Abstract
Purpose
To investigate the role and related mechanisms of miR-106a in sepsis-induced AKI.
Methods
Serum from sepsis and healthy patients was collected, sepsis mouse model was established by cecal ligation and puncture (CLP). TCMK-1 cells were treated with lipopolysaccharide (LPS) and transfected with THBS2-small interfering RNA (siTHBS2), miR-106a inhibitor, miR-106a mimics and their negative controls (NCs). The expression of miR-106a, thrombospondin 2 (THBS2), Bax, cleaved caspase-3 and Bcl-2, cell viability, relative caspase-3 activity and TNF-α, IL-1β, IL-6 content were respectively detected by quantitative real-time polymerase chain reaction (qRT-PCR), western blotting, Cell Counting Kit-8 (CCK-8) and enzyme linked immunosorbent assay (ELISA). The relationship between miR-106a and THBS2 was confirmed by dual luciferase reporter assay.
Results
MiR-106a was up-regulated in serum of sepsis patients, CLP-induced mice models and LPS-induced TCMK-1 cells. LPS reduced cell viability and Bcl-2 expression, and increased caspase-3 activity, Bax expression, the content of TNF-α, IL-1β, IL-6. THBS2 was a target of miR-106a. The decreases of caspase-3 activity, TNF-α, IL-1β, IL-6, Bax expression and the increases of cell viability, Bcl-2 expression caused by miR-106a knockdown were reversed when THBS2 silencing in LPS-stimulated TCMK-1 cells.
Conclusion
MiR-106a aggravated LPS-induced inflammation and apoptosis of TCMK-1 cells via regulating THBS2 expression.
Keywords: Sepsis, Acute Kidney Injury, Lipopolysaccharides, Mice
Introduction
Sepsis is a systemic inflammatory response syndrome caused by infection or highly suspicious infection, and is also a major cause of multiple organ failure and septic shock until death1,2. The uncontrolled inflammatory response and immune dysfunction of the body are the main pathophysiological basis of sepsis2. The degree of excessive inflammation and immune dysfunction directly affect the occurrence and development of sepsis2. Moreover, acute kidney injury (AKI) is one of the most common organ failure symptoms in sepsis3. The pathways of AKI induced by sepsis may include coagulopathy, inflammation, oxidative stress and changes in renal tubular epithelial cells to injury4. However, the mechanism of these effects is complex and there is no clear research report at present.
MicroRNAs (miRNAs) are non-coding endogenous RNAs that exert translational inhibition by targeting messenger RNA (mRNA), and have been shown to regulate various stages of sepsis5-7. A number of studies have shown that miRNAs disorders are associated with clinical symptoms of inflammation and sepsis6-8. For example, the up-regulation of miR-21-3p promotes sepsis-induced cardiac dysfunction6. MiR-15a/16 is up-regulated in serum of neonatal sepsis patients, and inhibits LPS-induced inflammatory signaling pathway activity7. MiR-128 expression decreased, while miR-21 levels increased in serum and LPS-induced podocytes of patients with sepsis, leading to podocyte damage in sepsis8. The above studies show that miRNAs play a crucial role in regulating immune response and sepsis. Therefore, understanding the potential mechanism of miRNAs in sepsis will help to better treat this disease. In addition, it has been reported that miR-106a expression is up-regulated in the serum of sepsis mice and inflammatory bowel disease mice model, and then the absence of miR-106a can alleviate the inflammatory response9,10. However, the role of miRNA-106 in sepsis-induced AKI remains unclear.
Thrombospondins (THBSs) is a protein family with a minimum of five members that participate in a variety of biological processes by combining some target proteins, such as cell motility, apoptosis, cytoskeletal formation, and can also serve as a response platform for extracellular matrices11,12. THBS2 is a relatively special member of THBSs family that has anti-angiogenic effects and interacts with various cellular receptors and growth factors to regulate cell proliferation, apoptosis and adhesion13. THBS2 has also been reported to act as important downstream targeting gene of miRNAs to form a miRNAs/THBS2 network for various diseases, such as cervical cancer, human myxoid liposarcoma and renal cancer14-16.
In this study, we collected serum from sepsis and healthy patients, constructed sepsis mouse models by cecal ligation and puncture (CLP) and stimulated mouse kidney epithelial TCMK-1 cells with LPS in vitro to investigate the role of miR-106a in sepsis-induced AKI and its related mechanisms, as well as the possible regulatory relationship with THBS2.
Methods
Between March, 2018 and January, 2019, a total of 50 patients (34 males and 16 females, mean age: 48±7.21 years) with sepsis who lived in Intensive Care Unit (ICU) and 30 healthy controls (21 males and 9 females, mean age: 42±5.46 years) were recruited in this study from The Affiliated Hospital of Hangzhou Normal University. The diagnosis of sepsis complied with the American College of Chest Physicians (ACCP) and the Society of Critical Care Medicine (SCCM) joint definition of sepsis diagnostic criteria in US17.
The venous blood (5 mL) was collected from patients with sepsis and healthy people, and then the whole blood was preserved by anticoagulation with 1 mL of Edathamil (EDTA-K2, Solarbio, Beijing, China). Serum was obtained by centrifugation after 4 mL EDTA-K2 coagulation and stored at -20°C. All patients signed the informed consent, and the experiment was approved by the Ethics Committee of The Affiliated Hospital of Hangzhou Normal University.
The establishment of the sepsis model
All animal experiences were approved by the Laboratory Animal Management Committee of Hangzhou Normal University Laboratory Animal Center.
