Abstract
Salt is a fundamental nutrient that is required for many physiological processes, including electrolyte homeostasis and neuronal activity. In mammals and Drosophila, the detection of NaCl induces two different behaviors: low-salt concentrations provide an attractive stimulus, whereas high-salt concentrations are avoided. We identified the gene called serrano (sano) as being expressed in the sensory organs of Drosophila larvae. A transgenic reporter line showed that sano was coexpressed with Gr66a in a subset of gustatory neurons in the terminal organ of third-instar larvae. The disruption of sano gene expression in gustatory neurons led to the specific loss of high-salt concentration avoidance in larvae, whereas the detection of other attractive or aversive substances was unaffected. Moreover, using a cellular marker sensitive to calcium levels, Sano function was shown to be required for neuronal activity in response to high-salt concentrations. In these neurons, the loss of the DEG/ENaC channel PPK19 function also eliminated the cellular response to high-salt concentrations. Our study revealed that PPK19 and Sano are required in the neurons of the larval gustatory organs for the detection of high-salt concentrations.
Keywords: behavior, chemosensory system, Drosophila melanogaster, larva, salt, taste
Introduction
Gustatory information is considered to be important for the control of animal behaviors, such as searching for food or sexual partners. In Drosophila, larvae sense olfactory and gustatory cues with the three major chemosensory organs located on the head surface, dorsal organ (DO), terminal organ (TO) and ventral organ (VO), and three pharyngeal organs (Vosshall and Stocker, 2007). Olfactory receptor neurons located in the DO project into glomeruli of the antennal lobe, whereas gustatory receptor neurons (GRNs) project via four different nerves to the suboesophageal ganglion (Vosshall and Stocker, 2007).
Insects, like mammals, are able to detect and discriminate among different gustatory stimuli, such as sugars, bitter substances, and salts, that can induce an attractive or a repulsive response in behavioral tests. Electrophysiological studies performed on Drosophila adult taste sensilla have revealed that low- and high-NaCl concentrations are detected by two distinct gustatory neurons (Hiroi et al., 2004; Amrein and Thorne, 2005; Ishimoto and Tanimura, 2004). Larvae are also able to display an attractive behavior in response to low-salt concentrations and an aversive response in the presence of high-salt concentrations (Heimbeck et al., 1999; Balakireva et al., 2000; Gerber and Stocker, 2007; Niewalda et al., 2008; Russell et al., 2011). The dpr locus (for defective proboscis extension response), a member of the Ig superfamily, is required for the aversive response to high-salt concentrations in adult flies (Nakamura et al., 2002). Two members of the pickpocket (ppk) gene family (DEG/ENaC channels, ppk19 and ppk11) have been implicated in the responses to low- and high-NaCl concentrations (Liu et al., 2003). In mammals, the detection of NaCl also requires the expression of ENaC protein in a subset of the taste receptor cells in the taste buds (Chandrashekar et al., 2010). More recently, Zhang et al. (2013) suggested that NaCl perception in Drosophila adults is determined by a bimodal switch system operating in GRNs that allows for detecting separately low- and high-NaCl concentrations. Moreover, they demonstrated that the ionotropic channel IR76b is selectively involved in the attractive pathway.
In this study, we describe the serrano (sano) locus, which we identified by the specific loss of the aversive response to high-NaCl concentrations in the mutant larvae; the responses to other attractive (sucrose or low-NaCl concentrations) and aversive (caffeine) stimuli remained unaltered. The sano locus encodes a putative cytoplasmic protein of 778 aa, which is related to the Themis gene family (Chung et al., 2009; Johnson et al., 2009). In our study, we show that Sano function is specifically required in the larval peripheral nervous system for the detection of high-NaCl concentrations and the subsequent induction of aversive behavior. Moreover, we show that Sano is necessary for neuronal activity of GRNs present in the TO of the larvae in response to high salt. Thus, Sano plays a key role in the detection of high-NaCl concentrations through the gustatory neurons of the TO.
Materials and Methods
Fly strains.
The fly strains used in this study were the wild-type strain CantonS, the sanoGal4 strain (identified from lines generated in a previous screen and provided by Prof. Denise Busson, Université Pierre et Marie Curie, Paris, France), the sanoGE12233 and sanoGE15762 strains (purchased from Genexel), and the UAS–TNT (tetanic toxin light chain), 10XUAS–IVS–mCD8::GFP, 10XUAS–IVS–mCD8::RFP, tubP–Gal80ts, and UAS–Cameleon2.1 (Cam2.1) strains (from the Bloomington Stock Center). The UAS–ppk19dsRNA strain was kindly provided by Dr. Michael J. Welsh (Howard Hugues Medical Institute, University of Iowa, Iowa City, IA). The Gr66a–GFP and Gr66aGal4 strains were kindly provided by Dr. Kristin Scott (Howard Hugues Medical Institute, University of California, Berkeley, Berkeley, CA) and Dr. Hubert Amrein (Texas A&M Health Science Center, College Station, TX), respectively. The strains used in behavioral tests have been outcrossed to a w1118 strain for five generations.
The generation of transgenic flies.
The sano ORF was amplified from the cDNA clone RE56731 (Berkeley Drosophila Genome Project), which contained the full RB cDNA, and was cloned into the pGEM-T vector for sequencing and into the pUAST vector to generate the UAS–sano lines.
A fragment of 575 bp from the exon 7 sequence was amplified from the cDNA clone RE56731 with the primers 5′-tctagaaactgcgactgctgggctgc and 5′-tctagacggaggtggtggcctgctgg and cloned into the pGEM-T vector for sequencing. Then, the fragment was cloned into the pWIZ vector to generate the UAS–sanodsRNA strain (protocol as in the study by Lee and Carthew, 2003). The construct was introduced into the Drosophila germ line by injection into w1118 embryos of either sex, as described previously (Rubin and Spradling, 1982).
