Streptococcus suis is one of the most important pathogens affecting the swine industry and is also an emerging zoonotic agent for humans. Two-component signaling systems (TCSs) play important roles in the adaptation of pathogenic bacteria to host environments. In this study, we identified a novel TCS, named TCS09HKRR, which facilitated Streptococcus suis serotype 2 (SS2) resistance to clearance by the host immune system and contributed to bacterial pathogenicity.
KEYWORDS: Streptococcus suis, TCS09HKRR, autorepression, capsular biosynthesis, pathogenicity
ABSTRACT
Streptococcus suis is one of the most important pathogens affecting the swine industry and is also an emerging zoonotic agent for humans. Two-component signaling systems (TCSs) play important roles in the adaptation of pathogenic bacteria to host environments. In this study, we identified a novel TCS, named TCS09HKRR, which facilitated Streptococcus suis serotype 2 (SS2) resistance to clearance by the host immune system and contributed to bacterial pathogenicity. Furthermore, RNA-sequencing analyses identified 79 genes that were differentially expressed between the wild-type (WT) and ΔTCS09HKRR strains, among which half of the 39 downregulated genes belonged to the capsular biosynthesis clusters. Transmission electron microscopy confirmed that the capsule of the ΔTCS09HKRR strain was thinner than that of the WT strain. Electrophoretic mobility shift assays (EMSA) showed that the regulator of TCS09HKRR (TCS09RR) could not bind the promoter regions of cps and neu clusters, which suggested that TCS09HKRR regulates capsule biosynthesis by indirect pathways. Unexpectedly, the TCS09HKRR operon was upregulated when TCS09HKRR was deleted. A specific region, ATGACATTTGTCAC, which extends from positions −193 to −206 upstream of the TCS09HKRR operon, was further identified as the TCS09RR-binding site using EMSA. These results suggested the involvement of a negative feedback loop in this regulation. In addition, TCS09RR was significantly upregulated by more than 18-fold when coincubated with RAW264.7 macrophages. Our data suggested that autorepression renders TCS09HKRR more sensitive to host stimuli, which optimizes the regulatory network of capsular biosynthesis in SS2.
INTRODUCTION
To survive in diverse environments, bacteria must sense and respond to external stimuli. Two-component signaling systems (TCSs) are ubiquitous in prokaryotes and represent a major mechanism through which bacterial cells adapt to environmental changes (1, 2). A typical TCS consists of a membrane-bound sensor histidine kinase (HK) and a cytoplasmic response regulator (RR) (1). Upon encountering certain external signals, HK protein autophosphorylates at a conserved histidine residue and subsequently transfers the phosphate group to a conserved aspartate residue of the cognate RR protein (1, 3). After phosphorylation, the RR protein is activated and in turn modulates downstream gene expression to respond to the initial stimulus.
Streptococcus suis is a Gram-positive, facultative anaerobic pathogen responsible for important economic losses in the swine industry worldwide (4, 5). Among the described serotypes based on capsular antigens, S. suis serotype 2 (SS2) is considered the most prevalent in most countries (6). Additionally, with the increased number of human cases of S. suis infections, this species is recognized as an important zoonotic bacterium (5–7). In China, two large-scale outbreaks of SS2 associated with meningitis and streptococcal toxic shock-like syndrome occurred in 1998 and 2005, respectively, which resulted in 229 human cases and 52 deaths (7). Although a variety of virulence factors have been proposed in recent years, the pathogenesis of S. suis infection is still unclear.
SS2 strains enter the host mainly through the upper respiratory tract, as well as by the oral route or skin injury (8). Once the mucosal barriers are breached, the bacteria disseminate to different organs and tissues by the bloodstream. TCSs are usually employed to sense and respond to extracellular signals during the infection process (2, 9). Bioinformatics analysis of the genome of SS2 strains identified 15 TCSs (10), among which RevS, Salk/SalR, CovR, CiaRH, Ihk/Irr, VirR/VirS, NisK/NisR, 1910HK/RR, and VraSRSS have been reported to regulate the expression of putative virulence factors, mediate multidrug resistance, or alter bacterial metabolism (11–20), thereby contributing to the adaption of SS2 strains to environmental stimuli. In bacteria, RRs often regulate the transcription of their own operons, which contain cognate sensor HK genes. Such autoregulation can be useful if the pathogen needs to respond to environmental changes rapidly (21, 22). VraSRSS exhibits positive autoregulation ability, as its own transcription is activated by the regulator protein VraRSS (20). Autorepression is less common than autoactivation among TCSs and appears to be inherent in areas of metabolism for maintaining metabolic homeostasis (22). In Escherichia coli, 14 TCSs were reported to be autoregulated, and only three of them function by autorepression (22). These TCSs can restrict the duration and intensity of stress responses and thereby improve sensitivity to external signals (23–25).
When invading the host, SS2 strains encounter phagocytes of the immune system. However, the mechanism through which they resist phagocytosis and persist in the host bloodstream is not yet understood. In this study, a novel TCS designated TCS09HKRR was found to contribute to bacterial resistance to phagocytosis and survival in swine blood, thereby enhancing the virulence of SS2. RNA-sequencing (RNA-Seq) analyses and transmission electron microscopy (TEM) revealed that TCS09HKRR regulates capsule biosynthesis in SS2. Further work identified an autorepressing process in TCS09HKRR that might render bacterial cells more sensitive to host stimuli and optimize the regulatory network of capsule biosynthesis in SS2.
RESULTS
Macrophage phagocytosis activates TCS genes.
The genome of the ZY05719 strain encodes 15 putative TCSs, among which RevS, Salk/SalR, CovR, CiaRH, Ihk/Irr, VirR/VirS, NisK/NisR, 1910HK/RR, and VraSRSS have already been identified to contribute to SS2 virulence (11–20). To explore the potential roles of the remaining six uncharacterized TCSs in bacterial pathogenicity, murine macrophage-like RAW264.7 cells were applied to coincubate with the SS2 strain ZY05719. Unexpectedly, the expression of these TCS RR genes, particularly zy05719_06495 and zy05719_08020, were all significantly higher after macrophage stimulation than when cultured in Dulbecco’s modified Eagle’s medium (DMEM). As shown in Fig. 1, when mRNA levels in DMEM were set to 1, the expression of zy05719_06495 was significantly increased by more than 18-fold, whereas zy05719_08020 showed a 15.7-fold increase. These results suggested that the two TCSs, zy05719_06490-zy05719_06495 and zy05719_08020-zy05719_08025, play important roles in the SS2 response to macrophage phagocytosis.
FIG 1.
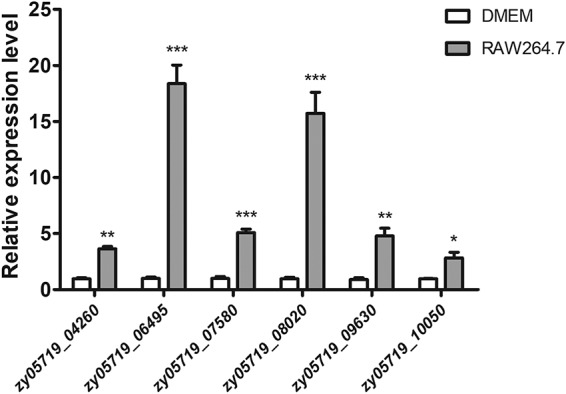
Transcription of the six uncharacterized TCS RRs of SS2 strain ZY05719 upon coincubation with RAW264.7 macrophages. Bacteria incubated in DMEM were used as the negative control. The results of qRT-PCR are expressed as means ± standard deviations (SD) from three independent experiments. An unpaired two-tailed Student's t test was used for statistical analysis. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Identification of zy05719_06490-zy05719_06495 and zy05719_08020-zy05719_08025 TCSs.
