Abstract
Heterotrophic protists play pivotal roles in aquatic ecosystems by transferring matter and energy, including lipids, from primary producers to higher trophic predators. Using Oxyrrhis marina as a model organism, changes to the non-saponifiable protist lipids were investigated under satiation and starvation conditions. During active feeding on the alga Cryptomonas sp., the O. marina hexane soluble non-saponifiable fraction lipid profile reflected its food source with the observed presence of long chain mono-unsaturated fatty alcohols up to C25:1. Evidence of trophic upgrading in O. marina was observed with long chain mono-unsaturated fatty alcohol accumulation of up to C35:1. To the best of our knowledge, this is the first evidence that heterotrophic dinoflagellates are capable of producing ester derived alcohols and that dinoflagellates like O. marina are capable of synthesizing fatty alcohols up to C35. Additionally, we show evidence of trophic upgrading of lipids. During a 20-day resource deprivation, the lipid profile remained constant. During starvation, the mobilization of wax esters as energy stores was observed with long chain fatty alcohols mobilized first. Changes in lipid class profile and utilization of wax esters in O. marina provides insight into the types of lipids available for energy demand, the transfer of lipids through the base of marine food webs, and the catabolic response induced by resource deprivation.
Keywords: Oxyrrhis marina, Wax ester, Resource deprivation, Trophic upgrading, Catabolism, Microzooplankton
Introduction
Heterotrophic dinoflagellates are ubiquitous, important components of the pelagic protozoan community. They are significant consumers of bacterial and phytoplankton biomass, and contribute to the cycling of organic matter and nutrients, serving as important trophic links in marine microbial food webs (Strom, 1991; Sherr & Sherr, 1994; Steinberg & Landry, 2017). Trophic interactions within complex marine food webs can strongly influence pathways and efficiencies of material and energy transfer to higher level consumers (Anderson & Menden-Deuer, 2017; Mitra & Flynn, 2005; Rose et al., 2011). Heterotrophic protists, like dinoflagellates, add biochemical value during this transfer through the production and chemical elaboration of compounds (Klein Breteler et al., 1999). Thus changes to the diet of heterotrophic dinoflagellates (i.e., through starvation) can alter the biomass and cellular composition of herbivores. Although heterotrophic protists add biochemical value during trophic transfer, little is known about how cellular composition changes in response to food availability. These changes in biomass can impact higher trophic levels through changes in cellular composition. It has been shown that heterotrophic dinoflagellates such as Oxyhrris marina, Gyrodinium dominans, and G. spirale can survive long periods (>10 days) without algal prey. For example, starvation of O. marina for up to 3 weeks resulted in a reduction in cell volume of 17–57% with some cells deformed and transparent (Menden-Deuer et al., 2005). It has been puzzling how a single celled heterotrophic organism can sustain survival in the absence of substantive organic matter, particularly over such extended periods.
Lipids are important energy stores that can be used in times of resource deprivation. Many of the studies on lipids of dinoflagellates fed on algal prey have focused on fatty acid and sterol composition (Klein Breteler et al., 1999; Veloza, Chu & Tang, 2006; Park et al., 2016). These studies have suggested that the fatty acid composition of O. marina may not be dependent on its prey and have highlighted this organism’s ability to upgrade lipids acquired from its diet. A subclass of the neutral lipids, wax esters, have traditionally only been found in marine animal phyla (Sargent, Gatten & McIntosh, 1977; Bauermeister & Sargent, 1979), but some examples have been reported in zooplankton species. Wax esters have been observed in the chlorophyte Chlorella kessleri (Sargent, Gatten & Henderson, 1981), the cryptomonad Chroomonas salina (Antia Naval et al., 1974; Henderson & Sargent, 1989), and the euglenoid Euglena gracilis (Furuhashi et al., 2015). In Chroomonas salina ester derived alcohols are almost exclusively saturated with the most predominant species C13 and C15 while in E. gracilis ester alcohol moieties of up to C22 have been observed.
The mechanism of trophic upgrading by heterotrophic protists may bridge the gap and deliver essential nutrients between higher trophic levels (Klein Breteler et al., 1999; Veloza, Chu & Tang, 2006). Given the importance of heterotrophic dinoflagellates in marine food webs by providing essential nutrients to higher trophic levels, an understanding of the changes to the lipid profile under varying availability of prey can provide insight into the nutritional quality available to higher trophic levels. Here, we report the changes to the non-saponifiable fraction (NSF) lipid composition of the representative dinoflagellate O. marina, as our model organisms during active feeding and in response to long term starvation. O. marina is a free living, cosmopolitan, and phagotrophic alveolate that feeds on a variety of algae and bacteria (Landry et al., 2000) and has been recognized for its unique starvation ability lasting for several months (Menden-Deuer et al., 2005; Calbet et al., 2013).
