Abstract
Pancreatic cancer (PanCa) is a major cause of cancer-related death due to limited therapeutic options. As pancreatic tumors are highly desmoplastic, they prevent appropriate uptake of therapeutic payloads. Thus, our objective is to develop a next-generation nanoparticle system for treating PanCa. We generated a multi-layered Pluronic F127 and polyvinyl alcohol stabilized and poly-L-lysine coated paclitaxel loaded poly(lactic-co-glycolic acid) nanoparticle formulation (PPNPs). This formulation exhibited optimal size (~160 nm) and negative Zeta potential (−6.02 mV), efficient lipid raft mediated internalization, pronounced inhibition in growth and metastasis in vitro, and in chemo-naive and chemo-exposed orthotopic xenograft mouse models. Additionally, PPNPs altered nanomechanical properties of PanCa cells as suggested by the increased elastic modulus in nanoindentation analyses. Immunohistochemistry of orthotopic tumors demonstrated decreased expression of tumorigenic and metastasis associated proteins (ki67, vimentin and slug) in PPNPs treated mice. These results suggest that PPNPs represent a viable and robust platform for (PanCa).
Keywords: Paclitaxel nanoformulation
Graphical abstract

In this study, we demonstrate the therapeutic efficacy of a novel PLGA-PTX nanoformulation (PPNPs). This system shows efficacy both in vitro and in vivo. Specifically, nanoindentation studies show an increased rigidity of PPNPs-treated pancreatic cancer cells, indicating reduced metastatic potential as confirmed with additional results herein (proliferation, colony formation, invasion, migration, cell cycle arrest, apoptosis induction). In vivo results indicate successful reduction in tumor growth, metastasis, and EMT markers in chemo-naïve and chemo-exposed orthotopic xenograft mice, suggesting that PPNPs may be a next-generation paclitaxel nanoformulation for PanCa treatment.
Background
Pancreatic cancer (PanCa) ranks as the fourth highest cause of cancer-related mortality in the US mainly due to difficulties in detection and chemoresistance(1). Hoff et al recently showed that gemcitabine with Abraxane (albumin-bound paclitaxel) only led to approximately 8.5 months survival increase as compared to 6.7 months for gemcitabine alone (2). Many new drugs and combinations have led to poor tolerance, low half-life, low cellular uptake, and high systemic toxicity (3–6). Such options largely fail by Phase II or III trials, causing huge financial burdens. PanCa exhibits several features leading to disorganized, leaky, nonfunctional vasculature (7–9), dense stroma (10), and deregulated cellular transport proteins (11). This leads to ineffective drug delivery and chemoresistance via high interstitial fluid pressure (12), stopping drugs from reaching vasculature to the extracellular compartment.
Paclitaxel (PTX), a natural product derived from Pacific Yew trees, is a second-line therapy in PanCa (13). PTX has shown therapeutic efficacy in many cancer types including PanCa (14). More recently, Abraxane (albumin-bound paclitaxel) has improved the efficacy of gemcitabine; since PTX reduced levels of cytidine deaminase (an enzyme responsible for gemcitabine degradation), allowing for greater concentrations of intracellular gemcitabine(15). Studies have shown that PTX alters TME and desmoplasia to improve GEM uptake in PanCa (2). In this study, we developed a next-generation PLGA-PTX nano-formulation (PPNPs) by incorporating Pluronic F127 to improve PTX therapy in PanCa patients. Poly(lactic-co-glycolic acid) (PLGA) is an FDA-approved biodegradable and biocompatible polymer employed for drug delivery applications including anti-cancer drug delivery (16, 17) and Pluronic F127 has shown potential in the reversing drug resistance (18, 19). Herein, we have shown that PPNPs inhibit PanCa growth and metastasis in chemo-naïve and chemo-exposed orthotopic xenograft mouse models. Moreover, nanoindentation analyses demonstrate altered nanomechanical properties of PanCa cells by PPNPs treatment.
Methods
Cell culture
PanCa cells (AsPCl, Panc-1, MIA PaCa-2 and HPAF-II) were purchased from the ATCC and were grown according to manufacturer’s protocol.
PLGA-PTX nanoparticle (PPNPs) preparation
PPNPs were generated using a modified procedure as described previously (20). First, an aqueous solution of 1% PVA (Sigma, P8136) was made. Then, a solution of PLGA (Lactel Polymers, B6010–4) in acetone (90 mg/5ml) was made in duplicate (PLGA control and PPNPs). Once both solutions were dissolved, 10 mg of PTX (Advanced ChemBlocks, F-4194) was added to one PLGA-acetone mixture to generate PPNPs. The PLGA-acetone mixture was added dropwise to the PVA solution then mixed overnight at 800 rpm while loosely covered in foil to allow for acetone evaporation. The next day, 5 mg/ml of PLL (Sigma, P2636) and F127 (Sigma, P2443) was made in Milli-Q water and was added dropwise to each beaker (2 ml each). This solution was mixed for seven hrs, then the volume was brought to 20 ml using Milli-Q. Samples were aliquoted into 1.5 ml tubes and stored at −20°C. One tube of control and loaded particles were taken and briefly sonicated using a probe sonicator (MISONIX Inc.) to ensure proper homogeneity for DLS analysis.
Particles size and zeta potential
Size, distribution and zeta potential of PPNPs were determined using DLS (ZetaSizer Nano ZS, Malvern Instruments, Malvern, UK). Briefly, 10 μl. of the particles was mixed into 990 μl of the dispersant (PBS, cell culture media, or 10% filtered human serum) and run using DLS measurements. Both particle size and zeta potential were collected in all three media.
HPLC-based drug loading assay
One ml of PPNPs aliquot was placed at −80°C for 4 hrs with loosely closed lids. The particles were lyophilized using a FreeZone 2.5 (LabConco) lyophilizer system and were mixed with 1 ml of acetonitrile with constant shaking for 24 hrs, then sonicated in a water bath for 2–3 hrs. The vials were then centrifuged at 10,000 rpm for 15 minutes and 500 μl of supernatant was collected for PTX loading by HPLC analysis. A mobile phase of 50/50 acetonitrile/water was used with a flow rate of 1 ml/minute, and PTX loading was determined at wavelength of 227 nm. The samples were run through a 50×2.1 mm column (ThermoFisher) with a 5 minute run time. A standard curve of PTX was generated using varying concentrations (1–1000 μg/ml). Encapsulation efficiency was calculated by taking the amount of drug determined to be held in the particles divided by the total amount of drug added, and drug loading was calculated by dividing the encapsulated drug by the total nanoparticle weight.