Homology analysis of ZY05719_06490 and ZY05719_06495 proteins revealed that their amino acid sequences shared 70% and 67% identity with the VicK and VicR proteins of Streptococcus pneumoniae (26), whereas ZY05719_08020 and ZY05719_08025 exhibited 44% and 30% amino acid sequence identity with DesR and DesK of Bacillus subtilis (27). To analyze their potential roles in the pathogenesis of SS2, we constructed homologous suicide plasmids to create zy05719_06490-zy05719_06495 and zy05719_08020-zy05719_08025 deletion mutants. However, we were not able to recover any deletion mutant of the putative TCS ZY05719_06490-ZY05719_06495 in the SS2 strain ZY05719, as this TCS is essential for bacterial viability, similar to that found for its homolog VicKR, which is essential for many Gram-positive pathogens (28–31). However, the zy05719_08020-zy05719_08025 double gene deletion mutant was successfully constructed. Previous studies demonstrated that DesKR is responsible for the cold adaptation of B. subtilis (27). Hence, we performed growth kinetics assays under normal (37°C) and low (25°C) temperatures to assess whether ZY05719_08020-ZY05719_08025 plays a similar role in the SS2 strain ZY05719. However, the zy05719_08020-zy05719_08025 mutant did not exhibit any growth difference compared to the wild-type (WT) strain at both normal and low temperatures (see Fig. S1 in the supplemental material), indicating that ZY05719_08020-ZY05719_08025 has unknown functions; hence, it was renamed TCS09RR/TCS09HK (for the ninth TCS located in the ZY05719 genome).
TCS09HKRR contributes to resistance to macrophage phagocytosis and survival in swine blood.
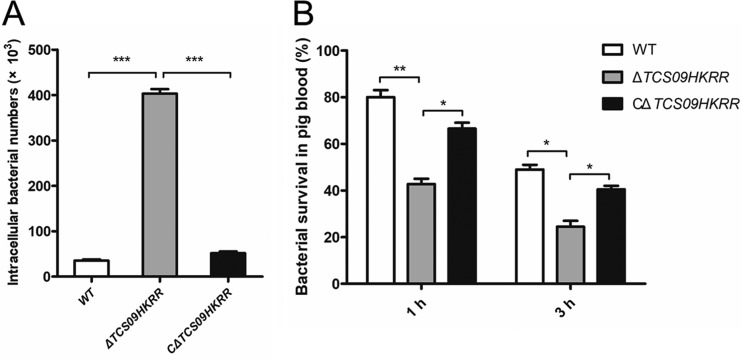
As the mRNA level of TCS09RR increased significantly when the ZY05719 strain was incubated with RAW264.7 macrophages, we compared the susceptibility to phagocytosis of ΔTCS09HKRR and WT strains. We observed that the ΔTCS09HKRR mutant was more susceptible to phagocytosis by RAW264.7 macrophages than the WT or complemented ΔTCS09HKRR (CΔTCS09HKRR) strains (Fig. 2A). In addition, a porcine alveolar macrophage cell line, 3D4/21, was used for phagocytosis assays, and the results were consistent with those obtained using RAW264.7 macrophages (Fig. S2). These results showed that TCS09HKRR is required for SS2 resistance to macrophage phagocytosis. It is well known that pathogenic bacteria must survive in the bloodstream during systemic infection. Hence, we evaluated the effect of TCS09HKRR on survival of the SS2 strain in whole swine blood. As shown in Fig. 2B, the percent survival rates of the ΔTCS09HKRR strain were significantly lower than those of the WT and CΔTCS09HKRR strains at both 1 and 3 h. These results indicated that TCS09HKRR plays important roles in resisting elimination by the host defense system.
FIG 2.
Coincubation of RAW264.7 macrophages or swine whole blood with S. suis wild-type (WT), ΔTCS09HKRR, and CΔTCS09HKRR strains. (A) Effect of the TCS09HKRR deletion on the ability of SS2 to resist phagocytosis by RAW264.7 macrophages. At 1.5 h postinfection, the cells were washed and incubated in DMEM containing antibiotics (penicillin and gentamicin) for another 1.5 h. The cells then were washed again and lysed to determine CFU numbers. (B) Effect of the TCS09HKRR deletion on the ability of SS2 to survive in swine whole blood. Survival rates were calculated as CFU per ml at 1 or 3 h relative to that at 0 h. The experiments were repeated three times. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
TCS09HKRR is important in a mouse model of bacteremia.
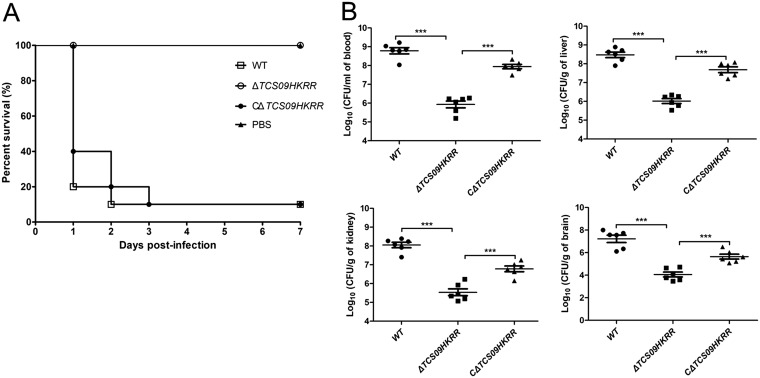
To evaluate the role of TCS09HKRR in bacterial pathogenicity, groups of 10 BALB/c mice were injected intraperitoneally with 5 × 108 CFU of different bacterial strains. We observed that mice in groups challenged by the WT and CΔTCS09HKRR strains developed severe clinical signs at 24 h postinfection, including shivering, rough hair coat, and depression, and the survival rates were 10%, whereas mice challenged by the ΔTCS09HKRR strain exhibited only slight clinical signs and had a 100% survival rate (Fig. 3A). To better understand the effects of TCS09HKRR during systemic infection, we determined the bacterial load in infected mice. As shown in Fig. 3B, the bacterial loads in the blood, livers, kidneys, and brains from ΔTCS09HKRR strain-challenged mice were all significantly reduced compared to those with the WT or CΔTCS09HKRR strain. Hence, TCS09HKRR is important for in vivo bacterial proliferation and contributes to the pathogenicity of SS2.
FIG 3.
TCS09HKRR contributes to SS2 pathogenicity in a mouse infection model. (A) Survival curves of mice infected with WT, ΔTCS09HKRR, and CΔTCS09HKRR strains. Groups containing 10 mice each were challenged with the indicated strains at a dose of 5 × 108 CFU/mouse and monitored until 7 days postinfection. The negative-control group was challenged with an equal volume of sterile PBS. (B) Bacterial loads in different mouse organs at 6 h postinfection. Groups containing six mice each were challenged with the indicated strains at a dose of 3 × 108 CFU/mouse. Blood, livers, kidneys, and brains were harvested to determine CFU numbers. Asterisks indicate significant differences (***, P < 0.001).
Transcriptomic analysis identifies genes that are differentially expressed between WT and ΔTCS09HKRR strains.
