Abstract
During neural development, even in seemingly homogeneous cell populations, individual neurons acquire diverse morphology and behavior, and even adjacent neurons can establish synaptic connections with distinct targets. Although diversity among individual neurons is essential for a functional nervous system, the underlying molecular mechanism of establishing heterogeneity in a population of neuronal precursors has been poorly addressed at the single-cell level. We focused on the development of the zebrafish posterior lateral line (PLL) and revealed a molecular mechanism that differentiates a homogenous neuronal population in the ganglion into the two types of neurons, leaders and followers. We developed a method to analyze gene expression levels in leaders and followers at the single-cell resolution, and found that leaders expressed significantly higher levels of neurod compared with followers. Furthermore, neurod expression was found to be heterogeneous among neurons before the onset of phenotypic differentiation of leaders and followers, and neurod overexpression in single PLL neurons promoted differentiation into leaders. These results suggest that the quantity, rather than quality (i.e., the ON/OFF states), of neurod expression directly or indirectly determines the two subtypes of PLL neurons.
Introduction
The vertebrate nervous system is composed of an enormous number of neurons that exhibit diversity in their morphology, gene expression, neuronal connections, responsivity and other properties. Such cell-to-cell variety is observed even in seemingly identical neuronal cell populations. For example, it is frequently observed that immediately adjacent neurons extend their axons in different tracts and connect with discrete targets (Macagno et al., 1973; Livet et al., 2007). The diversity among individual neurons driving this process must be essential for establishing a functional nervous system. However, the underlying mechanisms are poorly understood due to the lack of knowledge about phenotypes and gene expression profiles in individual developing neurons; the complexity of the nervous system often hampers single-cell analysis. For this reason, the zebrafish lateral line system is potentially one of the best models with which to address individuality of developing neurons.
The lateral line is a simple sensory system that responds to water movement in fish and amphibians. This system comprises a set of discrete sensory organs, called neuromasts, and sensory neurons (Metcalfe et al., 1985). These neurons form ganglia near a otic vesicle, and extend central and peripheral projections to the hindbrain and neuromasts, respectively (Ghysen and Dambly-Chaudière, 2004). The early-stage ganglion of the posterior lateral line (PLL) system consists of only 20–30 neurons that are superficially identical. However, two subpopulations of neurons soon become evident at the time of axon projection but remain intermingled within the ganglion (Sato et al., 2010). One is leading PLL neurons that are first to extend their axons. Leaders (almost one third of PLL neurons at this stage) contact with a PLL primordium, a precursor of neuromasts, which appear immediately posterior to the ganglion and comigrates to the tip of the tail along the midline. During primordium migration, the leaders' growth cones maintain contact with the primordium to the posterior end. The other neurons are defined as followers, which extend their axons along the axon tract of leaders. These two subpopulations display significantly different axonal elongation dynamics and growth cone morphology (Sato et al., 2010). Furthermore, the patterns of their projection are distinct; followers establish a topographic organization, that is, the relative position of cell bodies in the ganglion represents that of the neuromast they innervate (Alexandre and Ghysen, 1999), whereas leaders connect with posterior neuromasts regardless of their position in a ganglion (Sato et al., 2010).
To gain molecular insight into individuality created in a neuronal precursor population, we adopted the single-cell gene expression profiling approach combined with live-imaging in the PLL ganglion, and found that heterogeneity in neurod expression levels before the onset of axon projection plays a pivotal role in subtype differentiation.
Materials and Methods
Zebrafish strains and husbandry.
Zebrafish were maintained and raised under standard conditions. We used a transgenic strain, Tol047, which was generated by using the Tol2 transposon-mediated enhancer trap technique (Kawakami, 2005), and express GFP in the lateral line neurons and a subset of neurons. Males and females of this strain were mated to generate embryos.
Single-cell labeling and cDNA amplification.
To obtain mosaic embryos expressing GFP in a single PLL neuron, embryos were injected and screened as previously described (Sato et al., 2010). Single-cell cDNA of the labeled neurons were amplified following previously reported protocols (Tang et al., 2010) with slight modifications. PLL ganglions were extirpated from agarose-mounted embryos with fine injection needles, and incubated for 10 min at 37°C in 10–15 U/ml papain solution containing 0.1 mg/ml DNase I. The labeled PLL neurons were manually isolated and transferred into lysis buffer with a mouth pipette. The lysis buffer contains mRNAs of external control genes (ERCC RNA Spike-In Mix, Life Technologies) and other spike genes (Kurimoto et al., 2007). cDNA libraries were synthesized and amplified as described in the previous study (Tang et al., 2010).
qRT-PCR.
qRT-PCR was performed with THUNDERBIRD SYBR qPCR Mix (TOYOBO) according to the manufacturer's protocol. Gene names and primer sequences are listed in Table 1.
