Abstract
Neurobeachin (Nbea) is implicated in vesicle trafficking in the regulatory secretory pathway, but details on its molecular function are currently unknown. We have used Drosophila melanogaster mutants for rugose (rg), the Drosophila homolog of Nbea, to further elucidate the function of this multidomain protein. Rg is expressed in a granular pattern reminiscent of the Golgi network in neuronal cell bodies and colocalizes with transgenic Nbea, suggesting a function in secretory regulation. In contrast to Nbea−/− mice, rg null mutants are viable and fertile and exhibit aberrant associative odor learning, changes in gross brain morphology, and synaptic architecture as determined at the larval neuromuscular junction. At the same time, basal synaptic transmission is essentially unaffected, suggesting that structural and functional aspects are separable. Rg phenotypes can be rescued by a Drosophila rg+ transgene, whereas a mouse Nbea transgene rescues aversive odor learning and synaptic architecture; it fails to rescue brain morphology and appetitive odor learning. This dissociation between the functional redundancy of either the mouse or the fly transgene suggests that their complex composition of numerous functional and highly conserved domains support independent functions. We propose that the detailed compendium of phenotypes exhibited by the Drosophila rg null mutant provided here will serve as a test bed for dissecting the different functional domains of BEACH (for beige and human Chediak–Higashi syndrome) proteins, such as Rugose, mouse Nbea, or Nbea orthologs in other species, such as human.
Introduction
BEACH proteins are evolutionarily ancient, with one family member in the yeast Saccharomyces cerevisiae and varying numbers in all eukaryotes (Wang et al., 2000). Disruption of the yeast Bph1 (BEACH protein homolog) results in defective vacuolar protein sorting and aberrant cell wall formation without affecting vacuolar morphology (Shiflett et al., 2004). Neurobeachin (NBEA) codes for a brain enriched member of the BEACH protein family, which is defined by the BEACH–WD40 domains named after the mouse beige and the human Chediak–Higashi syndrome (Nagle et al., 1996; Wang et al., 2000). In the latter syndrome, the LYST (lysosomal trafficking regulator) protein is mutated and phenotypically manifests in enlarged lysosomes and lysosome-related organelles (Shiflett et al., 2002; Ward et al., 2002). Overall, it appears that mutations in BEACH proteins result in defective vesicle formation, fusion, or trafficking (De Lozanne, 2003; Kaplan et al., 2008; Albers et al., 2011; Volders et al., 2011). However, despite their abundance among eukaryotes and successful crystallization of the BEACH domain from NBEA (Jogl et al., 2002) and LRBA (LPS-responsive vesicle trafficking, beige-like anchor protein) (Gebauer et al., 2004), very little is known about BEACH protein function in general, and no molecular mechanisms have been discovered.
NBEA is a multidomain scaffolding protein with an AKAP (A-kinase anchor protein) domain and is localized near the trans-Golgi network (TGN) (Wang et al., 2000), supporting a function in vesicle sorting or secretion as demonstrated by experiments performed in Nbea knock-out mice and in vitro (Wang et al., 2000; Su et al., 2004; Medrihan et al., 2009; Castermans et al., 2010; Niesmann et al., 2011). Whereas NBEA is known for its neuronal function and its implication in autism spectrum disorders (Castermans et al., 2003; Volders et al., 2011), the human homolog LRBA has a broad expression pattern, and its function is currently unknown (Wang et al., 2001). Both NBEA and LRBA have been linked to cancer (Wang et al., 2001, 2004; O'Neal et al., 2009). In Drosophila, these proteins have a common ortholog, namely Rugose (Rg). Rg mutants display a rough eye phenotype primarily resulting from cone cell loss, defective ommatidial organization, absence of photoreceptors, and disorganized pigment cell lattice (Shamloula et al., 2002; Shamloula, 2003; Wech and Nagel, 2005). The typical organized structure of an ommatidium results from spatially restricted cell death depending on the balance between epidermal growth factor receptor (EGFR) and Notch signaling (Doroquez and Rebay, 2006). Rugose has been shown to interact genetically with components of the EGFR and Delta–Notch pathway (Schreiber et al., 2002; Shamloula et al., 2002; Wech and Nagel, 2005). In the past, human neurological disorders have been successfully modeled in Drosophila, providing valuable insights into disease mechanisms and protein functions (Bier, 2005; Bilen and Bonini, 2005).
Here, we introduce a Drosophila rg null mutant and characterize phenotypes affecting associative odor learning, brain morphology, and synaptic architecture/function. We used this phenotype compendium to dissociate the functional homology between Drosophila Rugose and mouse Nbea and thus provide a viable model to study molecular mechanisms of Nbea/Rugose in vivo.
Materials and Methods
Drosophila stocks.
The following stocks were used: rugose mutants rgγ1, rgγ5, rgγ6, rgP2, rgP4, and rgP6 (Shamloula et al., 2002); PBac(WH)rgf02898 and PBac(WH)f02325 (Exelixis Collection, Harvard Medical School, Boston, MA) for generation of rgFDD; and for galactosidase-4 (Gal4) lines, OK107–Gal4 (Connolly et al., 1996), GH298 (Stocker et al., 1997), GH146 (Heimbeck et al., 2001), MB247 (Zars et al., 2000), and N-synaptobrevin (Nsyb)–Gal4 (Pauli et al., 2008). rg1, Df(1)ovo41/FM6, elav–Gal4, and tub–Gal80ts were obtained from the Bloomington Drosophila Stock Center (Indiana University, Bloomington, IN). Stocks were rebalanced over fluorescent balancers whenever necessary.
Generation of a rugose null allele.
A null allele of rugose (hereafter called rgFDD) was generated with the flippase recognition target (FRT)-derived deletion (FDD) system (Parks et al., 2004). The stocks used were PBac(WH)rgf02898 and PBac(WH)f02325, respectively, located in rg and 3′ of the gene. Because rg is located on the X chromosome and deletion of the gene might be lethal in males, both male and female progeny (157 in total) were balanced again after activation of the flippase recombinase. Of 128 surviving stocks, one line carried the rg deletion. Recombination of the FRT sites was confirmed using inverse PCR with primers 5′-caccccactcacatgccatc-3′ and 5′-tccaagcggcgactgagatg-3′, 5′-acagaccgataaaacacatgcgtc-3′ and 5′-ggcatgtcggccattccaag-3′. DNA was isolated with the single fly genomic DNA preparation (Dr. Barry Dickson's Laboratory, Research Institute of Molecular Pathology, Vienne, Austria) with squishing buffer containing 10 mm Tris-HCl, pH 8, 1 mm EDTA, 25 mm NaCl, and 200 g/ml proteinase K (Roche). After a 30 min incubation at 37°C, proteinase K was inactivated by heating to 95°C for 3 min. After centrifugation, the supernatant was used for PCR applications.
cDNA synthesis and mRNA quantification.
RNA was isolated from whole flies using a phenol–chloroform extraction. cDNA was generated with the iScript Select cDNA Synthesis kit (qRT-PCR, random primers; Bio-Rad) or Superscript III First Strand Synthesis kit (rg cloning, oligo-dT primer, or a rg specific primer 5′ctatcgccagcgagagga-3′; Invitrogen) starting from 1 μg of RNA according to the protocol of the manufacturer. qRT-PCRs were performed using IQ SYBR Green Supermix (Bio-Rad) on a MyIQ single color real-time PCR detection system (Bio-Rad). The primers were designed with the web-based Roche Probefinder software. Primers used for the detection of rugose were 5′-atcgcgcgacagtagcat-3′ and 5′-aactacgataagggcaatgtgg-3′. Housekeeping genes used were RpL32 and αTub84B, respectively, with primers 5′-gctaagctgtcgcacaaatgg-3′, 5′-cggcgacgcactctgtt-3′ and 5′-caatatgcgtgaatgtatctctatcca-3′, 5′-ccagcaggcgtttccaat-3′.
Cloning of rugose and Nbea and generation of transgenic flies.
The full-length 10.5 kb cDNA of rugose [Rg forward; National Center for Biotechnology Information (NCBI) accession number NM_001144687, with additional in-frame insertion of 39 nucleotides “ttctaccagttgttctgtttctcgtttctcacgtggcag ” after nucleotide 1635 of the coding sequence, short form described by Han et al. (1997)] was cloned in three parts, i.e., a 5′-terminal part and two parts of ∼5 kb, starting from the cDNA of Canton-S Drosophila heads using AccuPrime Taq polymerase (Invitrogen). Primers used were, respectively, 5′-gaagcggccgccaccatggcggacatcatgcgt-3′, 5′-ggaaccaggtggtaaaggtg-3′ and 5′-cgccgtacagtgagatcaaa-3′, 5′-gaactcgagacgcgttcgctccttgca-3′ and 5′-gtgacttgaacgcgttacaatgtgggc-3′, 5′-gtgtctagattagtagcgctgctggta-3′. Restriction sites used are underlined or bold. Start and stop codons are in italics and bold. Overlap extension PCR was used to combine the first two fragments. Mutations were removed by flipping parts between clones and with the Quickchange Site-Directed Mutagenesis kit (Stratagene) according to the protocol of the manufacturer. The 5′ and 3′ fragments of ∼5 kb of the rugose cDNA were cloned in pUAST with NotI, XhoI and MluI, XbaI, respectively. To clone the full-length cDNA of mouse Nbea in pUAST, a new multiple cloning site was generated by annealing and ligation of oligos 5′-aattcgttaacagatctgcggccgcttgtatctgaccggtagtggctctgactatgtcctgatttggaggc3′ and 5′-tcgagcctccaaatcaggacatagtcagagccactaccggtcagatacaagcggccgcagatctgttaacg3′in pUAST digested with EcoRI and XhoI. The 5′ terminus of Nbea was generated from the construct described by Castermans et al. (2010) with primers 5′-ctaataaccggtatggcgagcgacaagccgggcccggggctag-3′ and 5′-gggaggcaatgcaattgccgcagcactacacccagg-3′ and cloned in pUAST with PinAI and AspI. The remaining part of Nbea was cloned with AspI and XhoI. Randomly inserted transgenic lines were generated by standard embryo injections (Duke University Model System Genomics, Durham, NC). Expression of the transgenes was referred to as the presence of rg+ and Nbea cDNA.
