Abstract
Background
Osteoarthritis (OA) affects the health and wellbeing of the elderly. Shaoyao Gancao decoction (SGD) is used in traditional Chinese medicine (TCM) for the treatment of OA and has two active components, shaoyao (SY) and gancao (GC). This study aimed to undertake a network pharmacology analysis of the mechanism of the effects of SGD in OA.
Material/Methods
The active compounds and candidates of SGD were obtained from the Traditional Chinese Medicine (TCM) Databases@Taiwan, the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database, the STITCH database, the ChEMBL database, and PubChem. The network pharmacology approach involved network construction, target prediction, and module analysis. Significant signaling pathways of the cluster networks for SGD and OA were identified using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
Results
Twenty-three bioactive compounds were identified, corresponding to 226 targets for SGD. Also, 187 genes were closely associated with OA, of which 161 overlapped with the targets of SGD and were considered to be therapeutically relevant. Functional enrichment analysis suggested that SGD exerted its pharmacological effects in OA by modulating multiple pathways, including cell cycle, cell apoptosis, drug metabolism, inflammation, and immune modulation.
Conclusions
A novel approach was developed to systematically identify the mechanisms of the TCM, SGD in OA using network pharmacology analysis.
MeSH Keywords: Health Care Evaluation Mechanisms; Medicine, Chinese Traditional; Osteoarthritis, Knee
Background
Osteoarthritis (OA) is an age-related degenerative disease that is characterized by the degradation of joint cartilage and inflammation of the synovium [1–3]. The typical clinical signs and symptoms of OA are pain, swelling, and stiffness, usually associated with reduced activity and limitation of movement [4]. Chronic OA results in the formation of osteophytes, and deformation and narrowing of the joint space. OA significantly reduces the quality of life for patients and can result in physical disability, which has an increasing socioeconomic and healthcare burden [5,6]. Severe OA commonly results in joint replacement, particularly in elderly individuals [7]. Currently, pharmacological treatments for OA primarily include the use of oral pain medication, including opioid analgesics, nonsteroidal anti-inflammatory drugs (NSAIDs), intra-articular injection of corticosteroids, and surgical treatment including osteotomy, arthroplasty, and arthrodesis [8–10]. However, pharmacological treatments for OA are aimed at alleviating the symptoms of the disease rather than treating the underlying causes, and have several side effects, including an increased risk of cardiovascular events and infection [11,12]. Therefore, more effective and safer therapeutic approaches are required for treating patients with OA.
Traditional Chinese medicine (TCM) has been used widely for several decades for the treatment of a range of diseases and has the advantage of being inexpensive and widely available, and because many medicines are derived from natural sources such as herbs, they have fewer side effects [13]. Several TCMs have been used to treat OA and are both effective and safe [14,15]. Therefore, for the treatment of OA, it would be helpful to identify the most effective TCM compounds. Previous studies have shown that Shaoyao Gancao decoction (SGD) is effective in reducing the clinical symptoms of OA by improving joint function and movement. SGD is an effective formula that has been described in the Treatise on Febrile and Miscellaneous Diseases (Shang Han Za Bing Lun) by the third-century Chinese physician Zhang Zhognjing. SGD contains two Chinese herbal medicines, shaoyao (SY) derived from Radix Paeoniae Alba, and gancao (GC) derived from Glycyrrhizae Radix et Rhizoma, in a 1: 1 ratio [16]. Pharmacological studies have shown that the two compounds in the SGD formulation have a synergistic effect in reducing inflammation, pain, and swelling and improving joint function in patients with OA [17]. However, the underlying pharmacological mechanisms of action of SGD and its components in the treatment of OA remain unclear, and the pharmacodynamic properties of its components and key targets remain to be identified.
Network pharmacology is a new and powerful method that integrates chemo-informatics, bio-informatics, network biology, network analysis and traditional pharmacology [18]. The method of network pharmacology conforms to the systemic or holistic view of TCM theory and is a novel strategy to elucidate the active compounds and potential mechanisms of TCM formulas. Therefore, this study aimed to use network pharmacology to identify the bioactive components and targets of SGD, to search for common targets for SGD in the treatment of OA, to understand the underlying mechanisms of action of the disease targets, and to mine for disease-related genes.
Material and Methods
Construction of a database of the components of Shaoyao Gancao decoction (SGD)
Figure 1 shows a schematic representation of the network pharmacology study of Shaoyao Gancao decoction (SGD) in the treatment of osteoarthritis (OA), including the two active components, shaoyao (SY) and gancao (GC). The data relating to the chemical compounds, SY and GC were derived from the Traditional Chinese Medicine (TCM) Databases@Taiwan (http://tcm.cmu.edu.tw/) [19], and the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database (http://lsp.nwu.edu.cn/tcmsp.php) [20]. In total, 365 compounds were identified in SGD after removing the duplicate data, including 280 compounds in GC and 85 compounds in SY.
Figure 1.
Schematic representation of network pharmacology study of Shaoyao Gancao decoction (SGD) in the treatment of osteoarthritis (OA). SGD – Shaoyao Gancao decoction; OA – osteoarthritis.
Screening of the active ingredients in SGD
The 365 potential compounds from SY and GC were filtered using two adsorption, distribution, metabolism, and excretion (ADME)-related models, integrating drug-likeness (DL) and oral bioavailability (OB). Drug-likeliness is a qualitative concept used in drug design to determine how drug-like a prospective compound is to describe and optimize pharmacokinetic and pharmaceutical properties [19,21]. Oral bioavailability indicates the drug-like nature of molecules as therapeutic agents and represents the relative amount of orally administered drug that reaches the blood circulation, shown by the convergence of the ADME process [22]. To identify the active components of SGD, the ingredients conforming to the requirements of both OB ≥30% and DL ≥0.18, based on the published literature and the information from the TCMSP database, were identified for further analysis [23]. Also, putative targets of potential compounds in SY and GC were identified from the STITCH, ChEMBL and PubChem databases, and those without target information were excluded.
Target genes related to the identified compounds
To identify the relevant targets of the potential compounds in SY and GC, the STITCH (http://stitch.embl.de/) [24], ChEMBL (http://www.ebi.ac.uk/chembl/) [25], and PubChem (http://pubchem.ncbi.nih.gov/) databases were used [26]. A final list of genes associated with compounds, with a confidence score of >0.7, was obtained that suggested a high confidence score according to STITCH. The ChEMBL is a manually curated database for storing standardized bioactivity, molecules, targets, and drug data, which are abstracted regularly from the primary medicinal chemistry literature [27]. The PubChem database is a resource for biological activities of small molecules, including substance information, compound structure, and bioactivity, and the data are experimentally validated. All the active ingredients identified in the present study were entered into the STITCH, ChEMBL and PubChem databases with the Homo sapiens species setting. The gene information, including the name, gene ID, and organism, was confirmed using the UniProt protein sequence resource (https://www.uniprot.org) [28]. After removing duplicates, the detailed information of targets obtained is described in Supplementary Table 1.
Related targets of osteoarthritis (OA)
Information on OA-associated target genes was collected from the following resources. DrugBank (https://www.drugbank.ca/) [29] is a comprehensive online database that provides extensive biochemical and pharmacological information on drugs and their mechanisms of action and targets, and 78 genes related to OA were identified from this database. GeneCards (http://www.genecards.org/) is a comprehensive database incorporating information on all annotated and predicted genes [30], which was searched using the keyword “osteoarthritis,” which identified 46 genes. The Online Mendelian Inheritance in Man® (OMIM) database (http://www.omim.org/) [31] is a comprehensive research resource of human genes and genetic phenotypes, from which 65 genes associated with OA were selected. There were 187 targets linked with OA after deleting redundant targets, and the information regarding these targets is provided in Supplementary Table 2.
Construction of the pharmacological networks
Network construction was established using four main steps. First, a compound-compound target network was established by linking compounds and predicted targets with a degree of >3. Second, a protein-protein interaction (PPI) network of compounds and targets was developed by linking the compound targets and predicted targets of other human proteins. Third, a PPI network of OA targets was constructed by linking the known OA-related targets and predicted targets of other human proteins. Fourth, a PPI network of targets for SGD and OA was developed by intersecting the PPI network of compounds and the PPI network of OA targets. The graphical and diagrammatic visualized networks were constructed using Cytoscape version 3.7.0 (http://www.cytoscape.org/) [32], which is a software package for visualizing network analysis.
Cluster analysis
Cluster analysis is a classification method that involves interconnected regions showing the inherent laws in the network [13]. The Molecular Complex Detection (MCODE) plug-in was used to detect densely connected regions and cluster analysis in the PPI network [33]. In this study, we selected significant cluster modules from the constructed PPI network using MCODE. The criteria settings were set as follows: node score cutoff=0.2; K-core=2; and degree of cutoff=2.
Gene Ontology (GO) and pathway enrichment analysis
The Gene Ontology (GO) database (http://geneontology.org/), including biological process, cell component, and molecular function terms, was used to identify the possible biological mechanisms using high-throughput genome or transcriptome data [34]. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.kegg.jp/) is a knowledge database for identifying the systematic functions and biological relevance of candidate targets [35]. In this study, GO functional annotation and KEGG pathway analysis were performed using Bioconductor clusterProfiler, an R package used for enrichment analysis of gene clusters [36].
Results
Screening for the active compounds of Shaoyao Gancao decoction (SGN) involved in osteoarthritis (OA)
From the two active components of Shaoyao Gancao decoction (SGD), shaoyao (SY) and gancao (GC), 365 compounds were obtained from the Traditional Chinese Medicine Systems Pharmacology(TCMSP) database and the traditional Chinese medicine (TCM) Databases@Taiwan, with 280 compounds from GC and 85 from SY. The values of oral bioavailability (OB) and drug-likeness (DL) (OB ≥30% and DL ≥0.18) were used to screen potential active compounds from GC and SY, and a total 23 active compounds met the screening standards. The properties of the compounds are shown in Table 1.
Table 1.
The active ingredients of the two components of Shaoyao Gancao decoction (SGD), shaoyao (SY) and gancao (GC).
| Molecule ID | Molecule name | Structure | OB | DL | Herb |
|---|---|---|---|---|---|
| MOL000359 | Sitosterol |

|
36.91 | 0.75 | SY |
| MOL000358 | beta-Sitosterol |

|
36.91 | 0.75 | SY |
| MOL000422 | Kaempferol |

|
41.88 | 0.24 | SY |
| MOL001924 | Paeoniflorin |
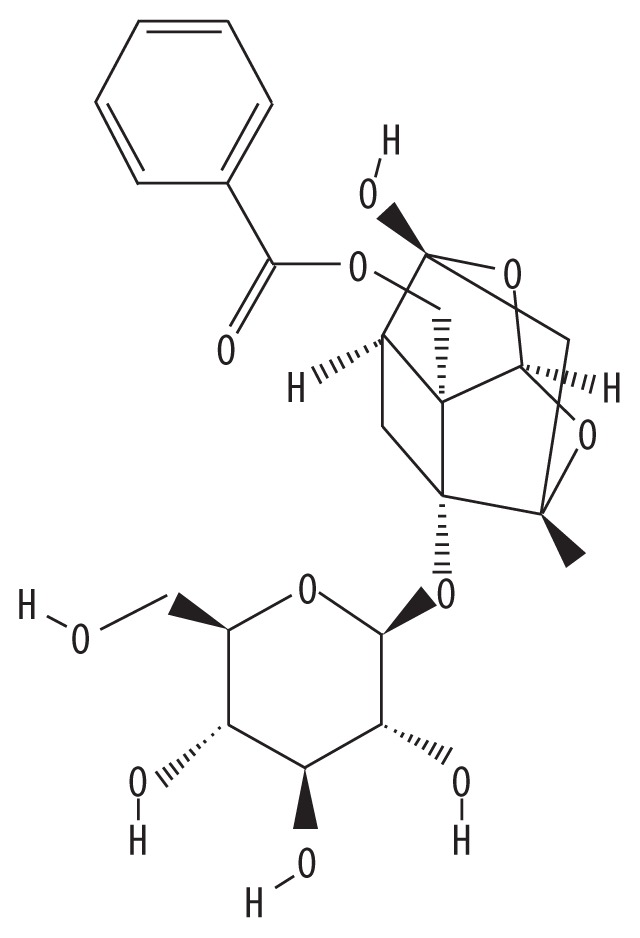
|
53.87 | 0.79 | SY |
| MOL000492 | (+)-Catechin |
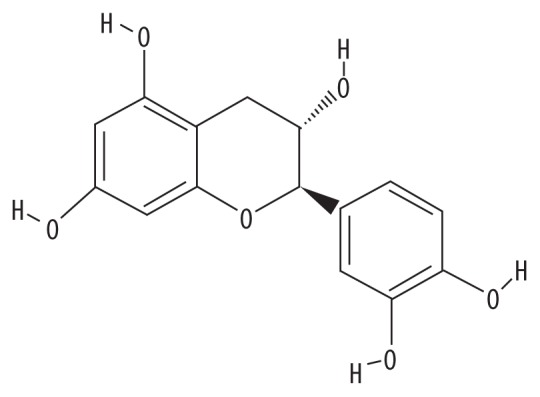
|
54.83 | 0.24 | SY |
| MOL000211 | Mairin |
|
55.38 | 0.78 | BS |
| MOL001792 | Liquiritigenin |

|
32.76 | 0.18 | GC |
| MOL000500 | Vestitol |
|
74.66 | 0.21 | GC |
| MOL004328 | Naringenin |

|
59.29 | 0.21 | GC |
| MOL000392 | Formononetin |

|
69.67 | 0.21 | GC |
| MOL000417 | Calycosin |
|
47.75 | 0.24 | GC |
| MOL004991 | 7-Acetoxy-2-methylisoflavone |
|
83.71 | 0.27 | GC |
| MOL000098 | Quercetin |

|
46.43 | 0.28 | GC |
| MOL000354 | Isorhamnetin |

|
49.6 | 0.31 | GC |
| MOL004910 | Glabranin |
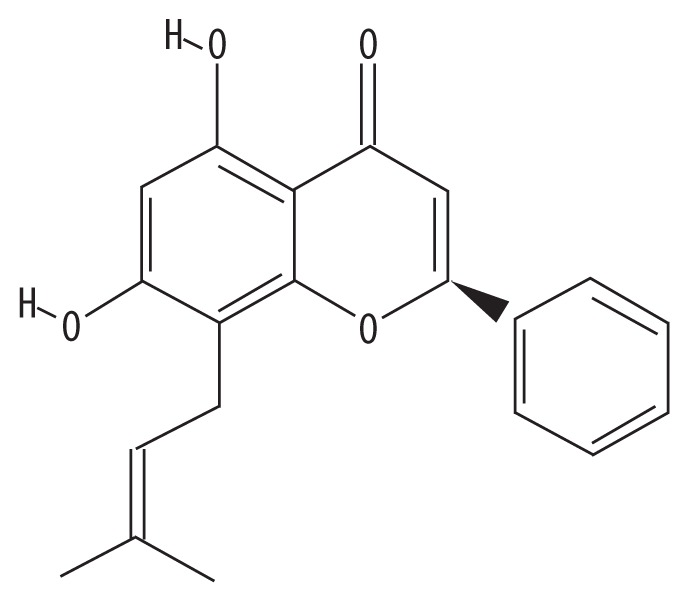
|
52.9 | 0.31 | GC |
| MOL002565 | Medicarpin |
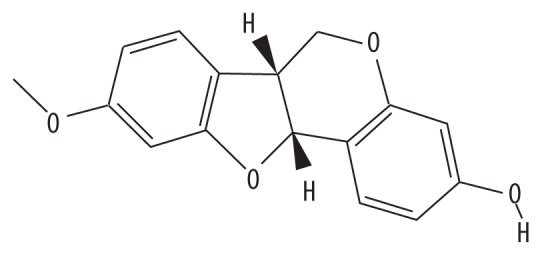
|
49.22 | 0.34 | GC |
| MOL004949 | Isolicoflavonol |

|
45.17 | 0.42 | GC |
| MOL004908 | Glabridin |

|
53.25 | 0.47 | GC |
| MOL001484 | Inermine |
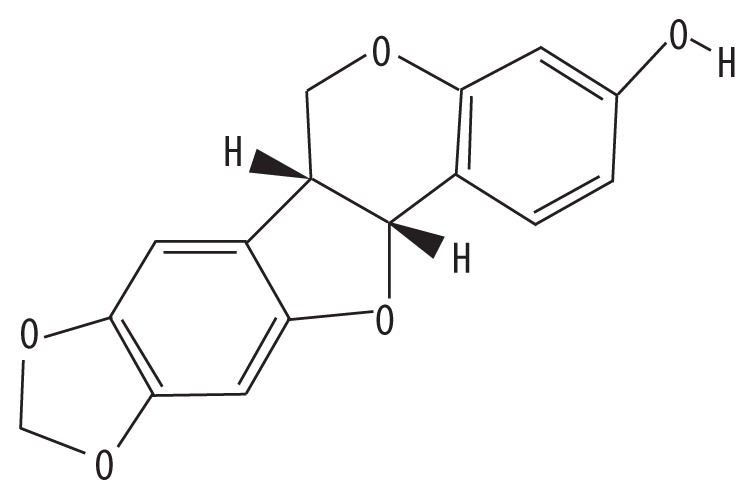
|
75.18 | 0.54 | GC |
| MOL004827 | Semilicoisoflavone B |

|
48.78 | 0.55 | GC |
| MOL004959 | 1-Methoxyphaseollidin |

|
69.98 | 0.64 | GC |
| MOL004903 | Liquiritin |

|
65.69 | 0.74 | GC |
| MOL004948 | Isoglycyrol |

|
44.7 | 0.84 | GC |
Target screening of SGD in the treatment of osteoarthritis
In the present study, the STITCH, ChEMBL, and PubChem databases were used to screen 226 targets corresponding to the active ingredients in SGD, with 188 targets for SY, 146 targets for GC, and 108 for SY and GC. These gene targets included cellular tumor antigen p53 (TP53), chlorotoxin derivative (CA4), estrogen receptor beta (ESR2), and multidrug resistance protein 1 (ABCB1), which are involved in inflammation [37], cell proliferation [38], and angiogenesis [39]. DrugBank, GeneCards, and the Online Mendelian Inheritance in Man® (OMIM) databases were also used to screen 187 targets associated with OA, removing compounds with duplication targets (Supplementary Table 3). The obtained compounds and targets were used to construct the pharmacology network.