Larval behavioral assay.
Petri dishes (9.5 cm diameter) were divided into two compartments. Each compartment was filled with either 2% agarose/water (control) or 2% agarose/test solution mixed in water (sucrose; S0389; Sigma-Aldrich), quinine (quinine hydrochloride; Q1125; Sigma-Aldrich), caffeine (C0750; Sigma-Aldrich), and sodium chloride (S9625; Sigma-Aldrich). At t = 0 min, 50 early-to-mid L3 larvae of either sex were placed on the dividing line between the two zones and allowed to move freely. The number of larvae found on the control (Nc) and test (Nt) areas was counted at 10 min. The larvae found at a distance of <0.5 cm on either side of the dividing line were not included in the index calculation. A gustatory preference index (GPI) was calculated using the following formula: GPI = (Nt − Nc)/(Nt + Nc). Positive preference indices indicate an attractive behavior, whereas negative ones indicate avoidance behavior toward tested substances. For each test, n = 10.
RT-PCR.
RNA was extracted from various portions of the dissected larvae of either sex using the TRIzol reagent (Invitrogen) and treated with RNase-free DNase to eliminate contamination by genomic DNA. Total RNA (1 μg) was reverse transcribed using the iScript cDNA Synthesis kit (Bio-Rad).
PCR reactions were conducted using a thermocycler (Bio-Rad). PCR primers were designed for different exons of the sano coding region (see Fig. 2): forward 1 (exon 1), 5′-ctgcgtctctttgcgtgttgg; forward 2 (exon 2), 5′-gaagccggtttcgtttctgtgtc; forward 3 (exons 5a and 5b), 5′-cggttcggtgtgtcccattgc; forward 4 (exon 5b), 5′-ggtatcagcgtgtgtttcgtcac; forward 5 (exon 8), 5′-acaacaattgctcctctgtgacg; reverse 1 (exon 6), 5′-ctcctttcgattccctttgacgc; reverse 2 (exon 8), 5′-atggcagctagaaacgagttggc; and reverse 3 (exon 9), 5′-cgattgcgagcagatggg.
Figure 2.
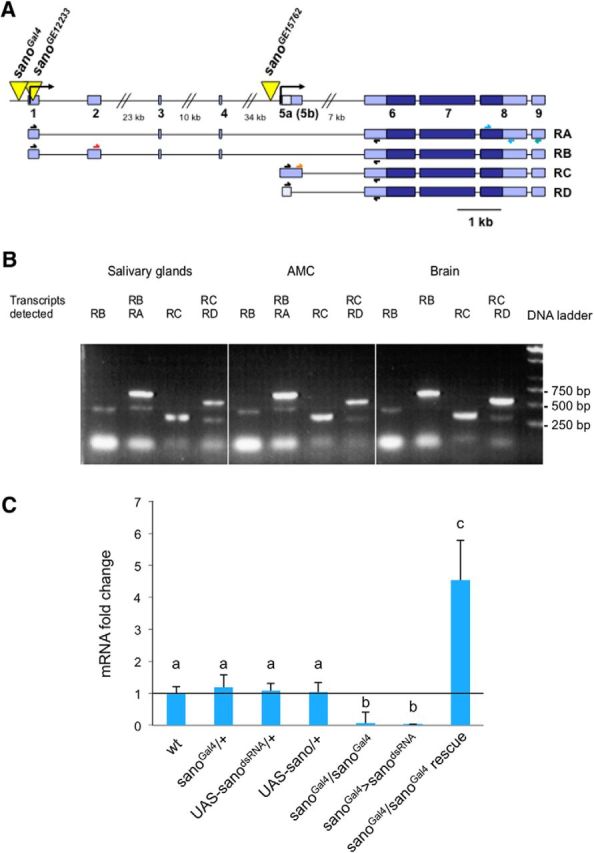
Map and transcript analysis of sano gene. A, Structure of the sano gene. Yellow triangles represent the P-element insertions [sanoGal4 is a P{GawB} line, and sanoGE12233 and sanoGE15762 are P{UAS} lines]. Exons are shown in blue. Light blue boxes represent the 5′ and 3′ untranslated regions. Scale bar, 1 kb. The arrows above the transcripts indicate the primers used for the RT-PCR analysis. B, sano expression analyzed by RT-PCR. RNAs were extracted from the dissected structures of the CantonS larvae. A PCR with a primer specific to the sano sequence was performed to obtain cDNAs by reverse transcription. All of the four transcripts were present in the different organs tested. AMC, Antenno-maxillary complex. C, The data represent the fold change in the amount of sano transcripts determined by qPCR. The CantonS strain was used as the wild-type reference, and the level of wild-type transcripts was defined as 1.0. The error bars represent the SEM, n = 3–10. The same letter over error bars indicates values that are not significantly different (ANOVA, p < 0.05).
Real-time PCR.
RNA from the various genotypes was extracted from 80 anterior portions of the third-instar larvae of either sex and treated with DNase to eliminate contamination by genomic DNA. The standard protocol was used for real-time PCR (Applied Biosystems, Roche). PCR primers were designed for the region spanning from exon 6 to exon 7: sano exon 6 forward, 5′-ggtgtccacaccgtcaaga; and sano exon 7 reverse, 5′-cactgccgtgaacgagtct.
Fluorescence and calcium imaging.
For GFP and RFP expression imaging, the anterior part of the larvae was dissected and incubated in a glycerol/1× PBS (50:50) mix for 1 h and mounted in Vectashield mounting medium (Kwon et al., 2011). Then direct observation of the fluorescence was performed using a confocal microscope (Leica TCS SP2).