Table 1.
Primers for qRT-PCR
| Gene | 5′ primer (5′ to 3′) | 3′ primer (5′ to 3′) |
|---|---|---|
| Ppia | TTTGTGAAGTCACCTCCCTGGCAC | ACTGAGGATGAGGTTGAGAGGGTGG |
| Rps18 | ACTGAGGATGAGGTTGAGAGGGTGG | AGTGACGGAGACCACGGTGAGC |
| Actb1 | TCACCTCTCTTGCTCCTTCCACC | GACAGGGAGGCCAAGATGGAGC |
| Egfp | TCAAGATCCGCCACAACATC | TCTCGTTGGGGTCTTTGCTC |
| Stathmin 1b | GGCATCTAGTGGTTGTGAGTTTG | CTGTTCCTGCAACAAAGGATG |
| Nestin | ACAGCCCAGCATAGTATGGCAGC | GGAATCCTTTCTGCCAGCGGCT |
| Neurog1 | CAACTCAGACCCGGGCAGCC | TGCTAGGCACGAAGTTGCGGT |
| Neurod | CCCTGCGTCATTCCATGCTACCC | AGGACGGGAATTGTGCAACTCTGC |
| Elavl3 | GGGCCTCTTGGATTTGTCACCCTTG | GCGACTTGCTTGGAAGGGCCT |
Immunohistochemistry.
Whole-mount immunohistochemistry was performed as previously described (Bae et al., 2009) with slight modifications. Embryos were fixed in 4% PFA/PBS for 1 h at 4°C, and permeabilized in acetone for 15 min at −30°C. Then, samples were incubated with the primary antibody solution (1:500, mouse monoclonal anti-Neurod antibody (Kani et al., 2010), a kind gift from Dr. M. Hibi, Nagoya University, Nagoya, Japan) at 4°C overnight, and incubated with secondary antibody (1:250, Alexa Fluor 488 anti-mouse IgG) at 4°C overnight.
Plasmid DNA construction.
To generate a neurod-2A-mCherry construct, the full-length neurod cDNA was cloned into pCS2+ vector. The primers contain a BamHI site (forward primer) and a ClaI site (reverse primer which mutates the stop codon) as follows: 5′-CTAGGGATCCGACATGACGAAGTCATAC-3′ and 5′-AGGATCGATAGTC GTGAAATATCGC-3′. This plasmid was used as a template for a PCR amplification of pCS2neurod fragment (forward, 5′-GAATTCAAGGCCTCT CGAGC-3′; reverse, 5′-CGAGTCGTGAAATATCGCGTTC-3′). The 2A-mCherry fragment was synthesized by PCR with primers as follows: forward containing P2A sequence (Kim et al., 2011), 5′-ATATTTCACGACTCGGGAAGCGGAGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGAGACGTGGAGGAGAACCCTGGACCTATGGTGAGCAAGGGCGAGGAG-3′ and reverse, 5′-AGAGGCCTTGAATTCTTACTTGTACAGCTCGTCCATG-3′. This fragment was cloned into the pCS2neurod using In-Fusion HD cloning kit (Clontech).
Results
Method for single-cell gene expression analysis in the developing zebrafish
To compare gene expression levels in leading PLL neurons with those in followers, a single-cell expression analysis was required. We adopted the previously reported method of single-cell cDNA amplification with slight modifications and combined it with live-imaging to link gene expression profiles to the neuronal subtypes (Fig. 1A). First, we generated mosaic embryos by plasmid DNA injection into an enhancer trap strain, Tol047, in which all PLL neurons express GFP, to randomly label individual neurons with another reporter gene (DsRed). Then we selected embryos in which a single PLL neuron was labeled (almost 1∼2% of injected embryos). We observed the labeled neuron to determine its subtype, as leader or follower. When the subtype was evident, the neuron was isolated and subjected to cDNA library amplification.