Fourier Transform infra-red (FT-IR) and X-ray diffraction
FT-IR spectra of PTX, PLGA, and PPNPs were obtained using Fourier Transform infra-red (FT-IR) microscope (Smiths Detection, Danbury, CT). The spectra data, 4,000–1000 cm−1, was acquired at a resolution of 4 cm−1 for 32 scans by placing PTX or lyophilized PLGA/PPNPs powder on the attenuated total reflection objective. The X-ray diffractograms measurements were acquired using Rigaku D/Max-B diffractometer at a scanning speed of 1° per minute (Rigaku Americas Corp, Woodlands, TX).
PPNPs characterization by atomic force microscopy (AFM)
AFM studies were conducted to determine the physical properties of PLGA and PPNPs. Nanoparticles were diluted in Milli-Q (1:1000) and placed on a glass slide to air dry overnight. AFM images were taken (2×2 square microns) using a ScanAsyst Air probe. To assess the physical characteristics of these nanoparticles, an RTESPA-300 probe was used (nominal spring constant 40N/m, tip radius ~8nm). The probe was calibrated by engaging onto a clean glass slide and ramping to calculate the deflection sensitivity. The spring constant was then calculated using the Sader method. A 2×2 square micron region was selected, and a force scan was conducted (128×128 pixels, ramp speed 60 Hz). Ten particles were selected at random from multiple scans and their modulus and adhesion data were analyzed and averaged.
Cellular uptake studies
Six-well plates were seeded (200,000/well). Once 75% confluent, treatment groups (PLGA or PPNPs with additional coumarin-6) were added for three hrs (2.5, 5, and 10 nM). The cells were rinsed with PBS and phenol red-free media was added. Microscopic images were taken to observe fluorescence of intracellular coumarin-6. Then cells were collected in PBS and run on the flow cytometer using channel FL1. Plates were seeded in the same way for uptake studies: HPAF-II and Panc-1 were treated with pathway inhibitors (genistein, amiloride, nocodazole, chlorpromazine, MBCD, incubation at 4°C) for one hr. The media was replaced and treated with coumarin-6 labelled PPNPs for one hr. The cells were then washed with PBS, collected in phenol red-free media, and analyzed for internalized coumarin-6 fluorescence in PanCa cells by flow cytometry using channel FL1.
MTT assay
The effect of cell proliferation on PanCa cells was assessed by MTT assay as described previously (21). For this experiment, at ~75% confluency, cells were treated with PPNPs (1.25–40 nM). for 48 hrs. Effect of PPNPs on cell proliferation was also determined by real time xCELLigence system as described previously (21).
Colony formation assay
The cells were seeded into 12-well plates (300/well) and attached overnight. Treatment was initiated once 50-cell colonies were seen. After two weeks, the media was aspirated, and colonies were fixed using cold methanol for 2–3 minutes. Cells were stained using 0.5% crystal violet and the colonies were counted.
Cell cycle analysis
A six-well plate was seeded (300,000/well) and attached overnight then treated with PPNPs for 24 hrs. Cells were fixed using cold 70% EtOH with gentle vortexing, then kept at 4°C for at least one hr, centrifuged, washed with PBS and re-suspended in 1 ml of Telford reagent with RNAse. The cells were then incubated in the dark for 4 hrs at 4°C, then run using a BD Accuri Flow Cytometer using channel FL2.
Apoptosis assessment by flow cytometry
Cells (300,000/well) were seeded in six well plates, attached overnight, then treated for 48 hrs. Cells were washed with PBS and diluted in Annexin-V binding buffer (1×106 cells/ml). One sample was left unstained as a gate control, the other samples were treated with 5 μl Annexin-V and 7-AAD and incubated for 20 minutes in the dark at room temperature and cells were analyzed using channels FL2 and FL3.
Western blot analysis
Cells were seeded into 150 mm dishes and grown to 75% confluency. PPNPs treatment was then given for 48 hrs. Cell lysates were prepared and quantified using the Bradford assay. Forty micrograms of protein were added to a 10% SDS-PAGE gel. The protein was then transferred to a PVDF membrane and blocked in 5% milk for 30 minutes at room temperature. Blots were incubated in primary antibody overnight at 4°C, followed by HRP-conjugated secondary antibody for 1 hr and developed with ECL reagent using a UVP gel documentation system.
Chemoinvasion assay
Cell invasion assays were conducted to determine if PPNPs could inhibit invasive potential of PanCa cells using our previously published protocol (21). Real-time assays on exCELLigence were also conducted (21).
Cell migration assay
Cell migration assays were performed using Boyden chambers (BD Biosciences) according to the manufacturer’s protocol as described previously(21). Effect of PPNPs on migration assay was also performed using the xCELLigence system as previously mentioned (21).
Nanoindentation and AFM imaging analysis
Cells were seeded in 60 mm dishes (500,000 cells) and attached overnight then treated with PLGA and PPNPs (10 nM) for 24 hrs. The cells were washed with PBS three times and fresh media was added. The dish was taken to the AFM and placed on a heated stage (37°C) and equilibrated for 15 minutes. Using PFQNM-LC probes, the deflection sensitivity was calculated via thermal tune. High-resolution images (256×256 resolution, 0.3Hz, 100 pN peak force set point) were taken to assess cellular morphology. A force curve was then generated on the center of a cell to confirm contact, then a force volume scan was conducted (128×128 resolution, 512 ramps per pixel, 15Hz ramp rate) to generate a force map of the entire cell. After collecting data on multiple cells, the average physical parameters (modulus and adhesion) were calculated at the nuclear, cytoplasmic, and peripheral regions and averaged.