For calcium imaging, living larvae of either sex were placed in 25 μl of distilled water between a cover slide and a perforated slide. The preparation was placed on an inverted confocal spectral Nikon C1Si microscope and observed with a 40× objective. Then, 25 μl of distilled water, 25 μl of 20 mm NaCl (to obtain a final concentration of 10 mm NaCl), or 25 μl of 600 mm NaCl (to obtain a final concentration of 300 mm NaCl) was injected through the hole in the slide. The fluorescence images (2 frames/s, 96 × 96 pixels) were acquired simultaneously using the Nikon EZ-C1 acquisition software and were analyzed using the Nikon EZ-C1 FreeViewer software. The FRET changes were measured as the ratio (R) of the donor over the acceptor emission intensities (427 nm/527 nm) and expressed as a percentage increase over the mean of the R values obtained during the measurement (ΔR/R0%). Data are expressed as the mean ± SEM.
Statistical analysis.
For each experiment, data were presented as mean ± SEM. All statistical analyses were performed with Prism 5 software for Mac OSX (GraphPad Software). Statistical analysis was generally made using either Student's t test or, for multiple comparisons, one-way ANOVA, followed by Bonferroni's post hoc test. p values < 0.05 were considered to be statistically significant.
Results
Identification of the sano gene
We identified the sano gene from a genetic screen of a P-Gal4 insertion collection based on both expression in larval chemosensory organs and the gustatory choice behavior defects observed in viable homozygous mutants. Among several selected lines, one P-Gal4 line (later called sanoGal4), when combined with 10XUAS–IVS–mCD8::GFP, showed expression in a few GRNs (5.5 ± 0.24; n = 10) in the TO and in three neurons in the ventral pharyngeal sense organ (VPS) of third-instar larvae (Fig. 1Aa,Ab). We also observed expression in a few neurons along the body. Then, we studied gustatory choice behavior on early third-instar sano mutant larvae with different attractive (100 mm sucrose) or aversive (10 mm caffeine and 10 mm quinine) substances and NaCl at different concentrations ranging from 10 to 500 mm. In a dose–response behavioral analysis toward NaCl, we observed that the homozygous larvae from the three mutant strains (sanoGal4, sanoGE12233, and sanoGE15762) showed a response to low-salt concentrations (at 10 and 50 mm NaCl) similar to that observed for wild-type or sanoGal4/+ (Fig. 1B). At 100 mm NaCl, sano mutant larvae displayed an attractive response, whereas wild-type larvae showed no preference. For higher NaCl concentrations (200, 300, and 500 mm), mutant larvae showed strong defects in the aversion to NaCl. To verify that the behavioral defect was related to the reduction of sano expression, we generated a UAS–sano construct containing the cDNA of the RB transcript and introduced it into a sanoGal4 mutant background (Fig. 2A). The defects in aversion could then be rescued; therefore, these defects are likely attributable to sano misexpression (Fig. 1B). sano mutant larvae, such as wild-type larvae, were attracted by 100 mm sucrose and were repelled by 10 mm caffeine or 10 mm quinine, showing that the GRNs of the mutant larvae were functional and could detect other aversive and attractive substances (Fig. 1C).
Figure 1.
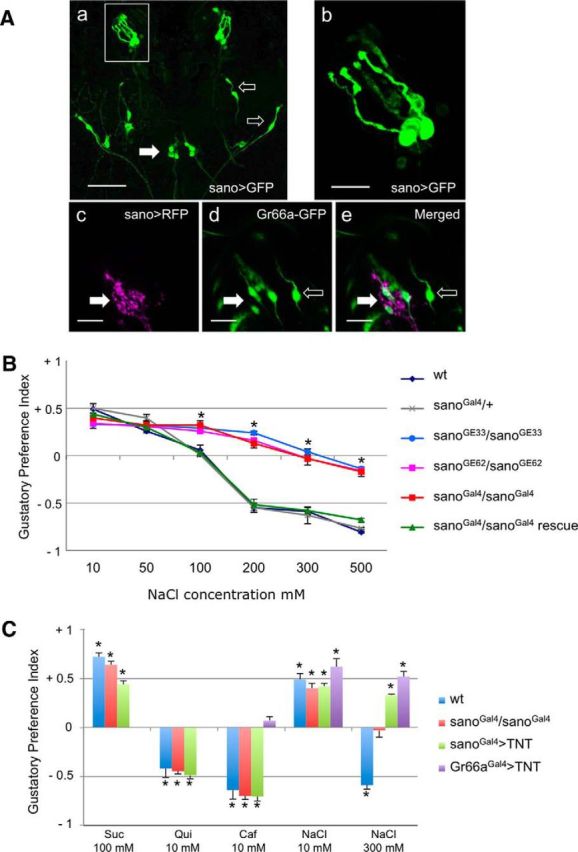
Role of sano in taste perception. Aa, Ab, Ventral view of the anterior part of a third-instar sanoGal4/sanoGal4;10XUAS–IVS–mCD8::GFP/10XUAS–IVS-mCD8::GFP larva. Aa, Expression is observed in gustatory neurons: five neurons in the TO (box) and three neurons in the ventral pharyngeal sense organ (filled arrow). Expression is also present in few neurons along the body (open arrows). Scale bar, 50 μm. Ab, Detailed view of the expression in five neurons of the TO. Scale bar, 10 μm. Ac–Ae, Ventral view of the anterior part of a third-instar sanoGal4/+;10XUAS–IVS–mCD8::RFP/Gr66a–GFP larva. Ac, sanoGal4 drives expression of RFP in six neurons of the TO ganglion. Ad, Gr66a–GFP drives expression of GFP in four neurons of the TO ganglion (filled arrow) and in two adjacent neurons of the TO dorsolateral group (open arrow). Ae, sanoGal4 and Gr66a–GFP are coexpressed in four neurons of the TO ganglion (filled arrow). B, Behavioral responses of third-instar larvae to NaCl concentrations. The GPIs for indicated NaCl concentrations versus water were calculated. For each condition, 10 trials with 50 larvae were performed. Control lines (wild-type; sanoGal4/+), sano mutants (sanoGal4/sanoGal4; sanoGE12233/sanoGE12233; sanoGE15762/sanoGE15762), and the sano rescue line (sanoGal4/sanoGal4;UAS–sano/+) were used in this experiment. Each bar represents a mean ± SEM of GPI (n = 10). Asterisks above error bars indicate that results were significantly different (ANOVA, p < 0.05). C, Behavioral responses of third-instar larvae to attractive [100 mm sucrose (Suc) or 10 mm NaCl] or repulsive [10 mm caffeine (Caf), 10 mm quinine (Qui), or 300 mm NaCl] tastants. Control larvae (wild-type), sano mutant larvae (sanoGal4/sanoGal4), and larvae with impaired gustatory neurons (sanoGal4>TNT or Gr66aGal4>TNT) were tested. Each point represents the mean ± SEM of GPI obtained from 10 trials of 50 larvae. A Student's t test comparing with the theoretical value 0 was performed. Asterisks above error bars indicate that results were significantly different (p < 0.05).
