Abstract
Background
Carbon ion (12C) radiotherapy is becoming very promising to kill highly metastatic cancer cells keeping adjacent normal cells least affected. Our previous study shows that combined PARP-1 inhibition with 12C ion reduces MMP-2,-9 synergistically in HeLa cells but detailed mechanism are not clear. To understand this mechanism and the rationale of using PARP-1 inhibitor with 12C ion radiotherapy for better outcome in controlling metastasis, we investigated metastatic potential in two non-small cell lung cancer (NSCLC) A549 and H1299 (p53-deficient) cells exposed with 12C ion in presence and absence of PARP-1 inhibition using siRNA or olaparib.
Methods
We monitored cell proliferation, in-vitro cell migration, wound healing, expression and activity of MMP-2, − 9 in A549 and p53-deficient H1299 cell lines exposed with 12C ion with and without PARP-1 inhibitor olaparib/DPQ. Expression and phosphorylation of NF-kB, EGFR, Akt, p38, ERK was also observed in A549 and H1299 cells exposed with 12C ion with and without PARP-1 inhibition using siRNA or olaparib. We also checked expression of few marker genes involved in epithelial-mesenchymal transition (EMT) pathways like N-cadherin, vimentin, anillin, claudin-1, − 2 in both NSCLC. To determine the generalized effect of 12C ion and olaparib in inhibition of cell’s metastatic potential, wound healing and activity of MMP-2, − 9 was also studied in HeLa and MCF7 cell lines after 12C ion exposure and in combination with PARP-1 inhibitor olaparib.
Results
Our experiments show that 12C ion and PARP-1 inhibition separately reduces cell proliferation, cell migration, wound healing, phosphorylation of EGFR, Akt, p38, ERK resulting inactivation of NF-kB. Combined treatment abolishes NF-kB expression and hence synergistically reduces MMP-2, − 9 expressions. Each single treatment reduces N-cadherin, vimentin, anillin but increases claudin-1, − 2 leading to suppression of EMT process. However, combined treatment synergistically alters these proteins to suppress EMT pathways significantly.
Conclusion
The activation pathways of transcription of MMP-2,-9 via NF-kB and key marker proteins in EMT pathways are targeted by both 12C ion and olaparib/siRNA. Hence, 12C ion radiotherapy could potentially be combined with olaparib as chemotherapeutic agent for better control of cancer metastasis.
Electronic supplementary material
The online version of this article (10.1186/s12885-019-6015-4) contains supplementary material, which is available to authorized users.
Keywords: Non-small-cell lung cancer, Carbon ion exposure, PARP-1, Matrix metalloproteinases (MMPs), Epithelial-mesenchymal transition (EMT), Metastatic potential
Background
Lung cancer is one of the leading causes of death of cancer patient worldwide [1]. Most of the newly diagnosed lung cancers are non-small-cell lung cancer (NSCLC) whose prognosis is very poor. Unfortunately, its survival rate is extremely low because of developing chemo-resistance and metastasis [2]. Although ionizing radiation is widely established standard radiotherapy against NSCLC, but a number of reports show the increase of malignant trait after gamma irradiation [3, 4]. However, these limitations are overcome in hadrontherapy especially using carbon ion (12C) which has been established as a promising modality to treat cancer [5, 6]. Epithelial-mesenchymal transition (EMT) is a key event in metastasis - the major cause of death of cancer patients. EMT is normally found during embryogenesis and is taken by cancer cells resulting proliferation of multiple malignant trait with alteration of marker genes characteristic to this process [7]. During EMT, there is secretion and activation of Matrix metalloproteinases (MMPs) that cleave extracellular matrix (ECM) to help these transformed cells to invade or migrate [8]. Unlike to gamma, 12C ion radiotherapy reduces invasiveness and metastatic potential although the detailed mechanisms are still unresolved [9–11].
Now-a-days, combined therapy is found to be more efficient than single mode treatment and radio-chemotherapy is becoming very promising for its better outcomes. Since Poly(ADP-ribose) polymerase-1 (PARP-1) is well-known active candidate in DNA repair, pharmacological inhibitors of PARP (iPARP) has long been used as chemosensitizer or radiosensitizer after gamma radiation [12–14]. The iPARP alone or in combination with other chemotherapeutic drugs is showing good results in clinical trials for treatment of various cancers [15–17]. Although, role of PARP-1 in metastasis is poorly understood but few reports show that PARP-1 can modulate marker genes of EMT pathway like vimentin and E-cadherin [18]. Again, iPARP alone is effective in cancer-types having defective homologous recombination (HR) pathways of repair. Can olaparib (an inhibitor of PARP-1 and PARP-2, approved after clinical trial) in combination with 12C ion radiotherapy be a better modality to control cancer metastasis irrespective of its efficiency in repair pathways? Actually, very few studies have been done so far combining chemotherapy with 12C ion radiotherapy [19, 20].
Notably, our previous report shows that combined 12C ion and PARP-1 inhibition induces apoptosis, reduces MMP-2/− 9 activity in a synergistic manner in HeLa cells although the detailed mechanism is not clear [20, 21]. To extend this work, we looked into cell migration, wound healing, MMP-2/− 9 activities in several human cancer cells A549, H1299, HeLa and MCF7 cells. Detailed signaling pathways of transcriptional regulation of MMP-2/− 9 and expression of key marker genes in EMT pathways were investigated in NSCLC A549 and H1299 cells after exposure with 12C ion in presence and absence of PARP-1 inhibition using iPARP (olaparib or DPQ) or siRNA.
Methods
Chemicals
Trypan blue, bovine serum albumine (BSA) and gelatin were purchased from Sigma-Aldrich, USA. Olaparib and 3, 4-dihydro-5-[4-(1-piperidinyl) butoxy]-1(2H)-isoquinolinone (DPQ) from Selleckchem, UK. Trypsin, fetal bovine serum (FBS), antibiotic solution and DMEM were purchased from HiMedia, India. The detail information about antibodies used is given in Additional file 1. Other reagents and bio-chemicals were purchased locally.
Cell culture, treatment with iPARP and siRNA against PARP-1
Human NSCLC A549, human breast adenocarcinoma MCF7 and human cervical cancer cell HeLa, normal lung cell line L-132 were obtained from National Centre for Cell Sciences, Pune, India. H1299 (ATCC CRL-5803) cells were obtained from ATCC. All the cells were grown in DMEM medium supplemented with 10% fetal bovine serum (FBS), (complete medium) at 37 °C in a humidified incubator (5% CO2) [20]. The cell lines were directly used after purchase. ATCC and NCCS performs authentication of cell lines via short tandem repeat (STR) profiling analysis. So, we did not carry out additional testing to authenticate the cell lines, but its morphology and behavior were consistent with ATCC and NCCS descriptions. The cell lines were found to be without mycoplasma contamination after testing in our laboratory.
