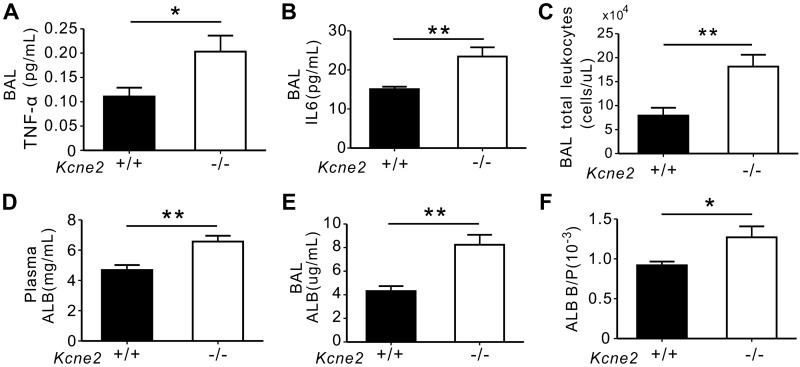
TNF-α and IL-6 serve as major inflammatory cytokines, the elevation of which may amplify lung injury and stimulate the inflammatory and autoimmune processes (34). The production of these 2 cytokines was up-regulated in Kcne2−/− lungs (TNF-α: P = 0.025, IL-6: P = 0.006 vs. Kcne2+/+ mice; Fig. 4A, B). Consistent with this, a marked increase in the total number of leukocytes was noted in the BAL fluid in Kcne2−/− mice compared with their wild-type littermates (P = 0.004; Fig. 4C). Moreover, we found that albumin concentrations were significantly increased by Kcne2 deletion in plasma (40% increase, P = 0.003; Fig. 4D) and BAL fluid (2-fold higher, P = 0.001; Fig. 4E), with a 44% higher BAL:plasma albumin concentration ratio in Kcne2−/− mice compared with that in Kcne2+/+ mice (P = 0.032; Fig. 4F), suggestive of relatively more pulmonary vascular leakage in Kcne2−/− mice at baseline.
Figure 4.
Kcne2 deletion increases the production of inflammatory mediators at baseline. A) Mouse TNF-α content in BAL fluids from Kcne2+/+ and Kcne2−/− mice was determined using ELISA; n = 5–7 mice/genotype. B) IL-6 content in BAL fluids from Kcne2+/+ and Kcne2−/− mice; n = 6 mice/genotype. C) Total leukocytes were quantitated in BAL fluids from Kcne2+/+ and Kcne2−/− mice; n = 6 mice/genotype. D) Albumin (ALB) levels in plasma from Kcne2+/+ and Kcne2−/− mice; n = 7 mice/genotype. E) ALB in BAL fluids from Kcne2+/+ and Kcne2−/− mice; n = 7 mice/genotype. F) The ratio of BAL ALB content:plasma albumin content (ALB B/P) in Kcne2+/+ and Kcne2−/− mice; n = 7 mice/genotype. *P < 0.05, **P < 0.01 between genotypes.
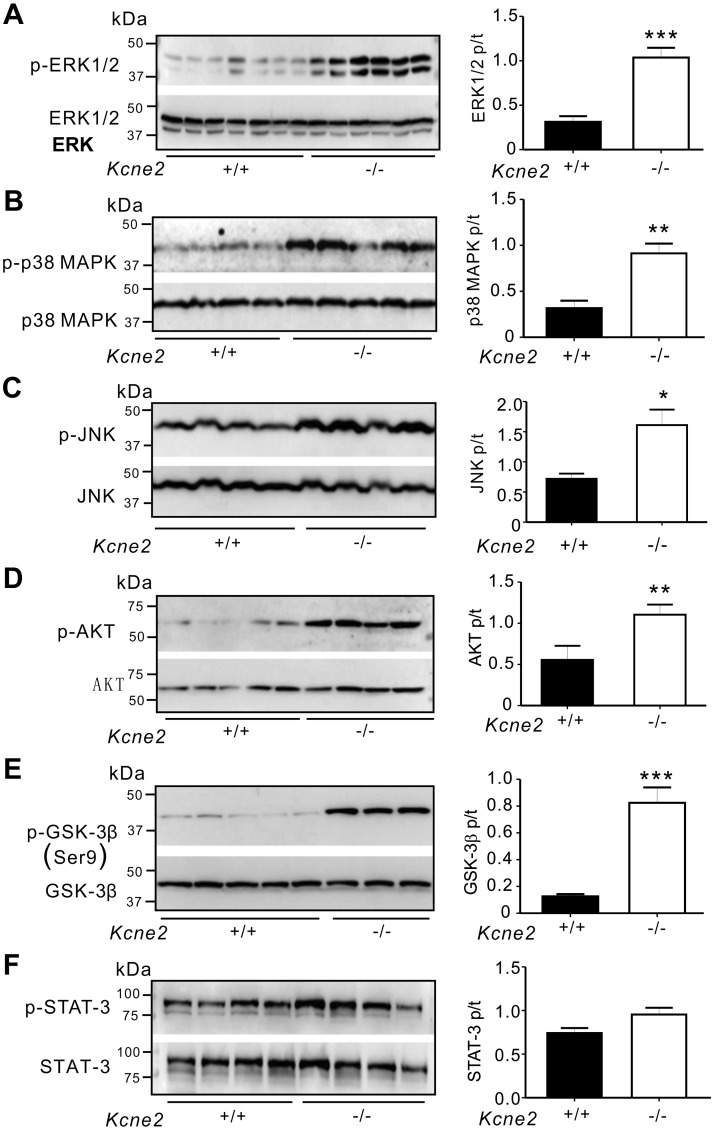
Kcne2 deletion stimulates baseline protein phosphorylation
We also detected differences arising from Kcne2 deletion in the phosphorylation of proteins known to be involved in signaling cascades activated in response to ischemic damage (35, 36). We found no differences in tERK1/2, tp38 MAPK, tJNK, tAKT, tGSK-3β, or tSTAT-3 protein levels in all groups. pERK1/2, pp38 MAPK, pJNK, pAKT, pGSK-3β, or pSTAT-3 levels were normalized by their total protein levels. We observed a 2.3-fold increase in ERK1/2 phosphorylation in Kcne2−/− lungs (P = 0.0001; Fig. 5A), whereas the ratio of pp38 MAPK:tp38 MAPK in Kcne2−/− mice post ischemia/reperfusion (I/R) injury (IRI) was double that of Kcne2+/+ mice (P = 0.004; Fig. 5B). JNK phosphorylation was increased by 30% in Kcne2−/− lungs (P = 0.013; Fig. 5C). Similarly, we also observed a 55% increase in AKT phosphorylation in Kcne2−/− lung lysates (P = 0.031; Fig. 5D). Strikingly, the ratio of pGSK-3β:tGSK-3β at baseline was increased by 5-fold in Kcne2−/− lungs (P = 0.0003; Fig. 5E). However, pSTAT-3 levels remained unchanged between genotypes (P = 0.071; Fig. 5F).