Using PCR, we located the P-Gal4 insertion 104 bp upstream of a transcription start site for CG12758 (serrano). Five transcripts (RA, RB, RC, RD, and RE) were predicted, with at least two transcription start sites separated by 70 kb (Fig. 2A). All of the transcripts code for the same predicted protein of 778 aa. The Sano protein is a member of the uncharacterized Themis protein family (Chung et al., 2009; Johnson et al., 2009). Members of this family are present in animals from cnidarians to mammals and contain at least one cysteine-rich CABIT domain and a low-complexity proline-rich stretch (Chung et al., 2009; Johnson et al., 2009). In Drosophila, Sano interacts with Dishevelled, a planar cell polarity regulator, to control tracheal tube length during development (Chung et al., 2009).
The sano coding region spans over 80 kb and contains a second predicted transcription start site. To determine whether some of the putative transcripts were specifically present in the larval chemosensory organs, we examined sano expression by RT-PCR in several larval tissues. To detect the different transcripts, we selected specific primers located in exons spanning other introns (Fig. 2A). The combination of different primers allowed for the amplification of the cDNA fragments corresponding to each transcript. At least four sano transcripts (RA, RB, RC, and RD were detected, but RE was not) were expressed and detectable in several parts of the larvae, including the most anterior part of the larvae, which encompasses the chemosensory organs, the brain, and the salivary glands (Fig. 2B). This result indicated that both of the transcription start sites were activated in these different structures, resulting in the presence of all of the transcripts. To confirm that the P-Gal4 element insertion in the sano locus caused the gustatory choice behavior defects observed in the sanoGal4 larvae, we isolated two other transgenic lines containing P-UAS element insertions, which were located 147 bp downstream of the first transcription start site (sanoGE12233) and 352 bp upstream of the second transcription start site (sanoGE15762). Homozygous larvae for these P-UAS insertions were viable and showed salt perception defects similar to those observed for the sanoGal4 mutants (Fig. 1B). Because all of these insertions were located close to the sano gene or in the coding region, they should have disturbed and reduced sano transcription. Accordingly, we measured a significant 3.94-fold decrease of sano expression by qPCR in the sanoGal4 mutant compared with the wild-type Drosophila (Fig. 2C). When we introduced an UAS–sano construct in a sanoGal4 mutant background, we induced a significant 4.54-fold increase in the sano transcript levels (Fig. 2B). Therefore, the sanoGal4 mutant behavioral defects are attributable to the reduction of sano expression and can be rescued inducing sano expression in the sanoGal4-expressing GRNs.
sanoGal4 is expressed in the gustatory neurons required for the aversive response to salt
Several chemosensory organs are located in the anterior part of the larval body (DO, TO, and VO) and along the larval pharynx (dorsal, ventral, and posterior pharyngeal sense organs; Colomb et al., 2007; Gerber et al., 2009). Previous studies have associated salt detection with the TO (Heimbeck et al., 1999; Liu et al., 2003). When combined with a 10XUAS–IVS–mCD8::GFP transgene, sanoGal4 expression was detectable in 5.5 neurons of the TO and 3 neurons in the VPS (Fig. 1Aa,Ab). To better characterize sano-expressing neurons in the TO, we compared sanoGal4>RFP and Gr66a–GFP expression in larvae. We observed that sano and Gr66a are coexpressed in four neurons, and sano is expressed alone in two neurons in the TO ganglion. Gr66a is expressed alone in two neurons that project their dendrites to the terminal dome but located in the DO ganglion (TO dorsolateral group; Kwon et al., 2011; Fig. 1Ac–Ae).