Compound-compound network targets
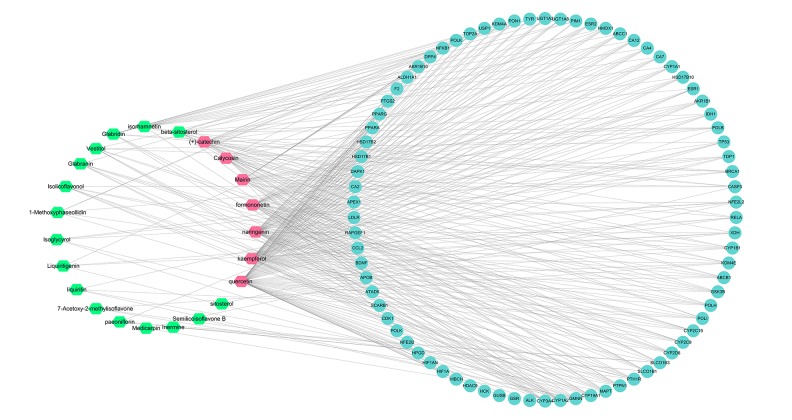
A compound-compound target network was developed to identify the relationship between the compounds of SGD and their candidate targets (Figure 2). The compound-compound target network consisted of 101 nodes (23 compounds and 78 compound targets) and 338 edges (degree >3). The average degree of 14.69 per compound in such a network was based on the network analysis, demonstrating the multitarget treatment characteristics of SGD. In this network, the values of the degree for quercetin (degree=63) and kaempferol (degree=54) were considerably higher than that of the other components, suggesting that two chemicals probably were served as significant therapeutic compounds in OA.
Figure 2.
The compound-compound target network of Shaoyao Gancao decoction (SGD) in the treatment of osteoarthritis (OA). Blue represents the compound targets, green represents the compounds of Shaoyao Gancao decoction (SGD), and red hexagons represent the central compounds of SGD.
Protein-protein interaction (PPI) network targets
The PPI networks of compound targets were developed to identify the interactions between SGD-related proteins and other relative proteins with 448 nodes (45 compound targets, 26 OA targets, 19 compound/OA targets, and other relevant proteins) and 1,869 edges (Figure 3) were constructed to determine the interactive effects of compounds modulated by SGD. About 19 intersection targets between compound targets and OA-related targets were identified in this network including, multidrug resistance protein 1 (ABCB1), multidrug resistance-associated protein 1(ABCC1), carbonic anhydrase 2, C-C motif chemokine 2, cytochrome P450 1A1 (CYP1A1), cytochrome P450 1A2 (CYP1A2), cytochrome P450 2C19, cytochrome P450 2C9 (CYP2C9), cytochrome P450 2D6 (CYP2D6), cytochrome P450 3A4 (CYP3A4), estrogen receptor (ER), estrogen receptor beta (ESR2), peroxisome proliferator-activated receptor alpha, peroxisome proliferator-activated receptor gamma, prostaglandin G/H synthase 2 (PTGS2), solute carrier organic anion transporter family member 1B1, TP53, UDP-glucuronosyltransferase 1–3 (UGT1A3), and UDP-glucuronosyltransferase 1–8.
Figure 3.
The protein-protein interaction (PPI) network of compound targets of Shaoyao Gancao decoction (SGD) in the treatment of osteoarthritis (OA). Incarnadine (crimson) represent other proteins, purple represent compound targets, yellow represent osteoarthritis (OA) targets, and green represent compound/OA targets).
PPI network of OA targets
The PPI network of OA targets was developed to identify the relationship between the OA-related targets and other proteins, with 394 nodes (123 OA targets and 271 other proteins that interacted with OA targets) and 2,184 edges (Figure 4). Considering the median values for degree (10), betweenness centrality (81.71), and closeness centrality (104.63), 27 highly connected nodes with degree >20, betweenness centrality >81.71, and closeness centrality >104.63 were identified as significant OA-related targets. These targets included collagen alpha-2(V) chain, collagen alpha-1(XII) chain, cytochrome P450 3A5 (CYP3A5), CYP2C9, collagen alpha-1(XI) chain, collagen alpha-1(VI) chain, collagen alpha-1 (III) chain, collagen alpha-1(I) chain, collagen alpha-1(IX) chain, CYP1A2, collagen alpha-1(II) chain, nuclear receptor coactivator 1 (NCOA1), collagen alpha-1(X) chain, nuclear factor NF-kappa-B p105 subunit, UDP-glucuronosyltransferase 1-1 (UGT1A1), vascular endothelial growth factor A, C-C motif chemokine 5, CYP3A4, collagen alpha-2 (I) chain, IL-8, thrombospondin-1, plasminogen activator inhibitor 1, parathyroid hormone, plasminogen, transforming growth factor beta-1 proprotein, IL-6, and transcription factor AP-1 (JUN).
Figure 4.
The protein-protein interaction (PPI) network of osteoarthritis (OA) targets. Green ovals represent osteoarthritis (OA) targets and purple ovals represent other human proteins that interacted with OA targets.
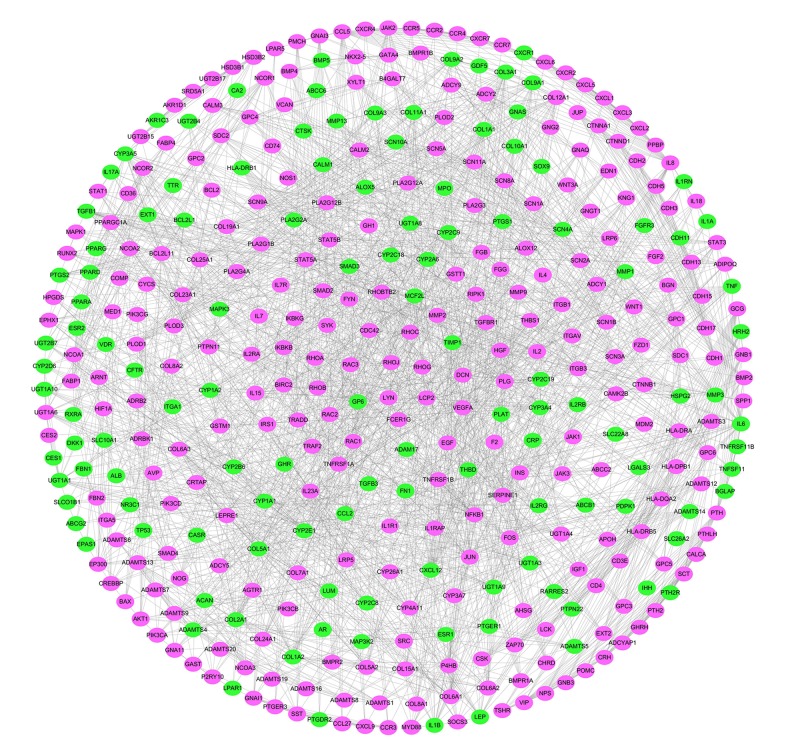
PPI network of targets for SGD in OA
To further identify the functional mechanisms of SGD in OA, the PPI network of targets for SGD in the treatment of OA was established by intersecting the two networks described above (Figure 5). The network was composed of 161 nodes (21 compound targets, 17 OA targets, 27 compound/OA targets, and 96 other proteins) and 546 edges (Figure 5). Based on the median values for degree, betweenness centrality, and closeness centrality, which were 6, 9.93, and 44.68, respectively, nodes with the degree, betweenness centrality, and closeness centrality values that were higher than the corresponding median values (degree >20, betweenness centrality >81.71, and closeness centrality >104.63) were considered as significant targets. The identified nodes included CYP3A4, nuclear receptor corepressor 1, TP53, JUN, CYP2C9, UGT1A1, CYP1A1, CYP1A2, NCOA1, nuclear receptor coactivator 2, UGT1A3, CYP3A5, CYP2D6, peroxisome proliferator-activated receptor gamma coactivator 1-alpha, IL-6, and tyrosine-protein kinase JAK2.
Figure 5.
The protein-protein interaction (PPI) network of targets for Shaoyao Gancao decoction (SGD) in osteoarthritis (OA). Yellow ovals represent osteoarthritis (OA) targets, incarnadine (crimson) ovals represent compound targets, green ovals represent compound/OA targets, and blue ovals represent other human proteins that interacted with OA targets or compound targets.
Cluster analysis
The PPI network of targets for SGD in OA was analyzed by using Molecular Complex Detection (MCODE), and five modules were obtained (Figure 6A). The biological processes, molecular functions, and signaling pathways enriched by the targets in the cluster modules were used to clarify the integral regulation of SGD for the treatment of OA (Figure 6B, 6C). In Gene Ontology (GO) terms, we discovered that (i) fatty acid binding, hormone receptor binding, and microtubules; (ii) regulation of lipid metabolism, hormone receptor binding, and nuclear chromatin; (iii) protein phosphatase activator activity, adenylate cyclase binding, and negative regulation of ryanodine-sensitive calcium-release channel activity; (iv) chemokine receptor activity, C-C chemokine receptor activity, and caveola; and (v) X chromosome, cyclin-dependent protein kinase holoenzyme complex, and cyclin-dependent protein serine, were enriched in clusters, supporting the role of SGD in the treatment of OA. The KEGG enrichment analysis showed that the signaling pathways were enriched in different modules (Figure 6C) [40]. Module 1 was highly associated with drug metabolism, including cytochrome P450; Module 2 was highly associated with the 5′AMP-activated protein kinase (AMPK) signaling pathway; Module 3 was related to gastric acid secretion; Module 4 was associated with the tumor necrosis factor (TNF) signaling pathway and chemokine signaling pathway; Module 5 was associated with the p53 signaling pathway.
Figure 6.
Enrichment analysis of the targets for Shaoyao Gancao decoction (SGD) in osteoarthritis (OA). (A) Clusters of the merged protein-protein interaction (PPI) network. Yellow ovals represent osteoarthritis (OA) targets, incarnadine (crimson) ovals represent compound targets, green ovals represent compound/OA targets, and blue ovals represent other human proteins that interacted with OA targets or compound targets. (B) The Gene Ontology (GO) pathway enrichment analysis of each cluster. (C) The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of each cluster.
Discussion
Osteoarthritis (OA) is a common form of chronic arthritis that is associated with painful symptoms that affect the quality of life for patients [41,42]. Currently, the therapeutic strategies for OA are mainly symptomatic and do not treat the underlying causes. Herbal traditional Chinese medicines (TCMs) contain several compounds that will have multiple targets, pathways, and modes of action but have been shown to treat the in OA [43]. Although Shaoyao Gancao decoction (SGD) has been used for centuries as an effective TCM for OA, its pharmacological mechanisms of action have been unclear. In this study, a network pharmacology approach was applied to determine the underlying mechanisms of SGD in OA.
After screening SGD for oral bioavailability (OB) (≥30%) and drug-likeness (DL) (≥0.18), 23 bioactive compounds were retrieved, including quercetin (OB=46.43; DL=0.28) and kaempferol (OB=41.88; DL=0.24) as potential bioactive compounds. Quercetin, one of the most abundant bioflavonoids, is known for its anti-oxidative [44], anti-inflammatory [45], antimicrobial [46], and antiviral activities [47] and its active role in promoting apoptosis in arthritic fibroblast-like synoviocytes and in protecting chondrocytes against oxidative stress [48]. Qiu et al. showed that quercetin reduced the symptoms of OA by reducing the level of reactive oxygen species (ROS), reversing mitochondrial dysfunction, and maintaining the integrity of the extracellular matrix (ECM) of the joint cartilage [49]. Kaempferol, a dietary element and an important bioflavonoid in vegetables and fruits [50], has a variety of pharmacological effects and acts as an anti-oxidant, anti-inflammatory, anti-apoptotic, anti-estrogenic, and neuroprotective agent [51]. Studies have shown that kaempferol significantly reduced in IL-1β-stimulated pro-inflammatory mediators in rat OA chondrocytes by inhibiting the NF-κB pathway [52]. Paeoniflorin (OB=53.87; DL=0.79) plays an important role in immune regulation [53], and hepatic protection [54]. Several studies have reported that liquirtin (OB=65.69; DL=0.74) has multiple pharmacological effects, as an immunomodulating agent, with anti-inflammatory, anti-allergic, anti-oxidant, and antiviral properties [55].
