Abstract
This study describes a technical platform that allows us to measure the pore-forming activity of antimicrobial peptides (AMPs) in the lipid bilayer and estimate antimicrobial activity. We selected six different AMPs of diverse species from urochordata to vertebrata and measured the channel current signals using a microfabricated lipid bilayer system. As a result of the electrophysiological measurements, we were able to estimate the pore-forming activity and roughly predict the antimicrobial activity although there was not a strong correlation between the pore-forming activity and the variety of species. Our method will be a unique tool for analyzing a wide variety of diverse AMPs.
1. Introduction
Antimicrobial peptides (AMPs) are short, immunity-related peptides that act against bacteria, viruses, and parasites and also have demonstrated an ability to act against transformed or cancerous cells.1 AMPs are ubiquitous among eukaryotes, and the modes of action by which these peptides interact with malignant cells and microbes are diverse, such as follows: (1) interference with protein folding and DNA, protein, and cell wall synthesis as endogenous activities2,3 and (2) pore formation in the cell membrane as an exogenous activity.4,5 Among them, pore formation in cell membranes has broad-spectrum antibiotics with the membrane disruption. This physical activity will be hard to make tolerance. A short (<50 a.a.) peptide that has cationic amino acids initially binds to the bacterial surface because it consists of an anionic lipid membrane. The binding peptide forms an α-helical structure that is amphiphilic and constructs a transmembrane structure; then, the peptide monomers are assembled and form pores using “barrel-stave”,6−11 “carpet”,12,13 or “toroidal pore”14−18 models.19 The AMP structure and mode of action are conserved in large animals, for instance, those from urochordata to vertebrata.20 In this study, we attempted to understand the difference of pore-forming activity depending on the species. To investigate this subject, we used electrophysiological analysis to examine the pore-forming activity of AMPs.
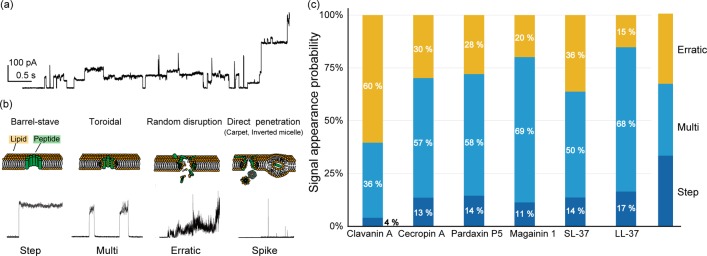
We have previously developed a method of directly observing and analyzing pore formation using electrophysiological measurement in real time with microfabricated lipid bilayer system (Figure 2).21−23 We were able to assign between the channel current signals and the pore-forming models proposed previously.24 The barrel-stave model pore in which the rigid-ring pore exhibits step-like signals. In contrast, the toroidal-model pore exhibits multilevel current signals. The current signal, in this case, fluctuates after jumping up and returns to the initial state. In the carpet model, this model was assigned as an erratic signal, which exhibits random, fluctuating current signals, whereas it could not assign perfectly. Based on this method, we investigate the pore-forming activities of a wide range of species from ascidiacea to mammalia. We select six different AMPs:
-
(1)
Clavanin A (Cla, VFQFLGKIIHHVGNFVHGFSHVF, 23 a.a.) is from Styela clava (a sea squirt).25
-
(2)
Cecropin A (Cec, KWKLFKKIEKVGQNIRDGIIKAGPAVAVVGQATQIAK, 37 a.a.) is from Hyalophora cecropia (a moth).26
-
(3)
Pardaxin P5 (Par, GFFALIPKIISSPLFKTLLSAVGSALSSSGDQE, 33 a.a.) is from Pardachirus marmoratus (a red sea sole).27
-
(4)
Magainin 1 (Mag, GIGKFLHSAGKFGKAFVGEIMKS, 23 a.a.) is from Xenopus laevis (an African clawed frog).28
-
(5)
SL-37 (SLGNFFRKARKKIGEEFKRIVQRIKDFLQHLIPRTEA, 37 a.a.) is from Hylobates moloch (a gibbon).29
-
(6)
LL-37 (LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES, 37 a.a.) is from Homo sapiens (a human).30
Figure 2.