Rg antibody generation, sample preparation, and Western blot analysis.
For antisera production, a genomic DNA fragment overlapping the region encoding for amino acids 962–1445 from Rg forward (NCBI accession number NP_001138159) was PCR amplified using the primers:5′-accaatcaaacggcagctgttaaag-3′ and 5′-caaggttcggggtctttggttctac-3′ and cloned into the TOPO TA cloning vector (Invitrogen). By using the flanking EcoRI restriction sites, the PCR product was fused to the glutathione S-transferase coding sequence. A rabbit and rat were immunized with the resulting fusion protein (Pineda ABservice). Adult, 1- to 4-d-old flies were lysed (Mammalian Cell Lysis kit; Sigma), and protein concentrations were determined with a BCA protein assay (Pierce). β-Mercaptoethanol (0.04%) was added, and samples were heated for 10 min at 65°C. NuPAGE Novex 3–8% Tris-Acetate gels, Novex Tris-Acetate SDS running buffer, and HiMark prestained protein standard were used (all from Invitrogen). Blocking buffer [100 mm Tris/HCl, pH 7.4, 150 mm NaCl, 0.5% blocking reagent (Roche), and 0.2% Triton X-100] was used for blocking and antibody dilution. Primary antibodies used are rabbit anti-Rugose (1:2000; this study) and rabbit anti-Nbea (1:3000) (Castermans et al., 2010); mouse anti-actin (JLA20, 1:100; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) was used as a loading control. Secondary peroxidase-conjugated antibodies (1:2500) were from Dako. The antibodies directed against Rg or Nbea did not cross-react with Nbea or Rg, respectively, as assessed with immunohistochemistry or Western blot.
Immunohistochemistry.
Tissues were dissected in ice-cold PBS and fixed in 3.6% formaldehyde. After several washes with PBS, the tissues were incubated in PAXD (5% BSA, 0.3% sodium deoxycholate, and 0.3% Triton X-100 in PBS) for 20 min. Primary antibodies were diluted in PAXD and incubated overnight at 4°C. After washing multiple times with PAXD, tissues were incubated with secondary antibodies in PAXD for 2 h at room temperature. After several washes with PAXD, tissues were mounted in Vectashield mounting medium (Vector Laboratories). Larval filets for the visualization of the neuromuscular junction (NMJ) were dissected in HL3 (in mm: 110 NaCl, 5 KCl, 10 NaHCO3, 5 HEPES, 30 sucrose, 5 trehalose, and 10 MgCl2, pH 7.2), fixed in HL3 with 3.6% formaldehyde, and washed in PBS with 0.4% Triton X-100. Subsequently, filets were blocked in 10% normal goat serum, 0.25% BSA, and 0.4% Triton X-100 in PBS. Antibody incubations and subsequent washes were in 0.25% BSA and 0.4% Triton X-100 in PBS. Stainings were analyzed using a confocal microscope (Olympus Fluoview FV1000). Only tissues of male origin were dissected, with the exception of the larval filets for the bouton number. The following primary antibodies were used: mouse anti-fasciclin II (1:200; 1D4; Developmental Studies Hybridoma Bank), rabbit anti-GM130 (1:100; ab30637; Abcam), rabbit anti-HRP (1:500; Jackson ImmunoResearch), rabbit anti-Nbea (1:1000; Castermans et al., 2010), rat anti-Rugose, and rabbit anti-Rugose (1:1000; this study). Secondary Alexa Fluor 488-conjugated (1:500; larval filets; Invitrogen), FITC-conjugated, and Cy3-conjugated antibodies were diluted 1:200 (Jackson ImmunoResearch). Rhodamine phalloidin (1:2000; Invitrogen) was used to stain the muscles of larval filets.
Analysis of Gal4 expression patterns.
Whole-mount immunofluorescent preparation of the brain was described in a previous study (Thum et al., 2006). Microdissection was performed in Ringer's solution to remove cuticle and connective tissues. Brains were fixed in PBS (Sigma) containing 4% formaldehyde for 2 h on ice and subsequently rinsed with PBT (PBS, BSA, and Triton X-100) three times at room temperature. Blocking of samples was performed in 5% normal goat serum (Sigma) in PBT for 2 h; afterward, brains were incubated with the primary antibodies in blocking solution at 4°C for 2 d. The following primary antibodies were used: mouse monoclonal anti-synapsin 3C11 (1:10; Developmental Studies Hybridoma Bank) and rabbit polyclonal anti-GFP (1:1000; A6455; Invitrogen).Samples were washed three times for 10 min and once for 2 h with PBT and incubated with secondary antibodies in blocking solution at 4°C for 2 d. Goat anti-rabbit Alexa Fluor-488 (1:200; Invitrogen) and goat anti-mouse Alexa Fluor-647 (1:200; Invitrogen) were used for detection of the primary antibodies. After five 10-min rinses with PBT, brains were mounted in Vectashield (Vector Laboratories), and confocal image stacks were taken with a LeicaTCS SP5 at 1 μm steps with a 20× glycerol objective. The resulting image stacks were projected and analyzed with NIH ImageJ software; adjustments to contrast and brightness as well as rotations and organization of images were performed in Photoshop (Adobe Systems).
Quantification of the bouton number.
Flies were cultured at 25°C. Boutons were stained with anti-HRP and muscles were visualized with rhodamine phalloidin. Confocal z-stack images were taken from the synapse of NMJ 6/7 at abdominal segment A2 with a confocal microscope (Olympus Fluoview FV1000). Boutons were manually counted on projected images, and muscle surface area was measured with the NIH ImageJ software.
Electrophysiology.
Two-electrode voltage-clamp experiments were performed using sharp electrodes (10–15 MΩ) filled with 3 m KCl and fileted larvae bathed in modified HL3 (in mm 110 NaCl, 5 KCl, 10 NaHCO3, 5 HEPES, 30 sucrose, 5 trehalose, 10 MgCl2, and 0.5 CaCl2, pH 7.2). For excitatory junctional current (EJC) recordings, axons innervating segments A2–A3 of dissected larvae were electrically stimulated 50% above threshold, and for miniature EJCs (mEJCs) recordings were performed in the presence of 1 μm TTX. All recordings were made with an Axoclamp 900A (Molecular Devices) from muscle 6. Input resistances were ≥4 MΩ, and data were filtered using a 1 kHz Bessel filter and digitized at 10 kHz using a Digidata 1440A (Molecular Devices). Membrane resting potentials were between −65 and −75 mV and were clamped at −70 mV. EJC amplitudes were analyzed in Clampfit 10.3(Molecular Devices) and calculated by averaging the peak amplitude of 60 responses recorded at 0.2 Hz. mEJCs were analyzed using the Clampfit 10.3 (Molecular Devices) event-detection algorithm. All error bars indicate SEM.
Fly care and behavioral experiments.
All flies were raised at 24°C and 60% relative humidity with a 14/10 h light/dark cycle on cornmeal-based food (Guo et al., 1996). Genetic crosses were performed according to standard procedures, and we used male F1 progeny for behavioral experiments. Genotypes, including the Gal80ts repressor (McGuire et al., 2003), were raised at permissive temperature (19°C), placed at 30°C for 36 h to inactivate the Gal80ts repressor, and tested at room temperature after a 2 h rest period. A detailed description of the site-by-site comparison between aversive and appetitive odor memory can be found in the study by Schwaerzel et al. (2003). Specific modifications of that protocol applied to the olfactory cues used 3-octanole (1:100 dilution in mineral oil presented in a 14 mm cup) and 4-methylcyclohexanole (1:120 dilution in mineral oil presented in a 14 mm cup). Increased performance of appetitive memory is attributable to adopting an improved training apparatus (Colomb et al., 2009).
Statistical analyses.
Statistical analyses were performed with the Statistica 9.0 software package (StatSoft). For the quantification of mRNA levels, unpaired t tests were performed on ΔCt cycle threshold values to compare statistical difference with controls. For the quantification of bouton numbers, when appropriate, an unpaired t test or Mann–Whitney U test was performed for the comparison of two groups. One-way ANOVA with post hoc Tukey's tests or Kruskal–Wallis test with the multiple comparisons of mean ranks for all groups was used if more groups were compared. For morphological data on the mushroom bodies (MBs), Fisher's exact test was used on the raw data to assess statistical significance. Electrophysiological characteristics were compared using a Student's t test. Behavioral data were analyzed using one-way ANOVA with a post hoc test, including the Bonferroni's correction for multiple comparisons. The level of statistical significance was depicted as follows: *p < 0.05, **p < 0.01, and ***p < 0.001.
