Abstract
The present study investigated the effects of iron, iron chelators, and mutations of tonB or iroN fepA genes on the growth and virulence of Salmonella Typhimurium. Results indicated that organic iron (ferric citrate and ferrous-l-ascorbate) supported better growth of Salmonella compared to inorganic iron. Among tested chelators, 2,2′-bipyridyl at 500 μM showed the highest inhibition of Salmonella growth with 5 μM ferrous sulfate. Deletion of genes (tonB– and iroN– fepA–) in the iron uptake system attenuated Salmonella invasion of Caco-2 cells and its ability to damage the epithelial monolayer. The expression of all tested host genes in Caco-2 was not affected under the iron-poor condition. However, claudin 3, tight junction protein 1, tumor necrosis factor α (TNF-α), and interleukin-8 (IL-8) were altered under the iron-rich condition depending on individual mutations. In Caenorhabditis elegans, a significant down-regulation of ferritin 1 expression was observed when the nematode was infected by the wild-type (WT) strain.
1. Introduction
Nontyphi Salmonella (NTS) are invasive pathogens that cause nontyphoidal salmonellosis, leading to significant public health issues and economic losses.1 In Salmonella enterica and other pathogens, the acquisition of iron determines multiple pathogenic characters and is required for their full virulence.2 The NTS possess a complicated iron uptake system including the use of various siderophores to acquire enough iron from the environment.3 Previous studies have shown that the iron-siderophore system of S. enterica consists of the ferric-enterobactin (Fep) transporter system and ferrichrome-iron (Fhu) transporter system.4,5 Importantly, the siderophore receptors of the two iron transporter systems are powered by TonB, an energy transduction protein that mediates the active transport of ferric-siderophore complex across the outer membrane of Gram-negative bacteria.6 Some outer membrane proteins of Salmonella Typhimurium not only function as catecholate siderophore receptors but also play a role in bacterial pathogenesis. A previous study has shown that the virulence of an iron uptake-defective mutant (iroN– fepA– cirA–) was attenuated in a systemic infection in mice supplemented with l-norepinephrine.7 In addition, Tsolis et al.8 reported that the mutation of tonB attenuated the infection of Salmonella Typhimurium in mice by an intragastric route. The effects of several iron sources on Salmonella pathogenesis have been well studied. Ferric citrate and ferric chloride were shown to increase the virulence of Salmonella in in vitro studies with Caco-2 cells.9,10 Similarly, Lin et al.11 reported that supplementation of ferrous sulfate enhanced the virulence of Salmonella in mice. Ferric EDTA demonstrated a high iron bioavailability in human studies and has been proposed to be used as a fortificant in breakfast cereal and cereal bars.12 However, little is known about the effect of ferric EDTA on the growth and pathogenesis of Salmonella. Also, Kamdi and Palkar13 reported that ferrous-l-ascorbate had a high iron bioavailability in pregnant women through an oral supplement. The effect of ferrous-l-ascorbate on Salmonella pathogenesis is still largely unknown.
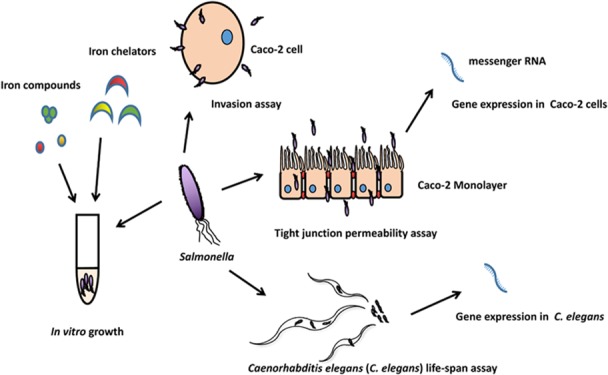
Massive and dynamic microbial communities including pathogens inhabit in the animal gut. Epithelial cells are lined up and form mucosal surfaces that provide a barrier between hostile external environments and the internal milieu.14 Colonization or invasion of gastrointestinal mucosal surfaces is the first step for enteric pathogens to cause systematic infection.15 As an intracellular pathogen, Salmonella species possess the ability to infect a wide variety of cell types, from kidney epithelial cells to macrophages.16 Caco-2, a human colon carcinoma cell line that expresses and organizes brush border membrane components as enterocytes, could mimic the differentiation of normal intestines under in vitro conditions.17,18 Well-differentiated Caco-2 cell monolayers could be adhered and invaded by Salmonella Typhimurium, providing a valuable in vitro model for pathogenic studies.16 The tight junction of the epithelial barrier is highly relevant to the health of animal guts. A previous study showed that tight junction proteins (TJP1 and CLDN3) were potential targets of many pathogens including Salmonella.(19) The intestinal oligopeptide transporter, PepT1, is responsible for transportation of dipeptides and tripeptides.20 It has also been reported to transport peptidomimetic drugs such as β-lactam antibiotics cefadroxil and valacyclovir. Cytokines play an indispensable role in the host–pathogen interaction.21Salmonella infection often induces the expression of multiple chemokines and proinflammatory cytokines including TNF-α and IL-8.21,22 In the present study, the expression of these genes was therefore examined for the host response to Salmonella infection.
Caenorhabditis elegans has become a popular model for studying animal development and behavior as well as bacterium and host interactions since the 1960s.23,24 Labrousse et al.24 reported that Salmonella Typhimurium, a highly adapted strain with a broad range of target hosts,25 was capable of infecting and causing the death of C. elegans. In addition, it has been reported that acid-sensitive mutants (UK1, fur-1, and ompR) of Salmonella Typhimurium presented a reduced virulence not only in mammals but also in C. elegans.(24) The DAF/insulin-like growth factor (DAF/IGF), p38 mitogen-activated protein kinase (p38 MAPK), and transforming growth factor-β (TGF-β) signaling pathways that have remained essentially unchanged throughout evolution are critical components in immune defense mechanisms of C. elegans.(26,27) Genes nsy-1 and pmk-1 encode two major components of the p38 MAPK signaling pathway, which is essential for the innate immune system of C. elegans against pathogens.28 Gene daf-16 encodes a FOXO-family transcription factor that is a member in the DAF/IGF signaling pathway.29 Murphy et al.26 reported that daf-16 regulates a set of genes with functions in adulthood, aging, and lifespan of C. elegans. Genes clec-85, lys-7, and spp-1 encodes a C-type lectin, lysozyme-like protein, and saposin-like protein, respectively.30,31 These proteins possess antimicrobial activities and are essential for the immunity of C. elegans.(32,33) Zhou et al.33 recently described the host response of the nematode to Escherichia coli infection and probiotic protection by activating the production of antimicrobial peptides (including genes clec-85, lys-7, and spp-1) by regulating its cell signaling, the p38 MAPK (e.g., Nsy-1 and Pmk-1) and DAF/IGF pathways in particular, to combat the bacterial infection. C. elegans expresses two different kinds of ferritin to regulate intracellular iron, which are ferritin 1 (encoded by ftn-1) and ferritin 2 (encoded by ftn-2).34 Simonsen et al.30 reported that ferritin 2 was involved in the immune response against the infection by Gram-positive (Staphylococcus aureus) and Gram-negative pathogens (E. coli). However, the functionality of ftn-1 on C. elegans against Salmonella infection remains unknown. Gene fgt-1 encodes a major glucose transporter that plays an indispensable role in energy supply to C. elegans.(35,36) It has also been reported to have a role in the longevity and lifespan of C. elegans through the DAF/IGF signaling pathway.35 Gene sod-3 encodes a mitochondrial superoxide dismutase (MnSOD) that is involved in the breakdown of reactive oxygen species.37 Previous studies have shown that MnSOD is related to the longevity of many animals including the lifespan of C. elegans.(37,38) In the present study, the expression of these genes in response to Salmonella infection, both the wild type (WT) and mutants, was examined to reveal the host response of C. elegans about iron requirement.
In the present study, Salmonella Typhimurium from broiler chicken was used to understand the role of specific iron uptake regulated genes in its pathogenesis, aiming for improving the effectiveness of Salmonella control in poultry production. Different iron uptake-defective mutant strains of Salmonella as well as their complemented strains were generated and compared. We examined four aspects:1 the effect of different iron sources on Salmonella growth (both the WT and mutants);2 the effect of iron chelators including EDTA, citric acid, and 2,2′-bipyridyl on inhibiting Salmonella growth;3 the invasion of Caco-2 cells by Salmonella grown in the presence of different iron concentrations and the effect on their transepithelial electrical resistance and their gene expression profile;4 and the ability of the Salmonella strains to infect C. elegans and the response of the nematode to the infection.
2. Results
2.1. Effect of Different Iron Forms on Salmonella Growth
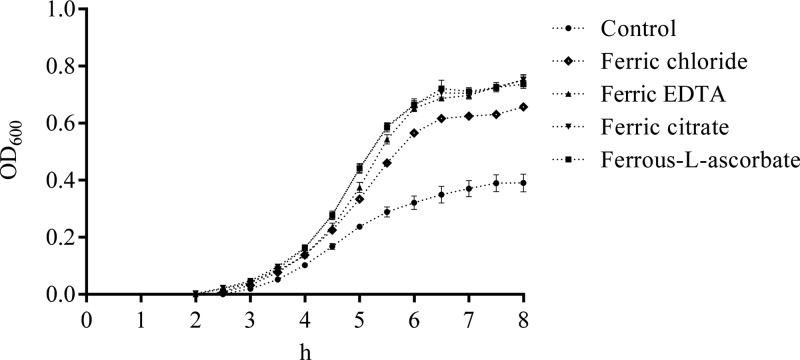
The effect of five different forms of iron including ferric chloride, ferrous sulfate, ferric EDTA, ferric citrate, and ferrous-L-ascorbate on the growth of Salmonella Typhimurium WT and its mutant strains were investigated. As shown in Figure 1, regardless of the forms of iron, the WT had much better growth than the two mutants (tonB– and iroN– fepA–; P < 0.05) after 4 h incubation and the growth of the WT in IMDM only (control) showed the slowest growth among all the treatments (P < 0.05). The growth of the WT with ferrous-l-ascorbate or ferric citrate was improved compared to ferric EDTA or ferric chloride (P < 0.05). However, the WT treated with ferrous sulfate showed comparable growth (P ≥ 0.05) with ferrous-l-ascorbate or ferric citrate. Both mutants hardly grew in IMDM only and had little growth when the medium was supplemented with 0.2 μM iron regardless of the forms. No statistical analysis was therefore applied.
Figure 1.