Materials and Methods
Materials
Long chain fatty alcohol standards were obtained from Millipore Sigma (Burlington, MA, USA). High pressure liquid chromatography (HPLC) lipid standards included a phospholipid mixture and mono-, di-, and tri-acylglycerol mixtures (Millipore Sigma, Burlington, MA, USA). Nile Red was purchased from Invitrogen (Carlsbad, CA, USA). All solvents used were of HPLC or spectroscopic grade. All HPLC mobile phases were filtered through a 0.22 μm membrane prior to use.
Cell culture
Non-axenic cultures of the cryptophyte alga Cryptomonas sp. were maintained in triplicate two-L, transparent polycarbonate (PC) bottles to serve as prey under culture conditions that included a 12:12 h light-dark cycle at 14 °C, salinity of approximately 30 practical salinity units (PSU), and a light intensity of 70–80 µmol photons · m−2 · s−1. The culture medium was prepared from sterile autoclaved 0.2 µm filtered seawater amended with nutrients following the f/2 medium without silica recipe of Guillard (1975). The seawater was collected at high tide from Narragansett Bay, Rhode Island, USA.
Non-axenic, clonal cultures of the heterotrophic dinoflagellate O. marina (CCMP3375; Om), were established via single-cell isolation and grown in two-L transparent PC bottles on a 12 h:12 h light–dark cycle at 14.5 °C, salinity of approximately 30 psu, and a light intensity of 8–15 µmol photons · m−2 · s−1. Grazers were maintained in exponential growth phase by feeding them once a week with Cryptomonas sp. prey and diluted with autoclaved filtered seawater.
Estimating cell abundance, size, and biomass
Grazer and phytoplankton prey abundance and cell size were monitored using a Multisizer TM 3 Coulter counter (version 3.53; Beckman Coulter, Indianapolis, IN, USA). The Coulter counter provided a more rapid and reliable sampling approach than microscopy and allowed convenient monitoring of the cultures over the course of the experiments (Kim & Menden-Deuer, 2013). Grazer and phytoplankton prey were easily distinguishable on the Coulter counter based on their respective size distributions. Grazer volume was determined using the equivalent spherical diameter measurements from the Coulter counter, and converted to carbon biomass (pg C. cell−1) using previously established conversion equations (Menden-Deuer & Lessard, 2000).
Starvation and re-feeding experiments were set-up using established methods (Anderson & Menden-Deuer, 2017). Briefly, grazers fed with prey were transferred to triplicate four-L bottles and starved for 1–3 weeks until a reduction in predator abundance or cell size was detected, which indicated a negative impact of algal prey deprivation and marked the initiation of grazer starvation (Anderson & Menden-Deuer, 2017). Grazers were starved for 20 days, received a fresh pulse of phytoplankton prey after starvation, and were monitored for 3–7 days after re-feeding. Samples were taken at regular intervals (0, 8, 10, 15, 18, 20 days) during the starvation for measurements of grazer abundance and cell size. Grazer and prey abundances obtained using the coulter counter and flow cytometer were verified by light microscopy.
Lipid isolation
Filter membranes containing Cryptomonas sp. or O. marina cells were extracted using the Bligh-Dyer procedure (Bligh & Dyer, 1959). Briefly, 105 to 107 cells adhered to membrane were suspended in five mL of 1:2 (v/v) CHCl3:MeOH and sonicated for 5 min at 65% amplitude, (10 s on, 20 s off). Lipids were extracted with the addition of three mL of 2:1 MeOH:CHCl3 and mixed by vortexing. The sample was then converted to a two phase Bligh-Dyer by the addition of one mL CHCl3 and 1.8 mL dH2O. The resulting biphasic samples were centrifuged (4,000 rpm, 2 min) and the bottom layer was recovered and transferred to a clean sample vial. The organic layer was dried under a stream on N2. Samples were then analyzed by HPLC. Portions of the lipid extracts were subjected to saponification (methanolic KOH) (Haubrich et al., 2015) for analysis of neutral lipids and analyzed by gas chromatography-mass spectrometry (GCMS) without further derivatization. Negative controls of solvent extracted filter membranes were incorporated into the experiment to control for contaminants arising from the filter membranes.
Lipid staining and flow cytometry
Samples were stained with Nile Red as described by De la Jara et al. (2003). The optimal fluorescence of Nile Red can be highly selective and variable based on dye concentration and cell type stained (Rumin et al., 2015), and given the novelty of Nile Red staining with marine heterotrophic dinoflagellates, we followed an optimization protocol. The effect of several parameters on dye permeation and fluorescence were tested such as final dye concentration (0.5–5 µg mL−1), incubation time (5–30 min), solvents (e.g., DMSO and acetone), and temperature (Rumin et al., 2015). An optimum Nile Red concentration of two µg mL−1 dissolved in acetone was determined based on fluorescence profiles (via flow cytometry), and thus represented the dye concentration used in starvation experiments. Triplicate five mL samples were spiked with Nile Red, gently vortexed, and incubated for 10 min at room temperature in the dark to ensure dye permeation while avoiding quenching effects. Nile Red samples were then fixed using glutaraldehyde (0.5% final conc. v/v), flash frozen in liquid N2, and stored at −80 °C until flow cytometry analysis (measured within 1–2 months).