Figure 5.
Kcne2 deletion affects protein activation at baseline. Left, representative Western blots of pERK1/2 and tERK1/2 (A), pp38 MAPK and tp38 MAPK (B), pJNK and tJNK (C), pAKT and tAKT (D), pGSK3β and tGSK3β (E), and pSTAT-3 and tSTAT-3 (F) from baseline Kcne2+/+ and Kcne2−/− lungs; 1 mouse/lane. Right, mean ratios of pERK:tERK (n = 6–7) (A), pp38 MAPK:tp38 MAPK (n = 5) (B), pJNK:tJNK (n = 5) (C), pAKT:tAKT (n = 7–8) (D), pGSK3β:tGSK3β (n = 5) (E), and pSTAT-3:tSTAT-3 (n = 5) (F) band densities from blots as in left. *P < 0.05, **P < 0.01, ***P < 0.001 between genotypes.
Kcne2 deletion exacerbates I/R-induced pulmonary injury
We next quantified W/D in lungs from Kcne2+/+ and Kcne2−/− mice subjected to 1 h of left lung ischemia followed by 2 h of reperfusion (IRI). Sham treatment did not cause alternation of the W/D when compared with baseline values (P = 0.291), and there were no genotype-dependent differences at baseline or in sham-treated mice (Fig. 6A). However, after pulmonary IRI, the W/D was significantly larger in Kcne2−/− mice (6.3 ± 0.4) than in Kcne2+/+ mice (5.4 ± 0.3%, P < 0.05, n = 6–9; Fig. 6B). Pulmonary damage was also evaluated by H&E staining, and quantification was evaluated using a previously described histologic scoring system (32). After IRI, both genotypes suffered various degrees of pulmonary injury (all P < 0.001 vs. sham-treated mice). Compared with Kcne2+/+ mice, the degree of pulmonary injury was more severe in Kcne2-deleted lungs (P < 0.001). In the lungs of Kcne2−/− mice, the I/R-induced pulmonary damage or inflammatory response was considerably more intense, as evidenced by a more enhanced alveolar accumulation of neutrophils and lymphocytes, thickened alveolar walls, interstitial edema, and hemorrhage (Fig. 6C). Consistent with this, after 2 h of reperfusion, TUNEL-positive cells were observed within the left lungs in both genotypes (all P < 0.001 vs. sham-operated ones). Furthermore, the number of apoptotic cells was significantly larger in Kcne2−/− lungs compared with their wild-type littermates (P < 0.01; Fig. 6D).
Figure 6.
Kcne2 deletion increases pulmonary injury after experimentally imposed pulmonary I/R. A) W/D of left lung sections from baseline and sham Kcne2+/+ and Kcne2−/− mice; n = 7–10 mice/genotype. NS, no difference between genotypes. B) W/D of left lung sections from Kcne2+/+ and Kcne2−/− mice post I/R. No difference was found between sham Kcne2+/+ and Kcne2−/− mice (P > 0.05). *P < 0.05, ***P < 0.001 compared with sham Kcne2+/+ mice; ##P < 0.01 compared with sham Kcne2−/− mice; †P < 0.05 compared with Kcne2+/+ mice post I/R (by 1-way ANOVA). C) Left: representative histologic H&E-stained micrographs of lung sections from Kcne2+/+ and Kcne2−/− mice post I/R. Representative of n = 5 mice/genotype. Scale bars, 20 µm. Right: quantitative morphologic evaluation of pulmonary damage after reperfusion in ischemic left lungs of both genotypes (n = 5 mice/genotype). ***P < 0.001 compared with sham Kcne2+/+ mice; ###P < 0.001 compared with sham Kcne2−/− mice; †††P < 0.001 compared with Kcne2+/+ mice post I/R (by 1-way ANOVA). D) Left: representative images from TUNEL-stained lung tissue sections from Kcne2+/+ and Kcne2−/− mice post I/R. Apoptotic nuclei were stained green, and nuclear staining DAPI is shown in red. Right: bar graph showing numbers of TUNEL-positive nuclei in tissue sections from each group. n = 5–6 hearts in each group. ***P < 0.001 compared with sham Kcne2+/+ mice; ###P < 0.001 compared with sham Kcne2−/− mice; ††P < 0.01 compared with Kcne2+/+ mice post I/R (by 1-way ANOVA). Scale bars, 20 µm.