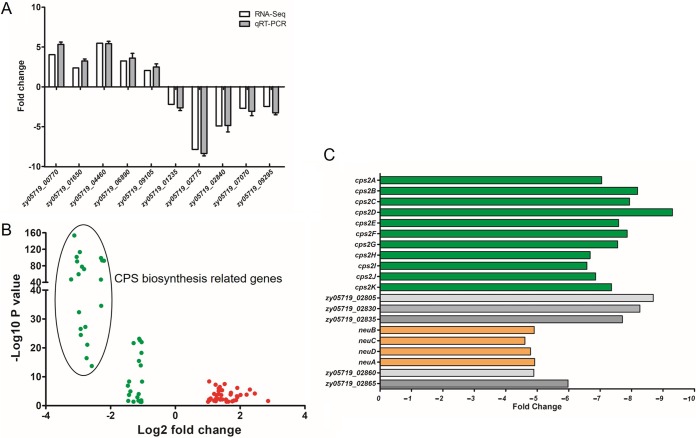
To further investigate the molecular mechanisms by which TCS09HKRR contributes to pathogenicity of the SS2 strains, RNA-Seq was performed and the transcriptomes of the WT and ΔTCS09HKRR strains were compared. As shown in Table S1, compared to gene expression in the WT strain, 79 genes were significantly differentially expressed in the ΔTCS09HKRR strain, among which 40 genes were upregulated and 39 were downregulated. We randomly selected 10 of these 79 genes (five upregulated and five downregulated genes) to perform quantitative reverse transcription-PCR (qRT-PCR) for further verification. The qRT-PCR results were consistent with the transcriptomic data (Fig. 4A), which confirmed the results of RNA-Seq analyses. Interestingly, we found that at least half of the downregulated genes were involved in the biosynthesis of capsular polysaccharide (CPS) (Fig. 4B and C), which is a proven critical virulence factor for SS2 (32–34). Meanwhile, the transcriptional levels of many well-known virulence factors of SS2, such as suilysin, enolase, and muramidase-released protein, were unaltered. These results suggested that the pathogenic role of TCS09HKRR is mediated by its potential regulation of CPS biosynthesis.
FIG 4.
RNA-Seq analysis of TCS09HKRR-regulated genes. (A) Validation of gene regulation by qRT-PCR. Ten genes were randomly selected and amplified by qRT-PCR to validate the expression level change observed by RNA-Seq analysis. The qRT-PCR results were expressed as means ± SD from three independent experiments. (B) Volcano plot showing gene expression in ΔTCS09HKRR versus WT strains determined based on RNA-Seq analysis. The x axis represents the log2 fold change value, while the y axis displays the −log10(P) value. Red dots represent genes with 2-fold higher expression in the ΔTCS09HKRR strain than in the WT strain with a P value of <0.05, and the green dots represent genes with 2-fold lower expression in ΔTCS09HKRR than in the WT strain with a P value of <0.05. (C) TCS09HKRR deletion significantly downregulated the CPS biosynthesis clusters, as observed based on RNA-Seq analysis. The x axis represents the fold change.
TCS09HKRR controls CPS biosynthesis in SS2.
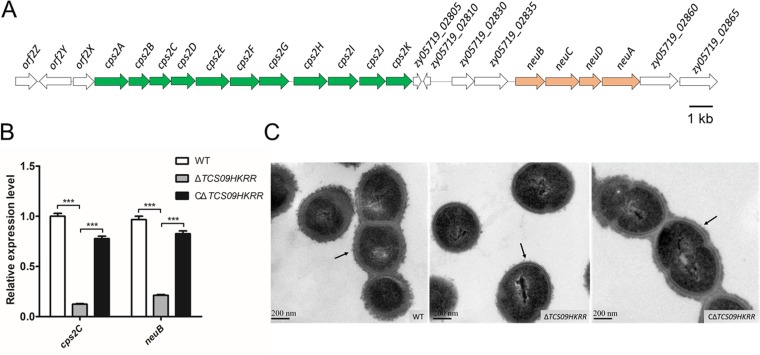
Capsules of the SS2 strains are composed of glucose, galactose, N-acetylglucosamine, rhamnose, and sialic acid, which are synthesized by the capsule synthesis clusters, mainly including cps2A to cps2K and neuBCDA (32–37) (Fig. 5A). To confirm that deletion of the TCS09HKRR genes was responsible for the downregulation of the capsule biosynthesis cluster in the mutant, we checked the mRNA levels of cps2C and neuB in the complemented strain. As shown in Fig. 5B, the expression of cps2C and neuB was restored in the CΔTCS09HKRR strain. TEM was performed to visualize the bacterial capsule structures. We observed that the capsule of the ΔTCS09HKRR strain was thinner than that of the WT and CΔTCS09HKRR strains (Fig. 5C). In addition, the surface proteins of the WT, ΔTCS09HKRR, and CΔTCS09HKRR strains were extracted and compared using SDS-PAGE, which showed no obvious differences (Fig. S3). These results indicated that TCS09HKRR regulates CPS biosynthesis.
FIG 5.
TCS09HKRR controls CPS biosynthesis in SS2. (A) Schematic diagram showing the genetic organization of the CPS synthesis clusters in SS2. The green cluster is cps2A to cps2K, while the orange cluster is neuBCDA. The arrows indicate the direction of transcription. (B) Expression of cps2C and neuB in WT, ΔTCS09HKRR, and CΔTCS09HKRR strains was determined using qRT-PCR. qRT-PCR results are represented as means ± SD from three independent experiments. Asterisks indicate significant differences (***, P < 0.001). (C) TEM images of WT, ΔTCS09HKRR, and CΔTCS09HKRR strains. Black arrows show the capsule, and the scale bars indicate the magnification size.
The TCS09HKRR promoter is autorepressed via a negative feedback loop by TCS09RR to regulate the cps gene cluster.
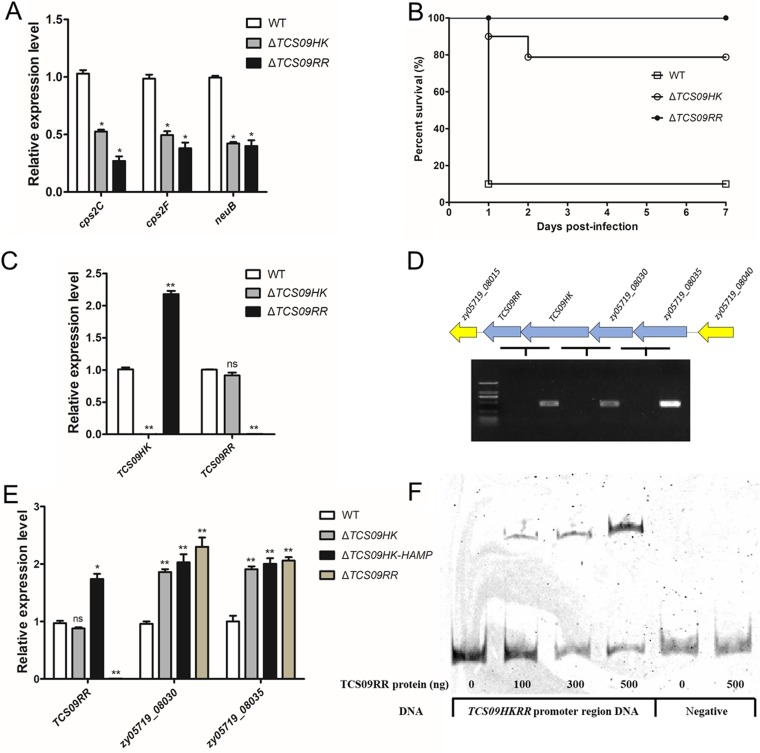
To investigate the relationship between TCS09HK and TCS09RR during the regulation of CPS biosynthesis in SS2, we created TCS09HK and TCS09RR single deletion mutants. As shown in Fig. 6A, the mRNA levels of cps2C, cps2F, and neuB in both ΔTCS09HK and ΔTCS09RR strains were all significantly lower than those in the WT strain. These results were consistent with the mouse survival assay, as the survival rates of both mutants were significantly higher than those with the WT strain (Fig. 6B). The mechanism underlying this regulation was subsequently investigated using electrophoretic mobility shift assay (EMSA).
FIG 6.