A total of 12 adult clean Kunming mice aged 6-8 weeks and weighing 18-22g were provided by the Laboratory Animal Center of Chongqing Medical University (Chongqing, China). All mice were raised independently at 20-25°C and 50% humidity, and then were randomly divided into two groups: the sham group and the CLP group. Animals were fasted one day before the experiment and anesthetized with 0.3% sodium pentobarbital (30 mg/kg, Solarbio, Beijing, China). In the midline of the mouse, we cut the mouth about 1cm long, found the cecum and ligatured the root, retained the blood supply, and then used the suture to puncture the ligated cecum through the serosal surface of the blind end of the intestine opposite to the mesentery. Two holes were made once to extrude a little intestinal content, close the abdominal cavity layer by layer with 1-0 silk thread, and immediately subcutaneously inject 1 mL of normal saline to supplement the intraoperative fluid loss18 Animals in the sham group were treated in the same manner, but the cecum was not perforated and not ligated.
Cell culture and transfection
TCMK-1 mouse kidney epithelial cell line was purchased from BioVector NTCC Inc. (Beijing, China) and cultured in 90% high-glucose Dulbecco’s modified Eagle medium (DMEM, Solarbio, Beijing, China) and 10% fetal bovine serum (FBS, Solarbio, Beijing, China) at 37°C. After incubation for 24 h, cells were divided into two groups: one group was added with 5 mg/L of lipopolysaccharide (LPS, Solarbio, Beijing, China) to mimic inflammation, and the other group was added with equal amount of DMEM for 2 h at 37°C as control group.
HEK293 cell line was purchased from American Type Culture Collection (ATCC; Manassas, USA) and cultured in Eagle’s Minimum Essential Medium (EMEM, Solarbio, Beijing, China) with 10% FBS at 37°C for dual luciferase reporter assay.
THBS2-small interfering RNA (siTHBS2), miR-106a inhibitor, miR-106a mimics or their negative control (NC), siNC, NC inhibitor and NC mimics were added to cells cultured in 96-well plate to transfect for 48h at 37°C by Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, USA).
Cell viability
Cell viability was detected by Cell Counting Kit-8 (CCK-8, MedChemExpress, New Jersey, USA). Briefly, cells (100 μL/well) were seeded in 96-well plate for 24 h, added to 10 μL of CCK-8 solution and incubated in an incubator at 37°C for 3h. The absorbance at 450 nm (OD450) was detected by a microplate reader (Bio-Rad, Hercules, USA).
Enzyme linked immunosorbent assay (ELISA)
Cultured cells were centrifuged for 10 min to obtain the culture supernatant. The contents of tumor necrosis factor (TNF-α), interleukin 1 beta (IL-1β) and interleukin 6 (IL-6) were detected by ELISA Kit (Abcam, Cambridge, USA) following the instructions. The absorbance at 450 nm (OD450) was measured by a microplate reader.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Cells were lysed with Biozol Reagent (Biobee, Beijing, China), extracted with chloroform and isopropanol (Sustgreen, Nanjing, China), precipitated in 75% ethanol and washed to obtain total RNA. The synthesis of cDNA was performed using SYBR primeScriptTM RT-PCR Kit (TaKaRa, Dalian, China) according to the Kit instructions. The primers were designed by Primer 5.0 software (PREMIER Biosoft International, California, USA) and synthesized by TaKaRa Biotechnology Co., Ltd (Dalian, China). The specific sequences of primers were shown in Table 1. qRT-PCR was performed by SYBR Green I dye method on the PCR instrument according to the operation instructions of ExScriptTM RT-PCR Kit (TaKaRa, Dalian, China). Reaction conditions of PCR were as follow: pre-denaturation 95°C for 10s, denaturation 95°C for 5s, fluorescence detection 72°C for 10s, in total of 35 cycles. The data were calculated by 2- method 19.
Table 1. The sequence of primers.
| Gene | F/R | Primer Sequence (5’ to 3’) |
|---|---|---|
| miR-106a | F | AAAAGUGCUUACAGUGCAGGUAG |
| R | ACCUGCACUGUAAGCACUUUUUU | |
| TNF-α | F | ACCACGCTCTTCAGCCTACTG |
| R | ACGGGCATATCTGAGGTATGAG | |
| IL-1β | F | AGCTGACCCTAAACAGATGA |
| R | GATCTACACTCTCCAGCTGTAGC | |
| IL-6 | F | CCTCCAGGAACCCAGGTATGAA |
| R | TCAGGTGCCCCAGCTACATTAT | |
| THBS2 | F | CGTGGACAATGACCTTGTTG |
| R | GCCATCGTTGTCATCATCAG |
F: Forward; R: Reward
Western blotting
The total protein was obtained by cells lysis with RIPA Lysis Buffer (Haigene, Harbin, China), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose (NC) membrane. Then, the membrane was placed in 5% milk for 2 h at room temperature, washed in phosphate buffer saline (PBS). The B-cell lymphoma-2 (Bcl-2, ab32124), Bcl-2 associated X protein (Bax, ab182733), cleaved caspase-3 (ab2302), β-actin (ab179467) and THBS2 (ab84469) antibodies from Abcam were diluted 1,000 times and added into membranes to incubate at 4°C overnight. And then, anti-rabbit IgG antibody (1:2,000; Abcam) was added to incubate for 30 min at room temperature. Finally, the proteins on the membrane were detected by an imaging system (Bio-Rad, Hercules, USA).