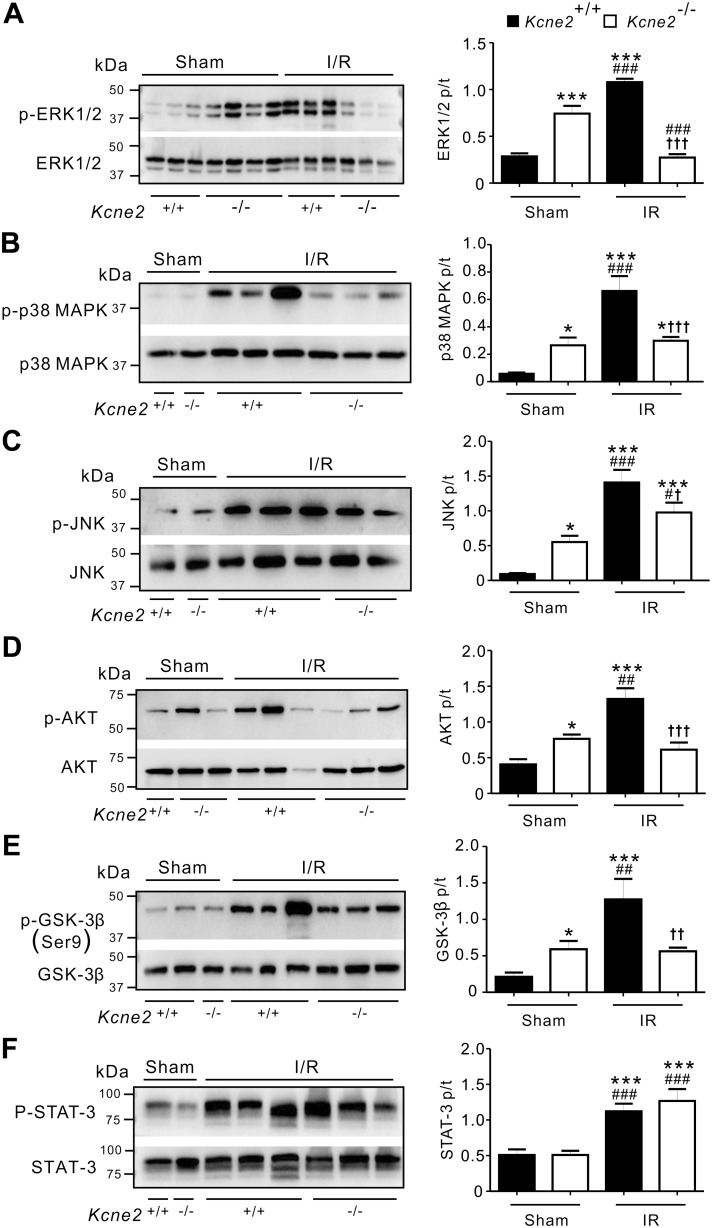
Effects of Kcne2 deletion on pulmonary protein phosphorylation post I/R
Consistent with data in Fig. 4, the levels of baseline phosphorylation in lung tissue of ERK, p38 MAPK, JNK, AKT, and GSK-3β but not STAT-3 were increased in sham-treated Kcne2−/− mice (P < 0.05 or P < 0.001) compared with sham-treated Kcne2+/+ mice (Fig. 7). At the end of 2 h of reperfusion, I/R stimuli led to a significant elevation of protein phosphorylation; that is, a 3.7-fold increase in ERK1/2 phosphorylation in Kcne2+/+ lungs (P < 0.001 vs. sham-treated Kcne2+/+ mice; Fig. 7A), a 10-fold increase in p38 MAPK phosphorylation (P < 0.001; Fig. 7B), a 15-fold increase in JNK phosphorylation (P < 0.001; Fig. 7C), and 3.2- and 6-fold increases in AKT and GSK-3β phosphorylation, respectively (P < 0.001; Fig. 7D, E). We also observed a 2-fold increase in STAT-3 phosphorylation in Kcne2+/+ lungs post I/R compared with sham-treated Kcne2+/+ lungs (P < 0.001; Fig. 7F).
Figure 7.
Kcne2 deletion impairs pulmonary protein phosphorylation post I/R. Left: representative Western blots of pERK1/2 and tERK1/2 (A), pp38 MAPK and tp38 MAPK (B), pJNK and tJNK (C), pAKT and tAKT (D), pGSK3β and tGSK3β (E), and pSTAT-3 and tSTAT-3 (F) in Kcne2+/+ and Kcne2−/− lungs post I/R injury; 1 mouse/lane. Right: mean ratio of pERK:tERK (n = 7–8) (A), pp38 MAPK:tp38 MAPK (n = 5) (B), pJNK/tJNK (n = 5–6) (C), pAKT/tAKT (n = 5–6) (D), pGSK3β/tGSK3β (n = 5–9) (E), and pSTAT-3/tSTAT-3 (n = 5) (F) band densities from blots as in left. *P < 0.05, **P < 0.01, ***P < 0.001 compared with sham-treated Kcne2+/+ mice; #P < 0.05, ##P < 0.01, ###P < 0.001 compared with sham-treated Kcne2−/− mice; †P < 0.05, ††P < 0.01, †††P < 0.001 compared with Kcne2+/+ mice post I/R (by 1-way ANOVA).
Kcne2 deletion dramatically impaired the pulmonary signaling cascade response to IRI. Thus, pulmonary IRI in Kcne2−/− mice resulted in only a 2.5-fold elevation in STAT-3 phosphorylation (P < 0.001 vs. sham-treated Kcne2−/− mice) and a 1.8-fold increase in JNK phosphorylation (P < 0.05). Furthermore, Kcne2 deletion eliminated the IRI-induced elevation in phosphorylation of p38 MAPK, AKT, or GSK-3β (P > 0.05 vs. sham-treated Kcne2−/− mice). Most strikingly, we observed a 2.7-fold decrease in ERK1/2 phosphorylation in Kcne2−/− lungs post I/R compared with sham-treated Kcne2−/− lungs (P < 0.001) (Fig. 7A–F).
