Abstract
Glioblastoma multiforme (GBM) is the most malignant primary brain tumor with a median survival of approximately 14 months. Despite aggressive treatment of surgical resection, chemotherapy and radiation therapy, only 3–5% of GBM patients survive more than 3 years. Contributing to this poor therapeutic response, it is believed that GBM contains both intrinsic and acquired mechanisms of resistance, including resistance to radiation therapy. In order to define novel mediators of radiation resistance, we conducted a functional knockdown screen, and identified the immunoglobulin superfamily protein, PTGFRN. In GBM, PTGFRN is found to be overexpressed and to correlate with poor survival. Reducing PTGFRN expression radiosensitizes GBM cells and potently decreases the rate of cell proliferation and tumor growth. Further, PTGFRN inhibition results in significant reduction of PI3K p110β and phosphorylated AKT, due to instability of p110β. Additionally, PTGFRN inhibition decreases nuclear p110β leading to decreased DNA damage sensing and DNA damage repair. Therefore overexpression of PTGFRN in glioblastoma promotes AKT-driven survival signaling and tumor growth, as well as increased DNA repair signaling. These findings suggest PTGFRN is a potential signaling hub for aggressiveness in GBM.
Keywords: PTGFRN, glioblastoma multiforme, AKT, PI3K p110β, radiation
1. Introduction
Glioblastoma multiforme (GBM) is a common and aggressive primary malignant brain tumor, and has one of the worst 5-year survival rates among all cancers despite aggressive treatment (1). Current therapies, including surgical resection followed by concurrent treatment with temozolomide and radiation therapy, have demonstrated limited effectiveness due to the anatomic location of GBM, the blood brain barrier, and intrinsic and acquired mechanisms of resistance (2–4). Notably, GBM has been shown to exhibit radioresistance, especially in recurrent tumors. Despite identification of several mechanisms that contribute to the radioresistant phenotype (5–7), the molecular basis of radioresistance remains incompletely defined. Through a recently published functional knockdown screen (8), we identified PTGFRN, Prostaglandin F2 receptor negative regulator, as a novel mediator of radioresistance in GBM cells.
PTGFRN (EWI-F, FPRP, CD9P-1) is a cell surface transmembrane protein in the immunoglobulin superfamily (9). To date, the role of PTGFRN has been little studied in cancer. Published studies show that PTGFRN expression is essential for angiogenesis, and its expression at both transcriptional and translational levels correlates with the metastatic status of lung cancer (10, 11). PTGFRN has also been shown to be associated with lipid accumulation in preadipocytes (12), involved in cell migration (13), and has been found to directly interact with ezrin-radixin-moesin proteins (9). PTGFRN self-associates but is also a component of tetraspanin-enriched microdomains (TEMs), which can include tetraspanins, growth factors, integrins, complement regulatory proteins, signaling enzymes and proteoglycans (9, 14, 15). As a member of TEMs, PTGFRN is primed to act as a scaffolding protein and associate with other proteins within the TEM to regulate downstream signaling events.
The AKT/PI3K signaling pathway is critical survival pathway in cells. PI3K contains a catalytic subunit p110 (α, β, and γ) and a regulatory subunit p85. Upon ligand binding to cell surface receptor tyrosine kinases, the phosphorylation of tyrosine residues creates binding sites for p85, resulting in a conformation change that releases the catalytic subunit p110. Activated p110 phosphorylates phosphatidy-linositol-3,4-bisphosphate (PIP2) into phosphatidy-linositol-3,4,5-bisphosphate (PIP3). PIP3 then recruits downstream AKT to the inner membrane for phosphorylation and activation (16). Studies have demonstrated increased activation of the PI3K/AKT pathway in GBM, among several other cancers, by a variety of mechanisms including homozygous deletion of the negative regulator of PI3K, PTEN (17). Crucially, pathway activation is significantly associated with radiation resistance (18, 19). Additionally, studies have also shown PI3K/AKT signaling to be altered by proteins found in TEMs, suggesting that multiple mechanisms exist for cancer cells to employ (20, 21).
In the present work, we demonstrate that PTGFRN supports tumorigenesis and survival signaling through increased PI3K/AKT signaling and p110β stability. We find PTGFRN to be elevated in primary GBM compared to normal brain tissue, and to significantly associate with poorer survival among patients under standard treatment paradigms. Our results indicate the inhibition of PTGFRN decreases tumor growth and sensitizes GBM cells to radiation treatment by affecting the turnover of the PI3K catalytic subunit p110β, hindering AKT signaling and DNA damage sensing. The findings support a novel role for PTGFRN in GBM tumorigenesis and survival signaling, and provide a novel therapeutic target expressed on the cell surface.
2. Materials and Methods
2.1. Cell lines and reagents
U87MG and A172 cells were obtained from ATCC and authenticated by STR profiling by Genetica DNA Laboratories. The pLKO.1 Tet-ON construct was given by Dr. Eli Bar (Case Western Reserve University, Cleveland, OH). Neurosphere stem cell lines GBM0821 and 0913 were a gift of Dr. Angelo Vescovi (University of Bicocca, Milan) (22–24). D-luciferin came from Gold Biotechnology. Stable PTGFRN inhibition was performed with lentiviral shRNA pLKO.1 clones: TRCN0000057452 and TRCN 0000057451 (Sigma). shRNA GFP was used as control. The PIK3CB overexpression plasmid was purchased from Addgene (plasmid #116555). Quantitative real-time PCR (qRT-PCR) was performed using Power SYBR Green PCR Master Mix from Thermo Fisher Scientific and normalized to either β-actin or GAPDH. Primer sequences:
PTGFRN F-5’ACAACAGCTGGGTGAAAAGC-3’, PTGFRN R-5’ TTTCATTGGGACTGGAGAGG-3’; Actin F-5’CATGTACGTTGCTATCCAGGC-3’, R-5’CTCCTTAATGTCACGCACGAT-3’; GAPDH F-5’AAGGTGAAGGTCGGAGTCAAC-3’, R-5’GGGGTCATTGATGGCAACAATA-3’; p85α F-5’AGTGGTTGGGCAATGAAAAC-3’, R-5’GAAAAAGTGCCATCTCGCTTC-3’; p11β F-5’GGGAAAGCTCATCGTAGCTG-3’, R-5’CTACTCTCCCGCTGACTTGC-3’; BRCA1 F-5’TGGAAGAAACCACCAAGGTC-3’, R-5’ACCACAGAAGCACCACACAG-3’.