Results
rugose is a Drosophila homolog of mammalian Neurobeachin and LRBA
Drosophila rg is a large gene located on the X chromosome consisting of 30 exons distributed over 163 kb of genomic DNA. The locus codes for multiple isoforms and is organized in two translational units with their translation start sites located 134 kb apart (Fig. 1A). Multiple hypomorphic alleles of the Drosophila rg gene have been isolated that phenotypically manifest in a rough surface of the compound eye attributable to perturbation of the reiterated pattern of individual ommatidia (Shamloula et al., 2002). Here, we report the generation of an rg null allele by use of the FDD system (hereafter called rgFDD; Parks et al., 2004) (for details, see Materials and Methods). Starting from FRT-containing P-elements located within the rg gene 5′ of the second translational unit and 3′ of the rugose locus (Fig. 1A), the complete coding sequence of the second translational unit of rg including the 3′UTR was deleted. qRT-PCR (Fig. 1B), Western blot (Fig. 1C), and immunofluorescent staining of adult brains (Fig. 1D,E) confirmed that rgFDD is a null allele. Immunofluorescent staining of Drosophila brains revealed widespread distribution of Rg in neuronal tissue restricted to the cortical region of cell bodies sparing synaptic connections that are mainly located within neuropil regions in the adult (Fig. 1D) and larval stage (Fig. 1F). Costaining of Rg with the neuronal marker elav or the glial marker repo clearly demonstrated Rg expression in neurons but not in glia (Fig. 1G,H). Anti-Rg staining also displayed a granular pattern reminiscent of the Golgi network (Fig. 1I), a cellular structure that is dispersed throughout the cell in fruit flies (Kametaka et al., 2010). Using the cis-Golgi marker GM130, we demonstrated that Drosophila Rg resides near or within the Golgi complex (Fig. 1I). Rugose protein showed the same location by expression of rg+ cDNA in an rgFDD mutant background under control of the neuron-specific Nsyb–Gal4 driver (data not shown). Moreover, mouse Nbea is expressed more diffusely but is enriched at a site colocalized with endogenous Rugose when expressed under control of Nsyb–Gal4 (Fig. 1J).
Figure 1.
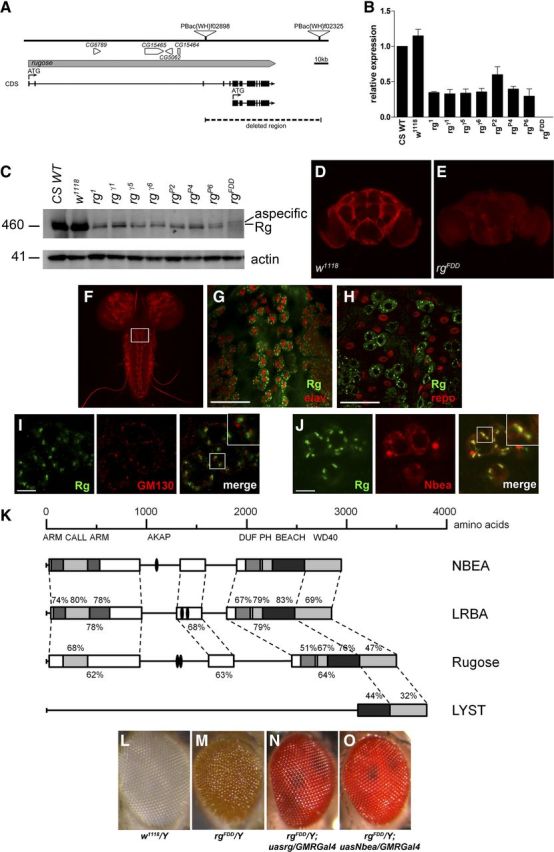
Generation of a Drosophila rugose −/− mutation. The rg locus is located on the X chromosome, and a precise deletion could be generated by remobilization of P-elements. A, Genomic structure of the rg locus. The rg gene is depicted in gray, and the coding sequence (CDS) is shown underneath. Translational start sites of the multiple isoforms are indicated with an arrow and “ATG. ” Other genes in this region are indicated in white, and CG numbers are mentioned. The P-elements, PBac(WH)f02898 and PBac(WH)f02325, used to generate the FDD allele of rg, are visualized as triangles and are located within the rg gene and 3′ of the gene, deleting the complete second translational unit, generating the rgFDD null allele. B, Relative rg mRNA levels extracted from adult male fly lysates, measured with qRT-PCR, confirm that the rg alleles generated by Shamloula et al. (2002) are hypomorphic alleles [p < 0.05, t < −10.109, df = 4, Student's t test for all mutants except rgP2 compared with w1118 or Canton-S wild type (CS WT)]. The rgFDD allele is a clear null (p < 0.001, t = −9.689, df = 4, Student's t test compared with w1118 or CS WT). The expression of each sample was normalized to αTub84B and relative to the mRNA expression in CS WT, which were arbitrarily set as 1. Data represent mean ± SEM; n = 3. C, Western blot of adult head extracts of male rg mutants, Canton-S wild-type (CS WT), and w1118 controls probed with an anti-Rugose antibody. Rugose expression is reduced in all rg mutants but is only absent in the rg null allele generated in this study. Actin was used as a loading control. D, E, Staining of adult brains with anti-Rg antibodies showing that endogenous Rg is localized in neuronal cell bodies and not in the neuropil regions. The antibody is specific because the staining is absent in rgFDD adult brains. F, Rugose staining of w1118 larval brain shows staining in the neuronal cell bodies, whereas the neuropil regions do not demonstrate Rg expression. G, H, Costaining of larval brain with antibodies against Rugose and the pan-neuronal marker elav (G) or the glial marker repo (H). Rugose is localized in neurons and not in glia. Scale bars: G, H, 40 μm. I, Endogenous Rugose in w1118 flies is located opposite of the cis-Golgi marker GM130 in the cell bodies of the dorsal medial cluster neurons. J, Confocal image of endogenous Rugose colocalizing with transgenic Nbea in UASNbea/+;NsybGal4/+ flies. Insets in I and J show a magnification of the boxed region in these images. Scale bars: I, J, 10 μm. K, Domain structure of NBEA, LRBA, Rugose, and LYST. The conserved regions are boxed, and domains are indicated in gray. Percentages represent the amino acid sequence identity. Concanavalin A-like lectin (CALL) domain, Armadillo (ARM) repeats, and AKAP domains were based on Burgess et al. (2009), Han et al. (1997), and Wang et al. (2000), respectively. NCBI reference sequences used are NP_056493.3 (NBEA), NP_006717.1 (LRBA), NP_001138159 [Rugose isoform forward, short isoform as described by Han et al. (1997)] and NP_000072.2 (LYST). DUF, Domain of unknown function. This schematic representation was derived and adapted from Wang et al. (2000) and Volders et al. (2011). L–O, The rough eye phenotype of rugose mutants can be rescued by expression of either transgenic rugose or Nbea in the Drosophila eye. w1118/Y eyes (L) show a smooth eye phenotype compared with the rough eye of rgFDD/Y (M). rgFDD/Y; UASrg/GMRGal4 (N) and rgFDD/Y; UASNbea/GMRGal4 (O) flies expressing the wild-type rg+ cDNA or Nbea cDNA in the eye of rgFDD mutants eye show an eye phenotype similar to wild type.
The Drosophila rugose gene encodes a protein with high homology to mouse Neurobeachin and LRBA. Specifically, Rg shares high sequence conservations within the BEACH, pleckstrin homology-like, WD40 repeats, armadillo repeats, and Concanavalin A-like lectin domains, suggesting that Drosophila Rg might be an ortholog or functional homolog of mammalian NBEA and/or LRBA (Fig. 1K). To test for that hypothesis, we aimed to generate appropriate transgenes for expression under control of the GAL/UAS system, but, because of the very large size and high GC content of part of the cDNA, we failed to generate a stable, full-length cDNA required to generate transgenic flies bearing the UAS–LRBA transgene. At the same time, targeting expression of UAS–rg+ or UAS–Nbea transgenes to the Drosophila compound eye rescued the rough eye phenotype of rugose mutant when ectopically expressed under control of eye-specific glass multiple reporter (GMR)–Gal4 (Fig. 1L–O). This result established functional homology between mouse Nbea and fly Rg at the level of retinal architecture.