DISCUSSION
In this first examination of the role of KCNE2 in the lungs, we found that Kcne2 deletion in mice down-regulates KCNQ1 and another known partner of KCNE2, KCNB1 (37). In this and a previous study (33), we readily detected Kcne2 transcript expression in mouse lung tissues (verified using knockout tissue as a negative control). In addition, here, we were able to detect Kcne2 protein in mouse lungs and verify the specificity of the signal using 2 approaches: Kcne2−/− tissue as a negative control and Kcne2RR tissue as an additional positive control. KCNQ1 is expressed in human airway smooth muscle cells and may play a role in regulating airway diameter (38); Kcnq1 mRNA was also previously detected in epithelial cells lining the bronchioles (39), and Kcnq1 protein was detected in tracheal epithelial cells (40) in adult mice. We speculate that deletion of Kcne2 may perturb the function of pulmonary KCNQ1-KCNE2 complexes (and we showed here that Kcne2 deletion results in pulmonary Kcnq1 down-regulation in mice) in epithelial cells lining the bronchi and bronchioles, perturbing fluid and ion secretion.
Regulation of ionic currents is crucial to all aspects of epithelial cell function and physiology and is achieved primarily by ion channels and transporters. The KCNQ1 Kv channel is essential for the normal function of many cell types, including both excitable and nonexcitable cells (41). This raises the question of how a voltage-gated ion channel activated by membrane depolarization can function effectively in both excitable and nonexcitable cells. KCNQ1 achieves this versatility largely by interaction with members of the KCNE family of single transmembrane domain ancillary or β subunits (1, 42, 43).
In human cardiac myocytes, KCNQ1 interacts with KCNE1, generating a β-adrenergic–stimulated, slow-activating current perfect for facilitating phase 3 repolarization and responding appropriately to increased demands during exercise, for example (44, 45). In contrast, in epithelial cells of the lower intestine, KCNQ1 interacts with KCNE3, forming a channel that is constitutively active because KCNE3 locks open the KCNQ1 voltage sensor, effectively mimicking membrane depolarization so that the channel can be active at resting membrane potential (46–50). KCNE3 also confers an additional mode of sensitivity to PKC and is regulated by estrogen (51, 52). These properties make KCNQ1-KCNE3 ideal for regulating cAMP-stimulated Cl− secretion in the intestine.
Kcne1 was previously reported to be absent from epithelial cells lining the trachea and bronchioles in adult mice (39, 40), whereas Kcne3 was suggested to regulate Kcnq1 in the tracheal epithelium (40). In agreement with others (24, 39, 40), we detected Kcnq1, Kcne2, Kcne3, and Kcne4 in mouse lungs, and in addition, we found evidence that Kcnq1 and Kcne2 coassemble with one another in epithelial cells of the bronchi and bronchioles. Disruption of lung epithelial Kcnq1-Kcne2 complexes is therefore a plausible explanation for at least some of the pulmonary defects we found in Kcne2−/− mice, although it remains possible that there is also contribution from other aspects of the multisystem syndrome that arises from Kcne2 deletion (including hypothyroidism, atherosclerosis, achlorhydria, and nonalcoholic fatty liver disease) (15, 18, 33, 53–55).
We were able to ascertain that Kcne2 is required for normal lung function in mice, and its deletion perturbs blood gases, increases pulmonary apoptosis, and increases the presence of inflammatory mediators in BAL fluid and plasma. Notably, Kcne2 deletion also increased the phosphorylation of several proteins in the reperfusion injury salvage kinase (RISK) protective protein signaling cascade, also indicative of baseline damage in the lungs due to Kcne2 deletion. Furthermore, Kcne2 deletion prevented the normal RISK and STAT-3 responses to pulmonary IRI and increased the level of IRI to the lungs. This is opposite to the effects of Kcne2 deletion in the context of cardiac IRI in which Kcne2 deletion preconditions the heart and lessens the extent of myocardial infarction produced by an imposed IRI (56).
As we also found here for Kcne2 protein localization in mouse lungs, the KCNE2 transcript was previously detected in human bronchial epithelial cells (30). A single-nucleotide polymorphism (SNP) upstream of human KCNE2 (rs9978142) is associated with reduced pulmonary function (30), as is another SNP close to KCNE2 (rs973754/rs9982601); both these SNPs are also associated with increased risk of coronary artery disease and early myocardial infarction (29, 57, 58). Although the functional effects that SNPs exert on KCNE2 are unknown (they are outside the coding region), there are clear parallels between lung and coronary artery disease occurring both in people with KCNE2 SNPs and in Kcne2−/− mice (54).
Together with the human population data, our mouse data support a causal link between disruption of the KCNE2 gene and altered lung function, with altered response to lung IRI (which can become especially important during lung transplant surgery). Future studies will investigate the precise mechanisms by which KCNE2 regulates lung physiology and how its perturbation leads to pulmonary dysfunction.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
The authors thank Dr. Siegfried Breitling (Charité – Universitätsmedizin, Berlin), Angele de Silva (University of California–Irvine), and Dr. Maria Papanikolaou (University of California–Irvine) for technical assistance. This study was supported by Grants 81700074 (to L.Z.) and 81670300 (to Z.H.) from the National Natural Science Foundation of China and GM115189 and GM130377 from the U.S. National Institutes of Health (NIH) National Institute of General Medical Sciences (to G.W.A.). The authors declare no conflicts of interest.
