Salmonella enterica serovar Typhimurium (S. Typhimurium) induces inflammatory changes in the ceca of streptomycin-pretreated mice. In this mouse model of colitis, the type III secretion system 1 (T3SS-1) has been shown to induce rapid inflammatory change in the cecum at early points, 10 to 24 h after infection. Five proteins, SipA, SopA, SopB, SopD, and SopE2, have been identified as effectors involved in eliciting intestinal inflammation within this time range.
KEYWORDS: Salmonella, cytotoxicity, inflammation, type III effectors
ABSTRACT
Salmonella enterica serovar Typhimurium (S. Typhimurium) induces inflammatory changes in the ceca of streptomycin-pretreated mice. In this mouse model of colitis, the type III secretion system 1 (T3SS-1) has been shown to induce rapid inflammatory change in the cecum at early points, 10 to 24 h after infection. Five proteins, SipA, SopA, SopB, SopD, and SopE2, have been identified as effectors involved in eliciting intestinal inflammation within this time range. In contrast, a T3SS-1-deficient strain was shown to exhibit inflammatory changes in the cecum at 72 to 120 h postinfection. However, the effectors eliciting T3SS-1-independent inflammation remain to be clarified. In this study, we focused on two T3SS-2 phenotypes, macrophage proliferation and cytotoxicity, to identify the T3SS-2 effectors involved in T3SS-1-independent inflammation. We identified a mutant strain that could not induce cytotoxicity in a macrophage-like cell line and that reduced intestinal inflammation in streptomycin-pretreated mice. We also identified five T3SS-2 effectors, SifA, SpvB, SseF, SseJ, and SteA, associated with T3SS-1-independent macrophage cytotoxicity. We then constructed a strain lacking T3SS-1 and all the five T3SS-2 effectors, termed T1S5. The S. Typhimurium T1S5 strain significantly reduced cytotoxicity in macrophages in the same manner as a mutant invA spiB strain (T1T2). Finally, the T1S5 strain elicited no inflammatory changes in the ceca of streptomycin-pretreated mice. We conclude that these five T3SS-2 effectors contribute to T3SS-1-independent inflammation.
INTRODUCTION
Salmonella enterica serovar Typhimurium (S. Typhimurium) is an important foodborne pathogen causing gastroenteritis. Two distinct type III secretion systems (T3SSs), T3SS-1 and T3SS-2, encoded by Salmonella pathogenicity islands 1 and 2, respectively, play central roles in Salmonella pathogenesis. It has been reported that T3SS-1 and T3SS-2 are pivotal in inducing intestinal inflammation in several animal models (1–3). Five T3SS-1 effectors, SipA, SopA, SopB, SopD, and SopE2, contribute to intestinal inflammation during the early stage of infection (4, 5). In a streptomycin-pretreated mouse model of colitis, a T3SS-1-deficient (invA::kan or ΔinvG) mutant is unable to elicit intestinal inflammation in the early stages of infection (within the first 48 h after infection), but severe inflammation is observed in the cecum in the late stages of infection (72 to 120 h after infection) (6, 7). Since the induction of T3SS-1-independent inflammation cannot be elicited in mice infected with a mutant deficient in both T3SS-1 and T3SS-2 (invA::kan ΔssaR or ΔinvG sseG::aphT) (6, 7), the T3SS-2 effectors seem to be important for intestinal inflammation during the late stage of infection. However, the mechanisms by which the T3SS-1-independent inflammation is induced are poorly understood, and it is still unknown which effector proteins are involved in T3SS-1-independent inflammatory changes in the streptomycin-pretreated mouse model.
T3SS-2 is associated with the survival and replication of S. Typhimurium in the host and is necessary for systemic infection in a mouse typhoid model (8–10). Approximately 30 T3SS-2 effectors have been identified so far, and many of these effectors are associated with maturation of Salmonella-containing vacuoles (SCVs) and required for survival of S. Typhimurium in SCVs (11). SifA has been studied most extensively and was originally identified as the T3SS-2 effector required for formation of Salmonella-induced filaments (SIFs) in S. Typhimurium-infected epithelial cells (12–14). SifA was also shown to contribute to proliferation in macrophages and S. Typhimurium virulence in a mouse typhoid model (12–14).
Some T3SS-2-deficient (ssaT::mTn5, ssaV::mTn5, and ssaJ::mTn5) mutants lead to reduced cytotoxicity in RAW 264.7 (RAW) cells, a mouse macrophage-like cell line (15). S. Typhimurium has also been shown to induce T3SS-1-independent cytotoxicity in RAW cells and another murine macrophage cell line, J774.A1, from 12 h after infection onwards (16). Only two proteins, SpvB and SseL, have been identified as T3SS-2 effectors that induce cytotoxicity in macrophages. SpvB ADP-ribosylates actin, which is required for the macrophage cytotoxicity (17) and contributes to virulence in mice (18). SseL also induces cytotoxicity in macrophages and plays a role in Salmonella virulence through its deubiquitinase activity (19). Although some targets of SseL have been observed (20, 21), the mechanisms by which SpvB and SseL contribute to macrophage cytotoxicity are not understood.
In this study, we focused on two T3SS-2 phenotypes for Salmonella pathogenesis, bacterial proliferation and cytotoxicity in S. Typhimurium-infected macrophages, and tried to identify effectors involved in T3SS-1-independent inflammation. Our results show that an S. Typhimurium mutant that is unable to cause cytotoxicity phenocopies a mutant lacking T3SS-1 and T3SS-2 in the streptomycin-pretreated mouse model. In addition, we identified T3SS-2 effectors that were required for induction of T3SS-1-independent inflammation in streptomycin-pretreated mice.
RESULTS
An S. Typhimurium macrophage proliferation-deficient mutant induces T3SS-1-independent inflammation in streptomycin-pretreated mice.
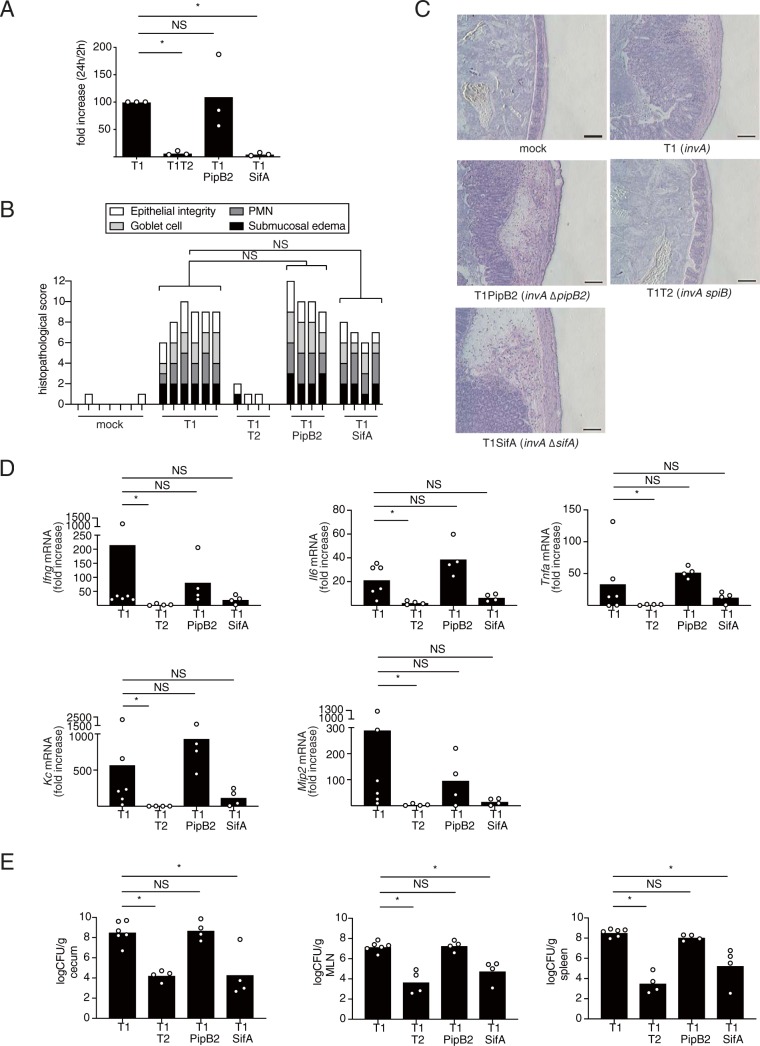
The T3SS-2 system contributes to S. Typhimurium-induced T3SS-1-independent colitis in streptomycin-pretreated mice (6, 7). We therefore hypothesized that T3SS-2 effectors play an important role in T3SS-1-independent inflammation. It is well known that T3SS-2 confers a survival advantage to S. Typhimurium within macrophages (15). To test whether proliferation of S. Typhimurium within macrophages is necessary for the induction of Salmonella T3SS-1-independent colitis, streptomycin-pretreated mice were infected with a macrophage proliferation-proficient ΔpipB2 mutant (22) or a macrophage proliferation-deficient ΔsifA mutant (23). Intracellular survival of a mutant lacking a functional T3SS-1 and pipB2 (invA ΔpipB2 mutant, T1PipB2) was similar to that of the T3SS-1-deficient invA mutant (T1) (Fig. 1A). In contrast, intercellular survival of a mutant lacking a functional T3SS-1 and sifA (invA ΔsifA mutant, T1SifA) in RAW cells was reduced compared to that of T1 (Fig. 1A). To determine whether a pipB2 or a sifA mutation reduces T3SS-1-independent inflammation, streptomycin-pretreated mice were infected with the T1PipB2 or the T1SifA strain. Histopathological analysis showed that inactivation of the pipB2 or the sifA gene had no effect on the ability of the S. Typhimurium T1 strain to induce inflammation in the cecum (Fig. 1B and C). Similarly, no statistically significant differences in mRNA expressions of the proinflammatory cytokine genes Ifng, Il6, and Tnfa and the chemokine genes Kc and Mip2 were detected in the ceca of mice infected with the T1PipB2 or T1SifA strain compared to those of mice infected with the T1 strain (Fig. 1D). The number of bacteria recovered from the ceca, mesenteric lymph nodes, and spleens of mice infected with T1PipB2 were similar to those in mice infected with T1, but those in mice infected with T1SifA were decreased (Fig. 1E). These results suggested that survival or growth of bacteria in macrophages is not required for the induction of T3SS-1-independent inflammation.