Xenograft study
Six-week-old athymic nude mice were purchased from Jackson Laboratory, housed under pathogen-free environment with a 12-h light/12-h dark schedule, and fed an autoclaved diet and water ad libitum. This study was conducted using protocols approved by UTHSC Institutional Animal Care and Use Committee (IACUC ID #16–049.0-C). Sixteen mice were used to establish orthotopic xenograft tumors with human PanCa cells (HPAF-II-luciferase). Cells were harvested from subconfluent cultures and washed once in serum-free medium and suspended in HBSS. Only suspensions with >90% viability were used. The pancreas of anaesthetized mice was exposed through a midline laparotomy incision and by splenic retraction. Cells (2.0×106) in 50 μl of HBSS containing 10% (v/v) Matrigel was injected into pancreatic parenchyma. Incisions were closed by suture followed by clipping. Ten days later, mice were imaged via IVIS Spectrum bioluminescence scanner (PerkinElmer, Waltham, MA): Five minutes prior, 100 μl of D-luciferin substrate (30 mg/ml) was injected i.p. Quantification was conducted using vendor software (Living Image® 4.0) by measuring the total photons over the pancreatic region and the average photon flux within regions-of-interest (ROI) represented as photons/second/cm2/sr (sr denotes steradian). Mice with equal tumor volumes were divided into control (n=6) and PPNPs (n=6) groups and rest (n=4) were excluded. For dose-escalation studies, we first administered PPNPs (1 mg/kg body weight; twice a week for two weeks, then 10 mg/kg twice a week for three weeks). Control mice were administered unloaded PLGA. Mice were imaged for tumor growth at 10, 21, 30, and 37 days. At the time of sacrifice, tumors were removed, fixed in formalin, embedded in paraffin, and sliced into 5 mm sections for further analysis. To determine the effect of PPNPs on PanCa metastasis, distant organs (liver, lungs and lymph nodes) were excised, imaged, and analyzed for histopathology. We further evaluated the effect of PPNPs on PTX resistance. In this experiment, HPAF-II-luciferase labelled cells derived orthotopic xenograft tumor mice (n=6) were treated with PTX (10 mg/kg) i.p twice weekly for 5 weeks. At six weeks, half of the mice received PPNPs (10 mg/kg) i.p and the other half received PLGA. Tumor growth in these mice was monitored by bioluminescence imaging as described previously (22).
Immunohistochemistry analysis
Immunohistochemistry (IHC) was performed to determine the expression of ki67, vimentin, and slug in excised tumors of control and PPNPs treated mice. Our IHC protocol can be seen in previously published data (21).
Statistical analysis
Statistical analyses were performed using unpaired two-tailed Student t tests employed to assess the statistical significance between the control and PPNPs treated groups. P values < 0.05 were considered significant.
Results
Optimization of PPNPs formulation for PanCa therapeutics
Our nano-formulation (Fig. 1A) has several unique properties: (i) the FDA-approved PLGA core is capable of storage and controlled release of PTX; (ii) PVA, a widely used NP stabilizer, supports stability for over 6 month(s) and avoids non-specific adsorption of serum proteins in vivo; (iii) PLL promotes cellular internalization and is less toxic compared to other polycationic polymers; (iv) amine functional groups (PLL) assist in antibody conjugation, and (v) F127 polymer has shown potential in overcoming drug resistance (19, 23). DLS characterization yielded an average size of 140 nm for PLGA and 160 nm for PPNPs (Fig. 1B) and exhibited moderately negative zeta potential (Fig. 1C). To confirm PTX uptake in PPNPs, we performed FT-IR and XRD analyses. In the FT-IR spectrum of PLGA (Fig. 1E, black line), the intense peaks were found at 1000 cm−1, 1281 cm−1, and 1010 cm−1 due to C=O stretching of ester, C-O stretching of ester, and glycosidic (C–O–C/C–C/C–O) stretch vibrations. District PTX FT-IR and XRD peaks were noticed (Fig. 1D/E, red line), but after encapsulation these sharp peaks disappeared in PPNPs (Fig. 1D,E, green line), which closely resembled control PLGA (Fig. 1D,E, black line). XRD analysis showed similar results – PTX crystalline peaks disappeared upon encapsulation in PPNPs (Fig. 1D/E).
Figure 1: Synthesis and characterization of PLGA-PTX nanoparticles (PPNPs).
(A) Schematic overview of PPNPs synthesis. (B) DLS characterization of PLGA and PPNPs. (C) DLS characterization of zeta potential for PLGA and PPNPs. (D) FTIR spectra for free PTX, PLGA, and PPNPs. (E) XRD spectra for free PTX, PLGA, and PPNPs. (F) Topographical and physical analysis of PLGA and PPNPs by atomic force microscopy (AFM). Representative 2D and 3D images (2×2 μm2) of PLGA and PPNPs (i-ii). Modulus data for two separate samples (iii-iv).
Characterization of PPNPs by atomic force microscopy
With AFM imaging, particles were noted to be somewhat smaller in air (between 90–120 nm). This may be due to swelling in a liquid environment seen with DLS (Fig. 1Fi–ii). For both PLGA and PPNPs, there was no significant difference in the modulus. However, the effect of the other polymers in this particle system are not yet fully understood.
PPNPs internalize in PanCa cells primarily through lipid-raft mediated endocytosis
Qualitative analysis of coumarin-6 loaded nanoparticles showed a dose-dependent increase in PPNPs uptake. The uptake was noticeably higher in Panc-1 compared to HPAF-II (Fig. 2A–B). Results revealed a strong uptake by both HPAF-II (Fig. 2Aii) and Panc-1 cells (Fig. 2Bii) which was inhibited by MBCD. However, Panc-1 cells showed energy-dependent uptake, while HPAF-II cells showed energy-independent uptake (Fig. 2Aiii, Biii). Although this indicates overall differences, both cell lines demonstrated similar inhibition of lipid raft mediated endocytosis (24).
Figure 2: Cellular uptake of PPNPs in PanCa cells.
(A,B) Representative images of HPAF-II (Ai) and Panc-1 cells (Bi). Quantification of uptake by flow cytometry for HPAF-II (Aii) and Panc-1 (Bii). Uptake inhibitor assay for HPAF-II (Aiii) and Panc-1 (Biii) cells. Values in bar graphs represent mean±SE value of three sample in each group. *P<0.05.