2.2. Colony formation assay
Radiation was performed with a137Cs irradiator (Shepherd). A total of 500 to 10,000 cells per plate were stained after 10 days with 0.1% crystal violet. Assays were done ≥ 3 times with individual samples in triplicate. Clonogenic assays of neurosphere lines were plated in 1 mL of neuro stem cell (NSA) media (22) containing 1.5% methylcellulose and fed every 3 days. Sphere formation was monitored and scored using GelCount (Oxford Optronix) after 10–12 days.
2.3. Cell Proliferation Assay
20,000 cells/well were plated into a 12 well dish and allowed to grow for 3 days. On day 3, cells were counted, and 20,000 cells were replated. The process was repeated up to day 9 or day 10.
2.4. Tumor Formation Assay
Animal studies were performed in accordance with CWRU institutional guidelines. Subcutaneous U87MG knockdown tumors were produced by injection of 1×106 cells in the flanks of eight-week-old nude mice, and measured with calipers twice weekly. Eight-week-old nude mice were injected intracranially with 105 luciferase-expressing GBM0913 neurosphere stem cells into the right cerebral cortex at a depth of 3 mm. Two weeks following injection, Tet-on-shRNA injected mice were given doxycycline in their food for 6 days, and half of the mice were irradiated at 12 Gy using GammaKnife. Tumor growth was monitored and quantified using bioluminescent imaging. Animal appearance, behavior, and weight were monitored to evaluate tumor progression as per a Case Western Reserve University approved IACUC protocol.
2.5. Cellular Fractionation
GBM shPTGFRN and shGFP neurosphere stem cell lysates were fractionated as described(25).
2.6. Western Blots
Western blots were performed using standard procedures. Antibodies against PTGFRN (ab97567 Abcam), PI3K p110beta (Cell Signaling #3011), PI3K p110alpha (Cell Signaling #4249), PI3K p85alpha (Cell Signaling #4257), β-actin (Sigma A1987), Phospho-AKT Ser473 (Cell signaling #9271), AKT (Cell signaling #9272), Histone H3 (Santa Cruz sc-8654), α-Tubulin (Sigma T9026), Phospho-Histone H2A.X (Ser139) (Millipore 05–636), Phospho-DNA-PKcs (PA5–78130, Invitrogen), DNA-PKcs (MA5–13238, Invitrogen) or Na/K-ATPase (Cell signaling #3010) were used to decorate the membranes, and developed by chemiluminescence on an Azure c300 imaging system. GBM0913 shPTGFRN or shGFP cells were pretreated with MG132 (20μM) or control vehicle (DMSO) for 30 mins. After 30 min, cycloheximide (50mg/ml) was added to each sample to block protein synthesis. Cells were collected at indicated time points.
2.7. Proximity Ligation Assay
The Sigma Duolink® In Situ Fluorescence (DUO92101) was used according to the manufacturer’s protocol. Antibodies: Anti-PIK3CB (SAB1404204–100UG), Anti-PTGFRN (SAB2700379), monoclonal Anti-CD9 (SAB1402143–100UG), polyclonal Anti-CD9 (SAB4503606). Positive cells were detected by flow cytometry.
2.8. HR reporter assay
U87 DRGFP cells were transfected with siRNA using DharmaFECT. After 24 hours, cells were infected at 10 MOI with I-SceI-expressing adenovirus or control (empty) virus (gift of Dr. Junran Zhang). Cells were incubated for an additional 48 hours and then analyzed by flow cytometry as described previously (26).
2.9. Immunofluorescent Staining
Cells were fixed in 3% paraformaldehyde and stained and analyzed as described previously (8). Antibody: Phospho-Histone H2A.X (Ser139) (Millipore 05–636), secondary Alexa-Fluor 594 anti-mouse (A11032; Invitrogen), Anti-PIK3CB (SAB1404204–100UG; Sigma), Anti-PTGFRN (SAB2700379; Sigma), secondary Alexa-Fluor 488 anti-rabbit (A11070; Invitrogen).
2.10. EdU pulse chase
Cells were treated with EdU (25uM) for 8 hours, cells were washed with HBSS and NSA complete media to remove the EdU and put back into culture to allow the cells to continue proliferating. A portion of cells were taken at indicated time points, fixed, permeabilized and stained according to Invitrogen’s Click-iT Plus EdU flow cytometry assay (C10636) protocol. Cells were analyzed on a CytoFLEX flow cytometer. Analysis of the cell cycle progression was completed as previously described (27).
2.11. Statistical Analysis
Student t tests and one or two-way ANOVA were used throughout the study to test the significance of differences between samples as indicated. Survival analyses were performed by log-rank tests in GraphPad Prism.