FIG 1.
The proliferation of S. Typhimurium in macrophages is not related to intestinal inflammation in streptomycin-pretreated mice. (A) Proliferation rates of the indicated S. Typhimurium mutant strains in RAW cells. The proliferation rate of the S. Typhimurium invA (T1) strain in RAW cells was taken as 100. (B and C) Histopathology of the murine cecum 5 days after infection with the indicated S. Typhimurium mutant strains. Representative images of hematoxylin and eosin-stained cecal sections are shown. Scale bars, 100 μm (B). Histopathology scores were determined by blinded examination of cecal sections. Each bar represents an individual mouse (C). (D) Transcript levels of Ifng, Il6, Tnfa, Kc, and Mip2 in the ceca of mice infected with the indicated S. Typhimurium strains at 5 days after infection measured by qPCR. (E) Numbers of bacteria recovered from the ceca, the mesenteric lymph nodes, and the spleens of mice 5 days after infection. A sample t test (A) and the two-tailed Mann-Whitney U test (B, D, and E) were performed for statistical analysis. Individual data are shown as a scatter plot, and bars indicate the means. Asterisks indicate differences that were statistically significant (P < 0.05). NS, not significant.
A macrophage cytotoxicity-deficient mutant of S. Typhimurium reduces T3SS-1-independent inflammation.
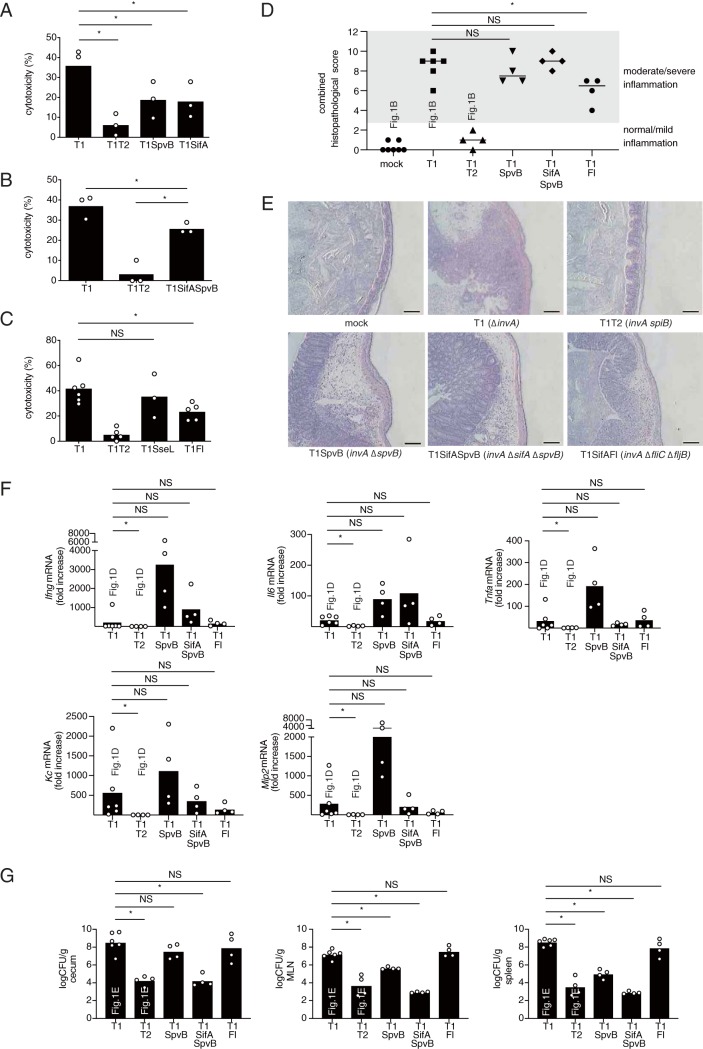
S. Typhimurium can induce cytotoxicity in macrophages in a manner dependent on T3SS-2 (16). Next, we investigated whether cytotoxicity in S. Typhimurium-infected macrophages is required for T3SS-1-independent inflammation. For these experiments, we selected a mutant lacking a functional T3SS-1 and spvB (invA ΔspvB mutant, T1SpvB) as a positive control and T1SifA as a negative control. The cytotoxicity of T1SpvB for RAW cells was lower than that of T1 (Fig. 2A). Unexpectedly, however, the cytotoxicity caused by T1SifA was similar to that triggered by T1SpvB and significantly lower than that of the parental strain, T1 (Fig. 2A). To determine whether an spvB or sifA mutation reduces T3SS-1-independent inflammation, the streptomycin-pretreated mice were infected with the T1SpvB or the T1SifA strain. No statistically significant differences in histopathological score or in proinflammatory cytokine or chemokine expression were detected in the ceca of mice infected with the T1SpvB or T1SifA strain compared to mice infected with the T1 strain (Fig. 1B, 1C, 1D, 2D, 2E, and 2F). However, we noticed that the cytotoxicity of T1SpvB or T1SifA in RAW cells was still higher than that of the T1T2 strain (Fig. 2A). Therefore, we hypothesized that T3SS-1-independent inflammation required macrophage cytotoxicity. An invA ΔsifA ΔspvB mutant (T1SifASpvB) induced more cytotoxicity in RAW cells and inflammation in the ceca of the streptomycin-pretreated mice than T1T2 (Fig. 2B, D, E, and F), suggesting that additional T3SS-2 effector proteins might contribute to T3SS-1-independent inflammation. Thus, we tested the contribution of other T3SS-2 effectors to cytotoxicity in macrophages and cytokine expression in the murine cecum.
FIG 2.