Triplicate samples were analyzed using flow cytometry (BD-Influx flow cytometer, Becton Dickinson Instruments) with an excitation wavelength of 488 nm. A minimum of 200 cells were counted for each sample. Populations of cells were identified based on fluorescence vs forward and side scatter. Chlorophyll autofluorescence was determined using a 692/40 nm filter and differentiated autotrophic prey from grazer lipid fluorescence. Lipid content, measured as fluorescence intensity per cell, was estimated from the fluorescence emission of NR-stained cells using 580/30 nm (neutral lipid), and 610/20 nm (polar lipid) filters (Alonzo & Mayzaud, 1999; De la Jara et al., 2003). Non-stained samples were used to control for NR autofluorescence; fluorescence of unstained cells was consistently less than 10% of stained samples. Lipid content for each grazer species was measured as a function of fluorescence, and expressed as relative fluorescence units, rather than as equivalent lipid concentrations.
Non-saponifiable lipid extracts
Lipid extracts were treated with 10:10:80 (v/w/v) of dH2O:KOH:MeOH and refluxed for 30 min (Haubrich et al., 2015). After cooling to room temperature, water and hexanes were added. Saponified lipids were extracted three times with hexanes and pooled. The combined hexane extracts were dried over anhydrous Na2SO4 and evaporated to dryness under a stream of N2. Prior to analysis samples were dissolved in equal volumes of CHCl3.
Chromatographic analysis
RP-HPLC
Chromatographic separation was performed on a Prominence uHPLC system (Shimadzu, Columbia, MD, USA) equipped with a COSMOSIL 5 μ C18-MS II (4.6 × 150 mm) column (Nacalai Tesque Inc., Kyoto, Japan). Lipid classes were separated employing a binary gradient system described by Knittelfelder et al. (2014) with detection at 205 nm (Guarrasi et al., 2010). Mobile phase A consisted of water:methanol (1:1 v/v), mobile phase B was 2-propanol. Both solvents contained phosphoric acid (eight μM) and formic acid (0.1% v/v). A linear gradient with initial conditions starting at 45% mobile phase B was increased to 90% B over 30 min. Mobile phase B was then increased to 100% over 2 min and was held at 100% for 10 min. The column was re-equilibrated for 15 min between injections. Retention time regions for polar, and neutral lipids were established using commercial lipid standards. Data was analyzed using LabSolutions (Shimadzu, Columbia, MD, USA) and statistical analysis (ANOVA) performed using GraphPad Prism version 7.0. All samples were run in biological triplicate and technical duplicate.
GCMS
NSF lipid samples were analyzed GCMS with an Agilent 7890A gas chromatograph equipped with a 5975C electron impact mass spectrometer set to 70 eV using a Restek Rtx-5 column (30 m × 25 μm diameter). The GC flow rate of He was set at 1.2 mL/min, injector port set to 250 °C, and the initial temperature set at 170 °C, held for 1 min, then increased at 20 °C/min to a final temperature of 280 °C (Haubrich et al., 2015). Chromatograms were processed using ChemStation (Agilent, Santa Clara, CA, USA) and analyte identification of the resulting chromatograms was performed via interrogation of resulting electron impact mass spectra with the NIST database and manual analysis. Octadecanol (RT = 5.001 min) was the standard for determination of relative retention time. All samples were run in biological triplicate and technical duplicate.