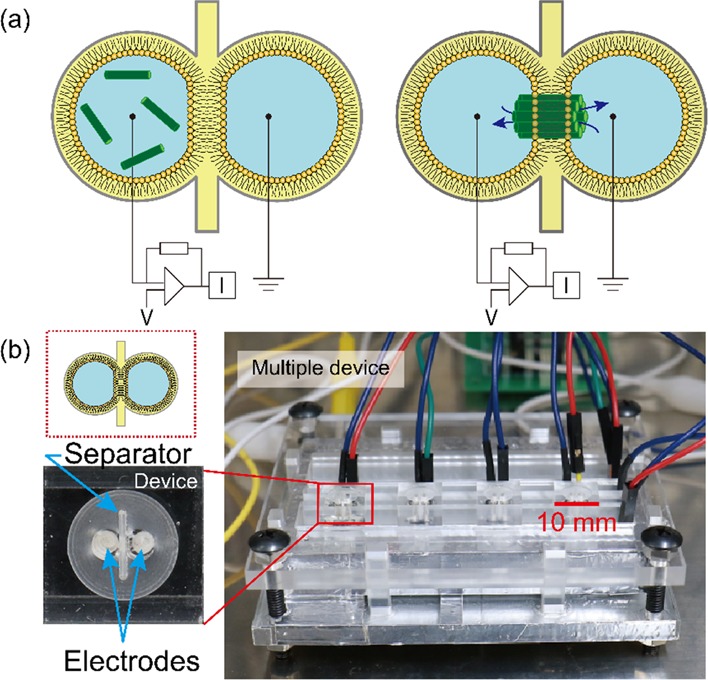
Method and setup for the electrophysiological measurements. (a) Formation of the lipid bilayer using the droplet contact method in the chamber, and the pore formation of antimicrobial peptides with assembling the monomers. (b) Microfabricated device used for electrophysiological measurements; this device has four individual chambers for the droplet contact method.
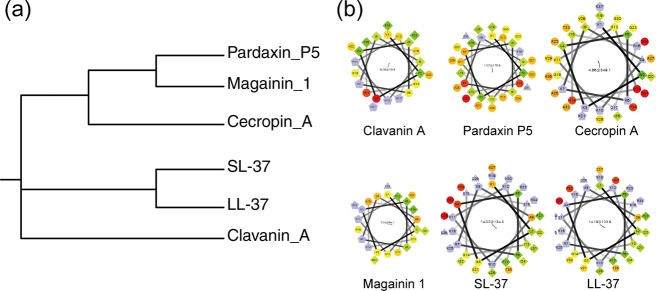
These peptides are genetically distant except for SL-37 and LL-37, and the antimicrobial activity with the α-helical structure and pore formation are preserved in all AMPs. To estimate the relatedness of these peptides, we describe the molecular dendrogram prepared by ClustalW as shown in Figure 1a. This diagram was created using peptide sequences because the genetic relationships among them are unknown. Par, Mag, and Cec are relatively closely related. Cla is isolated, and SL-37 and LL-37 are close, as was predicted due to their inclusion in the same gene family (cathelicidin antimicrobial peptide, CAMP family). These results mirror the tendency of the evolution of a biological species. These AMPs have an amphiphilic structure in the helices, commonly known as the helical wheel structure that is shown in Figure 1b. Based on this molecular information, we investigate the molecular basis of the pore-forming activity and its relations with antimicrobial activity using electrophysiological methods in the context of biological diversity.31
Figure 1.
(a) Molecular dendrogram of AMPs calculated with ClustalW. (b) Helical wheel structures of AMPs.
2. Results
2.1. Current Signal Classification and Estimation of Pore Formation
The AMPs used in this study form pores or defects in lipid bilayer membranes. The typical current and time traces of Mag are shown in Figure 3a. Several shapes of the current signals were observed as previously reported. We have already proposed the assignment of these current signals into the molecular mechanism of pore or defect formation in the lipid membrane. We are still trying to improve this assignment, and they are updated in this study, as described below.24,32
-
(1)
Step signal: In this type of current signal, the current jumps up orthogonally and maintains a plateau state. This signal can apply to the barrel-stave model wherein transmembrane peptides are tightly assembled and form a rigid circular pore (Figure 3b).
-
(2)
Multilevel signal: In this type of current signal, the current fluctuates after jumping up and returns to the initial state. This current may indicate a toroidal model wherein transmembrane peptides form a pore with lipids. In this model, the size of the pore can change dynamically with or without the participation of the lipid between the monomers (Figure 3b).
-
(3)
Erratic signal: In this type of current signal, the current randomly increases with fluctuation. We considered that this signal might indicate random disruption behavior33 because there is not a suitable pore-forming model. The random current behavior of the erratic signal might be caused by the random size of the pore or cluster (Figure 3b).
-
(4)
Spike signal: In this type of current signal, the current suddenly rises and then returns to the baseline over a period of less than 20 ms. This signal indicates an instantaneous membrane defect. We applied this signal to peptide permeation through the membrane. Three different models for direct penetration have been reported: pore formation, inverted micelle formation, and carpet models.34 The pore formation was already assigned to other current signals. Therefore, we assign the spike signal as the inverted micelle and/or carpet model. The inverted micelle model suggests forming the inverted micelle structure in the lipid membrane that encapsulates the peptides and releases them into the cytosol by the inversion (Figure 3b). The carpet model has been described as peptides that bind parallel to the lipid bilayer surface and, after reaching sufficient coverage, unwrap lipids as a peptide–lipid cluster from the membrane (Figure 3b).
The definition of the shape of signals is shown in Figure S2.
Figure 3.
Results of the channel current measurements. (a) Typical current and time traces of AMP (magainin) in the bacterial model membrane. (b) Classification of AMP channel current signals and possible models of the peptide mechanisms (these typical signals are from magainin). (c) Current signal analysis and classification of six AMPs. (n = 243–681).