The cell lines used in this study did not require any ethical approval for their use.
The cells were treated with 1 μM of iPARP (DPQ or olaparib) 4 h before irradiation. After irradiation the cells were grown in presence of iPARP.
Equal number of cells were counted and seeded (appox. 70%) in 35 mm petridish for transfection experiment. After overnight incubation, the media were discarded and replaced with Opti-MEM, a reduced serum media (Invitrogen, life technologies). Using lipofectamine RNAiMAX Reagent (Invitrogen, life technologies) cells were transfected with Silencer Select siRNA (Ambion, life technologies) specific for PARP-1 and also with a negative control (scrambled version) according to manufacturer’s protocol. Irradiation was performed for transfected cells 48 h post transfection.
Carbon ion (12C) irradiation
The cells were irradiated with 12C ion using the 15UD Pelletron. 12C ion with energy 85 MeV (equivalent to 7.08 MeV/nucleon) from the accelerator was used but the energy of 12C ion on the cell surface was 62 MeV (equivalent 5.16 MeV/nucleon). The corresponding entrance LET from the accelerator was 290 keV/μm, which was calculated using SRIM software. Irradiation was done following the protocol described in [19] and dose in Gray (Gy) was calculated from the particle fluence using the standard relation
Clonogenic cell survival
Clonogenic cell survival was done following the protocol described in [19].
Cell proliferation assay
About 106 cells were seeded in 35 mm plate and allowed to grow for 24 h. Then the cells were exposed with 12C ion with and without iPARP. The cells were trypsinized after 24 h of 12C ion exposure. Then, 3 × 103 cells were seeded in 96 well plates with culture medium having 10% FBS. The plates were then incubated for 24 h, 48 h, 72 h and 96 h at 37 °C in a humidified incubator (5% CO2) in presence or absence of iPARP. After incubation MTT assay were performed and absorbance was measured using a microplate reader (Victor X5).
Wound healing assay
About 1X 106 number of cells were seeded on 35 mm plates and allowed to grow for 24 h in CO2 incubator. A scratch was made by a tip in each plate in a similar way after cells were irradiated with 12C ion with and without iPARP. To study the wound healing property of the cells after such treatment, the plates were photographed immediately after treatment (0 h) and after 24 h and 48 h under light microscope (Carl Zeiss, Germany). Wound healing assay was done in A549, H1299, MCF7 and in HeLa cells after treatment.
Cell migration assay
Cell migration assay was done in A549 and H1299 cells using the protocol described in Entschladen et al. 2005 [22]. Briefly, 0.3X106 cells were seeded in 35 mm petri plates and grown in complete medium for 22 h. After incubation, cells were treated with iPARP olaparib at a dose of 1 μM in serum free medium. The cells were exposed to 12C ion after 4 h treatment of iPARP. After irradiation, plates were incubated for 24 h in presence of iPARP. Post Incubation, medium was discarded and cells were trypsinized and counted using hemocytometer. 2 × 105 cells were seeded in the upper chamber of Thin Cert™ multi-well plate having pore size of 8 μm, in serum free medium. Then, 2 ml medium containing 10% serum was added in lower chamber of the multi well plates and the cells were incubated for 19 h at at 37 °C in a humidified incubator. After incubation, medium from both the lower and upper chamber of the multi well plates was discarded and the cells in lower chamber were trypsinized and counted using hemocytometer to determine the number of cells migrated to the lower chamber from the upper porous membrane. The experiment was done in triplicates and was repeated thrice.
Gelatin Zymography
Gelatin zymography was done in A549, H1299 and MCF7 cells according to our previously described protocol in [21]. Briefly, 0.3 × 106 cells were seeded in complete medium and incubated at 37 °C for 22 h. After incubation, medium was discarded and cells were treated with 1 μM olaparib in serum free medium. Cells were irradiated with 12C ion 4 h post treatment of iPARP and then incubated for 24 h at 37 °C in presence of iPARP. Then, the medium was collected and centrifuged at 3000 g at 4 °C for 10 min to remove cells and other debris from the collected medium. Then the supernatant was further cleared by centrifugation with same speed. The supernatant was collected without disturbing pellet and concentrated using Amicon ultra cenrifugal protein concentrator (UFC901024, cut off 10 kDa) by centrifugation at 4600 g for 10 min at 4 °C and the total protein concentration was estimated using Victor X5 (Invitrogen). Equal amount (15 μg) of protein from each sample was resolved in 10% polyacrilamide gel containing 0.1% gelatin as a substrate. Then the gel was washed twice with 2.5x triton-X and was incubated for 19 h at 37 °C in developing buffer. The gel was then stained with Coomassie Brilliant Blue followed by overnight destaining. The gels were photographed and intensity of bands was measured using imageJ software.
RNA extraction and Q RT- PCR
Total RNA was isolated from the treated and untreated cells using TRIzol Reagent (Invitrogen; life technologies) following standard protocol [23]. Relative expression of the genes involved in EMT pathway was studied using SYBR green master mix. The primer list is given in S1 (See Additional file 1).
Western blot
Western blot was done following the protocol as described in [21]. Expression of MMP-2 and MMP-9 was measured both from the whole cell lysate and from the culture medium (secreted). All the secondary antibodies are HRP-conjugated and bands are developed in Luminata Crescendo western HRP substrate (Millipore) and image was taken in Chemidoc (Chemidoc XRS+ system, Biorad). The concentrations of various antibodies used are given in S2 and S3 in Additional file 1.
Image analysis
The intensities of the gel images were determined using imageJ software [21].
Statistical analysis
The statistical significance of the treated samples was evaluated with respect to the untreated one using ANOVA with Dunnett’s test from IBM SPSS Statistics Version 21 software. The values of significance were denoted as ‘*’ (0.01 < p ≤ 0.05), ‘**’ (0.001 < p ≤ 0.01) and ‘***’ (p ≤ 0.001). All the experiments were performed at least in triplicates.
In all figures, ‘D’ or ‘O’ means only 1 μM of DPQ or olaparib respectively; 0.5, 1, 2 represent dose of 12C ion in Gy; DPQ or olaparib combined with various doses of 12C are represented as D + 1, D + 2 etc. or O + 1, O + 2 etc.