Drosophila associative odor learning requires Rg/NBEA function
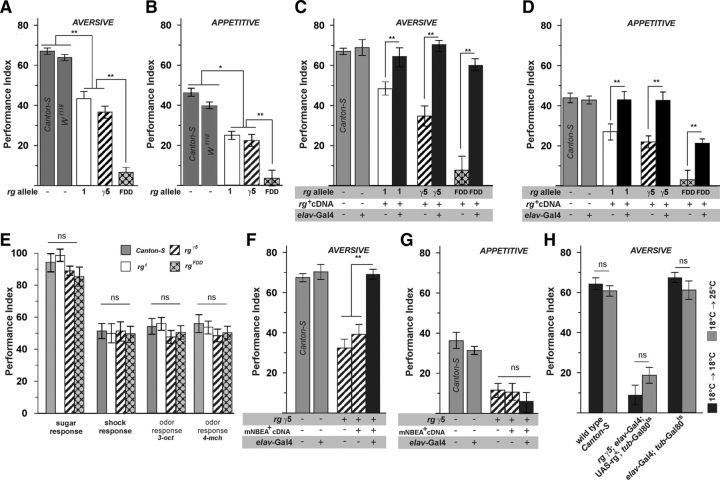
In mouse, Nbea is highly expressed within the hippocampus, a brain region functionally assigned to associative learning and memory (Wang et al., 2000). However, Nbea-deficient mice die shortly after birth because of respiratory problems; thus, the role of Nbea in associative learning is as yet untested (Su et al., 2004; Medrihan et al., 2009). To test whether Drosophila rugose functions in learning and memory, we used the well-established olfactory conditioning paradigm (Tully and Quinn, 1985). Here, aversive and appetitive memories can be compared in a site-by-site manner (Schwaerzel et al., 2003). The quality of the behavioral reinforcer used during the training procedure, i.e., either sugar reward for appetitive learning or electric shock punishment for aversive learning, influences the neuronal circuits (Krashes et al., 2007, 2009; Pitman et al., 2011) required to establish an appropriate memory at the level of the Drosophila MBs (Schwaerzel et al., 2003; Thum et al., 2007). When assayed for immediate aversive or appetitive memory performance, rg1 and rgγ5 animals show a significant reduction compared with wild-type Canton-S controls. At the same time, rgFDD mutants exhibit memory performance close to zero, thus significantly lower than w1118 controls (Fig. 2A,B). Decreased performance of rugose mutants was rescued by pan-neuronal expression of rg+ cDNA in appropriate rg mutant backgrounds (Fig. 2C,D). Aversive memory was rescued up to wild-type levels within background of either rg1, rgγ5, or rgFDD mutants (Fig. 2C). In contrast, appetitive memory was rescued completely within rg1 and rgγ5, whereas rgFDD yielded only partial but significant rescue (Fig. 2D). At the same time, we did not detect significant differences between sensory acuity of rg mutants and wild-type animals, thus excluding the trivial explanation that mutants are anosmic or unable to perceive either punitive or rewarding stimuli (Fig. 2E). Rather, these data show that Rg function is essential for associative odor learning per se rather than differentially required for either aversive or appetitive memory.
Figure 2.
Rugose mutants are defective in associative odor learning. The requirement of Rugose for associative learning was assayed in an olfactory conditioning paradigm, in which flies associate an odor cue with either aversive electric shock or appetitive sugar reward. Appropriate associative memories were immediately assayed after conditioning, and the performance index indicates percentage of correct choice, e.g., PI = 100 indicates that all animals made a correct choice; PI = 0 indicates that animals distributed randomly. We used rg1 and rgγ5 compared with its appropriate wild-type Canton-S reference, as well as rgFDD compared to w1118. A, B, Rugose mutant alleles rg1 and rgγ5 reduce immediate odor memory to approximately half the scores of Canton-S or w1118. rgFDD results in a complete loss of performance that reflects equal distribution of animals during the test situation. Rg mutants were equally impaired in either aversive (A: p < 0.01, F = 41.750, df = 51, ANOVA) or (B: p < 0.01, F = 39.709, df = 73, ANOVA) appetitive odor memory. C, D, Memory performance of rg mutants can be restored by neuron-specific expression of a wild-type UAS–rg+ cDNA within an otherwise rg mutant animal. Performance of aversive memory is completely restored within either allele of rg to levels indistinguishable from appropriate reference strains (p < 0.01, F = 29.589, df = 71, ANOVA when comparing appropriate rescues to Canton-S and w1118 shown in A). At the same time, appetitive memory is only partially rescued within an rgFDD background (D; p < 0.01, F = 15.682, df = 101, ANOVA). E, Sensory acuity of shock and sugar response, as well as odor response of rg mutants was indistinguishable from wild-type Canton-S (p > 0.05, F = 0.250, df = 45, ANOVA). F, G, Reduced performance of aversive, but not appetitive, odor memory, of rgγ5 mutants can be rescued by pan-neuronal expression of mammalian Nbea cDNA (F: p < 0.01, F = 26.127, df = 29, ANOVA; G: p < 0.01, F = 10.217, df = 36, ANOVA). H, Acute expression of rg+ cDNA at the adult stage prohibits the rescue of aversive odor memory (p > 0.05, F = 26.397, df = 59, ANOVA). All data represent mean ± SEM; n = 8–10 for associative behavior and n = 12–20 for non-associative controls (E).
To test whether mouse Nbea and fly Rg exhibit functional homology at the level of associative learning, we used pan-neuronal expression of Nbea cDNA in an rgγ5 mutant background because this is a strong hypomorph but yields many progeny that facilitates the behavioral experiments. Memory performance after aversive conditioning was fully rescued by mouse Nbea (Fig. 2F). In contrast, the rescue of appetitive conditioning failed (Fig. 2G). This result restricts a functional homology between mammalian Nbea and Drosophila Rugose to aversive but not to appetitive associative odor learning and directly suggests that different functions of those proteins might be engaged for either type of learning.
To test whether Rg is involved in the actual association process or alternatively required during development, we restricted expression of rescuing UAS–rg+ cDNA to the adult stage by use of the temperature-sensitive Gal80ts repressor (McGuire et al., 2003). Acute expression of rg+ cDNA at the adult stage 36 h before a behavioral experiment abolished rescue of aversive memory in rgγ5 animals (Fig. 2H). Although this result does not exclude a function of Rg throughout the associative process, it clearly indicates its requirement during development of the CNS.
Rugose supports odor learning at the level of the MBs
Olfactory information about the surrounding environment is internally represented by neural activity within the olfactory pathway of the brain. In Drosophila, this pathway follows a stereotyped organization orthologous to the mammalian olfactory bulb (Vosshall and Stocker, 2007). Airborne chemicals result in activation of olfactory receptor neurons, which project their axons to the antennal lobes. At this level, information is computed within the layers of local neurons (LNs) and projection neurons (PNs) and conveyed to higher olfactory centers, i.e., the lateral horn and the MBs (Fiala et al., 2002; Wang et al., 2003; Shang et al., 2007; Chou et al., 2010).
To probe which layer(s) of this circuitry require Rugose function for support of odor learning, we used a set of Gal4 lines, each driving expression in a particular neural layer, i.e., LNs (GH298–Gal4), PNs (GH146–Gal4), or MBs (MB247–Gal4, OK107–Gal4) (Fig. 3, see A for a schematic overview and B–E for expression patterns). When rg+ cDNA was expressed in the rgγ5 mutant background, we observed the rescue of aversive and appetitive odor learning on targeting expression to MBs but not to LNs or PNs (Fig. 3F,G). Thus, Rg supports odor learning at the level of MB intrinsic neurons, the Kenyon cells (KCs), regardless of the behavioral reinforcer used throughout the training procedure. This result suggests that the learning defect observed in rg mutants is not attributable to impaired computation of appropriate behavioral reinforcement but rather linked to memory formation at the level of KCs. Given the mandatory requirement of Rg function throughout development of the CNS, we sought to test for morphology of the MB structure.
Figure 3.
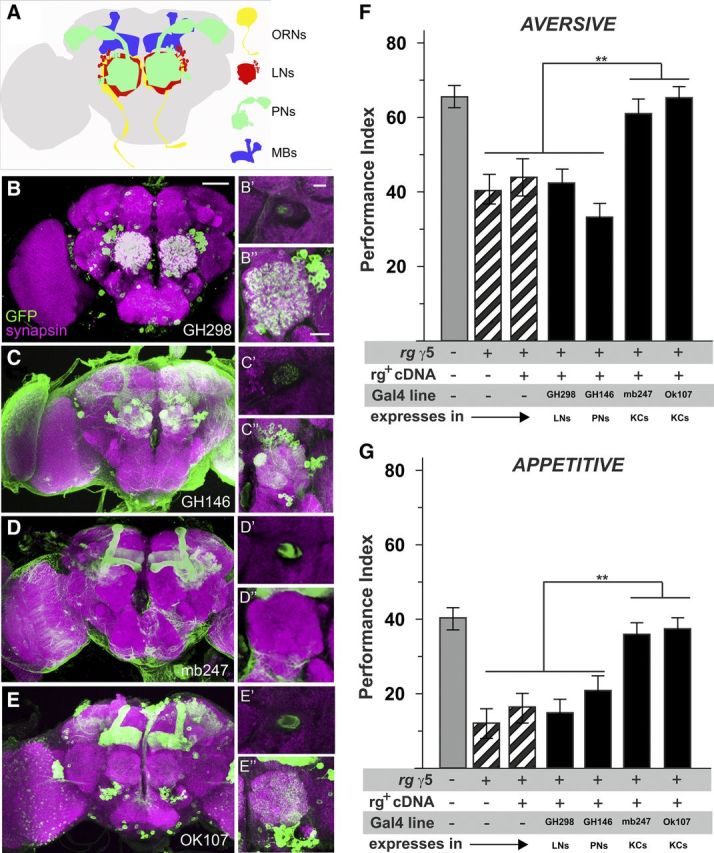
Rugose is required within KCs of the MBs to support immediate odor memory. Several Gal4 lines specific for different layers of the olfactory pathway were crossed to UAS–mCD8::GFP and double-stained by anti-GFP (green) and anti-synapsin (magenta) as a neuronal marker. Each main panel shows a frontal view of z-projections of 1 μm sections revealing the expression pattern of the driver line in the brain. For the insets, on the top right side, a single section focusing on the peduncle is shown, followed by a single section of the antennal lobe underneath. A, Schematic representation of the olfactory pathway: first-order olfactory receptor neurons (ORNs) convey odor information to the antennal lobe, which is innervated by GABAergic and cholinergic local interneurons. Second-order PNs distribute the olfactory information to higher brain centers, MB calyx, and lateral horn. MB intrinsic neurons, the KCs, represent third-order olfactory neurons and are presumed to be the site of a memory trace for aversive associative odor memory. B, GH298 shows staining mainly restricted to a set of local interneurons; limited MB expression is also seen at the center of the penduncle. C, GH146 specifically labels approximately two-thirds of the PNs. D, E, mb247 and OK107 Gal4 drives GFP expression particularly in a comprehensive set of the intrinsic MB KCs. F, G, Successful rescue of aversive and appetitive odor memory within rgγ5 background requires expression of rg+ cDNA at the level of MBs. At the same time, expression at the level of local interneurons (GH298–Gal4) or PNs (GH146–Gal4) was not successful (aversive: p < 0.01, F = 24.059, df = 34, ANOVA; appetitive: p < 0.01, F = 28.029, df = 44, ANOVA). All data represent mean ± SEM; n = 8–10.
