Abstract
MicroRNAs (miRNAs) have been shown to play important roles in both brain development and the regulation of adult neural cell functions. However, a systematic analysis of brain miRNA functions has been hindered by a lack of comprehensive information regarding the distribution of miRNAs in neuronal versus glial cells. To address this issue, we performed microarray analyses of miRNA expression in the four principal cell types of the CNS (neurons, astrocytes, oligodendrocytes, and microglia) using primary cultures from postnatal d 1 rat cortex. These analyses revealed that neural miRNA expression is highly cell-type specific, with 116 of the 351 miRNAs examined being differentially expressed fivefold or more across the four cell types. We also demonstrate that individual neuron-enriched or neuron-diminished RNAs had a significant impact on the specification of neuronal phenotype: overexpression of the neuron-enriched miRNAs miR-376a and miR-434 increased the differentiation of neural stem cells into neurons, whereas the opposite effect was observed for the glia-enriched miRNAs miR-223, miR-146a, miR-19, and miR-32. In addition, glia-enriched miRNAs were shown to inhibit aberrant glial expression of neuronal proteins and phenotypes, as exemplified by miR-146a, which inhibited neuroligin 1-dependent synaptogenesis. This study identifies new nervous system functions of specific miRNAs, reveals the global extent to which the brain may use differential miRNA expression to regulate neural cell-type-specific phenotypes, and provides an important data resource that defines the compartmentalization of brain miRNAs across different cell types.
Introduction
MicroRNAs (miRNAs) are 19- to 24-nucleotide (nt) noncoding RNAs that act as important regulators of posttranscriptional gene expression (Ambros, 2004; Kim, 2005). miRNAs bind to messenger RNAs (mRNAs) based on sequence complementarity and direct the degradation or repression of translation of the mRNAs to which they are bound (known as their targets). Typically, a miRNA can bind to many targets and each target may be regulated by multiple miRNAs.
Recent studies have shown that numerous miRNAs exist in mammalian systems, where they play important roles in development, are responsible for regulating cell-type-specific functions in the adult organism, and are involved in disease processes (Bartel, 2009). Interestingly, miRNAs show varying levels in different organs, which is consistent with their anticipated role in regulating tissue-specific protein expression (Lagos-Quintana et al., 2002). Compared with other organs, the brain has a particularly high percentage of tissue-specific and tissue-enriched miRNAs (Lagos-Quintana et al., 2002; Kim et al., 2004; Sempere et al., 2004; Smirnova et al., 2005). The physiological importance of miRNAs in nervous system functions and disease states has also been suggested by previous studies of a small number of brain-enriched miRNAs (Leucht et al., 2008; Mellios et al., 2008; Packer et al., 2008; Cheng et al., 2009; Schratt, 2009). However, the full scope of miRNA-mediated regulation of brain functions is largely unknown. Contributing to the limitations in current knowledge was the lack of data on brain miRNA expression at the cellular level.
Neural tissue is highly heterogeneous, being composed of different types of neurons, astrocytes, and oligodendrocytes, which develop from a common pool of neural progenitor cells (Gage, 2000), and microglia, which develop from the hematopoietic lineage (Ritter et al., 2006). The heterogeneous phenotypes of the various neural cells must be established during cell specification and be maintained throughout the life of the adult organism. To better understand the extent to which brain miRNAs might govern specific cellular phenotypes, we sought to establish and quantify differences in miRNA expression across the four neural cell subtypes. We then proceeded to test the hypothesis that cell-type-specific miRNA expression contributes to neural cell specification and maintenance.
Consistent with our hypothesis, our analyses showed that neural cell subtypes differed extensively in their miRNA expression patterns. Functional testing of the newly identified cell-type-specific miRNAs also indicated that cell-type-specific miRNAs participate in the specification of neuronal versus glial fates. Moreover, we have implicated a number of new miRNAs in the regulation of cell type specification by showing that neuron-specific miRNAs promoted and glia-specific miRNAs inhibited neuronal differentiation. In addition, we show that glial miRNAs are capable of targeting neuron-specific mRNAs and may thereby prevent inappropriate glial expression of neuronal proteins and phenotypes.
In addition to identifying new roles of specific brain miRNAs, our data represent a valuable resource that delineates the relative levels of miRNA expression in each of the four neural cell types. These data and analyses support further study of brain miRNAs that may have important nervous system functions.
Materials and Methods
Cell cultures.
Primary cultures were prepared in accordance with the European Community directive for the care and use of laboratory animals (86/-609/-EEC) and the Swiss Academy of Medical Science and with the authorization (1667.2) of the veterinary office of the canton of Vaud. Dissociated neuronal and glial cultures were prepared from cortexes of postnatal d 1 (P1) rats (of both sexes). To obtain neuronal cultures, cells were grown in neurobasal medium supplemented with B-27 (Invitrogen) and cytosine arabinoside. To obtain glial cells, mixed cortical cultures were grown in basal minimum Eagle's medium (BMEM, Invitrogen) supplemented with 10% fetal calf serum, 0.36% glucose, 0.5 mm glutamine, and 1× penicillin-streptomycin (Invitrogen). After 10–14 d in vitro, microglial cells were collected from the medium of the mixed culture. To eliminate loosely adherent microglia and neurons, mixed cultures were shaken on a rotary shaker at 300 rpm for 30 min, after which the medium was replaced. Oligodendrocyte progenitor cells were collected from the medium of cultures shaken overnight. These were plated onto poly-l-lysine-coated plates and grown in differentiation medium comprising DMEM-Ham's F12 (DMEM-F12) supplemented with 10% fetal calf serum, 0.36% glucose, 0.5 mm glutamine, and 1× penicillin-streptomycin (Invitrogen). The remaining cells attached to the original culture dish shaken overnight comprised astrocytes. Additional procedural details and additional cellular characterization data can be found in Kuhn et al. (2011).
Rat cortical stem cells were obtained from D&B Biosciences (catalog #NSC001) and handled according to the supplier's instructions. Upon reception, cells were plated in DMEM/F-12 with N-2 supplement, basic FGF (20 ng/ml), and l-glutamine (1 mm) on poly-l-ornithine- and fibronectin-coated plates. Fresh FGF was added to the culture daily. One day after plating, cells were infected with lentiviral expression vectors. Three days after lentiviral exposure, differentiation was induced by FGF withdrawal, changing the medium to neurobasal with B-27 supplement (Invitrogen), 1× penicillin-streptomycin (Invitrogen), and 0.5 mm l-glutamine.
RNA isolation and qPCR analysis.
Total RNA used for the qPCR analyses was isolated using the Ambion mirVana system. Reverse transcription for miRNA analyses in primary cultures and mouse brain was conducted using TaqMan miRNA assays and TaqMan miRNA reverse transcription kits (Applied Biosystems). Reverse transcription for mRNA analysis was performed using the High-Capacity cDNA Archive Kit (Applied Biosystems). The resultant cDNA was stored at −20°C.
Gene expression measures were recorded from triplicate culture samples, each of which were analyzed by triplicate qPCR assays. TaqMan assays were performed on a 7900HT Real-Time PCR System and analyzed with SDS 2.3 software (Applied Biosystems). Measurement of miRNAs in cell lines (PC12 or C6) were performed with Quantimir assays and reagents (System Biosciences). Relative expression (V) was calculated by δCt corrected for amplification efficiencies as described in Zucker et al. (2005), using normalization to sno-U6, β-3 tubulin (Tubb3), or β-actin (Actb) expression, as indicated. PCR amplification efficiencies were calculated from standard curves with total RNA input amounts ranging between 10 ng and 10 μg using the equation Ex = 10(−1 slope) − 1.
miRNA microarray sample processing.
Samples were prepared according to the miRNA Microarray System protocol (Agilent Technologies). Total RNA (100 ng) from neurons (N), oligodendrocytes (O), astrocytes (A), and microglia M (three biological replicates for N, O, and M samples and four replicates for A samples) were dephosphorylated with calf intestine alkaline phosphatase (GE Healthcare Europe), denatured with dimethyl sulfoxide, and labeled with pCp-Cy3 using T4 RNA ligase (GE Healthcare Europe). The labeled RNAs were hybridized to Agilent Technologies rat miRNA microarrays (Design ID 019159) for 20 h at 55°C with rotation. After hybridization and washing, the arrays were scanned with an Agilent Technologies microarray scanner using high-dynamic-range settings as specified by the manufacturer. Agilent Technologies's feature extraction software was used to collect the data. The microarray signals were then quantile normalized using GeneSpring software (Agilent Technologies).
Identification of differentially expressed miRNAs.