Results
Clonogenic cell survival and cell proliferation
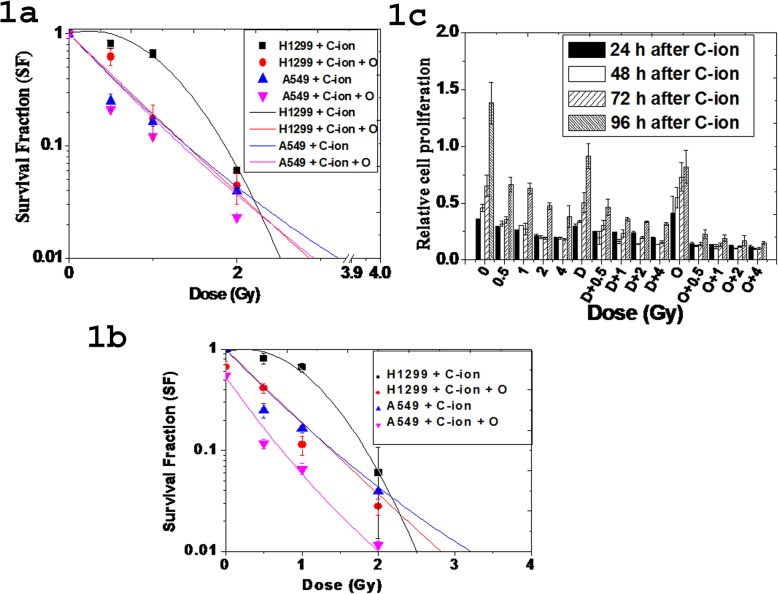
Cell survival fraction was reduced in a dose-dependent manner in both A549 and H1299 cells treated with 12C ion with or without olaparib as shown in Fig. 1a. Notably, A549 cells (p53 wild-type) were more sensitive compared with H1299 (p53 mutant) cells below 2 Gy. 12C ion produces DNA breaks in both A549 and H1299 cells. However, wild-type p53 drives the A549 cells to cell cycle arrest and apoptosis. So, cell survival fraction (SF) is low in A549 than that of H1299 for 12C ion treatment only. H1299 cells escapes from cell cycle arrest and apoptosis due to absence of p53. So, the colony forming ability of H1299 is higher than that of A549 cells after 12C treatment only. The H1299 cells escaped from cell cycle arrest or apoptosis, fail to repair after 12C irradiation due to olaparib treatment and thus olaparib treatment makes H1299 cells sensitive to 12C ion. In Fig. 1b, we represented the data where SF of untreated control is 1 and all other treated cells (either 12C ion or olaparib or combined) show SF values less than 1 because each treatment gives some toxicity to the each type of cells (A549 and H1299). Only olaparib treatment reduced SF to about 35–40% in both the cell-types. But, olaparib combined with 12C ion reduced more than that obtained by 12C ion alone.
Fig. 1.
a Survival fraction (SF) of A549 and H1299 cells exposed with 12C ion with and without olaparib (O). The survival fraction (SF) of olaparib treated cells at 0 Gy was corrected to 1 and all the olaparib treated cells were normalized accordingly. b represent the same data where survival fraction of untreated control was assumed as 1 and of all other treated cells (either 12C ion or olaparib or combined) normalized accordingly. c Time course cell proliferation in A549 cells after treatment with PARP-1 inhibitor DPQ (D) and olaparib (O) separately and in combination with 12C ion exposure (0–4 Gy). Each bar represents mean cell proliferation ± standard deviation obtained from three independent experiments. All the differences with respect to control are significant and p-values at each dose were p ≤ 0.001 in each cell-type
DPQ treatment alone reduced SF in a dose-dependent manner as shown in S4(B) (Additional file 1). However, DPQ combined with 12C ion always reduced SF more compared with single treatment. Either iPARP (olaparib or DPQ) or 12C ion exposure alone significantly reduced cell proliferation in a dose-dependent manner at different time interval (24 h – 96 h) as shown in Fig. 1c. Combined treatment significantly (p < 0.001) reduced cell proliferation at all time intervals (24 h – 96 h). Notably, olaparib sensitized A549 cells more than that obtained by DPQ within the dose range used in our experimental condition.
Wound healing, in-vitro cell migration and activity of MMP-2,-9 by gelatin zymography assay
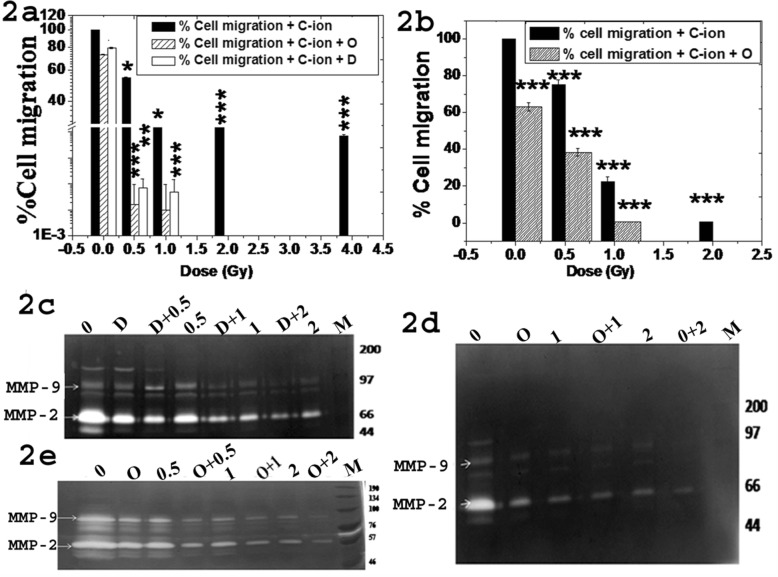
Both 12C ion and olaparib treatment alone significantly reduced wound healing in A549 (Additional file 1: S5), H1299 (Additional file 1: S6), HeLa (Additional file 1: S7) and MCF7 cells (Additional file 1: S8). Combined treatment showed synergistic effect. In-vitro cell migration was reduced dose-dependently at all doses (p < 0.001) in both A549 (Fig. 2a) and H1299 (Fig. 2b) cells after exposure with 12C ion alone. However, the in-vitro cell migration was drastically reduced to below 10% of untreated control at as low as 0.5 Gy 12C ion combined with 1 μM of iPARP and the cell migration was further reduced to almost nil at higher dose of 12C ion in presence of iPARP in both cell lines. So, combined iPARP and 12C ion exposure reduces wound healing and cell migration synergistically in various human cancer cells.
Fig. 2.
Cell migration and MMP-2, − 9 activity in A549 and H1299 cells. Percent cell migration after PARP-1 inhibition with olaparib (O), DPQ (D) and combined with 12C ion in A549 (a) and H1299 (b) cells. Each bar with different pattern represents mean percent cell migration with standard deviation obtained from three independent experiments in triplicates. c-d-e Typical photograph of gelatin zymogram to determine MMP-2 (72 kDa) and MMP-9 (92 kDa) activity after exposure with 12C in presence and absence of DPQ (d) (c) or olaparib (O) (d) in A549 cells and H1299 (e) cells
Activity of MMP-2 and MMP-9 was reduced to more than 45% after 1 μM of iPARP treatment alone in A549, H1299 and MCF7 cells (Additional file 1: S9). Combined treatment further reduced MMPs activity in all three cell-types as shown in Fig. 2c-e. MMP-2/− 9 activity as determined from the densitometry analysis from three independent zymogram and the loading control for each zymogram is shown in (Additional file 1: S10). This data implicates that olaparib or DPQ alone significantly reduces MMP-2, − 9 activities and combined treatment further reduces MMPs activity in all cell-types. So, reduction of MMP-2,-9 activity by single or combined treatment is not cell-specific but a generalized phenomenon irrespective of p53 status in cells.