The signal classification of all AMPs is presented in Figure 3b,c. In this classification, we eliminated the spike signals to estimate the pore formation and to compare the pore formation versus penetration. The step and multilevel signals reflected relatively stable pore formation in the barrel-stave and the toroidal models. In contrast, the erratic signals were assigned to the random disruption model that elicited detergent-like membrane disruption. Cla yielded notably unstable pore formation, whereas stable pore formation was observed in more than 60% in the other AMPs. Next, we analyzed AMP pore-forming capability by comparing stable pore formation and penetration using the spike signals, which can be assigned to the direct penetration of peptide molecules. The appearance ratio of [the penetration (spike signals)]/[the pore formation (step + multi + erratic signals)] was calculated to estimate the capability of pore formation. The order of the ratio was Cec > Par > LL-37 > SL-37 > Mag > Cla, as listed in Table 1. These results may imply the leakage activity of AMPs.
Table 1. Pore-Forming Activity by Estimating the Comparison between the Current Signals of Pore Formation and Membrane Penetration.
| parameter | clavanin A | cecropin A | pardaxin P5 | magainin 1 | SL-37 | LL-37 |
|---|---|---|---|---|---|---|
| pore (n) | 366 | 309 | 681 | 529 | 505 | 243 |
| spike (n) | 17212 | 371 | 922 | 5967 | 4908 | 1386 |
| penetration/pore form | 47.0 | 1.2 | 1.4 | 11.3 | 9.7 | 5.7 |
2.2. Analysis of the Pore Size and the Ion Flux Via the Pore
The size of the pore can be theoretically estimated using the channel conductance of the step signals and Hille’s equation35 (Figure 4a).
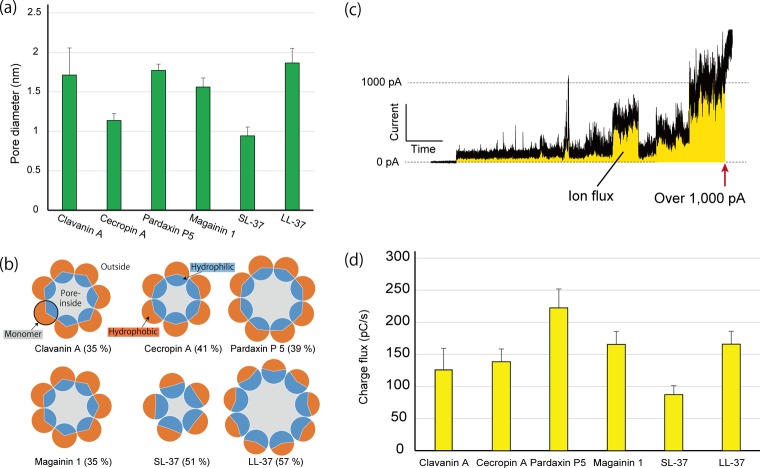
Here, r is the pore radius, ρ is the resistivity of the buffered solution in 25 °C, l is the length of the pore (7 nm from the thickness of the lipid bilayer), and R is the resistivity of the pore. R is calculated as V/I, where I is the current through the pore, and V is the applied voltage between two chambers. The diameter of the pores ranges from 0.9 to 1.9 nm (Table S1), and the tendency is divided into two groups: (LL-37 ≥ Par ≥ Cla ≥ Mag) > (Cec ≥ SL-37). The number of assembling monomers for constructing pores is mathematically calculated using the pore diameter and width of the peptides’ α-helical structure. Figure 4b shows a schematic illustration of the assembling monomers and the hydrophilic region of each monomer that is estimated by examining the number of hydrophilic residues compared to all residues (the number of hydrophilic/entire residues). Larger pore size seems to correlate with increasing hydrophilic region. However, mammalian AMPs have a large hydrophilic region but exhibit a notable pore size difference even in similar peptide sequences.
Figure 4.
Analysis of pore-forming activity. (a) Pore diameter and (b) number of assembling monomers with hydrophobicity percentage (hydrophobic residues/all residues) of AMPs calculated from step signals. (c) Diagram of charge flux estimation. The yellow area indicates the total charge flux through the membrane pore or defect. (d) Results of the charge flux in AMPs.
Charge flux (CF) through the nanopore reflected the ion flux (Figure 4c) and exhibited similar behavior to that of lipid bilayer leakage.32 Therefore, it is strongly correlated with membrane dysfunction-related antimicrobial activity. In these measurements, we set the threshold value at 1000 pA; this value was established in reference to our previous study of CF measurements.24 The order of CF was Par > LL-37 ≥ Mag > Cec ≥ Cla > SL-37 (Figure 4d and Table S1). The results indicate a similar tendency of pore stability.