The S. Typhimurium strains, T1SpvB, T1SifA, and T1Fl, that reduce the cytotoxicity of macrophages elicit T3SS-1-independent inflammation in mice. (A to C) LDH release from RAW cells infected with the indicated S. Typhimurium mutant strains at 20 h after infection. (D and E) Histopathology of the murine cecum 5 days after infection with the indicated S. Typhimurium mutant strains. The histopathology score was determined by the blinded examination of cecal sections. Each plot represents an individual mouse (D). Representative images of hematoxylin and eosin-stained cecal sections are shown. Scale bars, 100 μm (E). (F) Transcript levels of Ifng, Il6, Tnfa, Kc, and Mip2 in the ceca of mice infected with the indicated S. Typhimurium strains at 5 days after infection, as measured by qPCR. (G) Numbers of bacteria recovered from the ceca, mesenteric lymph nodes and spleen of mice 5 days after infection. The combined histopathological score, the expression levels of genes, and bacterial numbers in the tissue of mice infected with invA (T1) or invA spiB (T1T2) were determined as shown in Fig. 1B, D, and E, respectively. The two-tailed Student t test (A to C) and the Mann-Whitney U test (E to G) were performed for statistical analysis. Individual data are shown as a scatter plot, and bars indicate the means. Asterisks indicate differences that were statistically significant (P < 0.05). NS, not significant.
We first investigated the T3SS-2 effector SseL, which has been shown to be required for the T3SS-1-independent cytotoxicity of macrophages (19). We also included the flagellar proteins FliC and FljB, because flagella induce pyroptotic cell death of macrophages via caspase-1-dependent processing (24), although it is uncertain whether these proteins are translocated through T3SS-2. An invA ΔsseL mutant (T1SseL) caused cytotoxicity at levels similar to those of T1, whereas an invA fliC ΔfljB mutant (T1Fl) triggered a reduction in cytotoxicity compared to that of the T1 strain (Fig. 2C). However, although the T1Fl strain significantly reduced the inflammation, compared to T1, T1Fl induced moderate inflammation in the ceca of the streptomycin-pretreated mice (Fig. 2C to E). The mRNA levels of proinflammatory cytokines and chemokines in the ceca of mice infected with T1Fl were similar to those infected with T1 (Fig. 2F). The numbers of bacteria recovered from the ceca of mice infected with T1SpvB or T1Fl were similar to those recovered from mice infected with T1, but the numbers in mice infected with T1T2 or T1SifASpvB were decreased (Fig. 2G). Consistent with previous reports (12, 14, 18, 25), mutation of spvB and/or sifA resulted in decreased bacterial numbers at the systemic sites, i.e., the mesenteric lymph nodes and spleen (Fig. 2G).
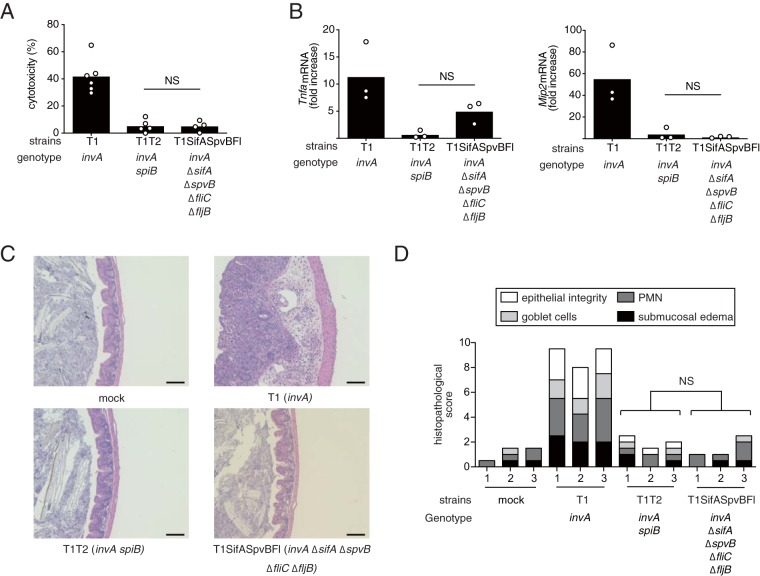
Next, we constructed an invA ΔsifA ΔspvB fliC ΔfljB mutant (T1SifASpvBFl) and performed a lactate dehydrogenase (LDH) release assay. Interestingly, the level of cytotoxicity of RAW cells infected with the S. Typhimurium strain having mutations of the five genes was markedly lower than that of cells infected with T1 and was the same as that of cells infected with the strains lacking T3SS-1 and T3SS-2 (T1T2) (Fig. 3A). Notably, T1SifASpvBFl elicited significantly lower levels of inflammatory genes in the ceca compared to T1 (Fig. 3B). Furthermore, the analysis of histopathological changes indicated that infection with T1 caused marked inflammation, but these responses were blunted during infection with T1SifASpvBFl or T1T2 (Fig. 3C and D). Collectively, these data suggested that macrophage cytotoxicity is required for T3SS-1-independent inflammation.
FIG 3.
Effect of different combinations of the sifA, spvB, fliC, and fljB mutations on T3SS-1-independent inflammation in mice. (A) LDH release from RAW cells infected with the indicated S. Typhimurium mutant strains at 20 h after infection. (B) The levels of Tnfa and Mip2 in the ceca of mice infected with the indicated S. Typhimurium strains at 5 days after infection. (C and D) Histopathology of the murine cecum 5 days after infection with the indicated S. Typhimurium mutant strains. Representative images of hematoxylin and eosin-stained cecal sections are shown. Scale bars, 100 μm (C). The histopathology score was determined by blinded examination of the cecal sections. Each bar represents an individual mouse (D). A two-tailed Student t test (A) and the Mann-Whitney U test (B and D) were performed for statistical analysis. Individual data are shown as a scatter plot, and bars indicate the means. Asterisks indicate differences that were statistically significant (P < 0.05). NS, not significant.
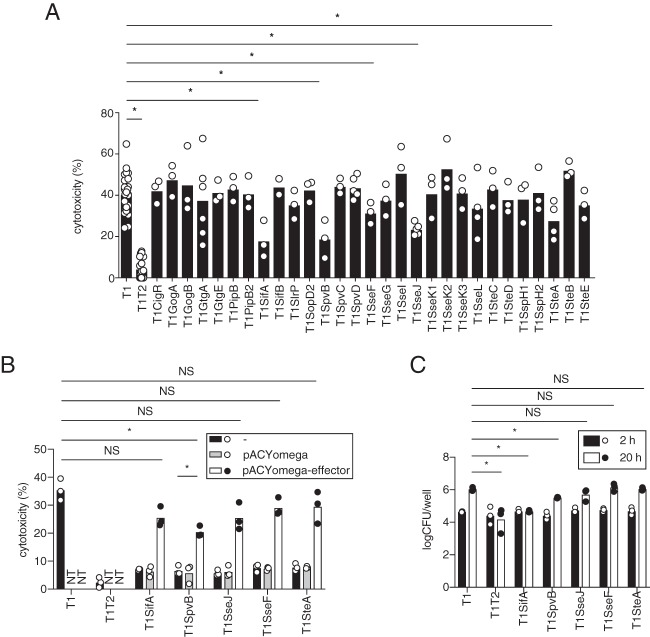
The S. Typhimurium T3SS-2 effectors SifA, SpvB, SseF, SseJ, and SteA contribute to T3SS-1-independent macrophage cytotoxicity.
As mentioned above, the invA ΔsifA ΔspvB fliC ΔfljB mutant (T1SifASpvBFl) induced less macrophage cytotoxicity and less inflammation in the ceca of the streptomycin-pretreated, S. Typhimurium-infected mice, in the same manner as the invA spiB mutant (T1T2). Of note, however, the invA spiB mutant has both a fliC gene and a fljB gene. These facts may indicate that T3SS-2 effectors other than SifA and SpvB also contribute to T3SS-1-independent macrophage cytotoxicity and inflammation. To identify these effectors, we introduced mutations in each of the 29 T3SS-2 effector genes into strain T1 and tested each resulting strain for macrophage cytotoxicity. Three mutant strains, the invA ΔsseF (T1SseF), invA ΔsseJ (T1SseJ), and invA ΔsteA (T1SteA) mutants, exhibited a reduction in cytotoxicity similar to those in the invA ΔsifA (T1SifA) and invA ΔspvB (T1SpvB) mutant strains (Fig. 2A and 4A). The expression of the effector from the plasmids was sufficient for the complementation of effector mutants; the single exception was SpvB, which showed partial complementation (Fig. 4B). In addition, to rule out an effect of bacterial proliferation in RAW cells, the numbers of bacteria in the cells infected with T1SifA, T1SpvB, T1SseF, T1SseJ, or T1SteA after 2 or 20 h were compared to those of the cells infected with T1. At 20 h after infection, the numbers of T1SifA or T1SpvB in RAW cells were reduced, but those of T1SseF, T1SseJ, or T1SteA were not (Fig. 4C). These results suggested that the growth of bacteria in macrophages is not necessarily correlated with the induction of T3SS-1-independent cytotoxicity.