Signaling pathways of EGFR/Akt/p38/ERK involved in the transcriptional regulation of MMP-2 and MMP-9
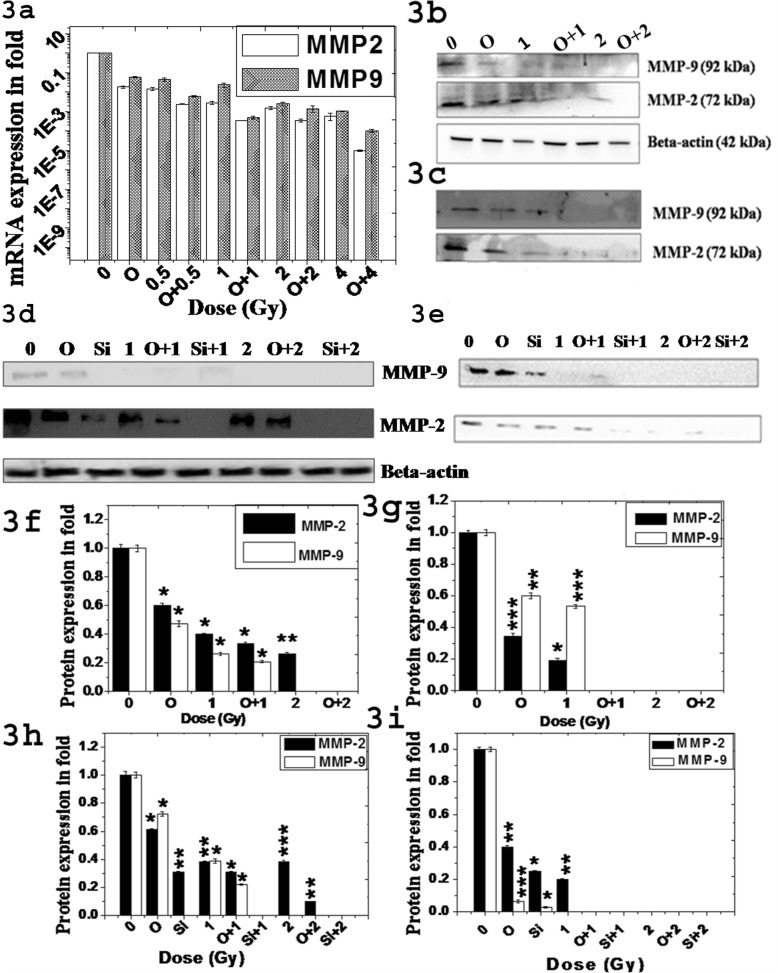
We checked expression of these two MMPs and their secretion in culture medium during cell growth of A549 and H1299. Dose-dependent decrease of these two MMPs expression in RNA level was detected by real time PCR after single and combined treatment in A549 cells as shown in Fig. 3a. Notably, only olaparib or DPQ treatment reduced almost 40–50% expressions of these MMPs. Reduction of these MMPs were more than 80% in siPARP-1 cells (PARP-1 knockdown). We further confirmed such reduction of MMPs expression using western blot in both A549 and H1299 cells as shown in Fig. 3b and d respectively and their relative expression as measured in fold as shown in Fig. 3f and h respectively. We used total proteins secreted in serum free medium in western blot to check MMPs secretion as shown in Fig. 3c (A549) & e (H1299) respectively and their relative expression as measured in fold as shown in Fig. 3g and i respectively, which followed the same pattern as that of Fig. 3b & d. Thus, the secretion of these MMPs remained unaltered after single or combined treatment in each cell-type. However reduction of MMPs observed in medium was due to reduction of expression of those proteins inside the cells.
Fig. 3.
Expression of MMP-2 and MMP-9 in A549 and H1299 cells treated with 12C with and without olaparib (O). a Relative mRNA expression of MMP-2 and MMP-9 in A549 cells after 24 h treatment as determined by Real Time PCR. Each bar represents mean expression ± standard deviation obtained from three independent experiments. All the differences with respect to control are significant and p-values at each dose was p ≤ 0.001 in each cell-type. b-c & d-e Typical western blot to determine expression of MMP-2 and MMP-9 from whole cell lysate in b (A549) & d (H1299) and their respective secretion in culture medium in c (A549) & e (H1299). f,g, h & i Relative expression as measured in fold of MMP-2 and MMP-9 from whole cell lysate f (A549) & h (H1299) and their respective secretion in culture medium in g (A549) & i (H1299)
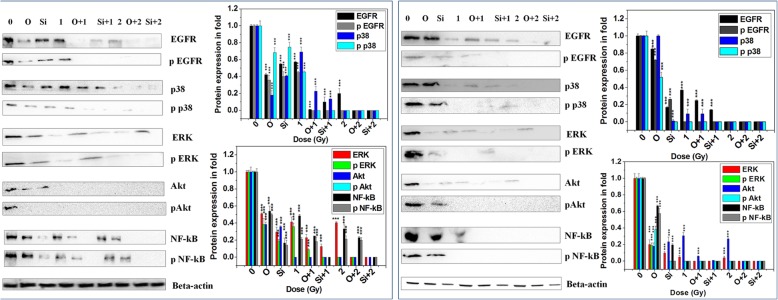
We checked the signaling pathways of up-stream of transcriptional regulation of these MMPs in both A549 and H1299 cells as shown in Fig. 4. Treatment with iPARP or siRNA significantly reduced NF-kB expression (well-known transcription factor of MMPs) but reduction was more for siRNA. 12C ion exposure alone also reduced NF-kB and combined treatment just abolished NF-kB expression. Hence phospho-NF-kB was also reduced after either single treatment or combined treatment. Olaparib/siRNA or 12C ion treatment alone reduced all four proteins (except p38 in A549 after olaparib treatment) EGFR, ERK1/2, Akt and p38 in both A549 and H1299 cells but extent of reduction was varied in two cell-types as shown in Fig. 4. However, combined treatment always reduced synergistically all the proteins. This data implicates that PARP-1 inhibition or 12C ion exposure interferes with the EGFR/Akt, EGFR/p38 and EGFR/ERK1/2 signaling pathways and combined treatment completely inhibit all these pathways to inactivate NF-kB in both p53 wild-type and p53 mutant cells. Notably, olaparib treatment did not alter expression of NF-kB in normal lung cells L-132 as shown in Additional file 1. This data implicates that olaparib sensitizes cancer cells but not normal cells against 12C ion exposure.
Fig. 4.