Results and discussion
O. marina lipid class profile
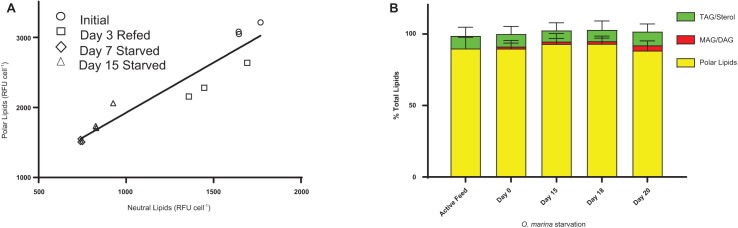
The lipidome of protist predators can change rapidly in response to environmental conditions. Nile Red lipid staining of O. marina demonstrated significant changes in the concentration of neutral and polar lipids in satiated and starved cells (Fig. 1A). A linear relationship (r2 = 0.9044) in the decrease in polar and neutral lipids during starvation was observed via flow cytometry (Fig. 1A). The apparent differences in total polar and neutral lipids after 7 and 15 days respectively are non-significant (p > 0.05). The change in polar and neutral lipid concentration during starvation was further investigated by RP-HPLC. While total polar and neutral lipids decreased during starvation the relative abundance of the subclasses of these lipids showed a consistent ratio of polar to neutral lipids of 92.1 ± 3.2:3.0 ± 2.2:5.7 ± 2.6 (PL:MAG/DAG:TAG) was maintained (Fig. 1B). The lipid class composition of O. marina is comparable to what has been previously observed in dinoflagellates and phytoplankton (Harvey et al., 1988; Bourdier & Amblard, 1989; Yoon et al., 2017). Nutrient deprived O. marina have been shown to decrease cell volume by 17–57% (Anderson & Menden-Deuer, 2017) and increase expression of genes involved in the degradation of lipids (Rubin et al., 2019) suggesting a homeostatic requirement for O. marina to maintain relative amounts of each lipid class as cell volume decreases and stress-induced catabolism progresses.
Figure 1. O. marina maintains a constant ratio of polar:neutral lipids during a 20-day starvation period.
(A) Linear relationship between depletion of polar and neutral lipids during starvation measured using flow cytometry. O. marina feeding on Cryptomonas sp. polar vs neutral lipids is characterized by a significant, linear relationship (model II regression, p < 0.0001, r2 = 0.903). (B) RP-HPLC analysis of lipid class. O. marina were fed Cryptomonas sp. prior to start of starvation. O. marina maintains a balance of 92.1 ± 3.2:3.0 ± 2.2:5.7 ± 2.6 (phospholipid (PL):monoacyl/diacylglyerols (MAG/DAG):triacylglycerol/sterol/wax ester (TAG/sterol/WE) as cell volume decreases. Lipid extracts were analyzed in biological triplicate and technical duplicate. Statistical analysis (ANOVA, p < 0.01) of lipid class abundance indicates no significant difference during the starvation period.
GCMS analysis of hexane soluble non-saponifiable fraction
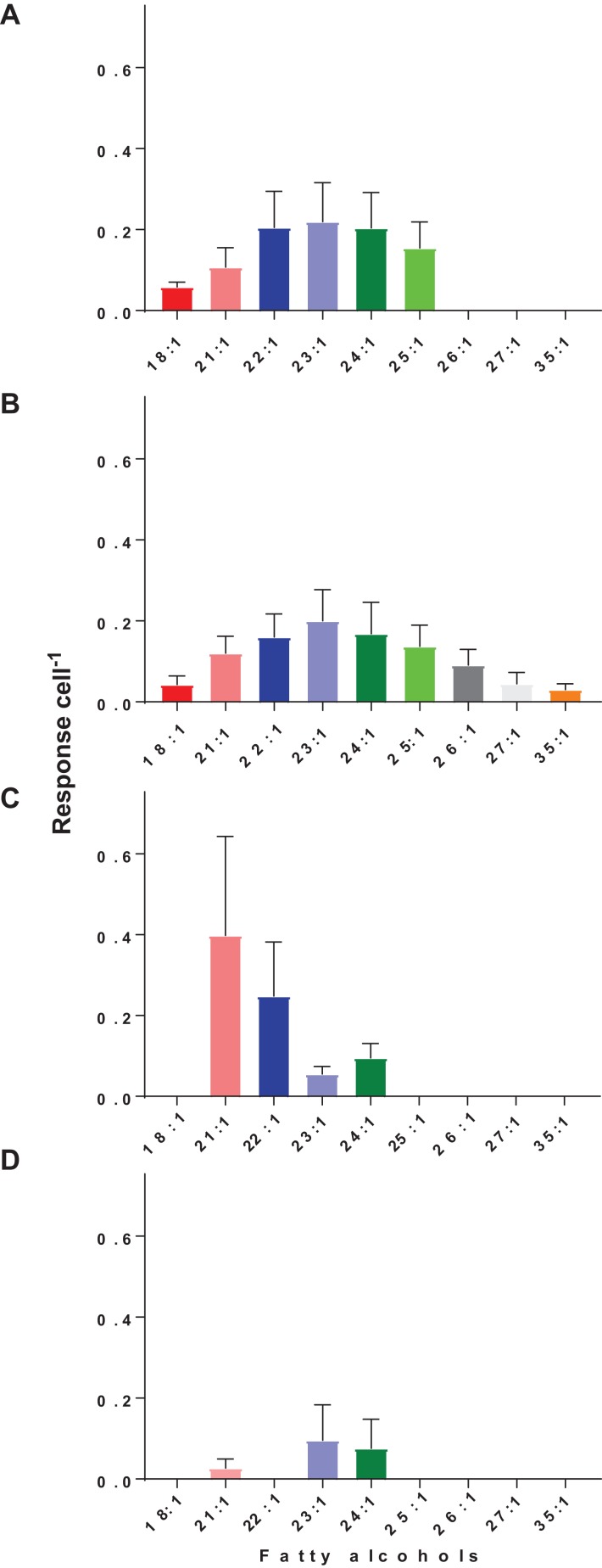
The prey Cryptomonas sp. contained wax ester-derived mono-unsaturated fatty alcohols ranging in size from C18:1 to C25:1 (Fig. 2A; Fig. S1; Table S1). These results are in contrast to observations in other phytoplankton in which fatty alcohols up to C22 have been observed (Henderson & Sargent, 1989; Furuhashi et al., 2015). Our results obtained here for Cryptomonas sp. indicate the presence of both even and odd chain alcohols.