Glossary
- AKT
protein kinase B
- BAL
bronchoalveolar lavage
- Cas9
CRISPR-associated protein 9
- CFTR
cystic fibrosis transmembrane conductance regulator
- CRISPR
clustered regularly interspaced short palindromic repeats
- GSK
glycogen synthase kinase
- H&E
hematoxylin and eosin
- I/R
ischemia/reperfusion
- IRI
I/R injury
- JNK
c-Jun N-terminal kinase
- Kv
voltage-gated potassium
- PBS-T
PBS containing 0.025% Tween-20
- qPCR
quantitative PCR
- RISK
reperfusion injury salvage kinase
- SNP
single-nucleotide polymorphism
- STAT-3
signal transducer and activator of transcription 3
- W/D
wet:dry lung weight ratio
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
Z. Hu, T. K. Roepke, and G. W. Abbott designed research; L. Zhou, C. Köhncke, Z. Hu and G. W. Abbott performed research; L. Zhou, C. Köhncke, Z. Hu, T. K. Roepke, and G. W. Abbott analyzed data; and Z. Hu and G. W. Abbott wrote the manuscript.
REFERENCES
- 1.Abbott G. W. (2015) The KCNE2 K+ channel regulatory subunit: ubiquitous influence, complex pathobiology. Gene 569, 162–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abbott G. W., Sesti F., Splawski I., Buck M. E., Lehmann M. H., Timothy K. W., Keating M. T., Goldstein S. A. (1999) MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell 97, 175–187 [DOI] [PubMed] [Google Scholar]
- 3.Finley M. R., Li Y., Hua F., Lillich J., Mitchell K. E., Ganta S., Gilmour R. F., Jr., Freeman L. C. (2002) Expression and coassociation of ERG1, KCNQ1, and KCNE1 potassium channel proteins in horse heart. Am. J. Physiol. Heart Circ. Physiol. 283, H126–H138 [DOI] [PubMed] [Google Scholar]
- 4.Franco D., Demolombe S., Kupershmidt S., Dumaine R., Dominguez J. N., Roden D., Antzelevitch C., Escande D., Moorman A. F. (2001) Divergent expression of delayed rectifier K(+) channel subunits during mouse heart development. Cardiovasc. Res. 52, 65–75 [DOI] [PubMed] [Google Scholar]
- 5.Jiang M., Zhang M., Tang D. G., Clemo H. F., Liu J., Holwitt D., Kasirajan V., Pond A. L., Wettwer E., Tseng G. N. (2004) KCNE2 protein is expressed in ventricles of different species, and changes in its expression contribute to electrical remodeling in diseased hearts. Circulation 109, 1783–1788 [DOI] [PubMed] [Google Scholar]
- 6.Kundu P., Ciobotaru A., Foroughi S., Toro L., Stefani E., Eghbali M. (2008) Hormonal regulation of cardiac KCNE2 gene expression. Mol. Cell. Endocrinol. 292, 50–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anantharam A., Abbott GW. (2005) Does hERG coassemble with a beta subunit? Evidence for roles of MinK and MiRP1. Novartis Found Symp. 266:100-12; discussion 112-7, 155-8 [Google Scholar]
- 8.Lundquist A. L., Manderfield L. J., Vanoye C. G., Rogers C. S., Donahue B. S., Chang P. A., Drinkwater D. C., Murray K. T., George A. L., Jr (2005) Expression of multiple KCNE genes in human heart may enable variable modulation of I(Ks). J. Mol. Cell. Cardiol. 38, 277–287 [DOI] [PubMed] [Google Scholar]
- 9.Gordon E., Panaghie G., Deng L., Bee K. J., Roepke T. K., Krogh-Madsen T., Christini D. J., Ostrer H., Basson C. T., Chung W., Abbott G. W. (2008) A KCNE2 mutation in a patient with cardiac arrhythmia induced by auditory stimuli and serum electrolyte imbalance. Cardiovasc. Res. 77, 98–106 [DOI] [PubMed] [Google Scholar]
- 10.Roepke T. K., Kontogeorgis A., Ovanez C., Xu X., Young J. B., Purtell K., Goldstein P. A., Christini D. J., Peters N. S., Akar F. G., Gutstein D. E., Lerner D. J., Abbott G. W. (2008) Targeted deletion of kcne2 impairs ventricular repolarization via disruption of I(K,slow1) and I(to,f). FASEB J. 22, 3648–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dedek K., Waldegger S. (2001) Colocalization of KCNQ1/KCNE channel subunits in the mouse gastrointestinal tract. Pflugers Arch. 442, 896–902 [DOI] [PubMed] [Google Scholar]
- 12.Grahammer F., Herling A. W., Lang H. J., Schmitt-Gräff A., Wittekindt O. H., Nitschke R., Bleich M., Barhanin J., Warth R. (2001) The cardiac K+ channel KCNQ1 is essential for gastric acid secretion. Gastroenterology 120, 1363–1371 [DOI] [PubMed] [Google Scholar]
- 13.Heitzmann D., Grahammer F., von Hahn T., Schmitt-Gräff A., Romeo E., Nitschke R., Gerlach U., Lang H. J., Verrey F., Barhanin J., Warth R. (2004) Heteromeric KCNE2/KCNQ1 potassium channels in the luminal membrane of gastric parietal cells. J. Physiol. 561, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee M. P., Ravenel J. D., Hu R. J., Lustig L. R., Tomaselli G., Berger R. D., Brandenburg S. A., Litzi T. J., Bunton T. E., Limb C., Francis H., Gorelikow M., Gu H., Washington K., Argani P., Goldenring J. R., Coffey R. J., Feinberg A. P. (2000) Targeted disruption of the Kvlqt1 gene causes deafness and gastric hyperplasia in mice. J. Clin. Invest. 106, 1447–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roepke T. K., Anantharam A., Kirchhoff P., Busque S. M., Young J. B., Geibel J. P., Lerner D. J., Abbott G. W. (2006) The KCNE2 potassium channel ancillary subunit is essential for gastric acid secretion. J. Biol. Chem. 281, 23740–23747 [DOI] [PubMed] [Google Scholar]