To confirm that sanoGal4 is specifically expressed in gustatory neurons involved in high-salt detection, we used an UAS–TNT transgene to block synaptic transmission and investigate taste perception defects in the presence of attractive or repellent substances (Sweeney et al., 1995; Heimbeck et al., 1999). We tested the behavioral responses of larvae from the sanoGal4 × UAS–TNT cross for 100 mm sucrose, bitter compounds (10 mm caffeine and 10 mm quinine), and two NaCl concentrations (10 and 300 mm). Mutant larvae showed responses similar to those observed with wild-type larvae toward sucrose, caffeine, quinine, and 10 mm NaCl (Fig. 1C). However, mutant larvae showed an avoidance defects toward 300 mm NaCl (Fig. 1C). More intriguingly, sanoGal4 × UAS–TNT larvae showed the same response toward low- and high-NaCl concentrations; they were also attracted by high-salt concentration. We also tested the behavioral responses of larvae from the Gr66aGal4 × UAS–TNT cross for 10 mm caffeine and low- and high-NaCl concentrations. Mutant larvae show no preference for caffeine as expected and attraction to low-salt concentration. Surprisingly, they also showed an attraction to high-NaCl concentration (Fig. 1C). Our results showed that the inactivation of neural transmission in the Gr66aGal4-expressing neurons can block the avoidance to caffeine and high-salt concentrations, whereas blocking neural transmission in the sanoGal4-expressing neurons specifically blocked the avoidance toward high-NaCl concentration.
sano is involved in high-salt perception
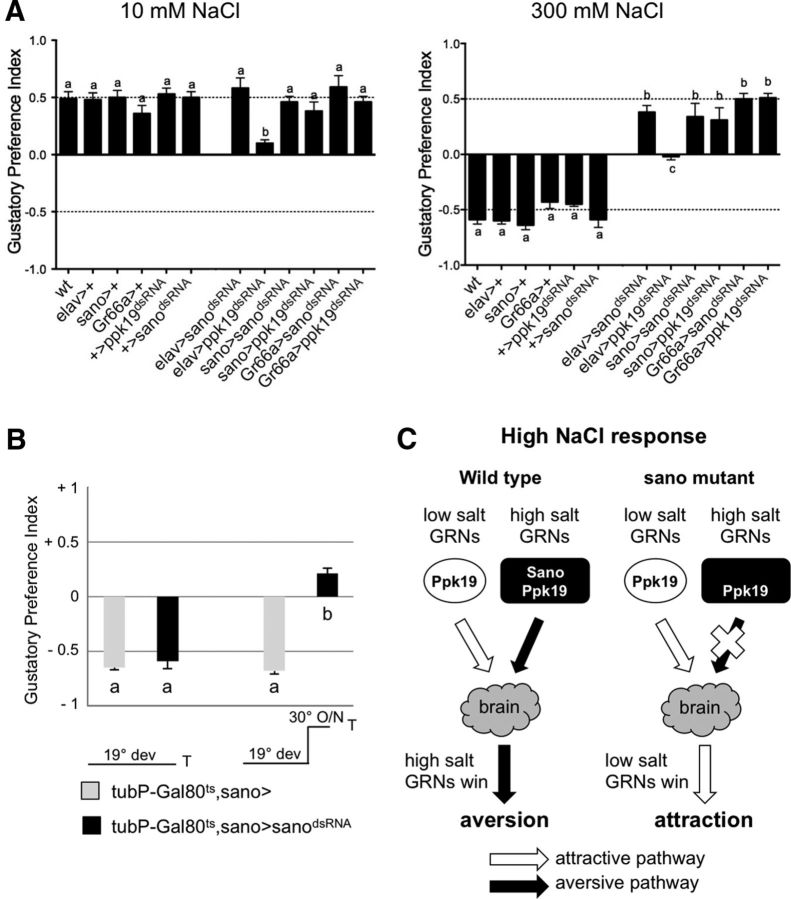
To confirm the role of sano in salt perception, we generated transgenic lines containing a dsRNA construct that included part of exon 7. This construct was intended to induce the degradation of all the sano transcripts. When we activated the expression of this sanodsRNA construct using sanoGal4 as a driver, we observed a fivefold decrease of sano mRNA levels detected by a qPCR analysis on dissected anterior parts of larvae (Fig. 2C). We then tested the ability of larvae expressing sanodsRNA to discriminate between attractive or repellent compounds. As predicted, larvae from the sanoGal4 × UAS–sanodsRNA cross showed responses similar to the wild-type larvae toward 10 mm NaCl, 10 mm sucrose, and 10 mm caffeine (Fig. 3A; data not shown). Similarly, larvae from the elavGal4 × UAS–sanodsRNA cross and Gr66aGal4 × UAS–sanodsRNA cross also showed responses similar to the wild-type larvae toward 10 mm NaCl, 10 mm sucrose, and 10 mm caffeine (Fig. 3A; data not shown). In the presence of 300 mm NaCl, larvae expressing UAS–sanodsRNA in sanoGal4 or Gr66aGal4 neurons or all the neurons showed an attractive response, whereas wild-type larvae showed an aversive response (Fig. 3A). These results are similar to the response obtained when neural transmission is blocked by expressing TNT in sanoGal4- or Gr66aGal4-expressing neurons.
Figure 3.
Disruption of sano and ppk19 by dsRNA. A, Behavioral assay for 10 mm NaCl and 300 mm NaCl. The effects of sano or ppk19 disruption in all larval neurons (elavGal4 crosses), sano-expressing neurons (sanoGal4 crosses), or Gr66a-expressing neurons (Gr66aGal4 crosses) were tested in these experiments. Each histogram (mean ± SEM of GPI) was calculated from 10 trials. The same letter over error bars indicates values that were not significantly different (ANOVA, p < 0.05). B, The effects of the temporal disruption of Sano on aversive behavior at 300 mm NaCl. Two temperature conditions were used. The offspring from the crosses were raised at 19°C (19° dev). At 19°C, GAL4 activity is repressed by GAL80ts such that sano transcripts are not impaired in tubP–Gal80ts,sano>sanodsRNA larvae. When tubP–Gal80ts,sano>sanodsRNA larvae are transferred to a permissive temperature of 30°C overnight (O/N), sanoGal4 drives expression of sanodsRNA. Each histogram (mean ± SEM of GPI) was calculated from 10 trials. The same letter over the error bars indicates values that were not significantly different (ANOVA, p < 0.05). C, Schematic model showing that the loss of Sano selectively impairs the aversive salt taste pathway. ppk19 is required in both low- and high-salt GRNs to detect NaCl. sano is required in high-salt GRNs to detect high-NaCl concentrations. In wild-type context, both low- and high-salt GRNs are activated by high-NaCl concentration, leading to aversion. Loss of sano selectively disrupts the aversive pathway, leading to attraction in the presence of high-NaCl concentration.