Figure 2. Changes in hexane soluble NSF lipid extracts during active feeding and prolonged starvation of O. marina.
(A) Prey Cryptomonas sp., (B) O. marina during active feeding on Cryptomonas sp., (C) O. marina after 15 days starvation, (D) O. marina after 18 days starvation. Day zero of starvation commenced when prey were not detectable by Coulter Counter and microscopy. Evidence of trophic upgrading of observed fatty alcohols in actively feeding O. marina. During a 20-day starvation, O. marina mobilized wax esters as an energy source.
Oxyrrhis marina actively feeding on Cryptomonas sp. showed the same base fatty alcohol profile as its prey. In addition to the Cryptomonas sp. derived fatty alcohols, there was evidence of trophic upgrading in that derived fatty alcohols were upgraded to chain lengths of up to C35, which were not detected in the prey fatty alcohol profiles (Fig. 2B). To the best of our knowledge, this is the first evidence that heterotrophic dinoflagellates are capable of producing ester derived alcohols and that dinoflagellates like O. marina are capable of synthesizing fatty alcohols up to C35, compared to zooplankton species from arctic waters where mono-unsaturated fatty alcohols up to C22 have been observed (Sargent, Gatten & McIntosh, 1977; Wakeham, 1982).
Over a 20 day starvation period, O. marina appeared to mobilize the fatty alcohols as energy reserves with longer chain fatty alcohols utilized first (Figs. 2C and 2D). After 20 days near complete depletion of fatty alcohols was observed, consistent with observations of increased expression of genes involved in lipid degradation in starved O. marina (Rubin et al., 2019).
Wax ester production in dinoflagellates has been suggested to be involved in buoyancy regulation and as a deposit of an energy rich food reserve during periods of low prey abundance (Sargent, Gatten & Henderson, 1981). Wax esters and TAGs are commonly found in lipid bodies within dinoflagellates. These compounds have been the focus of investigations during nitrogen stress (Dagenais Bellefeuille et al., 2014) and in coral-dinoflagellate symbiont relationships (Chen et al., 2012). This analysis of changes in lipid class profile and utilization of wax esters in O. marina provides insight into the catabolic response induced by general resource deprivation. We have provided evidence that during starvation in O. marina that wax esters are mobilized as energy stores while the ratio of polar:non-polar lipids remain constant as cell volume decreases. These data provide information on the changes in lipid content, in particular the NSF in O. marina during prolonged resource deprivation.
Conclusions
Here, we evaluated the lipid profile of a marine herbivorous zooplankton during starvation and contrasted this with the NSF lipid profile of its phytoplankton prey. We found evidence both of direct trophic transfer of lipids from the algal source, as well as trophic upgrade of neutral lipids. While diet deprivation did not seem to affect ratios of polar to neutral lipids in starved O. marina, starvation was accompanied by a time-dependent depletion of longer-chain fatty alcohols from energy stores. In addition, the presence of remarkably long fatty alcohols was noted in saponified lipids of a heterotrophic dinoflagellate. Characterization and quantification of catabolic responses to resource stress in marine herbivores provides opportunities to use lipids as biomarkers for energy demand and assessment of energy status in marine microbial food webs and improve coastal ecosystem models.
Supplemental Information
Compounds were identified through NIST library search and manual analysis of associated EIMS spectra.
(A) Cryptomonas sp. and (B) O. marina during active feeding and (C) 15 and (D) 20 days of starvation, respectively. Day zero of starvation commenced when prey were not detectable by Coulter counter and microscopy. During a 20 day starvation, O. marina mobilized wax esters as energy source.
Each file chromatographic file (GCMS, HPLC) has been converted to the corresponding Analytical Data Interchange format (.cdf). Flow cytometry raw data is provided as an excel spreadsheet.
Acknowledgments
We would like to thank Krystyna Kula for technical assistance.