FIG 4.
Screening of 29 T3SS-2 effectors for cytotoxicity of RAW cells. (A) LDH release from RAW cells infected with the indicated S. Typhimurium mutant strains at 20 h after infection. The cytotoxicities of RAW cells infected with the invA ΔsifA, invA ΔspvB, or invA ΔsseL strain were determined as shown in Fig. 1A, 2A, or 2C. Asterisks indicate that the cytotoxicities of RAW cells infected with the T3SS-1-deficient and T3SS-2 effector mutants (invA Δeffector) were significantly lower than those of cells infected with the T3SS-1-deficient strain (invA). (B) LDH release from RAW cells infected with the indicated S. Typhimurium mutant strains at 20 h after infection. NT, not tested. (C) Numbers of bacteria recovered from RAW cells infected with the indicated S. Typhimurium mutant strains at 2 and 20 h after infection. A two-tailed Student t test (A to C) was performed for statistical analysis. Individual data are shown as a scatter plot, and bars indicate the means. Asterisks indicate differences that were statistically significant (P < 0.05). NS, not significant.
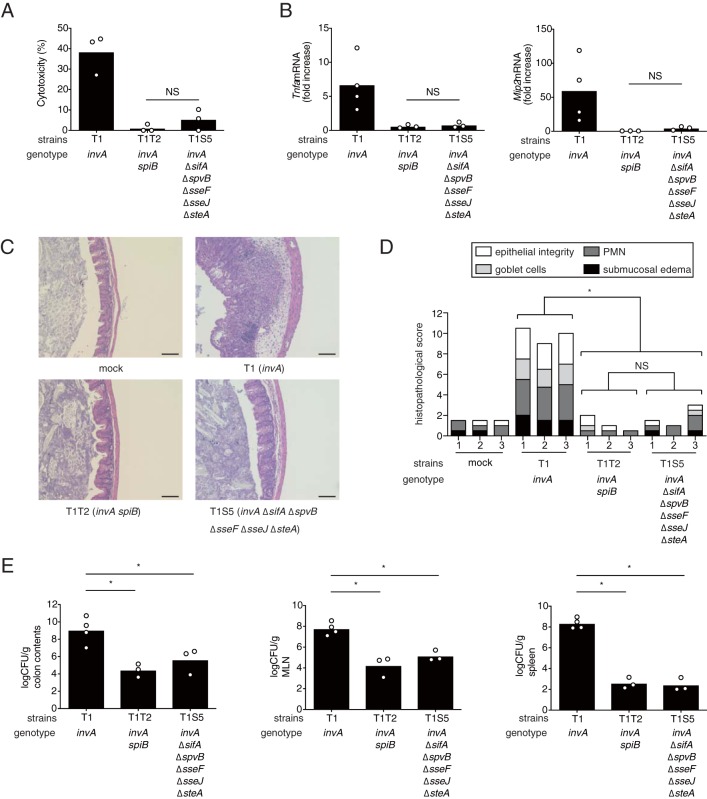
The S. Typhimurium invA ΔsifA ΔspvB ΔsseF ΔsseJ ΔsteA mutant does not elicit T3SS-1-independent cytotoxicity in macrophages or inflammation in mice.
Next, we examined whether the S. Typhimurium mutant with deletions of all five of the above-described genes impaired the cytotoxicity of macrophages. Intriguingly, we found that the cytotoxicity of RAW cells infected with the S. Typhimurium invA ΔsifA ΔspvB ΔsseJ ΔsseF ΔsteA mutant (T1S5) strain was dramatically reduced compared to that of the invA mutant (T1) and was similar to that of the invA spiB mutant (T1T2) (Fig. 5A). To confirm that the T1S5 strain is capable of translocating effectors other than these five into host cells, RAW cells were infected with S. Typhimurium T1, T1T2, or T1S5, each carrying a plasmid expressing the SseJ-CyaA protein. The levels of cyclic AMP (cAMP) in RAW cells infected with T1 or T1S5 were dramatically increased, but cAMP was not detected in RAW cells infected with T1T2 (see Fig. S1 in the supplemental material). In addition, we investigated whether these five deletions collectively attenuate intestinal inflammation elicited by an S. Typhimurium invA mutant. Compared to the invA mutant (T1), the invA ΔsifA ΔspvB ΔsseJ ΔsseF ΔsteA mutant (T1S5) elicited significantly lower levels of Tnfa and Mip2 in the ceca of the streptomycin-pretreated mice 5 days after infection (Fig. 5B). Moreover, the deletion of all five genes in the S. Typhimurium invA strain significantly reduced the inflammation observed in the cecum, as shown by histopathological analysis (Fig. 5C and D). Similarly, the bacterial numbers from the colon contents and tissues of mice infected with the T1T2 or the T1S5 strain were significantly lower than those of mice infected with the T1 strain (Fig. 5E).
FIG 5.
Effect of different combinations of the sifA, spvB, sseF, sseJ, and steA mutations on T3SS-1-independent inflammation in mice (A) LDH release from RAW cells infected with the indicated S. Typhimurium mutant strains at 20 h after infection. (B) Levels of Tnfa and Mip2 in the ceca of mice infected with the indicated S. Typhimurium strains at 5 days after infection. (C and D) Histopathology of the murine cecum 5 days after infection with the indicated S. Typhimurium mutant strains. Representative images of hematoxylin and eosin-stained cecal sections are shown. Scale bars, 100 μm. (C) Histopathological scoring of cecal sections. (D) Each bar represents an individual mouse. (E) Numbers of bacteria recovered from colon contents, mesenteric lymph nodes (MLN), and spleens of mice. A two-tailed Student t test (A) and Mann-Whitney U test (B, D, and E) were performed for statistical analysis. Individual data are shown as a scatter plot, and bars indicate the means (A, B, and E). Asterisks indicate differences that were statistically significant (P < 0.05). NS, not significant.
Collectively, these data suggested that SifA, SpvB, SseJ, SseF, and SteA cooperate to induce macrophage cytotoxicity and T3SS-1-independent inflammation.
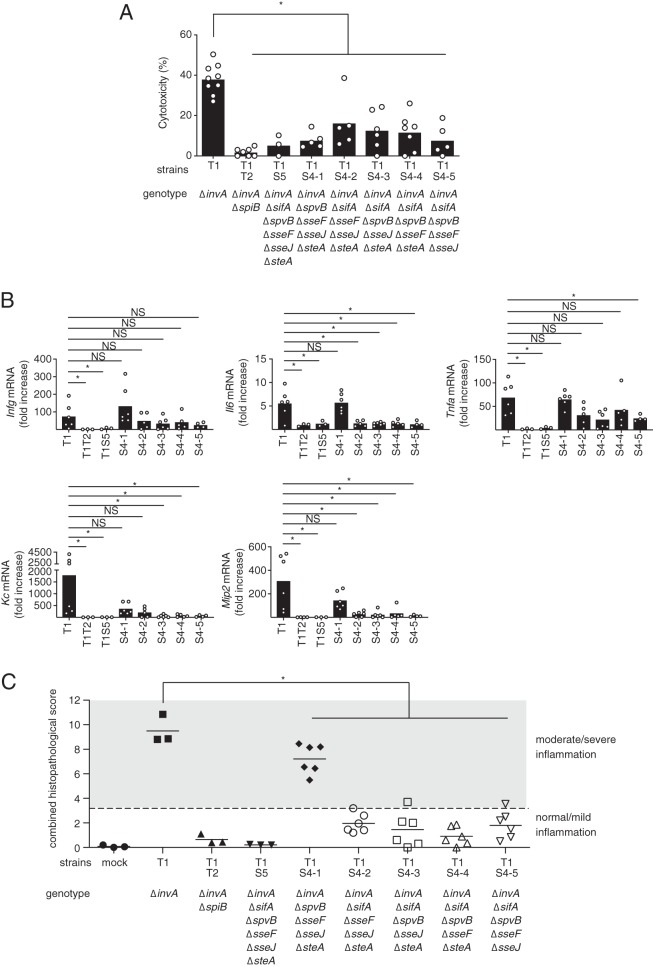
SifA is essential for the induction of intestinal inflammatory responses by an S. Typhimurium invA mutant.