Dual luciferase reporter assay
miR-106a mimics, NC-mimics, empty pmiR-GLO-NC, THBS2-wild type (WT) and THBS2-mutant (MUT) were transfected HEK293 cells by Lipofectamine 3000, and relative luciferase activity was tested by Dual-Luciferase® Reporter Assay System Protocol (Promega, Madison, USA).
Statistical analysis
All data were analyzed by SPSS 19.0 software, and students’ t-test was used for comparison between the two groups. The difference was significant when p<0.05, and then the results were represented by mean ± standard error (SE).
Results
MiR-106a was up-regulated in serum of sepsis patients and CLP-induced mice models
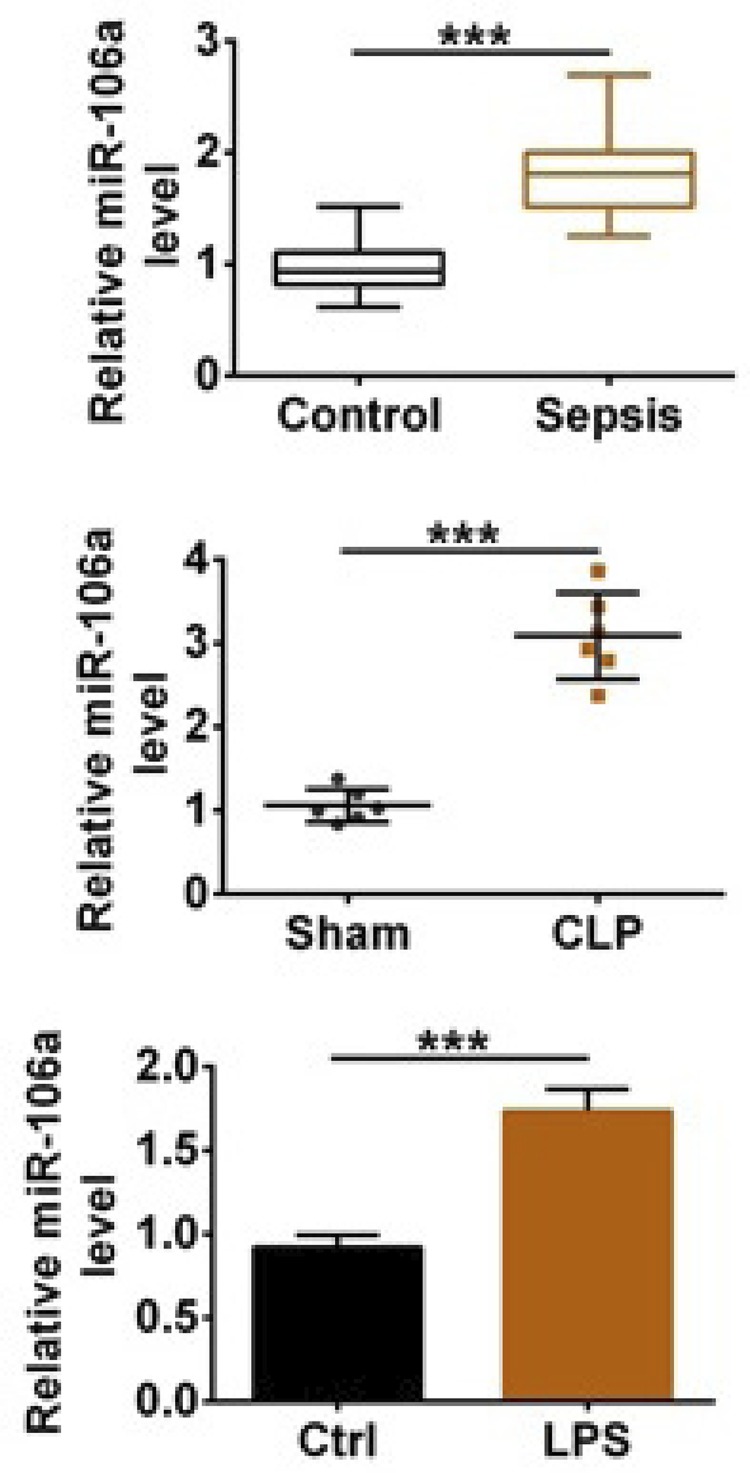
In order to study the expression level of miR-106a, we collected serum from sepsis patients and healthy patients, established CLP-induced sepsis model in mice and treated TCMK-1 cells with LPS. The results in Figure 1 showed that miR-106a expression level in the sepsis, CLP and LPS group was higher than that in the control or sham group (p<0.001), illustrating that sepsis and sepsis-induced inflammation promoted miR-106a up-regulation in vivo and in vitro.
Figure 1. MiR-106a was up-regulated in serum of sepsis patients and CLP-induced mice models. The relative miR-106a level was tested by qRT-PCR in the serum from sepsis and healthy patients, CLP-induced sepsis mouse models and TCMK-1 cells treated with LPS. *** p<0.001.