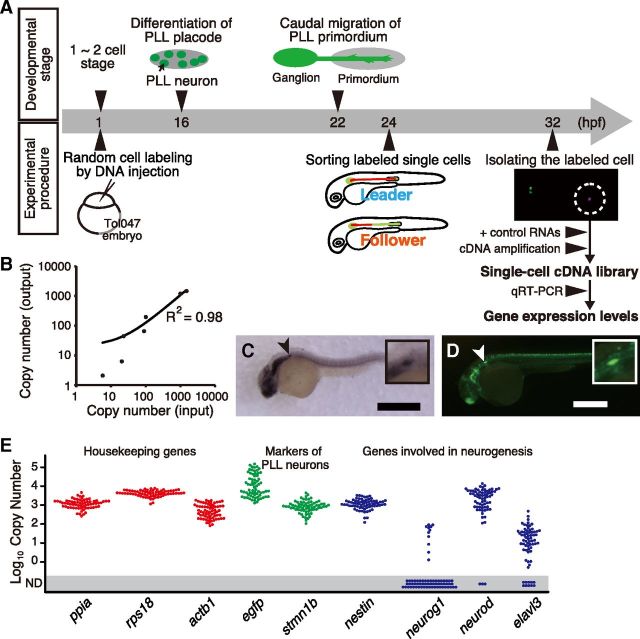
Figure 1.
Single-cell expression analysis in PLL neurons. A, The schedule of zebrafish PLL development and our experimental procedure. B, Quantitative representation of initial mRNA abundance of external control genes (on the horizontal axis) after cDNA amplification (on the vertical axis). We examined four control RNAs of ERCC RNA Spike-In Mix (ERCC00035, 00042, 00136, 00130) and three synthesized mRNAs (Lys, Dap, and Phe, derived from Bacillus subtilis). C, The expression pattern of stmn1b at 24 hpf. stmn1b is expressed in the PLL ganglion (arrowhead). D, GFP is expressed in the PLL ganglion (arrowhead) in the Tol047 embryo at 3 dpf. Scale bars, 0.5 mm. Insets show higher magnifications of the PLL ganglion. E, The expression levels of three housekeeping genes (red dots), two PLL neuron markers (green dots), and four neurogenesis markers (blue dots), in single PLL neurons (n = 57). ND, Not detected.
For quantitative expression analysis, an amplified cDNA library must faithfully represent the relative amount of mRNAs expressed in an intact single cell. We thus quantified amounts of externally added control mRNAs to control for consistency and accuracy in the amplification process. The results of qRT-PCR followed by normalization after cDNA amplification were indeed proportional to the copy number of control genes before amplification (Fig. 1B).
In the present study, the expression levels of the following nine genes were quantified: peptidylprolyl isomerase A (ppia), ribosomal protein S18 (rps18), actin β 1 (actb1), egfp, stathmin 1b (stmn1b), nestin (nes), neurogenin 1 (neurog1), neurogenic differentiation (neurod), ELAV-like 3 (elavl3). ppia, rps18 and actb1 are housekeeping genes generally used for controls, and nes, neurog1, neurod and elavl3 are well known genes that regulate neurogenesis. stmn1b, a member of the Stathmin family that regulates microtubule dynamics (Belmont and Mitchison, 1996), is expressed in all PLL neurons (Fig. 1C). stmn1b was chosen because it is the gene trapped in the enhancer trap line Tol047 expressing GFP in all PLL neurons (Fig. 1D). Thus, stmn1b and gfp were used as PLL neuron markers. Expectedly, the housekeeping genes and PLL neuron markers were detected in all cDNA libraries (n = 57), demonstrating that our method works properly for endogenous genes (Fig. 1E). By contrast, three of four other marker genes (neurog1, neurod, elavl3) were not detected in some neurons. In these cells, the marker genes were not expressed or expressed at undetectable levels.
neurod and actb1 are strongly expressed in leaders
We next quantified gene expression levels in leaders (n = 8) and followers (n = 21), respectively, at 32 hours postfertilization (hpf) (Fig. 1A). qRT-PCR revealed that the expression level of neurod, which is a transcription factor involved in neuronal differentiation, and actb1 in leaders were significantly higher than those in followers (Fig. 2A). In leaders, the copy number of neurod and actb1 mRNA calculated by using the amount of external control genes was 3.3 and 2.3 times higher than in followers, respectively. This result shows that the expression levels of these genes are correlated with the subtype-differentiation of PLL neurons.
Figure 2.