Expression and phosphorylation status of the proteins involved in the upstream signaling pathway in the transcriptional regulation of MMP-2,-9 in H1299 (left panel) and A549 (right panel) cells after 12C ion exposure with and without olaparib (O) or siRNA against PARP-1. The bar graphs in each panel represents the quantitative analysis of the proteins as obtained from densitometric analysis of the bands from three independent experiments using imageJ
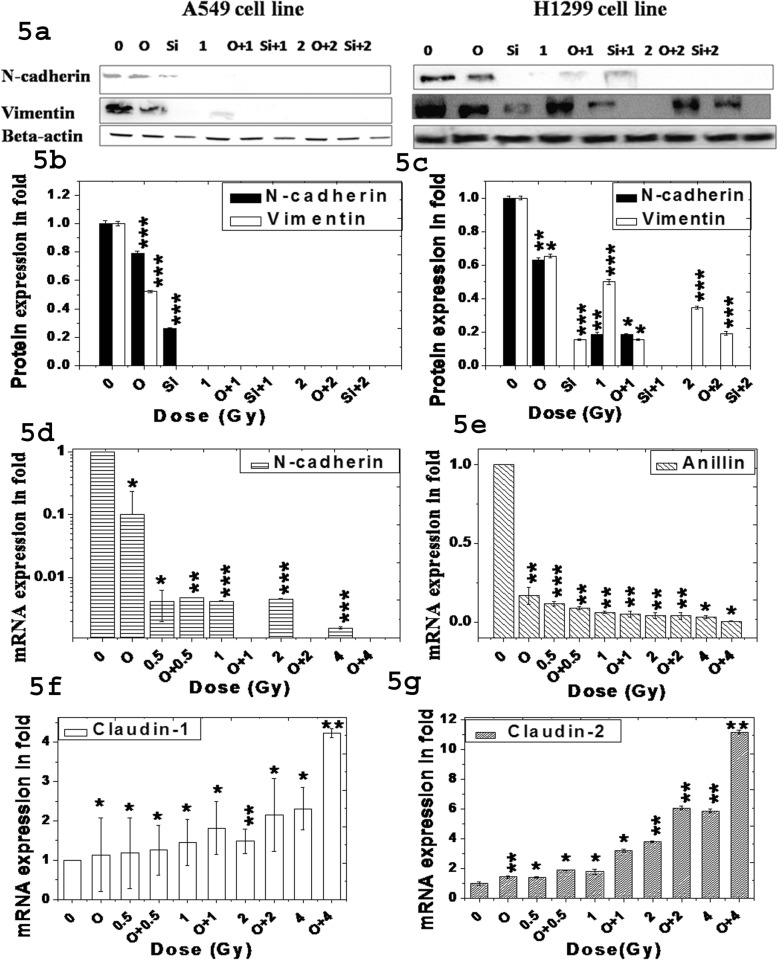
Expression of key proteins in the EMT pathways
We checked expression of key proteins involved in the EMT pathways such as N-cadherin, vimentin, anillin, claudin-1 and claudin-2 by western blot and real time PCR after single and combined treatment. N-cadherin and vimentin was decreased in A549 and H1299 cells after single or combined treatment as shown in Fig. 5a and their relative expression as measured in fold as shown in Fig. 5b and c respectively. This data was further supported by real time PCR data which indicated decrease of N-cadherin and increase of claudin-1 in RNA level after single or combined treatment as shown in Fig. 5d and f. Claudin-2 was enhanced and anillin was reduced significantly (p < 0.001) after treatment with olaparib or 12C ion separately or in combination as shown in Fig. 5g and e. This data implicates that single or combined treatment inhibits EMT pathway by altering these genes in both p53 wild-type A549 and p53 mutant H1299 cells.
Fig. 5.
Expression of few EMT markers after 12C ion exposure with and without olaparib or siRNA. a Typical western blots of N-cadherin and vimentin in A549 (left) and H1299 (right) cells. b & c Relative expression of N-cadherin and vimentin in A549 and H1299 cells as measured in fold. d, e, f & g Expression of the EMT markers N-cadherin, anillin, claudin-1 and claudin-2 respectively by Real Time PCR after normalizing with GAPDH in A549 cell line. All the differences with respect to control are significant and p-values at each dose were p ≤ 0.001
Discussion
Combined chemotherapy with radiotherapy has proven better outcomes than single mode but sufficient radiobiological information are lacking in radio-chemotherapy. Until recently, the research on iPARP has been concentrated on its role in DNA repair [24–26]. However, recent literature reports are indicating other novel functions of PARP-1 for which some of the iPARP are very much potent in vivo [27]. Incidentally, 12C ion reduces cell invasion and MMPs activity of various cancer cells [9, 21]. Here, we observe that PARP-1 inhibition (siRNA or iPARP) alone reduces expression and activity of MMP-2/− 9 in three cell-types and combined treatment with 12C ion further reduces their expression/activity. Earlier we reported that siPARP-1 HeLa cells showed reduced expression/activity of MMPs [21]. Thus reduction of MMPs expression/activity by single or combined treatment is not cell type specific. Although 12C ion induces cluster DNA damage and olaparib sensitizes cells to 12C ion. But, synergistic reduction of metastatic potential by combined treatment is not due to cytotoxicity. We observed that MMP-2, − 9 expressions were reduced about 60% and 80% after treatment with only 1 μM olaparib and 1 Gy 12C ion respectively after 2 h as shown in Additional file 1. However, there is no induction of apoptosis after either single treatment as shown upper panel of this file S13 in Additional file 1. Combined treatment almost abolished MMPs expression with little induction of apoptosis in terms of pro-caspase-3 activation. This data implicates that reduction of markers of metastasis is not dependent upon cytotoxicity in our experimental condition. Furthermore, Fig. 2a in our manuscript clearly shows that only olaparib treatment for 24 h reduces cell migration about 27%. But, at the same dose of olaparib the cell viability is about 90%. This data implicates that inhibition of in-vitro cell migration by single or combined treatment was not due to cytotoxicity.
Nuclear factor kappa B (NF-kB), AP1/AP2 and SP1 are known Transcription Factors (TFs) of MMP-2/− 9 [28–31]. We did not check status of AP1/AP2, SP1 here. Our data suggests that 12C ion exposure or PARP-1 inhibition reduces MMPs expression via reducing NF-kB in both p53 wild-type and p53 mutant cells. Functional modulation of NF-kB by PARP-1 is very complex in presence and absence of ionizing radiation and it is stimulus or cell-specific [32–34]. Other than cell proliferation and MMP’s activity, NF-kB can induce angiogenesis and anti-apoptotic genes making the cancer cells resistant to treatments [35] and hence targeting NF-kB has long been one of the good strategies for controlling cancer progression. Notably, olaparib treatment does not alter expression of NF-kB in normal lung cells L-132. In this respect, our proposed strategy of combining olaparib with 12C ion would target multiple pathways including NF-kB signaling to control metastatic potential of cancer cells without affecting normal cells.