Differentially expressed miRNAs were identified using the Bioconductor package “limma” (Wettenhall and Smyth, 2004). A statistical model specifying the four cell types (A, N, O, and M) as factors was used. All pairwise comparisons between the four groups were extracted from the model as contrasts (O vs N, O vs A, N vs A, O vs M, A vs M, N vs M). These six contrasts were combined into one F test (analogous to a one-way ANOVA for each gene). P values from the F test were adjusted for multiple testing with Benjamini and Hochberg's method to control the false discovery rate (FDR). To be called differentially expressed, a gene had to meet the following four criteria: (1) a FDR < 1% (F test), (2) a minimum of fivefold change versus the average expression in the other groups, (3) a minimum of twofold change versus each of the other groups taken individually, and (4) an miRNA called enriched in a particular cell type was called present in all replicates of this group and if a gene was called diminished in a particular cell type, it should be called present in all replicates of the other cell type groups. Gene Ontology (GO) analyses of differentially expressed miRNAs were conducted using the NCBI's online DAVID bioinformatics interface (Huang et al., 2008, 2009).
Antagomir experiments.
Locked nucleic modified antagomirs were used to knock down expression of miRNAs (anti-miR-223: GGGGTATTTGACAAACTGACA, anti-miR-19a: (TCAGTTTTGCATAGATTTGCACA, anti-miR-376a: ACGTGGATTTTCCTCTACGAT, anti-miR-434: AGGAGTCGAGTGATGGTTCAAA and anti-miR-124: GGCATTCACCGCGTGCCTTA). The modified positions are indicated in bold letters.
The antagomirs, at a 40 nm concentration, were transfected into PC12 (neuronal) and C6 (glial-like) cell lines to target neuronal (miR-376a, miR-434, miR-124) and glial (miR-223, miR-19a) miRNAs, respectively. Both cell lines were also treated with a control antagomir (ACCAATCGACCAAC). In cotransfection experiments, luciferase–3′-UTR clones were transfected along with respective anti-miRNAs. Twenty-four hours after transfection, cells were lysed in 20 μl of lysis buffer (Promega). The luciferase assay was performed using a dual-luciferase assay kit (catalog #E1960; Promega) and Renilla luciferase was normalized to the untargeted firefly luciferase.
Fluorescence-activated cell sorting (FACS) of Thy1+ and Gfap+ cells from mouse brain.
Brain tissue was collected from 2 male and 2 female 3-week-old C57BL/6-Tg (Thy1-EGFP) mice and 3 male 3-week-old C57BL/6-Tg (Gfap-EGFP) mice. Dissociation of cortex and FACS were performed as described previously (Bélanger et al., 2011). As we have shown in recent cell-resolved analyses of brain tissue, brain parenchyma is principally composed of the 4 neural cell types (neurons, astrocytes, oligodendrocytes, and microglia), with insignificant contributions from endothelial cells, pericytes, fibroblasts, smooth muscle cells, and erythrocytes (Kuhn et al., 2011).
Synapse formation assay.
A synapse formation assay originally developed by Scheiffele et al. (Scheiffele et al., 2000) was used here. This assay examines the synaptogenic activity of an individual gene (or protein). In our implementation of the assay, the cDNA encoding the postsynaptic protein neuroligin 1 was transfected into astrocytes and the effect of miR-146a-mediated regulation of the neuroligin 1-dependent activity was tested. Astrocytes were prepared from P1 rat cortex and plated on poly-l-lysine-coated coverslips. After astrocytes reached confluence, lentiviral expression vectors were used to express neuroligin 1 within the cells. Ten days after lentiviral transduction, neurons prepared from E16 rat cortex were plated on astrocytes at a plating density of 100 cells per well in a 24-well plate. (This low density avoids neuron-to-neuron contacts and neuron-to-neuron synapse formation). Forty-eight hours after the plating of neurons, cells were fixed with 4% paraformaldehyde and further analyzed using immunolabeling and microscopic image analysis. Synapse formation was analyzed by counting neuronal presynapses, which were identified by Synapsin 1/2 immunolabeling.
Immunolabeling and microscopic image analyses.
Cell cultures were washed with cold PBS and fixed with 4% paraformaldehyde (Fluka) for 15 min at room temperature. Cultures were subsequently washed with PBS and incubated in a blocking solution of PBS with 10% normal goat serum (NGS) (DakoCytomation) and 0.1% Triton X-100 (Sigma). Cells were then incubated overnight at 4°C in blocking solution containing the following primary antibodies: mouse monoclonal anti-neuronal nuclei (NeuN) (1:1000, MAB377 clone A60; Millipore), mouse monoclonal anti-neuronal class III tubulin (1:500, TUJ1; Covance), rabbit polyclonal anti-Synapsin 1/2 (1:500, Synaptic Systems), or mouse monoclonal anti-nestin (1:100, BD Biosciences). Secondary antibodies coupled to fluorophores (goat anti-mouse IgG Alexa Fluor 488, goat anti-mouse IgG Alexa Fluor 594, goat anti-rabbit IgG Alexa Fluor 594, and goat anti-rabbit IgG Alexa Fluor 488, all 1:800; Invitrogen) were applied for 1 h at room temperature.
Images of differentiated stem cell cultures were acquired with a BD Biosciences Pathway 855 bioimaging system and cells were counted with ImageJ software (Rasband, 1997; Abramoff et al., 2004). Images for quantitative analyses were acquired under nonsaturating exposure conditions. Stem cells for Pathway 855 analyses were plated in 96-well plates, avoiding the wells in the outermost rows and columns. In stem cell differentiation experiments, 15 replicate wells were analyzed per condition, and the effect of a miRNA on stem cell differentiation was confirmed in a minimum of 3 independent experiments. The statistical significance of the differences between the miRNA overexpression condition and the control condition was tested using unpaired Student's t test.
Images of cell cultures for synapse formation assays were acquired using a Leica confocal microscope.
Immunoblotting.
For analysis of protein expression, cells were harvested in RIPA buffer (Sigma) containing protease inhibitor mixture (Sigma). Proteins were separated on 10% SDS-polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were blocked for 1 h in 30% Odyssey Blocking Buffer (LI-COR Biosciences) with 0.1% Tween 20 in PBS, followed by incubation in primary antibodies diluted in blocking buffer with 0.1% Tween 20 overnight at 4°C. Membranes were rinsed with 0.1% Tween 20/PBS, followed by incubation with secondary antibodies diluted in blocking solution for 1 h at room temperature. After final rinses, blots were scanned with an Odyssey infrared imager and measurements were obtained using Odyssey software (LI-COR Biosciences). Primary and secondary antibodies comprised: mouse monoclonal anti-synaptotagmin 1 (1:1000, clone 41.1; Synaptic Systems), mouse monoclonal anti-neuroligin 1 (1:1000, clone 4C12; Synaptic Systems), rabbit polyclonal anti-tubulin (1:1000; Sigma), donkey anti-mouse IRDye 800CW (1:15,000; LI-COR Biosciences), donkey anti-rabbit IRDye 680 (1:15,000; LI-COR Biosciences), and donkey anti-goat IRDye 800CW (1:15,000; LI-COR Biosciences,).
Luciferase-3′-UTR construct preparation.