3. Results
3.1. PTGFRN is overexpressed in GBM and correlates with poorer patient outcome
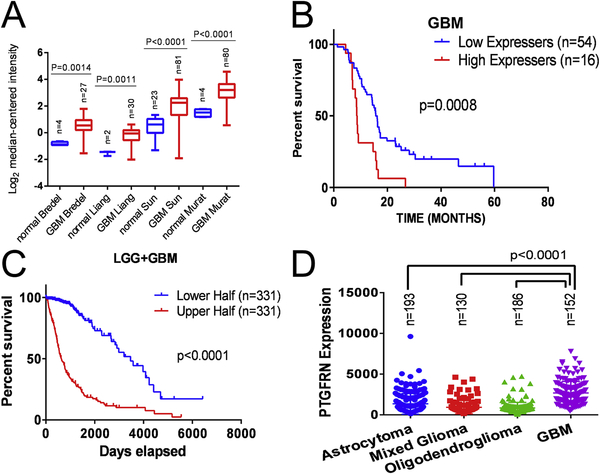
To identify novel mediators of radiation resistance in GBM, we performed a functional knockdown screen using a lentivirus-mediated shRNA library on two GBM cell lines (8) and filtered the expression levels of hits through in silico screens and patient data to determine relevance to GBM. In Oncomine, a public database of published gene expression studies (28), PTGFRN expression was found to be elevated in every study of GBM tumors versus normal brain tissue (Fig 1A). In the Bredel, Liang, Sun and Murat studies of GBM, PTGFRN was overexpressed by factors of 2.443 (P=0.0014), 2.513 (P=0.0011), 2.804 (P<0.0001) and 3.032 (P<0.0001), respectively. We next looked at GBM patient survival data to evaluate its effect by PTGFRN expression. We inquired Prognoscan, a public database for the meta-analysis of the prognostic value of genes (29), and found that GBM patients with elevated PTGFRN expression have a poorer outcome than patients with lower expression (15.66 months versus 8.67 months, P=0.0008, log-rank test) (Fig. 1B). We also queried TCGA database where PTGFRN expression significantly correlates with poor outcome in the low grade glioma and GBM cohort (Fig 1C). Additionally, PTGFRN was found to be significantly overexpressed in high grade GBM compared to subtypes of lower grade gliomas, including astrocytoma and oligodendroglioma (Fig 1D). Further, we queried TCGA to determine if known oncogenic mutations, such as PTEN or PI3K mutations, found in GBM tumors correlate with PTGFRN expression. We found no correlation of PTGFRN expression with the mutations analyzed (Supplemental Fig 1A). Finally, as GBM has been classified into four major molecular subtypes: classical, mesenchymal, proneural, and neural (30), we queried the GBM Bio Discovery Portal to determine how PTGFRN expression correlates with the molecular subtypes. PTGFRN expression was found to be higher in classical and mesenchymal GBM subtypes compared to proneural and neural subtypes (Supplemental Fig 1B). While a complete analysis of therapeutic response across the molecular subtypes is limited, literature has described mesenchymal GBM tumors to exhibit increased resistance to therapy resulting in a worse patient survival outcome compared to proneural GBM subtype tumors which have a better prognosis (31). Together, these data suggest that PTGFRN is a novel mediator of radiation resistance and may be pro-tumorigenic in GBM tumors and predictive of poor prognosis, independent of common tumor mutation status.
Figure 1:
Relevance of PTGFRN to GBM A, Oncomine data of PTGFRN expression in brain tumors versus normal brain tissue in four studies. B, PrognoScan survival curves of GBM patients whose tumors express high versus low levels of PTGFRN. C, From TCGA LGG+GBM database, Kaplan Maier survival plot for low and high grade gliomas dichotomized by PTGFRN expression. D, The expression of PTGFRN in low and high grade gliomas from the TCGA database. Pairwise statistical comparisons of each subtype of low grade glioma to GBM are indicated (student’s t-tests).
3.2. PTGFRN promotes cell proliferation and tumor growth
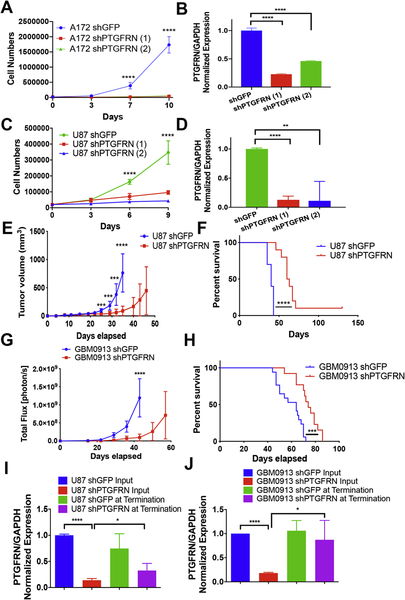
Next, we wished to determine the phenotype of PTGFRN in GBM cells by investigating the effects upon PTGFRN depletion. First, we performed cell proliferation assays using A172MG and U87MG GBM cell lines utilizing two different shRNAs to target PTGFRN. We found that reduction of PTGFRN expression significantly reduced cell proliferation in vitro (Fig 2A, B, C, D). Additionally, to access growth in vivo, we performed a subcutaneous tumor assay using PTGFRN knockdown U87MG cells. Tumors were measured twice weekly until the tumors reached greater than 1.5 cm3. Depleting PTGFRN drastically reduced tumor growth and increased mouse survival compared to shGFP control mice (Fig. 2 E, F).