2.3. Scoring on Pore-Forming Activities and the Minimum Inhibitory Concentration (MIC) of AMPs
To assess the pore-forming activity from several perspectives, we examined the activity on a scoring basis. Each AMP was scored from 1 to 6 in each pore-forming parameter, such as the pore size and CF, and the scores were determined as the sum of the score out of the total possible score. We considered that this value could predict the total pore-forming activity in the lipid bilayer, while the numerical values of the scores do not have quantitatively. The total scores are shown in Figure 5, and all values are listed in Table S1. The order of the total score was LL-37 > Par > Mag = Cec > Cla > SL-37. In order to elucidate the availability of this scoring, we compared the CF as the experimental value and the sum of the score to pore size and stability. According to our understanding, the value of CF should be strongly correlated with pore size and stability. If there were large and stable pores in the lipid bilayer, the CF would be large. The order of values were as follows: for scoring (size + stability), LL-37 > Par > Mag > Cec > Cla > SL-37; for charge flux (experimental), Par > LL-37 > Mag > Cec > Cla > SL-37. These orders were similar, suggesting the efficiency of this scoring method.
Figure 5.
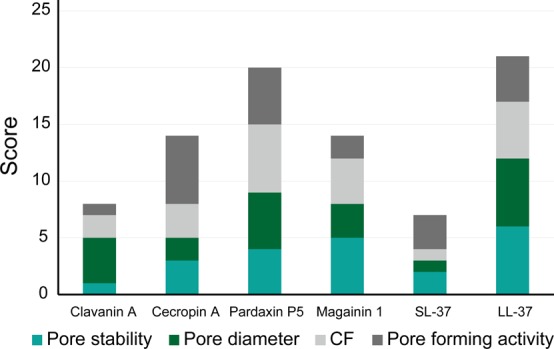
Total score of pore-forming activity parameters: pore stability, pore diameter, charge flux, and pore-forming activity. Each element was scored from 1 to 6.
Finally, we investigate the MICs of all AMPs against Escherichia coli. We categorize three MIC activity: high, middle, and low as MIC values exhibit intrinsic distribution due to the discontinuous changes in the sample concentration. The order of the antimicrobial activity from MIC measurements of each AMPs is Cec ≈ SL-37 ≈ LL-37 > Mag ≈ Par > Cla (Table 2). This order seems to somewhat reflect the order of the total scoring.
Table 2. MIC and the Classification of the Activity of each AMP.
3. Discussion
Our electrophysiological methods and analysis are useful tools for evaluating the mode of action of AMPs on pore formation and disruption in bacterial model membranes. The pore-forming activities and stability are different in each AMP (Table S1). The featured peptides and their characteristics are listed as follows:
Cla: pore-forming activity = low, stability = low
Cec: pore-forming activity = high, pore size = small
Par: pore-forming activity = high, pore size = large
Mag: pore-forming activity = low, stability = high
SL-37: pore-forming activity = low, stability = low, pore size = small
LL-37: pore-forming activity = high, stability = high, pore size = large
Considering these features, we found that the total pore-forming activities exhibit an obvious correlation with the species difference, such as the activity difference in young or old species.
The parameter scoring reflects roughly AMPs’ antimicrobial activity against E. coli, whereas the scores are not equal to the MIC values. The antimicrobial activity of these AMPs used in this study is mainly considered as membrane disruption. The score from electrophysiological measurements with the bacterial model membrane, which is the sum of the membrane disruption abilities, can predict the antimicrobial activity of the living bacteria. Similarly, the permeability induced by AMPs in the model membrane (DOPC) is quite similar to that in bacteria (spheroplast of E. coli).36 However, two unexplained results remain: (1) The total score of Cec is in the middle position, while the activity of MIC is the highest. (2) The pore-forming activity between LL-37 and SL-37 is notably different even though they are genetically close and have similar peptide sequences.
With respect to Cec behavior, the precise antibacterial mechanism of Cec is still argued. Recently, there is evidence showing that the membrane disruption is the main activity of Cec.37 However, another mechanism has also proposed that Cec induced apoptotic activity with inducing the imbalance of ions.38 This fact implies that there is some different mechanisms of antimicrobial activity, and they should be the reason for the difference between the scoring and MIC of Cec.
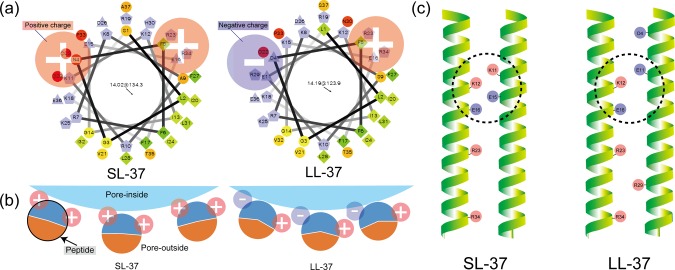
Next, SL-37 and LL-37 are from the same gene family (CAMP), and their sequences resemble each other. Although the value of their MICs seems to be similar, their scores are quite different. Among the scoring parameters, we focus on pore diameter because this parameter should reflect the assembling state of peptide monomers. The values between SL-37 (0.9 ± 0.1 nm) and LL-37 (1.9 ± 0.2 nm) are different; therefore, the number of assembling monomers is also different at 5 mers (SL-37) and 9 mers (LL-37). Both SL-37 and LL-37 exhibit an amphiphilic α-helical structure, and their monomers assemble in a lipid bilayer oriented so that the hydrophilic side faces the inside pore and the hydrophobic side is directed toward the lipid phase, as shown in Figure 4b. To explain this stoichiometry, we ascertain carefully the helical wheel structure and the position of amino acids. Figure 6 shows the schematic illustration of the geometry of amino acids in SL-37 and LL-37. The charges of the amino acids in the side region of the hydrophilic area are changed from positive to negative, resulting in an intensified electrostatic interaction between monomers in LL-37. This electrostatic change may enhance the oligomerization of LL-37 monomers.39 Besides, the F score (packing quality of helix) of LL-37 (F = 730) is two times higher than that of SL-37 (F = 321) simulated by a web tool of “Prediction tool for an ensemble of transmembrane α-helical dimer conformations” (PREDDIMER, https://preddimer.nmr.ru/preddimer/). These results also support the higher interaction in the LL-37 system.