The PPI network of candidate targets for SGD in the treatment of OA was established based on the component and OA target networks with 161 overlapping genes. Using the median values for the degree of betweenness centrality and closeness of centrality (degree >20, betweenness centrality >81.71, and closeness centrality >104.63), 16 targets were regarded as significant. It was apparent that most of these targets, including CYP3A4, CYP2C9, CYP1A1, CYP1A2, CYP3A5, and CYP2D6 in the cytochrome P450 family, were strongly associated with drug metabolism. For instance, CYP2D6 is involved in the metabolism of the dual opioid agonist and norepinephrine-serotonin re-uptake during OA therapy [56]. CYP2C9 is involved in the metabolism of several nonsteroidal anti-inflammatory drugs (NSAIDs), contributing to the wide variability in pharmacokinetics in the metabolism of drugs [56,57]. Some targets, such as TP53 and JAK2, are associated with cell growth. TP53 is associated with OA, and the SIRT1/TP53 signaling pathway modulates the pathogenesis of OA [58]. JAK2 has a role not only in mediating angiotensin-2-induced ARHGEF1 phosphorylation [59], but also cell in the cycle by phosphorylating CNKN1B [60]. Previous studies have shown that the TCM, danshen, reduces cartilage damage in OA by regulating the JAK2/STAT3 and the AKT signaling pathways [61]. Also, JAK2 is a direct target of miR-216a-5p, and long non-coding RNA (lncRNA) DANCR regulates the proliferation, inflammation, and apoptosis of chondrocytes in OA via the miR-216a-5p-JAK2-STAT3 axis [62]. Xiong et al. found that leptin levels significantly increased in the synovial fluid of patients with OA of the temporomandibular joint (TMJ), stimulating IL-6 expression mainly via the JAK2/STAT3, p38 MAPK, and PI3K/Akt pathways [63]. Previous studies have shown that lncRNA gastric cancer-associated transcript 3 affects cell proliferation in OA by the IL-6/STAT3 signaling pathway [64].
Because clustering modules can demonstrate the biological mechanisms of key targets in disease, we classified the PPI network into five clusters (Figure 6A), and performed the Gene Ontology (GO) analysis (Figure 6B) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis (Figure 6C). Based on the GO terms, it may be proposed that the pharmacological effects of SGD in OA occurred by simultaneously activating these biological processes, cell components, and molecular functions. For example, Zhang et al. found that in a mouse model, pharmaceutical inhibition of the fatty acid binding pathway reduced the symptoms of OA induced by a high-fat diet [65]. Also, lipid metabolism is a chemical reaction involving lipids, which are compounds soluble in organic solvents [66]. Park et al. showed that the functional integrity of ABCD2 in modulating lipid metabolism was through the dysregulation of miR-141, and through ACSL4 in OA [67].
From the findings of the present study, based on the KEGG terms, the potential targets for SGD in the treatment of OA were associated with the 5′AMP-activated protein kinase (AMPK) signaling pathway, the tumor necrosis factor (TNF) signaling pathway, and the p53 signaling pathway. In the AMPK signaling pathway, AMPK serves as an intracellular sensor that not only regulates protein synthesis related to inflammation but also modulates the energy balance within chondrocytes [68]. Previous studies have shown that several bioactive compounds protect against cartilage degeneration in an OA model via the AMPK signaling pathway, including increased mitochondrial biogenesis and reduced mitochondrial dysfunction [69,70]. Zhou et al. showed that AMPK activity in chondrocytes was involved in joint homeostasis and that OA developed by promoting chondrocyte apoptosis and enhancing catabolic activity [71]. As for the TNF signaling pathway, it includes apoptosis, cell survival, inflammation, and immune function [72]. The TNF signaling pathway is important in protecting against the effects of OA, and the correlation between TNF-α levels and the degree of OA has previously been shown [73,74]. Also, the p53 signaling pathway is involved in coordinating cellular responses to different types of stress and in promoting tumor progression. Yan et al. showed that microRNA-34a had a role in chondrocyte apoptosis and proliferation by modulating the SIRT1/p53 signaling pathway in OA [58]. However, we found that pharmacological studies on the mechanisms and targets of the effects of SGD in the treatment of OA were previously limited. Based on the findings from the present study, future studies should be undertaken to assess the relationship between agents used in TCM, including SGD in OA, and their effects in terms of specific targets at the molecular level to validate the results based on data analysis.
Conclusions
This study aimed to undertake a network pharmacology analysis of the mechanism of the effects of the traditional Chinese medicine (TCM), Shaoyao Gancao decoction (SGD), in osteoarthritis (OA). The findings showed that SGD exerted its pharmacological effects in OA by modulating multiple pathways, including the cell cycle, cell apoptosis, drug metabolism, inflammation, and immune modulation. This study also provided a theoretical basis to determine the synergistic effects of TCM in treating diseases and the role of systematic network pharmacology in elucidating the potential mechanisms of action of TCMs. However, as this study was based on data mining and data analysis, further clinical validation studies should be undertaken on the role of SGD in OA.
Supplementary Materials
Supplementary Table 1.
GSD-associated target genes.
| Gene symbol | Herb |
|---|---|
| ABCC2 | GC |
| APOB | GC |
| ATAD5 | GC |
| BAZ2B | GC |
| BDNF | GC |
| BRCA1 | GC |
| CALM1 | GC |
| CBR1 | GC |
| CBR3 | GC |
| CBX1 | GC |
| CCL2 | GC |
| GFER | GC |
| GSK3A | GC |
| HMOX1 | GC |
| HSPA5 | GC |
| LDLR | GC |
| MAPK8 | GC |
| MAPK9 | GC |
| MAZF | GC |
| MBNL1 | GC |
| MLLT3 | GC |
| MMP9 | GC |
| NOS2 | GC |
| PDE5A | GC |
| PLA2G7 | GC |
| PPME1 | GC |
| RAPGEF1 | GC |
| RAPGEF3 | GC |
| SHBG | GC |
| SLC5A1 | GC |
| SLC5A2 | GC |
| SLCO2B1 | GC |
| SMAD3 | GC |
| SMPD1 | GC |
| TIM23 | GC |
| UGT1A1 | GC |
| UGT1A10 | GC |
| UGT2B15 | GC |
| ABCB11 | SY |
| ABCG5 | SY |
| ABCG8 | SY |
| ADAM10 | SY |
| ADAM17 | SY |
| ALB | SY |
| ALPI | SY |
| ALPL | SY |
| APOBEC3F | SY |
| APOBEC3G | SY |
| APOE | SY |
| ARSA | SY |
| BIRC5 | SY |
| BLM | SY |
| CAT | SY |
| CDK1 | SY |
| CFTR | SY |
| CHRM1 | SY |
| CISD1 | SY |
| CTDSP1 | SY |
| CTSD | SY |
| CYCS | SY |
| CYP2A7 | SY |
| CYP7A1 | SY |
| DHCR24 | SY |
| DNMT1 | SY |
| DRD2 | SY |
| EHMT2 | SY |
| GAA | SY |
| GLI1 | SY |
| GLI3 | SY |
| GLS | SY |
| GPBAR1 | SY |
| GPT | SY |
| HSD11B2 | SY |
| HSF1 | SY |
| HSP90AA1 | SY |
| HSP90AB1 | SY |
| ICAM1 | SY |
| IL8 | SY |
| KCNA5 | SY |
| KCNH2 | SY |
| KCNMA1 | SY |
| LMNB1 | SY |
| NOS3 | SY |
| NPSR1 | SY |
| NQO1 | SY |
| NR1H2 | SY |
| NR1H3 | SY |
| NR1I2 | SY |
| NR1I3 | SY |
| PIM2 | SY |
| PLCG1 | SY |
| PLCG2 | SY |
| PMP22 | SY |
| PRKAA2 | SY |
| PRKCB | SY |
| PRKCE | SY |
| PTPRS | SY |
| PYGM | SY |
| RACGAP1 | SY |
| RARA | SY |
| RECQL | SY |
| RORC | SY |
| RPS6KA3 | SY |
| SAE1 | SY |
| SOD1 | SY |
| SP1 | SY |
| SREBF1 | SY |
| SREBF2 | SY |
| STK16 | SY |
| STK33 | SY |
| SYK | SY |
| TLR4 | SY |
| UBA2 | SY |
| UBE2I | SY |
| UGT1A7 | SY |
| UGT1A9 | SY |
| UGT3A1 | SY |
| XIAP | SY |
| ABCB1 | GC, SY |
| ABCC1 | GC, SY |
| ABCG2 | GC, SY |
| ACHE | GC, SY |
| AHR | GC, SY |
| AKR1B1 | GC, SY |
| AKR1B10 | GC, SY |
| AKT1 | GC, SY |
| ALDH1A1 | GC, SY |
| ALOX15 | GC, SY |
| ALOX15B | GC, SY |
| ALOX5 | GC, SY |
| AMY1A | GC, SY |
| APEX1 | GC, SY |
| APP | GC, SY |
| AR | GC, SY |
| ATXN2 | GC, SY |
| BACE1 | GC, SY |
| BCHE | GC, SY |
| CA1 | GC, SY |
| CA12 | GC, SY |
| CA2 | GC, SY |
| CA4 | GC, SY |
| CA7 | GC, SY |
| CASP3 | GC, SY |
| CDK6 | GC, SY |
| CLK1 | GC, SY |
| CYP19A1 | GC, SY |
| CYP1A1 | GC, SY |
| CYP1A2 | GC, SY |
| CYP1B1 | GC, SY |
| CYP2C19 | GC, SY |
| CYP2C9 | GC, SY |
| CYP2D6 | GC, SY |
| CYP3A4 | GC, SY |
| DAPK1 | GC, SY |
| DPP4 | GC, SY |
| DYRK1A | GC, SY |
| EGFR | GC, SY |
| ESR1 | GC, SY |
| ESR2 | GC, SY |
| ESRRA | GC, SY |
| F2 | GC, SY |
| FEN1 | GC, SY |
| FLT3 | GC, SY |
| GBA | GC, SY |
| GLO1 | GC, SY |
| GLP1R | GC, SY |
| GMNN | GC, SY |
| GSK3B | GC, SY |
| HDAC9 | GC, SY |
| HIF1A | GC, SY |
| HPGD | GC, SY |
| HSD17B1 | GC, SY |
| HSD17B10 | GC, SY |
| HSD17B2 | GC, SY |
| IDH1 | GC, SY |
| KDM4A | GC, SY |
| KDM4E | GC, SY |
| LMNA | GC, SY |
| MAPT | GC, SY |
| MDM2 | GC, SY |
| MDM4 | GC, SY |
| MPG | GC, SY |
| NEU2 | GC, SY |
| NFE2L2 | GC, SY |
| NFKB1 | GC, SY |
| NFKB2 | GC, SY |
| NOX4 | GC, SY |
| NR1H4 | GC, SY |
| NR3C1 | GC, SY |
| OPRD1 | GC, SY |
| OPRK1 | GC, SY |
| OPRM1 | GC, SY |
| PAFAH1B3 | GC, SY |
| PIM1 | GC, SY |
| PIP4K2A | GC, SY |
| PNLIP | GC, SY |
| POLB | GC, SY |
| POLH | GC, SY |
| POLI | GC, SY |
| POLK | GC, SY |
| PON1 | GC, SY |
| PPARA | GC, SY |
| PPARD | GC, SY |
| PPARG | GC, SY |
| PREP | GC, SY |
| PTGS1 | GC, SY |
| PTGS2 | GC, SY |
| PTH1R | GC, SY |
| PTPN1 | GC, SY |
| RAPGEF4 | GC, SY |
| RELA | GC, SY |
| RGS4 | GC, SY |
| RXRA | GC, SY |
| SIAE | GC, SY |
| SLCO1B1 | GC, SY |
| SLCO1B3 | GC, SY |
| SMN1 | GC, SY |
| TDP1 | GC, SY |
| TOP2A | GC, SY |
| TP53 | GC, SY |
| TYR | GC, SY |
| UGT1A3 | GC, SY |
| UGT1A4 | GC, SY |
| UGT1A8 | GC, SY |
| USP1 | GC, SY |
| XDH | GC, SY |
Supplementary Table 2.
Osteoarthritis-associated target genes.
