Abstract
Genetic or pharmacological activation of canonical Wnt/β-catenin signaling inhibits oligodendrocyte differentiation. Transcription factor 7-like 2 (TCF7l2), also known as TCF4, is a Wnt effector induced transiently in the oligodendroglial lineage. A well accepted dogma is that TCF7l2 inhibits oligodendrocyte differentiation through activation of Wnt/β-catenin signaling. We report that TCF7l2 is upregulated transiently in postmitotic, newly differentiated oligodendrocytes. Using in vivo gene conditional ablation, we found surprisingly that TCF7l2 positively regulates neonatal and postnatal mouse oligodendrocyte differentiation during developmental myelination and remyelination in a manner independent of the Wnt/β-catenin signaling pathway. We also reveal a novel role of TCF7l2 in repressing a bone morphogenetic protein signaling pathway that is known to inhibit oligodendrocyte differentiation. Thus, our study provides novel data justifying therapeutic attempts to enhance, rather than inhibit, TCF7l2 signaling to overcome arrested oligodendroglial differentiation in multiple sclerosis and other demyelinating diseases.
Keywords: BMP signaling, canonical Wnt/beta-catenin signaling, myelination, oligodendrocyte differentiation, remyelination, TCF7l2(TCF4)
Introduction
Transcription factor 7-like 2 (TCF7l2, also known as TCF4) is one of the four members of the TCF/LEF1 family (gene symbols, Tcf7, Tcf7l1, Tcf7l2, and Lef1 [lymphoid enhancer-binding factor 1]) that are essential for mediating canonical Wnt/β-catenin signaling in Wnt-activated cells (Fancy et al., 2010; Freese et al., 2010). During Wnt activation, TCF7l2 acts as an activator for Wnt target genes by which it regulates many Wnt-related biological processes. In the absence of Wnt activation, TCF7l2 interacts with co-repressors, such as Groucho, histone deacetylase, and other as yet unidentified factors, to repress gene expression in a context-dependent manner (Lien and Fuchs, 2014). TCF7l2 has a β-catenin-binding domain (encoded by exon 1) and a high mobility group–DNA binding domain (coded by exons 10 and 11), the latter of which is required for the function of TCF7l2 of both activating and repressing gene transcription (Lien and Fuchs, 2014). Among the four members of this group, only TCF7l2 is expressed specifically in oligodendroglial lineage cells (Fancy et al., 2009; Fu et al., 2009; Ye et al., 2009).
Enforced activation of Wnt/β-catenin signaling by genetic or pharmacologic manipulation inhibits or delays oligodendrocyte (OL) differentiation (Shimizu et al., 2005; Fancy et al., 2009, 2011, 2014; Feigenson et al., 2009; Ye et al., 2009; Lang et al., 2013; Dai et al., 2014). Because TCF7l2 is an essential effector for Wnt/β-catenin signaling, the current well accepted hypothesis is that TCF7l2 inhibits oligodendrogenesis (Ye et al., 2009) and OL differentiation (He et al., 2007; Fancy et al., 2009, 2011; Li et al., 2009; Swiss et al., 2011; Sabo and Cate, 2013; Wood et al., 2013) through Wnt/β-catenin activation. This hypothesis suggests that TCF7l2 expression in cells of the oligodendroglial lineage inhibits or delays OL differentiation and contributes to remyelination failure in the lesions of multiple sclerosis (Fancy et al., 2009; Pedre et al., 2011; Lürbke et al., 2013) and hypoxic ischemic encephalopathy (Fancy et al., 2011; Fu et al., 2012).
The specificity of TCF7l2 expression in oligodendroglial progenitor cells (OPCs) (Fancy et al., 2009, 2011, 2014) explained the specific activation of Wnt/β-catenin signaling in OPCs and the inhibition of OPC differentiation into OLs when Wnt/β-catenin is activated genetically. However, in a previous study, we reported that, when the Wnt negative regulator adenomatous polyposis coli (APC) is deleted conditionally, yielding Wnt/β-catenin activation and OPC differentiation block, TCF7l2 is downregulated (Lang et al., 2013). This finding suggests that TCF7l2 may play a role in oligodendroglial lineage cells that is not involved in Wnt/β-catenin signaling. Here we report that TCF7l2 is expressed intensely in myelin gene-expressing, newly differentiated OLs rather than in OPCs. Following up on these studies, we have used Cre–LoxP-mediated Tcf7l2 gene ablation to demonstrate that TCF7l2 is, in fact, a positive regulator of neonatal and postnatal OL differentiation during both normal myelination and remyelination in a manner independent of Wnt/β-catenin activity.
Materials and Methods
Transgenic mice.
The transgenic mice Olig2–CreERT2, Plp–CreERT2, Cnp–Cre, Apcfl/fl, and Ctnnb1fl/fl (loxP sites located in introns 1 and 6 of β-catenin gene, loss-of-function) and the Wnt reporter strains Bat–LacZ and Axin2–LacZ are described in our previous study (Lang et al., 2013). Tcf7l2fl/fl (exon 1 is flanked by loxP sites) mice were kindly provided by Dr. Melinda Angus-Hill and are described in her previous study (Angus-Hill et al., 2011). Both male and female mice were used in this study. All mice were maintained on the C57BL/6 background and covered by Institutional Animal Care and Use Committee protocols approved by University of California, Davis.
Tamoxifen-induced Tcf7l2 gene deletion.
Tamoxifen was dissolved in 90% sunflower seed oil and 10% ethanol and freshly prepared at 30 mg/ml solution.
For TCF7l2 conditional ablation in the early postnatal mice, tamoxifen was injected intraperitoneally with a dose of 100 μg/g body weight and with two injections at time points indicated in each figure according to our previous study (Lang et al., 2013). Control age-matched early postnatal mice (Tcf7l2fl/fl) were treated with tamoxifen in the same way as TCF7l2 conditional knock-out (cKO) mice. Our previous analysis, at 2 d after tamoxifen, indicated that OPCs were recombined predominantly in the spinal cord (Lang et al., 2013). Similarly, we have now found that, in the corpus callosum of Olig2–CreERT2, Rosa–EYFP mice that received tamoxifen at P6 and P7, almost all tamoxifen-induced recombined cells were OPCs when analysis at P8.
For TCF7l2 conditional ablation in the cuprizone-induced demyelination model, 2-month-old adult Oligo2–CreERT2, Tcf7l2fl/fl and Tcf7l2fl/fl control mice were injected intraperitoneally daily for 7 consecutive days from the end of the week 5 of cuprizone diet with a dose of 1.5 mg of tamoxifen per injection (for experimental design, see Fig. 9C).
Figure 9.
Conditional ablation of TCF7l2 inhibits OL differentiation in the cuprizone-induced demyelination model. A, Analysis of temporal dynamics of NG2+/Sox10+ OPCs and CC1+/Sox10+ OLs during cuprizone diet and return to normal diet (Recovery; n = 3 at each time point). We focus on the corpus callosum at the approximate coronal location of bregma −2.0 mm. Two-month-old mice are used at the initiation of cuprizone diet. B, Induction of TCF7l2 expression in APC+ cells in the corpus callosum at week 6 on cuprizone diet (left) and at 4 weeks after return to normal diet (right). C, Experimental design of TCF7l2 cKO and analysis for D–F. D, Plp mRNA ISH of forebrains at 10 d after the last tamoxifen injection. CC, Corpus callosum; Ctx, cortex. E, F, Representative confocal images of CC1 IHC (E) and quantification of CC1+ OLs and NG2+ OPCs of corpus callosum (F). Dotted areas in E refer to the corpus callosum. n = 3 for Olig2–CreERT2, Tcf7l2fl/fl, n = 6 for Tcf7l2fl/fl. Two-tailed Student's t test, **p < 0.01. n.s., Not significant. Scale bars: B, E, 50 μm; D, 500 μm.
Antibodies and qPCR primers.
Most antibodies used in this study have been described in our previous study (Lang et al., 2013). Four anti-TCF7l2 antibodies were used in this study: (1) rabbit, clone C48H11 (catalog #2569S; Cell Signaling Technology), recognizing DNA binding domain (Vacik et al., 2011); (2) rabbit, clone C9B9 (catalog #2566S; Cell Signaling Technology), recognizing an N-terminal epitope around Glu81 (Vacik et al., 2011); (3) goat, polyclonal, C19 (catalog #sc-8632; Santa Cruz Biotechnology), recognizing the C-terminal domain; and (4) mouse, clone 6H5-3 (catalog #05-511; Millipore), raised against a recombined protein corresponding to residues 31-331 of human TCF7l2 (Barker et al., 1999). The clone 6H5-3 antibody (lot #DAM1821079) gave a very high background when performing immunostaining on frozen sections without antigen retrieval and a clean background on paraffin sections; therefore, we used antigen retrieval when this antibody was used for immunohistochemistry (IHC) on frozen sections. All four TCF7l2 antibodies show the same immunostaining pattern, and the immunostaining signal was abolished in the TCF7l2 cKO mutants (see Fig. 5). Anti-phospho-Smad1/5 (Ser463/465) rabbit mAb 9516 was from Cell Signaling Technology. Mouse anti-BMP4 (MAB1049) was from Millipore. qPCR primers were all derived from the PrimerBank at pga.mgh.harvard.edu/primerbank/.
Figure 5.
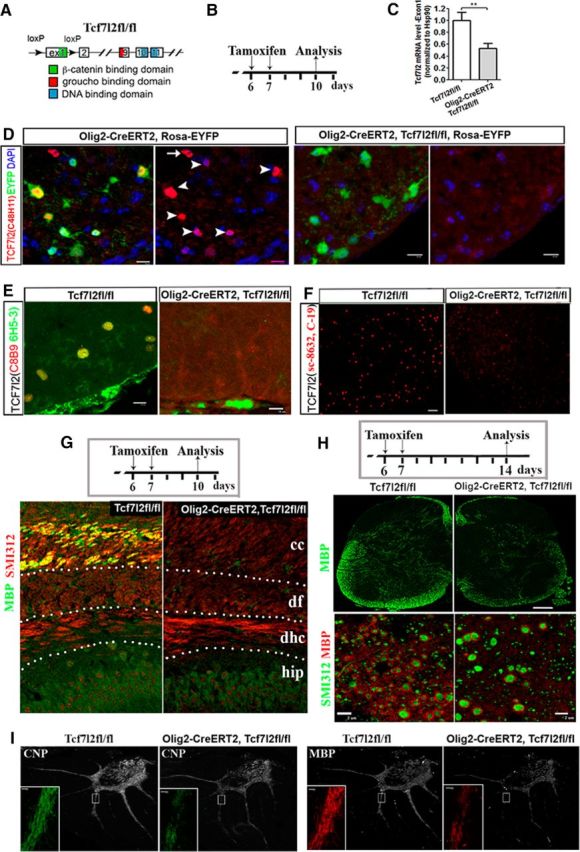
Conditional ablation of TCF7l2 results in hypomyelination. A, Structure of Tcf7l2fl/fl allele. B, Experimental design for C–F. C, Quantification of mRNA level of Tcf7l2 exon 1 by RT-qPCR. n = 5 in Tcf7l2fl/fl, n = 3 in Olig2–CreERT2, Tcf7l2fl/fl. Two-tailed Student's t test, ** p < 0.01. C, Western blot showing the ablation of the DNA-binding domain of TCF7l2. D–F, Representative confocal images of TCF7l2 immunostaining using antibodies against different regions of TCF7l2 (see Materials and Methods). Arrowheads and arrow in D point to EYFP+/TCF7l2+ and EYFP−/TCF7l2+ cells, respectively, in Olig2–CreERT2, Rosa–EYFP mice. G, Experimental design (top) and double IHC of MBP and pan-axonal marker SMI312 in the forebrain (bottom). Hip, Hippocampus; dhc, dorsal hippocampal commissure; df, dorsal fornix, cc, corpus callosum. H, Experimental design (top) for H and I and representative confocal images of MBP IHC at low magnification (middle) and of double IHC of MBP and SMI312 in the spinal cord dorsal columns at higher magnification (bottom). I, Representative confocal images of CNP and MBP in the cerebellum. Small boxed areas are shown at higher magnification at the left bottom corner of each panel. Scale bars: D, E, 10 μm; G, 20 μm; F, middle in H, 50 μm; bottom in H, 2 μm; I, 10 μm.