Funding Statement
This research was supported by the Rhode Island Science and Technology Advisory Council (to Susanne Menden-Deuer, Tatiana Rynearson, Christopher W. Reid) and NASA grant 80NSSC17K0716 (to Susanne Menden-Deuer, Tatiana Rynearson) as part of the Export Processes in the Global Ocean from Remote Sensing (EXPORTS) field campaign. This study was conducted using infrastructure supported by NSF EPSCoR research infrastructure improvement awards EPS-1004057 and OIA-1655221. There was no additional external funding received for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
Susanne Menden-Deuer is an Academic Editor for PeerJ.
Author Contributions
Keyana Roohani performed the experiments, analyzed the data, authored or reviewed drafts of the paper, approved the final draft.
Brad A. Haubrich conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Kai-Lou Yue performed the experiments, approved the final draft, prepared lipid extracts, saponified lipid samples, prepared and ran samples on GCMS.
Nigel D’Souza performed the experiments, approved the final draft.
Amanda Montalbano performed the experiments, analyzed the data, authored or reviewed drafts of the paper, approved the final draft.
Tatiana Rynearson conceived and designed the experiments, analyzed the data, authored or reviewed drafts of the paper, approved the final draft.
Susanne Menden-Deuer conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Christopher W. Reid conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Data Availability
The following information was supplied regarding data availability:
Raw data in the format of GCMS and HPLC chromatograms are provided as .netCDF files in the Supplemental Materials.
References
- Alonzo & Mayzaud (1999).Alonzo F, Mayzaud P. Spectrofluorometric quantification of neutral and polar lipids in zooplankton using Nile red. Marine Chemistry. 1999;67(3–4):289–301. doi: 10.1016/S0304-4203(99)00075-4. [DOI] [Google Scholar]
- Anderson & Menden-Deuer (2017).Anderson SR, Menden-Deuer S. Growth, grazing, and starvation survival in three heterotrophic dinoflagellate species. Journal of Eukaryotic Microbiology. 2017;64(2):213–225. doi: 10.1111/jeu.12353. [DOI] [PubMed] [Google Scholar]
- Antia Naval et al. (1974).Antia Naval J, Lee Richard F, Nevenzel Judd C, Cheng Joseph Y. Wax ester production by the marine Cryptomonad Chroomonas salina grown photoheterotrophically on glycerol. Journal of Protozoology. 1974;21(5):768–771. doi: 10.1111/j.1550-7408.1974.tb03749.x. [DOI] [PubMed] [Google Scholar]
- Bauermeister & Sargent (1979).Bauermeister A, Sargent JR. Wax esters: major metabolites in the marine environment. Trends in Biochemical Sciences. 1979;4(9):209–211. doi: 10.1016/0968-0004(79)90082-3. [DOI] [Google Scholar]
- Bligh & Dyer (1959).Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology. 1959;37(1):911–917. doi: 10.1139/y59-099. [DOI] [PubMed] [Google Scholar]
- Bourdier & Amblard (1989).Bourdier GG, Amblard CA. Lipids in Acanthodiaptomus denticomis during starvation and fed on three different algae. Journal of Plankton Research. 1989;11(6):1201–1212. doi: 10.1093/plankt/11.6.1201. [DOI] [Google Scholar]
- Calbet et al. (2013).Calbet A, Isari S, Martínez RA, Saiz E, Garrido S, Peters J, Borrat RM, Alcaraz M. Adaptations to feast and famine in different strains of the marine heterotrophic dinoflagellates Gyrodinium dominans and Oxyrrhis marina. Marine Ecology Progress Series. 2013;483:67–84. doi: 10.3354/meps10291. [DOI] [Google Scholar]
- Chen et al. (2012).Chen W-NU, Kang H-J, Weis VM, Mayfield AB, Jiang P-L, Fang L-S, Chen C-S. Diel rhythmicity of lipid-body formation in a coral-Symbiodinium endosymbiosis. Coral Reefs. 2012;31(2):521–534. doi: 10.1007/s00338-011-0868-6. [DOI] [Google Scholar]
- Dagenais Bellefeuille et al. (2014).Dagenais Bellefeuille S, Dorion S, Rivoal J, Morse D. The dinoflagellate Lingulodinium polyedrum responds to N depletion by a polarized deposition of starch and lipid bodies. PLOS ONE. 2014;9(11):e111067. doi: 10.1371/journal.pone.0111067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Jara et al. (2003).De la Jara A, Mendoza H, Martel A, Molina C, Nordstron L, De la Rosa V, Diaz R. Flow cytometric determination of lipid content in a marine dinoflagellate, Crypthecodinium cohnii. Journal of Applied Phycology. 2003;15(5):433–438. doi: 10.1023/A:1026007902078. [DOI] [Google Scholar]