| UniProt ID | Gene symbol | Description | Organism | Source |
|---|---|---|---|---|
| P43026 | GDF5 | Growth/differentiation factor 5 | Homo sapiens | OMIM |
| P02458 | COL2A1 | Collagen, type II, alpha-1 | Homo sapiens | OMIM |
| P16112 | ACAN | Aggrecan | Homo sapiens | OMIM |
| Q9BXN1 | ASPN | Asporin | Homo sapiens | OMIM |
| P84022 | SMAD3 | Mothers against decapentaplegic, drosophila, homolog OF, 3 | Homo sapiens | OMIM |
| P0DI81 | TRAPPC2 | Tracking protein particle complex, subunit 2 | Homo sapiens | OMIM |
| Q92765 | FRZB | Frizzled-related protein | Homo sapiens | OMIM |
| P20849 | COL9A1 | Collagen, type IX, alpha-1 | Homo sapiens | OMIM |
| Q99814 | EPAS1 | Endothelial pas domain protein 1 | Homo sapiens | OMIM |
| P49747 | COMP | Cartilage oligomeric matrix protein | Homo sapiens | OMIM |
| Q9UNA0 | ADAMTS5 | A disintegrin-like and metalloproteinase with thrombospondin type 1 Motif, 5 | Homo sapiens | OMIM |
| O15232 | MATN3 | Matrilin 3 | Homo sapiens | OMIM |
| Q14623 | IHH | Indian Hedgehog | Homo sapiens | OMIM |
| Q9NRR1 | CYTL1 | Cytokine-like protein 1 | Homo sapiens | OMIM |
| P49190 | PTH2R | Parathyroid hormone 2 receptor | Homo sapiens | OMIM |
| Q92731 | ESR2 | Estrogen receptor 2 | Homo sapiens | OMIM |
| P41159 | LEP | Leptin | Homo sapiens | OMIM |
| P0DP23 | CALM1 | Calmodulin 1 | Homo sapiens | OMIM |
| P41180 | CASR | Calcium-sensing receptor | Homo sapiens | OMIM |
| P98066 | TNFAIP6 | Tumor necrosis factor-apha-induced protein 6 | Homo sapiens | OMIM |
| Q92743 | HTRA1 | HTRA serine peptidase 1 | Homo sapiens | OMIM |
| P13942 | COL11A2 | Collagen, type XI, alpha-2 | Homo sapiens | OMIM |
| Q9UHF7 | TRPS1 | Trichorhinophalangeal syndrome, type I | Homo sapiens | OMIM |
| P11473 | VDR | Vitamin D receptor | Homo sapiens | OMIM |
| Q92633 | LPAR1 | Lysophosphatidic acid receptor 1 | Homo sapiens | OMIM |
| P30044 | PRDX5 | Peroxiredoxin 5 | Homo sapiens | OMIM |
| Q9HCJ1 | ANKH | ANK, mouse, Homolog OF | Homo sapiens | OMIM |
| Q9Y2L9 | LRCH1 | Leucine-rich repeats and calponin homology domain-containing 1 | Homo sapiens | OMIM |
| P98160 | HSPG2 | Heparan sulfate proteoglycan of basement membrane | Homo sapiens | OMIM |
| P56199 | ITGA1 | Integrin, alpha-1 | Homo sapiens | OMIM |
| P45452 | MMP13 | Matrix metalloproteinase 13 | Homo sapiens | OMIM |
| P02452 | COL1A1 | Collagen, type I, alpha-1 | Homo sapiens | OMIM |
| P03372 | ESR1 | Estrogen receptor 1 | Homo sapiens | OMIM |
| P13500 | CCL2 | Chemokine, CC Motif, ligand 2 | Homo sapiens | OMIM |
| Q16552 | IL17A | Interleukin 17A | Homo sapiens | OMIM |
| P78536 | ADAM17 | A disintegrin and metalloproteinase domain 17 | Homo sapiens | OMIM |
| P51884 | LUM | Lumican | Homo sapiens | OMIM |
| P48061 | CXCL12 | Chemokine, CXC Motif, ligand 12 | Homo sapiens | OMIM |
| P43235 | CTSK | Cathepsin K | Homo sapiens | OMIM |
| P11712 | CYP2C9 | Cytochrome P450, subfamily Iic, polypeptide 9 | Homo sapiens | OMIM |
| Q99969 | RARRES2 | Retinoic acid receptor responder 2 | Homo sapiens | OMIM |
| Q9NS15 | LTBP3 | Latent transforming growth factor-beta-binding protein 3 | Homo sapiens | OMIM |
| Q9HCN6 | GP6 | Glycoprotein VI, platelet | Homo sapiens | OMIM |
| P24001 | IL32 | Interleukin 32 | Homo sapiens | OMIM |
| Q92954 | PRG4 | Proteoglycan 4 | Homo sapiens | OMIM |
| O94907 | DKK1 | DICKKOPF, Xenopus, Homolog OF, 1 | Homo sapiens | OMIM |
| Q9H5V8 | CDCP1 | CUB domain-containing protein 1 | Homo sapiens | OMIM |
| Q9Y2U5 | MAP3K2 | Mitogen-activated protein kinase kinase kinase 2 | Homo sapiens | OMIM |
| Q8TCG1 | CIP2A | Cell proliferation-regulating inhibitor of protein phosphatase 2A | Homo sapiens | OMIM |
| P14784 | IL2RB | Interleukin 2 receptor, beta | Homo sapiens | OMIM |
| P17931 | LGALS3 | Lectin, galactoside-binding, soluble, 3 | Homo sapiens | OMIM |
| Q14050 | COL9A3 | Collagen, Type IX, alpha-3 | Homo sapiens | OMIM |
| P02751 | FN1 | Fibronectin 1 | Homo sapiens | OMIM |
| P30203 | CD6 | CD6 antigen | Homo sapiens | OMIM |
| Q96S44 | TP53 | Tumor protein P53 | Homo sapiens | OMIM |
| P31785 | IL2RG | Interleukin 2 receptor, gamma | Homo sapiens | OMIM |
| P55287 | CDH11 | Cadherin 11 | Homo sapiens | OMIM |
| Q8WVB3 | HEXDC | Hexosaminidase (glycosyl hydrolase family 20, catalytic domain)-containing protein | Homo sapiens | OMIM |
| O75711 | SCRG1 | Stimulator of chondrogenesis 1 | Homo sapiens | OMIM |
| P35354 | PTGS2 | Prostaglandin-endoperoxide synthase 2 | Homo sapiens | OMIM |
| Q12794 | HYAL1 | Hyaluronoglu-cosaminidase 1 | Homo sapiens | OMIM |
| Q8IUL8 | CILP2 | Cartilage intermediate layer protein 2 | Homo sapiens | OMIM |
| Q8WVQ1 | CANT1 | Calcium-activated nucleotidase 1 | Homo sapiens | OMIM |
| Q03692 | COL10A1 | Collagen, Type X, alpha-1 | Homo sapiens | OMIM |
| P10600 | TGFB3 | Transforming growth factor, beta-3 | Homo sapiens | OMIM |
| O15530 | PDPK1 | 3-phosphoinositide-dependent protein kinase 1 | Homo sapiens | Drugbank |
| P52209 | PGD | 6-phosphogluconate dehydrogenase, decarboxylating | Homo sapiens | Drugbank |
| Q9Y215 | COLQ | Acetylcholinesterase | Homo sapiens | Drugbank |
| P78348 | ASIC1 | Acid-sensing ion channel 1 | Homo sapiens | Drugbank |
| O60218 | AKR1B10 | Aldo-keto reductase family 1 member B10 | Homo sapiens | Drugbank |
| P42330 | AKR1C3 | Aldo-keto reductase family 1 member C3 | Homo sapiens | Drugbank |
| P10275 | AR | Androgen receptor | Homo sapiens | Drugbank |
| Q07817 | BCL2L1 | Apoptosis regulator Bcl-2 | Homo sapiens | Drugbank |
| P09917 | ALOX5 | Arachidonate 5-lipoxygenase | Homo sapiens | Drugbank |
| Q2M3GO | ABCB5 | ATP-binding cassette sub-family B member 5 | Homo sapiens | Drugbank |
| Q96J66 | ABCC11 | ATP-binding cassette sub-family C member 11 | Homo sapiens | Drugbank |
| Q9UNQ0 | ABCG2 | ATP-binding cassette sub-family G member 2 | Homo sapiens | Drugbank |
| Q92887 | ABCC2 | Canalicular multispecific organic anion transporter 1 | Homo sapiens | Drugbank |
| O15438 | ABCC3 | Canalicular multispecific organic anion transporter 2 | Homo sapiens | Drugbank |
| P00918 | CA2 | Carbonic anhydrase 2 | Homo sapiens | Drugbank |
| P07451 | CA3 | Carbonic anhydrase 3 | Homo sapiens | Drugbank |
| P06276 | BCHE | Cholinesterase | Homo sapiens | Drugbank |
| P08185 | SERPINA6 | Corticosteroid-binding globulin | Homo sapiens | Drugbank |
| P25024 | CXCR1 | C-X-C chemokine receptor type 1 | Homo sapiens | Drugbank |
| P13569 | CFTR | Cystic fibrosis transmembrane conductance regulator | Homo sapiens | Drugbank |
| P04798 | CYP1A1 | Cytochrome P450 1A1 | Homo sapiens | Drugbank |
| P05177 | CYP1A2 | Cytochrome P450 1A2 | Homo sapiens | Drugbank |
| P11509 | CYP2A6 | Cytochrome P450 2A6 | Homo sapiens | Drugbank |
| P20813 | CYP2B6 | Cytochrome P450 2B6 | Homo sapiens | Drugbank |
| P33260 | CYP2C18 | Cytochrome P450 2C18 | Homo sapiens | Drugbank |
| P33261 | CYP2C19 | Cytochrome P450 2C19 | Homo sapiens | Drugbank |
| P10632 | CYP2C8 | Cytochrome P450 2C8 | Homo sapiens | Drugbank |
| P10635 | CYP2D6 | Cytochrome P450 2D6 | Homo sapiens | Drugbank |
| P05182 | CYP2E1 | Cytochrome P450 2E1 | Homo sapiens | Drugbank |
| P08684 | CYP3A4 | PCytochrome P450 3A4 | Homo sapiens | Drugbank |
| P20815 | CYP3A5 | Cytochrome P450 3A5 | Homo sapiens | Drugbank |
| P02693 | FABP2 | Fatty acid-binding protein, intestinal | Homo sapiens | Drugbank |
| P04150 | NR3C1 | Glucocorticoid receptor | Homo sapiens | Drugbank |
| P25021 | HRH2 | Histamine H2 receptor | Homo sapiens | Drugbank |
| Q04760 | GLO1 | Lactoylglutathione lyase | Homo sapiens | Drugbank |
| P23141 | CES1 | Liver carboxylesterase 1 | Homo sapiens | Drugbank |
| P27361 | MAPK3 | Mitogen-activated protein kinase 3 | Homo sapiens | Drugbank |
| P08183 | ABCB1 | Multidrug resistance protein 1 | Homo sapiens | Drugbank |
| P33527 | ABCC1 | Multidrug resistance-associated protein 1 | Homo sapiens | Drugbank |
| O15439 | ABCC4 | Multidrug resistance-associated protein 4 | Homo sapiens | Drugbank |
| O95255 | ABCC6 | Multidrug resistance-associated protein 6 | Homo sapiens | Drugbank |
| P05164 | MPO | Myeloperoxidase | Homo sapiens | Drugbank |
| Q07869 | PPARA | Peroxisome proliferator-activated receptor alpha | Homo sapiens | Drugbank |
| Q03181 | PPARD | Peroxisome proliferator-activated receptor delta | Homo sapiens | Drugbank |
| P37231 | PPARG | Peroxisome proliferator-activated receptor gamma | Homo sapiens | Drugbank |
| P14555 | PLA2G2A | Phospholipase A2, membrane associated | Homo sapiens | Drugbank |
| O43526 | KCNQ2 | Potassium voltage-gated channel subfamily KQT member 2 | Homo sapiens | Drugbank |
| O43525 | KCNQ3 | Potassium voltage-gated channel subfamily KQT member 3 | Homo sapiens | Drugbank |
| Q9Y5Y4 | PTGDR2 | Prostaglandin D2 receptor 2 | Homo sapiens | Drugbank |
| P34995 | PTGER1 | Prostaglandin E2 receptor EP1 subtype | Homo sapiens | Drugbank |
| Q8VDQ1 | PTGR2 | Prostaglandin reductase 2 | Homo sapiens | Drugbank |
| P19793 | RXRA | Retinoic acid receptor RXR-alpha | Homo sapiens | Drugbank |
| Q9Y5Y9 | SCN10A | Sodium channel protein type 10 subunit alpha | Homo sapiens | Drugbank |
| P35499 | SCN4A | Sodium channel protein type 4 subunit alpha | Homo sapiens | Drugbank |
| Q14973 | SLC10A1 | Sodium/bile acid cotransporter | Homo sapiens | Drugbank |
| P46059 | SLC15A1 | Solute carrier family 15 member 1 | Homo sapiens | Drugbank |
| Q9NSA0 | SLC22A11 | Solute carrier family 22 member 11 | Homo sapiens | Drugbank |
| O15244 | SLC22A2 | Solute carrier family 22 member 2 | Homo sapiens | Drugbank |
| Q8VC69 | SLC22A6 | Solute carrier family 22 member 6 | Homo sapiens | Drugbank |
| Q9Y694 | SLC22A7 | Solute carrier family 22 member 7 | Homo sapiens | Drugbank |
| Q8TCC7 | SLC22A8 | Solute carrier family 22 member 8 | Homo sapiens | Drugbank |
| P46721 | SLCO1A2 | Solute carrier organic anion transporter family member 1A2 | Homo sapiens | Drugbank |
| Q9Y6L6 | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | Homo sapiens | Drugbank |
| Q9NYB5 | SLCO1C1 | Solute carrier organic anion transporter family member 1C1 | Homo sapiens | Drugbank |
| O94956 | SLCO2B1 | Solute carrier organic anion transporter family member 2B1 | Homo sapiens | Drugbank |
| P07204 | THBD | Thrombomodulin | Homo sapiens | Drugbank |
| P00750 | PLAT | Tissue-type plasminogen activator | Homo sapiens | Drugbank |
| P02766 | TTR | Transthyretin | Homo sapiens | Drugbank |
| P48775 | TDO2 | Tryptophan 2,3-dioxygenase | Homo sapiens | Drugbank |
| P22309 | UGT1A1 | UDP-glucuronosyltransferase 1-1 | Homo sapiens | Drugbank |
| Q9HAW8 | UGT1A10 | UDP-glucuronosyltransferase 1-10 | Homo sapiens | Drugbank |
| P35503 | UGT1A3 | UDP-glucuronosyltransferase 1-3 | Homo sapiens | Drugbank |
| Q9HAW9 | UGT1A8 | UDP-glucuronosyltransferase 1-8 | Homo sapiens | Drugbank |
| O60656 | UGT1A9 | UDP-glucuronosyltransferase 1-9 | Homo sapiens | Drugbank |
| P06133 | UGT2B4 | UDP-glucuronosyltransferase 2B4 | Homo sapiens | Drugbank |
| P16662 | UGT2B7 | UDP-glucuronosyltransferase 2B7 | Homo sapiens | Drugbank |
| P02768 | ALB | Serum albumin | Homo sapiens | Drugbank, Genecards |
| P23219 | PTGS1 | Prostaglandin G/H synthase 1 | Homo sapiens | Drugbank, Genecards |
| P01584 | IL1B | Interleukin 1 Beta | Homo sapiens | Genecards |
| P08123 | COL1A2 | Collagen Type I Alpha 2 Chain | Homo sapiens | Genecards |
| P08254 | MMP3 | Matrix Metallopeptidase 3 | Homo sapiens | Genecards |
| P01375 | TNF | Tumor Necrosis Factor | Homo sapiens | Genecards |
| P08887 | IL6 | Interleukin 6 | Homo sapiens | Genecards |
| P03956 | MMP1 | Matrix Metallopeptidase 1 | Homo sapiens | Genecards |
| P01137 | TGFB1 | Transforming Growth Factor Beta 1 | Homo sapiens | Genecards |
| O75339 | CILP | Cartilage Intermediate Layer Protein | Homo sapiens | Genecards |
| Q9NUQ7 | UFSP2 | UFM1 Specific Peptidase 2 | Homo sapiens | Genecards |
| Q14055 | COL9A2 | Collagen Type IX Alpha 2 Chain | Homo sapiens | Genecards |
| O14788 | TNFSF11 | TNF Superfamily Member 11 | Homo sapiens | Genecards |
| P10145 | CXCL8 | C-X-C Motif Chemokine Ligand 8 | Homo sapiens | Genecards |
| O00300 | TNFRSF11B | TNF Receptor Superfamily Member 11b | Homo sapiens | Genecards |
| P01033 | TIMP1 | TIMP Metallopeptidase Inhibitor 1 | Homo sapiens | Genecards |
| O75173 | ADAMTS4 | ADAM Metallopeptidase With Thrombospondin Type 1 Motif 4 | Homo sapiens | Genecards |
| P50443 | SLC26A2 | Solute Carrier Family 26 Member 2 | Homo sapiens | Genecards |
| Q16832 | DDR2 | Discoidin Domain Receptor Tyrosine Kinase 2 | Homo sapiens | Genecards |
| Q9HBA0 | TRPV4 | Transient Receptor Potential Cation Channel Subfamily V Member 4 | Homo sapiens | Genecards |
| P02741 | CRP | C-Reactive Protein | Homo sapiens | Genecards |
| P22301 | IL10 | Interleukin 10 | Homo sapiens | Genecards |
| P36222 | CHI3L1 | Chitinase 3 Like 1 | Homo sapiens | Genecards |
| O15068 | MCF2L | MCF.2 Cell Line Derived Transforming Sequence Like | Homo sapiens | Genecards |
| P12107 | COL11A1 | Collagen Type XI Alpha 1 Chain | Homo sapiens | Genecards |
| Q14807 | KIF22 | Kinesin Family Member 22 | Homo sapiens | Genecards |
| P22003 | BMP5 | Bone Morphogenetic Protein 5 | Homo sapiens | Genecards |
| P48436 | SOX9 | SRY-Box 9 | Homo sapiens | Genecards |
| P18510 | IL1RN | Interleukin 1 Receptor Antagonist | Homo sapiens | Genecards |
| P02461 | COL3A1 | Collagen Type III Alpha 1 Chain | Homo sapiens | Genecards |
| P21941 | MATN1 | Matrilin 1, Cartilage Matrix Protein | Homo sapiens | Genecards |
| Q8WXS8 | ADAMTS14 | ADAM Metallopeptidase With Thrombospondin Type 1 Motif 14 | Homo sapiens | Genecards |
| P22607 | FGFR3 | Fibroblast Growth Factor Receptor 3 | Homo sapiens | Genecards |
| P51798 | CLCN7 | Chloride Voltage-Gated Channel 7 | Homo sapiens | Genecards |
| P35555 | FBN1 | Fibrillin 1 | Homo sapiens | Genecards |
| P10912 | GHR | Growth Hormone Receptor | Homo sapiens | Genecards |
| P02818 | BGLAP | Bone Gamma-Carboxyglutamate Protein | Homo sapiens | Genecards |
| P63092 | GNAS | GNAS Complex Locus | Homo sapiens | Genecards |
| Q16394 | EXT1 | Exostosin Glycosyltransferase 1 | Homo sapiens | Genecards |
| P01583 | IL1A | Interleukin 1 Alpha | Homo sapiens | Genecards |
| Q86Y38 | XYLT1 | Xylosyltransferase 1 | Homo sapiens | Genecards |
| P208908 | COL5A1 | Collagen Type V Alpha 1 Chain | Homo sapiens | Genecards |
| Q9GIY3 | HLA-DRB1 | Major Histocompatibility Complex, Class II, DR Beta 1 | Homo sapiens | Genecards |
| Q9Y2R2 | PTPN22 | Protein Tyrosine Phosphatase, Non-Receptor Type 22 | Homo sapiens | Genecards |
| O15266 | SHOX | Short Stature Homeobox | Homo sapiens | Genecards |
| Q93099 | HGD | Homogentisate 1,2-Dioxygenase | Homo sapiens | Genecards |
Supplementary Table 3.