3′-UTRs were cloned into the multiple cloning site of psiCHECK-2 vector (catalog #C8021; Promega). 3′-UTR sequences were retrieved from TargetScan and primers were designed to amplify full-length 3′-UTRs. The following primers were used (F indicates forward; R, reverse): F-Syt1-3′-UTR (TTT ACT CGA GCT TTC TGC ATC TGC CCA CAT AG), R-Syt1-3′-UTR (TAA AGC GGC CGC AAT TCC TAA AGA TCA AGC ACG AGG); F-Nova1-3′-UTR (AATT CTCGAG TGCCCCGATTATACGTCAGATT), R-Nova1-3′-UTR (TTAA GCGGCCGC GATGCTACATGATGAACTAAGCAC); F-Nlgn1-3′-UTR (TAT ACT CGA GTT GGC TTT CAA CTT GGA AGA CTC), R-Nlgn1-3′-UTR (TTA TGC GGC CGC TAT GCC TTC CAA AAC CCA ACT C); R-Syt1-3′-UTR_mut (GGT CCT TAG GAA GTT TGT GCT AC), F-Syt1-3′-UTR_mut (GTA GCA CAA ACT TCC TAA GGA CC); R-Nova1_mut1 (GAC TCC ATG GAT AGA CCT TTG TTG), F-Nova1_mut1 (CAA CAA AGG TCT ATC CAT GGA GTC); R-Nova1_mut2 (TTT AGA GAT AGA TTT CTC TAA ATG), F-Nova1_mut2 (CAT TTA GAG AAA TCT ATC TCT AAA); F-Nlgn1-3′-UTR_mut (CAC TCT GAT TAC GTC GAT TGA TTT), R-Nlgn1-3′-UTR_mut (AAA TCA ATC GAC GTA ATC AGA GTG); F-Stat3-3′-UTR (TAA ACT CGA GGG AGC TGA AAG CGG AAA CTG C), R-Stat3 3′-UTR (ATT AGC GGC CGC CTG GAT GTT AAA GTA GTT ACA GCA); F-Erbb4-3′-UTR (TAT ACT CGA GCC AGT AGT TTT GAC ACT TCC G), R-Erbb4-3′-UTR (ATT AGC GGC CGC AGC ATT TGT TTC ATA TTT TAT TGA ATT); F-Hes5-3′-UTR (TAA ACT CGA GAC GGG TGC CTG GAG CTG AC), R-Hes5 3′-UTR (ATT AGC GGC CGC GAA GCC TTC AGA ACA GCT TGT G); F-Mef2C-3′-UTR (TAT TCT CGA GCT CTC TGA AGG ATG GGC AAC AT), R-Mef2C-3′-UTR (TTA TGC GGC CGC AAA ACA TTA CTA CAA GGC AGG CAG); F-Neurog1-3′-UTR (TAA ACT CGA GCC TTT GCA AGA CAA CGT TAA T), R-Neurog1-3′-UTR (ATT AGC GGC CGC CTT TGA TAG CTC ATA ATA ATA GAA).
Luciferase reporter assays.
The 3′-UTR sequences of the target mRNAs were cloned into the psiCHECK-2 vector (Promega). Five nanograms of the 3′-UTR–psi-CHECK-2 construct was cotransfected with 500 ng of a lentiviral vector encoding a miRNA gene into human embryonic kidney 293T (HEK293T). Two days after the transfection, luciferase activity was measured using the Dual-Luciferase assay kit (Promega) according to the manufacturer's instructions. Luminescence was measured on a Tecan Genios Pro plate reader in the linear range of the instrument.
Results
Individual neural cell types have distinct miRNA expression profiles
To address the question of which cell-type-specific miRNAs might play important roles in neural cell subtype specification and maintenance, we first comprehensively evaluated the extent to which the four distinct neural cell types differ in their expression of miRNAs. To our knowledge, full genome-wide profiling comparing miRNA expression in neurons, astrocytes, oligodendrocytes, and microglia had never before been reported. Previous studies had instead compared the miRNA expression in whole brain tissue with the expression in other tissues of the same organism (Sempere et al., 2004; Wienholds et al., 2005) or assessed differences in miRNA expression during different stages of development of individual neural cell types (Krichevsky et al., 2003; Smirnova et al., 2005; Zhao et al., 2010).
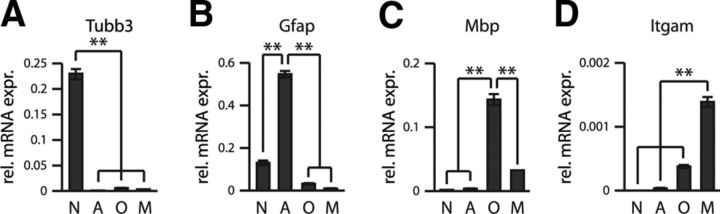
We therefore undertook neural cell-type-specific miRNA expression analyses in primary cultures derived from P1 rat cortex (see Materials and Methods). The cellular composition of these cultures was verified by immunocytochemistry using antibodies against well established cell-type-specific marker antigens: NeuN/Rbfox (neuronal nuclear antigen/RNA binding protein Fox) for mature neurons, GFAP (glial fibrillary acidic protein) for astrocytes, O4 antigen for oligodendrocytes, and integrin α M chain (Itgam) for microglia (Kuhn et al., 2011). In addition, we measured the relative expression levels of known cell-type-specific mRNAs for each of the 4 cell populations: β-3 tubulin (Tuj1) for neurons, Gfap for astrocytes, myelin basic protein (Mbp) for oligodendrocytes, and Itgam for microglia. These mRNA expression values indicated largely segregated expression of the cell-type-specific RNAs, further confirming that the primary cultures used for expression analyses represented the intended cell populations (Fig. 1). We subsequently performed expression profiling analyses of 351 miRNAs using Agilent Technologies Rat Genome V1 microarrays. These analyses showed that neural cell types have remarkably distinct miRNA expression profiles (Fig. 2), with 116 of the examined 351 miRNAs being at least fivefold differentially expressed across the four cell types.
Figure 1.
Distribution of neural cell-type-specific mRNAs in primary cultures. To verify the identity and purity of separated primary neural cells, levels of known cell-type-specific mRNAs were measured by qPCR normalized to β-actin. A, Tubb3. B, Gfap. C, Mbp. D, Itgam. Error bars represent SD. N, Neuronal cultures; A, astroglial cultures; O, oligodendroglial cultures; M, microglial cultures; and rel. mRNA abund., relative mRNA abundance.
Figure 2.
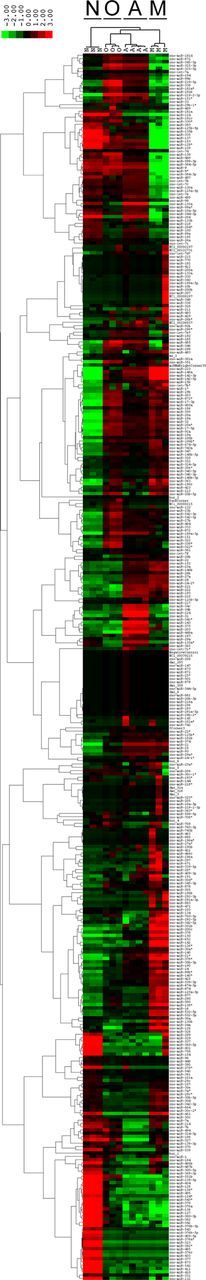
Differential miRNA expression across the four neural cell types. The four neural cell types differ extensively in their miRNA expression profiles. O indicates oligodendrocytes; M, microglia; N, neurons; and A, astrocytes. Scale bar shows the fold change in the miRNA expression in one sample compared with the average expression value in the others. A corresponding table with the means ± SD of microarray measures of expression in the four cell types is available upon request.
We then used the following criteria to identify individual miRNAs that were specifically enriched or diminished in individual cell types: the miRNAs had to have at least a fivefold difference in expression in one cell type compared with the average expression in the other three and at least a twofold difference compared with any other individual cell type, the expression change had to meet a statistical cutoff of false-discovery rate-adjusted (FDR) p ≤ 0.01, and miRNAs to be considered enriched in a cell type had to be reliably detected (called “present” by the Bioconductor package “limma” (Wettenhall and Smyth, 2004) in all of the replicate samples of that cell type. This analysis yielded multiple enriched or diminished miRNAs for neurons and microglia and relatively fewer miRNAs detected as uniquely enriched or diminished in astrocytes or oligodendrocytes (Table 1).
Table 1.
Cell-type-restricted miRNAs considering four cell subtypes
| Cell type | Specifically enriched miRNAs | Specifically diminished miRNAs |
|---|---|---|
| Neurons | miR-124, miR-127, miR-129, miR-129*, miR-136, miR-136*, miR-137, miR-154, miR-300-3p, miR-323, miR-329, miR-341, miR-369-5p, miR-376a, miR-376b-3p, miR-376c, miR-379, miR-382, miR-382*, miR-410, 411, miR-433, miR-434, miR-495, miR-541, miR-543*, miR-551b | miR-21, miR-142-3p, miR-142-5p, miR-146a, miR-150, miR-219-2-3p, miR-223, miR-449a |
| Astrocytes | miR-143, miR-449a | miR-327 |
| Oligodendrocytes | miR-219-2-3p | — |
| Microglia | miR-126, miR-126*, miR-141, miR-142-3p, miR-142-5p, miR-146a, miR-150, miR-200c, miR-223 | miR-9, miR-9*, miR-124, miR-127, miR-129, miR-129*, miR-135a, miR-135b, miR-136, miR-137, miR-153, miR-204, miR-325-3p, miR-335, miR-384-5p |
Shown are miRNAs specifically enriched or diminished in one cell type comparing neurons, astrocytes, oligodendrocytes, and microglia using the criteria of a minimum fivefold difference compared with the average expression in the other three types and a minimum twofold difference compared with any single cell type, differential expression using a statistical cutoff of FDR p < 0.01, and including a filter that enriched miRNAs be detected as present in all replicate samples of that cell type. For further details, please see Materials and Methods.