Figure 2:
PTGFRN is required for cell proliferation and tumor growth. A, C, Cell proliferation assay in A172MG and U87MG GBM knockdown cells. B, D, qRT-PCR for samples in A and C. E, Subcutaneous U87 shRNA tumor growth curve. F, Kaplan-Meier curve for mice in E. G, Bioluminescence of intracranial tumor growth of shRNA neurospheres. H, Kaplan-Meier curve of mice in G. I, PTGFRN expression from U87 cells before injection and from U87 subcutaneous tumors. J, PTGFRN expression from GBM0913 cells before injection and from GBM0913 intracranial tumors. ***, p<0.001; ****, p<0.0001 by two-way ANOVA (A, C, E, G) or log rank (F, H).
Further, as primary brain tumors are thought to be maintained by self-renewing, tumorigenic cells (22, 32–34), we also utilized neurosphere stem cell lines (22). An intracranial xenograft was performed after PTGFRN shRNA transduction into luciferase expressing GBM0913 neurosphere cells. Upon PTGFRN depletion, as determined by qRTPCR, cells were injected into the brains of nude mice and followed by bioluminescent imaging. We observed that similar to the established cell lines data, mice containing shPTGFRN neurospheres experienced decreased tumor growth rate (Fig 2G) and extended overall survival (Fig. 2H) compared to control shGFP neurospheres. Additionally, to determine whether the eventual tumor growth in the PTGFRN knockdown U87 subcutaneous and GBM0913 brain tumors correlated with regained expression of PTGFRN, tumors were excised and subjected to qRT-PCR. We found that the expression of PTGFRN was returning in the U87 cells (Fig 2I) and had returned in the GBM0913 cells (Fig 2J), compared to the expression of the cells injected suggesting a selective pressure exists for the expression of PTGFRN in the tumors.
Next, we wished to determine the cause of the significant decrease in cell proliferation seen in the GBM cell lines and GBM0913 neurospheres (Supplemental Fig 2A, B). In initial analyses of the cell proliferation, PTGFRN depletion increased the cell doubling time by roughly 20 hours compared to shGFP control cells (Supplemental Fig 2C). Also, upon performing an EdU pulse chase on GBM0913 shPTGFRN and shGFP neurosphere cells, we found that shPTGFRN cells are slower to progress through G2/M phases of the cell cycle compared to shGFP cells, as G1 peaks at 38 hours after release from growth factor starvation in shPTGFRN cells compared to shGFP cells where G1 peaks at 24 hours after release (Supplemental Fig 2D). These data suggest that the cause of decreased cell proliferation after depleting PTGFRN is due to an extension of cell cycle progression. Finally, as tumor stem cells are defined by their ability to self-renew, we wished to determine if PTGFRN affects self-renewal (35). We performed sphere formation assays using PTGFRN depleted GBM0821 and GBM0913 cells, and found no change in the number of spheres formed between shGFP and shPTGFRN neurospheres (Supplemental Fig 2E), but did find the colonies to be smaller (Supplemental Fig 2F), suggesting that PTGFRN does not affect the capability of the neurospheres to self-renew. Therefore, these results demonstrate that elevated expression of PTGFRN promotes cell proliferation and tumor growth by decreasing the time of cell cycle progression.
3.3. PTGFRN inhibition sensitizes GBM cells to radiation
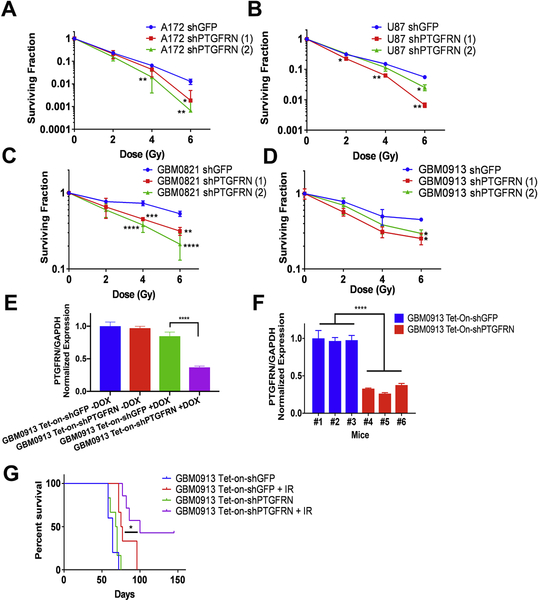
We next sought to verify the functional knockdown screen and determine whether PTGFRN reduction can sensitize GBM cells to radiation. Clonogenic survival assays were performed with varying doses of ionizing radiation (IR) targeting PTGFRN with two shRNAs in multiple GBM cell lines. qRT-PCR analysis was used to verify knockdown efficiency (Supplemental Fig 3). Both shRNAs, shPTGFRN (1) and shPTGFRN (2), sensitized A172 cells, with dose enhancement factors at 10% survival of 1.67 and 1.4 respectively (Fig 3A). Additionally, we observed that the depletion of PTGFRN also sensitized U87MG cells by a factor of 1.79 and 1.2 (Fig. 3B) (10% survival). Further, in GBM neurosphere lines, PTGFRN knockdown sensitized GBM0821 (Fig. 3C) by a factor of 1.77 and 3.28 (50% survival) and in GBM0913 (Fig. 3D) by 1.45 (35% survival) and 1.52 (45% survival). Notably, the neurosphere lines are significantly more radioresistant than the established cell lines (5). Together, these results show that inhibiting PTGFRN is capable of sensitizing multiple GBM tumor lines to radiation.