SGD compound targets.
| Gene symbol | Herb |
|---|---|
| ABCC2 | GC |
| APOB | GC |
| ATAD5 | GC |
| BAZ2B | GC |
| BDNF | GC |
| BRCA1 | GC |
| CALM1 | GC |
| CBR1 | GC |
| CBR3 | GC |
| CBX1 | GC |
| CCL2 | GC |
| GFER | GC |
| GSK3A | GC |
| HMOX1 | GC |
| HSPA5 | GC |
| LDLR | GC |
| MAPK8 | GC |
| MAPK9 | GC |
| MAZF | GC |
| MBNL1 | GC |
| MLLT3 | GC |
| MMP9 | GC |
| NOS2 | GC |
| PDE5A | GC |
| PLA2G7 | GC |
| PPME1 | GC |
| RAPGEF1 | GC |
| RAPGEF3 | GC |
| SHBG | GC |
| SLC5A1 | GC |
| SLC5A2 | GC |
| SLCO2B1 | GC |
| SMAD3 | GC |
| SMPD1 | GC |
| TIM23 | GC |
| UGT1A1 | GC |
| UGT1A10 | GC |
| UGT2B15 | GC |
| ABCB11 | SY |
| ABCG5 | SY |
| ABCG8 | SY |
| ADAM10 | SY |
| ADAM17 | SY |
| ALB | SY |
| ALPI | SY |
| ALPL | SY |
| APOBEC3F | SY |
| APOBEC3G | SY |
| APOE | SY |
| ARSA | SY |
| BIRC5 | SY |
| BLM | SY |
| CAT | SY |
| CDK1 | SY |
| CFTR | SY |
| CHRM1 | SY |
| CISD1 | SY |
| CTDSP1 | SY |
| CTSD | SY |
| CYCS | SY |
| CYP2A7 | SY |
| CYP7A1 | SY |
| DHCR24 | SY |
| DNMT1 | SY |
| DRD2 | SY |
| EHMT2 | SY |
| GAA | SY |
| GLI1 | SY |
| GLI3 | SY |
| GLS | SY |
| GPBAR1 | SY |
| GPT | SY |
| HSD11B2 | SY |
| HSF1 | SY |
| HSP90AA1 | SY |
| HSP90AB1 | SY |
| ICAM1 | SY |
| IL8 | SY |
| KCNA5 | SY |
| KCNH2 | SY |
| KCNMA1 | SY |
| LMNB1 | SY |
| NOS3 | SY |
| NPSR1 | SY |
| NQO1 | SY |
| NR1H2 | SY |
| NR1H3 | SY |
| NR1I2 | SY |
| NR1I3 | SY |
| PIM2 | SY |
| PLCG1 | SY |
| PLCG2 | SY |
| PMP22 | SY |
| PRKAA2 | SY |
| PRKCB | SY |
| PRKCE | SY |
| PTPRS | SY |
| PYGM | SY |
| RACGAP1 | SY |
| RARA | SY |
| RECQL | SY |
| RORC | SY |
| RPS6KA3 | SY |
| SAE1 | SY |
| SOD1 | SY |
| SP1 | SY |
| SREBF1 | SY |
| SREBF2 | SY |
| STK16 | SY |
| STK33 | SY |
| SYK | SY |
| TLR4 | SY |
| UBA2 | SY |
| UBE2I | SY |
| UGT1A7 | SY |
| UGT1A9 | SY |
| UGT3A1 | SY |
| XIAP | SY |
| ABCB1 | GC, SY |
| ABCC1 | GC, SY |
| ABCG2 | GC, SY |
| ACHE | GC, SY |
| AHR | GC, SY |
| AKR1B1 | GC, SY |
| AKR1B10 | GC, SY |
| AKT1 | GC, SY |
| ALDH1A1 | GC, SY |
| ALOX15 | GC, SY |
| ALOX15B | GC, SY |
| ALOX5 | GC, SY |
| AMY1A | GC, SY |
| APEX1 | GC, SY |
| APP | GC, SY |
| AR | GC, SY |
| ATXN2 | GC, SY |
| BACE1 | GC, SY |
| BCHE | GC, SY |
| CA1 | GC, SY |
| CA12 | GC, SY |
| CA2 | GC, SY |
| CA4 | GC, SY |
| CA7 | GC, SY |
| CASP3 | GC, SY |
| CDK6 | GC, SY |
| CLK1 | GC, SY |
| CYP19A1 | GC, SY |
| CYP1A1 | GC, SY |
| CYP1A2 | GC, SY |
| CYP1B1 | GC, SY |
| CYP2C19 | GC, SY |
| CYP2C9 | GC, SY |
| CYP2D6 | GC, SY |
| CYP3A4 | GC, SY |
| DAPK1 | GC, SY |
| DPP4 | GC, SY |
| DYRK1A | GC, SY |
| EGFR | GC, SY |
| ESR1 | GC, SY |
| ESR2 | GC, SY |
| ESRRA | GC, SY |
| F2 | GC, SY |
| FEN1 | GC, SY |
| FLT3 | GC, SY |
| GBA | GC, SY |
| GLO1 | GC, SY |
| GLP1R | GC, SY |
| GMNN | GC, SY |
| GSK3B | GC, SY |
| HDAC9 | GC, SY |
| HIF1A | GC, SY |
| HPGD | GC, SY |
| HSD17B1 | GC, SY |
| HSD17B10 | GC, SY |
| HSD17B2 | GC, SY |
| IDH1 | GC, SY |
| KDM4A | GC, SY |
| KDM4E | GC, SY |
| LMNA | GC, SY |
| MAPT | GC, SY |
| MDM2 | GC, SY |
| MDM4 | GC, SY |
| MPG | GC, SY |
| NEU2 | GC, SY |
| NFE2L2 | GC, SY |
| NFKB1 | GC, SY |
| NFKB2 | GC, SY |
| NOX4 | GC, SY |
| NR1H4 | GC, SY |
| NR3C1 | GC, SY |
| OPRD1 | GC, SY |
| OPRK1 | GC, SY |
| OPRM1 | GC, SY |
| PAFAH1B3 | GC, SY |
| PIM1 | GC, SY |
| PIP4K2A | GC, SY |
| PNLIP | GC, SY |
| POLB | GC, SY |
| POLH | GC, SY |
| POLI | GC, SY |
| POLK | GC, SY |
| PON1 | GC, SY |
| PPARA | GC, SY |
| PPARD | GC, SY |
| PPARG | GC, SY |
| PREP | GC, SY |
| PTGS1 | GC, SY |
| PTGS2 | GC, SY |
| PTH1R | GC, SY |
| PTPN1 | GC, SY |
| RAPGEF4 | GC, SY |
| RELA | GC, SY |
| RGS4 | GC, SY |
| RXRA | GC, SY |
| SIAE | GC, SY |
| SLCO1B1 | GC, SY |
| SLCO1B3 | GC, SY |
| SMN1 | GC, SY |
| TDP1 | GC, SY |
| TOP2A | GC, SY |
| TP53 | GC, SY |
| TYR | GC, SY |
| UGT1A3 | GC, SY |
| UGT1A4 | GC, SY |
| UGT1A8 | GC, SY |
| USP1 | GC, SY |
| XDH | GC, SY |
Abbreviations
- ABCB1
multidrug resistance protein 1
- ADME
adsorption, distribution, metabolism, and excretion
- BP
biological processes
- CC
cellular components
- CM
Chinese Medicine
- CYP1A1
cytochrome P450 1A1
- CYP1A2
cytochrome P450 1A2
- CYP2C9
cytochrome P450 2C9
- CYP2D6
cytochrome P450 2D6
- CYP3A4
cytochrome P450 3A4
- CYP3A5
cytochrome P450 3A5
- DL
drug-likeness
- ESR2
estrogen receptor beta
- GC
Gancao
- GO
Gene Ontology
- IL
interleukin
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- lncRNA
long non-coding RNA
- MCODE
Molecular Complex Detection
- NCOA1
nuclear receptor coactivator 1
- NSAID
nonsteroidal anti-inflammatory drug
- OA
osteoarthritis
- OB
oral bioavailability
- PTGS2
prostaglandin G/H synthase 2
- PPI
protein-protein interaction
- SGD
Shaoyao-Gancao decoction
- SY
shaoyao
- TCM
Traditional Chinese medicine
- TCMSP
Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform
- TNF
tumor necrosis factor
- TP53
cellular tumor antigen p53
- UGT1A1
UDP-glucuronosyltransferase 1-1
- UGT1A3
UDP-glucuronosyltranserase 1–3.
Footnotes
Source of support: The National Natural Science Foundation of China (Grant No. 81703659)
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request. The datasets generated and/or analyzed in this study included the Traditional Chinese Medicine Systems Pharmacology (TCMSP) repository, http://lsp.nwu.edu.cn/tcmsp.php; TCM@Taiwan, http://tcm.cmu.edu.tw/; STITCH, http://stitch.embl.de/; PubChem, http://pubchem.ncbi.nih.gov/; GeneCard, http://www.genecards.org/; ChEMBL, http://www.ebi.ac.uk/chembl/; the Kyoto Encyclopedia of Genes and Genomes (KEGG), https://www.kegg.jp/; OMIM, http://www.omim.org/; DrugBank, https://www.drugbank.ca/; Cytoscape, http://www.cytoscape.org/; and the Gene Ontology (GO) database, http://geneontology.org/.
Conflict of interest
None.
References
- 1.Glyn-Jones S, Palmer AJ, Agricola R, et al. Osteoarthritis. Lancet. 2015;386(9991):376–87. doi: 10.1016/S0140-6736(14)60802-3. [DOI] [PubMed] [Google Scholar]
- 2.Meulenbelt I, Kloppenburg M, Kroon HM, et al. Clusters of biochemical markers are associated with radiographic subtypes of osteoarthritis (OA) in subject with familial OA at multiple sites. The GARP study. Osteoarthritis Cartilage. 2007;15(4):379–85. doi: 10.1016/j.joca.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Pereira D, Ramos E, Branco J. Osteoarthritis. Acta Med Port. 2015;28(1):99–106. doi: 10.20344/amp.5477. [DOI] [PubMed] [Google Scholar]
- 4.Sinusas K. Osteoarthritis: Diagnosis and treatment. Am Fam Physician. 2012;85(1):49–56. [PubMed] [Google Scholar]
- 5.Hiligsmann M, Cooper C, Arden N, et al. Health economics in the field of osteoarthritis: an expert’s consensus paper from the European Society for Clinical and Economic Aspects of Osteoporosis and Osteoarthritis (ESCEO) Semin Arthritis Rheum. 2013;43(3):303–13. doi: 10.1016/j.semarthrit.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Clement ND, Howard TA, Immelman RJ, et al. Patellofemoral arthroplasty versus total knee arthroplasty for patients with patellofemoral osteoarthritis. Bone Joint J. 2019;101-B(1):41–46. doi: 10.1302/0301-620X.101B1.BJJ-2018-0654.R2. [DOI] [PubMed] [Google Scholar]
- 7.Woolf AD, Erwin J, March L. The need to address the burden of musculoskeletal conditions. Best Pract Res Clin Rheumatol. 2012;26(2):183–224. doi: 10.1016/j.berh.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Nelson AE. Osteoarthritis year in review 2017: Clinical. Osteoarthritis Cartilage. 2018;26(3):319–25. doi: 10.1016/j.joca.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W, Ouyang H, Dass CR, Xu J. Current research on pharmacologic and regenerative therapies for osteoarthritis. Bone Res. 2016;4:15040. doi: 10.1038/boneres.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newberry SJ, FitzGerald J, SooHoo NF, et al. Treatment of osteoarthritis of the knee: An update review. 2017. Rockville (MD): Agency for Healthcare Research and Quality (US); 2017. May, Report No.: 17-EHC011-EF. AHRQ Comparative Effectiveness Reviews. [PubMed] [Google Scholar]
- 11.Brosseau L, Wells GA, Kenny GP, et al. The implementation of a community-based aerobic walking program for mild to moderate knee osteoarthritis (OA): a knowledge translation (KT) randomized controlled trial (RCT): Part I: The Uptake of the Ottawa Panel clinical practice guidelines (CPGs) BMC Public Health. 2012;12:871. doi: 10.1186/1471-2458-12-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reginster JY, Neuprez A, Lecart MP, et al. Role of glucosamine in the treatment for osteoarthritis. Rheumatol Int. 2012;32(10):2959–67. doi: 10.1007/s00296-012-2416-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song W, Ni S, Fu Y, Wang Y. Uncovering the mechanism of Maxing Ganshi Decoction on asthma from a systematic perspective: A network pharmacology study. Sci Rep. 2018;8(1):17362. doi: 10.1038/s41598-018-35791-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen D, Su X, Wang N, et al. Chemical isotope labeling LC-MS for monitoring disease progression and treatment in animal models: Plasma metabolomics study of osteoarthritis rat model. Sci Rep. 2017;7:40543. doi: 10.1038/srep40543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Pan J, Wang Y, et al. Component analysis of Chinese medicine and advances in fuming-washing therapy for knee osteoarthritis via unsupervised data mining methods. J Tradit Chin Med. 2013;33(5):686–91. doi: 10.1016/s0254-6272(14)60043-1. [DOI] [PubMed] [Google Scholar]
- 16.Wang JX, Yang X, Zhang JJ, et al. [Effects of Shaoyao Gancao decoction on contents of amino acids and expressions of receptors in brains of spastic paralysis rats]. Zhongguo Zhong Yao Za Zhi. 2016;41(6):1100–6. doi: 10.4268/cjcmm20160621. [in Chinese] [DOI] [PubMed] [Google Scholar]
- 17.Wu G, Zhang J, Chen W, et al. Tougu Xiaotong capsule exerts a therapeutic effect on knee osteoarthritis by regulating subchondral bone remodeling. Mol Med Rep. 2019;19(3):1858–66. doi: 10.3892/mmr.2018.9778. [DOI] [PubMed] [Google Scholar]
- 18.Berger SI, Iyengar R. Network analyses in systems pharmacology. Bioinformatics. 2009;25(19):2466–72. doi: 10.1093/bioinformatics/btp465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CY. TCM Database@Taiwan: The world’s largest traditional Chinese medicine database for drug screening in silico. PLoS One. 2011;6(1):e15939. doi: 10.1371/journal.pone.0015939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ru J, Li P, Wang J, et al. 2014. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang F, Tang Q, Tian Y, et al. Network pharmacology-based prediction of the active ingredients and potential targets of Mahuang Fuzi Xixin decoction for application to allergic rhinitis. J Ethnopharmacol. 2015;176:402–12. doi: 10.1016/j.jep.2015.10.040. [DOI] [PubMed] [Google Scholar]
- 22.Xu X, Zhang W, Huang C, et al. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci. 2012;13(6):6964–82. doi: 10.3390/ijms13066964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yue SJ, Liu J, Feng WW, et al. System pharmacology-based dissection of the synergistic mechanism of huangqi and huanglian for diabetes mellitus. Front Pharmacol. 2017;8:694. doi: 10.3389/fphar.2017.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn M, Szklarczyk D, Franceschini A, et al. STITCH 3: Zooming in on protein-chemical interactions. Nucleic Acids Res. 2012;40(Database issue):D876–80. doi: 10.1093/nar/gkr1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaulton A, Bellis LJ, Bento AP, et al. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40(Database issue):D1100–7. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halladay CW, Trikalinos TA, Schmid IT, et al. Using data sources beyond PubMed has a modest impact on the results of systematic reviews of therapeutic interventions. J Clin Epidemiol. 2015;68(9):1076–84. doi: 10.1016/j.jclinepi.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 27.Davies M, Nowotka M, Papadatos G, et al. ChEMBL web services: Streamlining access to drug discovery data and utilities. Nucleic Acids Res. 2015;43(W1):W612–20. doi: 10.1093/nar/gkv352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandran U, Patwardhan B. Network ethnopharmacological evaluation of the immunomodulatory activity of Withania somnifera. J Ethnopharmacol. 2017;197:250–56. doi: 10.1016/j.jep.2016.07.080. [DOI] [PubMed] [Google Scholar]