To focus our analyses on the identification of miRNAs that could have important roles in neural cell type specification, we subsequently restricted our analyses to the differences in neurons, astrocytes, and oligodendrocytes, which develop from the same neural progenitor cells. Applying the same criteria as in the previous analyses (at least a fivefold difference in expression in one cell type compared with the average expression in the other two and a at least a twofold difference compared with any other individual cell type, expression change meeting a statistical cutoff of FDR p ≤ 0.01, and miRNAs to be considered enriched in a cell type being called “present” in all of the replicate samples of that cell type), we identified 53 enriched and 23 diminished miRNAs in neurons, 13 enriched and 9 diminished miRNAs in astrocytes, and 13 enriched and 4 diminished miRNAs in oligodendrocytes (Table 2).
Table 2.
Cell-type-restricted miRNAs considering neurons, astrocytes, and oligodendrocytes only
| Cell type | Specifically enriched miRNAs | Specifically diminished miRNAs |
|---|---|---|
| Neurons | miR-7b, miR-124, miR-124*, miR-127, miR-128, miR-129, miR-129*, miR-132, miR-135b, miR-136, miR-136*, miR-137, miR-139-5p, miR-154, miR-184, miR-188, miR-204, miR-299, miR-300-3p, miR-300-5p, miR-323, miR-329, miR-337, miR-335, miR-341, miR-369-3p, miR-369-5p, miR-376a, miR-376a*, miR-376b-3p, miR-376b-5p, miR-376c, miR-377, miR-379, miR-379*, miR-382, miR-382*, miR-409-5p, miR-410, miR-411, miR-431, miR-433, miR-434, miR-451, miR-466b, miR-485, miR-495, miR-539, miR-541, miR-543*, miR-551b, miR-758, miR-873 | miR-15b, miR-17-3p, 17-5p, miR-18a, miR-19a, miR-20a, miR-20a*, miR-21, miR-32, miR-92a, miR-93, miR-142-3p, miR-142-5p, miR-146a, miR-199a-3p, miR-219-2-3p, miR-219-5p, miR-223, miR-325, miR-338, miR-350, miR-484, miR-542-3p |
| Astrocytes | miR-21, miR-31, miR-34b, miR-34c, miR-135a, miR-143, miR-146a, miR-193, miR-210, miR-221, miR-222, miR-223, miR-449a | miR-128, miR-46b, miR-127, miR-551b, miR-188, miR-137, miR-327, miR-124, miR-494 |
| Oligodendrocytes | miR-17-3p, miR-20a, miR-20b-5p, miR-219-2-3p, miR-219-5p, miR-322*, miR-338, miR-338*, miR-346, miR-351, miR-450a, miR-503, miR-542-3p | miR-449a, miR-204, miR-222, miR-135a |
Shown are miRNAs specifically enriched or diminished in one cell type when comparing only neurons, astrocytes, and oligodendrocytes using the criteria of a minimum fivefold difference compared with the average expression in the other two types and a minimum twofold difference compared with any single cell type, differential expression using a statistical cutoff of FDR p < 0.01, and including a filter that enriched miRNAs be detected as present in all replicate samples of that cell type. For further details, please see Materials and Methods.
To address the issue that cell cultures derived from neonatal brain may still be in the process of development (and therefore not representative of mature phenotypes in the in vivo context), we further validated the differential expression of selected miRNAs in cells acutely isolated from mouse brain. By using FACS on brain samples of mice expressing EGFP under the control of the Thy1 promoter, we were able to successfully separate neurons from glial cells as confirmed by enrichment of neuronal marker Tubb3 and neuronal miR-124 in Thy1+ cells compared with Thy1− cells (Fig. 3A). These studies confirmed the neuron-enriched expression of miR-376a and miR-434 and the neuron-diminished expression of miR-223, miR-146a, and miR-19a (Fig. 3B). By implementing the same approach using Gfap-EGFP transgenic mice, we likewise confirmed the astrocyte-enriched expression of miR-143, miR-193, and miR-449a and the astrocyte-diminished expression of miR-327 and miR-494 (Fig. 3C,D). These data support the applicability of our results to intact brain tissues.
Figure 3.
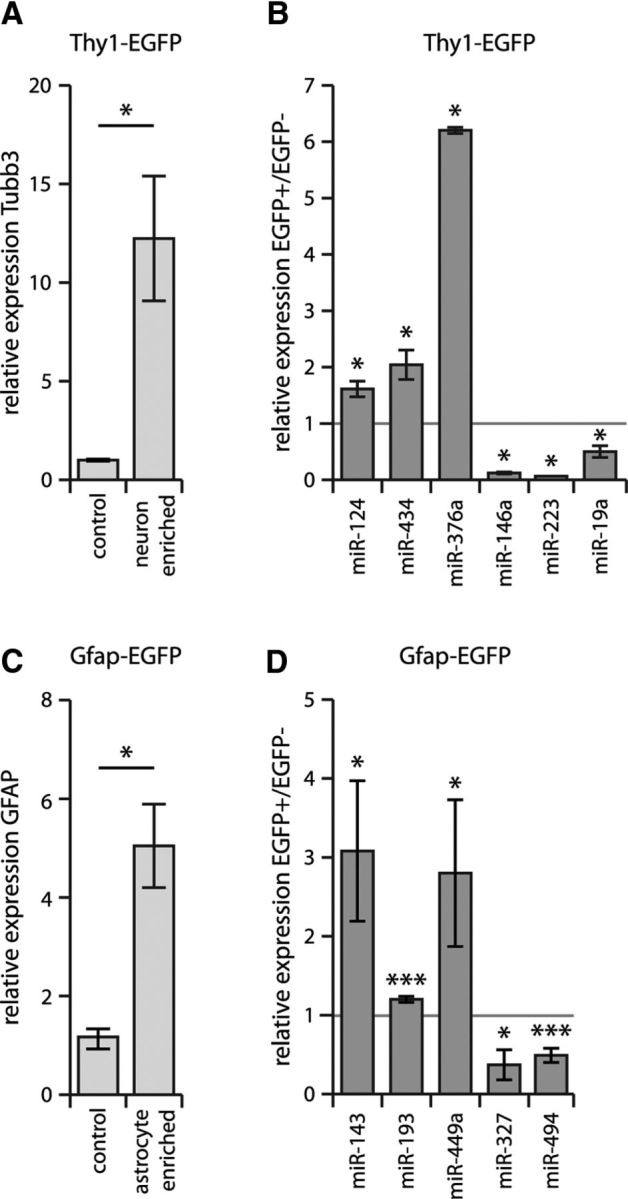
miRNAs identified as being differentially expressed in primary cultures demonstrate the same differential expression in vivo. A, B, Thy1+ neuronal cells (confirmed by Tubb3 enrichment, A) were isolated from mouse brain using FACS from Thy1-EGFP mice (see Materials and Methods). FACS-isolated cellular RNA from EGFP+ and EGFP− cells was used to confirm the neuron-enriched expression of miR-124 (positive control), miR-376a, and miR-434 and neuron-diminished expression of miR-146a, miR-19, and miR-223 (B). Relative expression values normalized to Actb (Tubb3 mRNA) or sno U6 RNA (miRNAs) are shown with respective SEM. *p < 0.05 by unpaired t test comparing EGFP+ and EGFP− samples. C, D, Gfap+ astrocytes (confirmed by Gfap enrichment, C) were isolated from mouse brain using FACS from Gfap-EGFP mice (see Materials and Methods); FACS-isolated cellular RNA confirmed the astrocyte-enriched expression of miR-143, miR-193, and miR-449a and the astrocyte-diminished expression of miR-327 and miR-494 (D). Relative expression values normalized to Actb (Gfap mRNA) or sno U6 RNA (miRNAs) are shown with respective SEM. *p < 0.05; ***p < 0.005 by unpaired t test comparing EGFP+ with EGFP− samples.