Primary OPC culture, tissue processing, IHC, mRNA in situ hybridization, co-immunoprecipitation, Western blotting, RT-qPCR, confocal imaging, microarray, and induction of experimental autoimmune encephalomyelitis- and cuprizone-demyelination models.
All these methods and procedures have been described in our previous studies (Guo et al., 2012; Lang et al., 2013).
Quantification and statistics.
For cell counting, we obtained confocal images with 10 μm optical thickness (12 μm physical thickness sections were used for immunostaining) and projected into one flattened image. Images were taken from the same anatomic location from cKO and control mice, and at least three histological sections were used from each animal. Cell counting and area calculation were conducted using confocal EZ-C1 viewer (Nikon). The cell density was calculated as the total number of cells counted divided by total area and expressed as cell number per square millimeter. For RT-qPCR quantification, the expression of genes of interest was normalized to that of internal control Hsp90, and the expression in controls was normalized to 1. The statistics used in this study are indicated in each figure. All data were expressed as mean ± SD.
Results
Wnt effector TCF7l2 is downregulated and LEF1 is upregulated in oligodendroglial lineage cells of APC cKO animals
We reported previously that disruption of Wnt negative regulator APC blocks OL differentiation and that this differentiation blockage persists in the adult (Lang et al., 2013), a result consistent with a recent study (Fancy et al., 2014). Additional analysis showed that OPCs were blocked at the postmitotic neuron–glial antigen 2-negative (NG2−)/PDGF receptor α-positive (PDGFR+) state and did not further differentiate into PDGFRα−/clone CC1+ OLs in the APC cKO spinal cord of Olig2–CreERT2, Apcfl/fl mice that received tamoxifen at P6 and P7 and were analyzed at P17 (data not shown).
APC is one of the components of the β-catenin destruction complex that is responsible for degrading cytoplasmic β-catenin, thus preventing activation of β-catenin-mediated canonical Wnt signaling (Aoki and Taketo, 2007). The Wnt target genes Sp5 (Fig. 1A; Boj et al., 2012; Fancy et al., 2014), Axin2, Naked1, and Notum, among other potential candidates of Wnt target genes (Table 1), were upregulated significantly in the spinal cord of APC cKO mice. Surprisingly, however, the Wnt effector TCF7l2 was downregulated at both mRNA and protein levels (Fig. 1B–D).
Figure 1.
Wnt effector TCF7l2 is downregulated and LEF1 is upregulated in APC cKO mutants. A, RT-qPCR of mRNA expression of generic Wnt target Sp5 in APC WT, APC heterozygous (Het) KO (Olig2–CreERT2, Apcfl/+), and APC homozygous (Homo) KO (Olig2–CreERT2, Apcfl/fl. n = 4 in each group. B, Representative confocal images of TCF7l2 and APC double IHC and density of TCF7l2+ cells. n = 4 in each group. C,-D, Western blot of TCF7l2 and RT-qPCR quantification of Tcf7l2 mRNA expression. n = 10 in WT, 6 in APC Het-KO, and 7 in APC Homo-KO. E, mRNA levels of TCF7, TCF7l1, and LEF1 quantified by RT-qPCR. n = 4 in each group. F, G, Representative confocal images of IHC of LEF1 and PECAM (F) and LEF1, PECAM, and Sox10 (G). Arrowheads in F point to LEF1+ cells that are PECAM+ endothelial cells in APC WT spinal cord. Note that all LEF1+ cells are positive for PECAM and negative for Sox10 in the APC WT spinal cord (G). H, Representative confocal images of IHC of LEF1, PECAM, and Sox10 in APC KO spinal cord and their quantification. Boxed area in H is shown at higher power at the right. Arrows point to LEF1+/Sox10+ cells, and arrowheads point to LEF1+/PECAM+ cells. n = 4 in each group. I, J, Dual ISH of Axin2 and IHC of TCF7l2 or LEF1 in the spinal cord of APC WT (I) and APC KO (J) at P14. Arrowheads in J point to double-positive cells. TCF7l2 protein is undetectable by IHC in APC KO attributable to the downregulation in APC KO. All data in Figure 1 were collected from spinal cord tissue. Tamoxifen was injected to each group at P7 and P8 daily, and the spinal cord was analyzed at P14. One-way ANOVA with Bonferroni's post hoc test (A, B, D, E). Two-tailed Student's t test (H). *p < 0.05, **p < 0.01, ***p < 0.001. Scale bars: B, 25 μm; F–H, 30 μm; I, J, 10 μm.
Table 1.
Significantly upregulated genes in the spinal cord of APC KO (Olig2–CreERT2, Apcfl/fl) compared with Apcfl/fl littermate controls from gene array analysis
| Gene symbol | Fold up |
|---|---|
| Mybpc1 | 3.3 |
| Nkd1 | 2.7 |
| Cxcr4 | 2.5 |
| Rpgr | 2.1 |
| Tnfrsf19 | 2.1 |
| E2f8 | 1.9 |
| Npy | 1.9 |
| Rnf43 | 1.9 |
| Sp5 | 1.9 |
| Gpr64 | 1.8 |
| Esyt3 | 1.8 |
| Axin2 | 1.7 |
| Nptx2 | 1.7 |
| C1qtnf1 | 1.6 |
| Wif1 | 1.5 |
| Kctd12 | 1.5 |
| Cdh3 | 1.5 |
| Gpr31b | 1.5 |
| Esr1 | 1.5 |
| Notum | 1.5 |
| Gpr31c | 1.5 |
| Hmgcs2 | 1.4 |
| Apcdd1 | 1.4 |
| Depdc2 | 1.4 |
| Abhd2 | 1.4 |
| Clic6 | 1.4 |
| Slc24a4 | 1.4 |
| Mr1 | 1.4 |
| Lef1 | 1.4 |
| Prmt8 | 1.3 |
| Uevld | 1.3 |
| Ptprm | 1.3 |
| Cyp26b1 | 1.3 |
| Abca1 | 1.3 |
| Fzd1 | 1.3 |
| Wwc1 | 1.3 |
| Akna | 1.3 |
| Olfm2 | 1.3 |
| Ndn | 1.3 |
| BC057079 | 1.3 |
| Slc1a3 | 1.3 |
| BC062109 | 1.3 |
| Nebl | 1.3 |
| Pde1a | 1.3 |
| Syngap1 | 1.3 |
| Hcn4 | 1.3 |
| Il16 | 1.3 |
| Gsg1l | 1.3 |
| Rnf144b | 1.3 |
| Egr1 | 1.3 |
These genes are potential candidates of Wnt-regulated genes in oligodendroglial lineage cells. Genes marked in bold are components of the Wnt/β-catenin signaling pathway or known Wnt target genes. APC was conditionally ablated in mice by tamoxifen injection at P6 and P7 (1 injection per day), and spinal cord tissue was harvested on P14. n = 4 in each group, p < 0.05 with false discovery rate correction. Only genes with a minimal upregulation of 1.3-fold in Olig2–CreERT2, Apcfl/fl mice are shown here.
These seemingly conflicting observations of upregulation of Wnt/β-catenin activity and downregulation of TCF7l2 prompted us to investigate what other Wnt effectors are upregulated to mediate Wnt/β-catenin signaling in APC cKO oligodendroglial lineage cells. Of mRNAs encoding the four members of the TCF/LEF1 family, only LEF1 mRNA was upregulated significantly in APC cKO mice (Fig. 1E). Consistent with this finding, LEF1 was upregulated significantly in Sox10+ oligodendroglial lineage cells in APC cKO mice (Fig. 1H), in line with a recent publication (Fancy et al., 2014), whereas in APC wild-type (WT) controls, LEF1 was absent in Sox10+ oligodendroglial lineage cells and only expressed in platelet endothelial cell adhesion molecule positive (PECAM+) endothelial cells (Fig. 1F,G). These data indicate that induction of LEF1 in oligodendroglial lineage cells may be responsible for the overactivation of Wnt/β-catenin signaling elicited by APC cKO. Supporting this notion, we found that Axin2 mRNA, a reliable readout of Wnt/β-catenin activation, was barely detectable in TCF7l2+ cells in APC WT (Fig. 1I) and upregulated in LEF1+/Sox10+ cells in APC cKO (Fig. 1J). The inhibition of OPC differentiation into OLs and downregulation of TCF7l2 in APC cKO led us to hypothesize that TCF7l2 is upregulated in cells that are developmentally downstream of “stalled” OPCs, and thus the downregulation of TCF7l2 in APC cKO simply reflects the blockage of OPC differentiation. We next investigated the cellular specificity of TCF7l2 expression during OL differentiation.
TCF7l2 is upregulated transiently in newly differentiated, premyelinating OLs during normal development
TCF7l2 was reported previously to be expressed more intensely in OPCs and then downregulated in OLs (Fancy et al., 2009, 2011). Using purified primary OPC cultures derived from neonatal rat forebrain, we found that TCF7l2 mRNA was low in OPCs, was sharply upregulated when purified OPCs were cultured in differentiation medium for 1 d, but then was downregulated after 4 d of differentiation to mature OLs (Fig. 2A). Western blot data confirmed this temporal pattern of TCF7l2 expression during OPC differentiation and maturation (Fig. 2B). (Note that two differently sized isoforms of TCF7l2 were detected, one at 58 kDa and one at 79 kDa.) In vivo, TCF7l2 was barely detectable in NG2+ or PDGFRα+/Sox10+ OPCs in the spinal cord (Fig. 2C, left) and forebrain (Fig. 2C, right) but intensely expressed in CC1+ differentiated OLs in spinal cord (Fig. 2D, left), forebrain (Fig. 2D, middle), and cerebellum (Fig. 2D, right). We validated the specificity of the TCF7l2 antibodies we used by demonstrating that their binding was abolished in TCF7l2 cKO tissue (see Fig. 5). Consistent with the temporal pattern in vitro (Fig. 2A,B), TCF7l2 expression was downregulated in the adult CNS (data not shown) in which most OLs are mature, thus resembling the temporal pattern we reported previously for APC expression (Lang et al., 2013).
Figure 2.
TCF7l2 is upregulated in OLs both in vitro and in vivo. A, B, TCF7l2 mRNA (RT-qPCR; A) and protein (Western blot; B) expression in purified primary OPCs (from neonatal rat cortex) and their differentiated progenies at days 1–4 (D1–D4). n = 4 at each time point. C, Representative confocal images of IHC of TCF7l2 and NG2 (left) and TCF7l2, PDGFRα, and Sox10 (right). Arrows point to NG2+ or PDGFRα+ OPCs with extremely low levels of TCF7l2. Arrowheads point to TCF7l2+ cells that are NG2− or PDGFRα−. D, IHC showing TCF7l2 expression in CC1+ differentiated OLs (arrowheads) in the spinal cord, corpus callosum, and cerebellar WM. One-way ANOVA with Bonferroni's post hoc test. ***p < 0.001. Scale bars: C, left, 5 μm; all others, 10 μm.