The purpose of this study was to identify the T3SS-2 effectors required for T3SS-1-independent inflammation in mice. To this end, we constructed five S. Typhimurium invA mutant strains, each containing deletions for four of the five effectors, sifA, spvB, sseF, sseJ, and steA, and designated them strains T1S4-1 to T1S4-5. For each of these mutant strains, the cytotoxicities of the infected RAW cells were weaker than those of the invA strain-infected RAW cells, but stronger than those of the invA spiB (T1T2) strain-infected RAW cells (Fig. 6A). This result indicated that all five effectors are required for the induction of T3SS-1-independent cytotoxicity in RAW cells. However, the expression of proinflammatory cytokines or chemokines did not correlate with the cytotoxicity in RAW cells (Fig. 6B). Interestingly, the expressions of proinflammatory cytokines and chemokines, except for Ifng, were elicited in the ceca of streptomycin-pretreated mice infected with the invA ΔspvB ΔsseF ΔsseJ ΔsteA mutant (T1S4-1) just as in the ceca of the streptomycin-pretreated mice infected with an S. Typhimurium invA mutant, T1 (Fig. 6B). Similarly, analysis of the histopathological changes showed that the invA ΔspvB ΔsseF ΔsseJ ΔsteA strain (T1S4-1), which retained only the sifA gene, caused intestinal inflammation similar to that of an S. Typhimurium invA mutant, T1 (Fig. 6C and see Fig. S2A in the supplemental material). Histopathological studies also revealed that two strains, the invA (T1) and invA ΔspvB ΔsseF ΔsseJ ΔsteA (T1S4-1) mutants, elicited moderate or severe inflammation in the cecal mucosa (Fig. 6C and Fig. S2A). The numbers of bacteria from the colon contents and tissue in mice infected with each T1S4 mutant were lower than the respective bacterial numbers in the mice infected with the T1 strain (Fig. S2B). However, there was no correlation between the bacterial numbers and inflammation in the ceca of mice. Collectively, these results suggested that SifA is the most important of the five effectors for the induction of T3SS-1-independent inflammation.
FIG 6.
Effect of deletions of four of the five effectors, sifA, spvB, sseF, sseJ, and steA, on T3SS-1-independent inflammation in mice. (A) LDH release from RAW cells infected with the indicated S. Typhimurium mutant strains at 20 h after infection. (B) Transcript levels of Ifng, Il6, Tnfa, Kc, and Mip2 in the ceca of mice infected with the indicated S. Typhimurium strains at 5 days after infection, as measured by qPCR. (C) Combined histopathological scoring of cecal sections. A two-tailed Student t test (A) and the Mann-Whitney U test (B and C) were performed for statistical analysis. Individual data are shown as a scatter plot, and bars indicate the means (A and B). Each plot represents an individual mouse (C). Asterisks indicate differences that were statistically significant (P < 0.05). NS, not significant.
SseF is the second most important effector for the induction of intestinal inflammation in streptomycin-pretreated mice.
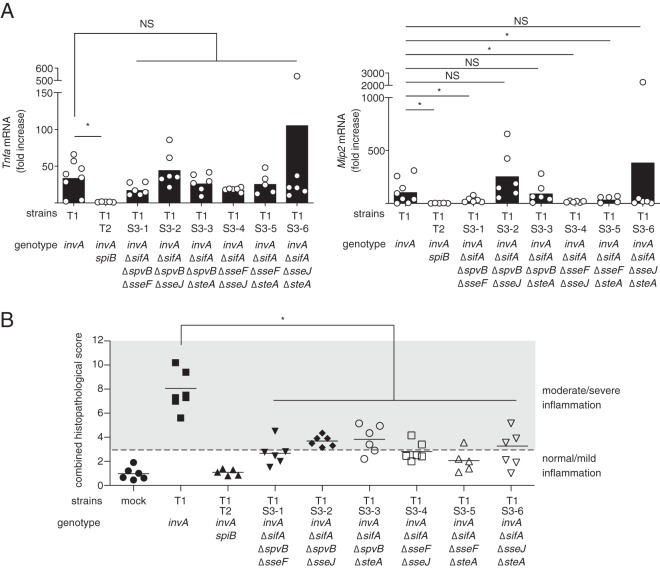
To further analyze the contribution of T3SS-2 effectors to T3SS-1-independent inflammation, we constructed six S. Typhimurium invA ΔsifA strains, each containing deletions of two of the four genes spvB, sseF, sseJ, and steA, and designated them strains T1S3-1 to T1S3-6. mRNA levels of tumor necrosis factor alpha (TNF-α) in the ceca from mice infected with S. Typhimurium strains were not significantly different between the invA (T1) and all T1S3 strains (Fig. 7A). Interestingly, higher expression levels of Mip-2 were observed in mice infected with an invA strain (T1) compared to mice infected with invA ΔsifA ΔspvB ΔsseF (T1S3-1), invA ΔsifA ΔsseF ΔsseJ (T1S3-4), and invA ΔsifA ΔsseF ΔsteA (T1S3-5) strains (Fig. 7A). Since deletion of the sseF gene was common to three of the mutants, T1S3-1, T1S3-4 and T1S3-5, it may be that SseF, which is adjacent to SifA, is important for T3SS-1-independent inflammation. To confirm this, the streptomycin-pretreated mice were infected with six strains, T1S3-1 to T1S3-6. In mice infected with all six strains, histopathological scores of ceca were significantly lower than in mice infected with T1 (invA::pEP185.2) (Fig. 7B and Fig. S3A). However, the average histopathological scores of the ceca in mice infected with the invA ΔsifA ΔspvB ΔsseF (T1S3-1), invA ΔsifA ΔsseF ΔsseJ (T1S3-4), or invA ΔsifA ΔsseF ΔsteA (T1S3-5) strain indicated normal/mild inflammation (Fig. 7B). In contrast to these three strains, the invA ΔsifA ΔspvB ΔsseJ (T1S3-2), invA ΔsifA ΔspvB ΔsteA (T1S3-3), and invA ΔsifA ΔsseJ ΔsteA (T1S3-6) strains tended to elicit moderate/severe inflammation in the ceca (Fig. 7B). These results suggested that, among the four effectors other than SifA, SseF is the most important for induction of T3SS-1-independent inflammation. Moreover, we observed no difference in the expressions of proinflammatory cytokines and chemokines in the ceca between the mice infected with the S. Typhimurium invA strain (T1) and those infected with the invA ΔsifA ΔsseF strain (T1SifASseF) (data not shown).
FIG 7.
Effect of different combinations of three of the four mutations—spvB, sseF, sseJ, and steA—in addition to the sifA mutation on T3SS-1-independent inflammation in mice. (A) Levels of Tnfa and Mip2 in the ceca of mice infected with the indicated S. Typhimurium strains at 5 days after infection, as measured by qPCR. Individual data are shown as a scatter plot, and bars indicate the means. (B) Combined histopathological scoring of cecal sections. Each bar represents an individual mouse. A two-tailed Mann-Whitney U test was performed for statistical analysis. Asterisks indicate differences that were statistically significant (P < 0.05). NS, not significant.
Taken together, our results suggested that SifA, SpvB, SseF, SseJ, and SteA are responsible for T3SS-1-independent cell cytotoxicity in RAW cells and inflammation in the streptomycin-pretreated mice infected with S. Typhimurium. Further, among these effectors, SifA and SseF trigger responses of a greater magnitude, but on their own they are not sufficient for the induction of intestinal inflammation during infection with an S. Typhimurium invA strain.