Figure 6.
Schematic illustration of pore-formation by assembling monomers of SL-37 and LL-37. (a) Characteristics of each peptide’s surface with the helical wheel structure. (b) Schematic illustration of oligomerization activities in SL-37 and LL-37 with the electrostatic interaction. In the hydrophilic region, 4th, 11th, and 29th amino acids exhibit different characteristics between SL-37 and LL-37. (c) Schematic illustration of the three-dimensional structures of SL-37 and LL-37. The circles indicate the probable electrostatic interaction region of each peptide.
4. Conclusions
In summary, we investigated the pore-forming activities of AMPs from diverse species using electrophysiological measurements. The activities were divided into four factors in terms of the stability and activity, and each peptide was scored. The order of the total scores is LL-37 > Par > Mag = Cec > Cla > SL-37. This order roughly reflects the order of antimicrobial activity from MIC measurements against E. coli: Cec ≈ SL-37 ≈ LL-37 > Mag ≈ Par > Cla. Although there is no distinct trend between pore-forming activity and evolution of the species, our method can estimate antimicrobial activity using electrophysiological measurements. In addition, it is found that the small difference of amino acid between SL-37 and LL-37 induces large differences in pore-forming activity, even in these genetically similar peptides. Moreover, our electrophysiological system and analysis would be a useful tool for selecting the pore-forming peptide in molecular evolution research.40,41
5. Materials and Methods
5.1. Reagents
In this study, we used the following reagents: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE; Avanti Polar Lipids, AL, USA); 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DOPG; Avanti Polar Lipids); n-decane (Wako Pure Chemical Industries, Ltd., Osaka, Japan); 3-morpholinopropane-1-sulfonic acid (MOPS, Nacalai Tesque, Kyoto, Japan); potassium chloride (KCl; Nacalai Tesque); clavanin A (GenScript); cecropin A (BACHEM); pardaxin (abcam); magainin 1 (AnaSpec Inc); SL-37 (Cosmo Bio Co., Ltd.); and LL-37 (AnaSpec Inc.). DOPE and DOPG were melted in n-decane at a concentration of 10 mg/mL, and we obtained a lipid mixture of DOPE/DOPG (3:1 mol/mol). The composition of the measurement buffer was regulated so that it became 200 mM KCl, 10 mM MOPS, and pH 7.0 in the Milli-Q system (Millipore, Billerica, MA, USA). The clavanin A, cecropin A, pardaxin, magainin 1, SL-37, and LL-37 powders were dissolved in the measurement buffer and preserved at 4 °C.
5.2. Fabrication of Multichannel Device
One of the multichannel devices measured 6 mm in height, 10 mm in length, and 10 in width and was constructed using poly(methyl methacrylate) (PMMA; Mitsubishi Rayon, Tokyo, Japan). We designed these devices using a RhinoCAM (3DS, Kanagawa, Japan) for computer-aided design and used MM-100 (Modia Systems, Saitama, Japan) for cutting. The devices have two chambers 2 mm in diameter and 4 mm in depth. There is a gap of 0.5 mm depth between the two chambers. Pores of 0.44 mm were cut at the bottom of each chamber for electrode interconnection. We inserted Ag/AgCl electrodes (Nilaco, Tokyo, Japan) in the pore and coated electrodes by Ag/AgCl ink for the reference electrode (BAS Inc., Tokyo, Japan). The parylene film was 5 μm thick, and a 100 μm pore was made using photolithography. We adhered it between two PMMA (0.2 mm thickness) plates that had a 1 mm pore and used it as the separator. We used Super X (Cemedine Co., Ltd., Tokyo, Japan) as the glue. The separator was fixed in the gap of the device using glue. The device is shown in Figure 2. The electrodes were connected to a breadboard, which was in turn connected to Jet patch-clamp amplifier (Tecella, CA, USA) with a jumper wire (E-Call Enterprise Co., Ltd., Taipei City, Taiwan).