PPNPs effectively inhibits growth, invasion and migratory potential of PanCa cells
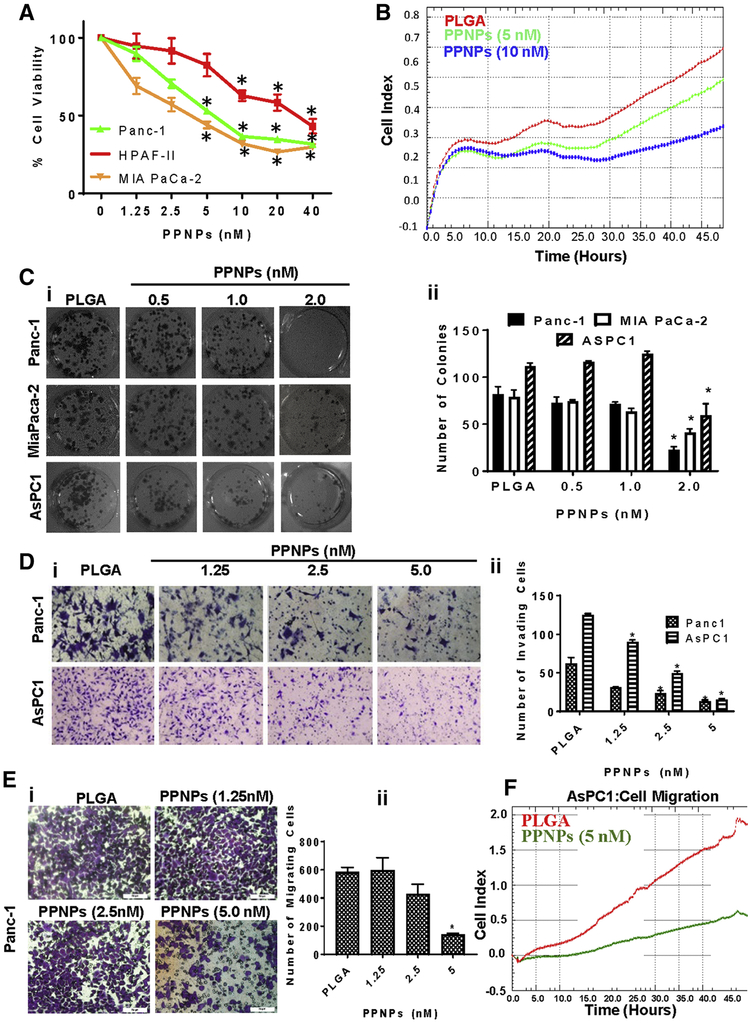
PPNPs inhibited cell viability of PanCa cells in a dose-dependent (1.25–40 nM) manner (Fig. 3A). IC50 of PPNPs in Panc-1 and MIA PaCa-2 cells was approximately 5 nM, while IC50 was 20 nM in HPAF-II cells at 48 hrs. A similar effect was also observed using xCELLigence assay (Fig. 3B) that monitors real-time inhibition of cell growth as described (21). PPNPs significantly (P<0.01) inhibited colony formation in all three PanCa cell lines (Panc-1 AsPC1 and MIA PaCa-2) at 2 nM (Figure 3, Ci–ii). We observed a more profound effects of PPNPs as compared to free PTX as shown in revised Supplementary Figure 4A–D. PPNPs have shown improved growth inhibitory effects on PanCa cells compared to free PTX. Moreover, PPNPs showed almost similar effects as abraxane in xCELLigence experiment in term of PanCa cells proliferation. In colony formation assays, PPNPs were shown to have a significantly higher therapeutic effect on GEM-resistant cells (Panc-1), however this effect was not significant in other non-Gemcitabine resistant cells. Overall, these data suggest that PPNPs nanoformulation has a superior efficacy compared to free PTX. PPNPs also dose-dependently (1.25–5.0 nM) inhibited invasion (Fig. 3D) and migration (Fig. 3E) of Panc-1 cells. This was further demonstrated by real-time inhibition of migration using AsPC1 cells with xCELLigence assay (Fig. 3F).
Figure 3: Effect of PPNPs on proliferation, invasion and migration of PanCa cells.
(A) Effect of PPNPs on cell viability of Panc-1, HPAF-II, and MIA PaCa-2. Values in bar graph represent mean ± SEM of 5 wells. (B) Real-time proliferation data using xCELLigence of AsPC1 cells. (C) Effect of PPNPs on clonogenic potential of PanCa cells. Representative colony images of control and PPNPs treated groups (i). Average colony counts in bar graph (ii). (D) Effect of PPNPs on invasive potential of PanCa cells. Representative images showing the effect of PPNPs on invasion of Panc-1 and AsPC1 cells through matrigel invasion assay (i). Bar graph quantification of invaded cells (ii). (E) Effect of PPNPs (24 hrs treatment) on migration of Panc-1 cells (i) Bar graph quantification of migrated cells (ii). (F) Real-time effect of PPNPs on AsPCl cells migration as shown using xCELLigence system.
PPNPs induce biophysical changes in PanCa cells as measured by nanoindentation analysis
After imaging the cells, differences were noted in the morphology – specifically, a notable shrinkage of the outer membrane (Fig. 4 Ai–ii, Bi–ii). After conducting nanoindentation analyses, the overall modulus was shown to increase in both cell lines (Fig. 4 Aiii, Biii). PPNPs treatment of AsPC1 for 24 hrs showed an increased modulus at the cytoplasmic and peripheral regions. Interestingly, the membrane adhesion was generally reduced with PPNPs treatment (Fig. 4 Aiv). Although cell modulus is a widely studied biophysical parameter, less evidence exists to explain the implications of reduced adhesion from a biochemical standpoint. The expected trends were also seen in Panc-1 (Fig. 4B), including an increased modulus across the cell. These results agree with the consensus that cancerous cells have a lower modulus compared to their healthy counterparts (25–27).
Figure 4: Effect of PPNPs on physical characteristics of PanCa cells.