To show that the gustatory choice behavior defects observed for the sano mutant could be attributable to a late disruption of sano expression during the third-larval instar, we investigated the effect of a temporal controlled inactivation of sano expression on NaCl perception. We performed temperature-shifting experiments on larvae bearing sanoGal4, a sanodsRNA construct, and a tubP–Gal80ts transgene. The Gal80ts protein can interact with and block Gal4 at low temperatures (19°C) but is inactivated at a higher temperature (30°C), allowing the Gal4 protein to activate sanodsRNA expression (McGuire et al., 2003). When the larvae were reared at 19°C, sanodsRNA was not expressed, and we observed an aversive response of the larvae to 300 mm NaCl similar to the wild-type response (Fig. 3B). When the larvae were transferred to 30°C overnight before the tests, sanodsRNA expression is activated and larvae showed behavioral defects toward high-salt concentrations (Fig. 3B). Larvae showed an attractive response similar to that observed for the larvae from the sanoGal4 × UAS–sanodsRNA cross. These results suggest that sano is expressed during the third-larval instar and is required for the activity of the gustatory neurons that detect high-salt concentrations.
Thus, the reduction of sano expression in sanoGal4- or Gr66aGal4-expressing gustatory neurons or in the whole nervous system led to a similar disruption of high-NaCl perception in larvae.
ppk19 is involved in low- and high-salt perception
ppk19 has been implicated in the perception of both low- and high-NaCl concentration at larval instar (Liu et al., 2003). In our behavioral tests, we showed that expression of a ppk19dsRNA construct in all the neurons using an elavGal4 driver led to the absence of a preference toward low- or high-NaCl concentrations (Fig. 3A). The expression of ppk19dsRNA using sanoGal4 or Gr66aGal4 did not affect the attraction toward 10 mm NaCl (Fig. 3A). However, disruption of ppk19 in sanoGal4 neurons induced an attractive response to 300 mm NaCl (Fig. 3A). Thus, loss of ppk19 in sanoGal4 or Gr66aGal4 neurons only impaired avoidance to high-NaCl concentration but not attraction attributable to low-salt perception.
Sano and PPK19 regulate the neuronal activity of taste neurons
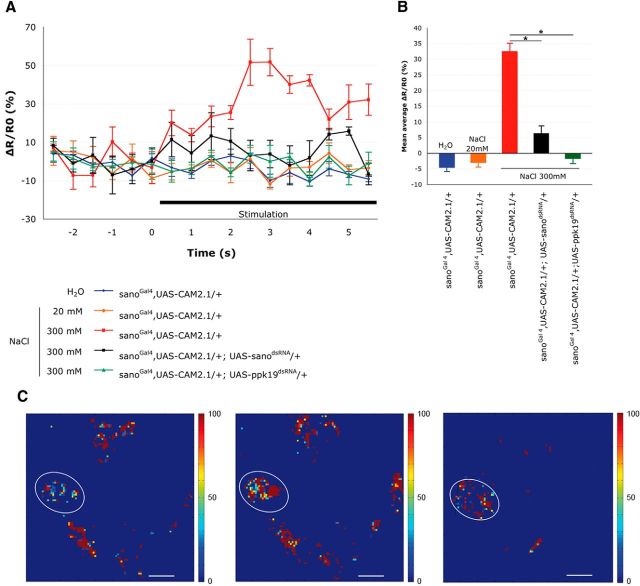
To provide physiological evidence showing that sano is required for NaCl detection, a calcium-sensitive protein, Cam2.1, was expressed in larval taste neurons to monitor Ca2+ levels in response to NaCl (Diegelmann et al., 2002). Expression of this protein was driven by the sanoGal4 line and was thus expressed in a subset of neurons in the TO (Fig. 1A). Fluorescence levels were analyzed in the cell bodies of these neurons in the TO that expressed the Cam2.1 protein. This allowed for the precise analysis of the calcium levels after stimulation with different concentrations of NaCl. In wild-type larvae, when a solution of 20 mm NaCl was applied, a variation of fluorescence was observed that was not significantly different from the variation observed in the controls without NaCl (Fig. 4). With the application of a 300 mm NaCl solution, a significant increase (50%) of the fluorescence was observed after 2 s of exposure. This result indicated that the observed neurons were sensitive to a high concentration of NaCl and respond to an increase of the Ca2+ level. To determine the role of sano and ppk19 in the activation of these neurons, an UAS–sanodsRNA or a UAS–ppk19dsRNA construct was then introduced into the sanoGal4;UAS–Cam2.1 background. In these two genetic contexts, the signal-to-noise level remained low after the addition of a 300 mm NaCl solution and close to the control levels in the neurons stimulated by high-NaCl concentrations. These data showed that PPK19 and Sano are required in sanoGal4 neurons to induce an increase of the intracellular calcium levels in response to high-NaCl concentrations. Therefore, PPK19 and Sano are both required for the activity of gustatory neurons in the TO that are involved in the detection of high-salt concentrations.
Figure 4.