DISCUSSION
The T3SS-1 mediates entry of S. Typhimurium into epithelial cells and induction of acute inflammation. Although the S. Typhimurium T3SS-1-deficient strain is incapable of translocating T3SS-1 effectors into the cells, bacteria invade the intestinal epithelial cells and elicit intestinal inflammation. Dendritic cell-mediated microbial sampling plays a crucial role in the invasiveness of the ΔinvG strain into the epithelial cells (3). After the invasion of epithelial cells, bacteria proliferate in the lamina propria and are phagocytosed by macrophages (7). It has also been shown that the S. Typhimurium ΔinvG and ΔaroA mutants cannot induce T3SS-1-independent inflammation (7). The ΔaroA mutant strain cannot replicate in macrophages, because an aroA gene is necessary for synthesizing aromatic amino acids (26). Thus, it could be speculated that bacterial proliferation in macrophages is relevant to the induction of T3SS-1-independent inflammation. However, our data indicated that bacterial replication in macrophages may not necessarily be responsible for inducing T3SS-1-independent inflammation. In the present study, we focused on the role of T3SS-2 effectors in cytotoxicity in macrophages to clarify the mechanism of T3SS-1-independent inflammation in the streptomycin-pretreated mice infected with S. Typhimurium.
S. Typhimurium induces caspase-1 activation, which is required for the canonical pyroptosis. Flagellin proteins, FliC and FljB, are detected by the inflammasomes NAIP5 and NAIP6 (27). Two other inflammasomes, NAIP1 and NAIP2, recognize needle and rod proteins of the T3SS, respectively, resulting in caspase-1 activation (28). As shown in Fig. 2C and 3A, cytotoxicity in RAW cells infected with an S. Typhimurium invA::pEP185.2 strain (T1) lacking fliC and fljB genes was reduced to about 20%, while the T1 strain lacking sifA, spvB, fliC, and fljB (T1SifASpvBFl) was not able to induce cytotoxicity in RAW cells at all, which was similar to the cytotoxicity induction ability of an invA and ssaV mutant (T1T2). These facts indicate that the T1SifASpvBFl strain is incapable of inducing cytotoxicity in RAW cells, although this strain possesses a normal T3SS-2 apparatus. Further, RAW cells are defective for NLRP3 activation because they lack the adaptor protein ASC (29). Thus, the death of RAW cells infected with the S. Typhimurium T1 strain could be not the canonical pyroptosis mediated through these inflammasomes. In this study, we showed that cytotoxicity in RAW cells was correlated with inflammation in streptomycin-pretreated mice. Our findings suggest that the induction of macrophage cytotoxicity by T3SS-2 contributes to the induction of an inflammatory response in mice during infection. Our group has shown that cell death genes are induced in the ceca of the streptomycin-pretreated mice infected with S. Typhimurium in a T3SS-2-dependent manner (unpublished data). However, to uncover the mechanism underlying the induction of macrophage cell death by S. Typhimurium infection, additional investigations are needed.
Previous studies have shown that the T3SS-2 effectors SpvB and SseL are associated with the induction of T3SS-1-independent cell death (16, 19, 30). As indicated in Fig. 2A and 3C, the cytotoxicities of RAW cells infected with the S. Typhimurium invA ΔspvB strain (T1SpvB), but not the invA ΔsseL strain (T1SseL), were reduced by about 50% compared to that of the cells infected with the invA strain. SseL was originally identified as deubiquitinase (DUB) and was shown to be required for macrophage cytotoxicity and virulence in mice (19). It has also been reported that SseL suppresses NF-κB activation by deubiquitination of IκBα, which functions as an inhibitor for NF-κB activation (31). However, the function of SseL is controversial, because other groups have shown that the DUB activity was absent on the NF-κB pathway, and an analysis using the proteomics approach suggested that the substrate of DUB was not a protein related to either the cell death pathway or the NF-κB pathway (21, 31). Our data showed for the first time that SseF, SseJ, and SteA contribute to the induction of cytotoxicity in the cells infected with S. Typhimurium. In addition to SifA and SpvB, these effectors are associated with the maturation of Salmonella-containing vacuoles (SCVs) and the biogenesis of Salmonella-induced filaments (SIFs) (32). Although SIFs are not detected in macrophage cell lines, little is known about the relationship between SCV maturation and cytotoxicity in the cells. Future studies are needed to reveal how T3SS-2 effectors induce macrophage cell death.
In the S. Typhimurium T1S5 strain, in which all five effectors associated with the induction of cytotoxicity in RAW cells were deleted, not only cytotoxicity in RAW cells but also intestinal inflammation in mice were markedly reduced compared to those of the parental strain T1. However, the T1S4-1 strain, in which four of the five effectors were deleted, with the retained effector being SifA, did not induce cytotoxicity in RAW cells like the T1S5 strain but did elicit severe intestinal inflammation in the streptomycin-pretreated mice. These findings may indicate that all five effectors are required for the induction of cytotoxicity in RAW cells and that one or more of the five effectors are shared between the two phenotypes, i.e., induction of cytotoxicity in RAW cells and inflammation in mice.
It has been reported that T3SS-2 is required for inflammatory change in the colon of streptomycin-pretreated mice, but the SifA and SpvB effectors are not (25). Consistent with this earlier report, the S. Typhimurium ΔinvA strains with deletion of sifA alone, spvB alone, or both sifA and spvB induced inflammation in the ceca of mice. We demonstrated that the S. Typhimurium invA strain lacking the sseF, sseJ, and steA genes, in addition to the sifA and spvB genes, dramatically reduced inflammation in mice, much like the invA spiB strain. This suggests that T3SS-1-independent inflammatory changes require the cooperative effects of these five effectors. Moreover, we showed that SifA is the most important of the five effectors, while SifA and SseF are required for the induction of T3SS-1-independent inflammation in the ceca of the streptomycin-pretreated mice.
In conclusion, our data suggest that five T3SS-2 effectors, SifA, SpvB, SseF, SseJ, and SteA, are associated with induction of T3SS-1-independent inflammation in mice. This is the first report showing that T3SS-2 effectors contribute to T3SS-1-independent, i.e., T3SS-2-dependent, inflammation in mice. Future studies are needed to determine the mechanism by which these effectors contribute to the inflammation of mice infected with S. Typhimurium.
MATERIALS AND METHODS
Animal experiments.
The protocols used in this study were approved by the Institutional Animal Care and Use Committee of Kitasato University (protocol J96-1 and 17-52). Eight- to ten-week-old female C57BL/6 mice were obtained from Japan SLC. The streptomycin-pretreated mouse model has been described by Barthel et al. (33). Mice were inoculated intragastrically with 0.1 ml of streptomycin (200 mg/ml solution in phosphate-buffered saline [PBS]) and were inoculated intragastrically 24 h later with either 0.1 ml of sterile LB broth or bacterial culture containing 5 × 108 CFU of S. Typhimurium bacteria. Groups of three to six mice were euthanized 5 days after infection. The ceca were collected, and the total weights were measured before division into the tip (for histopathological analysis), middle (RNA extraction), and proximal parts (protein extraction). Samples from the cecum, cecal contents, Peyer’s patches, mesenteric lymph nodes, and spleen were homogenized in PBS, and 10-fold serial dilutions were plated on LB agar plates containing the appropriate antibiotic to determine the bacterial count.
For histopathology, cecum samples were fixed with 10% buffered-formalin (Mildform 10N; Wako, Osaka, Japan), processed according to standard procedures for paraffin embedding, sectioned at 5 μm, and stained with hematoxylin and eosin solutions as described previously (34). The pathological changes were scored by a slight modification of the blinded examination method previously described by Barthel et al. (33).
RNA was extracted from the mouse cecum by using an RNeasy minikit (Qiagen, Hilden, Germany) or an RNA basic kit (Nippon Genetics, Tokyo, Japan). cDNA was synthesized using TaqMan reverse transcription reagent (Thermo, San Jose, CA). Real-time PCR was performed with the primer pairs listed in Table S2 in the supplemental material using KAPA SYBR FAST (Kapa Biosystems, Woburn, MA) and analyzed using the comparative CT method (Applied Biosystems, Foster City, CA). The levels of mRNA expression of target genes were normalized by the levels of Gapdh mRNA.
Bacterial strains and culture conditions.