Knockdown of miR-106a reduced LPS-induced apoptosis in TCMK-1 cells
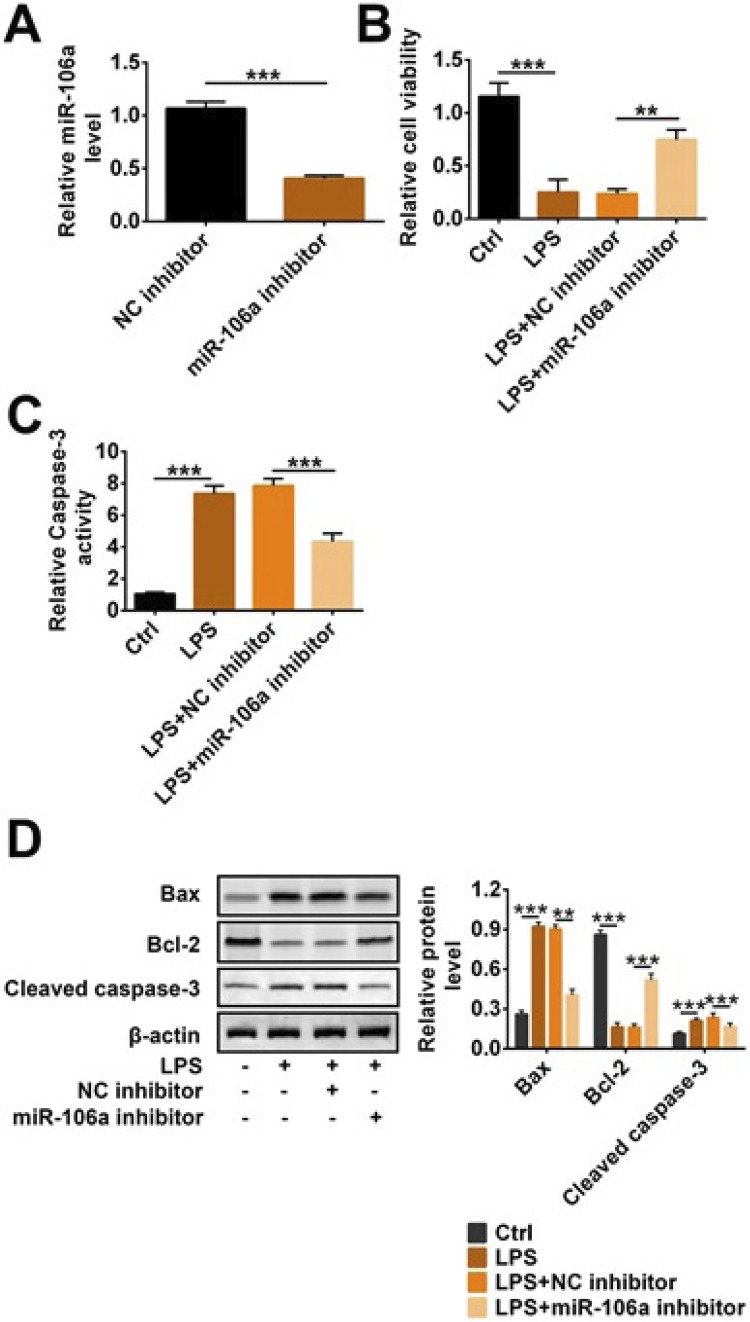
In order to investigate the mechanism and effect of miR-106a on sepsis-induced AKI, we used miR-106a inhibitor to transfect TCMK-1 cells treated with LPS. In Figure 2A, miR-106a expression level was significantly reduced in the miR-106a inhibitor group compared with the NC inhibitor group, indicating that miR-106a was efficiently transfected into cells (p<0.001). CCK-8 results in Figure 2B showed that cell viability was decreased in cells treated with only LPS, and this decrease was reversed after adding miR-106a (p<0.01 or p<0.001). On the contrary, caspase-3 activity in the LPS group was higher than that in the control group (p<0.001, Fig. 2C). But caspase-3 activity in the LPS+miR-106a inhibitor group was less than that in LPS+NC inhibitor (p<0.001, Fig. 2C). Moreover, western blotting results showed that the expression of Bax and cleaved caspase-3 were clearly increased and Bcl-2 expression was reduced when adding LPS; conversely, miR-106a inhibitor alleviated these increase and reduction (p<0.001, Fig. 2D). Above data indicated that miR-106a knockdown increased cell viability and decreased apoptosis-related protein expression in LPS-induced TCMK-1 cells.
Figure 2. Knockdown of miR-106a reduced LPS-induced apoptosis in TCMK-1 cells. miR-106a inhibitor, NC inhibitor and LPS were transfected into TCMK-1 cells. (A) Relative miR-106a level, (B) cell viability, (C) relative caspase-3 activity and (D) the protein level of Bax, cleaved caspase-3 and Bcl-2 were detected by qRT-PCR, CCK-8 and western blotting. ** p<0.01; *** p<0.001.
Knockdown of miR-106a reduced LPS-induced inflammatory factor levels in TCMK-1 cells
In order to further investigate the effects of miR-106a on sepsis, we selected three cellular inflammatory factors TNF-α, IL-1β, IL-6 as representatives for research. The results from qRT-PCR and ELISA in Figure 3A and B displayed that TNF-α, IL-1β, IL-6 expression and content were significantly increased in the LPS group compared with the control group (p<0.05, p<0.01 or p<0.001). After adding miR-106a inhibitor to LPS-induced cells, the expression and content of TNF-α, IL-1β, IL-6 were reduced, suggesting that miR-106a knockdown mitigated the increases of TNF-α, IL-1β, IL-6 caused by LPS.
Figure 3. Knockdown of miR-106a reduced LPS-induced inflammatory factor levels in TCMK-1 cells. miR-106a inhibitor, NC inhibitor and LPS were transfected into TCMK-1 cells. (A) The relative mRNA level and (B) content of TNF-α, IL-1β, IL-6 were detected by qRT-PCR and ELISA. * p<0.05; ** p<0.01; *** p<0.001.