- Furuhashi et al. (2015).Furuhashi T, Ogawa T, Nakai R, Nakazawa M, Okazawa A, Padermschoke A, Nishio K, Hirai MY, Arita M, Ohta D. Wax ester and lipophilic compound profiling of Euglena gracilis by gas chromatography-mass spectrometry: toward understanding of wax ester fermentation under hypoxia. Metabolomics. 2015;11(1):175–183. doi: 10.1007/s11306-014-0687-1. [DOI] [Google Scholar]
- Guarrasi et al. (2010).Guarrasi V, Mangione MR, Sanfratello V, Martorana V, Bulone D. Quantification of underivatized fatty acids from vegetable oils by HPLC with UV detection. Journal of Chromatographic Science. 2010;48(8):663–668. doi: 10.1093/chromsci/48.8.663. [DOI] [PubMed] [Google Scholar]
- Guillard (1975).Guillard RRL. Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH, editors. Culture of Marine Invertebrate Animals: Proceedings—1st Conference on Culture of Marine Invertebrate Animals Greenport. Boston: Springer; 1975. pp. 29–60. [Google Scholar]
- Harvey et al. (1988).Harvey HR, Bradshaw SA, O’Hara SCM, Eglinton G, Corner EDS. Lipid composition of the marine dinoflagellate Scrippsiella trochoidea. Phytochemistry. 1988;27(6):1723–1729. doi: 10.1016/0031-9422(88)80432-1. [DOI] [Google Scholar]
- Haubrich et al. (2015).Haubrich BA, Singha UK, Miller MB, Nes CR, Anyatonwu H, Lecordier L, Patkar P, Leaver DJ, Villalta F, Vanhollebeke B, Chaudhuri M, Nes WD. Discovery of an ergosterol-signaling factor that regulates Trypanosoma brucei growth. Journal of Lipid Research. 2015;56(2):331–341. doi: 10.1194/jlr.M054643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson & Sargent (1989).Henderson RJ, Sargent JR. Lipid composition and biosynthesis in ageing cultures of the marine cryptomonad, Chroomonas salina. Phytochemistry. 1989;28(5):1355–1361. doi: 10.1016/S0031-9422(00)97745-8. [DOI] [Google Scholar]
- Kim & Menden-Deuer (2013).Kim H, Menden-Deuer S. Reliability of rapid, semi-automated assessment of plankton abundance, biomass, and growth rate estimates: Coulter counter versus light microscope measurements. Limnology and Oceanography: Methods. 2013;11(7):382–393. doi: 10.4319/lom.2013.11.382. [DOI] [Google Scholar]
- Klein Breteler et al. (1999).Klein Breteler WCM, Schogt N, Baas M, Schouten S, Kraay GW. Trophic upgrading of food quality by protozoans enhancing copepod growth: role of essential lipids. Marine Biology. 1999;135(1):191–198. doi: 10.1007/s002270050616. [DOI] [Google Scholar]
- Knittelfelder et al. (2014).Knittelfelder OL, Weberhofer BP, Eichmann TO, Kohlwein SD, Rechberger GN. A versatile ultra-high performance LC-MS method for lipid profiling. Journal of Chromatography B. 2014;951–952:119–128. doi: 10.1016/j.jchromb.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry et al. (2000).Landry MR, Constantinou J, Latasa M, Brown SL, Bidigare RR, Ondrusek ME. Biological response to iron fertilization in the eastern equatorial Pacific (IronEx II). III. Dynamics of phytoplankton growth and microzooplankton grazing. Marine Ecology Progress Series. 2000;201:57–72. doi: 10.3354/meps201057. [DOI] [Google Scholar]
- Menden-Deuer & Lessard (2000).Menden-Deuer S, Lessard EJ. Carbon to volume relationships for dinoflagellates, diatoms, and other protist plankton. Limnology and Oceanography. 2000;45(3):569–579. doi: 10.4319/lo.2000.45.3.0569. [DOI] [Google Scholar]
- Menden-Deuer et al. (2005).Menden-Deuer S, Lessard EJ, Satterberg J, Grünbaum D. Growth rates and starvation survival of three species of the pallium-feeding, thecate dinoflagellate genus Protoperidinium. Aquatic Microbial Ecology. 2005;41:145–152. doi: 10.3354/ame041145. [DOI] [Google Scholar]
- Mitra & Flynn (2005).Mitra A, Flynn KJ. Predator–prey interactions: is ‘ecological stoichiometry’ sufficient when good food goes bad? Journal of Plankton Research. 2005;27(5):393–399. doi: 10.1093/plankt/fbi022. [DOI] [Google Scholar]