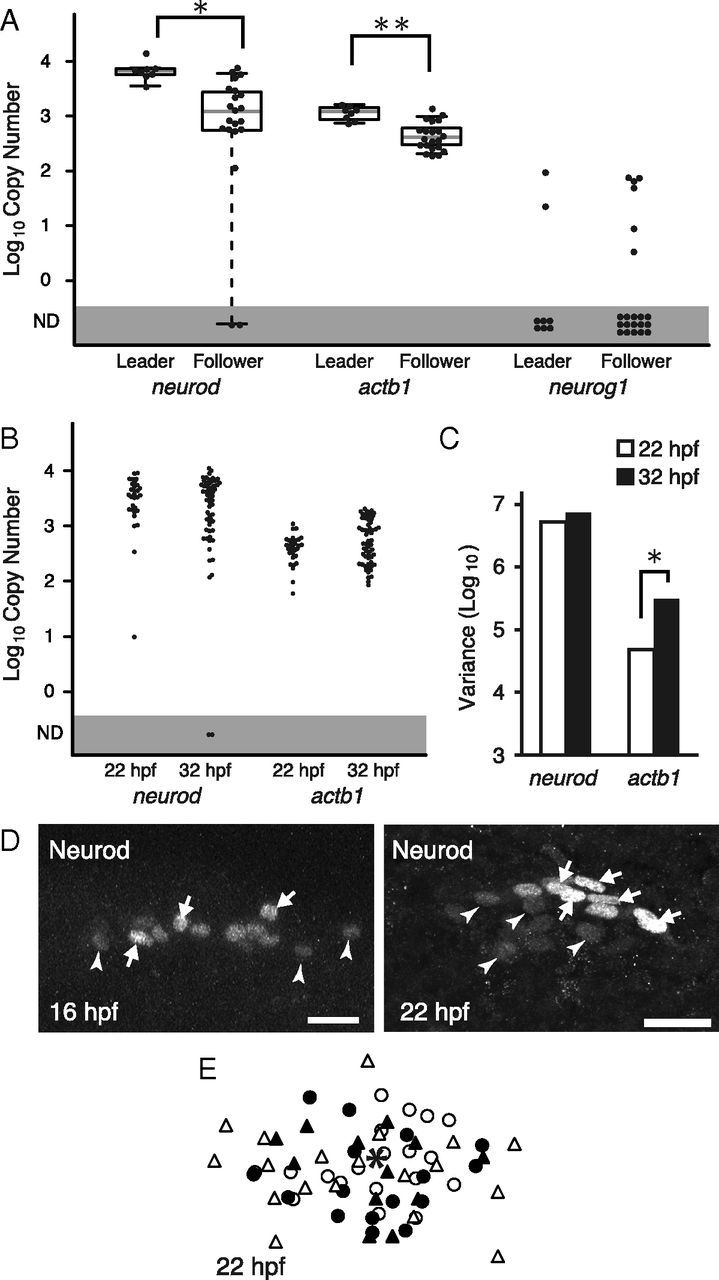
Expression of candidate genes involved in the neuronal specification. A, The expression levels of neurod and actb1 in leaders are significantly higher than those in followers (*p = 0.0007, **p = 0.0003, Wilcoxon rank sum test), while in neurog1, no significant differences are observed. B, The expression levels of neurod and actb1 at 22 and 32 hpf. The mean expression levels do not significantly change. C, The variance in neurod and actb1 expression levels of individual neurons. In neurod, no significant changes in expression variance between 22 and 32 hpf were observed (p = 0.14, Levene test). By contrast, in actb1, the variance is significantly increased at 32 hpf compared with 22 hpf (*p = 0.00016). D, Immunostaining of Neurod. Neurod level is heterogeneous at earlier stages, 16 and 22 hpf. Qualitatively, high-intensity cells (white arrows) and low-intensity cells (white arrowheads) are intermingled in a PLL ganglion. Scale bars, 20 μm. E, Superimposed image of NeuroD+ cells at 22 hpf. The centroid of the ganglion (indicated by an asterisk) was used to normalize their positions. Neurons in an individual ganglion are indicated by an identical symbol.
We then examined the expression of neurog1, a positive regulator of neurod. In contrast to neurod, no significant differences between leaders and followers were detected at 32 hpf (Fig. 2A). Indeed, in most PLL neurons, regardless of their subtypes, the expression of neurog1 was quite low or undetectable by our single-cell expression analysis. This is consistent with the result of in situ hybridization that neurog1 expression has disappeared by 20 hpf (Andermann et al., 2002). These results suggest that neurog1 has induced neurod expression at much earlier stages.
neurod expression level is heterogeneous from an earlier stage
If the expression level of neurod and/or actb1 is the determinant of the neuronal subtypes, there should be heterogeneous expression observed in a ganglion before the onset of their differentiation into leaders and followers (e.g., 16–22 hpf, Fig. 1A). We thus measured the expression abundance of these candidate genes in individual PLL neurons at 22 hpf, and compared their cell-to-cell variations at 22 hpf with those obtained at 32 hpf.