Axonal morphology of KCs and impaired odor learning are pleiotropic defects in rg mutants that are genetically separable
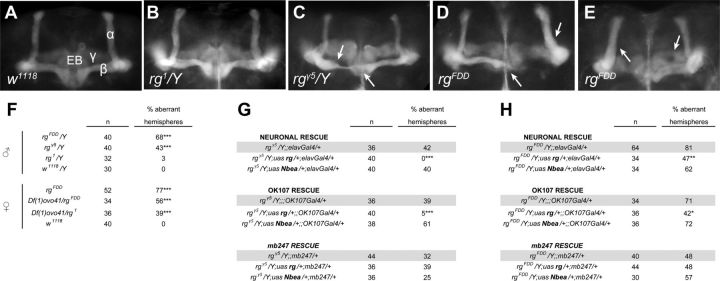
We visualized KCs with anti-fasciclin II staining (Fig. 4) and detected guidance defects within the fine structure of MB lobes, i.e., α/β, α′/β′, and γ lobes (Ito et al., 1997). Among the most prominent defects, we observed misguidance of α- or β-lobe axons, overextension of β-lobes, or β-lobe fusions as primary morphological abnormalities in rugose mutants (Fig. 4C--F). Additionally, a lower frequency of defects occurred, including lobe morphology, i.e., abnormal lobe thickness, aberrant morphology of the tips, or missing lobes, although the latter rather exceptionally.
Figure 4.
The morphological MB defects in rg mutants are genetically separable from the learning phenotype. Whole-mount anti-fasciclin II staining of the adult central brain labels α, β, γ MB lobes and the ellipsoid body (EB). A, Control w1118 MB lobes (α, β, γ) and EB. B, rg1 MBs show only minor defects compared with control brains. C, rgγ5 hemizygote with an aberrant morphology of the β lobe and β-lobe fusion. D, Overextension of the β lobe and full β-lobe misguidance in rgFDD brains. E, Full β misguidance and aberrant γ-lobe morphology of rgFDD MBs. MB defects mentioned for C–E are indicated with arrows. F–H, Quantification of the frequency of the morphological defects in rg mutants (F) and their respective rescues in rgγ5 (G) and rgFDD (H) background combined with a Gal4 driver. rgγ5 and rgFDD combined with the Gal4 driver were used as mutant controls (indicated in gray) to correct for effects of the Gal4 driver and genetic background. The rescues with the rugose transgene generally rescued the phenotypes completely or partially with, respectively, elav–Gal4 or the MB driver OK107–Gal4. Reintroducing rg+ cDNA with MB247–Gal4 had no effect. Nbea cDNA expression failed to rescue the phenotypes. Numbers represent the percentages of aberrant hemispheres. Fusion defects were counted as defects of two hemispheres. n is the number of brain hemispheres analyzed. Statistical significance between rgγ5 or rgFDD combined with a Gal4 driver and the respective rescues with transgenic rg+ or Nbea cDNA were assessed with the Fisher's exact test. For rugose mutants, statistical significance with w1118 brains was also assessed with the Fisher's exact test.
Overall, rugose mutants displayed defects with increasing numbers correlating with the severity of the rough eye phenotype, i.e., rgFDD > rgγ5 > rg1, but limited penetrance ranging from 3 to 68% in males. Remarkably, rg1 flies show hardly any defects with only one thin α lobe on a total of 32 hemispheres analyzed, thus directly raising the question of whether morphological defects might be causal for the associative learning defect. To address this issue, we introduced either rg+ or Nbea cDNA within either rgγ5 or rgFDD mutant background and quantified aberrant morphology of MB axons (Fig. 4G,H). Although rgγ5 phenotypes were rescued by rg+ cDNA expressed under control of either the pan-neuronal elav–Gal4 or the MB–Gal4 line OK107–Gal4 (Fig. 4G), rgFDD phenotypes showed only partial, albeit significant, rescue (Fig. 4H). In contrast, MB247–Gal4, which marks ∼700 KCs per hemisphere (Schwaerzel et al., 2002), failed to rescue in either combination. Rescue by Nbea failed completely (Fig. 4G,H).
These experiments support several conclusions. First, Rg is required for MB development and acts in a cell-autonomous manner at the level of KCs, as revealed by OK107–Gal4. The fact that MB247–Gal4 failed to rescue the morphological phenotype might be attributable to the fact that only approximately one-third of KCs is marked within this line. Moreover, expression levels of Rg might be critical as reflected by the difference between rgγ5 and rgFDD. Second, Rg and Nbea are not functional homologs at the level of MB morphology, similar to the situation in appetitive odor learning. In perspective to aversive odor learning, it seems to be a valid conclusion that rg mutants exhibit pleiotropic and genetically separable effects, because Nbea rescue is functional in terms of aversive learning but fails in terms of morphology. Thus, each phenotype might well be caused by different functional aspects of Rg, a hypothesis supported by the size of Rg protein and the multiple functional units present in this large scaffolding protein.
Rg mutants display aberrant morphology of the NMJ
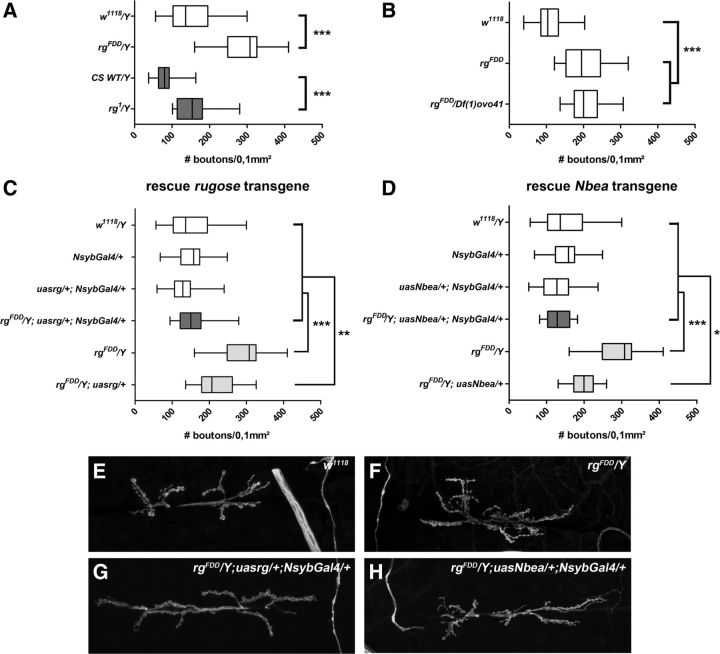
Although the allelic series of rg1 < rgγ5 < rgFDD exhibits gross morphological phenotypes with increasing severity at the level of MB axons, subtle changes at the level of synaptic architecture are neglected by this type of analysis. To address this issue, we quantified synaptic bouton numbers at the NMJ of third-instar larvae, a standard preparation used for investigating structural and functional aspects of synaptic function (Desai-Shah and Cooper, 2009). After anti-HRP staining, both, rgFDD and rg1 hemizygous male larvae showed increased bouton numbers per muscle area compared with w1118 and wild-type Canton-S controls (Fig. 5A). Similarly, homozygous rgFDD and trans-heterozygotes for rgFDD and Df(1)ovo41, a deficiency uncovering the rg locus, showed increased bouton numbers (Fig. 5B). This phenotype was restored to wild-type levels on ectopic expression of rg+ cDNA under control of Nsyb–Gal4 in an rgFDD mutant background. Genetic controls or overexpression of rg+ cDNA in a wild-type genetic background did not affect bouton numbers (Fig. 5C). Similarly, expression of Nbea cDNA rescued bouton numbers of rgFDD mutants to wild-type levels (Fig. 5D), thus establishing Rg and Nbea as functional homologs at the level of synaptic bouton number.