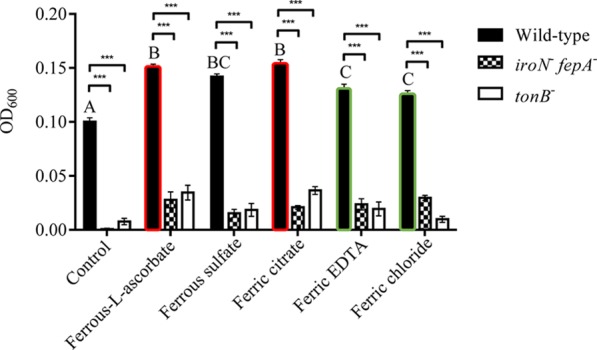
Effects of different iron forms on the growth of Salmonella Typhimurium. The optical density (OD600) of bacterial suspension of the wild type and iron uptake-defective mutants of Salmonella Typhimurium was measured after the strains were cultured in IMDM containing 0.2 μM iron for 4 h. Values are means ± standard error of the mean (SEM), n = 5. Control, no iron added; iroN– fepA–, mutant defective in both iroN and fepA; tonB–, mutant defective in tonB; EDTA, ethylenediaminetetraacetic acid. Symbol *** represents a significant difference between the wild-type and mutants within the control or each iron treatment (P < 0.05). A significant difference in the growth performance was detected between bars with the red edge and those with the green edge. Means marked with letters “A”, “B”, and “C” were significantly different (P < 0.05) for the wild type among different iron treatments.
2.2. Deletion of TonB, IroN, and FepA on Salmonella Growth
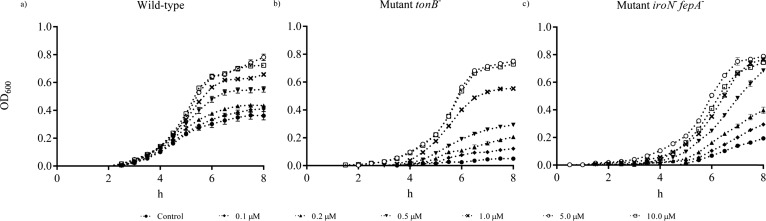
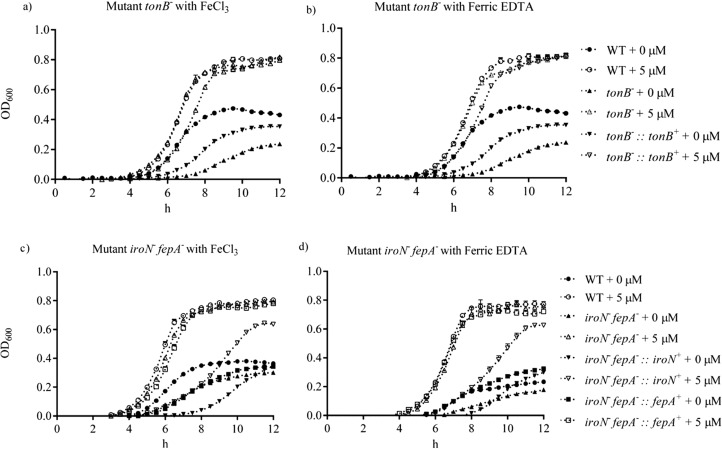
Figure 2 shows the growth of the WT, mutant tonB–, and mutant iroN– fepA– in IMDM containing ferric chloride from 0 to 10 μM. The growth of the mutants was retarded compared to the WT. A concentration-dependent growth was observed when ferric chloride was increased from 0.1 to 1 μM before 6 h. The growth curves of mutant tonB–, mutant iroN– fepA–, and the WT were comparable (P ≥ 0.05) when the supplementation of ferric chloride exceeded 5 μM. The growth of the WT, mutant, and complementary strains on ferric chloride and ferric EDTA is shown in Figure 3. Both ferric chloride and ferric EDTA supported similar growth of all tested strains. Under the iron-poor condition, the WT appeared to grow approximately 100 and 30% better than mutant tonB– and its complement (P < 0.05), respectively, but not under the iron-rich condition (Figure 3a,b). The growth of the WT, mutant iroN– fepA–, and its two partial complements were similar after 12 h incubation under the iron-poor condition (Figure 3c,d). The supplementation with 5 μM iron (iron-rich) generated full growth of the WT, mutant iroN– fepA–, and one of its partial complement except for the partial complement of iroN– fepA–::iroN+.
Figure 2.
Effects of different concentrations of ferric chloride on the growth of Salmonella Typhimurium. (a) In vitro growth of the wild type, (b) in vitro growth of mutant tonB–, and (c) in vitro growth of mutant iroN– fepA–. The interval of each measurement was 15 min for a total of 8 h. The concentrations of ferric chloride supplemented to the IMDM medium ranged from 0 to 10 μM. Values are means ± SEM; n = 5. Control, no iron added.
Figure 3.
Comparison in the growth of the wild-type, mutant, and complementary strains of Salmonella Typhimurium on ferric chloride and ferric EDTA. (a, b) Mutant tonB– and its complement cultured in the IMDM medium supplemented with ferric chloride and ferric EDTA, respectively. (c, d) Mutant iroN– fepA– and its partial complements cultured in the IMDM medium supplemented with ferric chloride and ferric EDTA, respectively. The wild type served as a control. The growth was evaluated in either iron-poor or iron-rich (5 μM) medium (IMDM). The interval of each measurement was 15 min for 12 h. Values are means ± SEM; n = 5. tonB–, mutant defective in tonB; tonB–::tonB+, complement of mutant tonB–; iroN– fepA–, mutant defective in both iroN and fepA; iroN– fepA–::iroN+, partial complement of mutant iroN– fepA– with iroN only; iroN– fepA–::fepA+, partial complement of mutant iroN– fepA– with fepA only.
2.3. Effect of Iron Chelators on the Growth of Salmonella
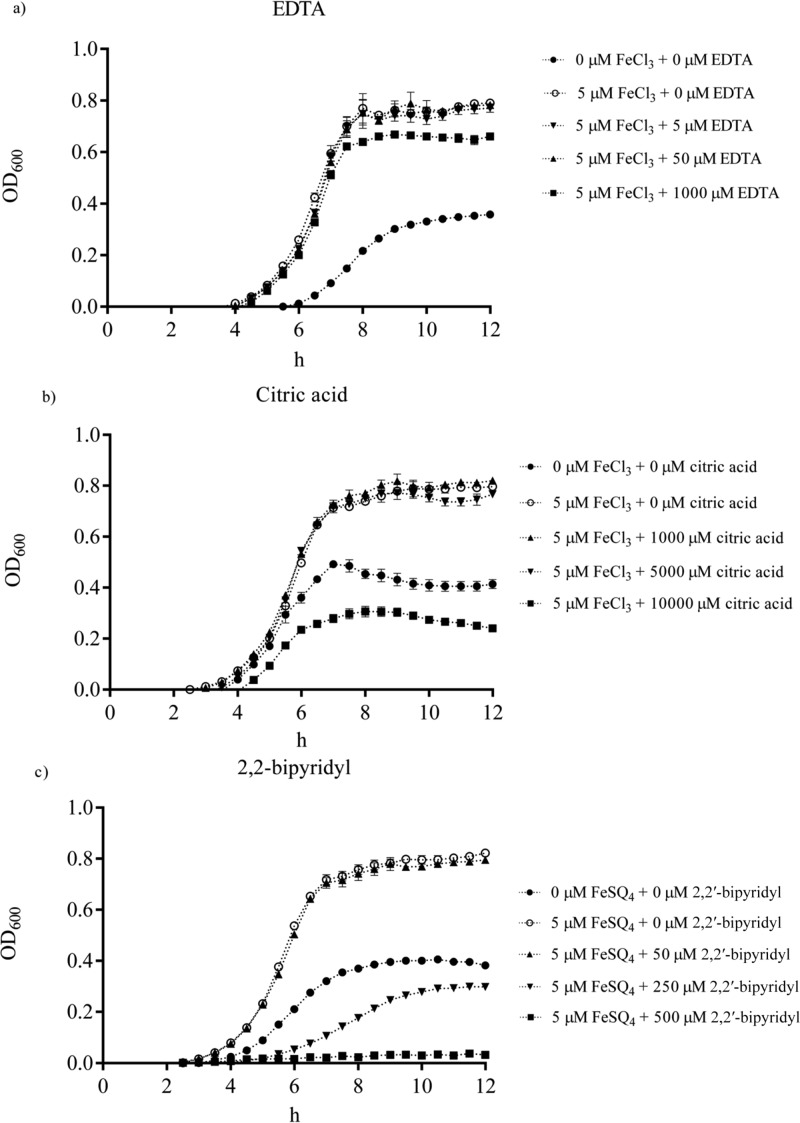
The growth of the WT was monitored under the iron-rich condition with either ferric chloride or ferrous sulfate in the presence of increased concentrations of EDTA, citric acid, or 2,2′-bipyridyl. EDTA at 1 mM showed significant inhibition of Salmonella growth (P < 0.05), reducing around 15% of growth (Figure 4a). Similarly, citric acid reduced Salmonella growth (P < 0.05, approximately 50%) at 1 mM (Figure 4b). In contrast, the growth of the pathogen was significantly inhibited (more than 50%) by 2,2′-bipyridyl at 250 μM (P < 0.05) and nearly abolished at 500 μM (Figure 4c).
Figure 4.
Effects of different iron chelators on the growth of Salmonella Typhimurium wild type. The IMDM contained no or 5 μM ferric chloride or 5 μM ferrous sulfate. (a) Growth in the presence of EDTA (0 to 1000 μM). (b) Growth in the presence of citric acid (0 to 10,000 μM). (c) Growth in the presence of 2,2′-bipyridyl (0 to 250 μM). The interval of each measurement was 15 min for 8 h. Values are means ± SEM; n = 5.
2.4. Deletion of TonB, IroN, and FepA on the Invasion of Caco-2 Cells
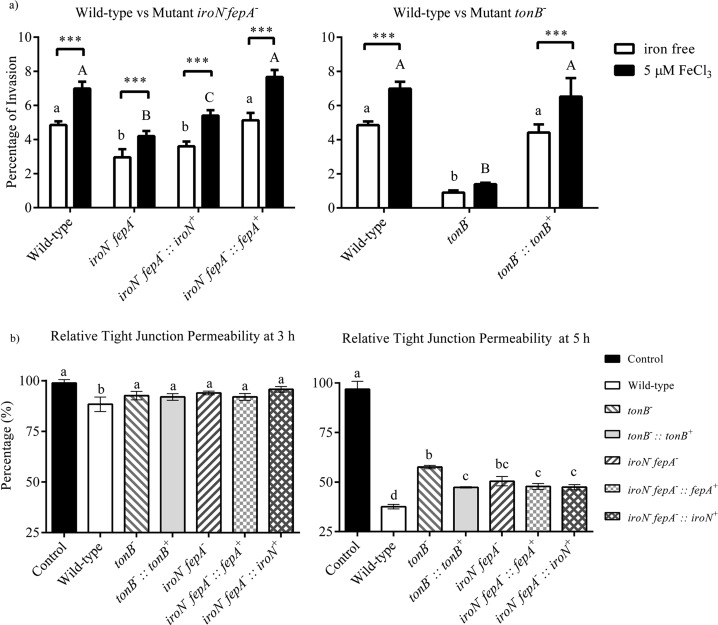
Caco-2 monolayers were used for the assay of Salmonella invasion into epithelial cells. As shown in Figure 5a, the percentages of invasion of mutant tonB– and mutant iroN– fepA– were significantly lower than that of the WT (P < 0.05) under both iron-rich and iron-poor conditions. The invasion ability of complement tonB–::tonB+ was fully restored under both conditions. However, while the invasion ability of complement iroN– fepA–::fepA+ was fully restored under both conditions, complement iroN– fepA–::iroN+ was not. Only a partial recovery of the ability was observed when the complement was under iron-rich condition. The invasion ability of all strains investigated was significantly improved (P < 0.05) expect mutant tonB– (P ≥ 0.05) under the iron-rich condition.