5.3. Preparation of Planar Lipid Bilayer
We prepared a planar lipid bilayer using a liquid contact method in a multichannel device, which was created using microprocessing.42 We obtained high-throughput data owing to multichannel devices’ ability to fabricate multiple lipid bilayers at the same time. We used a micropipette (Eppendorf, Hamburg, Germany) at fabrication for lipid bilayers in each chamber. After we added 2.3 μL of 10 mg/mL DOPE/DOPG (3:1 mol/mol) in n-decane solution to each chamber, we added 4.7 μL of 10 μM peptide liquid and 200 mM KCl liquid solution, allowing us to easily prepare the planar lipid bilayer. The peptide was added to only one chamber to which 100 mV voltage was applied to simulate the environment of living cells. A few minutes later, monolayers in each chamber were contacted, and a lipid bilayer was fabricated. When the bilayer was disrupted, we reconstructed the lipid bilayer by tracing between two chambers using a hydrophobic stick.43 We were able to alter the lipid ratio voluntarily and fabricate model cell membranes that imitated living cells. We used a bacterial model membrane (DOPE/DOPG (3:1 mol/mol)), which has a negative charge. This membrane has a disordered phase in this study because the phase transition temperatures of DOPE and DOPG are −16 and −18 °C, respectively, and the peptides can form the assembling structure in room temperature (23 ± 1 °C).
5.4. Channel Current Measurement
Channel current was observed using a JET patch-clamp amplifier. The measurement device had two electrodes. One side was connected to the Jet patch-clamp amplifier and applied 100 mV of constant voltage. Another side was grounded. As AMPs formed pores on the lipid bilayer, ions passed through the pores. Thus, we observed this process as a channel current. We measured current signals under the conditions of gain 1 G, 20 kHz of the sampling rate, and 4 kHz of low-pass filter in room temperature (23 ± 1 °C). Data were analyzed using pCLAMP ver. 10.5 (Molecular Devices, CA, USA) and Excel (Microsoft, Washington, USA). Data are described as mean ± SE unless otherwise stated. The classification of current signals obtained from several minutes to 2 h data was performed according to the previous definition (Figure S2)24,44 with using all data (n = 243–681, Figure 3b) and the same data numbers (n = 243, Figure S1). Both of these analyses showed similar results. The pore size of AMPs was calculated using the conductance of the step signal and Hille’s equation.35 Hille’s equation is based on simple Ohm’s law and the diffusion equation, and it is conventionally used to estimate the size of a pore or channel of ion channels,35 pore-forming toxin,45 and synthetic channels44 because of the validity.
5.5. MIC Measurement
MIC measurement of clavanin A, magainin 1, and LL-37 were conducted by the Hygiene & Microbiology Research Center. In the MIC measurements, the peptides were diluted as 100, 50, 25, 12.5, 6.3, 3.2, 1.6, 0.8, 0.4, and 0.2 μM and applied to E. coli (NBRC 3972). The E. coli with AMP was cultured at 35 °C for 24 h and the MIC was measured by the turbidity. After, the bacterial solution was prepared so that it became 104/mL. We mixed the bacterial solution with the Mueller-Hinton broth, and the potential bacterial increase was checked by examining the turbidity after 24 h of incubation at 35 °C. The value of cecropin A,46 pardaxin P5,47 and SL-3729 have previously reported and used the value in this study.
Acknowledgments
We thank H. Watanabe, Y. Sekiya, S. Shimizu, and M. Usami in our group for useful discussion on the channel current measurements and data analysis. This work was partially supported by the KAKENHI (grant nos. 17K19138 and 19H05382) from MEXT.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.9b01033.
Signal classification of six AMPs, definition of the current signal classification, and four parameters of pore-forming activities of each AMP (PDF).
The authors declare no competing financial interest.