THBS2 was a target of miR-106a
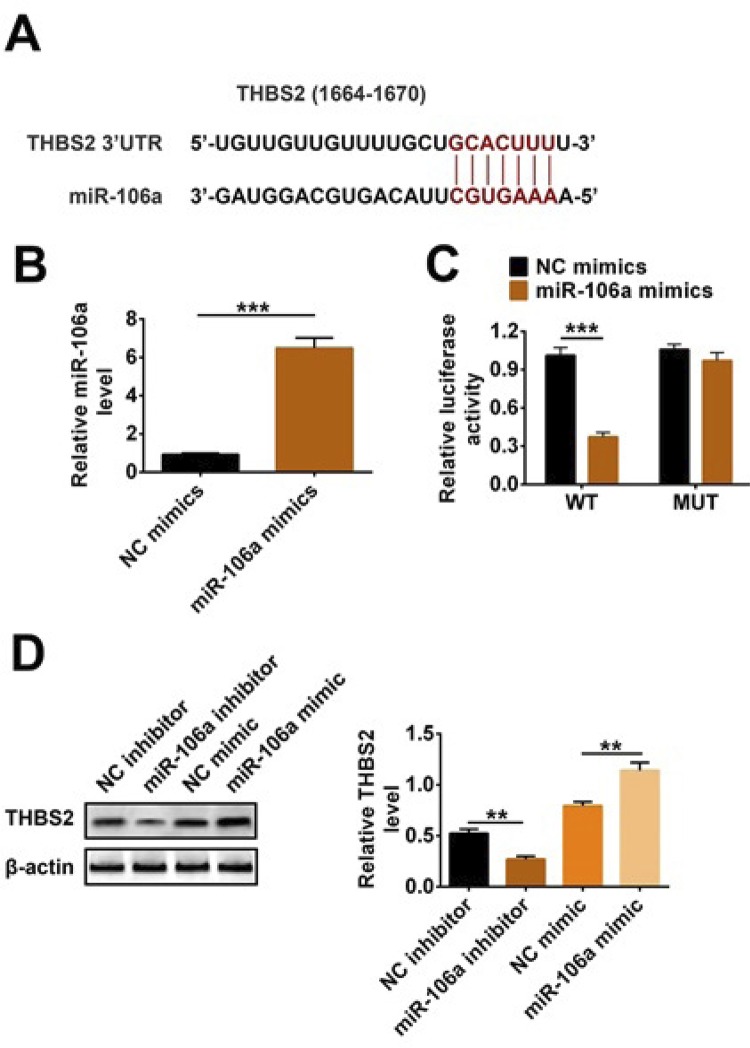
In this study, we used TargetScan website (http://www.targetscan.org) to find a potential target gene THBS2 downstream of miR-106a, and their binding site was shown in Figure 4A. Then, we also used double luciferase reporter assay to identify the targeting relationship between THBS2 and miR-106a. Figure 4B displayed that miR-106a mimics were successfully transfected into cells (p<0.001). In Figure 4C, relative luciferase activity in the miR-106a mimics+THBS2-WT group was less than that in the NC mimics+THBS2-WT group (p<0.001). However, relative luciferase activity did not change in the miR-106a mimics+THBS2-MUT group. The results in Figure 4C suggested that there was a targeting relationship between THBS2 and miR-106a. Furthermore, western blotting results showed that THBS2 expression level was up-regulated in the miR-106a inhibitor group in relation to the NC inhibitor group; conversely, it was clearly decreased when miR-106a overexpression in TCMK-1 cells (p<0.01). Hence, these results illustrated that THBS2 was a target of miR-106a and negatively regulated by miR-106a.
Figure 4. THBS2 was a target of miR-106a. MiR-106a inhibitor, NC inhibitor, and miR-106a mimics, NC mimics, THBS2-WT and THBS2-MUT were transfected into HEK293 and TCMK-1 cells. (A) The binding site was predicted by Targetscan website. (B) Relative miR-106a level was tested by qRT-PCR. (C) The target relationship between miR-106a and THBS2 was confirmed by dual luciferase reporter assay. (D) THBS2 expression was detected by western blotting. ** p<0.01; *** p<0.001.