We further demonstrated that TCF7l2 was upregulated transiently in postmitotic, newly differentiated myelin gene-expressing OLs (Fig. 3H). All TCF7l2+ cells were positive for myelin basic protein (MBP; Fig. 3A,B, arrowheads) and 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNP; Fig. 3C, arrowheads) in their cell bodies and proximal processes, and all MBP+ and CNP+ myelin gene-expressing OLs were positive for nuclear TCF7l2 in the spinal cord at late embryonic and very early postnatal stages when mature, myelinating OLs are sparse. Using a combination of mRNA in situ hybridization (ISH) and IHC, we demonstrated that all TCF7l2+ cells were positive for mRNA of Mbp (Fig. 3D, arrowheads) and proteolipid protein (Plp; Fig. 3E, arrowheads) in the spinal cord at P7 when active myelination occurs. However, a large proportion of Mbp or Plp mRNA+ cells were negative for TCF7l2 at this later time point (Fig. 3D,E, arrows), indicating that these cells were more mature, myelinating OLs that had downregulated TCF7l2. Consistent with their differentiated status, all TCF7l2+ cells were Ki67− and did not divide during 2 h pulse 5-ethynyl-2-deoxyuridine (EdU) labeling (Fig. 3F,G). Collectively, our data support the hypothesis that TCF7l2 is upregulated transiently in myelin gene-expressing, newly differentiated OLs rather than in OPCs.
Figure 3.
TCF7l2 is expressed in postmitotic newly differentiated premyelinating OLs. A–C, Double IHC of TCF7l2 and MBP or CNP in the spinal cord at E18.5 or P2. The boxed areas in B and C are shown as high-power orthogonal images at the right. Arrowheads point to TCF7l2+ cells that express MBP or CNP in the cell bodies and proximal processes, which are characteristic of newly differentiated, premyelinating OLs. D, E, IHC of TCF7l2 protein and ISH of Mbp (D) or Plp (E) mRNA in P7 spinal cord. Cells pointed by arrowheads 1–4 in D and boxed area in E are shown as high-power images at the right. Note that all TCF7l2+ cells are Mbp or Plp mRNA+ (arrowheads), whereas a large proportion of Plp or Mbp mRNA+ cells at this age are TCF7l2− (arrows). F, G, Double IHC of TCF7l2 and Ki67 or EdU (2 h pulse labeling) in the spinal cord. H, Schematic drawing depicting the temporal dynamics of TCF7l2 expression during OL development: low expression in OPCs, upregulation in postmitotic, newly differentiated, premyelinating OLs, and downregulation in mature, myelinating OLs. Scale bars: A–D, 10 μm; E–G, 50 μm.
Expression of TCF7l2 in oligodendroglial lineage cells is not associated with the activation of Wnt/β-catenin signaling
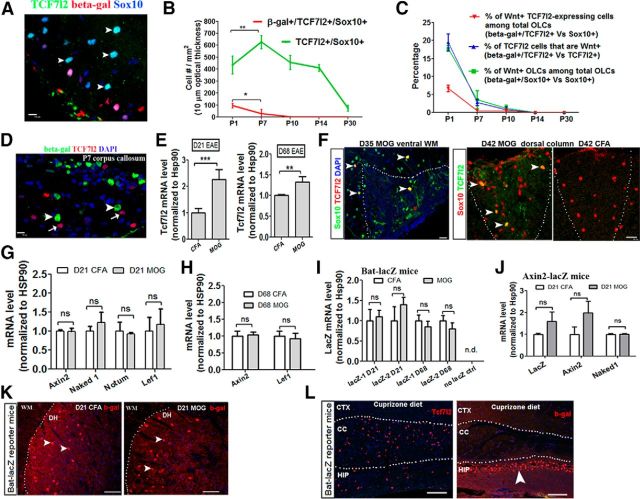
Next we asked whether TCF7l2 expression in OLs is an indicator of the activation of canonical Wnt/β-catenin signaling. To this end, we systematically analyzed the temporal dynamics of the Wnt reporter β-galactosidase (β-gal) and endogenous TCF7l2 within Sox10+ oligodendroglial lineage cells (Fig. 4A) in Bat–LacZ transgenic mice (Maretto et al., 2003) in which Wnt reporter β-gal expression is induced during the binding of the β-catenin/TCF/LEF1 complex to the binding site within the transgenic promoter region. The density of TCF7l2+/Sox10+ cells was increased significantly in the spinal cord of Wnt reporter mice from P1 (initiation of OPC differentiation) to P7 (peak of OPC differentiation and myelination; Fig. 4B). Conversely, the density of β-gal+/TCF7l2+/Sox10+ cells (Wnt-active cells) in the spinal cord of these mice was decreased significantly from P1 to P7 (Fig. 4B). Of note, TCF7l2+ cells were rarely β-gal+ in the spinal white matter (WM) by P10, although TCF7l2+ cells maintained a high density [456 ± 59/mm2 (mean ± SD) compared with 435 ± 75/mm2 in P1; Fig. 4B]. A total of 17.9% of total Sox10+ oligodendroglial lineage cells were β-gal+ in the spinal WM at P1 (Fig. 4C), consistent with a previous publication (Fancy et al., 2009). However, this percentage decreased to 2.5% by P7 and 0.7% by P10, and β-gal+ cells were virtually absent by P14 in spinal cord WM (Fig. 4C). Interestingly, many β-gal+ cells were NeuN+ neurons in the dorsal horn gray matter (GM) and ependymal cells surrounding the central canal at each time point (data not shown). In the forebrain of Bat–LacZ mice, few TCF7l2+ cells were β-gal+ (Fig. 4D, arrows), although β-gal+ cells were observed frequently in the P7 corpus callosum (Fig. 4D, arrowheads). As a positive control, we observed that many TCF7l2+ neurons in the thalamus, which are known to be subject to Wnt/β-catenin activation (Wisniewska et al., 2012), were β-gal+ (data not shown).
Figure 4.
TCF7l2 expression is not associated with canonical Wnt signaling during developmental myelination nor remyelination. A, Representative confocal images of triple IHC of TCF7l2, Wnt reporter β-gal, and Sox10 in P1 spinal cord of Bat–LacZ mice. Arrowheads point to TCF7l2+/Sox10+ cells that are β-gal−. B, Quantification of TCF7l2+/Sox10+ cells and β-gal+/TCF7l2+/Sox10+ cells at different time points. n = 4 at each time point of P1, P7, P10, and P14, except n = 3 at P30. C, Percentage of β-gal+ cells among different cell populations. n = 4 at each time point of P1, P7, P10, and P14, except n = 3 at P30. D, Representative confocal images showing that TCF7l2+ cells (arrows) are β-gal− (arrowheads) in the corpus callosum at P7. E, Quantification of Tcf7l2 mRNA expression in MOG35–55-peptide induced EAE and CFA control by RT-qPCR at day 21 (D21) and D68 after EAE. F, Representative confocal images of double IHC of TCF7l2 and Sox10 in the WM of D35 MOG (left) and in the dorsal columns of D42 MOG (middle) and CFA (right). Arrowheads point to double-positive cells. Dotted lines in F delineate areas of inflammatory infiltration in ventral WM (left image) and dorsal columns (right two images) of spinal cord. G, H, mRNA levels of Wnt target genes in MOG and CFA of D21 and D68 spinal cord. I, Quantification of LacZ mRNA by RT-qPCR in the spinal cord of D21 and D68 Bat–LacZ mice (2 primer sets LacZ-1 and LacZ-2 targeting different region of LacZ gene are used). Note that LacZ mRNA was not detected (n.d.) in nontransgene controls. J, Quantification of LacZ and Wnt target genes in D21 CFA and MOG spinal cord of Wnt reporter Axin2–LacZ mice. K, Representative confocal images of β-gal immunostaining at D21 after EAE of Bat–LacZ mice. Arrowheads point to β-gal+ neurons in the dorsal horn (DH). L, Representative confocal images of TCF7l2 (left) and β-gal (right) immunostaining in the forebrain of Bat–LacZ mice that had been maintained on cuprizone diet for 6 weeks. Note that many TCF7l2+ cells are present in the corpus callosum (CC), but no β-gal+ cells are present in the CC. Arrowhead points to β-gal+ cells in the hippocampus (HIP), a known area with Wnt signaling activation. CTX, Cortex. All data are collected from the spinal cord of Bat–LacZ Wnt reporter mice, except K is from the spinal cord of Axin2–LacZ Wnt reporter mice. Two-tailed Student's t test, *p < 0.05, **p < 0.01, ***p < 0.001. ns, Not significant. n = 4 in each group at D21, n = 5 at D68. Scale bars: A, D, F, 10 μm; K, 50 μm; L, 100 μm.
In an autoimmune model of multiple sclerosis, myelin oligodendrocyte glycoprotein (MOG)-peptide experimental autoimmune encephalomyelitis (EAE; Guo et al., 2011, 2012), we observed that TCF7l2 was significantly reinduced in Sox10+ newly differentiated OLs at both mRNA and protein levels on days 21 and 68 of post-MOG-peptide immunization (Fig. 4E,F), although the absolute density of TCF7l2+ cells was much lower than that during developmental myelination. Of note, most TCF7l2+ cells were distributed within and surrounding the inflammatory demyelinative lesions (Fig. 4F, dotted areas). However, mRNA expression of the endogenous Wnt target genes Axin2, Naked1, Notum1, and Lef1 were not increased at either of these time points (Fig. 4G,H). We further demonstrated that mRNA expression of Wnt reporter LacZ, which provides a more precise, real-time indicator for Wnt/β-catenin activation than β-gal protein (LacZ gene product; Fancy et al., 2011), was similar between Bat–LacZ mice treated with MOG and those treated with complete Freund's adjuvant (CFA) control on both days 21 and 68 after EAE (Fig. 4I). Using an independent Wnt reporter strain Axin2–LacZ, we showed that the mRNA expression of transgenic LacZ and endogenous Wnt target genes Axin2 and Naked1 were also similar between MOG-treated and CFA-treated mice on day 21 after EAE (Fig. 4J). At the histological level, β-gal+ cells were virtually absent from WM and mainly located in the dorsal horn GM (Fig. 4K) and ependymal cells (data not shown) at comparable levels in both MOG- and CFA-treated mice.
The conclusion that TCF7l2 induction was not associated with the activation of Wnt/β-catenin signaling was further supported by analyzing TCF7l2 and β-gal expression in the cuprizone demyelination model. Six weeks after initiating the cuprizone diet in Bat–LacZ mice, there was robust induction of TCF7l2 expression in the corpus callosum (Fig. 4L, left), with much higher numbers of TCF7l2+ cells than those in the spinal cord of MOG–EAE mice. However, few TCF7l2+ cells were β-gal+, although β-gal was strongly expressed in the hippocampus (Fig. 4L, right, arrowhead) in which Wnt/β-catenin signaling is active (Lie et al., 2005). Collectively, our in vivo data support the hypothesis that TCF7l2 expression in newly differentiated OLs is not associated with the activation of Wnt/β-catenin signaling in vivo during both developmental myelination and remyelination.
TCF7l2 cKO inhibits postnatal OL differentiation during normal myelination
Previous studies proposed that TCF7l2 may inhibit oligodendroglial differentiation (He et al., 2007; Fancy et al., 2009). However, this hypothesis is difficult to reconcile with the observation that there are fewer differentiated OLs in the spinal cord of Tcf7l2-null late embryos or newborns (Fu et al., 2009; Ye et al., 2009). Based on our data, we hypothesized that TCF7l2 promotes OL differentiation during postnatal myelination in the CNS. To test this hypothesis, we used the Cre–loxP system to conditionally ablate TCF7l2 in oligodendroglial lineage cells, thus circumventing the lethality of Tcf7l2-null newborns (Boj et al., 2012).