Figure 5.
The bouton number is increased in rg mutants and can be rescued by neuronal expression of Rg or Nbea. Because the motor terminal size is correlated with the muscle fiber size (Schuster et al., 1996), the bouton number was quantified at the synapse of muscle 6/7 of abdominal segment A2 and corrected for the muscle area. A, rgFDD (n = 12) and rg1 (n = 15) male larvae have a higher bouton number per muscle area compared with w1118 (n = 17) and Canton-S wild type (CS WT; n = 24), respectively. B, rgFDD (n = 22) female larvae and trans-heterozygote females for rgFDD and Df(1)ovo41 (n = 20), a deficiency deleting rg, also have an increased number of boutons compared with w1118 (n = 27) females. C, The morphological defect of male rgFDD third-instar larvae can be rescued by reintroducing Rg via the neuronal driver Nsyb–Gal4. The bouton number of rgFDD/Y (n = 12) and rgFDD/Y; UASrg/+ (n = 20) is significantly different from w1118 (n = 17), Nsyb–Gal4/+ (n = 31), UASrg/+; Nsyb–Gal4/+ (n = 19), and rgFDD/Y; UASrg/+; Nsyb–Gal4/+ (n = 32) male larvae. D, Neuronal expression Nbea with Nsyb–Gal4 can also rescue the increased number of boutons to wild-type levels. The number of boutons of rgFDD/Y (n = 12) and rgFDD/Y; UASNbea/+ (n = 20) is significantly different from w1118 (n = 17), Nsyb–Gal4/+ (n = 31), UASNbea/+; Nsyb–Gal4/+ (n = 19), and rgFDD/Y; UASNbea/+; Nsyb–Gal4/+ (n = 12) male larvae. In C and D, rescues are depicted with dark boxes, mutant (controls) in light gray boxes, and controls in white boxes. The data are depicted as box and whisker plots. The boxes represent the first quartile, the median and third quartile. The whiskers drawn at the left and right of the box represent the minimal and maximal scores. E–H, Examples of w1118 (E), rgFDD/Y (F), rgFDD/Y (G), and rgFDD/Y; UASNbea/+; Nsyb–Gal4/+ (H).
Rg mutants show no defects in basal synaptic function
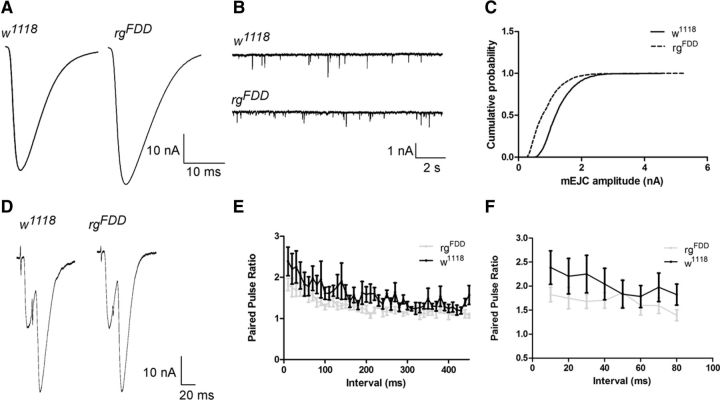
To test whether Drosophila rugose is involved in the regulation of synaptic transmission, we recorded EJCs elicited at 0.2 Hz and mEJCs at the larval NMJ (Fig. 6). EJC amplitudes in rgFDD were very similar to those measured during w1118 controls at 0.5 mm external Ca2+ (Fig. 6A; 49.63 ± 5.9 nA for rgFDD and 48.51 ± 6.39 nA for w1118). At the same time, spontaneous vesicle fusions (mEJCs) measured in 0.5 mm external Ca2+ and TTX were slightly but significantly decreased (Fig. 6B; 1.03 ± 0.07 nA for rgFDD and 1.35 ± 0.08 nA for w1118; p = 0.0179, t = 2.605, df = 18, Student's t test). However, calculation of the quantal content indicated that neuronal communication at NMJs of rgFDD mutants is primarily intact (48 ± 6 quanta for rgFDD and 40 ± 6 quanta for w1118; p = 0.305, t = 0.947, df = 15, Student's t test). Next we tested whether rg mutants affect paired-pulse facilitation, a measure for defective calcium handling at synapses. However, as shown in Figure 6, E and F, paired-pulse facilitation at different interpulse intervals was not different between rg mutants and controls. Together, these data indicate that basal synaptic transmission at third-instar larval NMJs in Drosophila rg mutants is not significantly affected.
Figure 6.
Basal synaptic function is not altered at the NMJ of rg mutants. A, Representative traces of EJCs amplitudes for w1118 and rgFDD mutants at 0.2 Hz stimulation. B, Representative traces for mEJCs recordings (B) and quantification of cumulative probability (C). The mEJC amplitude of rgFDD (n = 8) significantly differs from control w1118 (n = 12). D–F, Two-electrode voltage-clamp recordings of paired-pulse facilitation of control (w1118, n = 7) and rg mutants (rgFDD, n = 7). D, Representative traces in 0.5 mm CaCl2 for both w1118 and rgFDD. E, Quantification of facilitation as measured by the ratio between the EJC amplitudes at 10 ms interpulse intervals. F, Visualization of the first eight points of E. All error bars indicate SEM; n indicates the numbers of larvae tested per genotype. Statistical significance was determined by Student's t test, *p < 0.05.
Discussion
Here, we established functional homologies between Drosophila Rugose and mammalian NBEA that will help clarify the role of BEACH proteins in the development and function of the CNS in vivo. Drosophila Rg is neuronally expressed in the larval and adult CNS and localizes to the TGN and endomembranes near the TGN, as does NBEA in mammals (Wang et al., 2000). In contrast to Nbea knock-out mice that die shortly after birth (Su et al., 2004; Medrihan et al., 2009), loss of Rg function results in viable and fertile adults that display an increased number of synaptic boutons at the level of the larval NMJ without affecting basal synaptic transmission. At the level of CNS development, Drosophila rg mutants display gross morphological defects of the Drosophila MB, the fly “learning center,” and at the same time rg mutants are impaired in associative odor learning. However, genetic rescue experiments using either UAS–rg+ or UAS–NBEA—the mouse homolog of Drosophila rugose—indicate that functional and morphological aspects of Rg function are separable. Overall, this detailed compendium of phenotypes provides an experimental test bed to investigate systemic, developmental, and molecular aspects of NBEA function in a relatively simple but genetically well-developed model organism.
Morphological, structural, and functional aspects are separated in NBEA proteins
Rugose mutants display impaired odor learning, a defect that is attributable to Rg function within KCs of the MBs, the fly learning center (Davis, 2011). However, rgγ5 and rgFDD mutants exhibit aberrant MB morphology, and Rg function is required during development of the CNS, complicating the straightforward interpretation that Rg is functionally linked to memory. At the same time, it appears equally unlikely that solely the anatomical defects within the MB structure are causal of the learning defect.
We speculate that morphological and functional aspects of Rg are separable at the protein level, particularly given the overall size of the protein and the complex composition of numerous functional and highly conserved domains that might support independent functions. There are numerous arguments in favor of that hypothesis. First, all rg1 mutants—except for one brain with a minor defect—display normal gross morphology of MBs but impaired odor learning. Second, all learning defects are restored with a pan-neuronal Gal4 driver or a Gal4 driver expressing rg+ in the MBs. At the same time, global neuronal expression of rg+ cDNA or MB-specific expression with OK107 can only partially rescue the morphological phenotypes of rgγ5 and rgFDD flies. This is even more pronounced with the mouse Nbea transgene that restores the aversive, but not the appetitive, associative learning defects without affecting the morphological phenotype of rg mutants at all. In this respect, the temporally controlled expression of rg+ cDNA at the level of MBs clearly demonstrated a developmental contribution of Rg, without neglecting an additional contribution during the process of associative learning in the adult. Temporal control of rg-specific RNAi seems appropriate to resolve that dilemma, but we failed to phenocopy the rough eye phenotype characteristic of rg mutants by the multiple RNAi lines we tested.
The fact that Drosophila Rugose and mouse Nbea are functionally redundant in case of aversive but not appetitive odor learning is well in line with separable functions at the protein level. Similar dissociations have been reported previously for the Drosophila dunce mutant that affects immediate aversive, but not appetitive, memory (Tempel et al., 1983). In general, it is commonly accepted that aversive and appetitive odor learning depend on different neuronal circuits but share a requirement for MB KCs (Thum et al., 2007). The dissociation between the functional redundancies of either mouse Nbea or fly rg+ transgenes is well in line with the hypothesis that different functional domains are required to support either type of odor learning. Use of alternative putative homologs, e.g., human LRBA (Fig. 1K), or targeted mutagenesis of particular protein domains might aid in defining appropriate functions within future experiments.