Importantly, for neurod, there were no significant differences in expression variations between 22 (n = 30) and 32 hpf (n = 57) (Fig. 2B,C). The expression variance of neurod among neurons at both 22 and 32 hpf was significantly larger compared with that of a spike control RNA exhibiting similar mean expression abundance (data not shown), indicating that the variation observed was not due to technical reasons. The constant (or unchanged) variance in neurod expression level suggests that neurod is expressed heterogeneously within the ganglion before the onset of leader differentiation. By contrast, for actb1, the cell-to-cell variation in expression levels was significantly increased at 32 hpf (p = 0.00016, Levene test, Fig. 2B,C). This indicates the relatively uniform expression levels of actb1 within a ganglion before the determination of leaders. Therefore, neurod could be one of the candidate genes that play a role in leader differentiation.
Next, we confirmed the heterogeneous expression of Neurod proteins in individual cells at early stages by immunohistochemistry (Kani et al., 2010). At 16 hpf, a few Neurod+ cells were detected in the PLL placode (Fig. 2D). As previously reported (Mizoguchi et al., 2011), the number of Neurod+ cells was increased as development proceeds, and became broadly distributed in the PLL ganglion by 22 hpf (Fig. 2D). The signal of Neurod, a transcription factor, was localized in nuclei, which enabled us to roughly estimate the signal intensity of individual cells. Qualitatively, the signal intensity appeared to vary much among cells at 22 hpf (Fig. 2D), although this does not necessarily reflect the actual abundance of Neurod protein. Importantly, there was no fixed pattern in signal intensity and distribution within each ganglion (Fig. 2E). These results indicate the heterogeneous expression of Neurod within the early PLL ganglion, which is consistent with the above data of mRNA expression obtained by the single-cell qPCR.
neurod overexpression induces differentiation into leaders
Based on the above results, we hypothesized that cells expressing high levels of neurod at an early stage will later become leading PLL neurons. To test our hypothesis, we performed a mosaic overexpression analysis, and examined whether neurod-overexpressing neurons preferentially differentiate into leaders. For this purpose, embryos were injected with a plasmid that drives neurod-P2A-mCherry expression under the control of the CMV promoter (Fig. 3A). The translation product of this construct is self-cleaved at the site of 2A peptide, and Neurod and mCherry proteins are separately produced (Kim et al., 2011). First, neurod-P2A-mCherry mRNA synthesized in vitro was injected at the one- to two-cell stages, to confirm that this fusion product normally functions in vivo. The injected embryos displayed the typical known phenotypes caused by ubiquitous Neurod overexpression, that is, small eyes, poorly organized otic vesicles and short spinal cords (Fig. 3B) (Lee et al., 1995), indicating that this exogenous Neurod protein is indeed functional in vivo. Red fluorescence of mCherry was also ubiquitously detected (Fig. 3B).
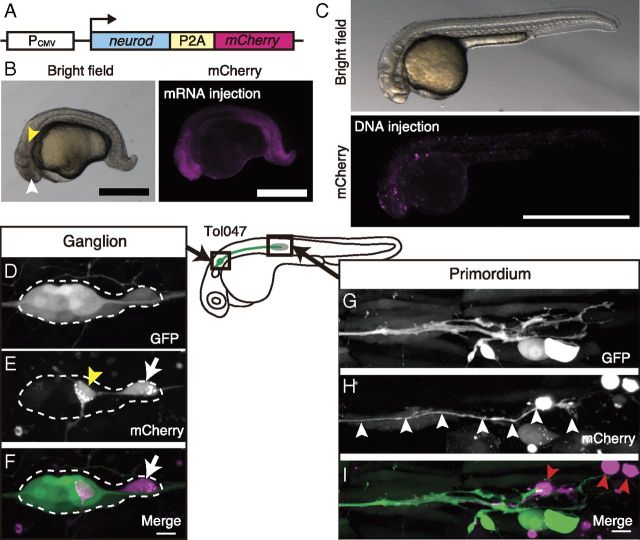
Figure 3.
High-level expression of neurod induces differentiation into leaders. A, DNA construct for the overexpression experiment. B, Phenotype of neurod-2A-mCherry mRNA injection at 32 hpf. The injected embryo displays a small eye (white arrowhead) and a poorly organized otic vesicle (yellow arrowhead). Scale bar, 0.5 mm. C, The overall development of mosaic embryos is normal at 32 hpf. Scale bar, 1 mm. D–F, The PLL ganglion of a Tol047 embryo in which the exogenous genes (neurod and mCherry) are expressed in a single PLL neuron (white arrow in E and F) at 32 hpf. The other labeled neuron (yellow arrowhead in E) is an abnormal neuron because its axon does not extend along the PLL nerve. Such abnormal PLL neurons were sometimes observed in neurod-overexpressing embryos. G–I, The tip of the PLL nerve in the same embryo as in D–F. The labeled axon reaches the tip of the PLL nerve (white arrowheads in H). Scale bars, 10 μm.