We first used inducible Olig2–CreERT2 (Lang et al., 2013) and Tcf7l2 exon 1 floxed (Angus-Hill et al., 2011) strains to delete the TCF7l2 exon 1 sequence in OPCs (Fig. 5A,C). Using Olig2–CreERT2, Rosa–EYFP reporter mice, we had reported previously that, in the spinal cord, 75% of EYFP+ recombined cells were OPCs when tamoxifen was injected at P6 and P7 and analysis was at P8 (Lang et al., 2013). These percentages of recombined OPCs were likely underestimated, because some of the initially recombined OPCs (at P6) probably would have differentiated into OLs by the time of tissue analysis (at P8) during this active oligodendroglial differentiation stage in spinal cord. Therefore, these data suggest that the deletion of Tcf7l2 exon 1 was initiated predominantly in OPCs in the Olig2–CreERT2, Tcf7l2fl/fl mutants. We showed that Cre-mediated deletion of Tcf7l2 exon 1 (Fig. 5C) abolished the potential truncated TCF7l2 protein in OLs that contains a DNA binding domain (Fig. 5D), an N-terminal domain (Fig. 5E), and a C-terminal domain (Fig. 5F; see antibodies in Materials and Methods), which is consistent with a previous publication (Angus-Hill et al., 2011), thus eliminating the dominant-negative effects of residual TCF7l2 in our study.
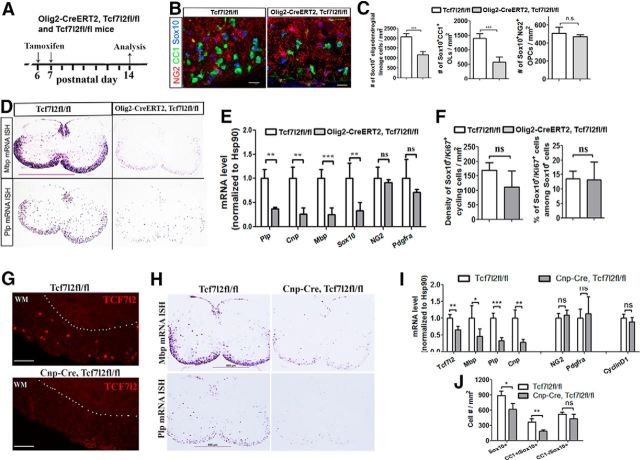
Deletion of the TCF7l2 exon 1 sequence initially in OPCs caused hypomyelination in the forebrain (Fig. 5G), spinal cord (Fig. 5H), and cerebellum (Fig. 5I), suggesting the hypothesis that TCF7l2 functions as a positive regulator of OL differentiation. Supporting this hypothesis, we found that conditional deletion of TCF7l2 starting at P6 (Fig. 6A) diminished the density of CC1+ OLs in the spinal cord of Olig2–CreERT2, Tcf7l2fl/fl mutants compared with Tcf7l2fl/fl controls at P14, whereas the density of NG2+ OPCs was not changed (Fig. 6B,C). Using mRNA ISH, we observed fewer Mbp mRNA+ and Plp mRNA+ differentiated OLs in the spinal cord of Olig2–CreERT2, Tcf7l2fl/fl mice compared with littermate controls (Fig. 6D). The mRNA levels of Sox10 and the myelin genes Plp, Cnp, and Mbp were all significantly diminished, whereas mRNAs coding the OPC markers NG2 and PDGFRα did not change in Olig2–CreERT2, Tcf7l2fl/fl mice by RT-qPCR quantification (Fig. 6E). Microarray analysis demonstrated that the expression of the myelin genes Opalin, myelin-associated glycoprotein (Mag), myelin-associated oligodendrocyte basic protein (Mobp), Mog, Cnp, Mbp, and Plp were all significantly decreased in Olig2–CreERT2, Tcf7l2fl/fl mice compared with those in Tcf7l2fl/fl littermate controls (Table 2, genes marked in bold). A cell proliferation assay revealed that the inhibition of OL differentiation was not attributable to a diminution in mitotic rate of TCF7l2-ablated OPCs (Fig. 6F), nor to increased apoptosis of differentiated OLs (data not shown). Together, these data suggest that TCF7l2 promotes OL differentiation during developmental myelination.
Figure 6.
Conditional ablation of TCF7l2 by two different Cre drivers inhibits OL differentiation. A, Experimental design for B–F. B, C, Representative confocal images of triple IHC of NG2, CC1, and Sox10 in the spinal cord (B) and quantification (C). D, mRNA ISH of Plp and Mbp in the spinal cord. E, mRNA expression level of oligodendroglial genes quantified by RT-qPCR in the spinal cord. F, Quantification of Ki67+/Sox10+ cells in the spinal cord. n = 5 in Tcf7l2fl/fl, n = 4 in Olig2–CreERT2, Tcf7l2fl/fl in C–F. Data from G–J are collected from P5 Cnp–Cre, Tcf7l2fl/fl and Tcf7l2fl/fl control mice. G, IHC of TCF7l2 (antibody against DNA-binding domain) in the spinal cord. H, mRNA ISH of Mbp and Plp in the spinal cord. I, RT-qPCR quantification of mRNA expression of Tcf7l2 exon 1, the myelin genes Mbp, Plp, and Cnp, the OPC markers NG2 and PDGFRα, and cell-cycle regulator Cyclin D1. J, Quantification of Sox10+ pan-oligodendroglial lineage cells, CC1+/Sox10+ differentiated OLs, and CC1−/Sox10+ undifferentiated OPCs. n = 4 in Tcf7l2fl/fl, n = 3 in Cnp–Cre, Tcf7l2fl/fl in I and J. Two-tailed Student's t test, *p < 0.05, **p < 0.01, ***p < 0.001. ns, Not significant. Scale bars: B, 10 μm; D, 1 mm; G, 50 μm; H, 500 μm.
Table 2.
Significantly downregulated genes in the spinal cord of TCF7l2 cKO (Olig2–CreERT2, Tcf7l2fl/fl) compared with Tcf7l2fl/fl controls from gene array analysis
| Gene symbol | Fold down |
|---|---|
| Opalin | 3.5 |
| Casr | 3.0 |
| 9630013A20Rik | 2.7 |
| Alox5 | 2.7 |
| Tmem141 | 2.5 |
| Gm3088 | 2.4 |
| Erbb3 | 2.4 |
| Mag | 2.3 |
| Tmem2 | 2.3 |
| Gm19439 | 2.2 |
| Parvb | 2.1 |
| Mobp | 2.1 |
| Trim59 | 2.1 |
| Gm98 | 2.0 |
| Mog | 2.0 |
| Fa2 h | 2.0 |
| Adamts4 | 2.0 |
| Ccnyl1 | 2.0 |
| Man1a | 2.0 |
| Tmem88b | 2.0 |
| Plekha1 | 1.9 |
| Il23a | 1.9 |
| Sytl2 | 1.9 |
| Aspa | 1.9 |
| Plekhh1 | 1.9 |
| Gsn | 1.9 |
| 1700047M11Rik | 1.9 |
| Cldn11 | 1.9 |
| Secisbp2l | 1.9 |
| Peli1 | 1.9 |
| Otud7b | 1.9 |
| Slc12a2 | 1.8 |
| 2810468N07Rik | 1.8 |
| Gjc2 | 1.8 |
| Anln | 1.8 |
| Plekhg3 | 1.8 |
| Ugt8a | 1.8 |
| Plat | 1.8 |
| Sh3gl3 | 1.8 |
| Tmem123 | 1.8 |
| Jam3 | 1.8 |
| Rasgef1b | 1.8 |
| C030030A07Rik | 1.7 |
| Ldlrap1 | 1.7 |
| Stxbp3a | 1.7 |
| Snx30 | 1.3 |
| Taf10 Ilk | 1.3 |
| Lgi3 | 1.3 |
| Pip4k2a | 1.3 |
| LOC100861831 | 1.3 |
| Ipo13 | 1.3 |
| Fbxo8 | 1.3 |
| Degs1 | 1.3 |
| Stk39 | 1.3 |
| Plekhb1 | 1.3 |
| Ube2g1 | 1.3 |
| Pop4 | 1.3 |
| Tbc1d5 | 1.3 |
| Pi4kb | 1.3 |
| Me1 | 1.2 |
| Mtap7 | 1.7 |
| Gltp | 1.7 |
| Elovl1 | 1.7 |
| Tprn | 1.7 |
| Cnp | 1.7 |
| Dhcr24 | 1.7 |
| Lass2 | 1.7 |
| Rhog | 1.7 |
| Ninj2 | 1.7 |
| Olig2 | 1.7 |
| Rap1a | 1.7 |
| Hspa2 | 1.7 |
| Tspan15 | 1.7 |
| Syde2 | 1.6 |
| Zdhhc9 | 1.6 |
| Sema4d | 1.6 |
| Tmem125 | 1.6 |
| Hapln2 | 1.6 |
| Ppp1r14a | 1.6 |
| 1500015L24Rik | 1.6 |
| Tspan2 | 1.6 |
| Klhl2 | 1.6 |
| Arhgef10 | 1.6 |
| Tns3 | 1.6 |
| Dock10 | 1.6 |
| Tmem182 | 1.6 |
| Piga | 1.6 |
| Plekhg1 | 1.6 |
| Strn | 1.6 |
| Carhsp1 | 1.6 |
| Atp8b1 | 1.6 |
| Idh1 | 1.6 |
| Tppp | 1.6 |
| Adipor2 | 1.6 |
| Nfasc | 1.6 |
| Il1rap | 1.6 |
| Ddc | 1.5 |
| Gss | 1.5 |
| Mal | 1.5 |
| Gab1 | 1.5 |
| Rab33a | 1.5 |
| Kndc1 | 1.5 |
| Stx2 | 1.5 |
| Gramd3 | 1.5 |
| Sorbs3 | 1.5 |
| Foxn3 | 1.2 |
| Hipk2 | 1.2 |
| Erf | 1.2 |
| Atp6v1g1 | 1.2 |
| D15Ertd621e | 1.2 |
| AW549877 | 1.2 |
| Lpar3 | 1.2 |
| Ltv1 | 1.2 |
| Aatk | 1.2 |
| Nudt4 | 1.2 |
| Pim3 | 1.2 |
| Nkain2 | 1.2 |
| Acsl3 Utp14b | 1.2 |
| Csrp1 | 1.2 |
| Txndc16 | 1.2 |
| St18 | 1.5 |
| Mcam | 1.5 |
| Nkx6–2 | 1.5 |
| Dbndd2 | 1.5 |
| Tmem63a | 1.5 |
| 6330503K22Rik | 1.5 |
| Klhl4 | 1.5 |
| Sc5d | 1.5 |
| Snx18 | 1.5 |
| Fbxo32 | 1.5 |
| Ppp1r16b | 1.5 |
| Dusp15 | 1.5 |
| Ypel2 | 1.5 |
| Rffl | 1.5 |
| Smad7 | 1.5 |
| Kif19a | 1.5 |
| G2e3 | 1.5 |
| Rcbtb1 | 1.5 |
| Apod | 1.5 |
| Enpp2 | 1.4 |
| Sox2 | 1.4 |
| Cyp51 | 1.4 |
| Ctnnal1 | 1.4 |
| Npc1 | 1.4 |
| Ephb1 | 1.4 |
| Pex5l | 1.4 |
| Fmnl2 | 1.4 |
| Il12rb1 | 1.4 |
| Hhip | 1.4 |
| Gm19500 | 1.4 |
| Slc44a1 | 1.4 |
| Sox10 | 1.4 |
| Map4k4 | 1.4 |
| Kat2b | 1.4 |
| Reep3 | 1.4 |
| Ick | 1.4 |
| Rab31 | 1.4 |
| Enpp4 | 1.4 |
| Zfp536 | 1.4 |
| Nde1 | 1.4 |
| Ankrd13a | 1.4 |
| Fntb LOC639541 | 1.4 |
| Ado | 1.4 |
| Acy3 | 1.4 |
| Chmp1b | 1.4 |
| Ctnna1 | 1.2 |
| Aplp1 | 1.2 |
| Lamp1 | 1.2 |
| Ktn1 | 1.2 |
| Creb5 | 1.2 |
| Prrg1 | 1.2 |
| Eps15 | 1.2 |
| Ankib1 | 1.2 |
| Picalm | 1.2 |
| Mmgt1 | 1.2 |
| Cap1 | 1.2 |
| Dock9 | 1.2 |
| Fth1 | 1.2 |
| Polr1d | 1.2 |
| Fkbp1a | 1.2 |
| Sirt2 | 1.4 |
| Creb3l2 | 1.4 |
| Ptpn11 | 1.4 |
| Prickle1 | 1.4 |
| Rtkn | 1.4 |
| Ptpdc1 | 1.4 |
| Npat | 1.4 |
| Cpd | 1.4 |
| Fgfr2 | 1.4 |
| Cdk19 | 1.4 |
| Acsl1 | 1.4 |
| Bace1 | 1.4 |
| Mbp | 1.4 |
| Sypl | 1.4 |
| Dram2 | 1.4 |
| Fam108b | 1.4 |
| Gm11492 | 1.4 |
| Eif2c3 | 1.4 |
| Npc2 | 1.4 |
| Bin1 | 1.4 |
| Gjb1 | 1.3 |