Generally EGFR phosphorylation activates NF-kB via Akt, p38 or ERK1/2 pathways. Here, the expressions/phosphorylation of EGFR, Akt, p38 and ERK1/2 are reduced by iPARP / 12C ion separately in A549 and H1299. Hence, synergistic reduction of phosphorylation of all four proteins is observed in combined treatment to completely inactivate NF-kB. Although reduction of expression/phosphorylation of Akt and ERK1/2 by 12C ion is reported earlier in various cells in the context other than metastasis [36, 37], but here we report such reduction of these proteins as a part of inhibiting metastatic potential in cancer cells independent of p53 status. PARP-1 inhibition is reported to decrease phosphorylation of EGFR, p38, Akt, ERK1/2 in different cell lines [38–41]. But, our new observation is the combining olaparib with 12C treatment that synergistically reduces phosphorylation/activation of all four molecules leading to drastic reduction of in-vitro cell migration.
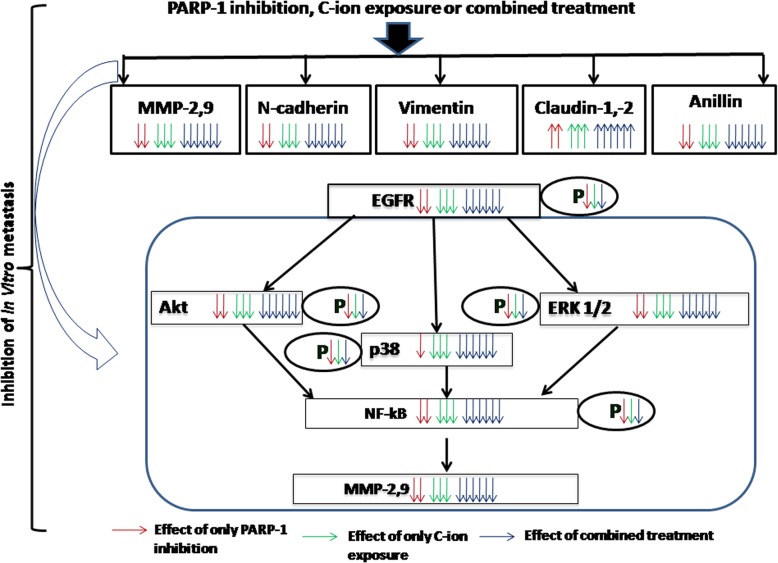
Role of 12C ion in EMT is largely an unexplored area. 12C ion reduces vimentin and anillin in different cells [9, 11]. Generally, N-cadherin, vimentin and anillin facilitates EMT whereas claudin-1, − 2 opposes EMT process. However, claudin may have complex role in metastasis [42]. Here, we report that 12C ion reduces N-cadherin, vimentin, anillin and increases claudin − 1, − 2 in NSCLC A549 and H1299 cells to reduce EMT process. In addition, combined 12C ion with PARP-1 inhibition (by siRNA or olaparib) synergistically increases claudin-1, − 2 expressions and decreases N-cadherin, vimentin, anillin. Role of PARP-1 as transcriptional modulator is well documented and it can alter expression of marker genes involved in EMT pathway [37, 43]. As such there is no report to show regulation of N-cadherin, claudin-1, − 2. Thus, decrease of N-cadherin, anillin and increase of claudin-1, − 2 by PARP-1 inhibition is our novel findings in the context of radio-chemotherapy using olaparib. So, the reduction of metastatic potential by 12C ion is potentiated by PARP-1 inhibition in cancer cells with wild-type or mutant p53 and the whole theme of our work is shown in Fig. 6.
Fig. 6.
The cartoon picture represents the mechanisms of inhibition of in-vitro metastatic potential after single and combined treatment. The downward or upward arrows with three different colours beside the protein represent down-regulation or up-regulation respectively of that protein for only PARP-1 inhibition (red), only 12C ion exposure (green) and combined treatment (blue). Phosphorylation of each protein is denoted by P within oval shape attached with the respective protein. The down arrow of different colours beside P denotes reduction of phosphorylation of the respective proteins after different treatments
Conclusions
12C ion with PARP-1 inhibition by olaparib/siRNA inhibits EGFR/Akt, EGFR/p38 and EGFR/ERK1/2 signaling pathways to reduce NF-kB-mediated MMP-2/− 9 expression. Some key markers in EMT pathway are also common target of 12C ion and iPARP/siRNA. That’s why combined treatment synergistically reduces metastatic potential in NSCLC irrespective of p53 status in it. Our radiobiological information establishes that combining olaparib as chemotherapeutic agent with carbon ion radiotherapy would be novel approach to treat highly metastatic lung cancer.
Additional file
Reduction of in-vitro cell migration and EMT pathway in non-small lung cancer cells treated with carbon ion alone and in combination with olaparib. (DOCX 9790 kb)
Acknowledgments
All the authors are thankful to IUAC for instrumental facility for carbon ion exposure. UG thanks DST-FIST for infrastructural facilities in the Department of Biochemistry & Biophysics, University of Kalyani.
Abbreviations
- DPQ
3, 4-dihydro-5-[4-(1-piperidinyl) butoxy]-1(2H)-isoquinolinone
- EMT
Epithelial-mesenchymal transition
- Gy
Gray
- MMPs
Matrix metalloproteinases
- PARP
Poly(ADP-ribose) polymerase
Authors’ contributions
UG conceived the whole study and framed all experiments. UG and PC critically analyzed the data. UG drafted the manuscript. PC carried out all the experiments. PD helped PC during cell exposure with carbon ion beam and also during western blot. SG helped PC during cell exposure with carbon ion beam. AS contributed his technical expertise in tuning the carbon ion beam with desired dose to irradiate the cells. All authors have read and approved the manuscript and ensure that this is the case.
Authors’ information
Priyanka Chowdhury, Payel Dey and Sourav Ghosh are Ph.D. scholars of the Department of Biochemistry & Biophysics, University of Kalyani, Kalyani- 741235, India.
Asitikantha Sarma, Ph.D., Scientist, Radiation Biology Laboratory, Inter-University Accelerator Centre, New Delhi-110067, India.