Using the DNA construct coding neurod-P2A-mCherry, we generated mosaic embryos in which cells expressing the transgene were sporadic. Those embryos were then screened so that mCherry was expressed in a single PLL neuron. In these selected mosaic embryos (almost 1∼2% of injected embryos), many transgene-positive cells in non-neuronal tissues underwent apoptosis (Fig. 3I, red arrowheads). This may be due to a general toxicity of ectopic Neurod expression in non-neuronal cells, because control-injected embryos expressing mCherry alone did not have such apoptotic cells (data not shown). Despite this effect, the overall development appeared normal (Fig. 3C). We found that in 6 of 7 embryos, the labeled PLL neurons extended their axons into the primordium, that is, they differentiated into leaders (Fig. 3D–I). The incidence of leaders in neurod-overexpressing PLL neurons was significantly higher than that in control neurons that express only a reporter gene (neurod-overexpressing; n = 6 of 7, control; n = 2 of 10, Fisher's exact test, p = 0.015). Hence, we conclude that the high-level of neurod expression induces the differentiation of leaders.
Discussion
Mechanism for determining the subtype of PLL neurons
In the current study, we first established the method of single-cell gene expression analysis targeting isolated individual PLL neurons. This single-cell analysis was combined with mosaic labeling to determine the subtype-specific expression profile. The data obtained demonstrated that the expression level of neurod and actb1 in leading PLL neurons is significantly higher than that in followers, suggesting that these two genes are candidates involved in determination of the neuronal subtypes in the ganglion. Importantly, the two genes are expressed in almost all PLL neurons, and thus quantitative differences, not qualitative differences (i.e., the ON/OFF states of expression), in expression levels among PLL neurons could influence the specification process of leaders and followers. The subsequent experiments support the idea that neurod is a regulator of subtype specification. The cell-to-cell variation of neurod expression level just before the subtype differentiation (at 22 hpf) was as large as that after the differentiation of leaders (at 32 hpf, Fig. 2C), suggesting that neurod is a potential regulatory gene whose expression level in an individual neuron determines the neuronal subtype at later stages. More directly, a neuron highly expressing neurod exhibits a strong tendency to become a leader (Fig. 3D–I). Indeed, neurod is a well known transcription factor that facilitates differentiation of neurons (Lee et al., 1995; Miyata et al., 1999; Roybon et al., 2009). At the present, due to technical difficulties, we are unable to determine by long-term live imaging whether precursor cells with high levels of Neurod indeed will later differentiate into leaders. Nonetheless, we propose that neurons expressing high levels of neurod at an early stage extend their axon first, and then come into contact with a PLL primordium at the time of the onset of its migration, which is characteristic behavior of the leader (Sato et al., 2010).
By contrast, the expression level of actb1 may not be a determinant of leader differentiation for the following reasons. The actb1 expression level at the early stage is homogeneous, and its variation increased as leaders and followers differentiate (Fig. 2C), indicating that the change in actb1 expression level occurs downstream of the subtype specification. Additionally, it has been reported that axonal growth upregulates β-actin mRNA expression in rats (Lund and McQuarrie, 1996). This is consistent with the fact that leaders expressing higher levels of actb1 actively extend their axons (Sato et al., 2010). It is thus likely that actb1 is upregulated in leaders by neural genes such as neurod and/or external signals present in the primordium. It is worth noting that in our experimental procedures, single-cell cDNA libraries were synthesized only from isolated cell bodies. Because β-actin mRNAs are known to be localized in growth cones and translated there (Lee and Hollenbeck, 2003; Rossoll et al., 2003), actb1 expression level in the present study may not precisely reflect the actual act1b level.
In many systems, cell-to-cell variation in gene expression quantity is known to play an important role in cell fate decision (Losick and Desplan, 2008; Enver et al., 2009; Huang, 2009; Eldar and Elowitz, 2010). However, no such phenomenon has been reported in development of nervous systems. Of course, the neurod heterogeneity alone cannot determine the entire process of subtype differentiation but could facilitate the process. Furthermore, the leaders may later be affected by signals from the primordium so that they maintain the characteristic axonal growth dynamics, growth cone morphology and projection pattern (Sato et al., 2010). The primordium, indeed, is known to express an axon guidance cue, semaphorin (Shoji et al., 1998).