A function for rugose in the modulation of presynaptic transmitter release could be consistent with its requirement for associative learning, because it has been shown that neurotransmission from KCs is critical for this type of associative paradigm (Schwaerzel et al., 2003). However, basal neurotransmission is essentially unaffected in rgFDD mutants with only a slight decrease in spontaneous vesicle fusions (mEJCs). Whether similar defects occur at the level of appropriate CNS neurons remains unknown.
Functional specifications in Rg, Nbea, and LRBA?
Nbea mainly localizes to endomembranes near the TGN, consistent with a possible function for Nbea upstream in a secretory pathway (Wang et al., 2000). A function for Nbea in trafficking is further suggested by the accumulation of actin and synaptopodin, a spine-associated protein with actin-bundling activity, near the Golgi of Nbea−/− neurons (Niesmann et al., 2011). In Drosophila, spine-like structures are abundant at lobular plate tangential cells of the visual system, thus directly providing a putative fly model for this aspect of Nbea/rg function. The observation that Rugose colocalizes with Nbea at a position opposite to the cis-Golgi marker GM130 provides evidence for a function in either sorting or maturation of secretory vesicles or granules more upstream. However, whether similar processes are affected in rugose mutants remains to be formally proven.
LRBA, the human homolog of NBEA, is expressed in many cell types that have an increased proliferative and survival capacity, and its expression is increased in multiple cancers (Wang et al., 2001, 2004). A dominant-negative form of LRBA (lacking BEACH–WD40 domains) reduces the phosphorylation of EGFR and MAPK and sensitizes cancer cells to apoptosis (Wang et al., 2004). This interaction with growth factor receptor signaling, more specifically EGFR, MAPK, and Notch signaling, has been shown previously for Rugose in eye development (Schreiber et al., 2002; Shamloula et al., 2002; Wech and Nagel, 2005). In Caenorhabditis elegans, loss of the Rugose ortholog sel-2 also results in aberrant patterning of vulvar precursor cells in a Notch-dependent manner (de Souza et al., 2007). At the same time, trafficking of synaptic receptors for glycine, the major inhibitory neurotransmitter in mammals, has been reported to require Nbea (del Pino et al., 2011), in line with a role of Nbea in the retrieval and post-Golgi trafficking of synaptic receptors. Mab21 genes, which are regulators of growth factor pathways, are nested within an intron of Nbea/LRBA in most organisms (Tsang et al., 2009). In Drosophila, however, Mab21 resides outside the rugose locus, so has no implication for the observed phenotypes.
Together, this study shows that Rugose is the functional homolog of Nbea in eye development, aversive odor learning, and NMJ morphology. The sophisticated genetic repertoire of Drosophila seems ideally suited to improving our understanding of the structure–function relationships in the neuronal roles of Neurobeach proteins, particularly given the overall size of those proteins and the complex composition of numerous functional and highly conserved domains that might support independent functions. Rg, NBEA, or LRBA—the later one remains to be established—would be predisposed to site-directed mutagenesis to test for the functional impact on the multiple phenotypes of rg null mutants. It will also help in understanding what the functional consequences are of the observed changes in synaptic structure and what its relation is with the learning and memory deficits observed in rugose mutants.
Footnotes
This work was supported by Flemish Agency for Innovation by Science and Technology Grant IDO/08/013 (K.V. and Catholic University Leuven) and German Research Foundation Grant SCHW1410/1-1 (M.S.). We thank Annelien Mestdagh and Sandra Meulemans for technical assistance. Andreas S. Thum (University of Konstanz, Konstanz, Germany) provided expression patterns of Gal4 lines.
References
- Albers CA, Cvejic A, Favier R, Bouwmans EE, Alessi MC, Bertone P, Jordan G, Kettleborough RN, Kiddle G, Kostadima M, Read RJ, Sipos B, Sivapalaratnam S, Smethurst PA, Stephens J, Voss K, Nurden A, Rendon A, Nurden P, Ouwehand WH. Exome sequencing identifies NBEAL2 as the causative gene for gray platelet syndrome. Nat Genet. 2011;43:735–737. doi: 10.1038/ng.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E. Drosophila, the golden bug, emerges as a tool for human genetics. Nat Rev Genet. 2005;6:9–23. doi: 10.1038/nrg1503. [DOI] [PubMed] [Google Scholar]
- Bilen J, Bonini NM. Drosophila as a model for human neurodegenerative disease. Annu Rev Genet. 2005;39:153–171. doi: 10.1146/annurev.genet.39.110304.095804. [DOI] [PubMed] [Google Scholar]
- Burgess A, Mornon JP, de Saint-Basile G, Callebaut I. A concanavalin A-like lectin domain in the CHS1/LYST protein, shared by members of the BEACH family. Bioinformatics. 2009;25:1219–1222. doi: 10.1093/bioinformatics/btp151. [DOI] [PubMed] [Google Scholar]
- Castermans D, Wilquet V, Parthoens E, Huysmans C, Steyaert J, Swinnen L, Fryns JP, Van de Ven W, Devriendt K. The neurobeachin gene is disrupted by a translocation in a patient with idiopathic autism. J Med Genet. 2003;40:352–356. doi: 10.1136/jmg.40.5.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castermans D, Volders K, Crepel A, Backx L, De Vos R, Freson K, Meulemans S, Vermeesch JR, Schrander-Stumpel CT, De Rijk P, Del-Favero J, Van Geet C, Van De Ven WJ, Steyaert JG, Devriendt K, Creemers JW. SCAMP5, NBEA and AMISYN: three candidate genes for autism involved in secretion of large dense-core vesicles. Hum Mol Genet. 2010;19:1368–1378. doi: 10.1093/hmg/ddq013. [DOI] [PubMed] [Google Scholar]
- Chou YH, Spletter ML, Yaksi E, Leong JC, Wilson RI, Luo L. Diversity and wiring variability of olfactory local interneurons in the Drosophila antennal lobe. Nat Neurosci. 2010;13:439–449. doi: 10.1038/nn.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomb J, Kaiser L, Chabaud MA, Preat T. Parametric and genetic analysis of Drosophila appetitive long-term memory and sugar motivation. Genes Brain Behav. 2009;8:407–415. doi: 10.1111/j.1601-183X.2009.00482.x. [DOI] [PubMed] [Google Scholar]
- Connolly JB, Roberts IJ, Armstrong JD, Kaiser K, Forte M, Tully T, O'Kane CJ. Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science. 1996;274:2104–2107. doi: 10.1126/science.274.5295.2104. [DOI] [PubMed] [Google Scholar]
- Davis RL. Traces of Drosophila memory. Neuron. 2011;70:8–19. doi: 10.1016/j.neuron.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lozanne A. The role of BEACH proteins in Dictyostelium. Traffic. 2003;4:6–12. doi: 10.1034/j.1600-0854.2003.40102.x. [DOI] [PubMed] [Google Scholar]
- del Pino I, Paarmann I, Karas M, Kilimann MW, Betz H. The trafficking proteins Vacuolar Protein Sorting 35 and Neurobeachin interact with the glycine receptor beta-subunit. Biochem Biophys Res Commun. 2011;412:435–440. doi: 10.1016/j.bbrc.2011.07.110. [DOI] [PubMed] [Google Scholar]
- Desai-Shah M, Cooper RL. Different mechanisms of Ca2+ regulation that influence synaptic transmission: comparison between crayfish and Drosophila neuromuscular junctions. Synapse. 2009;63:1100–1121. doi: 10.1002/syn.20695. [DOI] [PubMed] [Google Scholar]
- de Souza N, Vallier LG, Fares H, Greenwald I. SEL-2, the C. elegans neurobeachin/LRBA homolog, is a negative regulator of lin-12/Notch activity and affects endosomal traffic in polarized epithelial cells. Development. 2007;134:691–702. doi: 10.1242/dev.02767. [DOI] [PubMed] [Google Scholar]
- Doroquez DB, Rebay I. Signal integration during development: mechanisms of EGFR and Notch pathway function and cross-talk. Crit Rev Biochem Mol Biol. 2006;41:339–385. doi: 10.1080/10409230600914344. [DOI] [PubMed] [Google Scholar]
- Fiala A, Spall T, Diegelmann S, Eisermann B, Sachse S, Devaud JM, Buchner E, Galizia CG. Genetically expressed cameleon in Drosophila melanogaster is used to visualize olfactory information in projection neurons. Curr Biol. 2002;12:1877–1884. doi: 10.1016/s0960-9822(02)01239-3. [DOI] [PubMed] [Google Scholar]
- Gebauer D, Li J, Jogl G, Shen Y, Myszka DG, Tong L. Crystal structure of the PH-BEACH domains of human LRBA/BGL. Biochemistry. 2004;43:14873–14880. doi: 10.1021/bi049498y. [DOI] [PubMed] [Google Scholar]
- Guo A, Li L, Xia SZ, Feng CH, Wolf R, Heisenberg M. Conditioned visual flight orientation in Drosophila: dependence on age, practice, and diet. Learn Mem. 1996;3:49–59. doi: 10.1101/lm.3.1.49. [DOI] [PubMed] [Google Scholar]
- Han JD, Baker NE, Rubin CS. Molecular characterization of a novel A kinase anchor protein from Drosophila melanogaster. J Biol Chem. 1997;272:26611–26619. doi: 10.1074/jbc.272.42.26611. [DOI] [PubMed] [Google Scholar]