- 16.Roepke T. K., Purtell K., King E. C., La Perle K. M., Lerner D. J., Abbott G. W. (2010) Targeted deletion of Kcne2 causes gastritis cystica profunda and gastric neoplasia. PLoS One 5, e11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Q., Ma J., Zhou Q., Li J., Tang Y., Liu Y., Yang Y., Xiao J., Peng L., Li P., Liang D., Zhang H., Chen Y. H. (2010) KCNQ1 loss-of-function mutation impairs gastric acid secretion in mice. Mol. Biol. Rep. 37, 1329–1333 [DOI] [PubMed] [Google Scholar]
- 18.Roepke T. K., King E. C., Reyna-Neyra A., Paroder M., Purtell K., Koba W., Fine E., Lerner D. J., Carrasco N., Abbott G. W. (2009) Kcne2 deletion uncovers its crucial role in thyroid hormone biosynthesis. Nat. Med. 15, 1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abbott G. W., Tai K. K., Neverisky D. L., Hansler A., Hu Z., Roepke T. K., Lerner D. J., Chen Q., Liu L., Zupan B., Toth M., Haynes R., Huang X., Demirbas D., Buccafusca R., Gross S. S., Kanda V. A., Berry G. T. (2014) KCNQ1, KCNE2, and Na+-coupled solute transporters form reciprocally regulating complexes that affect neuronal excitability. Sci. Signal. 7, ra22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roepke T. K., Kanda V. A., Purtell K., King E. C., Lerner D. J., Abbott G. W. (2011) KCNE2 forms potassium channels with KCNA3 and KCNQ1 in the choroid plexus epithelium. FASEB J. 25, 4264–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boini K. M., Graf D., Hennige A. M., Koka S., Kempe D. S., Wang K., Ackermann T. F., Föller M., Vallon V., Pfeifer K., Schleicher E., Ullrich S., Häring H. U., Häussinger D., Lang F. (2009) Enhanced insulin sensitivity of gene-targeted mice lacking functional KCNQ1. Am. J. Physiol. Regul. Integr. Comp. Physiol. 296, R1695–R1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S. M., Baik J., Nguyen D., Nguyen V., Liu S., Hu Z., Abbott G. W. (2017) Kcne2 deletion impairs insulin secretion and causes type 2 diabetes mellitus. FASEB J. 31, 2674–2685 [DOI] [PubMed] [Google Scholar]
- 23.Cowley E. A., Linsdell P. (2002) Oxidant stress stimulates anion secretion from the human airway epithelial cell line Calu-3: implications for cystic fibrosis lung disease. J. Physiol. 543, 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowley E. A., Linsdell P. (2002) Characterization of basolateral K+ channels underlying anion secretion in the human airway cell line Calu-3. J. Physiol. 538, 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cuthbert A. W., MacVinish L. J. (2003) Mechanisms of anion secretion in Calu-3 human airway epithelial cells by 7,8-benzoquinoline. Br. J. Pharmacol. 140, 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moser S. L., Harron S. A., Crack J., Fawcett J. P., Cowley E. A. (2008) Multiple KCNQ potassium channel subtypes mediate basal anion secretion from the human airway epithelial cell line Calu-3. J. Membr. Biol. 221, 153–163 [DOI] [PubMed] [Google Scholar]
- 27.Tinel N., Diochot S., Borsotto M., Lazdunski M., Barhanin J. (2000) KCNE2 confers background current characteristics to the cardiac KCNQ1 potassium channel. EMBO J. 19, 6326–6330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heitzmann D., Koren V., Wagner M., Sterner C., Reichold M., Tegtmeier I., Volk T., Warth R. (2007) KCNE beta subunits determine pH sensitivity of KCNQ1 potassium channels. Cell. Physiol. Biochem. 19, 21–32 [DOI] [PubMed] [Google Scholar]
- 29.Sabater-Lleal M., Mälarstig A., Folkersen L., Soler Artigas M., Baldassarre D., Kavousi M., Almgren P., Veglia F., Brusselle G., Hofman A., Engström G., Franco O. H., Melander O., Paulsson-Berne G., Watkins H., Eriksson P., Humphries S. E., Tremoli E., de Faire U., Tobin M. D., Hamsten A. (2014) Common genetic determinants of lung function, subclinical atherosclerosis and risk of coronary artery disease. PLoS One 9, e104082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soler Artigas M., Loth D. W., Wain L. V., Gharib S. A., Obeidat M., Tang W., Zhai G., Zhao J. H., Smith A. V., Huffman J. E., Albrecht E., Jackson C. M., Evans D. M., Cadby G., Fornage M., Manichaikul A., Lopez L. M., Johnson T., Aldrich M. C., Aspelund T., Barroso I., Campbell H., Cassano P. A., Couper D. J., Eiriksdottir G., Franceschini N., Garcia M., Gieger C., Gislason G. K., Grkovic I., Hammond C. J., Hancock D. B., Harris T. B., Ramasamy A., Heckbert S. R., Heliovaara M., Homuth G., Hysi P. G., James A. L., Jankovic S., Joubert B. R., Karrasch S., Klopp N., Koch B., Kritchevsky S. B., Launer L. J., Liu Y., Loehr L. R., Lohman K., Loos R. J., Lumley T., Al Balushi K. A., Ang W. Q., Barr R. G., Beilby J., Blakey J. D., Boban M., Boraska V., Brisman J., Britton J. R., Brusselle G. G., Cooper C., Curjuric I., Dahgam S., Deary I. J., Ebrahim S., Eijgelsheim M., Francks C., Gaysina D., Granell R., Gu X., Hankinson J. L., Hardy R., Harris S. E., Henderson J., Henry A., Hingorani A. D., Hofman A., Holt P. G., Hui J., Hunter M. L., Imboden M., , Jameson K. A., Kerr S. M., Kolcic I., Kronenberg F., Liu J. Z., Marchini J., McKeever T., Morris A. D., Olin