Autorepression of TCS09HKRR in SS2. (A) Transcription levels of cps2C, cps2F, and neuB in WT, ΔTCS09HK, and ΔTCS09RR strains, as determined by qRT-PCR. (B) Survival curves of mice infected with WT, ΔTCS09HK, and ΔTCS09RR strains. Groups containing 10 mice each were challenged with the indicated strain at a dose of 5 × 108 CFU/mouse and monitored until 7 days postinfection. (C) Expression of TCS09HK and TCS09RR in WT, ΔTCS09HK, and ΔTCS09RR strains was determined by qRT-PCR. (D) RT-PCR showed that TCS09RR, TCS09HK, zy05719_08030, and zy05719_08035 formed a single operon. The RNA samples without reverse transcription served as the negative control. The DNA markers are 2,000, 1,000, 750, 500, 250, and 100 bp. (E) Expression of TCS09RR, zy05719_08030, and zy05719_08035 in WT, ΔTCS09HK, ΔTCS09HK-HAMP, and ΔTCS09RR strains was determined by qRT-PCR. (F) EMSA showing TCS09RR binding to the promoter region of the TCS09HKRR operon. The fragments from 16S rRNA served as the negative control. Asterisks indicate significant differences (ns, P > 0.05; *, P < 0.05; **, P < 0.01).
Unexpectedly, we also observed that TCS09HK expression was significantly higher in the ΔTCS09RR strain than in the WT strain, whereas the TCS09HK deletion had no significant effect on the expression of TCS09RR (Fig. 6C). As TCS09HK acts upstream of TCS09RR, we constructed a strain harboring a deletion in the N-terminal HAMP domain of TCS09HK to exclude the possibility that the entire TCS09HK deletion affects TCS09RR transcription. However, the expression level of TCS09RR was significantly higher in the ΔTCS09HK-HAMP strain than in the WT strain (Fig. 6E). These results suggested that TCS09HK and TCS09RR regulate their own expression and that this potential autoregulation of TCS09HKRR contributes to the optimal regulation of CPS biosynthesis.
The BPROM program (38) and reverse transcription-PCR analysis indicated that the four genes (TCS09RR, TCS09HK, zy05719_08030, and zy05719_08035) form a single operon and share one promoter (Fig. 6D). The expression levels of both zy05719_08030 and zy05719_08035 were significantly higher in ΔTCS09RR, ΔTCS09HK, and ΔTCS09HK-HAMP strains than in the WT strain (Fig. 6E), whereas these genes were not present in the ΔTCS09HKRR RNA-Seq data, as the false discovery rate was controlled by a log2 fold of ≥1 (0.9524 and 0.9161 for these two genes, respectively). Therefore, we further performed qRT-PCR analysis and showed that the expression levels of zy05719_08030 and zy05719_08035 were significantly increased in the ΔTCS09HKRR strain (Fig. S4). These results suggested that TCS09RR represses the transcription of its own promoter. Furthermore, the predicted promoter of this operon could be bound by the TCS09RR protein, as shown by EMSA (Fig. 6F), which confirmed that TCS09RR utilizes a negative feedback loop for autorepression.
As zy05719_08030 and zy05719_08035 are located in the TCS09HKRR operon and encode two ATP transport proteins, it is possible that they are the putative targets of TCS09HKRR regulation. Bioinformatics analyses showed that ZY05791_08030 and ZY05719_08035 possess putative conserved domains, YadH and CcmA, respectively (Fig. S5), and both have been predicted to be members of the ABC-type multidrug transport system in previous studies (39, 40). However, no difference in antibiotic resistance was observed between the ΔTCS09HKRR and WT strains. Thus, the identification of related phenotypes is required for further investigations in our future work.
Identification of a specific binding sequence for TCS09RR that directly regulates the TCS09HKRR promoter but not the cps cluster.
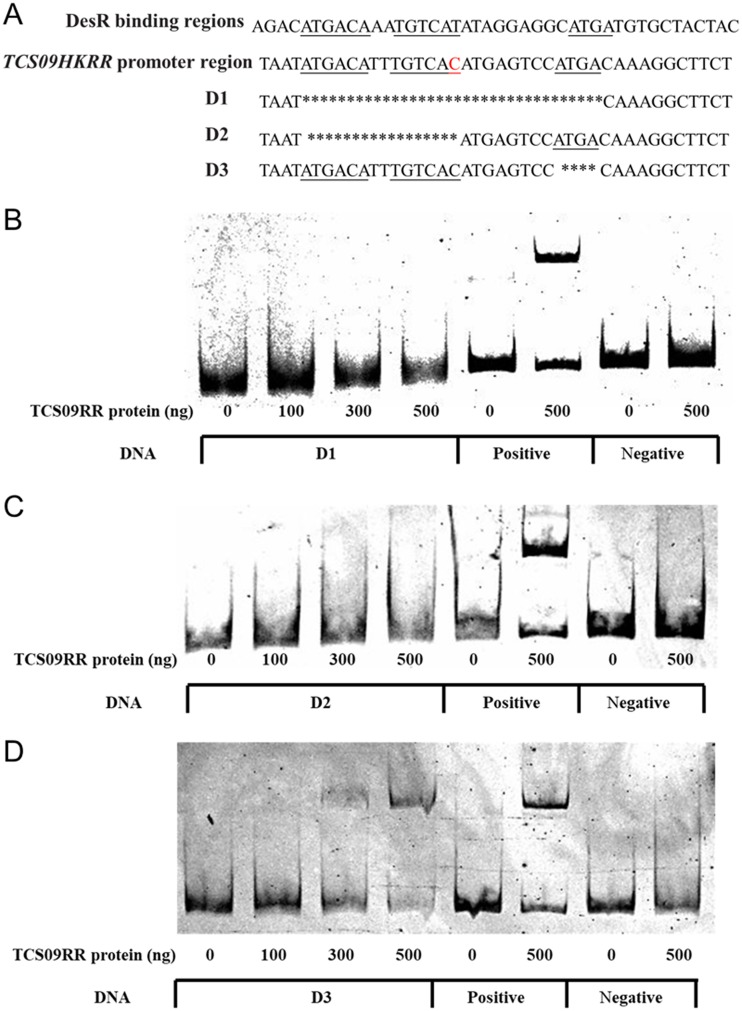
A previous study demonstrated that DesR regulates expression of the des gene, which encodes the Δ5-desaturase of B. subtilis, to alter membrane phospholipid compositions at low temperatures (27). The DesR binding sites within the des promoter harbor several inverted and direct repeats, including 5′-ATGACA-3′ and 5′-ATGA-3′ (Fig. 7A), and both repeats are necessary for DesR-DNA interactions (41, 42). Considering the homology between TCS09RR and DesR, we analyzed the sequence of the TCS09HKRR promoter region and identified similar repeats with only one base mutation (Fig. 7A). To determine whether the homologous binding motifs can also be recognized by TCS09RR, promoter variants (D1, D2, and D3) harboring deletions in these repeats were analyzed using EMSA. As shown in Fig. 7, D1 and D2 were unable to form complexes with TCS09RR, while D3 interacted with TCS09RR, similar to that observed in the positive control. These results suggested that TCS09RR binds a DNA sequence (5′-ATGACATTTGTCAC-3′) that extends from positions −193 to −206 upstream of the TCS09HKRR operon, thereby regulating its own expression. We next analyzed the promoter regions of cps2A to cps2K and neuBCDA but did not detect similar regions with the aforementioned binding sites. Furthermore, EMSAs did not show any significant shift in either of the promoters in the presence of recombinant TCS09RR (Fig. S6), suggesting that TCS09HKRR indirectly regulates CPS biosynthesis in SS2 strains.
FIG 7.