Source of the heterogeneous neurod expression
What is the source of the heterogeneity in neurod expression levels in early PLL ganglions? Since no reproducible pattern of the Neurod heterogeneity was observed within early ganglions, we exclude the possibility of positional cues provided by surrounding tissues.
One of the potential sources of heterogeneity is a low copy number of upstream regulatory factors. In general, biochemical processes driven by molecules expressed at a lower number are “noisy” because of stochastic fluctuations (Paulsson, 2004). This kind of spontaneous heterogeneity is also referred to as “intrinsic noise.” Neurog1, a bHLH transcription factor, could be a candidate upstream factor as it was reported to be prerequisite for neurod expression in the zebrafish sensory neurons (Andermann et al., 2002). In PLL neurons, the expression level of neurog1 was very low or undetectable at 32 and 22 hpf (Fig. 2A, and data not shown), and this is consistent with the previous study that shows neurog1 mRNA is decreased to undetectable by in situ hybridization in the PLL placode at 20 hpf (Andermann et al., 2002). Furthermore, even at earlier stages (14–18 hpf), neurog1 expression may be low, because the mRNA signal was very weak (Andermann et al., 2002). The variation in neurod expression level is thus likely to be explained by the low copy number of Neurog1. This idea is further supported by our preliminary data from a global expression analysis by next-generation sequencing (A. Sato, K. Yamaguchi, T. Tsukahara, S. Shigenobu, and H. Takeda, unpublished observations).
Another potential source for neurod heterogeneity is a temporal change in neurog1 expression levels. The gene expression level of neurogenin 2, another member of neurogenin family, is known to oscillate in neural progenitor cells of mice (Shimojo et al., 2008). If neurog1 expression also oscillates in individual PLL premature neurons without coupling, neurod expression level would then be heterogeneous.
Further analyses by quantitative live-imaging will be needed to distinguish these possibilities. In particular, a dual-reporter system, widely used to analyze the intrinsic noise, could answer the question (Hilfinger and Paulsson, 2011).
Regulating neuronal fates by quantity of regulatory genes
In most studies conducted thus far, neuronal diversification within precursor populations has been explained mainly by the ON/OFF state of regulatory genes, i.e., the combination of genes expressed (Tanabe and Jessell, 1996; Price and Briscoe, 2004; Guillemot et al., 2006). In the present study, however, we put more emphasis on the quantity of gene expression in determining subtype specification in neuronal precursor populations. The high levels of Neurod expression could turn on leader-specific genes or simply accelerate neuronal differentiation in PLL neurons. Indeed the timing of differentiation was reported to determine neuronal morphology and behavior in commissural neurons (Bak and Fraser, 2003). Thus, such quantitative regulation through differentiation timing would be a general mechanism for creating neural diversity. The nervous system consists of numerous types of neurons with complicated structure and network connectivity. The mechanisms relying only on the combination of regulatory genes alone may not fully explain the diversification of neurons, but instead the quantity of gene expression could play a leading role in production of the proper repertoire of neurons.
Footnotes
This work was supported in part by a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and Grants-in-Aid for JSPS (Japan Society for the Promotion of Science) Research Fellows (A. Sato). We thank Dr. K. Kawakami for collaborating in establishment of the zebrafish enhancer trap strain, and Dr. K. Horikawa for generating Tol047 strain. We also thank Dr. S. Higashijima for providing the plasmid for single-cell labeling, Dr. M. Saitou and Dr. K. Kurimoto for invaluable advice on the single-cell cDNA amplification, and Dr. M. Hibi for generating and providing the anti-Neurod antibody.
The authors declare no competing financial interests.