Genomic colocalization and coexpression of neural cell-type-specific miRNAs
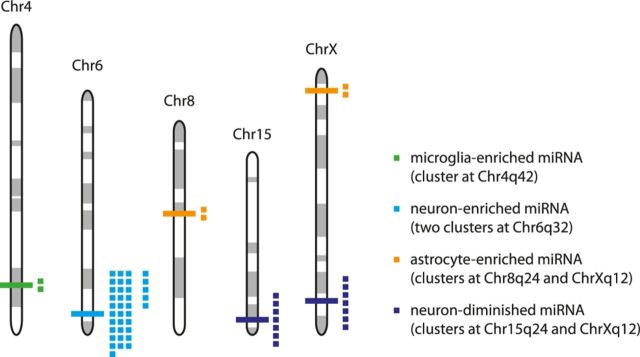
miRNAs are frequently found in genomic clusters (Bartel, 2009). The largest miRNA cluster discovered to date consists of >50 miRNAs in the human imprinted 14q32 domain (Seitz et al., 2004), which has orthologous clusters in the rat (6q32) and mouse (12qF1). Interestingly, a few miRNAs within this cluster have been implicated in regulating synapse-specific genes (Schratt et al., 2006) and shown to exhibit neuronal-activity-dependent expression (Fiore et al., 2009). Because miRNAs found in some genomic clusters have been reported previously to be subject to coordinated transcriptional control, we investigated whether miRNAs within clusters exhibited similar expression patterns across the four neural cell types. We therefore assessed the extent to which the coexpression of neural cell-type-specific miRNAs might reflect their genomic colocalization. Among the miRNAs exhibiting cell-type-specific expression among the four cell types, only 3 clusters were detected: a microglia-enriched cluster on Chr4q42 comprising miR-141 and miR-200c and two neuron-enriched clusters on Chr6q32 (the Rtl1 cluster comprising miR-127, miR-136, miR-337, miR-431, and miR-433 and the miR-379-410 cluster comprising miR-134, miR-136, miR-154, miR-300-3p, miR-300-5p, miR-323, miR-329, miR-369-3p, miR-369-5p, miR-376a, miR-376a*, miR-376b-3p, miR-376b-5p, miR-376c, miR-377, miR-379, miR-379*, miR-382, miR-382*, miR-409-5p, miR-410, miR-411, miR-434, miR-485, miR-487b, miR-494, miR-495, miR-539, miR-541, miR-543*, and miR-758; Fig. 4). Comparing differential miRNA expression only across neurons, astrocytes, and oligodendrocytes, four additional clusters were found: two neuron-diminished clusters on Chr15q24 comprising miR-17-3p, miR-17-5p, miR-18a, miR-19a, miR-20a, miR-20a*, and miR-92a and on ChrXq36 comprising miR-322, miR-322*, miR-351, miR-450a, miR-503, miR-542-3p, miR-542-5p, and two astrocyte-enriched clusters on ChrXq12 comprising miR-34b and miR-34c and on Chr8q24 comprising miR-221 and miR-222.
Figure 4.
Genomic colocalization and coexpression of neural cell-type-specific miRNAs. Clusters of miRNAs showing cell-type-specific expression (see also Results, Genomic colocalization and coexpression of neural cell-type-specific miRNAs). The microglia-enriched cluster on the Chr4q42 comprises rno-miR-141 and miR-200c. The neuron-enriched clusters on Chr6q32 consist of the Rtl1 cluster (comprising rno-miR-127, miR-136, miR-337, miR-431, and miR-433) and the miR-379-410 cluster (comprising miR-134, miR-136, miR-154, miR-300-3p, miR-300-5p, miR-323, miR-329, miR-369-3p, miR-369-5p, miR-376a, miR-376a*, miR-376b-3p, miR-376b-5p, miR-376c, miR-377, miR-379, miR-379*, miR-382, miR-382*, miR-409-5p, miR-410, miR-411, miR-434, miR-485, miR-487b, miR-494, miR-495, miR-539, miR-541, miR-543*, and miR-758). The astrocyte-enriched cluster on Chr8q24 comprises rno-miR-34b and miR-34c. The neuron-diminished cluster on Chr15q24 comprises rno-miR-17-3p, miR-17-5p, miR-18a, miR-19a, miR-20a, miR-20a, and miR-92a. The astrocyte-enriched cluster on ChrXq12 comprises rno-miR-221 and miR-222 and the neuron-diminished cluster on ChrXq36 comprises rno-miR-322, miR-322*, miR-351, miR-450a, miR-503, miR-542-3p, and miR-542-5p.
Evidence for alternate strand utilization among neural miRNAs
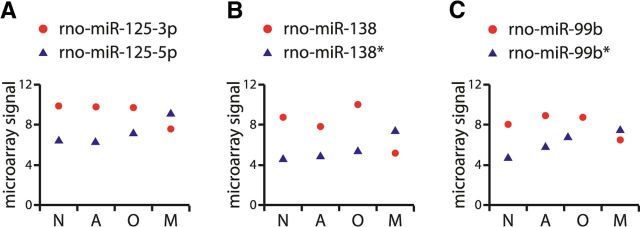
miRNA biogenesis is a complex process comprising several intermediate RNA products. It starts with transcription and genesis of a long primary miRNA, which typically consists of a 33 bp double stranded stem with a loop structure and long single stranded RNA flanking regions (Siomi and Siomi, 2009). Primary miRNAs are then recognized and cleaved by the Microprocessor complex to generate precursor miRNA hairpins (pre-miRNAs) that are ∼65–70 nt long. Pre-miRNAs are subsequently exported to the cytoplasm via exportin5-RanGTP and further processed in the cytoplasm by Dicer, which yields an ∼22 bp miRNA–miRNA* duplex. This miRNA duplex is then loaded onto the Argonaute (Ago) protein, which degrades one strand (the passenger strand) of the duplex while the other strand (the guide strand and mature miRNA) stays associated with Ago to generate RNA-induced silencing complexes. The small RNA cloning strategy used to identify mature miRNAs revealed that the process of the strand selection by Ago is not symmetrical (Lim et al., 2003). Studies investigating the relative thermodynamic stability of the two ends of the pre-miRNA duplex indicated that the strand in which the 5′ end is less tightly paired to its complement remains associated with Ago and performs the functions of the mature miRNA (Khvorova et al., 2003; Schwarz et al., 2003). By convention, the strand that is more frequently found to be the final product is referred to as miRNA and the rarer partner as miRNA*. Alternatively, when their relative persistence in the cell is unknown, they are denoted as miRNA-5p (5′-arm) and miRNA-3p (3′-arm). To assess the plausibility of whether “arm switching” may be used in neural miRNAs, we considered the relative distributions of individual members of miRNA–miRNA* and miRNA-5p–miRNA-3p miRNA duplex pairs. Of 43 duplex pairs we identified among our differentially expressed miRNAs, 39 showed correlated expression in which one of the two strands was always present at higher levels, but the relative expression of the two strands across neural cell types retained the same profile. Interestingly, however, there were three miRNA–miRNA* pairs that were found to have opposite expression patterns (i.e., they maintained a different strand in microglia compared with the other three cell types). These pairs are as follows: miR-125-5p/miR-125-3p, miR-138/miR-138*, and miR-99b/miR-99b* (Fig. 5). These data support the hypothesis that the relative strand utilization of some miRNAs are dependent on the cell type and not purely on the thermodynamic stability of their miRNA-miRNA* hairpin duplex (Chiang et al., 2010).
Figure 5.
miRNA strand utilization differs across neural cell subtypes. For each miRNA, the A values from the microarray analysis of miRNA expression in the four cell types are shown. N indicates neurons; A, astrocytes; O, oligodendrocytes; and M, microglia. Error bars representing SD are contained within the data symbols. Data are presented for miR-125-3p/-5p (A), miR-138/-138* (B), and miR-99b/-99b* (C).
Neuron- or glia-enriched miRNAs promote their respective differentiation programs in neural stem cells
Although neurons, astrocytes, and oligodendrocytes develop from the same stem cells, they fulfill highly specialized and distinct functions in the mature nervous system. Having observed that their miRNA profiles were also very divergent, we continued the pursuit of our hypothesis to determine whether these newly identified differentially expressed mRNAs might have important roles in neural cell specification. We tested this hypothesis for several miRNAs that showed either high enrichment or exclusion in neurons: neuron-enriched miRNAs comprised miR-124, miR-367a, and miR-434 and glial-enriched miRNAs comprised miR-223, miR-19, miR-32, and miR-146a. We overexpressed the selected miRNAs in rat cortical stem cells using lentiviral-vector-based gene delivery. To exclude the possibility that an absence of a miRNA effect on stem cell differentiation was due to its failure to be expressed by transduction with the lentiviral vector, we confirmed the overexpression of each miRNA using qPCR. In some cases, high overexpression levels led to a decrease in stem cell numbers and therefore confounded a definitive experimental conclusion. Therefore, we decreased the concentration of the applied vector to limit overexpression levels of each miRNA to a maximum of 100-fold compared with endogenous. In addition, we did not take into consideration any data from treatments that decreased the numbers of cells to <35% of the noninfected control.