TCS09RR binds a specific region of the TCS09HKRR promoter. (A) The TCS09HKRR promoter region contains similar sequence repeats, which have been identified as specific binding sites for DesR. The inverted and direct repeats are underlined, while the mutated base of the TCS09HKRR promoter region is marked in red. D1, D2, and D3 are promoter variants with deleted regions indicated by lines with asterisks. (B, C, and D) EMSA showing binding of the recombinant TCS09RR to D1, D2, and D3. The TCS09HKRR promoter region without deletion was used as the positive control, and fragments amplified from 16S rRNA served as the negative control.
DISCUSSION
TCSs are major mechanisms that allow bacteria to sense and respond to host environmental signals and are accompanied by coordinated changes in the expression of multiple genes (1–3, 9). In the present study, we demonstrated that autorepression via a negative feedback loop is mediated by TCS09HKRR to regulate capsule biosynthesis, thereby facilitating resistance to clearance by the host innate immune response in the SS2 strain and contributing to bacterial pathogenicity.
The SS2 strain ZY05719 is an encapsulated bacterium, and CPS is recognized as a critical virulence factor for highly virulent S. suis strains (32, 34). Previously, researchers have used isogenic unencapsulated mutant strains to demonstrate that the absence of CPS increases phagocytosis by phagocytes and promotes rapid clearance from circulation (32, 34, 43, 44). In addition, the S. suis transcriptional landscape has revealed that genes involved in CPS biosynthesis were upregulated in blood (45). Nevertheless, S. suis CPS-deficient mutants have been reported to adhere to epithelial cells better than the encapsulated parental strain (20, 46, 47), as CPS might cover adhesins on the bacterial surface (48). Therefore, it has been proposed that S. suis downregulates expression of the capsule for increased adhesion to epithelial cells during the early steps of infection, whereas it upregulates capsule production after entering the bloodstream to counter phagocytosis (49). However, the mechanisms regulating CPS expression during S. suis infection are complicated and remain unclear. In our study, we observed that TCS09HKRR can regulate transcription of the CPS biosynthesis cluster. Furthermore, TEM assays confirmed that capsule thickness in the ΔTCS09HKRR strain was decreased compared to that in the WT and complemented strains. The thinning capsule increased susceptibility to phagocytosis by macrophages and decreased survival in swine blood, thereby contributing to the diminished virulence of SS2 (32–34). Furthermore, the promoter probe of the cps cluster did not show any binding based on EMSA, suggesting that some unknown factors located downstream of TCS09HKRR are involved in this regulation.
In addition to TCS09HKRR, three other TCSs have already been identified to regulate CPS biosynthesis in the SS2 strains (13, 16, 20). The orphan response regulator CovR is a global virulence regulator that represses the expression of 193 genes, including that of cps2C and neuA (13), whereas proteomic analysis showed that VirR/VirS contribute to the expression of cps2B, cps2C, and neuC (16). Therefore, covR-defective mutants exhibit thicker capsules, enhanced resistance to phagocytosis, and increased bacterial lethality, whereas VirR/VirS contribute to capsule biosynthesis, resistance to host clearance, and full virulence in SS2 (13, 16). In addition, VraSRSS acted similarly to TCS09HKRR by indirectly upregulating transcription of the whole CPS biosynthesis cluster (cps2A-cps2K and neuBCDA) to improve capsule biosynthesis (20). Although TCS09HKRR is similar to VraSRSS regarding CPS regulation in SS2 strains, the two TCSs also show several differences. For example, VraSRSS can be positively autoregulated and plays critical roles in multidrug resistance in SS2 (20), whereas TCS09HKRR can be autorepressed and does not appear to affect the bacterial susceptibility profile against multiple antimicrobials. SS2 strains require a regulatory network to sensitively control CPS synthesis, and through this mechanism, it can better adapt to different external stimuli. Thus, the similarities between TCS09HKRR and VraSRSS, as well as the roles of CovR and VirR/VirS in the regulation of capsule gene expression in SS2, are reasonable and necessary. However, the interaction among the TCSs in SS2 during the regulation of CPS warrants further investigations.
Upon detecting external signals, certain TCSs can also regulate the transcription of their own genes in addition to specific downstream genes (21, 22). Autoregulation of TCSs is one of the common properties of bacterial regulatory circuits (22), which enhances sensitivity to input signals. For example, the autoactivation of PhoP/PhoQ promotes a transcriptional surge that initiates the regulation of Salmonella virulence (50), and the autorepression of CovR allows Streptococcus pyogenes to rapidly turn off the activated CovR regulatory circuit (51). In this study, we observed that the TCS09HKRR operon is upregulated when TCS09HKRR is deleted from the genome and that the predicted promoter of this operon can be specifically bound by the TCS09RR protein, suggesting that a negative feedback loop exists during this regulation. TCSs with autorepressing ability can derepress their own promoters, resulting in reduced RR production, which ensures the availability of adequate amounts of RR to respond to external signals (51). An analysis of the expression of TCS09RR and capsule genes in the host before and after the SS2 strain enters the blood circulation revealed significant upregulation in the blood compared to that in the abdominal cavity (Fig. S7). Such a system might increase the ability of the SS2 strain to respond to host environmental changes during the early steps of infection and downregulate the biosynthesis of CPS, which enhances contact between bacterial adhesins and host cells. When the SS2 strain enters the host circulation, the autorepression of TCS09HKRR might be relieved by other regulatory factors, such as VraSRSS, which form a positive autoregulation loop and exhibit significantly higher expression in host blood than in Todd-Hewitt broth (THB) culture (20). Therefore, SS2 strains can better persist and proliferate in the host.
In summary, a novel TCS09HKRR was identified as autorepressed by a negative feedback loop to regulate CPS biosynthesis in SS2. This mechanism allows SS2 strains to adapt to different host environmental stimuli, thereby contributing to bacterial pathogenicity. However, the external signals that activate HK and the CPS regulatory network require further investigations.
MATERIALS AND METHODS
Bacterial strains, plasmids, cells, and experimental animals.
The bacterial strains and plasmids used in this study are listed in Table S2 in the supplemental material. The wild-type SS2 strain ZY05719 was isolated from a diseased pig during an outbreak in the Sichuan province of China and grown at 37°C in THB (Becton, Dickinson) or Todd-Hewitt broth agar (THA). Escherichia coli strains were grown in Luria-Bertani (LB) broth or on LB agar plates at 37°C. When required, the following concentrations of antibiotics were added to the medium: for S. suis, spectinomycin (Sigma-Aldrich) at 100 μg/ml; for E. coli, spectinomycin (Sigma-Aldrich) at 50 μg/ml and ampicillin (Sigma-Aldrich) at 100 μg/ml.
The murine macrophage-like RAW264.7 cells and the porcine alveolar macrophage 3D4/21 cells were cultured in DMEM (Wisent, Canada) supplemented with 10% fetal bovine serum and maintained at 37°C with 5% CO2. Specific-pathogen-free BALB/c mice (female, 6 weeks old) were purchased from the Comparative Medicine Center of Yangzhou University (Jiangsu, China). All animal experiments were performed in the Laboratory Animal Center of the Nanjing Agricultural University with the approval of the Laboratory Animal Monitoring Committee of Jiangsu Province.
RNA isolation, RT-PCR, and qRT-PCR analysis.
The strains in logarithmic phase were washed three times with phosphate-buffered saline (PBS) and incubated with RAW264.7 macrophages or DMEM for 3 h at a bacterium-to-cell ratio of 100:1. Total RNA was isolated using TRIzol reagent (Vazyme, China) according to the manufacturer’s instructions. After removing contaminating DNA, cDNA was synthesized using the HiScriptII first-strand cDNA synthesis kit (Vazyme). qRT-PCR was performed to validate the transcript concentrations of the selected genes using the QuantStudio 6 Flex real-time PCR system and ChamQ Universal SYBR qPCR master mix (Vazyme). The housekeeping gene parC was used as the internal reference (45), and the relative fold change was calculated using the 2−ΔΔCT method (52). All primers used are listed in Table S3, and each sample procedure was repeated three times.