Figure 5.
Effects of gene mutation in iron uptake on the virulence of Salmonella Typhimurium. (a) Salmonella invasion into Caco-2 cells. Values are the percentage of invasion (mean ± SEM) of the pathogen into a monolayer of Caco-2 cells; n = 4. Percentage of the invasion was presented as the colony-forming units (CFU) of Salmonella that had invaded into Caco-2 cells divided by CFU of initial inoculation of Salmonella. Means marked with “a” and “b” and those without a common letter were significantly different (P < 0.05) for the iron-poor condition among different strains; means marked with “A” and “B” and those without a common letter were significantly different (P < 0.05) for the iron-supplemented condition among different strains. Symbol *** represents a significant difference between the iron-poor and iron-rich treatments within a strain (P < 0.05). (b) Ability of Salmonella to damage the epithelial monolayer of Caco-2 cells. Values are the percentage of relative tight junction permeability (mean ± SEM) that is presented by the measured transepithelial electrical resistance (TEER) values at 3 or 5 h of the assay divided by the initial TEER at the beginning (0 h); n = 4. Means without a common letter differ significantly (P < 0.05). tonB–, mutant defective in tonB; tonB–::tonB+, complement of tonB– mutant; iroN– fepA–, mutant defective in iroN and fepA; iroN– fepA–::iroN+, partial complement of mutant iroN– fepA– with iroN only; iroN– fepA–::fepA+, partial complement of mutant iroN– fepA– with fepA only.
2.5. Deletion of TonB, IroN, and FepA on Tight Junction Permeability of Caco-2 Cells
All the treatment groups of the Caco-2 monolayer shared a similar relative tight junction permeability after the first 2 h of incubation with different Salmonella strains, including the WT, mutants tonB– and iroN– fepA–, and their complements (data not shown). As shown in Figure 5b, the relative tight junction permeability of the Caco-2 monolayer was significantly lower (10% reduction, P < 0.05) only in the group treated with the WT compared with the control (uninfected group) after 3 h co-incubation. However, after 5 h co-incubation, significant damage to the monolayer occurred (Figure 5b). The WT showed the most severe damage to the monolayer among the different isolates. While both mutants caused less damage to the monolayer than the WT, the only complement of tonB– (tonB–::tonB+) significantly restored the damage to the level of WT (P < 0.05). At the end of co-incubation (sixth hour), the Caco-2 monolayer in all the treatment groups except for the one treated with mutant tonB– showed low relative TEER with no significant difference (P ≥ 0.05, data not shown).
2.6. Host Response of Caco-2 to Salmonella WT and Its TonB, IroN, and FepA Mutants
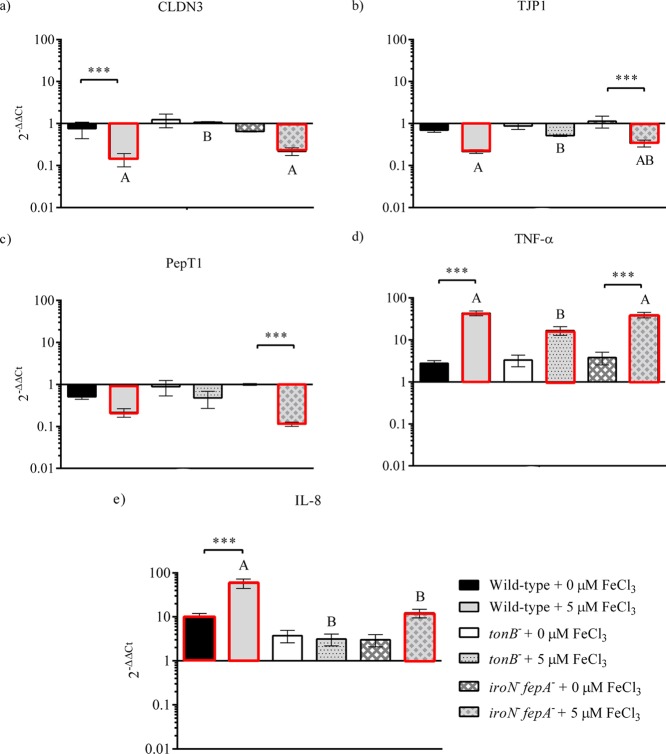
Tight junction proteins (CLDN3 and TJP1), nutrient transporter (PepT1), and proinflammatory cytokines (IL-8 and TNF-α) were used as indicators to investigate the host response of Caco-2 cells to the Salmonella infection. As shown in Figure 6, the gene expression in Caco-2 cells showed no significant changes for all examined genes under the iron-poor condition between the infected and uninfected Caco-2 except for IL-8 by the WT, as well as between the WT and mutants (tonB– and iroN–fepA–). Under the iron-rich condition, the transcription of both CLDN3 (5- to 9-fold changes) and TJP1 (3- to 4-fold changes) was down-regulated significantly when Caco-2 was invaded by the WT and mutant iroN–fepA– (P < 0.05). Compared to mutant tonB–, the WT and mutant iroN– fepA– significantly down-regulated the expression of CLDN3 (5- to 9-folds, P < 0.05) under the iron-rich condition (Figure 6a). A similar observation was also obtained with the expression of TJP1 (Figure 6b). Notably, mutant tonB– did not significantly suppress (P ≥ 0.05) the gene expression of tight junction proteins regardless of iron-rich or iron-deficient conditions (Figure 6d). No significant difference (P ≥ 0.05) in the gene expression of PepT1 was detected in Caco-2 cells treated with the WT and with the two mutants, respectively (Figure 6c), although the WT and mutant iroN– fepA– down-regulated the gene expression significantly compared with uninfected Caco-2 (5- to 8-folds, P < 0.05). The gene expression of TNF-α and IL-8 is shown in Figure 6d,e, respectively. All the strains significantly up-regulated the expression level of TNF-α in the infected Caco-2 cells under the iron-rich condition compared with uninfected (more than 10-folds, P < 0.05). In addition, the up-regulation was significantly higher in Caco-2 cells infected with the WT or mutant iroN– fepA– than with mutant tonB– (15- to 25-folds, P < 0.05) (Figure 6e). There was a significant up-regulation (more than 10-folds, P < 0.05) in the expression of IL-8 when Caco-2 cells were infected with the WT or mutant iroN– fepA– under the iron-rich condition (Figure 6e). The increase was larger by the WT than by the mutant (P < 0.05). No significant difference was detected in the gene expression between the treatments by the two mutants (P ≥ 0.05) (Figure 6e).
Figure 6.
Gene expression of (a) claudin 3 (CLDN3), (b) tight junction protein 1 (TJP1), (c) peptide transporter 1 (PepT1), (d) tumor necrosis factor α (TNF-α), and (e) interleukin 8 (IL-8) in Caco-2 cells as the response to Salmonella invasion. The Caco-2 cells were sampled on the second hour of the invasion assay. Relative expression was determined using the 2–ΔΔCt method. The ΔCt was presented as the comparison in the threshold cycle between the target genes and housekeeping genes (18S and GAPDH), and ΔΔCt represented the comparison between the Salmonella-infected Caco-2 cells and uninfected Caco-2 cells. The reference (=1) for the comparison was the gene expression in uninfected Caco-2 cells. The RNA sample of each treatment had three biological replicates in the qPCR assay; n = 3. Bars with the red edge indicate a significant difference in the gene expression between infected and uninfected Caco-2 cells. Symbol *** represents a significant difference in the gene expression within a strain between the iron-poor and iron-rich treatments (P < 0.05). Means marked with “A” and “B” and those without a common letter were significantly different (P < 0.05) for the iron-rich treatment among different strains. tonB–, mutant defective in tonB; iroN– fepA–, mutant defective in both iroN and fepA.
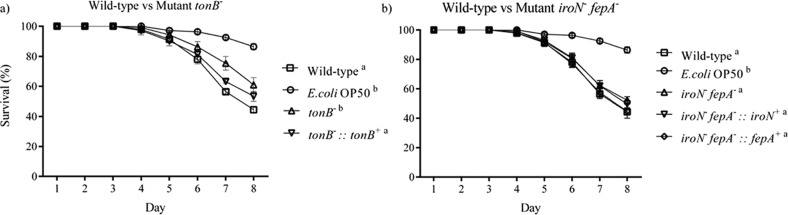
2.7. Deletion of TonB, IroN, and FepA on the Lifespan of C. elegans
The survival curves of C. elegans after infection by the WT, mutants tonB– and iroN– fepA–, and their complements of Salmonella Typhimurium are shown in Figure 7. The death of worms was first observed on day 4 of the assay, and viable worms dramatically decreased in the following 4 days. The worms infected by the WT, mutant iroN–fepA–, and its partial complements (iroN–fepA–::iroN+ and iroN–fepA–::fepA+) had a significantly reduced (P < 0.05) lifespan compared to the nematode fed E. coli OP50 only (negative control). In contrast, there was no significant difference (P ≥ 0.05) in the lifespan between the worms treated with mutant tonB– and with E. coli OP50 only (uninfected). Complement tonB–::tonB+ restored the ability to cause a similar level of worm death by the WT (P ≥ 0.05).
Figure 7.
Effects of gene mutation in iron uptake on the ability of Salmonella Typhimurium to infect C. elegans. (a) Lifespan of C. elegans treated with the wild type, mutant tonB–, or the complement of tonB–; (b) lifespan of C. elegans treated with the wild type, mutant iroN– fepA–, or partial complements of mutant iroN– fepA–. Each treatment group had about 50 worms that were incubated in the S medium containing 24 μM iron in the lifespan assay. Worms fed with E. coli OP50 (109 CFU/mL) only served as a reference. The final concentration of Salmonella in the assay mixture was 109 CFU/mL. tonB–, mutant defective in tonB; tonB–::tonB+, complement of mutant tonB–; iroN– fepA–, mutant defective in both iroN and fepA; iroN– fepA–::iroN+, partial complement of mutant iroN– fepA– with iroN only; iroN– fepA–::fepA+, partial complement of mutant iroN– fepA– with fepA only. Survival curves without a common letter (“a” and “b”) differ significantly (P < 0.05). The E. coli OP50 and Salmonella cultures used for the C. elegans lifespan assay were all in the early stationary phase.