A. C., Porteous D. J., Postma D. S., Rich S. S., Ring S. M., Rivadeneira F., Rochat T., Sayer A. A., Sayers I., Sly P. D., Smith G. D., Sood A., Starr J. M., Uitterlinden A. G., Vonk J. M., Wannamethee S. G., Whincup P. H., Wijmenga C., Williams O. D., Wong A., Mangino M., Marciante K. D., McArdle W. L., Meibohm B., Morrison A. C., North K. E., Omenaas E., Palmer L. J., Pietilainen K. H., Pin I., Pola Sbreve Ek O., Pouta A., Psaty B. M., Hartikainen A. L., Rantanen T., Ripatti S., Rotter J. I., Rudan I., Rudnicka A. R., Schulz H., Shin S. Y., Spector T. D., Surakka I., Vitart V., Volzke H., Wareham N. J., Warrington N. M., Wichmann H. E., Wild S. H., Wilk J. B., Wjst M., Wright A. F., Zgaga L., Zemunik T., Pennell C. E., Nyberg F., Kuh D., Holloway J. W., Boezen H. M., Lawlor D. A., Morris R. W., Probst-Hensch N., Kaprio J., Wilson J. F., Hayward C., Kahonen M., Heinrich J., Musk A. W., Jarvis D. L., Glaser S., Jarvelin M. R., Ch Stricker B. H., Elliott P., O'Connor G. T., Strachan D. P., London S. J., Hall I. P., Gudnason V., Tobin M. D. (2011) Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat. Genet. 43, 1082–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kredel S., Oswald F., Nienhaus K., Deuschle K., Röcker C., Wolff M., Heilker R., Nienhaus G. U., Wiedenmann J. (2009) mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures. PLoS One 4, e4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matute-Bello G., Downey G., Moore B. B., Groshong S. D., Matthay M. A., Slutsky A. S., Kuebler W. M.; Acute Lung Injury in Animals Study Group (2011) An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. Am. J. Respir. Cell Mol. Biol. 44, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Z., Kant R., Anand M., King E. C., Krogh-Madsen T., Christini D. J., Abbott G. W. (2014) Kcne2 deletion creates a multisystem syndrome predisposing to sudden cardiac death. Circ. Cardiovasc. Genet. 7, 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelley J. (1990) Cytokines of the lung. Am. Rev. Respir. Dis. 141, 765–788 [DOI] [PubMed] [Google Scholar]
- 35.Hausenloy D. J., Tsang A., Yellon D. M. (2005) The reperfusion injury salvage kinase pathway: a common target for both ischemic preconditioning and postconditioning. Trends Cardiovasc. Med. 15, 69–75 [DOI] [PubMed] [Google Scholar]
- 36.Lecour S. (2009) Activation of the protective Survivor Activating Factor Enhancement (SAFE) pathway against reperfusion injury: does it go beyond the RISK pathway? J. Mol. Cell. Cardiol. 47, 32–40 [DOI] [PubMed] [Google Scholar]
- 37.McCrossan Z. A., Roepke T. K., Lewis A., Panaghie G., Abbott G. W. (2009) Regulation of the Kv2.1 potassium channel by MinK and MiRP1. J. Membr. Biol. 228, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brueggemann L. I., Haick J. M., Neuburg S., Tate S., Randhawa D., Cribbs L. L., Byron K. L. (2014) KCNQ (Kv7) potassium channel activators as bronchodilators: combination with a β2-adrenergic agonist enhances relaxation of rat airways. Am. J. Physiol. Lung Cell. Mol. Physiol. 306, L476–L486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demolombe S., Franco D., de Boer P., Kuperschmidt S., Roden D., Pereon Y., Jarry A., Moorman A. F., Escande D. (2001) Differential expression of KvLQT1 and its regulator IsK in mouse epithelia. Am. J. Physiol. Cell Physiol. 280, C359–C372 [DOI] [PubMed] [Google Scholar]
- 40.Grahammer F., Warth R., Barhanin J., Bleich M., Hug M. J. (2001) The small conductance K+ channel, KCNQ1: expression, function, and subunit composition in murine trachea. J. Biol. Chem. 276, 42268–42275 [DOI] [PubMed] [Google Scholar]
- 41.Abbott G. W. (2014) Biology of the KCNQ1 potassium channel. New J. Sci. 2014, 1–26 [Google Scholar]
- 42.Abbott G. W. (2016) KCNE1 and KCNE3: the yin and yang of voltage-gated K(+) channel regulation. Gene 576, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abbott G. W. (2016) KCNE4 and KCNE5: K(+) channel regulation and cardiac arrhythmogenesis. Gene 593, 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barhanin J., Lesage F., Guillemare E., Fink M., Lazdunski M., Romey G. (1996) K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature 384, 78–80 [DOI] [PubMed] [Google Scholar]
- 45.Sanguinetti M. C., Curran M. E., Zou A., Shen J., Spector P. S., Atkinson D. L., Keating M. T. (1996) Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature 384, 80–83 [DOI] [PubMed] [Google Scholar]
- 46.Choi E., Abbott G. W. (2010) A shared mechanism for lipid- and beta-subunit-coordinated stabilization of the activated K+ channel voltage sensor. FASEB J. 24, 1518–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panaghie G., Abbott G. W. (2007) The role of S4 charges in voltage-dependent and voltage-independent KCNQ1 potassium channel complexes. J. Gen. Physiol. 129, 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schroeder B. C., Waldegger S., Fehr S., Bleich M., Warth R., Greger R., Jentsch T. J. (2000) A constitutively open potassium channel formed by KCNQ1 and KCNE3. Nature 403, 196–199 [DOI] [PubMed] [Google Scholar]