Recombinant DNA techniques.
The target gene deletion mutants were constructed using the pSET4S vector as described previously (53). Briefly, the upstream and downstream sequences surrounding the target gene were amplified from the chromosomal DNA of the ZY05719 strain using specific primers with restriction enzyme sites (Table S3). After the upstream and downstream sequences were ligated by overlap extension PCR, the fusion fragment was digested with respective endonucleases and inserted into the pSET4S vector with the same sites. The recombinant plasmid was electroporated into the WT strain, which then exchanged genetic fragments twice with the bacterial genome by intermolecular recombination. Putative deletion of the target gene was screened using PCR and verified by sequencing.
The pSET2 vector was used to construct the complemented strains (54). The open reading frame (ORF) of the target gene (including putative promoter sequences) from the genome was amplified and inserted into the pSET2 vector. After transformation into E. coli DH5α for propagation, the recombinant plasmid was electroporated into mutant competent cells.
To express the recombinant TCS09RR protein, the coding region of TCS09RR was cloned using PCR with the primers TCS09RRpColdF and TCS09RRpColdR (Table S3). After digestion with respective endonucleases, the sequence was inserted into the pCold II vector with the same sites. The recombinant pCold II-TCS09RR plasmid was verified by sequencing and transformed into BL21(DE3) cells.
Growth kinetics assay under different temperatures.
Strains in the logarithmic phase were diluted to an optical density at 600 nm (OD600) of 0.01 in THB. The cultures were then incubated at 37°C and 25°C with shaking, and the OD600 was determined at 1-h intervals for 12 h. Each growth curve was derived from at least three independent experiments.
Phagocytosis and swine blood killing assay.
Phagocytosis assays were performed as reported previously, with some modifications (55). The strains in logarithmic phase were washed three times with PBS and incubated with macrophages for 1.5 h at a bacterium-to-cell ratio of 100:1. After being washed three times with PBS, the infected cells were incubated in DMEM containing penicillin (5 μg/ml) and gentamicin (100 μg/ml) for another 1.5 h. The macrophages were then washed three times with PBS and lysed with water. Serial dilutions of the cell lysate were plated onto THA and incubated overnight at 37°C. All experiments were repeated three times.
A swine blood killing assay was performed to investigate bacterial survival in the presence of whole swine blood. The strains in logarithmic phase were washed three times with PBS and suspended in PBS to an OD600 of 0.1. After 0.1 ml of bacterial suspension was added to 0.9 ml of whole swine blood, the mixtures were incubated at 37°C with occasional moderate shaking. An aliquot of the sample was removed at 0, 1, and 3 h, and the serial dilutions were plated on THA. All experiments were repeated three times.
Murine infection assay.
Mouse survival assays were performed as described previously (20). Ten mice in each group were challenged by intraperitoneal injection with the strain at a dose of 5 × 108 CFU/mouse and monitored for signs until 7 days postinfection. The negative-control group was challenged with an equal volume of sterile PBS. To evaluate bacterial proliferation in vivo, a bacterial burden assay was conducted. Six mice in each group were inoculated with 3 × 108 CFU of the strain, and the blood, liver, kidney, and brain were harvested, weighed, and homogenized in PBS at 6 h postinfection. Subsequently, the homogenized samples were serially diluted and plated on THA to enumerate CFU.
To examine the expression of TCS09RR and capsule genes in the host before and after SS2 entered the blood circulation, six mice were challenged by intraperitoneal injection with 1 ml of the ZY05719 strain at a dose of 5 × 108 CFU/mouse. Ascitic fluid was collected from three mice at 2 h postinfection, while blood was harvested from another three mice at 6 h postinfection. RNA next was isolated and cDNA was synthesized for qRT-PCR.
Transcriptomic analysis.
RNA was extracted from the WT and mutant strains to generate the transcriptome library, which were sent to Novogene (Tianjin, China) for RNA-Seq analysis. The Illumina HiSeq 2000 platform was used as described previously (56). Each strain was analyzed based on three biological replicates. TopHat2 software was used to map the transcriptome reads against the reference sequence of ZY05719, and the Cuffdiff program was used to identify differentially expressed genes. To control the false discovery rate in the transcriptome data, comparisons with q values of ≤0.05 and estimated fold changes of ≥2 were declared significant.
Transmission electron microscopy.
TEM was utilized to analyze the morphological changes of the strains using a known method (57). In brief, the strains in logarithmic phase were centrifuged at 5,000 × g for 10 min and then fixed in 2.5% glutaraldehyde for more than 2 h. The specimens then were dehydrated in propylene oxide for 10 min and embedded in epoxy resin. Finally, the sections were examined with a transmission electron microscope (Hitachi H-7650) at an accelerating voltage of 200 kV. The experiments were performed three times independently.
Preparation of surface proteins and SDS-PAGE analysis.
The strains were centrifuged at 12,000 × g for 10 min at 4°C, washed twice with PBS, and then resuspended in 4 ml sample preparation solution (30 mM Tris-HCl [pH 7.5], 25% sucrose, 125 U/ml mutanolysin [Sigma], 3 mM MgCl2). After incubating the samples at 37°C for 1.5 h, the cell lysate was centrifuged at 12,000 × g for 10 min at 4°C and precipitated in 10% prechilled (4°C) trichloroacetic acid. The samples then were incubated in ice water for 0.5 h and centrifuged at 12,000 × g for 10 min at 4°C. Ten milliliters of chilled acetone was used to wash the pellet twice, which was then air dried. The proteins were then separated on a 12% SDS-PAGE gel.
Electrophoretic mobility shift assay.
E. coli BL21 with pCold II-TCS09RR was grown in 200 ml of LB overnight at 16°C, and protein expression was induced with the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma-Aldrich). Recombinant TCS09RR was purified using Ni-nitrilotriacetic acid spin columns (GE Healthcare) and dialyzed overnight at 4°C. The BProm program (SoftBerry) was used to predict the promoter regions, and a fragment from 16S rRNA was used as the negative control. The promoter variants were obtained by overlap extension PCR using the upstream and downstream sequences of the repeats. DNA probes were obtained by PCR amplification using specific primers (Table S3) and purified using a gel extraction kit (Vazyme). Increasing amounts of dialyzed TCS09RR protein (0 to 500 ng) were added to the DNA probe (100 ng) in binding buffer (10 mM Tris, 50 mM KCl, 50 mM MgCl2, 0.1 mM dithiothreitol, 10% glycerol, pH 7.4) at 37°C for 30 min. The reaction mixtures were loaded onto a 6% polyacrylamide gel in 0.5× TBE buffer (44.5 mM Tris base, 44.5 mM boric acid, 1 mM EDTA, pH 7.4) and electrophoresed at 200 V for 45 min. Finally, the gel was stained in 0.5× TBE buffer containing ethidium bromide for 15 min and the image was recorded.
Cotranscription assay.
Total RNA was extracted and reverse transcribed to prepare cDNA as described above. The primers were designed to span the ORFs of TCS09RR-TCS09HK, TCS09HK-zy05719_08030, and zy05719_08030-zy05719_08035, as listed in Table S3. One microgram of total RNA without reverse transcription served as the negative control to confirm that the samples were free of contaminating DNA. The matching cDNA samples were used as PCR templates for the cotranscription assay.
Statistical analysis.
All experiments were performed at least 3 times. The Prism 5 software package was used to perform statistical analyses. The unpaired two-tailed Student's t test and the log-rank (Mantel-Cox) test were used to analyze the data. For all tests, a P value of <0.05 was considered statistically significant.
Data availability.