2.8. Host Response of C. elegans to Salmonella WT and Its TonB, IroN and FepA Mutants
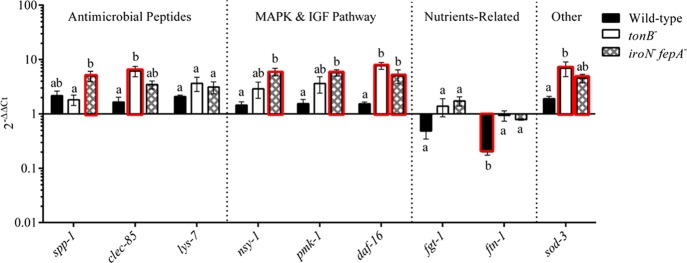
The gene expression of the defense molecule (sod-3), antimicrobial peptides (spp-1, clec-85, and lys-7), nutrient transporter (fgt-1), iron storage (ftn-1), and components in the IGF (daf-16) or p38 MAPK signaling pathway (nsy-1 and pmk-1) was investigated with the worms sampled on day 5 in the lifespan assay where iron was not limited. The treatment groups of nematodes were infected with the WT and mutants (tonB– and iroN–fepA–, respectively). As shown in Figure 8, no significant difference (P ≥ 0.05) was detected in the expression of all tested genes except for ftn-1 that was down-regulated (5-folds, P < 0.05) in the worms infected with WT compared to the uninfected worms. In contrast, more than 2-fold increase in the gene expression of clec-85, daf-16, and sod-3 (P < 0.05) was achieved by mutant tonB– (P < 0.05). Mutant iroN–fepA– also demonstrated a similar up-regulation (P < 0.05), including genes spp-1, nsy-1, pmk-1, daf-16, and sod-3. Compared with the WT, the two mutants enhanced the expression (4- to 7-folds, P < 0.05) of five genes in total (clec-85, nsy-1, pmk-1, daf-16, and sod-3), in which mutant tonB– altered clec-85, daf-16, and sod-3 and mutant iroN–fepA– up-regulated nsy-1, pmk-1, and daf-16. The ftn-1 was the only gene in the worms treated with the WT that was significantly down-regulated compared to the mutant-treated and uninfected worms (P < 0.05).
Figure 8.
Host response of C. elegans at the gene expression level to the infection of Salmonella Typhimurium wild type and mutants. Several genes related to antimicrobial peptide production, MAPK and IGF signaling pathways, and other molecules related to nutrient metabolism and defense in C. elegans were selected as the targets for the qPCR assay. The nematode was sampled on day 5 of the lifespan assay. Relative expression was determined using the 2–ΔΔCt method. The ΔCt was the comparison in the threshold cycle between target genes and housekeeping genes (snb and act-1). The ΔΔCt represented the comparison between Salmonella-infected C. elegans and C. elegans treated with E. coli OP50 only. The reference (=1) for the comparison was the gene expression in uninfected C. elegans. The RNA samples of each treatment had three biological replicates in the qPCR assay; n = 3. Means marked with “a” and “b” and those without a common letter were significantly different (P < 0.05) for the same gene among different strains. Bars with the red edge indicate a significant difference in the gene expression between infected and uninfected Caco-2 cells. tonB–, mutant defective in tonB; iroN– fepA–, mutant defective in both iroN and fepA; sod-3, superoxide dismutase 3; fgt-1, facilitated glucose transporter protein 1; ftn-1, ferritin; spp-1, saposin-like protein; clec-85, c-type lectin; lys-7, lysozyme-like protein 7; daf-16, forkhead-type transcription factor; pmk-1, mitogen-activated protein kinase pmk-1; nsy-1, mitogen-activated protein kinase kinase kinase nsy-1; IGF, insulin-like growth factor; MAPK, mitogen-activated protein kinases.
3. Discussion
Salmonella Typhimurium used in the present study was isolated from broiler chicken.39 The information generated from this study may be useful for developing a strategy to control the pathogen for poultry production.
One objective of the present study was to investigate the effect of different forms of iron on the growth of Salmonella Typhimurium. Organic iron (ferrous-l-ascorbate and ferric citrate) appeared to be favored by the pathogen for growth compared with inorganic iron (ferric EDTA and ferric chloride), in which Salmonella growth on organic iron showed a higher growth rate in the log phase than on inorganic iron (data not shown). A similar preference to organic iron for iron absorption has been reported previously in animals.40 Some studies proposed that animals had a high level of absorption of organic iron because organic acid and amino acid ligands of the organic iron can protect the ferrous iron from oxidation and/or interaction with other metal ions.40,41 There was also a study confirming that intestinal epithelial cells favored ferrous iron than ferric iron because DMT-1 was the major pathway for uptake of ferrous iron in both organic and inorganic forms.42 Even though Salmonella had an iron-porin-Feo (Feo) system that is similar to the DMT-1 pathway of a mammalian animal, a complicated iron-siderophore system is also used by Salmonella to acquire iron to grow and survive in iron-poor environments.43,44 Moreover, enteric bacteria including Salmonella possess both a citrate-dependent iron (III) transport and ferric dicitrate transport systems for uptaking ferric citrate as well as a wide variety of metal-free and metal-loaded tricarboxylic acids.45,46 An exclusive citrate-dependent iron transport system for uptaking ferric citrate might explain why ferric citrate showed a better enhancement on Salmonella growth compared with the inorganic iron investigated (FeSQ4 and FeCl3).47
In the present study, the function of IroN and FepA on the growth of Salmonella was evaluated. Results confirmed previous reports stating that TonB, FepA, and IroN proteins are important for Salmonella growth.48,49 Mutation of tonB impaired the functionality of all tonB-dependent (iron-siderophores) receptors on the outer membrane, as shown by growth curves. Similarly, mutation of iroN and fepA impaired the integrity and functionality of the Fep system, which retarded the growth of mutant iroN– fepA–. Interestingly, the growth curves of all mutant strains were similar to the WT under the iron-rich condition (> 5 μM), which suggested that Salmonella might have a Fe3+ iron uptake system that did not relate to the TonB protein nor to the investigated iron-siderophore system.
Among the three chelators tested in the present study, 2,2′-bipyridyl was the most effective in inhibiting Salmonella growth. Previous studies have reported that 2,2′-bipyridyl could increase the antimicrobial property of polymer-Cu(II) complexes by enhancing the lipophilic character of the central metal atom.50,51 However, most of the antimicrobial activities of 2,2′-bipyridyl were reported when it was combined with other metal ions (e.g., platinum, cobalt, and copper) or with metal complexes.52,53 It is possible that 2,2′-bipyridyl chelated Fe3+ and/or other metal ions supplemented in media, which retarded the growth of Salmonella. Unfortunately, a toxicity test on rat showed that LD50 of 2,2′-bipyridyl was 256 and 155 mg/kg through oral administration and vein injection, respectively.54 More studies are therefore required to determine its potential in application.
Our results clearly confirmed that iron increased the ability of Salmonella Typhimurium to invade a differentiated Caco-2 monolayer, in agreement with previous studies.55,56 Moreover, iron uptake system (Fep)-defective mutants of Salmonella Typhimurium significantly reduced their ability to invade the intestinal epithelial cells, suggesting that iron uptake systems are required for the virulence of Salmonella in animal guts. This is also confirmed by the evidence that defective iron uptake systems prevented an increase in permeability through tight junction stabilization (e.g., CLDN3) and blocking of proinflammatory response (e.g., reduced TNF-α and IL-8 gene expression). It is possible that, upon the down-regulation of the elevated expression of TNF-α and IL-8 by defective iron uptake systems (Fep), the expression of CLDN3 was stabilized; as a result, damage to the integrity of the enterocyte barrier was slowed down. A similar correlation was reported previously in the porcine jejunal epithelial IPEC-J2 cell line.57 In addition, IL-8 has been shown to be essential for Salmonella to pass through epithelial Caco-2 monolayers,58,22 and TNF-α contributes to the tissue pathology associated with Salmonella infection.59 Interestingly, the down-regulation of the PepT1 expression by Salmonella Typhimurium under iron-rich condition was prevented by the mutant tonB–, suggesting that Salmonella infection could reduce nutrient absorption (e.g., small peptides). TNF-α and IL-8 play important roles in the host defense mechanisms. Eckmann and Kagnoff21 reported that TNF-α and IL-8 are induced when the hosts are infected by Salmonella. One interesting finding is that the expression levels of TNF-α and IL-8 were altered in the Caco-2 cells infected by mutant iroN– fepA–. There might be some interaction between the iron receptor (iroN– fepA–) of Salmonella and the host immune systems. We also demonstrated that iron was necessary for observing the changes in the gene expression of tight junction protein and proinflammatory cytokines among the WT and iron uptake system-defective mutants of Salmonella Typhimurium. Taken together, our results suggest that iron and iron uptake systems are essential for the virulence of Salmonella in animal guts.
In the present study, the importance of TonB, IroN, and FepA in the virulence of Salmonella was evaluated in C. elegans with a lifespan assay followed by an investigation into the host gene expression and regulation of cell signaling and defense molecule production. The result of the lifespan assay (except for mutant iroN– fepA–) was consistent with the observation from the Caco-2 invasion experiment in the present study (Figure 5) as well as from mouse infection studies reported previously.8,60 It is interesting to note that the WT strain of Salmonella Typhimurium caused no changes in the expression of all tested genes related to cell signaling and defense molecule production (including antimicrobial peptides) in the nematode compared with uninfected worms (Figure 8). In contrast, either mutant tonB– or mutant iroN– fepA– was able to up-regulate some of the genes, for example, clec-85, sod-3, and daf-16 by mutant tonB– and spp-1, sod-3, nsy-1, pmk-1, and daf-16 by mutant iroN– fepA–, suggesting the involvement of both genes in Salmonella infection to C. elegans. Given the facts that only mutant tonB– (but not mutant iroN– fepA–) lost the ability to infect C. elegans and its complement restored the capacity in addition to the up-regulation of clec-85 and sod-3 expression by the mutant in particular, it appears that both clec-85 and sod-3 may play a substantial role in the defense of the nematode against Salmonella infection. It has been documented from previous studies that the p38 MAPK and DAF/IGF pathways control the expression of the antimicrobial peptides.61,62 Very recently, there was a report that a Lactobacillus isolate could regulate C. elegans signaling through the p38 MAPK and DAF/IGF pathways to control the production of antimicrobial peptides and defense molecules to combat E. coli infection.33 One significant finding from the C. elegans experiment in the present study was the down-regulation of ftn-1 by the WT of Salmonella Typhimurium only (Figure 8). The gene regulates the expression of ferritin 1 that has a role in iron storage of the host, and knocking out of ftn-1 would reduce the lifespan of C. elegans grown in the environment with excess iron.63 It may suggest that Salmonella could affect the expression of ftn-1 and create an intracellular environment with sufficient iron. Our observation on the inability of mutants tonB– and iroN– fepA– to alter the gene expression of ftn-1 supports the notion, providing further evidence for the importance of ftn-1 in the pathogen and host interaction during Salmonella infection.