MiR-106a increased LPS-induced inflammation and apoptosis of TCMK-1 cells via regulating THBS2 expression
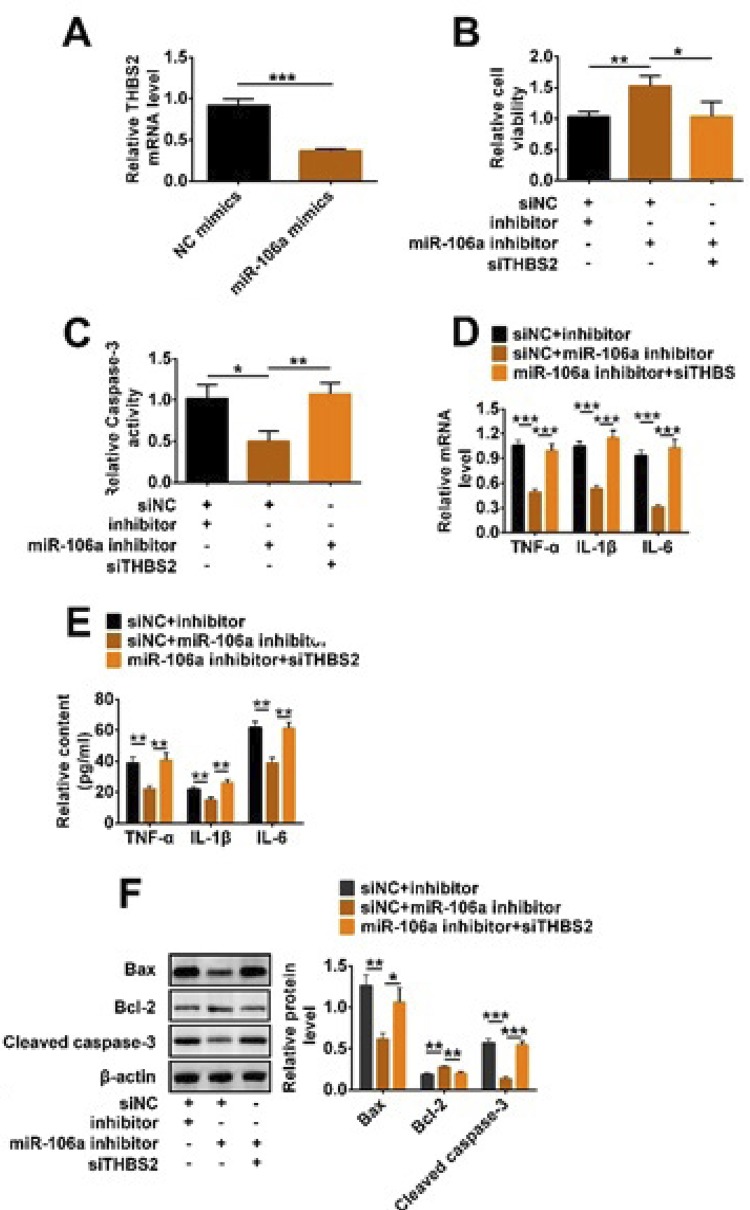
In order to study how miR-106a and THBS2 together affect sepsis-induced AKI in vitro, we used a complement experiment and simultaneously transfected siTHBS2 and miR-106a inhibitor into the LPS-treated TCMK-1 cells. qRT-PCR results in Figure 5A showed that THBS2 mRNA level was reduced in the miR-106a mimics group, indicating that there was a negative regulatory relationship between miR-106 and THBS2. Then, the increase of cell viability caused by miR-106a knockdown was decreased after adding siTHBS2 (p<0.05, Fig. 5B). On the contrary, siTHBS2 alleviated miR-106a knockdown-induced the reduction of caspase-3 activity (p<0.05, Fig. 5C). Furthermore, the mRNA level and content of TNF-α, IL-1β, IL-6 in the miR-106a inhibitor+siTHBS2 group were significantly more than that in the siNC+miR-106a inhibitor (p<0.01 or p<0.001, Fig. 5 D-E). Finally, western blotting results in Figure 5F showed that the down-regulations of Bax and cleaved caspase-3, and the up-regulation of Bcl-2 were reversed by siTHBS2 (p<0.01 or p<0.001). Sum up, miR-106a decreased cell viability and increased apoptosis by regulating THBS2 expression in LPS-induced TCMK-1 cells.
Figure 5. MiR-106a increased LPS-induced inflammation and apoptosis of TCMK-1 cells via regulating THBS2 expression. MiR-106a inhibitor, NC inhibitor, and miR-106a mimics, NC mimics, siTHBS2 and siNC were transfected into LPS-treated TCMK-1 cells. (A) Relative THBS2 mRNA level, (B) cell viability, (C) relative caspase-3 activity, (D-E) the relative mRNA level and content of TNF-α, IL-1β, IL-6, as well as (F) the protein level of Bax, cleaved caspase-3 and Bcl-2 were detected by qRT-PCR, CCK-8, ELISA and western blotting. * p<0.05; ** p<0.01; *** p<0.001.
Discussion
Sepsis is a disordered state in which an inflammatory response is out of control due to severe infection3. In recent years, it has been found that miRNAs can regulate the imbalanced inflammatory response by down-regulating inflammatory factors at the post-transcriptional level, suggesting that miRNAs play a very important role in inflammatory response and immune regulation of sepsis5. The latest findings confirm that the expression of miRNAs in serum of sepsis patients is also different from that of healthy people6,7,20,21. Vasilescu et al.20 used microarray detection technology to compare the differences in miRNA expression profiles in sepsis patients with normal human serum, and then the results showed that the expressions of miR-150, miR-182 and miR-486 were significantly different in patients with sepsis. Wang et al.21 reported that miR-146a and miR-233 expressions were reduced in serum of patients with sepsis. Moreover, Wu et al.9 displayed that the expression of miR-106a was up-regulated in the serum of sepsis mice and inflammatory bowel disease mice model. However, our results in this study were consistent with Wu’s report, that is, we found that the level of miR-106a was increased in the serum of sepsis patients, CLP-treated mice models and LPS-induced cells. These results suggested that miR-106a might be an ideal indicator for the detection and treatment of sepsis or sepsis-induced AKI. Furthermore, a research showed that the absence of miR-106a could alleviate the inflammatory response10. It has also been reported that miR-106a can inhibit cell proliferation and promote apoptosis in astrocytoma22. Hence, we detected LPS-stimulated TCMK-1 cell viability and apoptosis-related proteins expression, and found that miR-106a knockdown decreased apoptosis and increased cell viability, illustrating that miR-106a could involve in the process of sepsis-induced AKI.
During the recurrence and development of sepsis, the release of cytokines shows cascade effect, including the pro-inflammatory factors TNF-α, IL-1β and IL-623,24. Then, low levels of TNF-α, IL-1β, IL-8 and IL-6 could enhance cellular immune responses, activate defensive function and reduce the incidence of infection24-26.