- 29.Law V, Knox C, Djoumbou Y, et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2014;42(Database issue):D1091–97. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Safran M, Chalifa-Caspi V, Shmueli O, et al. Human gene-centric databases at the Weizmann institute of science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res. 2003;31(1):142–46. doi: 10.1093/nar/gkg050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amberger JS, Bocchini CA, Schiettecatte F, et al. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43(Database issue):D789–98. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics. 2014;47:8.13.1–24. doi: 10.1002/0471250953.bi0813s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J, Li C, Zhu Y, et al. Integrating GO and KEGG terms to characterize and predict acute myeloid leukemia-related genes. Hematology. 2015;20(6):336–42. doi: 10.1179/1607845414Y.0000000209. [DOI] [PubMed] [Google Scholar]
- 36.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–87. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ham SW, Jeon HY, Jin X, et al. TP53 gain-of-function mutation promotes inflammation in glioblastoma. Cell Death Differ. 2019;26(3):409–25. doi: 10.1038/s41418-018-0126-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu T, Fan Z, Li W, et al. Identification of two novel chlorotoxin derivatives CA4 and CTX-23 with chemotherapeutic and anti-angiogenic potential. Sci Rep. 2016;6:19799. doi: 10.1038/srep19799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park MN, Park KH, Lee JE, et al. The expression and activation of sex steroid receptors in the preeclamptic placenta. Int J Mol Med. 2018;41(5):2943–51. doi: 10.3892/ijmm.2018.3474. [DOI] [PubMed] [Google Scholar]
- 40.Liu X, Wu J, Zhang D, et al. Network pharmacology-based approach to investigate the mechanisms of Hedyotis diffusa Wild. in the treatment of gastric cancer. Evid Based Complement Alternat Med. 2018;2018 doi: 10.1155/2018/7802639. 7802639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy G, Nagase H. Reappraising metalloproteinases in rheumatoid arthritis and osteoarthritis: Destruction or repair? Nat Clin Pract Rheumatol. 2008;4(3):128–35. doi: 10.1038/ncprheum0727. [DOI] [PubMed] [Google Scholar]
- 42.Zhu N, Hou J, Wu Y, et al. Identification of key genes in rheumatoid arthritis and osteoarthritis based on bioinformatics analysis. Medicine (Baltimore) 2018;97(22):e10997. doi: 10.1097/MD.0000000000010997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng L, Yang K, Liu H, Zhang G. A network pharmacology approach to investigate the pharmacological effects of Guizhi Fuling Wan on uterine fibroids. Exp Ther Med. 2017;14(5):4697–710. doi: 10.3892/etm.2017.5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai HD, Su SL, Qian DW, et al. Renal protective effect and action mechanism of Huangkui capsule and its main five flavonoids. J Ethnopharmacol. 2017;206:152–59. doi: 10.1016/j.jep.2017.02.046. [DOI] [PubMed] [Google Scholar]
- 45.Meng LQ, Yang FY, Wang MS, et al. Quercetin protects against chronic prostatitis in rat model through NF-κB and MAPK signaling pathways. Prostate. 2018;78(11):790–800. doi: 10.1002/pros.23536. [DOI] [PubMed] [Google Scholar]
- 46.Şeker KG, Aydin G, Altinsoy B, et al. The effect of Pelargonium endlicherianum Fenzl. root extracts on formation of nanoparticles and their antimicrobial activities. Enzyme Microb Technol. 2017;97:21–26. doi: 10.1016/j.enzmictec.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 47.Terao J, Kawai Y, Murota K. Vegetable flavonoids and cardiovascular disease. Asia Pac J Clin Nutr. 2008;17(Suppl 1):291–93. [PubMed] [Google Scholar]
- 48.Na JY, Song K, Kim S, Kwon J. Rutin protects rat articular chondrocytes against oxidative stress induced by hydrogen peroxide through SIRT1 activation. Biochem Biophys Res Commun. 2016;473(4):1301–8. doi: 10.1016/j.bbrc.2016.04.064. [DOI] [PubMed] [Google Scholar]
- 49.Qiu L, Luo Y, Chen X. Quercetin attenuates mitochondrial dysfunction and biogenesis via upregulated AMPK/SIRT1 signaling pathway in OA rats. Biomed Pharmacother. 2018;103:1585–91. doi: 10.1016/j.biopha.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 50.Chen AY, Chen YC. A review of the dietary flavonoid, kaempferol on human health and cancer chemoprevention. Food Chem. 2013;138(4):2099–107. doi: 10.1016/j.foodchem.2012.11.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parveen Z, Deng Y, Saeed MK, et al. Anti-inflammatory and analgesic activities of Thesium Chinense Turcz extracts and its major flavonoids, kaempferol and kaempferol-3-O-glucoside. Yakugaku Zasshi. 2007;127(8):1275–79. doi: 10.1248/yakushi.127.1275. [DOI] [PubMed] [Google Scholar]
- 52.Zhuang Z, Ye G, Huang B. Kaempferol alleviates the interleukin-1β-induced inflammation in rat osteoarthritis chondrocytes via suppression of NF-κB. Med Sci Monit. 2017;23:3925–31. doi: 10.12659/MSM.902491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng YQ, Wei W, Zhu L, Liu JX. Effects and mechanisms of Paeoniflorin, a bioactive glucoside from paeony root, on adjuvant arthritis in rats. Inflamm Res. 2007;56(5):182–88. doi: 10.1007/s00011-006-6002-5. [DOI] [PubMed] [Google Scholar]
- 54.Ma Z, Chu L, Liu H, et al. Paeoniflorin alleviates non-alcoholic steatohepatitis in rats: Involvement with the ROCK/NF-κB pathway. Int Immunopharmacol. 2016;38:377–84. doi: 10.1016/j.intimp.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 55.Zhao WM, Jiang SW, Chen Y, et al. Laminaria japonica increases plasma exposure of glycyrrhetinic acid following oral administration of Liquorice extract in rats. Chin J Nat Med. 2015;13:540–49. doi: 10.1016/S1875-5364(15)30049-2. [DOI] [PubMed] [Google Scholar]
- 56.Balmaceda CM. The impact of ethnicity and cardiovascular risk on the pharmacologic management of osteoarthritis: A US perspective. Postgrad Med. 2015;127(1):51–56. doi: 10.1080/00325481.2015.998593. [DOI] [PubMed] [Google Scholar]
- 57.Dai DP, Wang SH, Li CB, et al. Identification and functional assessment of a new CYP2C9 allelic variant CYP2C9*59. Drug Metab Dispos. 2015;43(8):1246–49. doi: 10.1124/dmd.115.063412. [DOI] [PubMed] [Google Scholar]
- 58.Yan S, Wang M, Zhao J, et al. MicroRNA-34a affects chondrocyte apoptosis and proliferation by targeting the SIRT1/p53 signaling pathway during the pathogenesis of osteoarthritis. Int J Mol Med. 2016;38(1):201–9. doi: 10.3892/ijmm.2016.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guilluy C, Brégeon J, Toumaniantz G, et al. The Rho exchange factor Arhgef1 mediates the effects of angiotensin II on vascular tone and blood pressure. Nat Med. 2010;16(2):183–90. doi: 10.1038/nm.2079. [DOI] [PubMed] [Google Scholar]
- 60.Jäkel H, Weinl C, Hengst L. Phosphorylation of p27Kip1 by JAK2 directly links cytokine receptor signaling to cell cycle control. Oncogene. 2011;30(32):3502–12. doi: 10.1038/onc.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu X, Lv H, Li X, et al. Danshen attenuates cartilage injuries in osteoarthritis in vivo and in vitro by activating JAK2/STAT3 and AKT pathways. Exp Anim. 2018;67(2):127–37. doi: 10.1538/expanim.17-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang L, Zhang P, Sun X, et al. Long non-coding RNA DANCR regulates proliferation and apoptosis of chondrocytes in osteoarthritis via miR-216a-5p-JAK2-STAT3 axis. Biosci Rep. 2018;38(6) doi: 10.1042/BSR20181228. pii: BSR20181228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiong H, Li W, Li J, et al. Elevated leptin levels in temporomandibular joint osteoarthritis promote pro-inflammatory cytokine IL-6 expression in synovial fibroblasts. J Oral Pathol Med. 2019;48(3):251–59. doi: 10.1111/jop.12819. [DOI] [PubMed] [Google Scholar]
- 64.Li X, Ren W, Xiao ZY, et al. GACAT3 promoted proliferation of osteoarthritis synoviocytes by IL-6/STAT3 signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22(16):5114–20. doi: 10.26355/eurrev_201808_15705. [DOI] [PubMed] [Google Scholar]
- 65.Zhang C, Chiu KY, Chan BPM, et al. Knocking out or pharmaceutical inhibition of fatty acid binding protein 4 (FABP4) alleviates osteoarthritis induced by high-fat diet in mice. Osteoarthritis Cartilage. 2018;26(6):824–33. doi: 10.1016/j.joca.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 66.Hu G, Gu W, Sun P, et al. Transcriptome analyses reveal lipid metabolic process in liver related to the difference of carcass fat content in rainbow trout (Oncorhynchus mykiss) Int J Genomics. 2016;2016 doi: 10.1155/2016/7281585. 7281585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park S, Oh J, Kim YI, et al. Suppression of ABCD2 dysregulates lipid metabolism via dysregulation of miR-141: ACSL4 in human osteoarthritis. Cell Biochem Funct. 2018;36(7):366–76. doi: 10.1002/cbf.3356. [DOI] [PubMed] [Google Scholar]
- 68.Wang X, Jin J, Wan F, et al. AMPK promotes SPOP-mediated NANOG degradation to regulate prostate cancer cell stemness. Dev Cell. 2019;48(3):345–360.e7. doi: 10.1016/j.devcel.2018.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang L, Shan H, Wang B, et al. Puerarin attenuates osteoarthritis via upregulating AMP-activated protein kinase/proliferator-activated receptor-γ coactivator-1 signaling pathway in osteoarthritis rats. Pharmacology. 2018;102(3–4):117–25. doi: 10.1159/000490418. [DOI] [PubMed] [Google Scholar]
- 70.Zhou Y, Liu SQ, Yu L, et al. Berberine prevents nitric oxide-induced rat chondrocyte apoptosis and cartilage degeneration in a rat osteoarthritis model via AMPK and p38 MAPK signaling. Apoptosis. 2015;20(9):1187–99. doi: 10.1007/s10495-015-1152-y. [DOI] [PubMed] [Google Scholar]
- 71.Zhou S, Lu W, Chen L, et al. AMPK deficiency in chondrocytes accelerated the progression of instability-induced and ageing-associated osteoarthritis in adult mice. Sci Rep. 2017;7:43245. doi: 10.1038/srep43245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li M, Ren CX, Zhang JM, et al. The effects of miR-195-5p/MMP14 on proliferation and invasion of cervical carcinoma cells through TNF signaling pathway-based on bioinformatics analysis of microarray profiling. Cell Physiol Biochem. 2018;50(4):1398–413. doi: 10.1159/000494602. [DOI] [PubMed] [Google Scholar]
- 73.Zhao YP, Liu B, Tian QY, et al. Progranulin protects against osteoarthritis through interacting with TNF-α and β-catenin signalling. Ann Rheum Dis. 2015;74(12):2244–53. doi: 10.1136/annrheumdis-2014-205779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H, Xie S, Qi Y, et al. TNF-α increases the expression of inflammatory factors in synovial fibroblasts by inhibiting the PI3K/AKT pathway in a rat model of monosodium iodoacetate-induced osteoarthritis. Exp Ther Med. 2018;16(6):4737–44. doi: 10.3892/etm.2018.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
GSD-associated target genes.