- 49.Nakajo K., Kubo Y. (2007) KCNE1 and KCNE3 stabilize and/or slow voltage sensing S4 segment of KCNQ1 channel. J. Gen. Physiol. 130, 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kroncke B. M., Van Horn W. D., Smith J., Kang C., Welch R. C., Song Y., Nannemann D. P., Taylor K. C., Sisco N. J., George A. L., Jr., Meiler J., Vanoye C. G., Sanders C. R. (2016) Structural basis for KCNE3 modulation of potassium recycling in epithelia. Sci. Adv. 2, e1501228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alzamora R., O’Mahony F., Bustos V., Rapetti-Mauss R., Urbach V., Cid L. P., Sepúlveda F. V., Harvey B. J. (2011) Sexual dimorphism and oestrogen regulation of KCNE3 expression modulates the functional properties of KCNQ1 K+ channels. J. Physiol. 589, 5091–5107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rapetti-Mauss R., O’Mahony F., Sepulveda F. V., Urbach V., Harvey B. J. (2013) Oestrogen promotes KCNQ1 potassium channel endocytosis and postendocytic trafficking in colonic epithelium. J. Physiol. 591, 2813–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee S. M., Nguyen D., Anand M., Kant R., Köhncke C., Lisewski U., Roepke T. K., Hu Z., Abbott G. W. (2016) Kcne2 deletion causes early-onset nonalcoholic fatty liver disease via iron deficiency anemia. Sci. Rep. 6, 23118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee S. M., Nguyen D., Hu Z., Abbott G. W. (2015) Kcne2 deletion promotes atherosclerosis and diet-dependent sudden death. J. Mol. Cell. Cardiol. 87, 148–151 [DOI] [PubMed] [Google Scholar]
- 55.Roepke T. K., King E. C., Purtell K., Kanda V. A., Lerner D. J., Abbott G. W. (2011) Genetic dissection reveals unexpected influence of beta subunits on KCNQ1 K+ channel polarized trafficking in vivo. FASEB J. 25, 727–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu Z., Crump S. M., Zhang P., Abbott G. W. (2016) Kcne2 deletion attenuates acute post-ischaemia/reperfusion myocardial infarction. Cardiovasc. Res. 110, 227–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Myocardial Infarction Genetics Consortium ; Kathiresan S., Voight B. F., Purcell S., Musunuru K., Ardissino D., Mannucci P. M., Anand S., Engert J. C., Samani N. J., Schunkert H., Erdmann J., Reilly M. P., Rader D. J., Morgan T., Spertus J. A., Stoll M., Girelli D., McKeown P. P., Patterson C. C., Siscovick D. S., O’Donnell C. J., Elosua R., Peltonen L., Salomaa V., Schwartz S. M., Melander O., Altshuler D., Ardissino D., Merlini P. A., Berzuini C., Bernardinelli L., Peyvandi F., Tubaro M., Celli P., Ferrario M., Fetiveau R., Marziliano N., Casari G., Galli M., Ribichini F., Rossi M., Bernardi F., Zonzin P., Piazza A., Mannucci P. M., Schwartz S. M., Siscovick D. S., Yee J., Friedlander Y., Elosua R., Marrugat J., Lucas G., Subirana I., Sala J., Ramos R., Kathiresan S., Meigs J. B., Williams G., Nathan D. M., MacRae C. A., O’Donnell C. J., Salomaa V., Havulinna A. S., Peltonen L., Melander O., Berglund G., Voight B. F., Kathiresan S., Hirschhorn J. N., Asselta R., Duga S., Spreafico M., Musunuru K., Daly M. J., Purcell S., Voight B. F., Purcell S., Nemesh J., Korn J. M., McCarroll S. A., Schwartz S. M., Yee J., Kathiresan S., Lucas G., Subirana I., Elosua R., Surti A., Guiducci C., Gianniny L., Mirel D., Parkin M., Burtt N., Gabriel S. B., Samani N. J., Thompson J. R., Braund P. S., Wright B. J., Balmforth A. J., Ball S. G., Hall A., Wellcome Trust Case Control Consortium ; Schunkert H., Erdmann J., Linsel-Nitschke P., Lieb W., Ziegler A., König I., Hengstenberg C., Fischer M., Stark K., Grosshennig A., Preuss M., Wichmann H. E., Schreiber S., Schunkert H., Samani N. J., Erdmann J., Ouwehand W., Hengstenberg C., Deloukas P., Scholz M., Cambien F., Reilly M. P., Li M., Chen Z., Wilensky R., Matthai W., Qasim A., Hakonarson H. H., Devaney J., Burnett M. S., Pichard A. D., Kent K. M., Satler L., Lindsay J. M., Waksman R., Knouff C. W., Waterworth D. M., Walker M. C., Mooser V., Epstein S. E., Rader D. J., Scheffold T., Berger K., Stoll M., Huge A., Girelli D., Martinelli N., Olivieri O., Corrocher R., Morgan T., Spertus J. A., McKeown P., Patterson C. C., Schunkert H., Erdmann E., Linsel-Nitschke P., Lieb W., Ziegler A., König I. R., Hengstenberg C., Fischer M., Stark K., Grosshennig A., Preuss M., Wichmann H. E., Schreiber S., Hólm H., Thorleifsson G., Thorsteinsdottir U., Stefansson K., Engert J. C., Do R., Xie C., Anand S., Kathiresan S., Ardissino D., Mannucci P. M., Siscovick D., O’Donnell C. J., Samani N. J., Melander O., Elosua R., Peltonen L., Salomaa V., Schwartz S. M., Altshuler D. (2009) Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 41, 334–341; erratum: 762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wakil S. M., Ram R., Muiya N. P., Mehta M., Andres E., Mazhar N., Baz B., Hagos S., Alshahid M., Meyer B. F., Morahan G., Dzimiri N. (2016) A genome-wide association study reveals susceptibility loci for myocardial infarction/coronary artery disease in Saudi Arabs. Atherosclerosis 245, 62–70 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.