TNF-α has the characteristics of early rapid synthesis and release, and its content changes can reflect the changes of other inflammatory factors23. In addition, IL-6 in serum is a delayed cytokine, which can be used as a marker of cytokine cascade activation, reflecting the relationship between host inflammatory response and severity of disease, and also can be used as a prognostic indicator in sepsis27. In this study, the results showed that knockdown of miRNA-106a reduced LPS-induced the levels of inflammatory factor TNF-α, IL-1β and IL-6, suggesting that inhibiting the expression of miRNA-106a might inhibit the occurrence of AKI induced by sepsis.
THBS2 is considered to be a powerful angiogenesis inhibitor and a regulator of tissue remodeling after trauma. In recent years, studies have shown that THBS2 plays an important role in tumors, but no studies related to sepsis have been reported14-16. However, Wei et al.14 found that miR-221-3p could increase metastasis of cervical cancer by targeting THBS2. Then, we used TargetScan website to predict the potential target gene of miR-106a and found that there were seven binding sites between miR-106a and THBS2. Furthermore, we also confirmed THBS2 was a target gene of miR-106a and negatively regulated by miR-106a. In addition, our results showed that miR-106a increased LPS-induced inflammation and apoptosis of TCMK-1 cells via regulating THBS2 expression.
In conclusion, miR-106a aggravates sepsis-induced AKI by targeting THBS2 in mice model and cells. MiR-106a might be an ideal test and evaluation index, and further research on the downstream targeting gene THBS2 might provide a broader approach and method for the diagnosis and treatment of AKI caused by sepsis, the development of molecular targeted drugs and prognosis of sepsis patients.
Research performed at The Affiliated Hospital of Hangzhou Normal University, Hangzhou, Zhejiang Province, 310015, China.
Financial source: none
References
- 1.Mann EA, Baun MM, Meininger JC, Wade CE. Comparison of mortality associated with sepsis in the burn, trauma, and general intensive care unit patient: a systematic review of the literature. 10.1097/SHK.0b013e318237d6bfShock. 2012;37(1):4–16. doi: 10.1097/SHK.0b013e318237d6bf. Augusta, Ga. [DOI] [PubMed] [Google Scholar]
- 2.Wafaisade A, Lefering R, Bouillon B, Sakka SG, Thamm OC, Paffrath T, Neugebauer E, Maegele M. Epidemiology and risk factors of sepsis after multiple trauma: an analysis of 29,829 patients from the Trauma Registry of the German Society for Trauma Surgery. 10.1097/CCM.0b013e318206d3dfCriti Care med. 2011;39(4):621–628. doi: 10.1097/CCM.0b013e318206d3df. [DOI] [PubMed] [Google Scholar]
- 3.Zarjou A, Agarwal A. Sepsis and acute kidney injury. 10.1681/asn.2010050484J Amer Soci Nephrol. 2011;22(6):999–1006. doi: 10.1681/ASN.2010050484. [DOI] [PubMed] [Google Scholar]
- 4.Zarbock A, Gomez H, Kellum JA. Sepsis-induced acute kidney injury revisited: pathophysiology, prevention and future therapies. 10.1097/mcc.0000000000000153Curr Opin Criti Care. 2014;20(6):588–595. doi: 10.1097/MCC.0000000000000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts R, Steer CJ. Disease genes and gene regulation by microRNAs. 10.1007/s12265-010-9186-6J Cardiovasc Transl. 2010;3(3):169–172. doi: 10.1007/s12265-010-9186-6. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Bei Y, Shen S, Huang P, Shi J, Zhang J, Sun Q, Chen Y, Yang Y, Xu T, Kong X, Xiao J. miR-21-3p controls sepsis-associated cardiac dysfunction via regulating SORBS2. 10.1016/j.yjmcc.2016.03.014J Mol Cel Cardiol. 2016;94:43–53. doi: 10.1016/j.yjmcc.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Wang X, Liu X, Wang X, Xu J, Hou S, Zhang X, Ding Y. miR-15a/16 are upreuglated in the serum of neonatal sepsis patients and inhibit the LPS-induced inflammatory pathway. Int J Clin Exp Med. 2015;8(4):5683–5690. PMID: 26131152. [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S, Wang J, Zhang Z, Miao H. Decreased miR-128 and increased miR-21 synergistically cause podocyte injury in sepsis. 10.1007/s40620-017-0405-yJ Nephrol. 2017;30(4):543–550. doi: 10.1007/s40620-017-0405-y. [DOI] [PubMed] [Google Scholar]
- 9.Wu SC, Yang JC, Rau CS, Chen YC, Lu TH, Lin MW, Tzeng SL, Wu YC, Wu CJ, Hsieh CH. Profiling circulating microRNA expression in experimental sepsis using cecal ligation and puncture. 10.1371/journal.pone.0077936PLoS One. 2013;8(10):e77936. doi: 10.1371/journal.pone.0077936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma A, Kumar M, Ahmad T, Mabalirajan U, Aich J, Agrawal A, Ghosh B. Antagonism of mmu-mir-106a attenuates asthma features in allergic murine model. 10.1152/japplphysiol.00001.2012J Appl Physiol. 2012;113(3):459–464. doi: 10.1152/japplphysiol.00001.2012. [DOI] [PubMed] [Google Scholar]