Nanoindentation analyses of AsPCl (A) and PanC-1 cells. Three-dimensional images of untreated (i) and PPNPs-treated (ii) cells. Physical data collected by nanoindentation showing changes in modulus (iii) and adhesion (iv) with PPNPs. (B) Analysis of Panc-1 cells. Three-dimensional images of untreated (i) and PPNPs-treated (ii) cells. Physical data collected by nanoindentation shows changes in modulus (iii) and adhesion (iv) with PPNPs.
PPNPs arrests cell cycle in the G2/M phase and induces apoptosis in PanCa cells
PPNPs dose-dependently arrested cell cycle in G2/M phase (Fig. 5A). It is well documented that G2/M cell cycle arrest leads to the induction of apoptosis (28). Our data also shows that PPNPs (5–20 nM) of PanCa exhibited a dose-dependent increase in early-phase apoptosis (Fig. 5B). PPNPs dose-dependently increased the PARP cleavage in both Panc-1 and HPAF-II (Fig. 5C), which indicative of early apoptosis biomarker (29).
Figure 5: Effect of PPNPs on cell cycle distribution and apoptosis of PanCa cells.
(A) Flow cytometric analysis of cell cycle arrest by PPNPs on Panc-1 and HPAF-II. Representative histogram images of cell cycle analysis (i). Table showing percentage of Panc-1 and HPAF-II cells in each phase of cell cycle after PPNPs treatment (ii). (B) Flow cytometric analysis of Annexin-V/7-AAD positive Panc-1 cells after treatment with PPNPs (i). Quantification of early and late apoptosis induction by PPNPs in Panc-1 cells (ii). (C) Effect of PPNPs on PARP protein cleavage in Panc-1 and HPAF-II cells as determined by Western blot analysis (i). Quantification of cleaved PARP to PARP proteins ratio calculated by the band intensity using Image J software (ii).
PPNPs suppresses pancreatic tumor growth and metastasis in chemo-naïve and chemo-exposed orthotopic xenograft mice
In our in vivo studies, tumor volume was not affected at initial dosing of 1 mg/kg (Fig. 6 B,C). However, 10 mg/kg dosing showed a significant (P<0.01) difference from Day 30 onwards (Fig. 6 B,C). Bioluminescence results of excised pancreas demonstrated a significant (P<0.05) decrease of intensity in PPNPs treated mice (Fig. 6D). PPNPs treatment showed a significant (P<0.01) decrease in tumor weight compared to vehicle (Fig. 6E). These results agreed with the bioluminescence findings from excised pancreatic tumors. Histopathological analysis of excised tumor showed small necrotic areas in PPNPs treated mice (Fig. 6G). Immunohistochemistry results showed a decrease in ki67 in PPNPs treated mice (Fig. 6H). No apparent organ toxicity (heart and kidneys) was observed in PPNPs treated mice as determined by histopathological analysis (Supplementary Fig. 1). All control mice showed a high incidence of pancreatic metastasis in lymph nodes, liver, and lungs, which was significantly reduced with PPNPs (Fig.6F). Histopathological analysis results further confirmed decreased metastases in PPNPs treated mice – with only minimal foci in the lungs (Fig.7A). Immunohistochemistry results illustrated inhibition of EMT markers (vimentin and slug) in excised tumors of PPNPs treated mice (Fig.7B). PPNPs administration (10 mg/kg) significantly (P<0.05) inhibited tumor growth, even in pre-exposed mice as determined by significant (P<0.05) inhibition of bioluminescence counts (Fig. 7 E,F) and decrease in excised tumor weight (Fig. 7G).
Figure 6: Effect of PPNPs on pancreatic tumor growth in orthotopic xenograft mouse model.
(A) Schematic representation of schedule for dose escalation study of PPNPs in orthotopic xenograft mouse model. (B) Representative bioluminescence images of PLGA and PPNPs treated mice. (C) Quantification line graph of bioluminescence data in live mice. Values are shown as mean ± SEM with 6 mice per group. Inset images show a visual representation of tumor growth in indicated groups. (D) Representative pictures of ex vivo imaging of excised pancreatic tumor of PLGA and PPNPs treated mice. (E) Excised tumor weight of PLGA and PPNPs treated mice. Each dot represents one mouse. (F) Representative pictures of ex vivo imaging of liver, lymph nodes and lungs of PLGA and PPNPs treated mice. (G) Bar graphs indicating quantification of bioluminescence data for liver (i) and lymph nodes (ii) in PLGA and PPNPs treated mice. (H) H&E staining of pancreatic tumor of PLGA and PPNPs treated mice. Black arrows indicate pancreatic tumor. Red arrows indicate pancreatic acinar cells. Green arrows indicate necrotic area of pancreatic tumor. (I) Representative images of immunohistochemical analysis of Ki-67 in PLGA and PPNPs treated mice tissue sections.
Figure 7: Effect of PPNPs on pancreatic cancer metastasis and drug-resistant tumors.
(A) Representative H&E staining of liver and lung sections excised from PLGA and PPNPs treated mice. Black arrows indicate metastatic foci. (B-C) Representative immunohistochemical images of vimentin and slug in excised tissues. (D) Schematic representation of treatment schedule in orthotopic xenograft tumor-bearing mice pre-exposed to PTX. (E) Representative bioluminescence images showing tumor density in PLGA and PPNPs treated mice. (F) Quantification of bioluminescence data in PLGA and PPNPs treated mice. Values are shown as mean ± SEM with 3 mice per group. (G) Excised tumor weight in PLGA and PPNPs treated mice.
Discussion
Pancreatic cancer remains one of the deadliest cancers in the United States due to limited therapeutic options (30). Only 10–20% of patients diagnosed with localized disease are candidates for surgical resection; moreover, post-operative disease recurrence is very common (31). Nanoparticles (NPs) are known to preferentially accumulate in tumor tissue due to leaky vasculature, mainly via Enhanced Permeation and Retention (EPR) effect (32, 33). Abraxane® (albumin-bound paclitaxel) has been widely used in clinic for the treating various cancers, including PanCa (34–36), indicating that nano-drug delivery systems have superior therapeutic outcomes over free drug. In addition, nano-scale drug delivery has been shown by various studies to improve on-target uptake of drug, which can overcome drug resistance and lead to overall reduction in the administered dose and systemic toxicity (37). Therefore, the main objective of this study was to develop a new nano-formulation of PTX. Herein, we developed a unique formulation (PPNPs) using F127 polymer and optimized it against PanCa using in vitro and in vivo model systems.