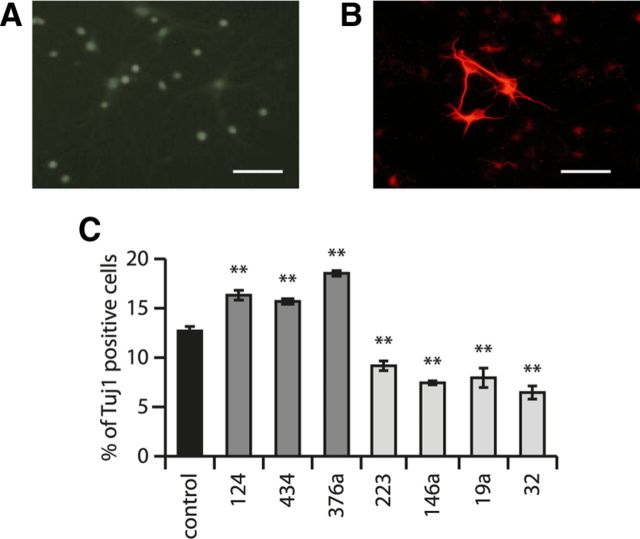
Three days after exposure of neural stem cells to the desired miRNA-expressing lentiviral vector (when the desired expression levels of miRNAs were attained), we induced stem cell differentiation by FGF withdrawal (see Materials and Methods) and, after an additional 3 d, evaluated the fraction of cells differentiating into neurons by comparing the number of Tuj1+ cells with the total number of cells assessed by Hoechst staining. Examples of cell morphology before and after FGF withdrawal are shown in Figure 6, A and B. Effects of the selected miRNAs on neural stem cell differentiation are shown in Figure 6C. These results show that several miRNAs had strong effects on neural stem cell differentiation. As a positive control, we showed that neuronal miR-124 increased the differentiation of stem cells toward neuronal cell fate, in agreement with previous observations (Krichevsky et al., 2003). Of our newly identified neuron-enriched miRNAs, miR-376a and miR-434 also increased the number of cells differentiating toward a neuronal phenotype, with an effect size similar to miR-124 (Fig. 6C).
Figure 6.
Effects of cell-type-selective miRNAs on neural stem cell differentiation. A, Neural stem cell cultures before differentiation labeled with anti-nestin antibody. Scale bar, 15 μm. B, Neural cultures after FGF withdrawal and differentiation labeled with anti-Tuj1 antibody (against Tubb3 protein). Scale bar, 15 μm. C, Overexpression of neuron-enriched miRNAs miR-124, miR-434, and miR-376a via lentivirus-mediated transduction in neural stem cells increases the number of Tuj1+ cells, whereas overexpression of glial-enriched miRNAs miR-223, miR-146a, miR-19a, and miR-32 in neural stem cells decreases the number of Tuj1+ cells. Control samples comprised parallel cultures not exposed to lentiviral vectors. The mean of 15 independent replicates is shown; error bars represent SEM. **p < 0.01 by unpaired t test compared with control.
Compared with neuron-enriched RNAs, glia-enriched miRNAs had the opposite effect on proneuronal differentiation. Several glia-enriched miRNAs decreased the number of stem cells differentiating into neurons, which was apparent by a decreased fraction of Tuj1+ cells (Fig. 6C).
To identify molecular mechanisms by which these miRNAs regulate cell specification, we considered which of the mRNA targets of these miRNAs would be involved in neuronal or glial differentiation. Using the TargetScan algorithm (Lewis et al., 2005) we found several target predictions for genes with a clear functional role in neural stem cell differentiation and tested these target predictions using 3′-UTR luciferase assays. For these experiments, we implemented neuron-like (PC12) and glia-like (C6) cell lines, which were determined to have the same relative expression of neuron-enriched and glia-enriched (neuron-diminished) miRNAs (Fig. 7A,B) and in which the corresponding miRNA levels could be diminished by transfection of LNA antagomirs (Fig. 7C,D). miR-434 negatively regulated the 3′-UTR of the Stat3 mRNA, the targeting of which has been shown previously to mediate miR-124-dependent proneuronal differentiation of neural stem cells (Krichevsky et al., 2003; Fig. 7E,G). Similarly, we demonstrated that miR-376a targets Hes5 mRNA, the negative regulation of which has also been found to promote neuronal differentiation in other studies (Ohtsuka et al., 1999; Fig. 7I,K).
Figure 7.
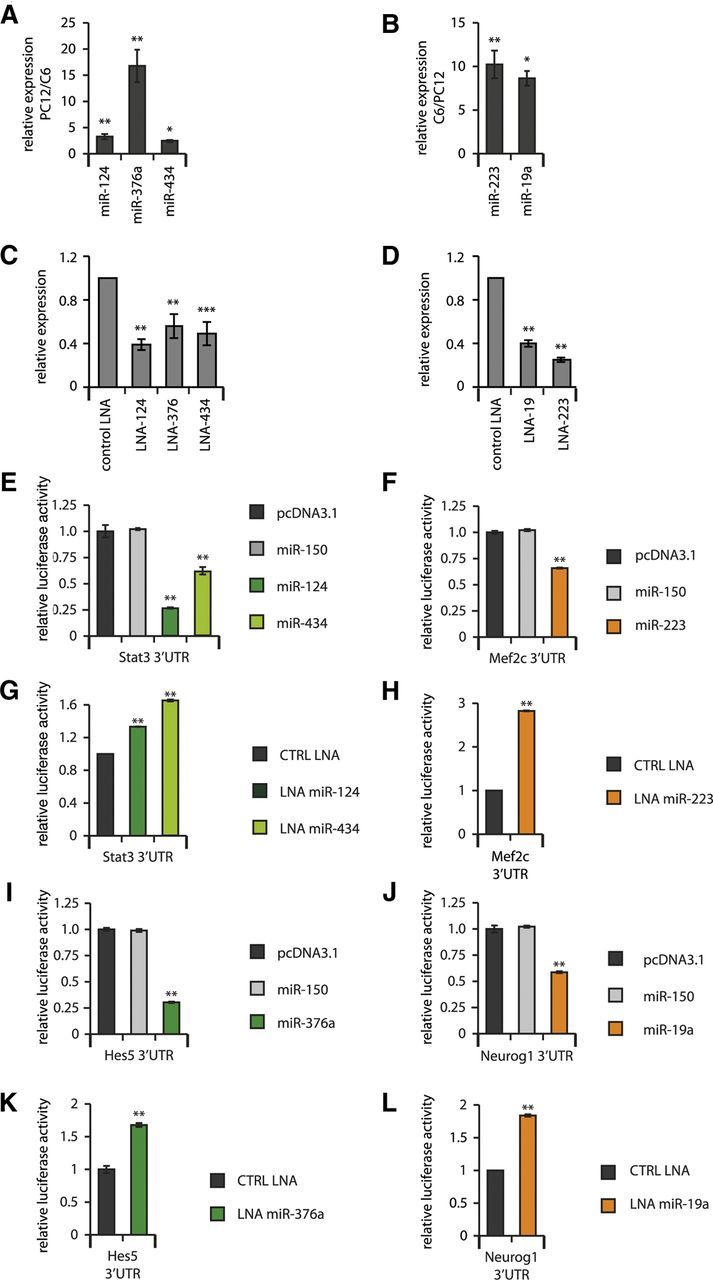
Neuron-enriched miRNAs negatively regulate 3′-UTRs of genes that would otherwise promote glial development, whereas glia-enriched miRNAs negatively regulate 3′-UTRs of genes that would otherwise promote neuronal development. A, Neuron-like PC12 cell line shows enriched expression of neuronal miRNAs compared with glia-like C6 cells. *p < 0.05; **p < 0.01 by unpaired t test comparing PC12 and C6 RNA samples. B, Glia-like cell line C6 shows enriched expression of neuronal miRNAs compared with glia-like C6 cells. *p < 0.05; **p < 0.01 by unpaired t test comparing C6 and PC12 RNA samples. C, Neuron-enriched miRNAs are successfully diminished by LNA antagomir transfection in PC12 cells. **p < 0.01; ***p < 0.001 by unpaired t test compared with control LNA-transfected cells. D, Glia-enriched miRNAs are successfully diminished by LNA antagomir transfection in C6 cells. **p < 0.01 by unpaired t test compared with control LNA-transfected cells. E, Overexpression of neuron-enriched miR-124 or miR-434 negatively regulates the 3′-UTR of the Stat3 mRNA. F, Overexpression of miR-223 negatively regulates the Mef2c 3′-UTR. G, Antagomirs against miR-124 or miR-434 positively regulate the 3′-UTR of the Stat3 mRNA. H, An antagomir against miR-223 positively regulates the Mef2c 3′-UTR. I, Overexpression of neuron-enriched miR-376a negatively regulates the 3′-UTR of the Hes5 mRNA. J, Overexpression of miR-19a negatively regulates the Neurog1 3′-UTR. K, Antagomir against miR-376a positively regulates the 3′-UTR of the Hes5 mRNA. L, Antagomir of miR-19a positively regulates the Neurog1 3′-UTR. For E–L, **p < 0.01 by unpaired t test compared with control miRNA- or control LNA-treated cells. Results shown are the mean of 6 independent replicates, error bars represent SD. The additional control shown in E, F, I, and J was transfected with empty pcDNA3.1 plasmid vector.