- 11.Stenina OI, Topol EJ, Plow EF. Thrombospondins, their polymorphisms, and cardiovascular disease. 10.1161/atvbaha.107.141713Arterioscler Thromb Vasc Biol. 2007;27(9):1886–1894. doi: 10.1161/ATVBAHA.107.141713. [DOI] [PubMed] [Google Scholar]
- 12.Adams JC, Lawler J. The thrombospondins. 10.1101/cshperspect.a009712Cold Spring Harbor Perspect Biol. 2011;3(10):a009712. doi: 10.1101/cshperspect.a009712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Rourke KM, Laherty CD, Dixit VM. Thrombospondin 1 and thrombospondin 2 are expressed as both homo- and heterotrimers. J Biol Chem. 1992;267(35):24921–24924. PMID: 1459996. [PubMed] [Google Scholar]
- 14.Wei WF, Zhou CF, Wu XG, He LN, Wu LF, Chen XJ, Yan RM, Zhong M, Yu YH, Liang L, Wang W. MicroRNA-221-3p, a TWIST2 target, promotes cervical cancer metastasis by directly targeting THBS2. 322010.1038/s41419-017-0077-5Cell Death Dis. 2017;8(12) doi: 10.1038/s41419-017-0077-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Nezu Y, Hagiwara K, Yamamoto Y, Fujiwara T, Matsuo K, Yoshida A, Kawai A, Saito T, Ochiya T. miR-135b, a key regulator of malignancy, is linked to poor prognosis in human myxoid liposarcoma. 10.1038/onc.2016.157Oncogene. 2016;35(48):6177–6188. doi: 10.1038/onc.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boguslawska J, Rodzik K, Poplawski P, Kedzierska H, Rybicka B, Sokol E, Tanski Z, Piekielko-Witkowska A. TGF-beta1 targets a microRNA network that regulates cellular adhesion and migration in renal cancer. 10.1016/j.canlet.2017.10.019Cancer Lett. 2018;412:155–169. doi: 10.1016/j.canlet.2017.10.019. [DOI] [PubMed] [Google Scholar]
- 17.Balc IC, Sungurtekin H, Gurses E, Sungurtekin U, Kaptanoglu B. Usefulness of procalcitonin for diagnosis of sepsis in the intensive care unit. Crit Care. 2003;7(1):85–90. doi: 10.1186/cc1843. PMID: 12617745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C, Hu J, Mao Z, Kang H, Liu H, Fu W, Lv Y, Zhou F. Acute kidney injury and inflammatory response of sepsis following cecal ligation and puncture in d-galactose-induced aging rats. 10.2147/cia.s132277Clin Int Aging. 2017;12:593–602. doi: 10.2147/CIA.S132277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Vasilescu C, Rossi S, Shimizu M, Tudor S, Veronese A, Ferracin M, Nicoloso MS, Barbarotto E, Popa M, Stanciulea O, Fernandez MH, Tulbure D, Bueso-Ramos CE, Negrini M, Calin GA. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. 10.1371/journal.pone.0007405PLoS One. 2009;4(10):e7405. doi: 10.1371/journal.pone.0007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang JF, Yu ML, Yu G, Bian JJ, Deng XM, Wan XJ, Zhu KM. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. 10.1016/j.bbrc.2010.02.145Biochem Biophys Res Commun. 2010;394(1):184–188. doi: 10.1016/j.bbrc.2010.02.145. [DOI] [PubMed] [Google Scholar]
- 22.Zhi F, Zhou G, Shao N, Xia X, Shi Y, Wang Q, Zhang Y, Wang R, Xue L, Wang S, Wu S, Peng Y, Yang Y. miR-106a-5p inhibits the proliferation and migration of astrocytoma cells and promotes apoptosis by targeting FASTK. 10.1371/journal.pone.0072390PLoS One. 2013;8(8):e72390. doi: 10.1371/journal.pone.0072390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurt AN, Aygun AD, Godekmerdan A, Kurt A, Dogan Y, Yilmaz E. Serum IL-1beta, IL-6, IL-8, and TNF-alpha levels in early diagnosis and management of neonatal sepsis. 31397Mediat Inflamm. 2007;2007 doi: 10.1155/2007/31397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moraes CA, Santos G, de Sampaio e Spohr TC, D’Avila JC, Lima FR, Benjamim CF, Bozza FA, Gomes FC. Activated microglia-induced deficits in excitatory synapses through IL-1beta: implications for cognitive impairment in sepsis. 10.1007/s12035-014-8868-5Mol Neurobiol. 2015;52(1):653–663. doi: 10.1007/s12035-014-8868-5. [DOI] [PubMed] [Google Scholar]
- 25.Cao YZ, Tu YY, Chen X, Wang BL, Zhong YX, Liu MH. Protective effect of Ulinastatin against murine models of sepsis: inhibition of TNF-alpha and IL-6 and augmentation of IL-10 and IL-13. 10.1016/j.etp.2010.11.011Exp Toxicol Pathol. 2012;64(6):543–547. doi: 10.1016/j.etp.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Veleminsky M, Jr, Stransky P, Veleminsky M, Sr, Tosner J. Relationship of IL-6, IL-8, TNF and sICAM-1 levels to PROM, pPROM, and the risk of early-onset neonatal sepsis. Neuro Endocrinol Lett. 2008;29(3):303–311. PMID: 18580842. [PubMed] [Google Scholar]
- 27.Leon LR, White AA, Kluger MJ. Role of IL-6 and TNF in thermoregulation and survival during sepsis in mice. 10.1152/ajpregu.1998.275.1.R269Am J Physiol. 1998;275(1):R269–R277. doi: 10.1152/ajpregu.1998.275.1.R269. [DOI] [PubMed] [Google Scholar]