Supplementary Material
References
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–95. 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- Yeaman M. R.; Yount N. Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- Hancock R. E.; Sahl H. G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–7. 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- Brogden K. A. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria?. Nat. Rev. Microbiol. 2005, 3, 238–50. 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- Melo M. N.; Ferre R.; Castanho M. A. Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 2009, 7, 245–50. 10.1038/nrmicro2095. [DOI] [PubMed] [Google Scholar]
- He K.; Ludtke S. J.; Worcester D. L.; Huang H. W. Neutron scattering in the plane of membranes: structure of alamethicin pores. Biophys. J. 1996, 70, 2659–66. 10.1016/S0006-3495(96)79835-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H.; Jähnig F. The structure of melittin in membranes. Biophys. J. 1986, 50, 573–82. 10.1016/S0006-3495(86)83497-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.; Harroun T. A.; Weiss T. M.; Ding L.; Huang H. W. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys. J. 2001, 81, 1475–85. 10.1016/S0006-3495(01)75802-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann G.; Mueller P. A molecular model of membrane excitability. J. Supramol. Struct. 1974, 2, 538–557. 10.1002/jss.400020504. [DOI] [PubMed] [Google Scholar]
- Mak D. O.; Webb W. W. Two classes of alamethicin transmembrane channels: Molecular models from single-channel properties. Biophys. J. 1995, 69, 2323–2336. 10.1016/S0006-3495(95)80102-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S.; Wang W.; Yang L.; Huang H. W. Structure of the alamethicin pore reconstructed by x-ray diffraction analysis. Biophys. J. 2008, 94, 3512–3522. 10.1529/biophysj.107.126474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit E.; Miller I. R.; Biggin P. C.; Sansom M. S.; Shai Y. Structure and orientation of the mammalian antibacterial peptide cecropin P1 within phospholipid membranes. J. Mol. Biol. 1996, 258, 860–70. 10.1006/jmbi.1996.0293. [DOI] [PubMed] [Google Scholar]
- Pouny Y.; Rapaport D.; Mor A.; Nicolas P.; Shai Y. Interaction of Antimicrobial Dermaseptin and Its Fluorescently Labeled Analogs with Phospholipid-Membranes. Biochemistry 2002, 31, 12416–12423. 10.1021/bi00164a017. [DOI] [PubMed] [Google Scholar]
- Matsuzaki K.; Murase O.; Fujii N.; Miyajima K. An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation. Biochemistry 1996, 35, 11361–8. 10.1021/bi960016v. [DOI] [PubMed] [Google Scholar]
- Matsuzaki K. Why and how are peptide–lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim. Biophys. Acta, Biomembr. 1999, 1462, 1–10. 10.1016/S0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- Ludtke S. J.; He K.; Heller W. T.; Harroun T. A.; Yang L.; Huang H. W. Membrane pores induced by magainin. Biophys. J. 1996, 35, 13723–13728. 10.1021/bi9620621. [DOI] [PubMed] [Google Scholar]
- Qian S.; Wang W.; Yang L.; Huang H. W. Structure of transmembrane pore induced by Bax-derived peptide: Evidence for lipidic pores. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 17379–17383. 10.1073/pnas.0807764105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclohier H.; Molle G.; Spach G. Antimicrobial Peptide Magainin-I From Xenopus Skin Forms Anion-Permeable Channels In Planar Lipid Bilayers. Biophys. J. 1989, 56, 1017–1021. 10.1016/S0006-3495(89)82746-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimley W. C. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol. 2010, 5, 905–17. 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J. A. Molecular evolution of animal antimicrobial peptides: widespread moderate positive selection. J. Evol. Biol. 2005, 18, 1387–1394. 10.1111/j.1420-9101.2005.00925.x. [DOI] [PubMed] [Google Scholar]
- Watanabe H.; Gubbiotti A.; Chinappi M.; Takai N.; Tanaka K.; Tsumoto K.; Kawano R. Analysis of Pore Formation and Protein Translocation Using Large Biological Nanopores. Anal. Chem. 2017, 89, 11269–11277. 10.1021/acs.analchem.7b01550. [DOI] [PubMed] [Google Scholar]
- Kunthic T.; Watanabe H.; Kawano R.; Tanaka Y.; Promdonkoy B.; Yao M.; Boonserm P. pH regulates pore formation of a protease activated Vip3Aa from Bacillus thuringiensis. Biochim. Biophys. Acta, Biomembr. 2017, 1859, 2234–2241. 10.1016/j.bbamem.2017.08.018. [DOI] [PubMed] [Google Scholar]
- Watanabe H.; Kawano R. Channel Current Analysis for Pore-forming Properties of an Antimicrobial Peptide, Magainin 1, Using the Droplet Contact Method. Anal. Sci. 2016, 32, 57–60. 10.2116/analsci.32.57. [DOI] [PubMed] [Google Scholar]
- Sekiya Y.; Sakashita S.; Shimizu K.; Usui K.; Kawano R. Channel current analysis estimates the pore-formation and the penetration of transmembrane peptides. Analyst 2018, 143, 3540–3543. 10.1039/C8AN00243F. [DOI] [PubMed] [Google Scholar]
- Zhao C.; Liaw L.; Hee Lee I.; Lehrer R. I. cDNA cloning of Clavanins: Antimicrobial peptides of tunicate hemocytes. FEBS Lett. 1997, 410, 490–492. 10.1016/S0014-5793(97)00646-7. [DOI] [PubMed] [Google Scholar]