Funding
UG and PD are thankful to IUAC (IUAC/XIII.7/UFR-60329) for financial support. PC thanks ICMR (BMS/FW/CMB/2015–24000/JUN-2016/05/WB/GOVT) for her senior research fellowship. UG is thankful to CSIR (37(1674)/16/EMR-II) and DBT (BT/PR 4809/BRB/10/1028/2012) for the partial financial assistance.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29 Available from: http://onlinelibrary.wiley.com/doi/10.3322/caac.21208/full%5Cnhttps://www.ncbi.nlm.nih.gov/pubmed/24399786. [DOI] [PubMed]
- 3.Zhou YC, Liu JY, Li J, Zhang J, Xu YQ, Zhang HW, et al. Ionizing radiation promotes migration and invasion of cancer cells through transforming growth factor-beta-mediated epithelial-mesenchymal transition. Int J Radiat Oncol Biol Phys. 2011;81(5):1530–1537. doi: 10.1016/j.ijrobp.2011.06.1956. [DOI] [PubMed] [Google Scholar]
- 4.Ho JN, Kang GY, Lee SS, Kim J, Bae IH, Hwang SG, et al. Bcl-XLand STAT3 mediate malignant actions of γ-irradiation in lung cancer cells. Cancer Sci. 2010;101(6):1417–1423. doi: 10.1111/j.1349-7006.2010.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loeffier JS, Durante M. Charged particle therapy- optimization, challenges and future directions. Nat Rev Clin Oncol. 2013;10:411–424. doi: 10.1038/nrclinonc.2013.79. [DOI] [PubMed] [Google Scholar]
- 6.Allen C, Borak TB, Tsujii H, Nickoloff JA. Heavy charged particle radiobiology: using enhanced biological effectiveness and improved beam focusing to advance cancer therapy. Mutat Res. 2011;711(1–2):150–157. doi: 10.1016/j.mrfmmm.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamouille Samy, Xu Jian, Derynck Rik. Molecular mechanisms of epithelial–mesenchymal transition. Nature Reviews Molecular Cell Biology. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radisky ES, Radisky DC. Matrix metalloproteinase-induced epithelial-mesenchymal transition in breast cancer. J Mammary Gland Biol Neoplasia. 2010;15(2):201–212. doi: 10.1007/s10911-010-9177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akino Y. Carbon-ion beam irradiation effectively suppresses migration and invasion of human non-small-cell lung cancer cells. Int J Radiat Oncol Biol Phys. 2009;75(9):475–481. doi: 10.1016/j.ijrobp.2008.12.090. [DOI] [PubMed] [Google Scholar]
- 10.Moncharmont C, Guy J, Wozny A, Battiston-montagne P, Ardail D, Beuve M. Carbon ion irradiation withstands cancer stem cells ’ migration / invasion process in Head and Neck Squamous Cell Carcinoma (HNSCC). Oncotarget. 2016;7:47738-49. 10.18632/oncotarget.10281. [DOI] [PMC free article] [PubMed]
- 11.Kim EH, Kim M-S, Furusawa Y, Uzawa A, Han S, Jung W-G, et al. Metformin enhances the radiosensitivity of human liver cancer cells to γ–rays and carbon ion beams. Oncotarget. 2016;7(49):80568–80578. doi: 10.18632/oncotarget.12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calabrese C. R., Almassy R., Barton S., Batey M. A., Calvert A. H., Canan-Koch S., Durkacz B. W., Hostomsky Z., Kumpf R. A., Kyle S., Li J., Maegley K., Newell D. R., Notarianni E., Stratford I. J., Skalitzky D., Thomas H. D., Wang L.-Z., Webber S. E., Williams K. J., Curtin N. J. Anticancer Chemosensitization and Radiosensitization by the Novel Poly(ADP-ribose) Polymerase-1 Inhibitor AG14361. JNCI Journal of the National Cancer Institute. 2004;96(1):56–67. doi: 10.1093/jnci/djh005. [DOI] [PubMed] [Google Scholar]
- 13.Bridges KA, Toniatti C, Buser CA, Liu H, Buchholz TA, Meyn RE. Niraparib (MK-4827), a novel poly(ADP-Ribose) polymerase inhibitor, radiosensitizes human lung and breast cancer cells. Oncotarget. 2014;5(13):5076–86. Available from: https://www.ncbi.nlm.nih.gov/pubmed/24970803. [cited 2018 Apr 5] [DOI] [PMC free article] [PubMed]
- 14.Güster Julian David, Weissleder Stephanie Valerie, Busch Chia-Jung, Kriegs Malte, Petersen Cordula, Knecht Rainald, Dikomey Ekkehard, Rieckmann Thorsten. The inhibition of PARP but not EGFR results in the radiosensitization of HPV/p16-positive HNSCC cell lines. Radiotherapy and Oncology. 2014;113(3):345–351. doi: 10.1016/j.radonc.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 15.Lee J.-m., Ledermann J. A., Kohn E. C. PARP Inhibitors for BRCA1/2 mutation-associated and BRCA-like malignancies. Annals of Oncology. 2013;25(1):32–40. doi: 10.1093/annonc/mdt384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hay Trevor, Matthews James R., Pietzka Lucie, Lau Alan, Cranston Aaron, Nygren Anders O.H., Douglas-Jones Anthony, Smith Graeme C.M., Martin Niall M.B., O’Connor Mark, Clarke Alan R. Poly(ADP-Ribose) Polymerase-1 Inhibitor Treatment Regresses Autochthonous Brca2/p53-Mutant Mammary Tumors In vivo and Delays Tumor Relapse in Combination with Carboplatin. Cancer Research. 2009;69(9):3850–3855. doi: 10.1158/0008-5472.CAN-08-2388. [DOI] [PubMed] [Google Scholar]
- 17.Fong Peter C., Boss David S., Yap Timothy A., Tutt Andrew, Wu Peijun, Mergui-Roelvink Marja, Mortimer Peter, Swaisland Helen, Lau Alan, O'Connor Mark J., Ashworth Alan, Carmichael James, Kaye Stan B., Schellens Jan H.M., de Bono Johann S. Inhibition of Poly(ADP-Ribose) Polymerase in Tumors fromBRCAMutation Carriers. New England Journal of Medicine. 2009;361(2):123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 18.Su S, Lin X, Ding N, Zhang H, Zhang Q, Ding Y, et al. Effects of PARP-1 inhibitor and ERK inhibitor on epithelial mesenchymal transitions of the ovarian cancer SKOV3 cells. Pharmacol Rep. 2016;68(6):1225–1229. doi: 10.1016/j.pharep.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Ghorai Atanu, Bhattacharyya Nitai P., Sarma Asitikantha, Ghosh Utpal. Radiosensitivity and Induction of Apoptosis by High LET Carbon Ion Beam and Low LET Gamma Radiation: A Comparative Study. Scientifica. 2014;2014:1–10. doi: 10.1155/2014/438030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghorai A, Sarma A, Bhattacharyya NP, Ghosh U. Carbon ion beam triggers both caspase-dependent and caspase-independent pathway of apoptosis in HeLa and status of PARP-1 controls intensity of apoptosis. Apoptosis. 2015;20:562–580. doi: 10.1007/s10495-015-1107-3. [DOI] [PubMed] [Google Scholar]