Our first step was to generate and characterize this formulation. DLS characterization yielded an average size of 160 nm and exhibited moderately negative zeta potential – both ideal for nano-scale drug delivery. In the FT-IR and XRD spectra, characteristic peaks of PLGA were not affected by PTX loading as seen PLGA and PPNPs peak similarities. It should be noted however that FTIR and XRD data only represents the overall composition and physical state of the formulation. To determine the PTX loading in our nanoformulation, we used an HPLC-based method to quantify the encapsulation efficiency. Our results indicated an encapsulation efficiency ranging from of 72–86%, and an experimentally calculated drug loading of 7.4–8.7% (Supplementary Figure 2). Our AFM results indicate no change in the physical properties of PLGA nanoparticles after PTX encapsulation which was suggested by a similar modulus in PLGA and PPNPs (~ 12 GPa). These results should be seen as semi-quantitative because the modulus of bulk PLGA is reported around 2 GPa (38), but the modulus of other components (PLL, PVA and F127) are not known. Our results indicate differing internalization of PPNPs in Panc-1 and HPAF-II cells and was noticeably higher in Panc-1. However, both cells showed inhibition with MBCD treatment (lipid raft mediated endocytosis). (24). Further study is required to determine whether PPNPs uptake is cell line specific.
In this study, we established that PPNPs demonstrate anti-cancer activity against PanCa. MTT and colony formation assays indicate that PPNPs inhibit growth and proliferation of PanCa cells. It is well documented that invasion and migration represent two critical components of metastatic cascades (39). We observed a significant reduction in the invasive and migratory properties of PanCa cells treated with PPNPs in vitro. Nanoindentation techniques have been successfully used to investigate cancer cells recently, however the literature on this field is still limited. Given the ability to differentiate between healthy and cancerous tissue, as well as observing the effects of various treatments can yield greater insights into phenotypical behaviors of cancer cells (40). Conventional nanoindentations have been conducted using large, spherical tips (40, 41). Although this can lead to the ability to quickly scan multiple cells, it also reduces the amount of data per cell. Using recently designed probes from Bruker custom-built for live cell applications (PFQNM-LC), we were able to generate force maps with significantly more data per cell, giving a greater understanding of the physical properties at different cellular regions. Based on our nanoindentation results, the overall modulus was increased in Panc-1 and AsPC1. These results clearly suggest that the increased rigidity caused by PPNPs will inhibit metastatic phenotype of PanCa cells.
It is well established that arresting cell cycle and inducing apoptosis are prime therapeutic targets for cancer treatment (42, 43). Various chemotherapeutics have been shown to induce apoptosis and arrest cell cycle (44, 45). Specifically, PTX has been shown to induce apoptosis and arrest the cell cycle during the G2/M phase in various cancer cells (46). Our results also suggest that PPNPs induce apoptosis and arrest cell cycle in G2/M phase in PanCa cells. This was further supported by cleavage in PARP protein, an early biomarker of apoptosis (29).
After accumulating significant in vitro data, we then assessed the in vivo efficacy of PPNPs. We used human HPAF-II-luciferase cells to generate an orthotopic xenograft mouse model. This model has previously been used to evaluate the therapeutic efficacy of chemotherapeutic agents against PanCa (47, 48). Histopathology results revealed no toxicity in the heart, kidney and liver of PPNPs treated mice, suggesting a 10 mg/kg dose is a safe dose in mice. Our imaging data of PPNPs treated mice (10 mg/kg twice a week for three weeks) showed a significant (P<0.01) inhibition in bioluminescence, suggesting that PPNPs inhibits pancreatic tumor growth. We also observed a significant (P<0.01) inhibition in the weight of excised tumors from PPNPs-treated mice. These findings demonstrate that PPNPs inhibits the progression of orthotopic xenograft tumors in athymic nude mice. In humans, lymph nodes, lungs, and the liver are frequent sites of PanCa metastasis. All control mice showed metastases in these organs with ex vivo imaging and histopathology. Our results showed a significant inhibition of HPAF-II-luciferase cells metastasis in PPNPs treated mice. PPNPs also inhibited tumor growth in PTX-exposed mice, suggesting that PPNPs are capable of overcoming drug resistance in PanCa.
Ki-67 is a well-known cell proliferation marker and has been used to determine the anti-proliferative effect of therapeutic agents (49). Our data indicated a decrease of nuclear ki-67 in PPNPs treated mice tissue. Our results show the growth inhibitory and anti-metastatic nature of PPNPs against human PanCa. EMT is a crucial step in metastasis (50). During EMT, phenotypic and molecular changes occur in cancer cells leading to loss of epithelial markers and an elevation of mesenchymal markers (vimentin, N-cadherin, Slug, and fibronectin) (51, 52). Our immunohistochemistry data of PPNPs treated mice indicated a decrease in vimentin and slug - indicating that PPNPs can suppress EMT in PanCa. These results suggest that PPNPs are a promising therapeutic modality for the treatment of PanCa growth and metastasis.
We characterized the properties and therapeutic efficacy of a novel paclitaxel nanoparticle system (PPNPs) which has shown ideal properties for nano-scale drug delivery, as well as significant effects on PanCa cells in vitro. Our in vivo data also suggests that PPNPs can reduce tumor growth and metastatic burden in both chemo-naïve and chemo-exposed orthotopic xenograft mouse models. Nanoindentation analyses demonstrate that PPNPs alter nanomechanical properties of PanCa cells. Therefore, we are thoroughly investigating molecular mechanisms and targeted delivery for this formulation using clinically relevant mouse model systems. Overall, our results suggest the anti-cancer potential of PPNPs which could be used as a novel therapeutic modality for PanCa treatment.
Supplementary Material
Acknowledgement
This work was supported by NIH R01CA210192, NIH R01CA206069 and College of Pharmacy/University of Tennessee Health Science Center Seed grant.