This study clearly demonstrated that EDTA, 2,2′-bipyridyl, and citric acid can effectively inhibit the growth of WT and iron uptake-defective mutants of Salmonella. 2,2′-Bipyridyl is not approved as a feed ingredient. Citric acid and EDTA can be used in animal feeds, but these chelators not only reduce iron availability to Salmonella but also decrease the bioavailability of minerals including iron to animal hosts. This makes it very difficult to include chelators into feeds to control Salmonella. However, these chelators could be included in poultry litters to reduce the survival of Salmonella in poultry farms.
4. Materials and Methods
4.1. Materials
Proteinase K was purchased from Qiagen (Germantown, Maryland, United States). Tryptic soy agar was purchased from BD Difco (Franklin Lakes, New Jersey, United States). All other chemical agents were purchased from Fisher Scientific (Ottawa, ON, Canada) and Sigma-Aldrich (Oakville, ON, Canada). Chemicals used in the present study were of analytical reagent grade.
4.2. Iron Content Analysis Using Inductively Coupled Plasma Optical Emission Spectrometry
Iscove’s modified Dulbecco’s medium (IMDM; Sigma-Aldrich) is a chemically defined iron-poor medium that is suitable for both tissue cultures and bacterial cultures. The iron concentration in the completed IMDM was determined using inductively coupled plasma optical emission spectrometry (ICP-OES). Briefly, the IMDM powder was dissolved in NERL high purity water (Fisher Scientific) and prepared according to the manufacturer’s instructions. The medium samples containing 2% (v/v) nitric acid were analyzed by the Manitoba Chemical Analysis Laboratory for iron content using ICP-OES. The results of ICP-OES analysis indicated that the iron concentration in the completed medium was approximately 0.8 μM. Although the form of iron in the IMDM medium is unknown, the contained iron level is negligible. Therefore, the IMDM medium supplemented with no iron is defined as an iron-poor medium, while that supplemented with iron at 5 μM or above is called an iron-rich medium in the present study.
4.3. Bacterial Strains and Growth Conditions
S. enterica serovar Typhimurium ABBSB1218-1 was isolated from broiler chicken as previously reported.43 The strains used in this study including both the WT and mutants as well as some complemented strains are shown in Table 1.39,64 IMDM was used as the culture medium in the in vitro experiments to investigate the effect of iron and iron chelators on bacterial growth and virulence. Lysogeny broth (LB, Fisher Scientific) was used for bacterial culture in the C. elegans experiments. The WT and mutant strains were cultured aerobically with shaking at 37 °C. Complemented strains were cultured and maintained under the same conditions as the WT strains except for the supplementation of 50 μg/mL kanamycin (Sigma-Aldrich) in the culture medium.
Table 1. Bacterial Strains Used in the Study.
| strain/plasmid of Salmonella Typhimurium | description and characteristic | reference |
|---|---|---|
| ABBSB1218-1 WT | wild type isolated from broiler chicken | (39) |
| ABBSB1218-1 ΔfepA | single deletion of FepA (enterobactin transporter) | (64) |
| ABBSB1218-1 ΔiroN | single deletion of iroN (catecholate transporter) | (64) |
| ABBSB1218-1 ΔtonB | single deletion of TonB (energy transducer) | (64) |
| ΔtonB + pSCA-tonB | ΔtonB carrying plasmid pSCA-tonB | (64) |
| ABBSB1218-1 ΔiroNΔfepA | double deletion of IroN and FepA | (64) |
| ΔiroNΔfepA + pSCA-fepA | ΔiroNΔfepA carrying plasmid pSCA-fepA | (64) |
| ΔiroNΔfepA + pSCA-iroN | ΔiroNΔfepA carrying plasmid pSCA-iroN | (64) |
4.4. Evaluation of Bacterial Growth on Different Forms of Iron
To determine the effect of different forms and different concentrations of iron on the growth of Salmonella Typhimurium, Bioscreen C MBR (Oy Growth Curves Ab Ltd., Helsinki, Vuorimiehenkatu, Finland) was used in this study to measure the optical density (OD) of bacterial suspension.
Salmonella Typhimurium was subcultured three times in IMDM to eliminate iron contamination from previous cultures. Bacteria were inoculated at 104 CFU/mL into IMDM media containing 0 to 50 μM of either ferric citrate, ferric chloride, ferric EDTA, ferrous-l-ascorbate, or ferrous sulfate. The inoculated IMDM (350 μL/well) from each treatment was transferred to Bioscreen honeycomb plates (Oy Growth Curves Ab Ltd.) with five wells of technical replicates. The optical density of the suspension in each well was measured every 15 min at 600 nm (OD600) for 16 h. Growth curves were analyzed with Bioscreen C MBR software.
4.5. Evaluation of Iron Chelators for Inhibiting Bacterial Growth
The growth of the WT of Salmonella Typhimurium was measured using Bioscreen C MBR (Oy Growth Curves Ab Ltd.) and the protocol described above. Ferric chloride or ferrous sulfate was used as the iron source in this study. Salmonella growth in IMDM only served as a negative control, while the positive control was the growth in IMDM supplemented with 5 μM ferric chloride or ferrous sulfate. Increasing concentrations of iron chelators, including EDTA (5 to 1000 μM), citric acid (1000 to 10,000 μM), and 2,2′-bipyridyl (50 to 500 μM), were supplemented to IMDM containing 5 μM iron to examine the effect of the iron chelators on the growth of Salmonella Typhimurium.
4.6. Cell Line and Growth Conditions
The colon adenocarcinoma cell line Caco-2 (ATCC HTB-37) was obtained from American Type Culture Collection (ATCC, Manassas, Virginia, United States). Six-well plates, 12-well plates, and 12-well Millicell membrane cell inserts were purchased from Corning Costar (Fisher Scientific).
Caco-2 cells were cultured at 37 °C under 5% CO2 in Dulbecco’s modified Eagle medium (DMEM) with 4.5 g/L glucose, 0.586 g/L l-glutamine, 1000 U/mL penicillin–streptomycin (Fisher Scientific), and 3.7 g/L sodium bicarbonate.65 The culture medium was changed every other day, and the antibiotics-free medium was applied at the last medium change before the experiments were performed. In the assay of Salmonella Typhimurium invasion into epithelial cells, Caco-2 cells were cultured in 12-well plates (Corning Costar, Fisher Scientific) with DMEM and 10% (v/v) FBS for 4 to 6 days to reach 100% cell confluency (5 × 104 cell/cm2). DMEM with 20% (v/v) FBS was used to cultivate the cells for the tight junction permeability assay.
4.7. Caco-2 Invasion Assay
The ability of Salmonella Typhimurium to invade epithelial cells was determined using Caco-2 cells at a multiplicity of infection (MOI) of 100:1.65 Caco-2 cells (1 × 105) were seeded in each well of 12-well plates. The DMEM medium was removed, and Caco-2 monolayers were washed twice with 0.5 mL/well of phosphate-buffered saline (PBS) prior to the invasion assay. Bacterial strains were inoculated into fresh IMDM containing 5 μM ferric chloride at a final concentration of 4 × 107 CFU/mL. The bacterial suspension was transferred to the wells (0.5 mL/well) containing Caco-2 monolayers and co-incubated in IMDM at 37 °C for 2 h. The bacterial cells were then killed by incubation with gentamicin (150 μg/mL, 0.5 mL) in PBS for 1 h.66,67 Epithelial monolayers were then washed twice with PBS and lysed with 200 μL of 0.1% (v/v) Triton X-100 (Sigma-Aldrich); the Salmonella count that had invaded Caco-2 cells was determined by 10-fold serial dilution and plating on tryptic soy agar (BD Difco). The percentage of bacterial invasion was presented by the following formula
4.8. Assay for Caco-2 Tight Junction Permeability
The effect of the WT, mutants, and complements of Salmonella Typhimurium on the tight junction permeability of epithelial cells was studied using Caco-2 cells. The Caco-2 cells were cultured in 12 mm Millicell cell culture inserts (12 wells, Corning Costar, Fisher Scientific) in DMEM for 12 to 16 days until the transepithelial electrical resistance (TEER) value of the monolayers in all wells became stable between 1200 and 1400 Ω cm2 prior to the assay for measuring TEER in IMDM after two washes of Caco-2 monolayers with PBS.68,69 The TEER was expressed after subtracting from the resistance reading of the supporting membrane and multiplying it by the surface area of the Caco-2 monolayer. The TEER value was measured every other day by a Millicell ERS-2 voltohmmeter (Millipore Co., Bedford, Massachusetts, United States). The MOI was 100:1, and the Salmonella Typhimurium suspension was prepared as the invasion assay stated above. The bacterial suspension was added to the wells (500 μL/well) containing Caco-2 monolayers and coculture for 6 h in IMDM at 37 °C under 5% CO2. The TEER value of the monolayers was measured and recorded immediately 1 h after inoculation. The permeability value was calculated following the formula
4.9. C elegans Lifespan Assay
C. elegans temperature-sensitive defect mutants (glp-4; SS104) and E. coli OP50 were obtained from the Caenorhabditis Genetics Center (Minneapolis, Minnesota, United States). C. elegans was maintained on a nematode growth medium (NGM) with E. coli OP50 lawn using standard protocols.70
The C. elegans lifespan assay with the treatment of various Salmonella Typhimurium strains was performed as described previously.71 Adult worms were collected by sterilized water from NGM plates to perform synchronization, as described by Stiernagle.70 Approximately 300 synchronized eggs were transferred on NGM agar with E. coli OP50 lawn and incubated at 25 °C for 72 h to grow to the L4 stage. After the L4 worms were collected in S basal solution and washed twice with S medium via centrifugation (1300g for 1 min) and resuspension, 20 to 25 worms were assigned to each well of a six-well titer plate (Costar) with 2 mL of S medium and then incubated at 25 °C for 8 days. The S medium was not changed during the assay. Each treatment had a total number of 45 to 50 worms. Worms fed with E. coli OP50 (109 CFU/mL) only served as the control, while worms treated with various Salmonella Typhimurium strains (109 CFU/mL) were regarded as the treatment groups. The E. coli OP50 culture and Salmonella Typhimurium cultures used for the lifespan assay were all in the early stationary phase. The bacteria were washed twice in S medium by centrifugation and resuspension before being fed to the nematode. To determine the survival of C. elegans, a worm was considered dead when it did not respond to touch. The number of dead worms was recorded daily, and the percentage of the worms’ survival was presented following the formula
4.10. Total RNA Extraction and cDNA Synthesis
The method for extracting RNA from Caco-2 was adapted from Cuadras et al.72 Briefly, Caco-2 cells infected with bacteria were stabilized at 4 °C overnight with RNAlater solution (Ambion, Fisher Scientific). The total RNA of Caco-2 cells was isolated using the RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. The RNA quality and yield were determined using NanoDrop 1000 (Fisher Scientific) and 1.5% agarose gel electrophoresis. Total RNA was purified using TURBO DNA-Free kit (Ambion) according to the manufacturer’s instructions. The complementary DNA (cDNA) synthesis was performed using qScript cDNA SuperMix (QuantaBio, Beverly, Massachusetts, USA). One microgram of purifying RNA from each sample was used for cDNA synthesis in 20 μL of a reaction mixture.