Figure 3:
PTGFRN reduction sensitizes GBM cells to IR. Clonogenic assays of A172 (A), U87MG (B), GBM0821 (C) and GBM0913 neurospheres (D) cells after indicated doses of IR. qRT-PCR of expression normalized to GAPDH can be found in supplemental figure 3. E, qRTPCR of GBM0913 Tet-on-shRNA cells with or without doxycycline in vitro. F, qRT-PCR of PTGFRN expression in animals injected with GBM0913 Tet-On-shRNA cells treated with doxycycline. G, Kaplan-Meier survival curve of GBM0913 Tet-onshRNA intracranial xenograft.*, p<0.05; **, p<0.01; ***, p<0.001 by student’s t tests (A, B, C, D) or log rank (E).
To specifically analyze radiosensitization in vivo upon PTGFRN depletion without the previously observed decreased cell proliferation as a variable, we utilized an inducible Tet-On shRNA tumor model. GBM0913 neurosphere stem cells, which overexpress the Tet repressor, were infected with lentiviruses carrying a doxycycline-inducible pLKO-Tet-On-shRNA (either shGFP or shPTGFRN) construct and implanted intracranially into nude mice. We validated the effect of doxycycline on expression of PTGFRN in vitro (Fig 3E) and on six animals in vivo and saw a significant reduction in PTGFRN expression (Fig. 3F). To test radiosensitization, the animals received either GBM0913 Tet-On-shGFP cells or GBM0913 Tet-On-shPTGFRN cells. Two weeks after implantation, tumors were measured by bioluminescence and mice were randomized into two treatment groups: doxycycline, or doxycycline plus 12 Gy IR. Radiation was delivered using the GammaKnife clinical irradiator as a single dose (36), and doxycycline was given for 6 days to obtain gene silencing during IR and then removed. As expected, control mice injected with GBM0913 Tet-On-shGFP and Tet-On-shPTGFRN showed no difference in overall survival (median survival: 64 days versus 69 days, respectively), given that PTGFRN was silenced for only 6 days. With the addition of IR, mice containing GBM0913 Tet-On-shPTGFRN neurosphere cells survived longer than the control, GBM0913 Tet-On-shGFP with IR (median survival: 100 days versus 76 days, respectively, P=0.0304) (Fig. 3G). In addition, three of animals were cured and sacrificed with no evidence of tumors at 150 days. Together, the data suggest that PTGFRN expression protects GBM cells from genotoxic IR in vitro as well as in vivo.
3.4. PTGFRN is necessary for P-AKT signaling by increasing stability of p110β
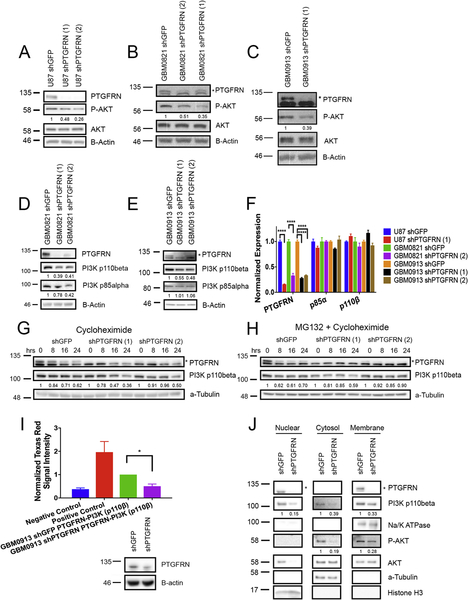
The molecular mechanisms of PTGFRN have been little studied. We found that PTGFRN inhibition decreases cell growth and sensitizes GBM cells to radiation, and as the PI3K/AKT pathway is a major survival signaling pathway found to be altered in GBM (18, 37), we hypothesized that PTGFRN modulates AKT signaling. We thus performed western blots to examine the levels of phospho-AKT and total AKT in PTGFRN-depleted GBM cells. We found that PTGFRN knockdown decreased basal phospho-AKT S473 levels compared to shGFP control cells in U87MG cells and both of the neurosphere lines (Fig 4A–C). Upstream of AKT, Phosphatidyl inositol 3 kinase (PI3K) is a major mediator of AKT phosphorylation, and we therefore assessed expression of the p110 and p85 subunits. We noted that PTGFRN knockdown-cells displayed decreased p110β protein levels compared to control, while p85α and p110α levels remained unchanged (Fig 4D, E, Supplemental Fig 4A,B). We next wished to see if we could rescue the decreased phospho-AKT by overexpressing p110β following PTGFRN depletion. Upon PTGFRN knockdown, we saw decreased p110β and phospho-AKT protein levels compared with shGFP control. Following overexpression of p110β (PIK3CB) utilizing an overexpression plasmid in the shPTGFRN cells, we noted increased levels of phospho-AKT compared to both shPTGFRN and shGFP (Supplemental Fig 4C). Additionally, overexpression of p110β following PTGFRN knockdown was able to partially rescue cell viability and proliferation (Supplemental Fig 4D), further supporting the proposed mechanism of PTGFRN signaling.
Figure 4:
PTGFRN depletion decreases p110β levels. A, B, C, Western blot evaluating basal P-AKT signaling in different GBM shPTGFRN cells compared to shGFP control. D, E, Western blot assessing p110β protein levels in GBM0821 and GBM0913 neurosphere cell lines. F, qRT-PCR for p85α and p110β in 3 different GBM shPTGFRN cell lines. Expression normalized to GAPDH. G, H, Immunoblot analyses of GBM0913 shRNA cells treated with cycloheximide alone (G) or in combination with proteasome inhibitor, MG132 (H). I, Proximity ligation assay of GBM0913 shPTGFRN or shGFP cells; negative control contained no primary antibody; positive control comprised two different antibodies targeting PTGFRN; * p<0.05 by student’s t test (above). Immunoblot of cells used for the PLA assay (below). J, Cellular fractionation assay in GBM0913 shRNA neurosphere cells using a-tubulin, Histone H3 and Na/K-ATPase as cellular fractionation internal and loading controls.