| Cyp3a13 | 1.3 |
| Rnf13 | 1.3 |
| Setd8 | 1.3 |
| Nkain1 | 1.3 |
| Evi2a Evi2b | 1.3 |
| Ankrd49 | 1.3 |
| Smurf1 | 1.3 |
| Erbb2ip | 1.3 |
| Phactr4 | 1.3 |
| Tmem163 | 1.3 |
| Mospd2 | 1.3 |
| Qk | 1.3 |
| 2810008M24Rik | 1.3 |
| Entpd5 | 1.3 |
| Rock1 | 1.3 |
| Ralgds | 1.3 |
| +Papss1 | 1.3 |
| Gm19897 | 1.3 |
| Myo6 | 1.3 |
| Mtmr2 | 1.3 |
| Rap2a | 1.3 |
| Arap2 | 1.3 |
| Snap23 | 1.3 |
| Twf1 Gm4887 | 1.3 |
| Tbc1d14 | 1.2 |
| Srd5a1 | 1.2 |
| Lrrn1 | 1.2 |
| Slc22a23 | 1.2 |
| Dhrs7 | 1.2 |
| Gclm | 1.2 |
| Rhbdl2 | 1.2 |
| Ncor2 | 1.2 |
| Kif5b | 1.2 |
| Rhoa | 1.2 |
| Mtmr6 | 1.2 |
| Gm428 | 1.2 |
| Add1 | 1.2 |
| Plp1 | 1.2 |
| Frmd4a | 1.2 |
TCF7l2 was conditionally ablated in mice by tamoxifen injection at P6 and P7 (1 injection per day), and spinal cord tissue was harvested on P14. n = 4 in each group, p < 0.05 with false discovery rate correction. Only genes with a minimal downregulation of 1.2-fold in Olig2–CreERT2, Tcf7l2fl/fl mice are shown here. Genes marked in bold are myelin genes or genes known to regulate OL differentiation.
To exclude any potential side-effects of tamoxifen on CNS development and to substantiate the conclusion derived from Olig2–CreERT2, Tcf7l2fl/fl transgenic mice, we used the constitutive Cre driver Cnp–Cre (Lappe-Siefke et al., 2003), which is activated initially in OPCs and remains activated persistently in differentiated OLs. Cnp–Cre, Tcf7l2fl/fl mutants were born in a normal Mendelian ratio and had no TCF7l2 expression in the spinal cord (Fig. 6G), cerebellum, and corpus callosum of the forebrain (data not shown). Histological and molecular quantification demonstrated that Cnp–Cre, Tcf7l2fl/fl mice contained fewer Plp mRNA+, Mbp mRNA+ cells (Fig. 6H,I) and fewer Sox10+/CC1+ differentiated OLs (Fig. 6J) in the spinal cord compared with Tcf7l2fl/fl littermate controls at P5 when active OL differentiation occurs. Consistent with the data from Olig2–CreERT2, Tcf7l2fl/fl mutants (Fig. 6A–F), numbers of Sox10+/CC1− undifferentiated OPCs and mRNA levels of the OPC markers NG2 and Pdgfrα and of the cell-cycle regulator Cyclin D1 were not altered in Cnp–Cre, Tcf7l2fl/fl mice (Fig. 6I,J). Collectively, these in vivo data from different Cre–loxP mutants provide compelling evidence indicating that the Wnt effector TCF7l2 is a required positive regulator for OL differentiation.
TCF7l2 expression in differentiated OLs does not regulate myelin gene expression
A previous study reported that Wnt/β-catenin signaling through TCF7l2 in Plp+ OLs directly activates myelin gene expression in zebrafish embryonic CNS (Tawk et al., 2011). The observation that TCF7l2 is upregulated transiently in newly differentiated, myelin gene-expressing OLs (Fig. 3) prompted us to determine whether the transcription factor TCF7l2 directly regulates myelin gene expression in Plp+ OLs. To this end, we used Plp–CreERT2 to delete TCF7l2 predominantly in Plp+ differentiated OLs by tamoxifen treatment. The cellular specificity of Plp–CreERT2 depends on the time of tamoxifen injection (Doerflinger et al., 2003; Guo et al., 2009, 2012; Harlow et al., 2014). Our previous studies showed that a small proportion of OPCs (∼10% among total OPCs) were recombined with EYFP in the spinal cord when tamoxifen was injected at very early postnatal ages (Guo et al., 2009). However, EYFP was predominantly expressed in differentiated OLs in the spinal cord when tamoxifen was injected at adult ages (Guo et al., 2012). Here we show that EYFP is predominantly expressed in CC1+ differentiated OLs in the spinal cord of Plp–CreERT2, Tcf7l2fl/fl, Rosa–EYFP triple transgenic mice (Fig. 7A2) when tamoxifen was injected at P9 (Fig. 7A1). TCF7l2 expression was ablated successfully in Plp–CreERT2, Tcf7l2fl/fl spinal cord 24 h after initial tamoxifen injection at P9 (Fig. 7A1), as indicated by the substantial diminution of TCF7l2 immunostaining signals (Fig. 7A3) and Tcf7l2 exon 1 mRNA (Fig. 7B, the first bar graph set) in Plp–CreERT2, Tcf7l2fl/fl mutants. However, the expressions of the major myelin genes Plp and Mbp, Sox10, and Olig2 and mature OL enzyme aspartoacylase (ASPA) were not altered during this “acute” short time window between Tcf7l2 deletion and analysis (Fig. 7B), suggesting that the myelin genes of Plp and Mbp are not immediate target genes of TCF7l2 in OLs in vivo. Furthermore, the expression of Yin Yang 1 (YY1), a transcription factor that has been shown to promote OL differentiation and to suppress TCF7l2 expression (He et al., 2007), did not change (Fig. 7B, right). Consistently, the intensities and the patterns of Plp and Mbp mRNAs in both forebrain (data not shown) and spinal cord (Fig. 7C) were similar in Plp–CreERT2, Tcf7l2fl/fl mice and littermate controls treated by the same tamoxifen paradigm. To evaluate the long-term effect of TCF7l2 deletion in differentiated OLs on myelin gene expression, we analyzed spinal cord tissue 1 week after TCF7l2 cKO in differentiated OLs (Fig. 7D). The expression of most myelin genes were not changed in Plp–CreERT2, Tcf7l2fl/fl mutants (Fig. 7E), nor was the density of CC1+ differentiated OLs (Fig. 7F), indicating that TCF7l2 expression in differentiated OLs neither directly nor indirectly regulates the expression of major myelin genes. Of note, however, the expressions of MAG and Elovl1, an elongase required for synthesis of very long-chain fatty acids, were significantly downregulated when TCF7l2 was ablated conditionally in differentiated OLs at both 24 h and 7 d (Fig. 7G,H).
Figure 7.
Conditional ablation of TCF7l2 in differentiated OLs does not affect myelin gene expression. A1, experimental design for A2, A3, B, and C. A2, Confocal images of immunostaining of EYFP and CC1 in the spinal cord of Plp–CreERT2, Tcf7l2fl/fl, Rosa–EYFP reporter mice. Arrowheads point to EYFP+/CC1+ differentiated OLs. A3, Confocal images of TCF7l2 in the spinal cord of Tcf7l2fl/fl (top) and Plp–CreERT2, Tcf7l2fl/fl (bottom) mice. B, mRNA levels of TCF7l2 exon 1, the oligodendroglial genes Sox10, Olig2, and ASPA, the myelin genes Plp and Mbp, and the transcription factor YY1 in the spinal cord. n = 6 in each group. C, mRNA ISH of Plp in the spinal cord. D, Experimental design (top) for E and F and confocal images of TCF7l2 IHC (bottom). E, Gene expression in the spinal cord quantified by RT-qPCR. n = 5 in Tcf7l2fl/fl, n = 5 in Plp–CreERT2, Tcf7l2fl/fl. Omgp, Oligodendrocyte myelin glycoprotein. F, Representative confocal images of CC1 IHC in the spinal cord and quantification. G, H, RT-qPCR quantification of MAG (G) and Elovl1 (H) in the spinal cord of PCE, Tcf7l2fl/fl and Tcf7l2fl/fl littermates at both 24 h and 7 d after Tcf7l2 conditional ablation. Two-tailed Student's t test, *p < 0.05, **p < 0.01, ***p < 0.001. Scale bars: A2, 10 μm; all others, 50 μm.
TCF7l2 regulates OL differentiation in a manner independent of Wnt/β-catenin signaling
The Wnt effector TCF7l2 mediates the activity of canonical Wnt/β-catenin signaling in Wnt-responsive cells, for example, in colorectal cancer cells (Anastas and Moon, 2013). In the absence of Wnt, the APC-containing destruction complex constitutively degrades cytoplasmic β-catenin, preventing it from binding nuclear TCF7l2, whereas in the presence of Wnt, β-catenin accumulates in the cytoplasm and translocates into the nucleus, in which it binds TCF7l2 to activate the expression of immediate target genes in Wnt-responsive cells (Fig. 8A), for example, Axin2, Naked1, Notum, Lef1 (Lang et al., 2013), and Sp5 (Fig. 1A; Fancy et al., 2014). Therefore, the altered expression of these immediate target genes during TCF7l2 deletion provides a reliable readout by which to evaluate the involvement of oligodendroglial TCF7l2 in Wnt/β-catenin signaling.