| Gene symbol | Herb |
|---|---|
| ABCC2 | GC |
| APOB | GC |
| ATAD5 | GC |
| BAZ2B | GC |
| BDNF | GC |
| BRCA1 | GC |
| CALM1 | GC |
| CBR1 | GC |
| CBR3 | GC |
| CBX1 | GC |
| CCL2 | GC |
| GFER | GC |
| GSK3A | GC |
| HMOX1 | GC |
| HSPA5 | GC |
| LDLR | GC |
| MAPK8 | GC |
| MAPK9 | GC |
| MAZF | GC |
| MBNL1 | GC |
| MLLT3 | GC |
| MMP9 | GC |
| NOS2 | GC |
| PDE5A | GC |
| PLA2G7 | GC |
| PPME1 | GC |
| RAPGEF1 | GC |
| RAPGEF3 | GC |
| SHBG | GC |
| SLC5A1 | GC |
| SLC5A2 | GC |
| SLCO2B1 | GC |
| SMAD3 | GC |
| SMPD1 | GC |
| TIM23 | GC |
| UGT1A1 | GC |
| UGT1A10 | GC |
| UGT2B15 | GC |
| ABCB11 | SY |
| ABCG5 | SY |
| ABCG8 | SY |
| ADAM10 | SY |
| ADAM17 | SY |
| ALB | SY |
| ALPI | SY |
| ALPL | SY |
| APOBEC3F | SY |
| APOBEC3G | SY |
| APOE | SY |
| ARSA | SY |
| BIRC5 | SY |
| BLM | SY |
| CAT | SY |
| CDK1 | SY |
| CFTR | SY |
| CHRM1 | SY |
| CISD1 | SY |
| CTDSP1 | SY |
| CTSD | SY |
| CYCS | SY |
| CYP2A7 | SY |
| CYP7A1 | SY |
| DHCR24 | SY |
| DNMT1 | SY |
| DRD2 | SY |
| EHMT2 | SY |
| GAA | SY |
| GLI1 | SY |
| GLI3 | SY |
| GLS | SY |
| GPBAR1 | SY |
| GPT | SY |
| HSD11B2 | SY |
| HSF1 | SY |
| HSP90AA1 | SY |
| HSP90AB1 | SY |
| ICAM1 | SY |
| IL8 | SY |
| KCNA5 | SY |
| KCNH2 | SY |
| KCNMA1 | SY |
| LMNB1 | SY |
| NOS3 | SY |
| NPSR1 | SY |
| NQO1 | SY |
| NR1H2 | SY |
| NR1H3 | SY |
| NR1I2 | SY |
| NR1I3 | SY |
| PIM2 | SY |
| PLCG1 | SY |
| PLCG2 | SY |
| PMP22 | SY |
| PRKAA2 | SY |
| PRKCB | SY |
| PRKCE | SY |
| PTPRS | SY |
| PYGM | SY |
| RACGAP1 | SY |
| RARA | SY |
| RECQL | SY |
| RORC | SY |
| RPS6KA3 | SY |
| SAE1 | SY |
| SOD1 | SY |
| SP1 | SY |
| SREBF1 | SY |
| SREBF2 | SY |
| STK16 | SY |
| STK33 | SY |
| SYK | SY |
| TLR4 | SY |
| UBA2 | SY |
| UBE2I | SY |
| UGT1A7 | SY |
| UGT1A9 | SY |
| UGT3A1 | SY |
| XIAP | SY |
| ABCB1 | GC, SY |
| ABCC1 | GC, SY |
| ABCG2 | GC, SY |
| ACHE | GC, SY |
| AHR | GC, SY |
| AKR1B1 | GC, SY |
| AKR1B10 | GC, SY |
| AKT1 | GC, SY |
| ALDH1A1 | GC, SY |
| ALOX15 | GC, SY |
| ALOX15B | GC, SY |
| ALOX5 | GC, SY |
| AMY1A | GC, SY |
| APEX1 | GC, SY |
| APP | GC, SY |
| AR | GC, SY |
| ATXN2 | GC, SY |
| BACE1 | GC, SY |
| BCHE | GC, SY |
| CA1 | GC, SY |
| CA12 | GC, SY |
| CA2 | GC, SY |
| CA4 | GC, SY |
| CA7 | GC, SY |
| CASP3 | GC, SY |
| CDK6 | GC, SY |
| CLK1 | GC, SY |
| CYP19A1 | GC, SY |
| CYP1A1 | GC, SY |
| CYP1A2 | GC, SY |
| CYP1B1 | GC, SY |
| CYP2C19 | GC, SY |
| CYP2C9 | GC, SY |
| CYP2D6 | GC, SY |
| CYP3A4 | GC, SY |
| DAPK1 | GC, SY |
| DPP4 | GC, SY |
| DYRK1A | GC, SY |
| EGFR | GC, SY |
| ESR1 | GC, SY |
| ESR2 | GC, SY |
| ESRRA | GC, SY |
| F2 | GC, SY |
| FEN1 | GC, SY |
| FLT3 | GC, SY |
| GBA | GC, SY |
| GLO1 | GC, SY |
| GLP1R | GC, SY |
| GMNN | GC, SY |
| GSK3B | GC, SY |
| HDAC9 | GC, SY |
| HIF1A | GC, SY |
| HPGD | GC, SY |
| HSD17B1 | GC, SY |
| HSD17B10 | GC, SY |
| HSD17B2 | GC, SY |
| IDH1 | GC, SY |
| KDM4A | GC, SY |
| KDM4E | GC, SY |
| LMNA | GC, SY |
| MAPT | GC, SY |
| MDM2 | GC, SY |
| MDM4 | GC, SY |
| MPG | GC, SY |
| NEU2 | GC, SY |
| NFE2L2 | GC, SY |
| NFKB1 | GC, SY |
| NFKB2 | GC, SY |
| NOX4 | GC, SY |
| NR1H4 | GC, SY |
| NR3C1 | GC, SY |
| OPRD1 | GC, SY |
| OPRK1 | GC, SY |
| OPRM1 | GC, SY |
| PAFAH1B3 | GC, SY |
| PIM1 | GC, SY |
| PIP4K2A | GC, SY |
| PNLIP | GC, SY |
| POLB | GC, SY |
| POLH | GC, SY |
| POLI | GC, SY |
| POLK | GC, SY |
| PON1 | GC, SY |
| PPARA | GC, SY |
| PPARD | GC, SY |
| PPARG | GC, SY |
| PREP | GC, SY |
| PTGS1 | GC, SY |
| PTGS2 | GC, SY |
| PTH1R | GC, SY |
| PTPN1 | GC, SY |
| RAPGEF4 | GC, SY |
| RELA | GC, SY |
| RGS4 | GC, SY |
| RXRA | GC, SY |
| SIAE | GC, SY |
| SLCO1B1 | GC, SY |
| SLCO1B3 | GC, SY |
| SMN1 | GC, SY |
| TDP1 | GC, SY |
| TOP2A | GC, SY |
| TP53 | GC, SY |
| TYR | GC, SY |
| UGT1A3 | GC, SY |
| UGT1A4 | GC, SY |
| UGT1A8 | GC, SY |
| USP1 | GC, SY |
| XDH | GC, SY |
Supplementary Table 2.
Osteoarthritis-associated target genes.
| UniProt ID | Gene symbol | Description | Organism | Source |
|---|---|---|---|---|
| P43026 | GDF5 | Growth/differentiation factor 5 | Homo sapiens | OMIM |
| P02458 | COL2A1 | Collagen, type II, alpha-1 | Homo sapiens | OMIM |
| P16112 | ACAN | Aggrecan | Homo sapiens | OMIM |
| Q9BXN1 | ASPN | Asporin | Homo sapiens | OMIM |
| P84022 | SMAD3 | Mothers against decapentaplegic, drosophila, homolog OF, 3 | Homo sapiens | OMIM |
| P0DI81 | TRAPPC2 | Tracking protein particle complex, subunit 2 | Homo sapiens | OMIM |
| Q92765 | FRZB | Frizzled-related protein | Homo sapiens | OMIM |
| P20849 | COL9A1 | Collagen, type IX, alpha-1 | Homo sapiens | OMIM |
| Q99814 | EPAS1 | Endothelial pas domain protein 1 | Homo sapiens | OMIM |
| P49747 | COMP | Cartilage oligomeric matrix protein | Homo sapiens | OMIM |
| Q9UNA0 | ADAMTS5 | A disintegrin-like and metalloproteinase with thrombospondin type 1 Motif, 5 | Homo sapiens | OMIM |
| O15232 | MATN3 | Matrilin 3 | Homo sapiens | OMIM |
| Q14623 | IHH | Indian Hedgehog | Homo sapiens | OMIM |
| Q9NRR1 | CYTL1 | Cytokine-like protein 1 | Homo sapiens | OMIM |
| P49190 | PTH2R | Parathyroid hormone 2 receptor | Homo sapiens | OMIM |
| Q92731 | ESR2 | Estrogen receptor 2 | Homo sapiens | OMIM |
| P41159 | LEP | Leptin | Homo sapiens | OMIM |
| P0DP23 | CALM1 | Calmodulin 1 | Homo sapiens | OMIM |
| P41180 | CASR | Calcium-sensing receptor | Homo sapiens | OMIM |
| P98066 | TNFAIP6 | Tumor necrosis factor-apha-induced protein 6 | Homo sapiens | OMIM |
| Q92743 | HTRA1 | HTRA serine peptidase 1 | Homo sapiens | OMIM |
| P13942 | COL11A2 | Collagen, type XI, alpha-2 | Homo sapiens | OMIM |
| Q9UHF7 | TRPS1 | Trichorhinophalangeal syndrome, type I | Homo sapiens | OMIM |
| P11473 | VDR | Vitamin D receptor | Homo sapiens | OMIM |
| Q92633 | LPAR1 | Lysophosphatidic acid receptor 1 | Homo sapiens | OMIM |
| P30044 | PRDX5 | Peroxiredoxin 5 | Homo sapiens | OMIM |
| Q9HCJ1 | ANKH | ANK, mouse, Homolog OF | Homo sapiens | OMIM |
| Q9Y2L9 | LRCH1 | Leucine-rich repeats and calponin homology domain-containing 1 | Homo sapiens | OMIM |
| P98160 | HSPG2 | Heparan sulfate proteoglycan of basement membrane | Homo sapiens | OMIM |
| P56199 | ITGA1 | Integrin, alpha-1 | Homo sapiens | OMIM |
| P45452 | MMP13 | Matrix metalloproteinase 13 | Homo sapiens | OMIM |
| P02452 | COL1A1 | Collagen, type I, alpha-1 | Homo sapiens | OMIM |
| P03372 | ESR1 | Estrogen receptor 1 | Homo sapiens | OMIM |
| P13500 | CCL2 | Chemokine, CC Motif, ligand 2 | Homo sapiens | OMIM |
| Q16552 | IL17A | Interleukin 17A | Homo sapiens | OMIM |
| P78536 | ADAM17 | A disintegrin and metalloproteinase domain 17 | Homo sapiens | OMIM |
| P51884 | LUM | Lumican | Homo sapiens | OMIM |
| P48061 | CXCL12 | Chemokine, CXC Motif, ligand 12 | Homo sapiens | OMIM |
| P43235 | CTSK | Cathepsin K | Homo sapiens | OMIM |
| P11712 | CYP2C9 | Cytochrome P450, subfamily Iic, polypeptide 9 | Homo sapiens | OMIM |
| Q99969 | RARRES2 | Retinoic acid receptor responder 2 | Homo sapiens | OMIM |
| Q9NS15 | LTBP3 | Latent transforming growth factor-beta-binding protein 3 | Homo sapiens | OMIM |
| Q9HCN6 | GP6 | Glycoprotein VI, platelet | Homo sapiens | OMIM |
| P24001 | IL32 | Interleukin 32 | Homo sapiens | OMIM |
| Q92954 | PRG4 | Proteoglycan 4 | Homo sapiens | OMIM |
| O94907 | DKK1 | DICKKOPF, Xenopus, Homolog OF, 1 | Homo sapiens | OMIM |
| Q9H5V8 | CDCP1 | CUB domain-containing protein 1 | Homo sapiens | OMIM |
| Q9Y2U5 | MAP3K2 | Mitogen-activated protein kinase kinase kinase 2 | Homo sapiens | OMIM |
| Q8TCG1 | CIP2A | Cell proliferation-regulating inhibitor of protein phosphatase 2A | Homo sapiens | OMIM |
| P14784 | IL2RB | Interleukin 2 receptor, beta | Homo sapiens | OMIM |
| P17931 | LGALS3 | Lectin, galactoside-binding, soluble, 3 | Homo sapiens | OMIM |
| Q14050 | COL9A3 | Collagen, Type IX, alpha-3 | Homo sapiens | OMIM |
| P02751 | FN1 | Fibronectin 1 | Homo sapiens | OMIM |
| P30203 | CD6 | CD6 antigen | Homo sapiens | OMIM |
| Q96S44 | TP53 | Tumor protein P53 | Homo sapiens | OMIM |
| P31785 | IL2RG | Interleukin 2 receptor, gamma | Homo sapiens | OMIM |
| P55287 | CDH11 | Cadherin 11 | Homo sapiens | OMIM |
| Q8WVB3 | HEXDC | Hexosaminidase (glycosyl hydrolase family 20, catalytic domain)-containing protein | Homo sapiens | OMIM |
| O75711 | SCRG1 | Stimulator of chondrogenesis 1 | Homo sapiens | OMIM |
| P35354 | PTGS2 | Prostaglandin-endoperoxide synthase 2 | Homo sapiens | OMIM |
| Q12794 | HYAL1 | Hyaluronoglu-cosaminidase 1 | Homo sapiens | OMIM |
| Q8IUL8 | CILP2 | Cartilage intermediate layer protein 2 | Homo sapiens | OMIM |
| Q8WVQ1 | CANT1 | Calcium-activated nucleotidase 1 | Homo sapiens | OMIM |
| Q03692 | COL10A1 | Collagen, Type X, alpha-1 | Homo sapiens | OMIM |
| P10600 | TGFB3 | Transforming growth factor, beta-3 | Homo sapiens | OMIM |
| O15530 | PDPK1 | 3-phosphoinositide-dependent protein kinase 1 | Homo sapiens | Drugbank |
| P52209 | PGD | 6-phosphogluconate dehydrogenase, decarboxylating | Homo sapiens | Drugbank |
| Q9Y215 | COLQ | Acetylcholinesterase | Homo sapiens | Drugbank |
| P78348 | ASIC1 | Acid-sensing ion channel 1 | Homo sapiens | Drugbank |
| O60218 | AKR1B10 | Aldo-keto reductase family 1 member B10 | Homo sapiens | Drugbank |
| P42330 | AKR1C3 | Aldo-keto reductase family 1 member C3 | Homo sapiens | Drugbank |
| P10275 | AR | Androgen receptor | Homo sapiens | Drugbank |
| Q07817 | BCL2L1 | Apoptosis regulator Bcl-2 | Homo sapiens | Drugbank |
| P09917 | ALOX5 | Arachidonate 5-lipoxygenase | Homo sapiens | Drugbank |
| Q2M3GO | ABCB5 | ATP-binding cassette sub-family B member 5 | Homo sapiens | Drugbank |
| Q96J66 | ABCC11 | ATP-binding cassette sub-family C member 11 | Homo sapiens | Drugbank |
| Q9UNQ0 | ABCG2 | ATP-binding cassette sub-family G member 2 | Homo sapiens | Drugbank |
| Q92887 | ABCC2 | Canalicular multispecific organic anion transporter 1 | Homo sapiens | Drugbank |
| O15438 | ABCC3 | Canalicular multispecific organic anion transporter 2 | Homo sapiens | Drugbank |
| P00918 | CA2 | Carbonic anhydrase 2 | Homo sapiens | Drugbank |
| P07451 | CA3 | Carbonic anhydrase 3 | Homo sapiens | Drugbank |
| P06276 | BCHE | Cholinesterase | Homo sapiens | Drugbank |
| P08185 | SERPINA6 | Corticosteroid-binding globulin | Homo sapiens | Drugbank |
| P25024 | CXCR1 | C-X-C chemokine receptor type 1 | Homo sapiens | Drugbank |
| P13569 | CFTR | Cystic fibrosis transmembrane conductance regulator | Homo sapiens | Drugbank |
| P04798 | CYP1A1 | Cytochrome P450 1A1 | Homo sapiens | Drugbank |
| P05177 | CYP1A2 | Cytochrome P450 1A2 | Homo sapiens | Drugbank |
| P11509 | CYP2A6 | Cytochrome P450 2A6 | Homo sapiens | Drugbank |
| P20813 | CYP2B6 | Cytochrome P450 2B6 | Homo sapiens | Drugbank |
| P33260 | CYP2C18 | Cytochrome P450 2C18 | Homo sapiens | Drugbank |
| P33261 | CYP2C19 | Cytochrome P450 2C19 | Homo sapiens | Drugbank |
| P10632 | CYP2C8 | Cytochrome P450 2C8 | Homo sapiens | Drugbank |
| P10635 | CYP2D6 | Cytochrome P450 2D6 | Homo sapiens | Drugbank |
| P05182 | CYP2E1 | Cytochrome P450 2E1 | Homo sapiens | Drugbank |
| P08684 | CYP3A4 | PCytochrome P450 3A4 | Homo sapiens | Drugbank |
| P20815 | CYP3A5 | Cytochrome P450 3A5 | Homo sapiens | Drugbank |
| P02693 | FABP2 | Fatty acid-binding protein, intestinal | Homo sapiens | Drugbank |