Sano and PPK19 are required for TO neuron activation by high-NaCl concentrations. A, Representative fluorescence change (ΔR/R0) of CAM2.1 expressed in sanoGal4 neurons in control and mutant larvae. Neuronal activity was measured before and after the addition of different stimuli: distilled water, 20 mm NaCl, or 300 mm NaCl. Responses to 300 mm NaCl were measured in larvae expressing either sanodsRNA or ppk19dsRNA in sanoGal4 neurons (n = 4–6). The error bars represent the SEM. B, Mean FRET ratio changes. Changes were measured during the first 5 s of the stimulation of different control and mutant larvae (n = 4–6). The error bars represent the SEM. Asterisks indicate that results are significantly different (Student's t test, p < 0.05). C, Peak responses are shown in false-color scale (ΔR/R0%, right of images). Left, sanoGAL4,UAS–CAM2.1/+ before stimulation. Middle, sanoGAL4,UAS–CAM2.1/+ at 2 s after stimulation with 300 mm NaCl. Right, sanoGAL4,UAS–CAM2.1/+;UAS--sanodsRNA/+ at 2 s after stimulation with 300 mm NaCl. The cell bodies of the neurons used for the FRET measurements are circled. Scale bar, 10 μm.
Discussion
sano is expressed in the neurons that detect high-salt concentrations
Our behavioral results on larvae showed that sanoGal4 is expressed in a group of GRNs in the TO that are specifically involved in the detection of high-NaCl concentrations. Because sano and Gr66a are coexpressed in four neurons in the TO, we propose that these neurons are specifically required for high-salt detection. The attractive behavioral response of larvae observed when these neurons are inactivated showed that other gustatory neurons are still able to detect NaCl. This result strongly suggests that two distinct types of neurons respond simultaneously to either low- or high-salt concentrations to generate an attractive or aversive response (Fig. 3C). Recently, Zhang et al. (2013) proposed a model in which competition between low-salt GRNs and high-salt GRNs in S- and L-type sensilla results in the bidirectional behavioral responses to salt in adults. They showed that loss of Ir76b selectively disrupted the attractive salt pathway and left the aversive salt pathway intact, leading to an aversive response to low-salt concentrations. In our experiments, we observed that inactivation of sano-expressing GRNs induced an attraction to high-salt concentrations, suggesting that the aversive salt pathway is disrupted and the attractive salt pathway remained unaffected. Thus, we conclude that, in larvae as in adults, a bimodal switch system determines the opposing behavioral responses to low- and high-salt; sano is only expressed in the neurons of the aversive salt pathway.
Previous results have demonstrated that the TO shows only weak electrophysiological responses to 100 mm NaCl (Oppliger et al., 2000). Here, using the FRET technique, we showed that the sano-expressing neurons are strongly stimulated by 300 mm NaCl but not by a low concentration of NaCl. Therefore, sanoGal4 neurons are involved in the detection of high-NaCl concentrations in larvae. Thus, the sanoGal4 line is characteristic of larval sensory neurons required for the perception of high-NaCl concentrations and provides an excellent genetic tool to specifically target these neurons for additional cellular and molecular study.
Disruption of sano impairs the aversive response to high-NaCl concentrations
Larvae with impaired sano function display behavioral defects in the presence of high-salt concentrations. Tests performed with caffeine and quinine provided evidence that the sano mutant larvae were able to detect and avoid other aversive molecules, suggesting that Sano does not play a role in the detection of aversive molecules in general. Attractive behaviors to sucrose or low-salt concentrations were also not affected. Similarly, the disruption of Sano function using UAS–sanodsRNA constructs under the control of sanoGal4, Gr66aGal4, or elavGal4 only caused the loss of high-salt detection in larvae. Thus, the disruption of Sano throughout the whole nervous system or in a subset of GRNs (including bitter GRNs) only affects the perception of high-NaCl concentrations.
Studies on the ppk gene family have shown that PPK19 is required for the detection of low- and high-salt concentrations (Liu et al., 2003). Our results highlight the particular role of Sano in the detection of high-NaCl concentrations; thus, the sano gene is the first characterized gene specifically required for an aversive response to salt.
Sano is involved in the activity of high-salt larval GRNs
Little is known about the cellular mechanisms of salt detection by GRNs. Here, we identified a protein that appears to be cytoplasmic and is part of the cellular mechanism required in high-salt detecting neurons. In mammals, intracellular calcium levels are modulated in response to chemosensory stimuli, such as salt, in taste-receptor cells (Chandrashekar et al., 2010). In Drosophila, calcium imaging has been used in adults to measure neuronal activity in response to sugars, bitter substances, and NaCl (Marella et al., 2006). Our results show that calcium levels are increased in sano-expressing neurons in the TO in response to high-salt concentrations. This increase in GRN activity requires PPK19 and Sano. Then Sano is involved in the detection of high salt by GRNs in the TO.
Previous studies have shown that Sano is a cytoplasmic protein that interacts with other proteins, such as Grb2 (a protein involved in the signaling pathways of tracheal and wing development; Chung et al., 2009). Moreover, a molecular interaction with Epac (exchange protein directly activated by cAMP) has been detected in a two-hybrid study (Giot et al., 2003). Epac is a member of the Rap1 signal transduction pathway and is involved in cell adhesion and differentiation, as well as in neuronal activity by regulating calcium levels or neurotransmitter release (Gloerich and Bos, 2010). The genetic and molecular interactions between Sano and Epac remain to be fully demonstrated by future in vitro and in vivo studies and may provide new insights into the cellular mechanisms taking place downstream of the DEG/ENaC channels in salt-detecting neurons.
Footnotes
This work was supported by grants from the National Center of Scientific Research and the Regional Council of Burgundy (PARI, Regional Action Plan for Innovation). We thank Isabelle Chauvel for technical assistance and Yann Roche and Christine Arnould from the Diamacell platform for spectral image analysis and technical assistance for the confocal microscopy.
The authors declare no competing financial interests.