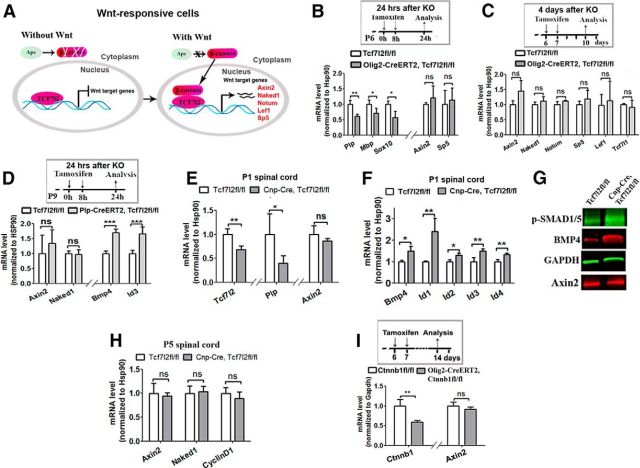
Figure 8.
Canonical Wnt signaling activity is not perturbed in the TCF7l2 cKO mutants driven by different Cre promoters at different time points. A, Schematic diagram depicting the role of Wnt negative regulator APC, β-catenin, TCF7l2, and Wnt target gene expression in Wnt-responsive cells. The binding of β-catenin to TCF7l2 in the presence of Wnt activation switches TCF7l2 from a transcriptional repressor to an activator. Gray circled area depicts the nucleus. B, Quantification of oligodendroglial genes and Wnt target genes in the spinal cord of Olig2–CreERT2, Tcf7l2fl/fl (n = 4) and Tcf7l2fl/fl (n = 5) controls at 24 h after conditional ablation of TCF7l2 in OPCs. C, Quantification of Wnt target genes and other members of Wnt effector LEF1 and TCF7l1 in the spinal cord of Olig2–CreERT2, Tcf7l2fl/fl (n = 4) and Tcf7l2fl/fl (n = 4) controls at 4 d conditional ablation of TCF7l2 in OPCs. D, RT-qPCR quantification of the Wnt target genes BMP4 and Id3 in the spinal cord of Plp–CreERT2, Tcf7l2fl/fl and Tcf7l2fl/fl controls at 24 h after conditional ablation of TCF7l2 in Plp+ OLs. n = 5 in each group. E, The level of gene expression quantified by RT-qPCR in the spinal cord of Cnp–Cre, Tcf7l2fl/fl and Tcf7l2fl/fl controls at P1 (f, n = 4 KOs, n = 3 controls). F, Quantification of Bmp4 and BMP target genes Id1, Id2, Id3, and Id4 by RT-qPCR in the spinal cord of Cnp–Cre, Tcf7l2fl/fl and Tcf7l2fl/fl controls at P1 (f, n = 4 KOs, n = 3 controls). G, Western blot of spinal cord protein extracts from P1 Cnp–Cre, Tcf7l2fl/fl and Tcf7l2fl/fl in OLs using antibodies against Axin2, BMP4, and phosphorylated SMAD1/5 at Ser463/467, the activated form of SMAD1/5 for BMP signaling activation. H, mRNA levels of Wnt targets Axin2 and Naked1 and the cell-cycle regulator Cyclin D1 quantified by RT-qPCR from the Cnp–Cre, Tcf7l2fl/fl and Tcf7l2fl/fl spinal cords at P5 (n = 4 each group). I, Quantification of β-catenin gene (Ctnnb1) and Wnt target Axin2 in the spinal cord of Olig2–CreERT2, Ctnnb1fl/fl (n = 3) and Ctnnb1fl/fl control (n = 4) 7 d after β-catenin cKO. Two-tailed Student's t test, applied to all, *p < 0.05, **p < 0.01, ***p < 0.001. ns, Not significant.
We used Olig2–CreERT2, Tcf7l2fl/fl transgenic mice to conditionally ablate TCF7l2 predominantly in OPCs at P6 and analyzed gene expression 24 h later (Fig. 8B, top of experimental design). In this acute cKO system, we observed significant decreases in Plp, Mbp, and Sox10 mRNAs (Fig. 8B, left part of bottom), again suggesting a positive role of TCF7l2 in the transition of OPCs into myelin gene-expressing OLs. However, the mRNA expressions of the Wnt target genes Axin2 and Sp5, which are upregulated significantly in APC cKO oligodendroglial lineage cells (Fig. 1A; Lang et al., 2013; Fancy et al., 2014), were unchanged in Olig2–CreERT2, Tcf7l2fl/fl mutants (Fig. 8B, right part of bottom). Furthermore, we also showed that the Wnt target genes Axin2, Naked1, Notum, Sp5, and Lef1 were all unchanged at 4 d after TCF7l2 deletion in Olig2–CreERT2, Tcf7l2fl/fl mutants (Fig. 8C), suggesting that TCF7l2 in oligodendroglial lineage cells does not mediate the activity of Wnt/β-catenin signaling. We further showed that another related member of the TCF/LEF family, TCF7l1, was not upregulated at the mRNA level (Fig. 8C, rightmost set of bar graph) or protein level (data not shown) to compensate for TCF7l2 deletion.
Does TCF7l2 disruption in Plp+ differentiated OLs perturb the Wnt/β-catenin activity in these OLs, as indicated in a previous study (Tawk et al., 2011)? To address this important question, we conditionally ablated Tcf7l2 in Plp+ differentiated OLs using Plp–CreERT2, Tcf7l2fl/fl transgenic mice by tamoxifen treatment (Fig. 8D, top; for TCF7l2 deletion in differentiated OLs, see Fig. 7A1–A3). In this TCF7l2 cKO system, we did not observe changes in mRNA expression of the Wnt target genes Axin2 and Naked1 (Fig. 8D, left half of bottom). Surprisingly, however, expression of mRNAs encoding BMP4, a ligand for BMP/SMAD signaling, and inhibitor of DNA binding 3 (Id3), a direct target of BMP/SMAD signaling, were upregulated significantly at 24 h after TCF7l2 ablation in Plp–CreERT2, Tcf7l2fl/fl mice compared with Tcf7l2fl/fl littermate controls (Fig. 8D, right half of bottom). These data suggest that TCF7l2 does not transcriptionally mediate upstream Wnt/β-catenin signaling in OLs and that BMP signaling may be an immediate target of Wnt effector TCF7l2 in the oligodendroglial lineage.
It was reported previously that TCF7l2 was colabeled with Wnt reporter β-gal in neonatal Bat–LacZ mice (Fancy et al., 2009; Fig. 4A–C), suggesting that TCF7l2 may mediate Wnt/β-catenin signaling in a temporally specific manner, i.e., transiently in neonatal oligodendroglial lineage cells before active OL differentiation and myelination. We found that tamoxifen had a toxic effect on postnatal murine development if injected into neonatal and very early postnatal pups. To avoid this complication, we used tamoxifen-independent, constitutive Cnp–Cre to ablate TCF7l2 in the oligodendroglial lineage. In the spinal cord of Cnp–Cre, Tcf7l2fl/fl neonates at P1 when TCF7l2 starts to appear and OL differentiation has just begun, TCF7l2 mRNA (Fig. 8E, left part) and protein (data not shown) were significantly decreased compared with Tcf7l2fl/fl littermate controls. Consistent with a positive role of TCF7l2 in OL differentiation, the expression of the myelin gene Plp was significantly decreased compared with Tcf7l2fl/fl controls (Fig. 8E, middle part). However, the expression of the Wnt target gene Axin2 was comparable between Cnp–Cre, Tcf7l2fl/fl mutant and controls (Fig. 8E, right part), suggesting that TCF7l2 does not mediate Wnt/β-catenin signaling in neonatal oligodendroglial lineage cells. Interestingly, BMP4 expression and BMP/SMAD signaling targets Id1, Id2, Id3, and Id4 were upregulated significantly in Cnp–Cre, Tcf7l2fl/f (Fig. 8F) at P1 compared with littermate controls, consistent with the data from Plp–CreERT2, Tcf7l2fl/f mutants (Fig. 8D, right half of bottom). Western blot confirmed the upregulation of BMP4 and also showed increased levels of phosphorylated-SMAD1/5 (Ser463/465), an active form of Smad1/5 required for BMP signaling activation, and comparable levels of Wnt target gene Axin2 at P1 (Fig. 8G). Collectively, these data suggest that TCF7l2 does not play a role in mediating Wnt/β-catenin signaling but, instead, represses BMP/SMAD signaling in neonatal and postnatal oligodendroglial lineage cells.
The nonperturbation of Wnt signaling in TCF7l2 cKO mutants was further confirmed by analyzing tamoxifen-independent Cnp–Cre, Tcf7l2fl/fl cKO mutants at P5 when substantial OL differentiation occurs, evidenced by comparable levels of Wnt target gene expression between cKO and controls (Fig. 8H). We further used gene microarray to screen for genes with altered expression in the TCF7l2 cKO of Olig2–CreERT2, Tcf7l2fl/fl mutants (Table 2). We reasoned that, if TCF7l2 regulates OL differentiation through mediating Wnt/β-catenin signaling activity, the genes of well known Wnt targets and potential candidates that are upregulated in APC cKO of Olig2–CreERT2, Apcfl/fl mutants (Table 1) will be significantly downregulated in TCF7l2 cKO. However, the results showed that none of the upregulated genes in APC cKO (Table 1) appeared in the downregulated gene list derived from TCF7l2 cKO microarray analysis (Table 2).
β-Catenin is a required molecule to mediate canonical Wnt/β-catenin signaling in Wnt-responsive cells. We had reported previously that β-catenin cKO did not affect the differentiation of neonatal and postnatal OLs (Lang et al., 2013) and now show that cKO of β-catenin (gene symbol, Ctnnb1) driven by Olig2–CreERT2 (Fig. 8I, top, and left part of bottom) did not affect the expression of Wnt target Axin2 (Fig. 8I, right part of bottom), nor of TCF7l2 expression (data not shown). Consistently, Wnt target gene Axin2 mRNA was barely detectable by mRNA ISH in TCF7l2+ OLs in the spinal cord (Fig. 1I). As a positive control, Axin2 mRNA was observed readily in LEF1+ but not TCF7l2+ oligodendroglia in APC cKO spinal cord of Olig2–CreERT2, Apcfl/fl mutants (Fig. 1J). ISH data indicate that TCF7l2 does not mediate Wnt/β-catenin activation in OLs under either physiological or Wnt genetic activated (APC cKO) conditions.
Together, our in vivo experimental data demonstrate that TCF7l2 positively regulates neonatal and postnatal OL differentiation and that this regulation does not involve the role of Wnt/β-catenin signaling, weakening the previous hypothesis that TCF7l2, through Wnt/β-catenin signaling activation, plays an inhibitory role in postnatal oligodendroglial differentiation. Our data also suggest that TCF7l2 positively regulates OL differentiation likely by suppressing BMP/SMAD signaling pathways in oligodendroglial lineage cells.
TCF7l2 promotes OL differentiation during remyelination after myelin damage
Previous studies proposed that TCF7l2 may inhibit OL differentiation during myelination (Fancy et al., 2009; Fancy et al., 2011; Pedre et al., 2011). However, no genetic data have thus far been available to support this hypothesis. We used the cuprizone demyelination/remyelination model to determine the role of TCF7l2 in remyelination. Analysis of the temporal dynamics of OPCs and OLs in the corpus callosum revealed that OL regeneration in this model occurred mainly between week 5 of cuprizone diet and week 2 after return to a normal diet (Fig. 9A). At the end of week 6 of cuprizone diet, there was robust reinduction of TCF7l2 in the corpus callosum, and TCF7l2 was downregulated by week 4 of recovery (Fig. 9B, red). TCF7l2 colabeled with Wnt negative regulator APC during and after remyelination (Fig. 9B, green).