The method for total RNA isolation from C. elegans was adapted from Ketting et al.73 Briefly, a total of 40 worms from each treatment condition were collected on the fifth day of the lifespan assay. The sampling on day 5 was chosen based on our observation that the worm’s survival decreased dramatically on days 6 and 7 postinfection with Salmonella. In each well, the worms were washed twice with RNase-free water to remove bacterial cells. The washed worms were transferred into a new RNase-free Eppendorf tube, mixed with 25 μL of lysis buffer, and then incubated at 65 °C for 10 min followed by 85 °C for 1 min. The lysis buffer consisted of 0.5% (v/v) Triton, 0.5% (v/v) Tween-20, 0.25 μM EDTA, 2.5 μM Tris–HCl buffer (pH 8.0), and 1 mg/mL proteinase K. The total RNA of C. elegans was isolated from the lysate using TRIzol RNA isolation reagents (Fisher Scientific) according to the manufacturer’s instructions. The RNA yield and RNA integrity were measured and determined, respectively, by the same method described above for Caco-2 RNA isolation. The isolated total RNA was purified, and cDNA was synthesized as previously described.33
4.11. Quantitative PCR Analysis
The mRNA abundance of various genes related to tight junction proteins, inflammation factors, and nutrient transporters of Caco-2 cells was analyzed using quantitative polymerase chain reaction (qPCR) assays. The qPCR was performed with 50 ng of cDNA constructed above using the fluorescent dye SYBR Green methodology and an ABI Prism 7500 Fast Real-Time PCR system (Applied Biosystems, Fisher Scientific). The qPCR conditions were as follows: a total of 40 PCR cycles, denaturing at 95 °C for 30 s, annealing at 60 °C for 1 min, extension at 72 °C for 30 s. The specificity of each gene amplification was verified at the end of each qPCR reaction by analysis of melting curves of the PCR products. The curves of amplification were read with ABI Prism 7500 software using the comparative cycle threshold. Relative quantification of the target mRNA levels was presented after normalization of the total amount of cDNA tested to endogenous references 18S RNA and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The primers for qPCR are listed in Table 2.74−79 The results of qPCR were analyzed using the 2–ΔΔCt method to determine the fold changes of target genes.33 The ΔCt was presented as the difference in threshold cycle between the target genes and housekeeping genes (18S and GAPDH), and ΔΔCt was the difference in ΔCt between the Salmonella-infected Caco-2 cells and uninfected Caco-2 cells. The gene expression in the uninfected Caco-2 cells was used as the baseline (reference = 1).
Table 2. Primers of qPCR Assay for Caco-2 Cellsa.
| gene | sequence 5′–3′ | product size (bp) | reference |
|---|---|---|---|
| PepT1 | FP: TTGGCCCAATGTCTCA | 120 | (74) |
| RP: GGCCCTGCTTGAAGTC | |||
| CLDN3 | FP: GGACTTCTACAACCCCGTGGT | 230 | (75) |
| RP: AGACGTAGTCCTTGCGGTCGT | |||
| TJP1 | FP: CCTTCAGCTGTGGAA GAG GATG | 287 | (75) |
| RP: AGCTCCACAGGCTTCAGGAAC | |||
| IL-8 | FP: AAGGAACCATCTCACTG | 357 | (76) |
| RP: GATTCTTGGATACCACAGAG | |||
| TNF-α | FP: GCCATTGGCCAG GAG GGC | 220 | (77) |
| RP: CGCCACCACGCT CTT CTG | |||
| GAPDH | FP: GGAGTCCACTGGCGTCTTCAC | 165 | (78) |
| RP: GAGGCATTGCTGATGATCTTGAGG | |||
| 18S | FP: CGCCGCTAGAGGTGAAATTC | 62 | (79) |
| RP: TTGGCAAATGCTTTCGCTC |
Note: TNF-α, tumor necrosis factor α; IL-8, interleukin 8; PepT1, peptide transporter 1; CLDN3, claudin 3; TJP1, tight junction protein 1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; 18S, 18S ribosomal RNA; bp, base pair; FP, forward primer; RP, reverse primer.
The mRNA abundance of several genes encoding for antimicrobial peptides and a defense molecule, components in the p38 MAPK and DAF/IGF signaling pathway, and nutrient utilization-related functions of C. elegans was analyzed by qPCR assays. A total of 10 ng of C. elegans cDNA was used for a qPCR assay using the same conditions stated above. Relative quantification of the target mRNA levels was presented after normalization of endogenous references (snb-1 and act-1). The primers for qPCR are listed in Table 3.33,35,63 The 2–ΔΔCt method was used to determine the fold changes of target genes.37 The ΔCt was presented as the difference in the threshold cycle between the target genes and housekeeping genes (act-1 and snb), and ΔΔCt was the difference in ΔCt between the Salmonella-infected C. elegans and uninfected C. elegans. The gene expression in C. elegans treated with E. coli OP50 only (uninfected worms) was used as the baseline (reference = 1).
Table 3. Primers of qPCR Assay for C. elegansa.
| gene | sequence 5′–3′ | product size (bp) | reference |
|---|---|---|---|
| sod-3 | FP: AAATGTCCGCCCAGACTATG | 124 | (33) |
| RP: TGGCAAATCTCTCGCTGA | |||
| fgt-1 | FP: GGCCAGCTACTCAGCCATC | 93 | (35) |
| RP: ATTTCGGAGACGAAGAACCA | |||
| ftn-1 | FP: GACGTGTTGCCATGCAGAACATT | 144 | (63) |
| RP: CATTGCGTTGTTCGGCGATT | |||
| spp-1 | FP: TGGACTATGCTGTTGCCGTT | 106 | (33) |
| RP: ACGCCTTGTCTGGAGAATCC | |||
| clec-85 | FP: CCAATGGGATGACGGAACCA | 121 | (33) |
| RP: CTTCTGTCCAGCCAACGTCT | |||
| lys-7 | FP: GTACAGCGGTGGAGTCACTG | 153 | (33) |
| RP: GCCTTGAGCACATTTCCAGC | |||
| daf-16 | FP: TCGTCTCGTGTTTCTCCAGC | 181 | (33) |
| RP: TAATCGGCTTCGACTCCTGC | |||
| pmk-1 | FP: CCAAAAATGACTCGCCGTGA | 115 | (33) |
| RP: CTTTTGCAGTTGGACGACGA | |||
| nsy-1 | FP: AGCGGCTCGATCAACAAGAA | 122 | (33) |
| RP: CCCATTCCACCGATATGCGA | |||
| act-1 | FP: CCCCACTCAATCCAAAGGCT | 121 | (33) |
| RP: GTACGTCCGGAAGCGTAGAG | |||
| snb | FP: CCGGATAAGACCATCTTGACG | 128 | (33) |
| RP: GACGACTTCATCAACCTGAGC |
Note: sod-3, superoxide dismutase 3; fgt-1, facilitated glucose transporter protein 1; ftn-1, ferritin 1; spp-1, saposin-like protein; clec-85, C-type lectin; lys-7, lysozyme-like protein 7; daf-16, forkhead-type transcription factor; pmk-1, mitogen-activated protein kinase pmk-1; nsy-1, mitogen-activated protein kinase kinase kinase nsy-1; act-1, actin; snb, synaptobrevin-1; bp, base pair; FP, forward primer; RP, reverse primer.
4.12. Statistical Analysis
All statistical analyses were performed using the GraphPad Prism 6 software (San Diego, United States) except that the survival curve analyses of C. elegans were performed using the Statistical Analysis System (SAS release 9.4, SAS Institute Inc., Cary, North Carolina, United States). Comparison of C. elegans survival curves was performed by Kaplan–Meier estimator with a log-rank test. In the bacterial invasion, tight junction permeability, and gene expression studies, Tukey’s multiple comparison tests were used to determine differences among treatment means. P < 0.05 was taken to indicate statistical significance.
Acknowledgments
This work was financially supported by Agriculture and Agri-Food Canada through the A-base Program (AAFC Project ID PSS # 1561, : J-001391) and the University of Manitoba Start-Up Grant (C. Yang, 46561). The authors are grateful to Emily Emmter, who was an NSERC summer undergraduate student in the Department of Animal Science at the University of Manitoba, for assistance.
Appendix
The effect of 1 μM of iron on the growth of Salmonella Typhimurium is shown in Figure A1.
Figure A1.
Effect of 1 μM of iron on the growth of Salmonella Typhimurium
Author Present Address
+ Present address: Food and Bioproduct Sciences, College of Agriculture and Bioresources, University of Saskatchewan, Saskatoon, Canada S7N 5A8.
The authors declare no competing financial interest.