As altered protein levels could be due to corresponding mRNA levels, we determined by qRT-PCR that p110β mRNA levels were unaffected upon knockdown of PTGFRN (Fig 4F). With mRNA levels unaltered, we questioned whether the decreased p110β protein could be due to increased turnover. We treated shPTGFRN and shGFP cells with cycloheximide to block translation and collected the cell lysate at multiple time points. Over 24 hours, we observed in shPTGFRN cells that p110β is less stable compared to shGFP control (Fig 4G). Further, we asked whether treatment with a proteasome inhibitor, MG132, could rescue p110β degradation in shPTGFRN cells. Cells were treated with cycloheximide in combination with MG132 and collected at specified time points (Fig 4H). The amount of degradation seen at 24 hours was rescued in shPTGFRN cells as well as shGFP control cells compared to cells treated with cycloheximide alone. The data thus suggest that PTGFRN plays a role in p110β stability.
Since both PTGFRN and p110β can be found at the plasma membrane and PTGFRN affects p110β turnover, we asked if PTGFRN is in close proximity with p110β. By performing a proximity ligation assay (PLA) in GBM0913 shGFP and shPTGFRN neurosphere cells, we found that PTGFRN and p110β are within close proximity to each other, as upon PTGFRN inhibition, the Texas Red signal decreased two-fold to levels similar to the negative control (Fig 4I). The close proximity of PTGFRN and p110β was further supported by evaluating the co-localization of PTGFRN and p110β by confocal imaging in GBM0913 shGFP and shPTGFRN cells (Supplemental Fig 4E). In GBM0913 shPTGFRN cells, the fluorescence intensity of both PTGFRN and p110β protein significantly decreased compared to shGFP (Supplemental Fig 4F). Furthermore, we determined that the majority of p110β co-localized with PTGFRN. Interestingly, not all of PTGFRN co-localized with p110β (Supplemental Fig 4G), suggesting that PTGFRN may have other roles in the cell.
Additionally, we hypothesized that if PTGFRN is necessary for p110β stability, then p110β expression should be decreased not just at the membrane, but also in the cytosol and nucleus of shPTGFRN GBM cells. When we fractionated proteins from shGFP and shPTGFRN neurosphere cells, we observed decreased levels of p110β at the membrane and in the cytosol and nucleus of shPTGFRN cells compared to control cells. Additionally, we found decreased phospho-AKT protein levels in the membrane and cytosol fractions suggesting that the reduced phospho-AKT may be due to the decreased availability of p110β to catalyze PIP2 to PIP3, allowing for AKT to translocate to the membrane for activation (Fig 4J). With these results, we posit that PTGFRN, acting as a scaffolding protein (9, 38), is important for p110β stability allowing for downstream AKT signaling.
3.5. PTGFRN promotes DNA damage sensing through p110β
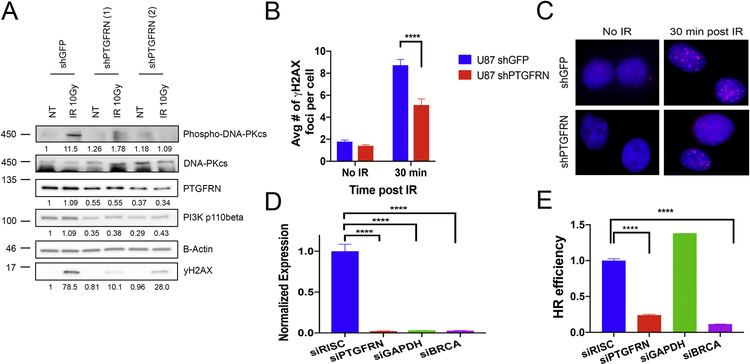
Radiation therapy is a major component of the treatment for GBM patients, and the main deleterious damage caused by IR is DNA double strand-breaks (DSBs) (39). Our results demonstrate that depleting PTGFRN radiosensitizes GBM cells in vitro and in vivo, which led us to question how PTGFRN depletion affects DNA damage repair. Interestingly, the literature has described a necessity for nuclear p110β in sensing DSBs and its depletion leads to a defect in DNA repair activation (40). Our cellular fractionation results also found decreased p110β in the nucleus (Fig 4J) suggesting that PTGFRN
depletion could impair DSB sensing through p110β. Therefore, we irradiated GBM0913 shRNA cells with 10 Gy and collected the cell lysates after 30 minutes to evaluate gH2AX activation, a canonical DSB marker, and DNA-PKcs activation, a canonical nonhomologous end joining (NHEJ) factor. Upon PTGFRN depletion, we find decreased gH2AX and phospho-DNA-PKcs following IR compared to shGFP control (Fig 5A). Additionally, we irradiated U87 shRNA cells and performed immunofluorescence to analyze formation of gH2AX foci. Similarly, we found decreased amounts of gH2AX foci in shPTGFRN cells compared to shGFP 30 min post IR suggesting a decrease in DSB recognition (Fig. 5B, C). To confirm that DNA damage repair was also hindered by PTGFRN depletion, we performed a homologous recombination (HR) DNA repair assay using a DR-GFP reporter in U87MG cells. Our results show that decreasing PTGFRN expression (Fig. 5D) also hindered the efficiency of HR DNA repair (Fig 5E), possibly due to decreased DSB sensing. Altogether, the data suggest that PTGFRN promotes DNA damage DSB sensing through p11β signaling.