References
- Amrein H, Thorne N. Gustatory perception and behavior in Drosophila melanogaster. Curr Biol. 2005;15:R673–R684. doi: 10.1016/j.sbi.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Balakireva M, Gendre N, Stocker RF, Ferveur JF. The genetic variant Voila causes gustatory defects during Drosophila development. J Neurosci. 2000;20:3425–3433. doi: 10.1523/JNEUROSCI.20-09-03425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J, Kuhn C, Oka Y, Yarmolinsky DA, Hummler E, Ryba NJ, Zuker CS. The cells and peripheral representation of sodium taste in mice. Nature. 2010;464:297–301. doi: 10.1038/nature08783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Vining MS, Bradley PL, Chan CC, Wharton KA, Jr, Andrew DJ. Serrano (sano) functions with the planar cell polarity genes to control tracheal tube length. PLoS Genet. 2009;5:e1000746. doi: 10.1371/journal.pgen.1000746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomb J, Grillenzoni N, Ramaekers A, Stocker RF. Architecture of the primary taste center of Drosophila melanogaster larvae. J Comp Neurol. 2007;502:834–847. doi: 10.1002/cne.21312. [DOI] [PubMed] [Google Scholar]
- Diegelmann S, Fiala A, Leibold C, Spall T, Buchner E. Transgenic flies expressing the fluorescence calcium sensor Cameleon 2.1 under UAS control. Genesis. 2002;34:95–98. doi: 10.1002/gene.10112. [DOI] [PubMed] [Google Scholar]
- Gerber B, Stocker RF. The Drosophila larva as a model for studying chemosensation and chemosensory learning: a review. Chem Senses. 2007;32:65–89. doi: 10.1093/chemse/bjl030. [DOI] [PubMed] [Google Scholar]
- Gerber B, Stocker RF, Tanimura T, Thum AS. Smelling, tasting, learning: Drosophila as a study case. Results Probl Cell Differ. 2009;47:139–185. doi: 10.1007/400_2008_9. [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Gloerich M, Bos JL. Epac: defining a new mechanism for cAMP action. Annu Rev Pharmacol Toxicol. 2010;50:355–375. doi: 10.1146/annurev.pharmtox.010909.105714. [DOI] [PubMed] [Google Scholar]
- Heimbeck G, Bugnon V, Gendre N, Häberlin C, Stocker RF. Smell and taste perception in Drosophila melanogaster larva: toxin expression studies in chemosensory neurons. J Neurosci. 1999;19:6599–6609. doi: 10.1523/JNEUROSCI.19-15-06599.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi M, Meunier N, Marion-Poll F, Tanimura T. Two antagonistic gustatory receptor neurons responding to sweet-salty and bitter taste in Drosophila. J Neurobiol. 2004;61:333–342. doi: 10.1002/neu.20063. [DOI] [PubMed] [Google Scholar]
- Ishimoto H, Tanimura T. Molecular neurophysiology of taste in Drosophila. Cell Mol Life Sci. 2004;61:10–18. doi: 10.1007/s00018-003-3182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AL, Aravind L, Shulzhenko N, Morgun A, Choi SY, Crockford TL, Lambe T, Domaschenz H, Kucharska EM, Zheng L, Vinuesa CG, Lenardo MJ, Goodnow CC, Cornall RJ, Schwartz RH. Themis is a member of a new metazoan gene family and is required for the completion of thymocyte positive selection. Nat Immunol. 2009;10:831–839. doi: 10.1038/ni.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon JY, Dahanukar A, Weiss LA, Carlson JR. Molecular and cellular organization of the taste system in the Drosophila larva. J Neurosci. 2011;31:15300–15309. doi: 10.1523/JNEUROSCI.3363-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/S1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Liu L, Leonard AS, Motto DG, Feller MA, Price MP, Johnson WA, Welsh MJ. Contribution of Drosophila DEG/ENaC genes to salt taste. Neuron. 2003;39:133–146. doi: 10.1016/S0896-6273(03)00394-5. [DOI] [PubMed] [Google Scholar]
- Marella S, Fischler W, Kong P, Asgarian S, Rueckert E, Scott K. Imaging taste responses in the fly brain reveals a functional map of taste category and behavior. Neuron. 2006;49:285–295. doi: 10.1016/j.neuron.2005.11.037. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Baldwin D, Hannaford S, Palka J, Montell C. Defective proboscis extension response (DPR), a member of the Ig superfamily required for the gustatory response to salt. J Neurosci. 2002;22:3463–3472. doi: 10.1523/JNEUROSCI.22-09-03463.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewalda T, Singhal N, Fiala A, Saumweber T, Wegener S, Gerber B. Salt processing in larval Drosophila: choice, feeding, and learning shift from appetitive to aversive in a concentration-dependent way. Chem Senses. 2008;33:685–692. doi: 10.1093/chemse/bjn037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppliger FY, Guerin PM, Vlimant M. Neurophysiological and behavioural evidence for an olfactory function for the dorsal organ and a gustatory one for the terminal organ in Drosophila melanogaster larvae. J Insect Physiol. 2000;46:135–144. doi: 10.1016/S0022-1910(99)00109-2. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Russell C, Wessnitzer J, Young JM, Armstrong JD, Webb B. Dietary salt levels affect salt preference and learning in larval Drosophila. PLoS One. 2011;6:e20100. doi: 10.1371/journal.pone.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney ST, Broadie K, Keane J, Niemann H, O'Kane CJ. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron. 1995;14:341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- Zhang YV, Ni J, Montell C. The molecular basis for attractive salt-taste coding in Drosophila. Science. 2013;340:1334–1338. doi: 10.1126/science.1234133. [DOI] [PMC free article] [PubMed] [Google Scholar]