- Heimbeck G, Bugnon V, Gendre N, Keller A, Stocker RF. A central neural circuit for experience-independent olfactory and courtship behavior in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2001;98:15336–15341. doi: 10.1073/pnas.011314898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Jogl G, Shen Y, Gebauer D, Li J, Wiegmann K, Kashkar H, Krönke M, Tong L. Crystal structure of the BEACH domain reveals an unusual fold and extensive association with a novel PH domain. EMBO J. 2002;21:4785–4795. doi: 10.1093/emboj/cdf502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka S, Sawada N, Bonifacino JS, Waguri S. Functional characterization of protein-sorting machineries at the trans-Golgi network in Drosophila melanogaster. J Cell Sci. 2010;123:460–471. doi: 10.1242/jcs.055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J, De Domenico I, Ward DM. Chediak-Higashi syndrome. Curr Opin Hematol. 2008;15:22–29. doi: 10.1097/MOH.0b013e3282f2bcce. [DOI] [PubMed] [Google Scholar]
- Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S. Sequential use of mushroom body neuron subsets during Drosophila odor memory processing. Neuron. 2007;53:103–115. doi: 10.1016/j.neuron.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes MJ, DasGupta S, Vreede A, White B, Armstrong JD, Waddell S. A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell. 2009;139:416–427. doi: 10.1016/j.cell.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Medrihan L, Rohlmann A, Fairless R, Andrae J, Doring M, Missler M, Zhang W, Kilimann MW. Neurobeachin, a protein implicated in membrane protein traffic and autism, is required for the formation and functioning of central synapses. J Physiol. 2009;587:5095–5106. doi: 10.1113/jphysiol.2009.178236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle DL, Karim MA, Woolf EA, Holmgren L, Bork P, Misumi DJ, McGrail SH, Dussault BJ, Jr, Perou CM, Boissy RE, Duyk GM, Spritz RA, Moore KJ. Identification and mutation analysis of the complete gene for Chediak-Higashi syndrome. Nat Genet. 1996;14:307–311. doi: 10.1038/ng1196-307. [DOI] [PubMed] [Google Scholar]
- Niesmann K, Breuer D, Brockhaus J, Born G, Wolff I, Reissner C, Kilimann MW, Rohlmann A, Missler M. Dendritic spine formation and synaptic function require neurobeachin. Nat Commun. 2011;2:557. doi: 10.1038/ncomms1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neal J, Gao F, Hassan A, Monahan R, Barrios S, Kilimann MW, Lee I, Chng WJ, Vij R, Tomasson MH. Neurobeachin (NBEA) is a target of recurrent interstitial deletions at 13q13 in patients with MGUS and multiple myeloma. Exp Hematol. 2009;37:234–244. doi: 10.1016/j.exphem.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell. 2008;14:239–251. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman JL, Huetteroth W, Burke CJ, Krashes MJ, Lai SL, Lee T, Waddell S. A pair of inhibitory neurons are required to sustain labile memory in the Drosophila mushroom body. Curr Biol. 2011;21:855–861. doi: 10.1016/j.cub.2011.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SL, Preiss A, Nagel AC, Wech I, Maier D. Genetic screen for modifiers of the rough eye phenotype resulting from overexpression of the Notch antagonist hairless in Drosophila. Genesis. 2002;33:141–152. doi: 10.1002/gene.10102. [DOI] [PubMed] [Google Scholar]
- Schuster CM, Davis GW, Fetter RD, Goodman CS. Genetic dissection of structural and functional components of synaptic plasticity. I. Fasciclin II controls synaptic stabilization and growth. Neuron. 1996;17:641–654. doi: 10.1016/s0896-6273(00)80197-x. [DOI] [PubMed] [Google Scholar]
- Schwaerzel M, Heisenberg M, Zars T. Extinction antagonizes olfactory memory at the subcellular level. Neuron. 2002;35:951–960. doi: 10.1016/s0896-6273(02)00832-2. [DOI] [PubMed] [Google Scholar]
- Schwaerzel M, Monastirioti M, Scholz H, Friggi-Grelin F, Birman S, Heisenberg M. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J Neurosci. 2003;23:10495–10502. doi: 10.1523/JNEUROSCI.23-33-10495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamloula HK. Mutation in rugose gene locus affects the pigment cell number and alters the optic lobe phenotype in the Drosophila visual system. J Egypt Soc Parasitol. 2003;33:955–967. [PubMed] [Google Scholar]
- Shamloula HK, Mbogho MP, Pimentel AC, Chrzanowska-Lightowlers ZM, Hyatt V, Okano H, Venkatesh TR. rugose (rg), a Drosophila A kinase anchor protein, is required for retinal pattern formation and interacts genetically with multiple signaling pathways. Genetics. 2002;161:693–710. doi: 10.1093/genetics/161.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Claridge-Chang A, Sjulson L, Pypaert M, Miesenböck G. Excitatory local circuits and their implications for olfactory processing in the fly antennal lobe. Cell. 2007;128:601–612. doi: 10.1016/j.cell.2006.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiflett SL, Kaplan J, Ward DM. Chediak-Higashi Syndrome: a rare disorder of lysosomes and lysosome related organelles. Pigment Cell Res. 2002;15:251–257. doi: 10.1034/j.1600-0749.2002.02038.x. [DOI] [PubMed] [Google Scholar]
- Shiflett SL, Vaughn MB, Huynh D, Kaplan J, Ward DM. Bph1p, the Saccharomyces cerevisiae homologue of CHS1/beige, functions in cell wall formation and protein sorting. Traffic. 2004;5:700–710. doi: 10.1111/j.1600-0854.2004.00213.x. [DOI] [PubMed] [Google Scholar]
- Stocker RF, Heimbeck G, Gendre N, de Belle JS. Neuroblast ablation in Drosophila P[GAL4] lines reveals origins of olfactory interneurons. J Neurobiol. 1997;32:443–456. doi: 10.1002/(sici)1097-4695(199705)32:5<443::aid-neu1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Su Y, Balice-Gordon RJ, Hess DM, Landsman DS, Minarcik J, Golden J, Hurwitz I, Liebhaber SA, Cooke NE. Neurobeachin is essential for neuromuscular synaptic transmission. J Neurosci. 2004;24:3627–3636. doi: 10.1523/JNEUROSCI.4644-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel BL, Bonini N, Dawson DR, Quinn WG. Reward learning in normal and mutant Drosophila. Proc Natl Acad Sci U S A. 1983;80:1482–1486. doi: 10.1073/pnas.80.5.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum AS, Knapek S, Rister J, Dierichs-Schmitt E, Heisenberg M, Tanimoto H. Differential potencies of effector genes in adult Drosophila. J Comp Neurol. 2006;498:194–203. doi: 10.1002/cne.21022. [DOI] [PubMed] [Google Scholar]
- Thum AS, Jenett A, Ito K, Heisenberg M, Tanimoto H. Multiple memory traces for olfactory reward learning in Drosophila. J Neurosci. 2007;27:11132–11138. doi: 10.1523/JNEUROSCI.2712-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WH, Shek KF, Lee TY, Chow KL. An evolutionarily conserved nested gene pair—Mab21 and Lrba/Nbea in metazoan. Genomics. 2009;94:177–187. doi: 10.1016/j.ygeno.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- Volders K, Nuytens K, Creemers JW. The autism candidate gene Neurobeachin encodes a scaffolding protein implicated in membrane trafficking and signaling. Curr Mol Med. 2011;11:204–217. doi: 10.2174/156652411795243432. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112:271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Wang JW, Howson J, Haller E, Kerr WG. Identification of a novel lipopolysaccharide-inducible gene with key features of both A kinase anchor proteins and chs1/beige proteins. J Immunol. 2001;166:4586–4595. doi: 10.4049/jimmunol.166.7.4586. [DOI] [PubMed] [Google Scholar]
- Wang JW, Gamsby JJ, Highfill SL, Mora LB, Bloom GC, Yeatman TJ, Pan TC, Ramne AL, Chodosh LA, Cress WD, Chen J, Kerr WG. Deregulated expression of LRBA facilitates cancer cell growth. Oncogene. 2004;23:4089–4097. doi: 10.1038/sj.onc.1207567. [DOI] [PubMed] [Google Scholar]
- Wang X, Herberg FW, Laue MM, Wullner C, Hu B, Petrasch-Parwez E, Kilimann MW. Neurobeachin: A protein kinase A-anchoring, beige/Chediak-higashi protein homolog implicated in neuronal membrane traffic. J Neurosci. 2000;20:8551–8565. doi: 10.1523/JNEUROSCI.20-23-08551.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DM, Shiflett SL, Kaplan J. Chediak-Higashi syndrome: a clinical and molecular view of a rare lysosomal storage disorder. Curr Mol Med. 2002;2:469–477. doi: 10.2174/1566524023362339. [DOI] [PubMed] [Google Scholar]
- Wech I, Nagel AC. Mutations in rugose promote cell type-specific apoptosis in the Drosophila eye. Cell Death Differ. 2005;12:145–152. doi: 10.1038/sj.cdd.4401538. [DOI] [PubMed] [Google Scholar]
- Zars T, Fischer M, Schulz R, Heisenberg M. Localization of a short-term memory in Drosophila. Science. 2000;288:672–675. doi: 10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]