Figure 5:
PTGFRN promotes DNA damage sensing. A, Western blot of GBM0913 shRNA cells that received either no treatment (NT) or radiation (10 Gy) and collected after 30 minutes. B, Quantification of γH2AX foci following IR (2 Gy) in U87 shRNA cells. C, Immunofluorescence of γH2AX (red) foci. D, qRTPCR of targeted genes in HR assay. Expression normalized to b-actin. E, DR-GFP HR reporter assay in siPTGFRN, siGAPDH (negative control), siBRCA1 (positive control), or siRISC (control) U87 cells.
4. Discussion
In the present study, we have defined a novel mediator of cell proliferation and radioprotection in GBM that associates with poor patient outcome. PTGFRN is overexpressed in GBM tumor samples compared to normal brain, as assessed bioinformatically in Oncomine and TCGA databases. Depletion of PTGFRN decreased cell proliferation and radiosensitized multiple cell lines, including neurosphere stem cell lines, and tumors in mice. Mechanistically, we found that depletion of PTGFRN decreases phospho-AKT signaling due to decreased stability of p110β at the cell membrane. Additionally, we found decreased nuclear p110β upon depletion of PTGFN, which decreased DNA damage sensing and reduced DNA repair. Together, our findings highlight a novel function for PTGFRN as a scaffolding protein in regulating survival signaling responses in tumors, and identify a potential therapeutic target.
PTGFRN was first discovered to inhibit the binding of prostaglandin F2α to the prostaglandin F2α receptor, a G-protein coupled receptor that could selectively activate p110β (41), by decreasing the receptor number rather than the receptor affinity (42). Increased levels of prostaglandin F2α, and subsequently increased prostaglandin F2α:prostaglandin F2α receptor signaling, have been found to increase cell proliferation and promote radiation resistance in prostate cancers (43, 44). As PTGFRN was found to decrease the receptor number, decreasing PTGFRN would cause increased levels of prostaglandin F2α receptor, allowing for prostaglandin F2α binding and signal transduction. However, in our study, decreasing PTGFRN affects cell proliferation and radiation resistance through survival signaling that does not affect protein levels of prostaglandin F2α receptor in the GBM cells tested (data not shown). Therefore, in our study and in the context of GBM, PTGFRN does not regulate prostaglandin signaling by depleting the number of prostaglandin F2α b1receptor molecules
Our results indicated that PTGFRN depletion leads to decreased cell proliferation and basal phospho-AKT protein levels, in agreement with other published studies completed in GBM cells and tumors (45, 46). In this study, the effects are at least partially due to the reduction of p110β at the membrane of shPTGFRN cells. Furthermore, we observed that PTGFRN and p110β are within close proximity, and shPTGFRN cells had more p110β degradation after 24 hours compared to control, thus suggesting that PTGFRN impacts p110β turnover. It is known that p110β is stabilized by dimerization with p85, which also regulates p110β catalytic activity (47). Our results thus suggest that tethering p110β to the membrane is the mechanistic reason for promoting stabilization and signaling.
At the same time, other functions for p110β have been discovered that relate to radioresistance. In the nucleus, p110β has been found to be involved in DNA damage sensing and cell survival by regulating recruitment of DNA damage proteins to DSB foci (40, 48). We found decreased p110β in the nuclear fraction of the shPTGFRN GBM cells compared to shGFP cells, suggesting that decreased p110β could also explain the observed radiation sensitivity upon PTGFRN depletion. After DNA damage by radiation, PTGFRN depleted cells have reduced DNA damage sensing and delayed protein recruitment to the site of damage as demonstrated by the reduced activation of γ H2AX and DNA-PKcs in shPTGFRN cells 30 minutes post IR. Furthermore, a decrease in DNA damage sensing could cause decreased DNA repair, as cells fail to detect the initial damage, agreeing with the decrease in HR repair efficiency seen in shPTGFRN GBM cells.
Targeting PTGFRN could be beneficial in decreasing cell survival signaling in GBM tumors. Thus far, a truncated form of PTGFRN, GS-168AT2, was developed and found to function as a dominant negative protein that interferes with normal PTGFRN extracellular interactions. GS-168AT2 decreased tumor growth in vivo in lung xenograft mice. The authors suggest that GS-168AT2 acts through preventing interactions with CD9 and CD151, two tetraspanin proteins implicated in tumorigenesis (10, 11, 49). Interestingly, CD9 has been found to promote tumor growth in GBM by stabilizing IL-6 receptor glycoprotein 130 from degradation (50), analogous to PTGFRN stabilizing p110β to promote survival signaling. Further, CD9 is a partner protein of PTGFRN (51) suggesting that CD9 and PTGFRN could regulate signaling as a complex, as well as individually. While not a focus of the present study, future analyses on the interaction between PTGFRN and CD9 in GBM and their possible dual implications to cell signaling would be attractive as both proteins, individually, seem to regulate turnover of signaling proteins.
Various PI3K inhibitors have been developed and studied in the last few decades since hyper-activation of the PI3K/AKT pathway confers rapid growth, tumor progression and multidrug resistance upon GBM cells. The first generation of pan-PI3K inhibitors, wortmannin and LY294002, are unable to be used clinically due to their high toxicities. The current PI3K inhibitors, which have improved safety, efficacy and pharmacokinetics, in clinical trials include BKM120, XL147, and PX-866 (52). BKM120 is a pan-PI3K inhibitor that impedes intracerebral U87 GBM cell xenograft growth in preclinical studies and inhibits AKT phosphorylation (53, 54). Currently, BKM120, which is well-tolerated and permeable to the blood-brain barrier, is the most-frequently used PI3K inhibitor in GBM clinical trials (52). Another promising PI3K inhibitor is Pilaralisib (XL147) that exhibits dose dependent decrease in AKT phosphorylation, pS6K1, and Ki67 expression, suggesting moderate blood brain barrier penetration and inhibition of proliferation (55, 56). Finally, PX-866 is an irreversible wortmannin analogue that inhibits intracranial xenograft growth by selectively blocking the PI3K/AKT pathway (57). As depleting PTGFRN decreases AKT phosphorylation and tumor growth, similar to the above PI3K inhibitors, targeting PTGFRN, which is positioned on the cell surface, has the potential to be beneficial to GBM patients.