Based on these temporal dynamics of OL differentiation and TCF7l2 reinduction, we decided to administer tamoxifen daily to 2-month-old Olig2–CreERT2, Tcf7l2fl/fl mice and Tcf7l2fl/fl controls between weeks 5 and 6 of cuprizone diet and analyze the corpus callosum 10 d after the last injection (for experimental design, see Fig. 9C). Plp mRNA ISH demonstrated substantially fewer Plp+ differentiated OLs in the corpus callosum of Olig2–CreERT2, Tcf7l2fl/fl mice compared with Tcf7l2fl/fl controls (Fig. 9D). We also used IHC of CC1 (Fig. 9E) and NG2 to quantify OLs and OPCs, respectively, and found that the density of differentiated OLs in the corpus callosum was significantly lower in Olig2–CreERT2, Tcf7l2fl/fl than that in Tcf7l2fl/fl control mice, whereas the density of OPCs was comparable between these two groups (Fig. 9F). These data suggest that Tcf7l2 reinduction promotes OL differentiation in the cuprizone-induced demyelination/remyelination model.
Discussion
There are several significant findings in this study: (1) in contrast to previous reports, TCF7l2 is upregulated transiently in postmitotic, newly differentiated OLs but not in OPCs; (2) TCF7l2 expression is not associated with the activation of canonical Wnt/β-catenin signaling during developmental myelination and remyelination; (3) TCF7l2 is an intrinsic positive regulator of OL differentiation, and its cKO inhibits OL differentiation; (4) TCF7l2 regulates OL differentiation without modulating Wnt/β-catenin signaling but instead may antagonize inhibition of OL differentiation mediated by BMP/Smad signaling (Grinspan et al., 2000; Samanta and Kessler, 2004; See et al., 2004); and (5) TCF7l2 reinduction after myelin damage promotes OL differentiation during remyelination. Results presented here weaken the hypothesis that TCF7l2 expression inhibits OL differentiation through activation of Wnt/β-catenin signaling. Our experimental data also indicate that the sparse expression of TCF7l2 in active lesions of multiple sclerosis (Fancy et al., 2009; Pedre et al., 2011; Lürbke et al., 2013) may play a Wnt-independent role, and TCF7l2 expression may be an indicator of the occurrence of OL differentiation rather than a sign of blockage of OL differentiation.
It is well accepted that oligodendroglial lineage cells express TCF7l2 among the four members of Wnt effector TCF/LEF1 family. One laboratory found that TCF7l2 is expressed specifically in PDGFRα+ OPCs but not in exon 3b-containing Plp mRNA+ mature OLs (Fancy et al., 2009, 2011). However, our data agree with the reports by Fu et al. (2009) who found that TCF7l2 is restricted to postmitotic premyelinating OLs and is absent in OPCs in the mixed glial cultures from neonate cortex and by Ye et al. (2009) who reported that TCF7l2 is upregulated in CC1+ differentiated OLs. A recent study in human tissue (Lürbke et al., 2013) also reported TCF7l2 expression in mature OLs. The issue of the temporal dynamics of TCF7l2 expression is important because it has been assumed that TCF7l2 is linked to the activation of Wnt/β-catenin signaling in OPCs. Our data do not support a link between TCF7l2 expression and Wnt/β-catenin signaling activation in oligodendroglial lineage cells (both OPCs and OLs). First, conditional disruption of β-catenin affected neither the OL differentiation (Lang et al., 2013) nor the expression of the Wnt target gene Axin2 (Fig. 8I) and TCF7l2 (data not shown). Second, conditional disruption of the Wnt inhibitor APC inhibited OL differentiation, activated Wnt/β-catenin signaling, and, conversely, downregulated TCF7l2 expression (Fig. 1B–D). Third, using the Wnt reporter mouse strains Bat–LacZ and Axin2–LacZ, we found that TCF7l2 expression was not correlated with activation of Wnt signaling (Fig. 4). Fourth, Axin2 mRNA, a reliable marker for Wnt activation, was not detected in TCF7l2+ OLs (Fig. 1I,J), consistent with a recent study (Fancy et al., 2014). Finally, conditional disruption of TCF7l2 did not affect Wnt signaling activity in either neonatal or postnatal oligodendroglial lineage cells (Fig. 8B–E,H). Collectively, our data suggest that neonatal and postnatal oligodendroglial lineage cells do not respond to Wnt signaling through TCF7l2 in vivo.
Wnt/β-catenin signaling has been shown to inhibit both oligodendrogenesis and OL differentiation. Based on the fact that TCF7l2 is the Wnt effector expressed in oligodendroglial lineage cells (Fancy et al., 2009; Fu et al., 2009; Ye et al., 2009), it has been long accepted that TCF7l2 inhibits OL differentiation through activation of Wnt/β-catenin signaling (He et al., 2007; Fancy et al., 2009, 2011; Li et al., 2009; Freese et al., 2010; Kotter et al., 2011). An early study suggested that TCF7l2 mediated the inhibition of OL differentiation caused by YY1 cKO (He et al., 2007; Li et al., 2009). In support of that suggestion, overexpression of TCF7l2 in an immortalized OPC cell line inhibited the luciferase activity driven by the Mbp promoter (He et al., 2007). However, it is possible that overexpression of full-length TCF7l2 in that progenitor cell line created an in vitro “pathological” environment (note that TCF7l2 expression is high in newly differentiated OLs in vivo) in which it inappropriately activated and potentiated Wnt/β-catenin activity, leading to the inhibition of transfected Mbp promoter activity (He et al., 2007). It has been reported that PDGF, which is included in the culture medium or is present in serum supplement to the culture medium, stimulates the activity of Wnt/β-catenin signaling in OPC culture systems (Chew et al., 2011). Subsequently, Ye et al. (2009) reported that expression of a dominant-negative TCF7l2 that lacks a β-catenin-binding domain (thus blocking the Wnt-related function of TCF7l2) accelerated oligodendrogenesis in chick embryos. However, it is not clear whether this precocious oligodendrogenesis was attributable to the inactivation of the Wnt-related function of TCF7l2 or the activation of the Wnt-independent role of TCF7l2, for example repressing BMP signaling activation as we reported here (Fig. 8D,F,G). Alternatively, precocious oligodendrogenesis may have been a consequence of an “ectopic” effect of dominant-negative TCF7l2 overexpression, because dominant-negative forms of TCF7l2 devoid of β-catenin-binding domain are, unlike in embryonic brain, normally absent in the embryonic spinal cord (Vacik et al., 2011; Nagalski et al., 2013). A previous study showed that TCF7l2 is also expressed in neural stem cells (NSCs), including Olig2+ OPC-generating NSCs in embryonic spinal cord before oligodendrogenesis (Wang et al., 2011). Thus, in the future, it would be interesting to assess whether TCF7l2 cKO affects embryonic oligodendrogenesis and, if yes, whether through modulating Wnt signaling, BMP signaling, or both.
Our study has focused on the neonatal and postnatal oligodendroglial lineage and demonstrates that the Wnt-related function of TCF7l2 plays a minor role, if any, in OL differentiation. Most importantly, we show that, contrary to the well accepted hypothesis, TCF7l2 functions as a positive regulator of OL differentiation during both developmental myelination and remyelination. This conclusion concerning the role of TCF7l2 in OL differentiation is compatible with data derived from Tcf7l2-null late embryos or newborns (Fu et al., 2009; Ye et al., 2009).
Our gene deletion data suggest, for the first time, a novel link between TCF7l2 and repression of BMP signaling in the context of the in vivo oligodendroglial lineage. BMP signaling is an inhibitory pathway for oligodendrogenesis and OL differentiation (Grinspan et al., 2000; Samanta and Kessler, 2004; See et al., 2004). The source of BMP ligands has not been established definitively. However, oligodendroglial lineage cells have been shown to secrete BMPs (Kondo and Raff, 2004; Ara et al., 2008; Choe et al., 2014), and BMPs may act in an autocrine manner to affect OL differentiation. Interestingly, BMP4, the most abundant BMP ligand in the CNS, is expressed in OPCs (Choe et al., 2014), significantly upregulated in newly formed OLs, and downregulated in myelinating OLs (Zhang et al., 2014). The genetic data presented here suggest that TCF7l2 antagonizes autocrine oligodendroglial BMP signaling to regulate the timing of OL differentiation. Our study also suggests that, when interpreting the data derived from ectopic expression of the dominant-negative TCF7l2 lacking the β-catenin-binding domain (Ye et al., 2009; Tawk et al., 2011), the previously unrecognized BMP-related role of TCF7l2, as shown here, should be considered. We are currently investigating how TCF7l2 antagonizes BMP signaling in oligodendroglial lineage cells and whether this antagonistic regulation of TCF7l2 and BMP signaling also plays a role in embryonic oligodendrogenesis.
Expression of the myelin genes Plp and Mbp was downregulated significantly in the Olig2–CreERT2, Tcf7l2fl/fl cKO system (Figs. 6D,E, 8B) in which TCF7l2 is initially ablated predominantly in OPCs but was unaltered in the Plp–CreERT2, Tcf7l2fl/fl cKO system in which TCF7l2 is initially ablated predominantly in OLs (Fig. 7B,E). This is quite unexpected considering that expression of dominant-negative TCF7l2 lacking a β-catenin-binding domain in differentiated OLs directly inactivates Plp and Mbp genes in vitro and in zebrafish embryonic spinal cord (Tawk et al., 2011). Our data suggest that TCF7l2 exerts a stage-dependent role during OL development. That is, in postmitotic OPCs that are committed to differentiation and immature OLs in which myelin genes are not fully activated, TCF7l2 may cooperate with other (transcription) factors to activate myelin gene expression to promote OL differentiation, whereas in newly differentiated, myelin gene-expressing OLs, high levels of TCF7l2 may promote terminal maturation and myelination by enhancing lipid synthesis (Chrast et al., 2011). In this context, we found that Elovl1, a fatty acid elongase essential for the synthesis of C24 fatty acids (Ohno et al., 2010) that are highly enriched in myelin lipid (Chrast et al., 2011), is significantly downregulated in the Plp-CreERT2 Tcf7l2fl/fl cKO system at both 24 h and 7 d after Tcf7l2 cKO in OLs (Fig. 7H). Of note, many genes involved in lipid metabolism are direct target genes of TCF7l2 in the liver, and their expression is impaired in Tcf7l2-null newborn livers (Boj et al., 2012). Among all the myelin genes we assessed (Fig. 7E) in the TCF7l2-disrupted OLs, only MAG, which serves as a signaling molecule between myelin and axons (Yin et al., 1998; Pernet et al., 2008), is significantly altered (Fig. 7G). Additional studies are needed to elucidate the function of TCF7l2 in differentiated OLs and its link to lipid metabolism and MAG expression for the terminal myelination event.
Footnotes
This work was supported by Shriners Hospitals for Children and the National Multiple Sclerosis Society.