Raw data for RNA-Seq analyses are available in the NCBI database under accession number SRP193660.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Development Program of the Jiangsu Modern Agricultural Industry Technology System (no. JSTS[2018]011), Postgraduate Research & Practice Innovation Program of Jiangsu Province (no. KYCX18_0717), Shanghai Agriculture Applied Technology Development Program (no. G2016060201), the Special Fund for Public Welfare Industry of Chinese Ministry of Agriculture (no. 201303041), and the project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
We have no competing financial interests to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00377-19.
REFERENCES
- 1.West AH, Stock AM. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci 26:369–376. doi: 10.1016/S0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen CT, Park SS, Rhee DK. 2015. Stress responses in Streptococcus species and their effects on the host. J Microbiol 53:741–749. doi: 10.1007/s12275-015-5432-6. [DOI] [PubMed] [Google Scholar]
- 3.Hoch JA. 2000. Two-component and phosphorelay signal transduction. Curr Opin Microbiol 3:165–170. doi: 10.1016/S1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 4.Bonifait L, Veillette M, Letourneau V, Grenier D, Duchaine C. 2014. Detection of Streptococcus suis in bioaerosols of swine confinement buildings. Appl Environ Microbiol 80:3296–3304. doi: 10.1128/AEM.04167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goyette-Desjardins G, Auger JP, Xu J, Segura M, Gottschalk M. 2014. Streptococcus suis, an important pig pathogen and emerging zoonotic agent—an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microbes Infect 3:e45. doi: 10.1038/emi.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fittipaldi N, Segura M, Grenier D, Gottschalk M. 2012. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol 7:259–279. doi: 10.2217/fmb.11.149. [DOI] [PubMed] [Google Scholar]
- 7.Feng Y, Zhang H, Ma Y, Gao GF. 2010. Uncovering newly emerging variants of Streptococcus suis, an important zoonotic agent. Trends Microbiol 18:124–131. doi: 10.1016/j.tim.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Segura M, Calzas C, Grenier D, Gottschalk M. 2016. Initial steps of the pathogenesis of the infection caused by Streptococcus suis: fighting against nonspecific defenses. FEBS Lett 590:3772–3799. doi: 10.1002/1873-3468.12364. [DOI] [PubMed] [Google Scholar]
- 9.Zheng C, Li L, Ge H, Meng H, Li Y, Bei W, Zhou X. 2018. Role of two-component regulatory systems in the virulence of Streptococcus suis. Microbiol Res 214:123–128. doi: 10.1016/j.micres.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Chen C, Tang J, Dong W, Wang C, Feng Y, Wang J, Zheng F, Pan X, Liu D, Li M, Song Y, Zhu X, Sun H, Feng T, Guo Z, Ju A, Ge J, Dong Y, Sun W, Jiang Y, Wang J, Yan J, Yang H, Wang X, Gao GF, Yang R, Wang J, Yu J. 2007. A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS One 2:e315. doi: 10.1371/journal.pone.0000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Greeff A, Buys H, van Alphen L, Smith HE. 2002. Response regulator important in pathogenesis of Streptococcus suis serotype 2. Microb Pathog 33:185–192. doi: 10.1016/S0882-4010(02)90526-7. [DOI] [PubMed] [Google Scholar]
- 12.Li M, Wang C, Feng Y, Pan X, Cheng G, Wang J, Ge J, Zheng F, Cao M, Dong Y, Liu D, Wang J, Lin Y, Du H, Gao GF, Wang X, Hu F, Tang J. 2008. SalK/SalR, a two-component signal transduction system, is essential for full virulence of highly invasive Streptococcus suis serotype 2. PLoS One 3:e2080. doi: 10.1371/journal.pone.0002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan X, Ge J, Li M, Wu B, Wang C, Wang J, Feng Y, Yin Z, Zheng F, Cheng G, Sun W, Ji H, Hu D, Shi P, Feng X, Hao X, Dong R, Hu F, Tang J. 2009. The orphan response regulator CovR: a globally negative modulator of virulence in Streptococcus suis serotype 2. J Bacteriol 191:2601–2612. doi: 10.1128/JB.01309-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Tan C, Zhou Y, Fu S, Hu L, Hu J, Chen H, Bei W. 2011. The two-component regulatory system CiaRH contributes to the virulence of Streptococcus suis 2. Vet Microbiol 148:99–104. doi: 10.1016/j.vetmic.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Han H, Liu C, Wang Q, Xuan C, Zheng B, Tang J, Yan J, Zhang J, Li M, Cheng H, Lu G, Gao GF. 2012. The two-component system Ihk/Irr contributes to the virulence of Streptococcus suis serotype 2 strain 05ZYH33 through alteration of the bacterial cell metabolism. Microbiology 158:1852–1866. doi: 10.1099/mic.0.057448-0. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Shen X, Zhao Y, Wang M, Zhong Q, Chen T, Hu F, Li M. 2012. Identification and proteome analysis of the two-component VirR/VirS system in epidemic Streptococcus suis serotype 2. FEMS Microbiol Lett 333:160–168. doi: 10.1111/j.1574-6968.2012.02611.x. [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Fu S, Liu M, Xu Q, Bei W, Chen H, Tan C. 2014. The two-component system NisK/NisR contributes to the virulence of Streptococcus suis serotype 2. Microbiol Res 169:541–546. doi: 10.1016/j.micres.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Yuan F, Tan C, Liu Z, Yang K, Zhou D, Liu W, Duan Z, Guo R, Chen H, Tian Y, Bei W. 2017. The 1910HK/RR two-component system is essential for the virulence of Streptococcus suis serotype 2. Microb Pathog 104:137–145. doi: 10.1016/j.micpath.2016.12.026. [DOI] [PubMed] [Google Scholar]
- 19.Chang P, Li W, Shi G, Li H, Yang X, Xia Z, Ren Y, Li Z, Chen H, Bei W. 2018. The VraSR regulatory system contributes to virulence in Streptococcus suis via resistance to innate immune defenses. Virulence 9:771–782. doi: 10.1080/21505594.2018.1428519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong X, Zhang Y, Zhu Y, Dong W, Ma J, Pan Z, Roy S, Lu C, Yao H. 2018. The two-component signaling system VraSRss is critical for multidrug resistance and full virulence in Streptococcus suis serotype 2. Infect Immun 86:e00096-18. doi: 10.1128/IAI.00096-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olivera BC, Ugalde E, Martinez-Antonio A. 2010. Regulatory dynamics of standard two-component systems in bacteria. J Theor Biol 264:560–569. doi: 10.1016/j.jtbi.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Hermsen R, Erickson DW, Hwa T. 2011. Speed, sensitivity, and bistability in auto-activating signaling circuits. PLoS Comput Biol 7:e1002265. doi: 10.1371/journal.pcbi.1002265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Kustu S, Stewart V. 1994. In vitro interaction of nitrate-responsive regulatory protein NarL with DNA target sequences in the fdnG, narG, narK and frdA operon control regions of Escherichia coli K-12. J Mol Biol 241:150–165. doi: 10.1006/jmbi.1994.1485. [DOI] [PubMed] [Google Scholar]