References
- Rabsch W.; Tschäpe H.; Bäumler A. J. Non-typhoidal salmonellosis: emerging problems. Microbes. Infect. 2001, 3, 237–247. 10.1016/S1286-4579(01)01375-2. [DOI] [PubMed] [Google Scholar]
- Frawley E. R.; Fang F. C. The ins and outs of bacterial iron metabolism. Mol. Microbiol. 2014, 93, 609–616. 10.1111/mmi.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. C.; Robinson A. K.; Rodríguez-Quiñones F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- Ferguson A. D.; Hofmann E.; Coulton J. W.; Diederichs K.; Welte W. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 1998, 282, 2215–2220. 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- Nagy T. A.; Moreland S. M.; Andrews-Polymenis H.; Detweiler C. S. The Ferric Enterobactin Transporter, Fep, is Required for Persistent Salmonella Infection. Infect. Immun. 2013, 81, 4063–4070. 10.1128/IAI.00412-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannavy K. C.; Barr G. C.; Dorman C. J.; Adamson J.; Mazengera L. R.; Gallagher M. P.; Evans J. S.; Levine B. A.; Trayer I. P.; Higgins C. F. TonB protein of Salmonella typhimurium: a model for signal transduction between membranes. J. Mol. Biol. 1990, 216, 897–910. 10.1016/S0022-2836(99)80009-6. [DOI] [PubMed] [Google Scholar]
- Williams P. H.; Rabsch W.; Methner U.; Voigt W.; Tschäpe H.; Reissbrodt R. Catecholate receptor proteins in Salmonella enterica: role in virulence and implications for vaccine development. Vaccine 2006, 24, 3840–3844. 10.1016/j.vaccine.2005.07.020. [DOI] [PubMed] [Google Scholar]
- Tsolis R. M.; Bäumler A. J.; Heffron F.; Stojiljkovic I. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect. Immun. 1996, 64, 4549–4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster S. L.; Richardson S. H.; Failla M. L. Elevated iron status increases bacterial invasion and survival and alters cytokine/chemokine mRNA expression in Caco-2 human intestinal cells. J. Nutr. 2001, 131, 1452–1458. 10.1093/jn/131.5.1452. [DOI] [PubMed] [Google Scholar]
- Kortman G. A. M.; Boleij A.; Swinkels D. W.; Tjalsma H. Iron availability increases the pathogenic potential of Salmonella typhimurium and other enteric pathogens at the intestinal epithelial interface. PLoS One 2012, 7, e29968 10.1371/journal.pone.0029968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F.; Wu H.; Zeng M.; Yu G.; Dong S.; Yang H. Probiotic/prebiotic correction for adverse effects of iron fortification on intestinal resistance to Salmonella infection in weaning mice. Food Funct. 2018, 9, 1070–1078. 10.1039/C7FO00990A. [DOI] [PubMed] [Google Scholar]
- Heimbach J.; Rieth S.; Mohamedshah F.; Slesinski R.; Samuel-Fernando P.; Sheehan T.; Dickmann R.; Borzelleca J. Safety assessment of iron EDTA [sodium iron (Fe3+) ethylenediaminetetraacetic acid]: summary of toxicological, fortification and exposure data. Food Chem. Toxicol. 2000, 38, 99–111. 10.1016/S0278-6915(99)00125-8. [DOI] [PubMed] [Google Scholar]
- Kamdi S. P.; Palkar P. J. Efficacy and safety of ferrous asparto glycinate in the management of iron deficiency anaemia in pregnant women. J. Obstet. Gynaecol. 2015, 35, 4–8. 10.3109/01443615.2014.930098. [DOI] [PubMed] [Google Scholar]
- Turner J. R. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 2009, 9, 799. 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- Giannasca K. T.; Giannasca P. J.; Neutra M. R. Adherence of Salmonella typhimurium to Caco-2 cells: identification of a glycoconjugate receptor. Infect. Immun. 1996, 64, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay B. B.; Falkow S. Salmonella interactions with polarized human intestinal Caco-2 epithelial cells. J. Infect. Dis. 1990, 162, 1096–1106. 10.1093/infdis/162.5.1096. [DOI] [PubMed] [Google Scholar]
- Neutra M. Differentiation of intestinal cells in vitro. Func. Epithelial Cells Cult. 1989, 363–398. [Google Scholar]
- Peterson M. D.; Mooseker M. S. Characterization of the enterocyte-like brush border cytoskeleton of the C2BBe clones of the human intestinal cell line, Caco-2. J. Cell Sci. 1992, 102, 581–600. [DOI] [PubMed] [Google Scholar]
- Guttman J. A.; Finlay B. B. Tight junctions as targets of infectious agents. Biochim. Biophys. Acta, Biomembr. 2009, 1788, 832–841. 10.1016/j.bbamem.2008.10.028. [DOI] [PubMed] [Google Scholar]
- Adibi S. A. The oligopeptide transporter (Pept-1) in human intestine: biology and function. Gastroenterology 1997, 113, 332–340. 10.1016/S0016-5085(97)70112-4. [DOI] [PubMed] [Google Scholar]
- Eckmann L.; Kagnoff M. F. Cytokines in host defense against Salmonella. Microbes Infect. 2001, 3, 1191–1200. 10.1016/S1286-4579(01)01479-4. [DOI] [PubMed] [Google Scholar]
- Gewirtz A. T.; Rao A. S.; Simon P. O. Jr.; Merlin D.; Carnes D.; Madara J. L.; Neish A. S. Salmonella typhimurium induces epithelial IL-8 expression via Ca 2+–mediated activation of the NF-κB pathway. J. Clin. Invest. 2000, 105, 79–92. 10.1172/JCI8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle D. L.; Blumenthal T.; Meyer B. J.; Priess J. R.. Introduction to C. elegans. C. elegans II, 2nd ed.; Riddle D. L.; Blumenthal T.; Meyer B. J.; Priess J. R., Ed.; Cold Spring Harbor Laboratory Press: New York, 1997; Chapter 1. [Google Scholar]
- Labrousse A.; Chauvet S.; Couillault C.; Léopold Kurz C.; Ewbank J. J. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr. Biol. 2000, 10, 1543–1545. 10.1016/S0960-9822(00)00833-2. [DOI] [PubMed] [Google Scholar]
- Jones B. D.; Falkow S. Salmonellosis: host immune responses and bacterial virulence determinants. Annu. Rev. Immunol. 1996, 14, 533–561. 10.1146/annurev.immunol.14.1.533. [DOI] [PubMed] [Google Scholar]
- Murphy C. T.; McCarroll S. A.; Bargmann C. I.; Fraser A.; Kamath R. S.; Ahringer J.; Li H.; Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 2003, 424, 277. 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Zugasti O.; Ewbank J. J. Neuroimmune regulation of antimicrobial peptide expression by a noncanonical TGF-β signaling pathway in Caenorhabditis elegans epidermis. Nat. Immunol. 2009, 10, 249. 10.1038/ni.1700. [DOI] [PubMed] [Google Scholar]
- Kim D. H.; Feinbaum R.; Alloing G.; Emerson F. E.; Garsin D. A.; Inoue H.; Tanaka-Hino M.; Hisamoto N.; Matsumoto K.; Tan M.-W.; Ausubel F. M. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 2002, 297, 623–626. 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- Dillin A.; Crawford D. K.; Kenyon C. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science 2002, 298, 830–834. 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- Simonsen K. T.; Møller-Jensen J.; Kristensen A. R.; Andersen J. S.; Riddle D. L.; Kallipolitis B. H. Quantitative proteomics identifies ferritin in the innate immune response of C. elegans. Virulence 2011, 2, 120–130. 10.4161/viru.2.2.15270. [DOI] [PubMed] [Google Scholar]
- Bányai L.; Patthy L. Amoebapore homologs of Caenorhabditis elegans. Biochim. Biophys. Acta. 1998, 1429, 259–264. 10.1016/S0167-4838(98)00237-4. [DOI] [PubMed] [Google Scholar]
- Kurz C. L.; Tan M. W. Regulation of aging and innate immunity in C. elegans. Aging Cell 2004, 3, 185–193. 10.1111/j.1474-9728.2004.00108.x. [DOI] [PubMed] [Google Scholar]
- Zhou M.; Liu X.; Yu H.; Yin X.; Nie S.-P.; Xie M.-Y.; Chen W.; Gong J. Cell signaling of Caenorhabditis elegans in response to enterotoxigenic Escherichia coli infection and Lactobacillus zeae protection. Front. Immunol. 2018, 9, 1745. 10.3389/fimmu.2018.01745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romney S. J.; Thacker C.; Leibold E. A. An iron enhancer element in the FTN-1 gene directs iron-dependent expression in Caenorhabditis elegans intestine. J. Biol. Chem. 2008, 283, 716–725. 10.1074/jbc.M707043200. [DOI] [PubMed] [Google Scholar]
- Feng Y.; Williams B. G.; Koumanov F.; Wolstenholme A. J.; Holman G. D. FGT-1 is the major glucose transporter in C. elegans and is central to aging pathways. Biochem. J. 2013, 456, 219–229. 10.1042/BJ20131101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka S.; Morielli A. D.; Zhao F. Q. FGT-1 is a mammalian GLUT2-like facilitative glucose transporter in Caenorhabditis elegans whose malfunction induces fat accumulation in intestinal cells. PLoS One 2013, 8, e68475 10.1371/journal.pone.0068475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogewijs D.; Houthoofd K.; Matthijssens F.; Vandesompele J.; Vanfleteren J. R. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 2008, 9, 9. 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk J. M.; Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 2009, 5, e1000361 10.1371/journal.pgen.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanani A. S.; Block G.; Dewar K.; Forgetta V.; Topp E.; Beiko R. G.; Diarra M. S. Genomic comparison of non-typhoidal Salmonella enterica serovars Typhimurium, Enteritidis, Heidelberg, Hadar and Kentucky isolates from broiler chickens. PLoS One 2015, 10, e0128773 10.1371/journal.pone.0128773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y. F.; Jiang M. M.; Sun J.; Shi R. B.; Liu D. S. Studies on Different Iron Source Absorption by in Situ Ligated Intestinal Loops of Broilers. Biol. Trace Elem. Res. 2015, 163, 154–161. 10.1007/s12011-014-0179-1. [DOI] [PubMed] [Google Scholar]
- Teucher B.; Olivares M.; Cori M. Enhancers of iron absorption: ascorbic acid and other organic acids. Int. J. Vitam. Nutr. Res. 2004, 74, 403–419. 10.1024/0300-9831.74.6.403. [DOI] [PubMed] [Google Scholar]
- Yeung C. K.; Glahn R. P.; Miller D. D. Inhibition of iron uptake from iron salts and chelates by divalent metal cations in intestinal epithelial cells. J. Agric. Food Chem. 2005, 53, 132–136. 10.1021/jf049255c. [DOI] [PubMed] [Google Scholar]