Financial Support: This work was supported by; NIH R01CA210192, NIH R01CA206069, and College of Pharmacy/University of Tennessee Health Science Center Seed Grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: No potential conflicts of interest were disclosed
References
- 1.Court CM, Ankeny JS, Sho S, Winograd P, Hou S, Song M, et al. Circulating Tumor Cells Predict Occult Metastatic Disease and Prognosis in Pancreatic Cancer. Ann Surg Oncol. 2018;25(4): 1000–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18): 1691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Marco M, Di Cicilia R, Macchini M, Nobili E, Vecchiarelli S, Brandi G, et al. Metastatic pancreatic cancer: is gemcitabine still the best standard treatment? (Review). Oncology reports. 2010;23(5): 1183–92. [DOI] [PubMed] [Google Scholar]
- 4.Kleynberg RL, Sofi AA, Chaudhary RT. Hand-foot hyperpigmentation skin lesions associated with combination gemcitabine-carboplatin (GemCarbo) therapy. American journal of therapeutics. 2011;18(6):e261–3. [DOI] [PubMed] [Google Scholar]
- 5.Pedersen AG. Phase I studies of gemcitabine combined with carboplatin or paclitaxel. Seminars in oncology. 1997;24(2 Suppl 7):S7–64–S7–8. [PubMed] [Google Scholar]
- 6.Rapoport N, Kennedy AM, Shea JE, Scaife CL, Nam KH. Ultrasonic nanotherapy of pancreatic cancer: lessons from ultrasound imaging. Molecular pharmaceutics. 2010;7(1):22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pries AR, Hopfner M, le Noble F, Dewhirst MW, Secomb TW. The shunt problem: control of functional shunting in normal and tumour vasculature. Nature reviews Cancer. 2010;10(8):587–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Provenzano PP, Cuevas C, Chang AE, Goel VK, Von Hoff DD, Hingorani SR. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer cell. 2012;21(3):418–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stylianopoulos T, Martin JD, Chauhan VP, Jain SR, Diop-Frimpong B, Bardeesy N, et al. Causes, consequences, and remedies for growth-induced solid stress in murine and human tumors. Proceedings of the National Academy of Sciences of the United States of America. 2012; 109(38): 15101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neesse A, Michl P, Frese KK, Feig C, Cook N, Jacobetz MA, et al. Stromal biology and therapy in pancreatic cancer. Gut. 2011;60(6):861–8. [DOI] [PubMed] [Google Scholar]
- 11.Farrell JJ, Elsaleh H, Garcia M, Lai R, Ammar A, Regine WF, et al. Human equilibrative nucleoside transporter 1 levels predict response to gemcitabine in patients with pancreatic cancer. Gastroenterology. 2009;136(1): 187–95. [DOI] [PubMed] [Google Scholar]
- 12.Jain RK. Transport of molecules across tumor vasculature. Cancer metastasis reviews. 1987;6(4):559–93. [DOI] [PubMed] [Google Scholar]
- 13.Maeda S, Motoi F, Onogawa T, Morikawa T, Shigeru O, Sakata N, et al. Paclitaxel as second-line chemotherapy in patients with gemcitabine-refractory pancreatic cancer: a retrospective study. International journal of clinical oncology. 2011;16(5):539–45. [DOI] [PubMed] [Google Scholar]
- 14.He L, Yang H, Zhou S, Zhu H, Mao H, Ma Z, et al. Synergistic antitumor effect of combined paclitaxel with FEN1 inhibitor in cervical cancer cells. DNA repair. 2018;63:1–9. [DOI] [PubMed] [Google Scholar]
- 15.Frese KK, Neesse A, Cook N, Bapiro TE, Lolkema MP, Jodrell DI, et al. nab-Paclitaxel potentiates gemcitabine activity by reducing cytidine deaminase levels in a mouse model of pancreatic cancer. Cancer discovery. 2012;2(3):260–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinarvand R, Sepehri N, Manoochehri S, Rouhani H, Atyabi F. Polylactide-co-glycolide nanoparticles for controlled delivery of anticancer agents. International journal of nanomedicine. 2011;6:877–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Preat V. PLGA-based nanoparticles: an overview of biomedical applications. Journal of controlled release : official journal of the Controlled Release Society. 2012;161(2):505–22. [DOI] [PubMed] [Google Scholar]
- 18.Sosnik A Reversal of multidrug resistance by the inhibition of ATP-binding cassette pumps employing “Generally Recognized As Safe” (GRAS) nanopharmaceuticals: A review. Advanced drug delivery reviews. 2013;65(13–14): 1828–51. [DOI] [PubMed] [Google Scholar]
- 19.Zhang W, Shi Y, Chen Y, Ye J, Sha X, Fang X. Multifunctional Pluronic P123/F127 mixed polymeric micelles loaded with paclitaxel for the treatment of multidrug resistant tumors. Biomaterials. 2011;32(11):2894–906. [DOI] [PubMed] [Google Scholar]
- 20.Khan S, Chauhan N, Yallapu MM, Ebeling MC, Balakrishna S, Ellis RT, et al. Nanoparticle formulation of ormeloxifene for pancreatic cancer. Biomaterials. 2015;53:731–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hafeez BB, Ganju A, Sikander M, Kashyap VK, Hafeez ZB, Chauhan N, et al. Ormeloxifene Suppresses Prostate Tumor Growth and Metastatic Phenotypes via Inhibition of Oncogenic beta-catenin Signaling and EMT Progression. Molecular cancer therapeutics. 2017;16(10):2267–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hafeez BB, Zhong W, Fischer JW, Mustafa A, Shi X, Meske L, et al. Plumbagin, a medicinal plant (Plumbago zeylanica)-derived 1,4-naphthoquinone, inhibits growth and metastasis of human prostate cancer PC-3M-luciferase cells in an orthotopic xenograft mouse model. Molecular oncology. 2013;7(3):428–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yallapu MM, Jaggi M, Chauhan SC. Poly(beta-cyclodextrin)/curcumin self-assembly: a novel approach to improve curcumin delivery and its therapeutic efficacy in prostate cancer cells. Macromolecular bioscience. 2010;10(10):1141–51. [DOI] [PubMed] [Google Scholar]
- 24.Wang M, Hajishengallis G. Lipid raft-dependent uptake, signalling and intracellular fate of Porphyromonas gingivalis in mouse macrophages. Cellular microbiology. 2008;10(10):2029–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo Q, Kuang D, Zhang B, Song G. Cell stiffness determined by atomic force microscopy and its correlation with cell motility. Biochimica et biophysica acta. 2016; 1860(9): 1953–60. [DOI] [PubMed] [Google Scholar]