- 24.Ansaldi M, Simon G, Lepelletier M, Mejean V. 2000. The TorR high-affinity binding site plays a key role in both torR autoregulation and torCAD operon expression in Escherichia coli. J Bacteriol 182:961–966. doi: 10.1128/jb.182.4.961-966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atkinson MR, Blauwkamp TA, Ninfa AJ. 2002. Context-dependent functions of the PII and GlnK signal transduction proteins in Escherichia coli. J Bacteriol 184:5364–5375. doi: 10.1128/jb.184.19.5364-5375.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wayne KJ, Li S, Kazmierczak KM, Tsui HC, Winkler ME. 2012. Involvement of WalK (VicK) phosphatase activity in setting WalR (VicR) response regulator phosphorylation level and limiting cross-talk in Streptococcus pneumoniae D39 cells. Mol Microbiol 86:645–660. doi: 10.1111/mmi.12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguilar PS, Hernandez-Arriaga AM, Cybulski LE, Erazo AC, de Mendoza D. 2001. Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J 20:1681–1691. doi: 10.1093/emboj/20.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fabret C, Hoch JA. 1998. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol 180:6375–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin PK, Li T, Sun D, Biek DP, Schmid MB. 1999. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J Bacteriol 181:3666–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Throup JP, Koretke KK, Bryant AP, Ingraham KA, Chalker AF, Ge Y, Marra A, Wallis NG, Brown JR, Holmes DJ, Rosenberg M, Burnham MK. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol Microbiol 35:566–576. doi: 10.1046/j.1365-2958.2000.01725.x. [DOI] [PubMed] [Google Scholar]
- 31.Senadheera MD, Guggenheim B, Spatafora GA, Huang YC, Choi J, Hung DC, Treglown JS, Goodman SD, Ellen RP, Cvitkovitch DG. 2005. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J Bacteriol 187:4064–4076. doi: 10.1128/JB.187.12.4064-4076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith HE, Damman M, van der Velde J, Wagenaar F, Wisselink HJ, Stockhofe-Zurwieden N, Smits MA. 1999. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect Immun 67:1750–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith HE, de Vries R, van't Slot R, Smits MA. 2000. The cps locus of Streptococcus suis serotype 2: genetic determinant for the synthesis of sialic acid. Microb Pathog 29:127–134. doi: 10.1006/mpat.2000.0372. [DOI] [PubMed] [Google Scholar]
- 34.Segura M, Gottschalk M, Olivier M. 2004. Encapsulated Streptococcus suis inhibits activation of signaling pathways involved in phagocytosis. Infect Immun 72:5322–5330. doi: 10.1128/IAI.72.9.5322-5330.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elliott SD, Tai JY. 1978. The type-specific polysaccharides of Streptococcus suis. J Exp Med 148:1699–1704. doi: 10.1084/jem.148.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson TL, Jeffers J, Rapp-Gabrielson VJ, Martin S, Klein LK, Lowery DE, Fuller TE. 2007. A novel signature-tagged mutagenesis system for Streptococcus suis serotype 2. Vet Microbiol 122:135–145. doi: 10.1016/j.vetmic.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 37.Van Calsteren MR, Gagnon F, Lacouture S, Fittipaldi N, Gottschalk M. 2010. Structure determination of Streptococcus suis serotype 2 capsular polysaccharide. Biochem Cell Biol 88:513–525. doi: 10.1139/o09-170. [DOI] [PubMed] [Google Scholar]
- 38.Salamov VSA, Solovyevand A. 2011. Automatic annotation of microbial genomes and metagenomic sequences, p 61–78. In Li RW, Metagenomics and its applications in agriculture. Nova Science Publishers, Hauppauge, NY. [Google Scholar]
- 39.Guo J, Wang Q, Wang X, Wang F, Yao J, Zhu H. 2015. Horizontal gene transfer in an acid mine drainage microbial community. BMC Genomics 16:496. doi: 10.1186/s12864-015-1720-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin M, Lu J, Chen Z, Nguyen SH, Mao L, Li J, Yuan Z, Guo J. 2018. Antidepressant fluoxetine induces multiple antibiotics resistance in Escherichia coli via ROS-mediated mutagenesis. Environ Int 120:421–430. doi: 10.1016/j.envint.2018.07.046. [DOI] [PubMed] [Google Scholar]
- 41.Cybulski LE, del Solar G, Craig PO, Espinosa M, de Mendoza D. 2004. Bacillus subtilis DesR functions as a phosphorylation-activated switch to control membrane lipid fluidity. J Biol Chem 279:39340–39347. doi: 10.1074/jbc.M405150200. [DOI] [PubMed] [Google Scholar]
- 42.Najle SR, Inda ME, de Mendoza D, Cybulski LE. 2009. Oligomerization of Bacillus subtilis DesR is required for fine tuning regulation of membrane fluidity. Biochim Biophys Acta 1790:1238–1243. doi: 10.1016/j.bbagen.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 43.Charland N, Harel J, Kobisch M, Lacasse S, Gottschalk M. 1998. Streptococcus suis serotype 2 mutants deficient in capsular expression. Microbiology 144:325–332. doi: 10.1099/00221287-144-2-325. [DOI] [PubMed] [Google Scholar]
- 44.Chabot-Roy G, Willson P, Segura M, Lacouture S, Gottschalk M. 2006. Phagocytosis and killing of Streptococcus suis by porcine neutrophils. Microb Pathog 41:21–32. doi: 10.1016/j.micpath.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 45.Wu Z, Wu C, Shao J, Zhu Z, Wang W, Zhang W, Tang M, Pei N, Fan H, Li J, Yao H, Gu H, Xu X, Lu C. 2014. The Streptococcus suis transcriptional landscape reveals adaptation mechanisms in pig blood and cerebrospinal fluid. RNA 20:882–898. doi: 10.1261/rna.041822.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lalonde M, Segura M, Lacouture S, Gottschalk M. 2000. Interactions between Streptococcus suis serotype 2 and different epithelial cell lines. Microbiology 146:1913–1921. doi: 10.1099/00221287-146-8-1913. [DOI] [PubMed] [Google Scholar]
- 47.Benga L, Goethe R, Rohde M, Valentin-Weigand P. 2004. Non-encapsulated strains reveal novel insights in invasion and survival of Streptococcus suis in epithelial cells. Cell Microbiol 6:867–881. doi: 10.1111/j.1462-5822.2004.00409.x. [DOI] [PubMed] [Google Scholar]
- 48.Talbot UM, Paton AW, Paton JC. 1996. Uptake of Streptococcus pneumoniae by respiratory epithelial cells. Infect Immun 64:3772–3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gottschalk M, Segura M. 2000. The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet Microbiol 76:259–272. doi: 10.1016/S0378-1135(00)00250-9. [DOI] [PubMed] [Google Scholar]
- 50.Shin D, Lee EJ, Huang H, Groisman EA. 2006. A positive feedback loop promotes transcription surge that jump-starts Salmonella virulence circuit. Science 314:1607–1609. doi: 10.1126/science.1134930. [DOI] [PubMed] [Google Scholar]
- 51.Federle MJ, McIver KS, Scott JR. 1999. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J Bacteriol 181:3649–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Takamatsu D, Osaki M, Sekizaki T. 2001. Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid 46:140–148. doi: 10.1006/plas.2001.1532. [DOI] [PubMed] [Google Scholar]
- 54.Takamatsu D, Osaki M, Sekizaki T. 2001. Construction and characterization of Streptococcus suis-Escherichia coli shuttle cloning vectors. Plasmid 45:101–113. doi: 10.1006/plas.2000.1510. [DOI] [PubMed] [Google Scholar]
- 55.Mitterstiller AM, Haschka D, Dichtl S, Nairz M, Demetz E, Talasz H, Soares MP, Einwallner E, Esterbauer H, Fang FC, Geley S, Weiss G. 2016. Heme oxygenase 1 controls early innate immune response of macrophages to Salmonella Typhimurium infection. Cell Microbiol 18:1374–1389. doi: 10.1111/cmi.12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilhelm BT, Landry JR. 2009. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods 48:249–257. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 57.Hyams C, Yuste J, Bax K, Camberlein E, Weiser JN, Brown JS. 2010. Streptococcus pneumoniae resistance to complement-mediated immunity is dependent on the capsular serotype. Infect Immun 78:716–725. doi: 10.1128/IAI.01056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data for RNA-Seq analyses are available in the NCBI database under accession number SRP193660.