- Park et al. (2016).Park J, Jeong HJ, Yoon EY, Moon SJ. Easy and rapid quantification of lipid contents of marine dinoflagellates using the sulpho-phospho-vanillin method. ALGAE. 2016;31(4):391–401. doi: 10.4490/algae.2016.31.12.7. [DOI] [Google Scholar]
- Rose et al. (2011).Rose KA, Allen JI, Artioli Y, Barange M, Blackford J, Carlotti F, Cropp R, Daewel U, Edwards K, Flynn K, Hill SL, HilleRisLambers R, Huse G, Mackinson S, Megrey B, Moll A, Rivkin R, Salihoglu B, Schrum C, Shannon L, Shin Y-J, Smith SL, Smith C, Solidoro C, St. John M, Zhou M. End-to-end models for the analysis of marine ecosystems: challenges, issues, and next steps. Marine and Coastal Fisheries. 2011;2(1):115–130. doi: 10.1577/C09-059.1. [DOI] [Google Scholar]
- Rubin et al. (2019).Rubin ET, Cheng S, Montalbano AL, Menden-Deuer S, Rynearson TA. Transcriptomic response to feeding and starvation in a herbivorous dinoflagellate. Frontiers in Marine Science. 2019;6:246. doi: 10.3389/fmars.2019.00246. [DOI] [Google Scholar]
- Rumin et al. (2015).Rumin J, Bonnefond H, Saint-Jean B, Rouxel C, Sciandra A, Bernard O, Cadoret J-P, Bougaran G. The use of fluorescent Nile red and BODIPY for lipid measurement in microalgae. Biotechnology for Biofuels. 2015;8(1):42. doi: 10.1186/s13068-015-0220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent, Gatten & Henderson (1981).Sargent JR, Gatten RR, Henderson RJ. Marine wax esters. Pure and Applied Chemistry. 1981;53(4):867–871. doi: 10.1351/pac198153040867. [DOI] [Google Scholar]
- Sargent, Gatten & McIntosh (1977).Sargent JR, Gatten RR, McIntosh RJ. Wax esters in the marine environment—their occurrence, formation, transformation and ultimate fates. Marine Chemistry. 1977;5(4–6):573–584. doi: 10.1016/0304-4203(77)90043-3. [DOI] [Google Scholar]
- Sherr & Sherr (1994).Sherr EB, Sherr BF. Bacterivory and herbivory: key roles of phagotrophic protists in pelagic food webs. Microbial Ecology. 1994;28(2):223–235. doi: 10.1007/BF00166812. [DOI] [PubMed] [Google Scholar]
- Steinberg & Landry (2017).Steinberg DK, Landry MR. Zooplankton and the ocean carbon cycle. Annual Review of Marine Science. 2017;9(1):413–444. doi: 10.1146/annurev-marine-010814-015924. [DOI] [PubMed] [Google Scholar]
- Strom (1991).Strom SL. Growth and grazing rates of the herbivorous dinoflagellate Gymnodinium sp. from the open subarctic Pacific Ocean. Marine Ecology Progress Series. 1991;78:103–113. doi: 10.3354/meps078103. [DOI] [Google Scholar]
- Veloza, Chu & Tang (2006).Veloza AJ, Chu F-LE, Tang KW. Trophic modification of essential fatty acids by heterotrophic protists and its effects on the fatty acid composition of the copepod Acartia tonsa. Marine Biology. 2006;148(4):779–788. doi: 10.1007/s00227-005-0123-1. [DOI] [Google Scholar]
- Wakeham (1982).Wakeham SG. Organic matter from a sediment trap experiment in the equatorial north Atlantic: wax esters, steryl esters, triacylglycerols and alkyldiacylglycerols. Geochimica et Cosmochimica Acta. 1982;46(11):2239–2257. doi: 10.1016/0016-7037(82)90198-3. [DOI] [Google Scholar]
- Yoon et al. (2017).Yoon EY, Park J, Jeong HJ, Rho J-R. Fatty acid composition and docosahexaenoic acid (DHA) content of the heterotrophic dinoflagellate Oxyrrhis marina fed on dried yeast: compared with algal prey. ALGAE. 2017;32(1):67–74. doi: 10.4490/algae.2017.32.3.5. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Compounds were identified through NIST library search and manual analysis of associated EIMS spectra.
(A) Cryptomonas sp. and (B) O. marina during active feeding and (C) 15 and (D) 20 days of starvation, respectively. Day zero of starvation commenced when prey were not detectable by Coulter counter and microscopy. During a 20 day starvation, O. marina mobilized wax esters as energy source.
Each file chromatographic file (GCMS, HPLC) has been converted to the corresponding Analytical Data Interchange format (.cdf). Flow cytometry raw data is provided as an excel spreadsheet.
Data Availability Statement
The following information was supplied regarding data availability:
Raw data in the format of GCMS and HPLC chromatograms are provided as .netCDF files in the Supplemental Materials.