Because glia-enriched miRNAs had the opposite effect on proneuronal differentiation, we also identified relevant neuronal targets. Among the predicted targets for miR-223 and miR-19a, we confirmed the targeting of Mef2c and Neurog1, which have been reported to stimulate neuronal differentiation (Fode et al., 2000; Li et al., 2008; Fig. 7F,H,J,L).
Glia-enriched miR-146a targets neuron-specific genes and functions
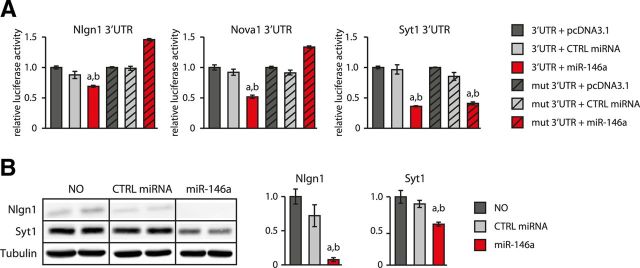
In considering the potential targets of various cell-type-specific miRNAs, we observed that some glia-enriched miRNAs were predicted to target genes known to have important functions in mature neurons. One such miRNA is miR-146a; GO analysis of the predicted miR-146a targets (Huang et al., 2008, 2009) showed significant overrepresentation of GO terms associated with neuron differentiation, axonogenesis, and synaptic transmission. We therefore sought to examine the functional significance of such miRNA target pairing. For this purpose, we chose to focus on three predicted miR-146a targets with neuron-specific expression and important, well characterized functional roles in neuronal cells: neuroligin 1 (Nlgn1), neuro-oncological ventral antigen 1 (Nova1), and synaptotagmin 1 (Syt1).
Because sequence-based target prediction alone is subject to false-positive identification, we first validated that Nlgn1, Nova1, and Syt1 mRNA 3′-UTRs indeed interact with miR-146a using 3′-UTR luciferase reporter assays (Fig. 8A). and confirmed for Nlgn1 and Syt1 that this interaction could lead to a decrease in endogenous protein levels (Fig. 8B). We also investigated whether this effect is due to the predicted binding events using deletion mutants of the identified miR-146a binding sites in the corresponding 3′-UTRs. As predicted, mutations in the Ngln1 and Nova1 3′-UTRs abrogated their regulation by miR-146a in the 3′-UTR-luciferase assay, confirming the functionality of the expected sites (Fig. 8A, first and second panels). Conversely, mutation of the predicted site in the 3′-UTR of Syt1 did not abolish its regulation by miR-146a. Combined with the protein expression data, this result implies that Syt1 is a target of miR-146a, but that the actual binding site is different from the predicted one (Fig. 8A, third panel); alternatively, Syt1 might be an indirect target of miR-146a.
Figure 8.
miR-146a inhibits the expression of neuron-specific targets Nlgn1, Nova1, and Syt1. A, miR-146a inhibits the expression of luciferase reporter constructs carrying the 3′-UTRs of Nlgn1, Nova1, and Syt1. Deleting the TargetScan-predicted miR-146a-binding sites in Nlgn1 and Nova1 3′-UTRs abolished miR-146a-mediated repression. Mean values of 6 independent replicates are shown; error bars represent SD. **p < 0.01 by unpaired t test compared with control samples transfected with empty pcDNA3.1 vector. B, Western blot experiments showing that miR-146a expression in neurons also inhibits accumulation of endogenous Nlgn1 and Syt1 proteins.
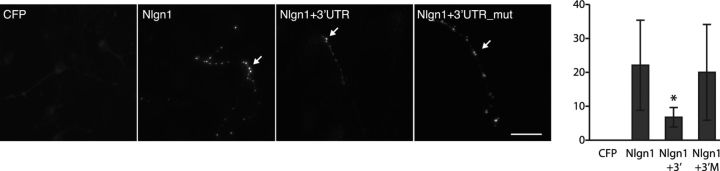
Given these results, we hypothesized that miR-146a might act to safeguard against the expression of neuronal genes in glial cells. It has been shown previously that heterologous expression of key synaptogenic proteins in non-neuronal cells allows them to receive projections from neurons and subsequently induce morphological and functional presynaptic differentiation in the contacting axons (Biederer and Scheiffele, 2007). Neuroligin 1 is among the neuronal proteins to possess such synaptogenic activity (Scheiffele et al., 2000; Biederer and Scheiffele, 2007), and we therefore postulated that the role of miR-146a expression in glia might be to prevent the formation of inappropriate connections with neuronal cells. To test this hypothesis, we transfected plasmids encoding Nlgn1 with its endogenous full-length 3′-UTR or artificially mutated 3′-UTR into primary cortical astrocytes and assessed the ability of cortical neurons to form synapses onto transfected astroglial cells. These experiments showed that the cDNA without the Nlgn1 3′-UTR allowed sufficient Nlgn1 expression to permit synaptogenic activity in adjacent neurons (as evaluated by quantifying numbers of Syn 1/2+ specializations) (Fig. 9). Conversely, adding back the 3′-UTR inhibited the Nlgn1 synaptogenic activity (Fig. 9). Moreover, selectively mutating the miR-146a-binding site restored synaptogenic activity, thereby supporting a specific role for miR-146a in regulating this phenotype. This result indicates that the expression of miR-146a by glial cells safeguards against their producing Nlgn1 and receiving inappropriate neuronal projections. By extension, this also implies that the targeting of other neuronal mRNAs by miR-146a and other glia-enriched miRNAs might likewise prevent glia from mistakenly adopting neuron-specific phenotypes.
Figure 9.
miR-146 prevents neurons from forming inappropriate synaptic contacts by targeting the Nlgn1 mRNA. Left panels show representative micrographs. Scale bar, 10 μm. The graph on the right represents the number of Syn 1/2+ punctae per field from 5 independent samples, 10 fields per sample. Error bars represent SD. *p < 0.05 by unpaired t test compared with Nlgn1 without its endogenous 3′-UTR sequence. CFP indicates cyan fluorescent protein. Scale bar, 10 μm.
Discussion
To define the role of an miRNA, it is crucial to determine both its molecular targets and its cellular expression. In a neurobiological context, this issue is important in comparing miRNA-mediated events in brain with those in other tissues and in identifying cell-type-specific miRNA functions in brain, which is composed of distinct cell types that fulfill very different roles. Previous efforts in this area had focused on brain- or neuron-enriched miRNAs (Lagos-Quintana et al., 2002; Kim et al., 2004; Seitz et al., 2004; Sempere et al., 2004; Smirnova et al., 2005) or on miRNAs in which the expression levels changed during neural cell differentiation (Krichevsky et al., 2003; Lau et al., 2008). However, a genome-wide investigation of miRNA expression in neurons, astrocytes, oligodendrocytes, and microglia had not been available previously.
Our miRNA microarray results are in agreement with previously published data on single miRNAs with neuron-associated functions. These include neuronal expression and/or functions for miR-124 (Lagos-Quintana et al., 2002; Smirnova et al., 2005), miR-128 (Smirnova et al., 2005), miR-132 (Vo et al., 2005; Klein et al., 2007; Wayman et al., 2008; Hansen et al., 2010; Impey et al., 2010), miR-137 (Siegel et al., 2009; Smrt et al., 2010; Willemsen et al., 2011), miR-184 (Nomura et al., 2008), and the miR379-410 miRNA cluster on chromosome 6 (Seitz et al., 2004). Similarly, we confirmed the glial expression of miR-219 and miR-338, which have been implicated previously in the differentiation of oligodendrocytes (Lau et al., 2008; Dugas et al., 2010; Zhao et al., 2010), and the astrocyte-enriched expression of miR-21, which has been reported to be under the transcriptional control of neuron-restrictive silence factor (NRSF/REST) (Jørgensen et al., 2009) and aberrantly induced in neurons after traumatic brain injury (Redell et al., 2011). These results, together with the confirmation of the differential expression of 10 newly identified cell-type-specific miRNAs by FACS-qPCR, support the applicability of our results to other contexts.