- Chu B. C.; Garcia-Herrero A.; Johanson T. H.; Krewulak K. D.; Lau C. K.; Peacock R. S.; Slavinskaya Z.; Vogel H. J. Siderophore uptake in bacteria and the battle for iron with the host; a bird’s eye view. BioMetals 2010, 23, 601–611. 10.1007/s10534-010-9361-x. [DOI] [PubMed] [Google Scholar]
- Lau C. K. Y.; Krewulak K. D.; Vogel H. J. Bacterial ferrous iron transport: the Feo system. FEMS Microbiol. Rev. 2016, 40, 273–298. 10.1093/femsre/fuv049. [DOI] [PubMed] [Google Scholar]
- Banerjee S.; Paul S.; Nguyen L. T.; Chu B. C. H.; Vogel H. J. FecB, a periplasmic ferric-citrate transporter from E. coli, can bind different forms of ferric-citrate as well as a wide variety of metal-free and metal-loaded tricarboxylic acids. Metallomics 2016, 8, 125–133. 10.1039/C5MT00218D. [DOI] [PubMed] [Google Scholar]
- Pressler U.; Staudenmaier H.; Zimmermann L.; Braun V. Genetics of the iron dicitrate transport system of Escherichia coli. J. Bacteriol. 1988, 170, 2716–2724. 10.1128/jb.170.6.2716-2724.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer A.; Braun V. Iron regulates transcription of the Escherichia coli ferric citrate transport genes directly and through the transcription initiation proteins. Arch. Microbiol. 1998, 169, 483–490. 10.1007/s002030050600. [DOI] [PubMed] [Google Scholar]
- Rabsch W.; Voigt W.; Reissbrodt R.; Tsolis R. M.; Bäumler A. J. Salmonella typhimurium IroN and FepA proteins mediate uptake of enterobactin but differ in their specificity for other siderophores. J. Bacteriol. 1999, 181, 3610–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mademidis A.; Köster W. Transport activity of FhuA, FhuC, FhuD, and FhuB derivatives in a system free of polar effects, and stoichiometry of components involved in ferrichrome uptake. Mol. Gen. Genet. 1998, 258, 156–165. 10.1007/s004380050718. [DOI] [PubMed] [Google Scholar]
- Chandraleka S.; Ramya K.; Chandramohan G.; Dhanasekaran D.; Priyadharshini A.; Panneerselvam A. Antimicrobial mechanism of copper (II) 1, 10-phenanthroline and 2, 2′-bipyridyl complex on bacterial and fungal pathogens. J. Saudi Chem. Soc. 2014, 18, 953–962. 10.1016/j.jscs.2011.11.020. [DOI] [Google Scholar]
- Kumar R. S.; Sasikala K.; Arunachalam S. DNA interaction of some polymer–copper (II) complexes containing 2, 2′-bipyridyl ligand and their antimicrobial activities. J. Inorg. Biochem. 2008, 102, 234–241. 10.1016/j.jinorgbio.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Srinivasan S.; Annaraj J.; Athappan P. Spectral and redox studies on mixed ligand complexes of cobalt (III) phenanthroline/bipyridyl and benzoylhydrazones, their DNA binding and antimicrobial activity. J. Inorg. Biochem. 2005, 99, 876–882. 10.1016/j.jinorgbio.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Egan T. J.; Koch K. R.; Swan P. L.; Clarkson C.; Van Schalkwyk D. A.; Smith P. J. In vitro antimalarial activity of a series of cationic 2,2‘-Bipyridyl- and 1,10-Phenanthrolineplatinum(II) benzoylthiourea complexes. J. Med. Chem. 2004, 47, 2926–2934. 10.1021/jm031132g. [DOI] [PubMed] [Google Scholar]
- Bodavari S.The Merck Index; 12th ed.; Bodavari S., Ed.; Whitehouse Station: New York, 2006. [Google Scholar]
- Altier C. Genetic and environmental control of Salmonella invasion. J. Microbiol. 2005, 43, 85–92. [PubMed] [Google Scholar]
- Cossart P.; Sansonetti P. J. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 2004, 304, 242–248. 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- Blais M.; Fortier M.; Pouliot Y.; Gauthier S. F.; Boutin Y.; Asselin C.; Lessard M. Colostrum whey down-regulates the expression of early and late inflammatory response genes induced by Escherichia coli and Salmonella enterica Typhimurium components in intestinal epithelial cells. Br. J. Nutr. 2015, 113, 200–211. 10.1017/S0007114514003481. [DOI] [PubMed] [Google Scholar]
- Coburn B.; Grassl G. A.; Finlay B. B. Salmonella, the host and disease: a brief review. Immunol. Cell Biol. 2007, 85, 112–118. 10.1038/sj.icb.7100007. [DOI] [PubMed] [Google Scholar]
- Arnold J. W.; Niesel D. W.; Annable C. R.; Hess C. B.; Asuncion M.; Cho Y. J.; Peterson J. W.; Klimpel G. R. Tumor necrosis factor-α mediates the early pathology in Salmonella infection of the gastrointestinal tract. Microb. Pathog. 1993, 14, 217–227. 10.1006/mpat.1993.1021. [DOI] [PubMed] [Google Scholar]
- Rabsch W.; Methner U.; Voigt W.; Tschäpe H.; Reissbrodt R.; Williams P. H. Role of receptor proteins for enterobactin and 2, 3-dihydroxybenzoylserine in virulence of Salmonella enterica. Infect. Immun. 2003, 71, 6953–6961. 10.1128/IAI.71.12.6953-6961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper S.; McBride S. J.; Lackford B.; Freedman J. H.; Schwartz D. A. Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol. Cell. Biol. 2007, 27, 5544–5553. 10.1128/MCB.02070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamaladevi A.; Balamurugan K. Lactobacillus casei triggers a TLR mediated RACK-1 dependent p38 MAPK pathway in Caenorhabditis elegans to resist Klebsiella pneumoniae infection. Food Funct. 2016, 7, 3211–3223. 10.1039/C6FO00510A. [DOI] [PubMed] [Google Scholar]
- Kim Y.-I.; Cho J. H.; Yoo O. J.; Ahnn J. Transcriptional regulation and life-span modulation of cytosolic aconitase and ferritin genes in C. elegans. J. Mol. Biol. 2004, 342, 421–433. 10.1016/j.jmb.2004.07.036. [DOI] [PubMed] [Google Scholar]
- Chekabab S. M.; Rehman M. A.; Yin X.; Carrillo C.; Mondor M.; Diarra M. S. Growth of Salmonella enterica Serovars Typhimurium and Enteritidis in iron-poor media and in meat: Role of catecholate and hydroxamate siderophore transporters. J. Food Prot. 2019, 82, 548–560. 10.4315/0362-028X.JFP-18-371. [DOI] [PubMed] [Google Scholar]
- Mickael C. S.; Lam P.-K. S.; Berberov E. M.; Allan B.; Potter A. A.; Köster W. Salmonella enterica serovar Enteritidis tatB and tatC mutants are impaired in Caco-2 cell invasion in vitro and show reduced systemic spread in chickens. Infect. Immun. 2010, 78, 3493–3505. 10.1128/IAI.00090-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon M.; Berner A. Z.; Chervet N.; Chassard C.; Lacroix C. Comparison of the Caco-2, HT-29 and the mucus-secreting HT29-MTX intestinal cell models to investigate Salmonella adhesion and invasion. J. Microbiol. Methods 2013, 94, 274–279. 10.1016/j.mimet.2013.06.027. [DOI] [PubMed] [Google Scholar]
- Hänel I.; Müller J.; Müller W.; Schulze F. Correlation between invasion of Caco-2 eukaryotic cells and colonization ability in the chick gut in Campylobacter jejuni. Vet. Microbiol. 2004, 101, 75–82. 10.1016/j.vetmic.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Ranaldi G.; Consalvo R.; Sambuy Y.; Scarino M. L. Permeability characteristics of parental and clonal human intestinal Caco-2 cell lines differentiated in serum-supplemented and serum-free media. Toxicol. In Vitro 2003, 17, 761–767. 10.1016/S0887-2333(03)00095-X. [DOI] [PubMed] [Google Scholar]
- Lu S.; Gough A. W.; Bobrowski W. F.; Stewart B. H. Transport properties are not altered across Caco-2 cells with heightened TEER despite underlying physiological and ultrastructural changes. J. Pharm. Sci. 1996, 85, 270–273. 10.1021/js950269u. [DOI] [PubMed] [Google Scholar]
- Stiernagle T.Maintenance of C. elegans. In The WormBook; Hobert O., Ed; The C. elegans Research Community: 2006. [Google Scholar]
- Wang C.; Wang J.; Gong J.; Yu H.; Pacan J. C.; Niu Z.; Si W.; Sabour P. M. Use of Caenorhabditis elegans for preselecting Lactobacillus isolates to control Salmonella Typhimurium. J. Food Prot. 2011, 74, 86–93. 10.4315/0362-028X.JFP-10-155. [DOI] [PubMed] [Google Scholar]
- Cuadras M. A.; Feigelstock D. A.; An S.; Greenberg H. B. Gene expression pattern in Caco-2 cells following rotavirus infection. J. Virol. 2002, 76, 4467–4482. 10.1128/JVI.76.9.4467-4482.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting R. F.; Tijsterman M.; Plasterk R. H. Isolation of RNA from C. elegans. CSH Protoc. 2010, 1, pdb-prot4321. 10.1101/pdb.prot4321. [DOI] [PubMed] [Google Scholar]
- Mooij M. G.; de Koning B. E.; Lindenbergh-Kortleve D. J.; Simons-Oosterhuis Y.; van Groen B. D.; Tibboel D.; Samsom J. N.; de Wildt S. N. Human intestinal PEPT1 transporter expression and localization in preterm and term infants. Drug Metab. Dispos. 2016, 44, 1014–1019. 10.1124/dmd.115.068809. [DOI] [PubMed] [Google Scholar]
- Ding C.; Cong X.; Zhang X. M.; Li S. L.; Wu L. L.; Yu G. Y. Decreased interaction between ZO-1 and occludin is involved in alteration of tight junctions in transplanted epiphora submandibular glands. J. Mol. Histol. 2017, 48, 225–234. 10.1007/s10735-017-9716-5. [DOI] [PubMed] [Google Scholar]
- Crabtree J. E.; Wyatt J. I.; Trejdosiewicz L. K.; Peichl P.; Nichols P. H.; Ramsay N.; Primrose J. N.; Lindley I. J. Interleukin-8 expression in Helicobacter pylori infected, normal, and neoplastic gastroduodenal mucosa. J. Clin. Pathol. 1994, 47, 61–66. 10.1136/jcp.47.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghizadeh M.; Ong J. M.; Garvey W. T.; Henry R. R.; Kern P. A. The expression of TNF alpha by human muscle. Relationship to insulin resistance. J. Clin. Invest. 1996, 97, 1111–1116. 10.1172/JCI118504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piana C.; Wirth M.; Gerbes S.; Viernstein H.; Gabor F.; Toegel S. Validation of reference genes for qPCR studies on Caco-2 cell differentiation. Eur. J. Pharm. Biopharm. 2008, 69, 1187–1192. 10.1016/j.ejpb.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Maubon N.; Le Vee M.; Fossati L.; Audry M.; Le Ferrec E.; Bolze S.; Fardel O. Analysis of drug transporter expression in human intestinal Caco-2 cells by real-time PCR. Fundam. Clin. Pharmacol. 2007, 21, 659–663. 10.1111/j.1472-8206.2007.00550.x. [DOI] [PubMed] [Google Scholar]