- 26.Cross SE, Jin YS, Rao J, Gimzewski JK. Nanomechanical analysis of cells from cancer patients. Nature nanotechnology. 2007;2(12):780–3. [DOI] [PubMed] [Google Scholar]
- 27.Swaminathan V, Mythreye K, O’Brien ET, Berchuck A, Blobe GC, Superfine R. Mechanical stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer research. 2011;71(15):5075–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmed W, Rahmani M, Dent P, Grant S. The cyclin-dependent kinase inhibitor p21(CIP1/WAF1) blocks paclitaxel-induced G2M arrest and attenuates mitochondrial injury and apoptosis in p53-null human leukemia cells. Cell cycle (Georgetown, Tex). 2004;3(10):1305–11. [DOI] [PubMed] [Google Scholar]
- 29.Chaitanya GV, Steven AJ, Babu PP. PARP-1 cleavage fragments: signatures of cell-death proteases in neurodegeneration. Cell communication and signaling : CCS. 2010;8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64(1):9–29. [DOI] [PubMed] [Google Scholar]
- 31.Hung JS, Yang CY, Hu RH, Lee PH, Tien YW. Surgical treatment of pancreatic serous cystadenoma: aggressive for operations but limited resections. Pancreas. 2007;35(4):358–60. [DOI] [PubMed] [Google Scholar]
- 32.Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Cancer nanotechnology: the impact of passive and active targeting in the era of modem cancer biology. Advanced drug delivery reviews. 2014;66:2–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Acharya S, Sahoo SK. PLGA nanoparticles containing various anticancer agents and tumour delivery by EPR effect. Advanced drug delivery reviews. 2011;63(3):170–83. [DOI] [PubMed] [Google Scholar]
- 34.Palacio S, Hosein PJ, Reis I, Akunyili II, Ernani V, Pollack T, et al. The nab-paclitaxel/gemcitabine regimen for patients with refractory advanced pancreatic adenocarcinoma. Journal of gastrointestinal oncology. 2018;9(1): 135–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuwayama T, Nakamura S, Hayashi N, Takano T, Tsugawa K, Sato T, et al. Randomized Multicenter Phase II Trial of Neoadjuvant Therapy Comparing Weekly Nab-paclitaxel Followed by FEC With Docetaxel Followed by FEC in HER2(−) Early-stage Breast Cancer. Clinical breast cancer. 2018. [DOI] [PubMed] [Google Scholar]
- 36.Cohen AL, Ray A, Van Brocklin M, Burnett DM, Bowen RC, Dyess DL, et al. A phase I trial of azacitidine and nanoparticle albumin bound paclitaxel in patients with advanced or metastatic solid tumors. Oncotarget. 2017;8(32):52413–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markman JL, Rekechenetskiy A, Holler E, Ljubimova JY. Nanomedicine therapeutic approaches to overcome cancer drug resistance. Advanced drug delivery reviews. 2013;65(13–14): 1866–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gentile P, Chiono V, Carmagnola I, Hatton PV. An overview of poly(lactic-co-glycolic) acid (PLGA)-based biomaterials for bone tissue engineering. International journal of molecular sciences. 2014;15(3):3640–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nature reviews Cancer. 2002;2(8):563–72. [DOI] [PubMed] [Google Scholar]
- 40.Yallapu MM, Katti KS, Katti DR, Mishra SR, Khan S, Jaggi M, et al. The roles of cellular nanomechanics in cancer. Medicinal research reviews. 2015;35(1): 198–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gavara N A beginner’s guide to atomic force microscopy probing for cell mechanics. Microscopy research and technique. 2017;80(1):75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fisher DE. Apoptosis in cancer therapy: crossing the threshold. Cell. 1994;78(4):539–42. [DOI] [PubMed] [Google Scholar]
- 43.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342–8. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Li L, Yang J, Clair PM, Glenn MJ, Stephens DM, et al. Drug-free macromolecular therapeutics induce apoptosis in cells isolated from patients with B cell malignancies with enhanced apoptosis induction by pretreatment with gemcitabine. Nanomedicine : nanotechnology, biology, and medicine. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karaca B, Degirmenci M, Ozveren A, Atmaca H, Bozkurt E, Karabulut B, et al. Docetaxel in combination with octreotide shows synergistic apoptotic effect by increasing SSTR2 and SSTR5 expression levels in prostate and breast cancer cell lines. Cancer chemotherapy and pharmacology. 2015;75(6): 1273–80. [DOI] [PubMed] [Google Scholar]
- 46.Jimenez-Guerrero R, Gasca J, Flores ML, Perez-Valderrama B, Tejera-Parrado C, Medina R, et al. Obatoclax and Paclitaxel Synergistically Induce Apoptosis and Overcome Paclitaxel Resistance in Urothelial Cancer Cells. Cancers. 2018;10(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shim IK, Yi HJ, Yi HG, Lee CM, Lee YN, Choi YJ, et al. Locally-applied 5-fluorouracil-loaded slow-release patch prevents pancreatic cancer growth in an orthotopic mouse model. Oncotarget. 2017;8(25):40140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kotopoulis S, Delalande A, Popa M, Mamaeva V, Dimcevski G, Gilja OH, et al. Sonoporation-enhanced chemotherapy significantly reduces primary tumour burden in an orthotopic pancreatic cancer xenograft. Molecular imaging and biology : MIB : the official publication of the Academy of Molecular Imaging. 2014;16(1):53–62. [DOI] [PubMed] [Google Scholar]
- 49.Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. Journal of cellular physiology. 2000; 182(3):311–22. [DOI] [PubMed] [Google Scholar]
- 50.Trager MM, Dhayat SA. Epigenetics of epithelial-to-mesenchymal transition in pancreatic carcinoma. International journal of cancer. 2017; 141 (1):24–32. [DOI] [PubMed] [Google Scholar]
- 51.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–90. [DOI] [PubMed] [Google Scholar]
- 52.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. The Journal of clinical investigation. 2009;119(6): 1420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.