- 21.Ghorai A, Sarma A, Chowdhury P, Ghosh U. PARP-1 depletion in combination with carbon ion exposure significantly reduces MMPs activity and overall increases TIMPs expression in cultured HeLa cells. Radiat Oncol. 2016;11:126. doi: 10.1186/s13014-016-0703-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Entschladen F, Drell TL, IV, Lang K, Masur K, Palm D, Bastian P, et al. Analysis methods of human cell migration. Exp Cell Res. 2005;307(2):418–426. doi: 10.1016/j.yexcr.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 23.Sadhukhan R, Chowdhury P, Ghosh S, Ghosh U. Expression of telomere-associated proteins is interdependent to stabilize native telomere structure and telomere dysfunction by G-Quadruplex ligand causes TERRA upregulation. Cell Biochem Biophys. 2017:1–9 Available from: https://www.ncbi.nlm.nih.gov/pubmed/218214750. [DOI] [PubMed]
- 24.Migwtl A, Alonso C, Quevedo C. Poly(ADP-ribose) polymerase-I (PARP-I) inhibitors in Cancer chemotherapy. Science (80- ) 2006;1(1):39–53. doi: 10.2174/157489206775246430. [DOI] [PubMed] [Google Scholar]
- 25.Malyuchenko NV, Kotova EY, Kulaeva OI, Kirpichnikov MP, Studitskiy VM. PARP1 Inhibitors: Antitumor drug design. Acta Nat. 2015;7(3):27–37. [PMC free article] [PubMed] [Google Scholar]
- 26.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol. 2011;5(4):387–393. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhen Y, Yu Y. Proteomic analysis of the downstream signaling network of PARP1. Biochemistry. 2018; acs.biochem.7b01022. Available from: https://www.ncbi.nlm.nih.gov/pubmed/29327913. [DOI] [PMC free article] [PubMed]
- 28.Mancini A, Di Battista JA. Transcriptional regulation of matrix metalloprotease gene expression in health and disease. Front Biosci. 2006;11(1):423–446. doi: 10.2741/1809. [DOI] [PubMed] [Google Scholar]
- 29.Aguilar-Quesada R, Muñoz-Gámez JA, Martín-Oliva D, Peralta-Leal A, Quiles-Pérez R, Rodríguez-Vargas JM, et al. Modulation of transcription by PARP-1: consequences in carcinogenesis and inflammation. Curr Med Chem. 2007;14(11):1179–1187. doi: 10.2174/092986707780597998. [DOI] [PubMed] [Google Scholar]
- 30.Hossain MB, Ji P, Anish R, Jacobson RH, Takada S. Poly(ADP-ribose) polymerase 1 interacts with nuclear respiratory factor 1 (NRF-1) and plays a role in NRF-1 transcriptional regulation. J Biol Chem. 2009;284(13):8621–8632. doi: 10.1074/jbc.M807198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaniolo Karine, Rufiange Anne, Leclerc Steeve, Desnoyers Serge, Guérin Sylvain L. Regulation of the poly(ADP-ribose) polymerase-1 gene expression by the transcription factors Sp1 and Sp3 is under the influence of cell density in primary cultured cells. Biochemical Journal. 2005;389(2):423–433. doi: 10.1042/BJ20041718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veuger SJ, Hunter JE, Durkacz BW. Ionizing radiation-induced NF-κB activation requires PARP-1 function to confer radioresistance. Oncogene. 2009;28(6):832–842. doi: 10.1038/onc.2008.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliver FJ, Ménissier-de Murcia J, Nacci C, Decker P, Andriantsitohaina R, Muller S, et al. Resistance to endotoxic shock as a consequence of defective NF-κB activation in poly (ADP-ribose) polymerase-1 deficient mice. EMBO J. 1999;18(16):4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, et al. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-κB-dependent transcription. J Biol Chem. 2005;280(49):40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- 35.Xia Y., Shen S., Verma I. M. NF- B, an Active Player in Human Cancers. Cancer Immunology Research. 2014;2(9):823–830. doi: 10.1158/2326-6066.CIR-14-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin X, Li F, Zheng X, Liu Y, Hirayama R, Liu X, et al. Carbon ions induce autophagy effectively through stimulating the unfolded protein response and subsequent inhibiting Akt phosphorylation in tumor cells. Sci Rep. 2015;5(September) Available from: 10.1038/srep13815. [DOI] [PMC free article] [PubMed]
- 37.Mitra AK, Bhat N, Sarma A, Krishna M. Alteration in the expression of signaling parameters following carbon ion irradiation. Mol Cell Biochem. 2005;276(1–2):169–173. doi: 10.1007/s11010-005-3903-5. [DOI] [PubMed] [Google Scholar]
- 38.Lai Y, Kong Z, Zeng T, Xu S, Duan X, Li S, et al. PARP1-siRNA suppresses human prostate cancer cell growth and progression. Oncol Rep. 2018;39(4):1901–1909. doi: 10.3892/or.2018.6238. [DOI] [PubMed] [Google Scholar]
- 39.Robaszkiewicz A, Valkó Z, Kovács K, Hegedus C, Bakondi E, Bai P, et al. The role of p38 signaling and poly(ADP-ribosyl)ation-induced metabolic collapse in the osteogenic differentiation-coupled cell death pathway. Free Radic Biol Med. 2014;76:69–79. doi: 10.1016/j.freeradbiomed.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 40.Wang D, Li C, Zhang Y, Wang M, Jiang N, Xiang L, Li T, Roberts TM, Zhao JJ, Cheng H, Liu P. Combined inhibition of PI3K and PARP is effective in the treatment of ovarian cancer cells with wild-type PIK3CA genes. Gynecol Oncol. 2016;142(3):548–556. doi: 10.1016/j.ygyno.2016.07.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Motta C, D’Angeli F, Scalia M, Satriano C, Barbagallo D, Naletova I, et al. PJ-34 inhibits PARP-1 expression and ERK phosphorylation in glioma-conditioned brain microvascular endothelial cells. Eur J Pharmacol. 2015;761:55–64. doi: 10.1016/j.ejphar.2015.04.026. [DOI] [PubMed] [Google Scholar]
- 42.Martin TA, Jiang WG. Loss of tight junction barrier function and its role in cancer metastasis. Biochim Biophys Acta Biomembr. 2009;1788(4):872–891. doi: 10.1016/j.bbamem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 43.Rodríguez María Isabel, Peralta-Leal Andreína, O'Valle Francisco, Rodriguez-Vargas José Manuel, Gonzalez-Flores Ariannys, Majuelos-Melguizo Jara, López Laura, Serrano Santiago, de Herreros Antonio García, Rodríguez-Manzaneque Juan Carlos, Fernández Rubén, del Moral Raimundo G., de Almodóvar José Mariano, Oliver F. Javier. PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant Transformation. PLoS Genetics. 2013;9(6):e1003531. doi: 10.1371/journal.pgen.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reduction of in-vitro cell migration and EMT pathway in non-small lung cancer cells treated with carbon ion alone and in combination with olaparib. (DOCX 9790 kb)
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.