S. Typhimurium strains used in this study are listed in Table S1. A T3SS-1-deficient mutant (T1) was used as the parental strain. The T1 strain was constructed by P22 phage transduction of invA::pEP185.2 from a derivative of strain IR715, AJB634 (35), to the S. Typhimurium wild-type strain SH100 (36). A mutant deficient in both T3SS-1 and T3SS-2 (T1T2) was also constructed by P22 transduction of spiB::KSAC from SPN452 (37) to the T3SS-1-deficient mutant (T1). To introduce flagellum-deficient mutations (ΔfliC and ΔfljB) in the T1 strain, a ΔfljB mutant strain was first constructed using the λ Red recombination system (38). Then, a DNA fragment was amplified by PCR with the specific primer pairs listed in Table S2, using pKD4 as a template. The PCR product was introduced to SH100 carrying plasmid pKD46. The ΔfljB::Km mutation was transferred by phage P22-mediated transduction into SH100. A kanamycin cassette was eliminated from the ΔfljB::Km strain using pCP20, which carries FLP recombinase. Next, the invA::pEP185.2 mutation was transferred by P22 transduction into a ΔfljB strain, and then a fliC::Tn10 mutation from the S. Typhimurium EHW26 strain (39) was transferred to yield the invA::pEP185.2 fliC::Tn10 ΔfljB (T1Fl) strain.
Deletions of T3SS-2 effectors were constructed using the λ Red recombination system mentioned above. The ΔspvB (HG203), ΔspvC (HG204), and ΔspvD (HG205) strains were from laboratory collections. To construct the invA::pEP185.2 and T3SS-2 effector double mutant, the invA::pEP185.2 mutation was introduced into each of the T3SS-2 effector mutants. Using the same method, the combinations of triple, fourth, or fifth T3SS-2 effector deletion mutants were constructed. All S. Typhimurium mutations were verified using PCR.
To construct T1S5 (pSseJ-CyaA-2HA), pSseJ-CyaA-2HA, which was reported previously (34, 40), was transformed into S. Typhimurium T1S5.
In order to construct the plasmids pACYomega-SseF and pACYomega-SseJ for the complementation analysis, pAC-ProSseJ-SseF-2HA or pAC-ProSseJ-SseJ-2HA was digested with SalI and BamHI, and the ProSseF-2HA fragment or ProSseF-2HA fragment was cloned in the same restriction sites on pACYomega (41). To construct pACYomega-SifA, pACyomega-SpvB, or pACyomega-SteA, sifA, spvB, or steA was amplified by PCR with the TH1151 and TH1152, TH1113 and TH1114, or TH1115 and TH1116 primers, respectively, and the genomic DNA extracted from S. Typhimurium SH100 as a template. The amplified DNAs were digested with the appropriate restriction enzymes and cloned in the same enzyme site on pFLAG-CTC (Sigma), yielding pFLAG-SifA, pFLAG-SpvB, and pFLAG-SteA. Next, SifA-FLAG, SpvB-FLAG, or SteA-FLAG was amplified by PCR with the TH1153 and TH1155 (SifA and SpvB) or TH1153 and TH1154 (SteA) primers, digested with SphI and EcoRV for SifA-FLAG and SpvB-FLAG and with SphI and BamHI for SteA-FLAG, and ligated with the same restriction sites of pACYomega-SseF, respectively.
S. Typhimurium strains were grown at 37°C in Luria-Bertani (LB) broth or on LB agar. Antibiotics were added to the media under the following conditions: ampicillin (100 μg/ml), chloramphenicol (25 μg/ml), kanamycin (25 μg/ml), nalidixic acid (25 μg/ml), and spectinomycin (25 μg/ml).
Cell culture experiments.
The mouse macrophage-like cell line RAW 264.7 was cultured in Dulbecco modified Eagle medium (DMEM; Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum (HyClone Laboratories, Inc., South Logan, UT). RAW cells were seeded in 24-well plates at a density of 5 × 105 cells per well and incubated for 18 h. S. Typhimurium strains were incubated for 18 h, washed two times with PBS, and used to infect RAW cells at a multiplicity of infection of 50 (cytotoxicity assays) or 10 (survival assays and CyaA translocation assays) for 1 h. Cells were washed three times with PBS, and DMEM containing gentamicin (100 μg/ml, Sigma) was added. After 60 min of incubation, the medium was replaced with DMEM containing gentamicin (10 μg/ml).
Cytotoxicity was assessed by LDH release into the culture medium using a CytoTox 96 nonradioactive cytotoxicity assay (Promega, Madison, WI). After 20 h of infection, the supernatants were collected from infected cells, and the optical density at 490 nm was measured using an iMark microplate reader (Bio-Rad, Hercules, CA). The relative LDH release was calculated by using a CytoTox 96 kit according to the manufacturer’s protocol.
For survival assays, the number of intracellular bacteria was determined at 2 and 20 h after infection. First, the cells were washed three times with PBS and lysed in 1% Triton X-100, and then the bacteria were quantified by spreading serial 10-fold dilutions of RAW cell lysates on LB agar plates to count the CFU. Proliferation rates were determined by calculating the ratio of CFU at 2 and 20 h after infection.
For CyaA translocation assays, the secretion of the different T3SS-2 effectors from S. Typhimurium into RAW cells was measured using a cAMP enzyme immunoassay (EIA) kit (Cayman Chemical, Ann Arbor, MI). After 20 h of infection, the cells were washed three times with PBS and lysed using the standard protocol recommended by the manufacturer. A cAMP EIA was performed using an iMark microplate reader.
Statistical analysis.
Unless otherwise indicated, data were expressed as means and standard errors of the means from at least three independent experiments. Statistical analyses were performed with Prism 8 software (GraphPad, La Jolla, CA). Levels of bacterial proliferation in RAW cells, cytotoxicity, and mRNA expression were compared using a Student t test. Histopathological scores were compared by Mann-Whitney U test. P values of <0.05 were considered statistically significant and are indicated by asterisks (*). “NS” is used in the figures to indicate not statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rina Hibino, Asuka Tsukada, Lisa Hatogai, and Nahoko Kubo for technical assistance. We also thank Andreas Bäumler for critical comments on the manuscript.
This study was supported in part by Japan Society for the Promotion of Science KAKENHI grants 15K08470 and 19K07543 (to T.H.) and 18K07119 (to N.O.) and by a Kitasato University Research Grant for Young Researchers (2012 to T.H.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00872-18.