- Steiner H.; Hultmark D.; Engström A.; Bennich H.; Boman H. G. Sequence and specificity of two antibacterial proteins involved in insect immunity. J. Immunol. 1981, 182, 6635–6637. 10.1038/292246a0. [DOI] [PubMed] [Google Scholar]
- Lazarovici P.; Primor N.; Loew L. M. Purification and pore-forming activity of 2 hydrophobic polypeptides from the secretion of the red-sea moses sole (pardachirus-marmoratus). J. Biol. Chem. 1986, 261, 6704–6713. [PubMed] [Google Scholar]
- Zasloff M. Magainins, a class of antimicrobial peptides from xenopus skin: isolation, characterization of two active forms, and partial cdna sequence of a precursor. Proc. Natl. Acad. Sci. U. S. A. 1987, 84, 5449–5453. 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelezetsky I.; Pontillo A.; Puzzi L.; Antcheva N.; Segat L.; Pacor S.; Crovella S.; Tossi A. Evolution of the primate cathelicidin. J. Biol. Chem. 2006, 281, 19861–19871. 10.1074/jbc.M511108200. [DOI] [PubMed] [Google Scholar]
- Gudmundsson G. H.; Agerberth B.; Odeberg J.; Bergman T.; Olsson B.; Salcedo R. The human gene FALL39 and processing of the cathelin precursor to the antibacterial peptide LL-37 in granulocytes. Eur. J. Biochem. 1996, 238, 325–332. 10.1111/j.1432-1033.1996.0325z.x. [DOI] [PubMed] [Google Scholar]
- Mylonakis E.; Podsiadlowski L.; Muhammed M.; Vilcinskas A. Diversity, evolution and medical applications of insect antimicrobial peptides. Philos. Trans. R. Soc., B 2016, 371, 20150290. 10.1098/rstb.2015.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya Y.; Shimizu K.; Kitahashi Y.; Ohyama A.; Kawamura I.; Kawano R. Electrophysiological Analysis of Membrane Disruption by Bombinin and Its Isomer Using the Lipid Bilayer System. ACS Appl. Bio Mater. 2019, 2, 1542. 10.1021/acsabm.8b00835. [DOI] [PubMed] [Google Scholar]
- Bechinger B.; Lohner K. Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochim. Biophys. Acta, Biomembr. 2006, 1758, 1529–1539. 10.1016/j.bbamem.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Guidotti G.; Brambilla L.; Rossi D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. 10.1016/j.tips.2017.01.003. [DOI] [PubMed] [Google Scholar]
- Hille B.Ion channels of excitable membranes. 3rd ed.; Sinauer: Sunderland, Mass., 2001; p xviii, 814 p. [Google Scholar]
- Faust J. E.; Yang P. Y.; Huang H. W. Action of Antimicrobial Peptides on Bacterial and Lipid Membranes: A Direct Comparison. Biophys. J. 2017, 112, 1663–1672. 10.1016/j.bpj.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H.; Felix J. B. Peptide-membrane interactions and mechanisms of membrane destruction by amphipathic α-helical antimicrobial peptides. Biochim. Biophys. Acta, Biomembr. 2006, 1758, 1245–1256. 10.1016/j.bbamem.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Yun J.; Lee D. G. Cecropin A-induced apoptosis is regulated by ion balance and glutathione antioxidant system in Candida albicans. IUBMB Life 2016, 68, 652–662. 10.1002/iub.1527. [DOI] [PubMed] [Google Scholar]
- Toniolo C.; Benedetti E. Intramolecularly hydrogen-bonded peptide Conformation. Crit. Rev. Biochem. 2008, 9, 1–44. 10.3109/10409238009105471. [DOI] [PubMed] [Google Scholar]
- Kumachi S.; Husimi Y.; Nemoto N. An RNA Binding Peptide Consisting of Four Types of Amino Acid by in Vitro Selection Using cDNA Display. ACS Omega 2016, 1, 52–57. 10.1021/acsomega.6b00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauson A. J.; Hall O. M.; Fuselier T.; Starr C. G.; Kauffman W. B.; Wimley W. C. Conformational Fine-Tuning of Pore-Forming Peptide Potency and Selectivity. J. Am. Chem. Soc. 2015, 137, 16144–16152. 10.1021/jacs.5b10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano R.; Tsuji Y.; Sato K.; Osaki T.; Kamiya K.; Hirano M.; Ide T.; Miki N.; Takeuchi S. Automated parallel recordings of topologically identified single ion channels. Sci. Rep. 2013, 3, 1995. 10.1038/srep01995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano R.; Tsuji Y.; Kamiya K.; Kodama T.; Osaki T.; Miki N.; Takeuchi S. A portable lipid bilayer system for environmental sensing with a transmembrane protein. PLoS One 2014, 9, e102427 10.1371/journal.pone.0102427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chui J. K.; Fyles T. M. Ionic conductance of synthetic channels: analysis, lessons, and recommendations. Chem. Soc. Rev. 2012, 41, 148–75. 10.1039/C1CS15099E. [DOI] [PubMed] [Google Scholar]
- Praper T.; Sonnen A.; Viero G.; Kladnik A.; Froelich C. J.; Anderluh G.; Dalla Serra M.; Gilbert R. J. Human perforin employs different avenues to damage membranes. J. Biol. Chem. 2011, 286, 2946–55. 10.1074/jbc.M110.169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A.; Weisshaar J. C. Effects of alterations of the E. coli lipopolysaccharide layer on membrane permeabilization events induced by Cecropin A. Biochim. Biophys. Acta, Biomembr. 2018, 1860, 1470–1479. 10.1016/j.bbamem.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren Z.; Shai Y. A class of highly potent antibacterial peptides derived from pardaxin, a pore-forming peptide isolated from Moses sole fish Pardachirus marmoratus. Eur. J. Biochem. 1996, 237, 303–310. 10.1111/j.1432-1033.1996.0303n.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.