References
- Alexandre D, Ghysen A. Somatotopy of the lateral line projection in larval zebrafish. Proc Natl Acad Sci U S A. 1999;96:7558–7562. doi: 10.1073/pnas.96.13.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andermann P, Ungos J, Raible DW. Neurogenin1 defines zebrafish cranial sensory ganglia precursors. Dev Biol. 2002;251:45–58. doi: 10.1006/dbio.2002.0820. [DOI] [PubMed] [Google Scholar]
- Bae YK, Kani S, Shimizu T, Tanabe K, Nojima H, Kimura Y, Higashijima S, Hibi M. Anatomy of zebrafish cerebellum and screen for mutations affecting its development. Dev Biol. 2009;330:406–426. doi: 10.1016/j.ydbio.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Bak M, Fraser SE. Axon fasciculation and differences in midline kinetics between pioneer and follower axons within commissural fascicles. Development. 2003;130:4999–5008. doi: 10.1242/dev.00713. [DOI] [PubMed] [Google Scholar]
- Belmont LD, Mitchison TJ. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84:623–631. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enver T, Pera M, Peterson C, Andrews PW. Stem cell states, fates, and the rules of attraction. Stem Cell. 2009;4:387–397. doi: 10.1016/j.stem.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Ghysen A, Dambly-Chaudière C. Development of the zebrafish lateral line. Curr Opin Neurobiol. 2004;14:67–73. doi: 10.1016/j.conb.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Molnár Z, Tarabykin V, Stoykova A. Molecular mechanisms of cortical differentiation. Eur J Neurosci. 2006;23:857–868. doi: 10.1111/j.1460-9568.2006.04626.x. [DOI] [PubMed] [Google Scholar]
- Hilfinger A, Paulsson J. Separating intrinsic from extrinsic fluctuations in dynamic biological systems. Proc Natl Acad Sci U S A. 2011;108:12167–12172. doi: 10.1073/pnas.1018832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. Non-genetic heterogeneity of cells in development: more than just noise. Development. 2009;136:3853–3862. doi: 10.1242/dev.035139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kani S, Bae YK, Shimizu T, Tanabe K, Satou C, Parsons MJ, Scott E, Higashijima S, Hibi M. Proneural gene-linked neurogenesis in zebrafish cerebellum. Dev Biol. 2010;343:1–17. doi: 10.1016/j.ydbio.2010.03.024. [DOI] [PubMed] [Google Scholar]
- Kawakami K. Transposon tools and methods in zebrafish. Dev Dyn. 2005;234:244–254. doi: 10.1002/dvdy.20516. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA, Choi SY. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Saitou M. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat Protoc. 2007;2:739–752. doi: 10.1038/nprot.2007.79. [DOI] [PubMed] [Google Scholar]
- Lee J, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by neurod, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Lee SK, Hollenbeck PJ. Organization and translation of mRNA in sympathetic axons. J Cell Sci. 2003;116:4467–4478. doi: 10.1242/jcs.00745. [DOI] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Losick R, Desplan C. Stochasticity and cell fate. Science. 2008;320:65–68. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund LM, McQuarrie IG. Axonal regrowth upregulates beta-actin and Jun D mRNA expression. J Neurobiol. 1996;31:476–486. doi: 10.1002/(SICI)1097-4695(199612)31:4<476::AID-NEU7>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Macagno ER, Lopresti V, Levinthal C. Structure and development of neuronal connections in isogenic organisms: variations and similarities in the optic system of Daphnia magna. Proc Natl Acad Sci U S A. 1973;70:57–61. doi: 10.1073/pnas.70.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe WK, Kimmel CB, Schabtach E. Anatomy of the posterior lateral line system in young larvae of the zebrafish. J Comp Neurol. 1985;233:377–389. doi: 10.1002/cne.902330307. [DOI] [PubMed] [Google Scholar]
- Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13:1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Togawa S, Kawakami K, Itoh M. Neuron and sensory epithelial cell fate is sequentially determined by notch signaling in zebrafish lateral line development. J Neurosci. 2011;31:15522–15530. doi: 10.1523/JNEUROSCI.3948-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsson J. Summing up the noise in gene networks. Nature. 2004;427:415–418. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- Price SR, Briscoe J. The generation and diversification of spinal motor neurons: signals and responses. Mech Dev. 2004;121:1103–1115. doi: 10.1016/j.mod.2004.04.019. [DOI] [PubMed] [Google Scholar]
- Rossoll W, Jablonka S, Andreassi C, Kröning AK, Karle K, Monani UR, Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybon L, Deierborg T, Brundin P, Li JY. Involvement of Ngn2, Tbr and NeuroD proteins during postnatal olfactory bulb neurogenesis. Eur J Neurosci. 2009;29:232–243. doi: 10.1111/j.1460-9568.2008.06595.x. [DOI] [PubMed] [Google Scholar]
- Sato A, Koshida S, Takeda H. Single-cell analysis of somatotopic map formation in the zebrafish lateral line system. Dev Dyn. 2010;239:2058–2065. doi: 10.1002/dvdy.22324. [DOI] [PubMed] [Google Scholar]
- Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Shoji W, Yee CS, Kuwada JY. Zebrafish semaphorin Z1a collapses specific growth cones and alters their pathway in vivo. Development. 1998;125:1275–1283. doi: 10.1242/dev.125.7.1275. [DOI] [PubMed] [Google Scholar]
- Tanabe Y, Jessell TM. Diversity and pattern in the developing spinal cord. Science. 1996;274:1115–1123. doi: 10.1126/science.274.5290.1115. [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Nordman E, Li B, Xu N, Bashkirov VI, Lao K, Surani MA. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc. 2010;5:516–535. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]