REFERENCES
- 1.Tsolis RM, Adams LG, Ficht TA, Bäumler AJ. 1999. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect Immun 67:4879–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coombes BK, Coburn BA, Potter AA, Gomis S, Mirakhur K, Li Y, Finlay BB. 2005. Analysis of the contribution of Salmonella pathogenicity islands 1 and 2 to enteric disease progression using a novel bovine ileal loop model and a murine model of infectious enterocolitis. Infect Immun 73:7161–7169. doi: 10.1128/IAI.73.11.7161-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hapfelmeier S, Müller AJ, Stecher B, Kaiser P, Barthel M, Endt K, Eberhard M, Robbiani R, Jacobi CA, Heikenwalder M, Kirschning C, Jung S, Stallmach T, Kremer M, Hardt WD. 2008. Microbe sampling by mucosal dendritic cells is a discrete, MyD88-independent step in ΔinvG S. Typhimurium colitis. J Exp Med 205:437–450. doi: 10.1084/jem.20070633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang S, Santos RL, Tsolis RM, Stender S, Hardt WD, Bäumler AJ, Adams LG. 2002. The Salmonella enterica serotype Typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infect Immun 70:3843–3855. doi: 10.1128/iai.70.7.3843-3855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marijke Keestra A, Winter MG, Klein-Douwel D, Xavier MN, Winter SE, Kim A, Tsolis RM, Bäumler AJ. 2011. A Salmonella virulence factor activates the NOD1/NOD2 signaling pathway. mBio 2:e00266-11. doi: 10.1128/mBio.00266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coburn B, Li Y, Owen D, Vallance BA, Finlay BB. 2005. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect Immun 73:3219–3227. doi: 10.1128/IAI.73.6.3219-3227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hapfelmeier S, Stecher B, Barthel M, Kremer M, Muller AJ, Heikenwalder M, Stallmach T, Hensel M, Pfeffer K, Akira S, Hardt WD. 2005. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar Typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol 174:1675–1685. doi: 10.4049/jimmunol.174.3.1675. [DOI] [PubMed] [Google Scholar]
- 8.Shea JE, Hensel M, Gleeson C, Holden DW. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci U S A 93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ochman H, Soncini FC, Solomon F, Groisman EA. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci U S A 93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 11.Jennings E, Thurston TLM, Holden DW. 2017. Salmonella SPI-2 type III secretion system effectors: molecular mechanisms and physiological consequences. Cell Host Microbe 22:217–231. doi: 10.1016/j.chom.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Stein MA, Leung KY, Zwick M, Portillo F-D, Finlay BB. 1996. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol Microbiol 20:151–164. doi: 10.1111/j.1365-2958.1996.tb02497.x. [DOI] [PubMed] [Google Scholar]
- 13.Guy RL, Gonias LA, Stein MA. 2000. Aggregation of host endosomes by Salmonella requires SPI2 translocation of SseFG and involves SpvR and the fms-aroE intragenic region. Mol Microbiol 37:1417–1435. doi: 10.1046/j.1365-2958.2000.02092.x. [DOI] [PubMed] [Google Scholar]
- 14.Beuzón CR, Méresse S, Unsworth KE, Ruíz-Albert J, Garvis S, Waterman SR, Ryder TA, Boucrot E, Holden DW. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J 19:3235–3249. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hensel M, Shea JE, Barbel R, Monack D, Falkow S, Gleeson C, Kubo T, Holden DW. 1997. Functional analysis of ssaJ and the ssaK/U operon, 13 genes encoding components of the type III secretion apparatus of Salmonella pathogenicity island 2. Mol Microbiol 24:155–167. doi: 10.1046/j.1365-2958.1997.3271699.x. [DOI] [PubMed] [Google Scholar]
- 16.van der Velden AW, Lindgren SW, Worley MJ, Heffron F. 2000. Salmonella pathogenicity island 1-independent induction of apoptosis in infected macrophages by Salmonella enterica serotype Typhimurium. Infect Immun 68:5702–5709. doi: 10.1128/iai.68.10.5702-5709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurita A, Gotoh H, Eguchi M, Okada N, Matsuura S, Matsui H, Danbara H, Kikuchi Y. 2003. Intracellular expression of the Salmonella plasmid virulence protein, SpvB, causes apoptotic cell death in eukaryotic cells. Microb Pathog 35:43–48. doi: 10.1016/S0882-4010(03)00066-4. [DOI] [PubMed] [Google Scholar]
- 18.Lesnick ML, Reiner NE, Fierer J, Guiney DG. 2001. The Salmonella spvB virulence gene encodes an enzyme that ADP-ribosylates actin and destabilizes the cytoskeleton of eukaryotic cells. Mol Microbiol 39:1464–1470. doi: 10.1046/j.1365-2958.2001.02360.x. [DOI] [PubMed] [Google Scholar]
- 19.Rytkönen A, Poh J, Garmendia J, Boyle C, Thompson A, Liu M, Freemont P, Hinton JCD, Holden DW. 2007. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc Natl Acad Sci U S A 104:3502–3507. doi: 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mesquita FS, Thomas M, Sachse M, Santos AJM, Figueira R, Holden DW. 2012. The Salmonella deubiquitinase SseL inhibits selective autophagy of cytosolic aggregates. PLoS Pathog 8:e1002743. doi: 10.1371/journal.ppat.1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakayasu ES, Sydor MA, Brown RN, Sontag RL, Sobreira TJP, Slysz GW, Humphrys DR, Skarina T, Onoprienko O, Di Leo R, Deatherage Kaiser BL, Li J, Ansong C, Cambronne ED, Smith RD, Savchenko A, Adkins JN. 2015. Identification of Salmonella Typhimurium deubiquitinase SseL substrates by immunoaffinity enrichment and quantitative proteomic analysis. J Proteome Res 14:4029–4038. doi: 10.1021/acs.jproteome.5b00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knodler LA, Vallance BA, Hensel M, Jäckel D, Finlay BB, Steele-Mortimer O. 2003. Salmonella type III effectors PipB and PipB2 are targeted to detergent-resistant microdomains on internal host cell membranes. Mol Microbiol 49:685–704. [DOI] [PubMed] [Google Scholar]
- 23.Brumell JH, Rosenberger CM, Gotto GT, Marcus SL, Finlay BB. 2001. SifA permits survival and replication of Salmonella typhimurium in murine macrophages. Cell Microbiol 3:75–84. doi: 10.1046/j.1462-5822.2001.00087.x. [DOI] [PubMed] [Google Scholar]
- 24.Miao EA, Leaf IA, Treuting PM, Mao DP, Dors M, Sarkar A, Warren SE, Wewers MD, Aderem A. 2010. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol 11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fierer J, Okamoto S, Banerjee A, Guiney DG. 2012. Diarrhea and colitis in mice require the Salmonella pathogenicity island-2 encoded secretion function but not SifA or Spv effectors. Infect Immun 80:3360–3370. doi: 10.1128/IAI.00404-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stocker BA, Hoiseth SK, Smith BP. 1983. Aromatic-dependent “Salmonella sp.” as live vaccine in mice and calves. Dev Biol Stand 53:47–54. [PubMed] [Google Scholar]
- 27.Zhao Y, Shi J, Shi X, Wang Y, Wang F, Shao F. 2016. Genetic functions of the NAIP family of inflammasome receptors for bacterial ligands in mice. J Exp Med 213:647–656. doi: 10.1084/jem.20160006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kofoed EM, Vance RE. 2011. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelegrin P, Barroso-Gutierrez C, Surprenant A. 2008. P2X7 receptor differentially couples to distinct release pathways for IL-1β in mouse macrophage. J Immunol 180:7147–7157. doi: 10.4049/jimmunol.180.11.7147. [DOI] [PubMed] [Google Scholar]
- 30.Monack DM, Detweiler CS, Falkow S. 2001. Salmonella pathogenicity island 2-dependent macrophage death is mediated in part by the host cysteine protease caspase-1. Cell Microbiol 3:825–837. doi: 10.1046/j.1462-5822.2001.00162.x. [DOI] [PubMed] [Google Scholar]
- 31.Le Negrate G, Faustin B, Welsh K, Loeffler M, Krajewska M, Hasegawa P, Mukherjee S, Orth K, Krajewski S, Godzik A, Guiney DG, Reed JC. 2008. Salmonella secreted factor L deubiquitinase of Salmonella typhimurium inhibits NF-κB, suppresses IκB ubiquitination, and modulates innate immune responses. J Immunol 180:5045–5056. doi: 10.4049/jimmunol.180.7.5045. [DOI] [PubMed] [Google Scholar]
- 32.Knuff K, Finlay BB. 2017. What the SIF is happening: the role of intracellular Salmonella-induced filaments. Front Cell Infect Microbiol 7:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Rüssmann H, Hardt WD. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71:2839–2858. doi: 10.1128/iai.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haneda T, Ishii Y, Shimizu H, Ohshima K, Iida N, Danbara H, Okada N. 2012. Salmonella type III effector SpvC, a phosphothreonine lyase, contributes to reduction in inflammatory response during intestinal phase of infection. Cell Microbiol 14:485–499. doi: 10.1111/j.1462-5822.2011.01733.x. [DOI] [PubMed] [Google Scholar]
- 35.Vazquez-Torres A, Jones-Carson J, Bäumler AJ, Falkow S, Valdivia R, Brown W, Le M, Berggren R, Parks WT, Fang FC. 1999. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- 36.Gotoh H, Okada N, Kim Y-G, Shiraishi K, Hirami N, Haneda T, Kurita A, Kikuchi Y, Danbara H. 2003. Extracellular secretion of the virulence plasmid-encoded ADP-ribosyltransferase SpvB in Salmonella. Microb Pathog 34:227–238. doi: 10.1016/S0882-4010(03)00034-2. [DOI] [PubMed] [Google Scholar]
- 37.Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio S-P, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, Bäumler AJ. 2009. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raffatellu M, Chessa D, Wilson RP, Dusold R, Rubino S, Bäumler AJ. 2005. The Vi capsular antigen of Salmonella enterica serotype Typhi reduces Toll-like receptor-dependent interleukin-8 expression in the intestinal mucosa. Infect Immun 73:3367–3374. doi: 10.1128/IAI.73.6.3367-3374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miki T, Shibagaki Y, Danbara H, Okada N. 2009. Functional characterization of SsaE, a novel chaperone protein of the type III secretion system encoded by Salmonella pathogenicity island 2. J Bacteriol 191:6843–6854. doi: 10.1128/JB.00863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, Libby SJ, Fang FC, Raffatellu M. 2013. Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.