Our microarray results showed that the four neural cell types differ extensively in their miRNA profiles. Moreover, hierarchical clustering of the expression profiles showed that cell types of common developmental origin differed least in their miRNA composition (astrocytes and oligodendrocytes), while neurons and microglia showed more unique miRNA expression profiles. These results indicate large differences in miRNA utilization between cells of neural and hematopoietic lineages. Our data also provide rationale for future studies of cell-type-specific regulation of miRNA expression and processing, given the interesting finding of alternate miRNA strand utilization in microglial cells.
Because neurons, astrocytes, and oligodendrocytes develop from the same precursor cells (Gage, 2000), we investigated whether our novel cell-type-specific miRNAs might play important roles in neural stem cell fate determination (Krichevsky et al., 2003; Le et al., 2009). While confirming that miR-124 is a potent determinant of neuronal identity (Krichevsky et al., 2003; Visvanathan et al., 2007), we were able to significantly expand the list of miRNAs involved in cell fate specification based on our newly identified cell-type-enriched miRNAs. We discovered that miR-434 and miR-376a drive stem cell differentiation toward a neuronal fate and identified Stat3, Erbb4, and Hes5 as targets in which negative regulation mediates this effect. Conversely, we demonstrated that the glia-enriched miRNAs miR-223, miR-146a, miR-19, and miR-32 were able to inhibit neuronal differentiation, with corresponding mRNA targets including Mef2c for miR-223 and Neurog1 for miR-19a. In addition to examining whether other miRNAs discovered to have cell-type-specific expression might also participate in embryonic neural stem cell differentiation, we intend to examine the roles of our cell-type-restricted miRNAs in adult brain neurogenesis in future studies.
The number of neural cell-type-specific miRNAs detected in our arrays is much larger than the set implicated previously in brain gene expression. Therefore, our study will be a valuable resource for future neuroscience miRNA research by delineating miRNAs expressed in each neural cell type and supporting future experiments to elucidate cell-type-specific miRNA functions.
In summary, we have found through our comprehensive analyses that levels of specific neural-cell-expressed miRNAs differ greatly across the four principal brain cell types. In addition to representing a valuable resource for cell-type-specific miRNA expression in brain, our study identifies new miRNA participants in neural stem cell differentiation and shows that miRNAs can prevent the inappropriate expression of cell-type-specific genes in fully differentiated cells. These studies therefore provide novel evidence that cell-type-specific miRNAs have important roles in both establishing and maintaining distinct neuronal and glial phenotypes.
Footnotes
Acknowledgements: Funding support was from the Roche Foundation and the Ecole Polytechnique Fédérale de Lausanne (to A.J. and R.L.-C.), Council of Scientific and Industrial Research of India (project #BSC0123, “Comparative Genomics of noncoding RNA,” to B.P. and R.R.), and the Indo Swiss Joint Research Programme (to B.P. and R.L.-C.). We thank Johann Weber, Floriane Consales, and the Lausanne Genomic Technologies Facility for assistance with the microarray experiments, Lely Feletti and Liliane Glauser for the preparation of primary cultures, Maria de Fatima Rey for the production of lentiviral expression vectors, Joel Gyger and Jean-Marie Petit for help with FACS experiments, Darren Moore and Hilal Lashuel for their collegial input, and Ralf Schneggenburger and Nicolas Toni for helpful comments on the manuscript.
The authors declare no competing financial interests.
References
- Abramoff M, Magalhaes P, Ram S. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bélanger M, Yang J, Petit JM, Laroche T, Magistretti PJ, Allaman I. Role of the glyoxalase system in astrocyte-mediated neuroprotection. J Neurosci. 2011;31:18338–18352. doi: 10.1523/JNEUROSCI.1249-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T, Scheiffele P. Mixed-culture assays for analyzing neuronal synapse formation. Nat Protocols. 2007;2:670–676. doi: 10.1038/nprot.2007.92. [DOI] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, Blelloch R, Schroth GP, Nusbaum C, Bartel DP. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT, Barres BA. Dicer1 and miR-219 are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65:597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, Greenberg ME, Schratt G. Mef2-mediated transcription of the miR379–410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- Hansen KF, Sakamoto K, Wayman GA, Impey S, Obrietan K. Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS One. 2010;5:e15497. doi: 10.1371/journal.pone.0015497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, Davare M, Lesiek A, Fortin D, Ando H, Varlamova O, Obrietan K, Soderling TR, Goodman RH, Wayman GA. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol Cell Neurosci. 2010;43:146–156. doi: 10.1016/j.mcn.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen HF, Terry A, Beretta C, Pereira CF, Leleu M, Chen ZF, Kelly C, Merkenschlager M, Fisher AG. REST selectively represses a subset of RE1-containing neuronal genes in mouse embryonic stem cells. Development. 2009;136:715–721. doi: 10.1242/dev.028548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci U S A. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Thu D, Waldvogel HJ, Faull RLM, Luthi-Carter R. Population-specific expression analysis (PSEA) reveals molecular changes in diseased brain. Nat Methods. 2011;8:945–947. doi: 10.1038/nmeth.1710. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28:11720–11730. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le MT, Xie H, Zhou B, Chia PH, Rizk P, Um M, Udolph G, Yang H, Lim B, Lodish HF. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol. 2009;29:5290–5305. doi: 10.1128/MCB.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucht C, Stigloher C, Wizenmann A, Klafke R, Folchert A, Bally-Cuif L. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat Neurosci. 2008;11:641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li H, Radford JC, Ragusa MJ, Shea KL, McKercher SR, Zaremba JD, Soussou W, Nie Z, Kang YJ, Nakanishi N, Okamoto S, Roberts AJ, Schwarz JJ, Lipton SA. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc Natl Acad Sci U S A. 2008;105:9397–9402. doi: 10.1073/pnas.0802876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. 2008;17:3030–3042. doi: 10.1093/hmg/ddn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Kimura M, Horii T, Morita S, Soejima H, Kudo S, Hatada I. MeCP2-dependent repression of an imprinted miR-184 released by depolarization. Hum Mol Genet. 2008;17:1192–1199. doi: 10.1093/hmg/ddn011. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as Notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS. ImageJ. Bethesda, MD: National Institutes of Health; 1997. [Google Scholar]
- Redell JB, Zhao J, Dash PK. Altered expression of miRNA-21 and its targets in the hippocampus after traumatic brain injury. J Neurosci Res. 2011;89:212–221. doi: 10.1002/jnr.22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter MR, Banin E, Moreno SK, Aguilar E, Dorrell MI, Friedlander M. Myeloid progenitors differentiate into microglia and promote vascular repair in a model of ischemic retinopathy. J Clin Invest. 2006;116:3266–3276. doi: 10.1172/JCI29683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaillé J. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, Hübel K, Dekker F, Hedberg C, Rengarajan B, Drepper C, Waldmann H, Kauppinen S, Greenberg ME, Draguhn A, Rehmsmeier M, Martinez J, Schratt GM. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- Smirnova L, Gräfe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, Teng ZQ, Luo Y, Peng J, Bordey A, Jin P, Zhao X. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase Mind Bomb-1. Stem Cells. 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Willemsen MH, Vallès A, Kirkels LAMH, Mastebroek M, Olde Loohuis N, Kos A, Wissink-Lindhout WM, de Brouwer APM, Nillesen WM, Pfundt R, Holder-Espinasse M, Vallée L, Andrieux J, Coppens-Hofman MC, Rensen H, Hamel BCJ, van Bokhoven H, Aschrafi A, Kleefstra T. Chromosome 1p21.3 microdeletions comprising DPYD and MIR137 are associated with intellectual disability. J Med Genet. 2011;48:810–818. doi: 10.1136/jmedgenet-2011-100294. [DOI] [PubMed] [Google Scholar]
- Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X, Mi QS, Xin M, Wang F, Appel B, Lu QR. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65:612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker B, Luthi-Carter R, Kama JA, Dunah AW, Stern EA, Fox JH, Standaert DG, Young AB, Augood SJ. Transcriptional dysregulation in striatal projectionand interneurons in a mouse model of Huntington's disease: neuronal selectivity and potential neuroprotective role of HAP1. Hum Mol Genet. 2005;14:179–189. doi: 10.1093/hmg/ddi014. [DOI] [PubMed] [Google Scholar]