| P04150 | NR3C1 | Glucocorticoid receptor | Homo sapiens | Drugbank |
| P25021 | HRH2 | Histamine H2 receptor | Homo sapiens | Drugbank |
| Q04760 | GLO1 | Lactoylglutathione lyase | Homo sapiens | Drugbank |
| P23141 | CES1 | Liver carboxylesterase 1 | Homo sapiens | Drugbank |
| P27361 | MAPK3 | Mitogen-activated protein kinase 3 | Homo sapiens | Drugbank |
| P08183 | ABCB1 | Multidrug resistance protein 1 | Homo sapiens | Drugbank |
| P33527 | ABCC1 | Multidrug resistance-associated protein 1 | Homo sapiens | Drugbank |
| O15439 | ABCC4 | Multidrug resistance-associated protein 4 | Homo sapiens | Drugbank |
| O95255 | ABCC6 | Multidrug resistance-associated protein 6 | Homo sapiens | Drugbank |
| P05164 | MPO | Myeloperoxidase | Homo sapiens | Drugbank |
| Q07869 | PPARA | Peroxisome proliferator-activated receptor alpha | Homo sapiens | Drugbank |
| Q03181 | PPARD | Peroxisome proliferator-activated receptor delta | Homo sapiens | Drugbank |
| P37231 | PPARG | Peroxisome proliferator-activated receptor gamma | Homo sapiens | Drugbank |
| P14555 | PLA2G2A | Phospholipase A2, membrane associated | Homo sapiens | Drugbank |
| O43526 | KCNQ2 | Potassium voltage-gated channel subfamily KQT member 2 | Homo sapiens | Drugbank |
| O43525 | KCNQ3 | Potassium voltage-gated channel subfamily KQT member 3 | Homo sapiens | Drugbank |
| Q9Y5Y4 | PTGDR2 | Prostaglandin D2 receptor 2 | Homo sapiens | Drugbank |
| P34995 | PTGER1 | Prostaglandin E2 receptor EP1 subtype | Homo sapiens | Drugbank |
| Q8VDQ1 | PTGR2 | Prostaglandin reductase 2 | Homo sapiens | Drugbank |
| P19793 | RXRA | Retinoic acid receptor RXR-alpha | Homo sapiens | Drugbank |
| Q9Y5Y9 | SCN10A | Sodium channel protein type 10 subunit alpha | Homo sapiens | Drugbank |
| P35499 | SCN4A | Sodium channel protein type 4 subunit alpha | Homo sapiens | Drugbank |
| Q14973 | SLC10A1 | Sodium/bile acid cotransporter | Homo sapiens | Drugbank |
| P46059 | SLC15A1 | Solute carrier family 15 member 1 | Homo sapiens | Drugbank |
| Q9NSA0 | SLC22A11 | Solute carrier family 22 member 11 | Homo sapiens | Drugbank |
| O15244 | SLC22A2 | Solute carrier family 22 member 2 | Homo sapiens | Drugbank |
| Q8VC69 | SLC22A6 | Solute carrier family 22 member 6 | Homo sapiens | Drugbank |
| Q9Y694 | SLC22A7 | Solute carrier family 22 member 7 | Homo sapiens | Drugbank |
| Q8TCC7 | SLC22A8 | Solute carrier family 22 member 8 | Homo sapiens | Drugbank |
| P46721 | SLCO1A2 | Solute carrier organic anion transporter family member 1A2 | Homo sapiens | Drugbank |
| Q9Y6L6 | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | Homo sapiens | Drugbank |
| Q9NYB5 | SLCO1C1 | Solute carrier organic anion transporter family member 1C1 | Homo sapiens | Drugbank |
| O94956 | SLCO2B1 | Solute carrier organic anion transporter family member 2B1 | Homo sapiens | Drugbank |
| P07204 | THBD | Thrombomodulin | Homo sapiens | Drugbank |
| P00750 | PLAT | Tissue-type plasminogen activator | Homo sapiens | Drugbank |
| P02766 | TTR | Transthyretin | Homo sapiens | Drugbank |
| P48775 | TDO2 | Tryptophan 2,3-dioxygenase | Homo sapiens | Drugbank |
| P22309 | UGT1A1 | UDP-glucuronosyltransferase 1-1 | Homo sapiens | Drugbank |
| Q9HAW8 | UGT1A10 | UDP-glucuronosyltransferase 1-10 | Homo sapiens | Drugbank |
| P35503 | UGT1A3 | UDP-glucuronosyltransferase 1-3 | Homo sapiens | Drugbank |
| Q9HAW9 | UGT1A8 | UDP-glucuronosyltransferase 1-8 | Homo sapiens | Drugbank |
| O60656 | UGT1A9 | UDP-glucuronosyltransferase 1-9 | Homo sapiens | Drugbank |
| P06133 | UGT2B4 | UDP-glucuronosyltransferase 2B4 | Homo sapiens | Drugbank |
| P16662 | UGT2B7 | UDP-glucuronosyltransferase 2B7 | Homo sapiens | Drugbank |
| P02768 | ALB | Serum albumin | Homo sapiens | Drugbank, Genecards |
| P23219 | PTGS1 | Prostaglandin G/H synthase 1 | Homo sapiens | Drugbank, Genecards |
| P01584 | IL1B | Interleukin 1 Beta | Homo sapiens | Genecards |
| P08123 | COL1A2 | Collagen Type I Alpha 2 Chain | Homo sapiens | Genecards |
| P08254 | MMP3 | Matrix Metallopeptidase 3 | Homo sapiens | Genecards |
| P01375 | TNF | Tumor Necrosis Factor | Homo sapiens | Genecards |
| P08887 | IL6 | Interleukin 6 | Homo sapiens | Genecards |
| P03956 | MMP1 | Matrix Metallopeptidase 1 | Homo sapiens | Genecards |
| P01137 | TGFB1 | Transforming Growth Factor Beta 1 | Homo sapiens | Genecards |
| O75339 | CILP | Cartilage Intermediate Layer Protein | Homo sapiens | Genecards |
| Q9NUQ7 | UFSP2 | UFM1 Specific Peptidase 2 | Homo sapiens | Genecards |
| Q14055 | COL9A2 | Collagen Type IX Alpha 2 Chain | Homo sapiens | Genecards |
| O14788 | TNFSF11 | TNF Superfamily Member 11 | Homo sapiens | Genecards |
| P10145 | CXCL8 | C-X-C Motif Chemokine Ligand 8 | Homo sapiens | Genecards |
| O00300 | TNFRSF11B | TNF Receptor Superfamily Member 11b | Homo sapiens | Genecards |
| P01033 | TIMP1 | TIMP Metallopeptidase Inhibitor 1 | Homo sapiens | Genecards |
| O75173 | ADAMTS4 | ADAM Metallopeptidase With Thrombospondin Type 1 Motif 4 | Homo sapiens | Genecards |
| P50443 | SLC26A2 | Solute Carrier Family 26 Member 2 | Homo sapiens | Genecards |
| Q16832 | DDR2 | Discoidin Domain Receptor Tyrosine Kinase 2 | Homo sapiens | Genecards |
| Q9HBA0 | TRPV4 | Transient Receptor Potential Cation Channel Subfamily V Member 4 | Homo sapiens | Genecards |
| P02741 | CRP | C-Reactive Protein | Homo sapiens | Genecards |
| P22301 | IL10 | Interleukin 10 | Homo sapiens | Genecards |
| P36222 | CHI3L1 | Chitinase 3 Like 1 | Homo sapiens | Genecards |
| O15068 | MCF2L | MCF.2 Cell Line Derived Transforming Sequence Like | Homo sapiens | Genecards |
| P12107 | COL11A1 | Collagen Type XI Alpha 1 Chain | Homo sapiens | Genecards |
| Q14807 | KIF22 | Kinesin Family Member 22 | Homo sapiens | Genecards |
| P22003 | BMP5 | Bone Morphogenetic Protein 5 | Homo sapiens | Genecards |
| P48436 | SOX9 | SRY-Box 9 | Homo sapiens | Genecards |
| P18510 | IL1RN | Interleukin 1 Receptor Antagonist | Homo sapiens | Genecards |
| P02461 | COL3A1 | Collagen Type III Alpha 1 Chain | Homo sapiens | Genecards |
| P21941 | MATN1 | Matrilin 1, Cartilage Matrix Protein | Homo sapiens | Genecards |
| Q8WXS8 | ADAMTS14 | ADAM Metallopeptidase With Thrombospondin Type 1 Motif 14 | Homo sapiens | Genecards |
| P22607 | FGFR3 | Fibroblast Growth Factor Receptor 3 | Homo sapiens | Genecards |
| P51798 | CLCN7 | Chloride Voltage-Gated Channel 7 | Homo sapiens | Genecards |
| P35555 | FBN1 | Fibrillin 1 | Homo sapiens | Genecards |
| P10912 | GHR | Growth Hormone Receptor | Homo sapiens | Genecards |
| P02818 | BGLAP | Bone Gamma-Carboxyglutamate Protein | Homo sapiens | Genecards |
| P63092 | GNAS | GNAS Complex Locus | Homo sapiens | Genecards |
| Q16394 | EXT1 | Exostosin Glycosyltransferase 1 | Homo sapiens | Genecards |
| P01583 | IL1A | Interleukin 1 Alpha | Homo sapiens | Genecards |
| Q86Y38 | XYLT1 | Xylosyltransferase 1 | Homo sapiens | Genecards |
| P208908 | COL5A1 | Collagen Type V Alpha 1 Chain | Homo sapiens | Genecards |
| Q9GIY3 | HLA-DRB1 | Major Histocompatibility Complex, Class II, DR Beta 1 | Homo sapiens | Genecards |
| Q9Y2R2 | PTPN22 | Protein Tyrosine Phosphatase, Non-Receptor Type 22 | Homo sapiens | Genecards |
| O15266 | SHOX | Short Stature Homeobox | Homo sapiens | Genecards |
| Q93099 | HGD | Homogentisate 1,2-Dioxygenase | Homo sapiens | Genecards |
Supplementary Table 3.
SGD compound targets.
| Gene symbol | Herb |
|---|---|
| ABCC2 | GC |
| APOB | GC |
| ATAD5 | GC |
| BAZ2B | GC |
| BDNF | GC |
| BRCA1 | GC |
| CALM1 | GC |
| CBR1 | GC |
| CBR3 | GC |
| CBX1 | GC |
| CCL2 | GC |
| GFER | GC |
| GSK3A | GC |
| HMOX1 | GC |
| HSPA5 | GC |
| LDLR | GC |
| MAPK8 | GC |
| MAPK9 | GC |
| MAZF | GC |
| MBNL1 | GC |
| MLLT3 | GC |
| MMP9 | GC |
| NOS2 | GC |
| PDE5A | GC |
| PLA2G7 | GC |
| PPME1 | GC |
| RAPGEF1 | GC |
| RAPGEF3 | GC |
| SHBG | GC |
| SLC5A1 | GC |
| SLC5A2 | GC |
| SLCO2B1 | GC |
| SMAD3 | GC |
| SMPD1 | GC |
| TIM23 | GC |
| UGT1A1 | GC |
| UGT1A10 | GC |
| UGT2B15 | GC |
| ABCB11 | SY |
| ABCG5 | SY |
| ABCG8 | SY |
| ADAM10 | SY |
| ADAM17 | SY |
| ALB | SY |
| ALPI | SY |
| ALPL | SY |
| APOBEC3F | SY |
| APOBEC3G | SY |
| APOE | SY |
| ARSA | SY |
| BIRC5 | SY |
| BLM | SY |
| CAT | SY |
| CDK1 | SY |
| CFTR | SY |
| CHRM1 | SY |
| CISD1 | SY |
| CTDSP1 | SY |
| CTSD | SY |
| CYCS | SY |
| CYP2A7 | SY |
| CYP7A1 | SY |
| DHCR24 | SY |
| DNMT1 | SY |
| DRD2 | SY |
| EHMT2 | SY |
| GAA | SY |
| GLI1 | SY |
| GLI3 | SY |
| GLS | SY |
| GPBAR1 | SY |
| GPT | SY |
| HSD11B2 | SY |
| HSF1 | SY |
| HSP90AA1 | SY |
| HSP90AB1 | SY |
| ICAM1 | SY |
| IL8 | SY |
| KCNA5 | SY |
| KCNH2 | SY |
| KCNMA1 | SY |
| LMNB1 | SY |
| NOS3 | SY |
| NPSR1 | SY |
| NQO1 | SY |
| NR1H2 | SY |
| NR1H3 | SY |
| NR1I2 | SY |
| NR1I3 | SY |
| PIM2 | SY |
| PLCG1 | SY |
| PLCG2 | SY |
| PMP22 | SY |
| PRKAA2 | SY |
| PRKCB | SY |
| PRKCE | SY |
| PTPRS | SY |
| PYGM | SY |
| RACGAP1 | SY |
| RARA | SY |
| RECQL | SY |
| RORC | SY |
| RPS6KA3 | SY |
| SAE1 | SY |
| SOD1 | SY |
| SP1 | SY |
| SREBF1 | SY |
| SREBF2 | SY |
| STK16 | SY |
| STK33 | SY |
| SYK | SY |
| TLR4 | SY |
| UBA2 | SY |
| UBE2I | SY |
| UGT1A7 | SY |
| UGT1A9 | SY |
| UGT3A1 | SY |
| XIAP | SY |
| ABCB1 | GC, SY |
| ABCC1 | GC, SY |
| ABCG2 | GC, SY |
| ACHE | GC, SY |
| AHR | GC, SY |
| AKR1B1 | GC, SY |
| AKR1B10 | GC, SY |
| AKT1 | GC, SY |
| ALDH1A1 | GC, SY |
| ALOX15 | GC, SY |
| ALOX15B | GC, SY |
| ALOX5 | GC, SY |
| AMY1A | GC, SY |
| APEX1 | GC, SY |
| APP | GC, SY |
| AR | GC, SY |
| ATXN2 | GC, SY |
| BACE1 | GC, SY |
| BCHE | GC, SY |
| CA1 | GC, SY |
| CA12 | GC, SY |
| CA2 | GC, SY |
| CA4 | GC, SY |
| CA7 | GC, SY |
| CASP3 | GC, SY |
| CDK6 | GC, SY |
| CLK1 | GC, SY |
| CYP19A1 | GC, SY |
| CYP1A1 | GC, SY |
| CYP1A2 | GC, SY |
| CYP1B1 | GC, SY |
| CYP2C19 | GC, SY |
| CYP2C9 | GC, SY |
| CYP2D6 | GC, SY |
| CYP3A4 | GC, SY |
| DAPK1 | GC, SY |
| DPP4 | GC, SY |
| DYRK1A | GC, SY |
| EGFR | GC, SY |
| ESR1 | GC, SY |
| ESR2 | GC, SY |
| ESRRA | GC, SY |
| F2 | GC, SY |
| FEN1 | GC, SY |
| FLT3 | GC, SY |
| GBA | GC, SY |
| GLO1 | GC, SY |
| GLP1R | GC, SY |
| GMNN | GC, SY |
| GSK3B | GC, SY |
| HDAC9 | GC, SY |
| HIF1A | GC, SY |
| HPGD | GC, SY |
| HSD17B1 | GC, SY |
| HSD17B10 | GC, SY |
| HSD17B2 | GC, SY |
| IDH1 | GC, SY |
| KDM4A | GC, SY |
| KDM4E | GC, SY |
| LMNA | GC, SY |
| MAPT | GC, SY |
| MDM2 | GC, SY |
| MDM4 | GC, SY |
| MPG | GC, SY |
| NEU2 | GC, SY |
| NFE2L2 | GC, SY |
| NFKB1 | GC, SY |
| NFKB2 | GC, SY |
| NOX4 | GC, SY |
| NR1H4 | GC, SY |
| NR3C1 | GC, SY |
| OPRD1 | GC, SY |
| OPRK1 | GC, SY |
| OPRM1 | GC, SY |
| PAFAH1B3 | GC, SY |
| PIM1 | GC, SY |
| PIP4K2A | GC, SY |
| PNLIP | GC, SY |
| POLB | GC, SY |
| POLH | GC, SY |
| POLI | GC, SY |
| POLK | GC, SY |
| PON1 | GC, SY |
| PPARA | GC, SY |
| PPARD | GC, SY |
| PPARG | GC, SY |
| PREP | GC, SY |
| PTGS1 | GC, SY |
| PTGS2 | GC, SY |
| PTH1R | GC, SY |
| PTPN1 | GC, SY |
| RAPGEF4 | GC, SY |
| RELA | GC, SY |
| RGS4 | GC, SY |
| RXRA | GC, SY |
| SIAE | GC, SY |
| SLCO1B1 | GC, SY |
| SLCO1B3 | GC, SY |
| SMN1 | GC, SY |
| TDP1 | GC, SY |
| TOP2A | GC, SY |
| TP53 | GC, SY |
| TYR | GC, SY |
| UGT1A3 | GC, SY |
| UGT1A4 | GC, SY |
| UGT1A8 | GC, SY |
| USP1 | GC, SY |
| XDH | GC, SY |