References
- Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- Angus-Hill ML, Elbert KM, Hidalgo J, Capecchi MR. T-cell factor 4 functions as a tumor suppressor whose disruption modulates colon cell proliferation and tumorigenesis. Proc Natl Acad Sci U S A. 2011;108:4914–4919. doi: 10.1073/pnas.1102300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Taketo MM. Adenomatous polyposis coli (APC): a multi-functional tumor suppressor gene. J Cell Sci. 2007;120:3327–3335. doi: 10.1242/jcs.03485. [DOI] [PubMed] [Google Scholar]
- Ara J, See J, Mamontov P, Hahn A, Bannerman P, Pleasure D, Grinspan JB. Bone morphogenetic proteins 4, 6, and 7 are up-regulated in mouse spinal cord during experimental autoimmune encephalomyelitis. J Neurosci Res. 2008;86:125–135. doi: 10.1002/jnr.21462. [DOI] [PubMed] [Google Scholar]
- Barker N, Huls G, Korinek V, Clevers H. Restricted high level expression of Tcf-4 protein in intestinal and mammary gland epithelium. Am J Pathol. 1999;154:29–35. doi: 10.1016/S0002-9440(10)65247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boj SF, van Es JH, Huch M, Li VS, José A, Hatzis P, Mokry M, Haegebarth A, van den Born M, Chambon P, Voshol P, Dor Y, Cuppen E, Fillat C, Clevers H. Diabetes risk gene and Wnt effector Tcf7l2/TCF4 controls hepatic response to perinatal and adult metabolic demand. Cell. 2012;151:1595–1607. doi: 10.1016/j.cell.2012.10.053. [DOI] [PubMed] [Google Scholar]
- Chew LJ, Shen W, Ming X, Senatorov VV, Jr, Chen HL, Cheng Y, Hong E, Knoblach S, Gallo V. SRY-box containing gene 17 regulates the Wnt/beta-catenin signaling pathway in oligodendrocyte progenitor cells. J Neurosci. 2011;31:13921–13935. doi: 10.1523/JNEUROSCI.3343-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe Y, Huynh T, Pleasure SJ. Migration of oligodendrocyte progenitor cells is controlled by transforming growth factor beta family proteins during corticogenesis. J Neurosci. 2014;34:14973–14983. doi: 10.1523/JNEUROSCI.1156-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrast R, Saher G, Nave KA, Verheijen MH. Lipid metabolism in myelinating glial cells: lessons from human inherited disorders and mouse models. J Lipid Res. 2011;52:419–434. doi: 10.1194/jlr.R009761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai ZM, Sun S, Wang C, Huang H, Hu X, Zhang Z, Lu QR, Qiu M. Stage-specific regulation of oligodendrocyte development by Wnt/beta-catenin signaling. J Neurosci. 2014;34:8467–8473. doi: 10.1523/JNEUROSCI.0311-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerflinger NH, Macklin WB, Popko B. Inducible site-specific recombination in myelinating cells. Genesis. 2003;35:63–72. doi: 10.1002/gene.10154. [DOI] [PubMed] [Google Scholar]
- Fancy SP, Baranzini SE, Zhao C, Yuk DI, Irvine KA, Kaing S, Sanai N, Franklin RJ, Rowitch DH. Dysregulation of the Wnt pathway inhibits timely myelination and remyelination in the mammalian CNS. Genes Dev. 2009;23:1571–1585. doi: 10.1101/gad.1806309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy SP, Kotter MR, Harrington EP, Huang JK, Zhao C, Rowitch DH, Franklin RJ. Overcoming remyelination failure in multiple sclerosis and other myelin disorders. Exp Neurol. 2010;225:18–23. doi: 10.1016/j.expneurol.2009.12.020. [DOI] [PubMed] [Google Scholar]
- Fancy SP, Harrington EP, Yuen TJ, Silbereis JC, Zhao C, Baranzini SE, Bruce CC, Otero JJ, Huang EJ, Nusse R, Franklin RJ, Rowitch DH. Axin2 as regulatory and therapeutic target in newborn brain injury and remyelination. Nat Neurosci. 2011;14:1009–1016. doi: 10.1038/nn.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy SP, Harrington EP, Baranzini SE, Silbereis JC, Shiow LR, Yuen TJ, Huang EJ, Lomvardas S, Rowitch DH. Parallel states of pathological Wnt signaling in neonatal brain injury and colon cancer. Nat Neurosci. 2014;17:506–512. doi: 10.1038/nn.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigenson K, Reid M, See J, Crenshaw EB, 3rd, Grinspan JB. Wnt signaling is sufficient to perturb oligodendrocyte maturation. Mol Cell Neurosci. 2009;42:255–265. doi: 10.1016/j.mcn.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Freese JL, Pino D, Pleasure SJ. Wnt signaling in development and disease. Neurobiol Dis. 2010;38:148–153. doi: 10.1016/j.nbd.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Cai J, Clevers H, Fast E, Gray S, Greenberg R, Jain MK, Ma Q, Qiu M, Rowitch DH, Taylor CM, Stiles CD. A genome-wide screen for spatially restricted expression patterns identifies transcription factors that regulate glial development. J Neurosci. 2009;29:11399–11408. doi: 10.1523/JNEUROSCI.0160-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Kesari S, Cai J. Tcf7l2 is tightly controlled during myelin formation. Cell Mol Neurobiol. 2012;32:345–352. doi: 10.1007/s10571-011-9778-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinspan JB, Edell E, Carpio DF, Beesley JS, Lavy L, Pleasure D, Golden JA. Stage-specific effects of bone morphogenetic proteins on the oligodendrocyte lineage. J Neurobiol. 2000;43:1–17. doi: 10.1002/(SICI)1097-4695(200004)43:1<1::AID-NEU1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Guo F, Ma J, McCauley E, Bannerman P, Pleasure D. Early postnatal proteolipid promoter-expressing progenitors produce multilineage cells in vivo. J Neurosci. 2009;29:7256–7270. doi: 10.1523/JNEUROSCI.5653-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Maeda Y, Ma J, Delgado M, Sohn J, Miers L, Ko EM, Bannerman P, Xu J, Wang Y, Zhou C, Takebayashi H, Pleasure D. Macroglial plasticity and the origins of reactive astroglia in experimental autoimmune encephalomyelitis. J Neurosci. 2011;31:11914–11928. doi: 10.1523/JNEUROSCI.1759-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Maeda Y, Ko EM, Delgado M, Horiuchi M, Soulika A, Miers L, Burns T, Itoh T, Shen H, Lee E, Sohn J, Pleasure D. Disruption of NMDA receptors in oligodendroglial lineage cells does not alter their susceptibility to experimental autoimmune encephalomyelitis or their normal development. J Neurosci. 2012;32:639–645. doi: 10.1523/JNEUROSCI.4073-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow DE, Saul KE, Culp CM, Vesely EM, Macklin WB. Expression of proteolipid protein gene in spinal cord stem cells and early oligodendrocyte progenitor cells is dispensable for normal cell migration and myelination. J Neurosci. 2014;34:1333–1343. doi: 10.1523/JNEUROSCI.2477-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi Y, Nave KA, Casaccia-Bonnefil P. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Raff MC. A role for Noggin in the development of oligodendrocyte precursor cells. Dev Biol. 2004;267:242–251. doi: 10.1016/j.ydbio.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Kotter MR, Stadelmann C, Hartung HP. Enhancing remyelination in disease—can we wrap it up? Brain. 2011;134:1882–1900. doi: 10.1093/brain/awr014. [DOI] [PubMed] [Google Scholar]
- Lang J, Maeda Y, Bannerman P, Xu J, Horiuchi M, Pleasure D, Guo F. Adenomatous polyposis coli regulates oligodendroglial development. J Neurosci. 2013;33:3113–3130. doi: 10.1523/JNEUROSCI.3467-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, Griffiths IR, Nave KA. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- Li H, He Y, Richardson WD, Casaccia P. Two-tier transcriptional control of oligodendrocyte differentiation. Curr Opin Neurobiol. 2009;19:479–485. doi: 10.1016/j.conb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie DC, Colamarino SA, Song HJ, Désiré L, Mira H, Consiglio A, Lein ES, Jessberger S, Lansford H, Dearie AR, Gage FH. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- Lien WH, Fuchs E. Wnt some lose some: transcriptional governance of stem cells by Wnt/beta-catenin signaling. Genes Dev. 2014;28:1517–1532. doi: 10.1101/gad.244772.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lürbke A, Hagemeier K, Cui QL, Metz I, Brück W, Antel J, Kuhlmann T. Limited TCF7L2 expression in MS lesions. PLoS One. 2013;8:e72822. doi: 10.1371/journal.pone.0072822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci U S A. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalski A, Irimia M, Szewczyk L, Ferran JL, Misztal K, Kuznicki J, Wisniewska MB. Postnatal isoform switch and protein localization of LEF1 and TCF7L2 transcription factors in cortical, thalamic, and mesencephalic regions of the adult mouse brain. Brain Struct Funct. 2013;218:1531–1549. doi: 10.1007/s00429-012-0474-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno Y, Suto S, Yamanaka M, Mizutani Y, Mitsutake S, Igarashi Y, Sassa T, Kihara A. ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc Natl Acad Sci U S A. 2010;107:18439–18444. doi: 10.1073/pnas.1005572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedre X, Mastronardi F, Bruck W, López-Rodas G, Kuhlmann T, Casaccia P. Changed histone acetylation patterns in normal-appearing white matter and early multiple sclerosis lesions. J Neurosci. 2011;31:3435–3445. doi: 10.1523/JNEUROSCI.4507-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernet V, Joly S, Christ F, Dimou L, Schwab ME. Nogo-A and myelin-associated glycoprotein differently regulate oligodendrocyte maturation and myelin formation. J Neurosci. 2008;28:7435–7444. doi: 10.1523/JNEUROSCI.0727-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo JK, Cate HS. Signalling pathways that inhibit the capacity of precursor cells for myelin repair. Int J Mol Sci. 2013;14:1031–1049. doi: 10.3390/ijms14011031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- See J, Zhang X, Eraydin N, Mun SB, Mamontov P, Golden JA, Grinspan JB. Oligodendrocyte maturation is inhibited by bone morphogenetic protein. Mol Cell Neurosci. 2004;26:481–492. doi: 10.1016/j.mcn.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Kagawa T, Wada T, Muroyama Y, Takada S, Ikenaka K. Wnt signaling controls the timing of oligodendrocyte development in the spinal cord. Dev Biol. 2005;282:397–410. doi: 10.1016/j.ydbio.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Swiss VA, Nguyen T, Dugas J, Ibrahim A, Barres B, Androulakis IP, Casaccia P. Identification of a gene regulatory network necessary for the initiation of oligodendrocyte differentiation. PLoS One. 2011;6:e18088. doi: 10.1371/journal.pone.0018088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawk M, Makoukji J, Belle M, Fonte C, Trousson A, Hawkins T, Li H, Ghandour S, Schumacher M, Massaad C. Wnt/β-Catenin signaling is an essential and direct driver of myelin gene expression and myelinogenesis. J Neurosci. 2011;31:3729–3742. doi: 10.1523/JNEUROSCI.4270-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacik T, Stubbs JL, Lemke G. A novel mechanism for the transcriptional regulation of Wnt signaling in development. Genes Dev. 2011;25:1783–1795. doi: 10.1101/gad.17227011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lei Q, Oosterveen T, Ericson J, Matise MP. Tcf/Lef repressors differentially regulate Shh-Gli target gene activation thresholds to generate progenitor patterning in the developing CNS. Development. 2011;138:3711–3721. doi: 10.1242/dev.068270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska MB, Nagalski A, Dabrowski M, Misztal K, Kuznicki J. Novel beta-catenin target genes identified in thalamic neurons encode modulators of neuronal excitability. BMC Genomics. 2012;13:635. doi: 10.1186/1471-2164-13-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TL, Bercury KK, Cifelli SE, Mursch LE, Min J, Dai J, Macklin WB. mTOR: a link from the extracellular milieu to transcriptional regulation of oligodendrocyte development. ASN Neuro. 2013;5:e00108. doi: 10.1042/AN20120092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, Hsieh J, Bassel-Duby R, Olson EN, Lu QR. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Crawford TO, Griffin JW, Tu Ph, Lee VM, Li C, Roder J, Trapp BD. Myelin-associated glycoprotein is a myelin signal that modulates the caliber of myelinated axons. J Neurosci. 1998;18:1953–1962. doi: 10.1523/JNEUROSCI.18-06-01953.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]