In summary, we have defined a novel function for PTGFRN in mediating survival signaling in brain tumors through regulation of p110β. Our findings describe a mechanism for PTGFRN to act as a scaffolding protein and regulate p110β protein stability, thus affecting cell proliferation and tumor growth, along with regulation of DNA DSB recognition and repair. Together, these findings contribute to our understanding of PTGFRN and the impact of scaffolding protein depletion on downstream signaling and suggest that inhibition of PTGFRN may prove to be a beneficial therapeutic target by affecting both cell proliferation and survival.
Supplementary Material
Highlights:
PTGFRN is widely overexpressed in glioblastoma and dictates poor survival.
PTGFRN depletion decreased tumor growth and radiosensitized GBM cells.
PTGFRN acts as a scaffolding protein to regulate p110β stability and signaling.
Inhibition of PTGFRN depletes nuclear p110β causing decreased DNA damage sensing.
Acknowledgments:
This work was supported by the National Cancer Institute (R01CA187053 to SMW, and R01CA187780 to EB), the Case Comprehensive Cancer Center at CWRU, and the Sylvester Comprehensive Cancer Center at the University of Miami. Core facilities of the Case Comprehensive Cancer Center were used: Radiation Resources, Cytometry and Imaging Microscopy, Small Animal Imaging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare no conflict of interest
Contributor Information
Brittany Aguila, Department of Biochemistry, Case Western Reserve University, Cleveland, OH 44106.
Adina Brett Morris, Department of Pharmacology, Case Western Reserve University, Cleveland, OH 44106.
Raffaella Spina, Department of Neurological Surgery, Case Western Reserve University, Cleveland, OH 44106.
Eli Bar, Department of Neurological Surgery, Case Western Reserve University, Cleveland, OH 44106.
Julie Schraner, Department of Radiation Onoclogy, University Hospitals Cleveland Medical Center, Seidman Cancer Center, Cleveland, OH 44106.
Robert Vinkler, Department of Radiation Onoclogy, University Hospitals Cleveland Medical Center, Seidman Cancer Center, Cleveland, OH 44106.
Jason W Sohn, Department of Radiation Oncology, Allegheny Health Network, Pittsburgh, PA 15212.
Scott M Welford, Department of Radiation Oncology, Miller School of Medicine, University of Miami, Miami, FL 33136; Sylvester Comprehensive Cancer Center, Miller School of Medicine, University of Miami, Miami, FL 33136.
References
- 1.Krex D, Klink B, Hartmann C, von Deimling A, Pietsch T, Simon M, Sabel M, Steinbach JP, Heese O, Reifenberger G, Weller M, Schackert G, German Glioma N. Long-term survival with glioblastoma multiforme. Brain : a journal of neurology. 2007;130(Pt 10):2596–606. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO, European Organisation for R, Treatment of Cancer Brain T, Radiotherapy G, National Cancer Institute of Canada Clinical Trials G. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England journal of medicine. 2005;352(10):987–96. [DOI] [PubMed] [Google Scholar]
- 3.Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF, Agarwalla PK, Chheda MG, Campos B, Wang A, Brennan C, Ligon KL, Furnari F, Cavenee WK, Depinho RA, Chin L, Hahn WC. Emerging insights into the molecular and cellular basis of glioblastoma. Genes & development. 2012;26(8):756–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen PY, Kesari S. Malignant gliomas in adults. The New England journal of medicine. 2008;359(5):492–507. [DOI] [PubMed] [Google Scholar]
- 5.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–60. [DOI] [PubMed] [Google Scholar]
- 6.Chakravarti A, Zhai GG, Zhang M, Malhotra R, Latham DE, Delaney MA, Robe P, Nestler U, Song Q, Loeffler J. Survivin enhances radiation resistance in primary human glioblastoma cells via caspase-independent mechanisms. Oncogene. 2004;23(45):7494–506. [DOI] [PubMed] [Google Scholar]
- 7.Hatanpaa KJ, Burma S, Zhao D, Habib AA. Epidermal growth factor receptor in glioma: signal transduction, neuropathology, imaging, and radioresistance. Neoplasia. 2010;12(9):675–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brett-Morris A, Wright BM, Seo Y, Pasupuleti V, Zhang J, Lu J, Spina R, Bar EE, Gujrati M, Schur R, Lu ZR, Welford SM. The polyamine catabolic enzyme SAT1 modulates tumorigenesis and radiation response in GBM. Cancer Res. 2014;74(23):6925–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sala-Valdes M, Ursa A, Charrin S, Rubinstein E, Hemler ME, Sanchez-Madrid F, Yanez-Mo M. EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. The Journal of biological chemistry. 2006;281(28):19665–75. [DOI] [PubMed] [Google Scholar]
- 10.Colin S, Guilmain W, Creoff E, Schneider C, Steverlynck C, Bongaerts M, Legrand E, Vannier JP, Muraine M, Vasse M, Al-Mahmood S. A truncated form of CD9-partner 1 (CD9P-1), GS-168AT2, potently inhibits in vivo tumour-induced angiogenesis and tumour growth. Br J Cancer. 2011;105(7):1002–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guilmain W, Colin S, Legrand E, Vannier JP, Steverlynck C, Bongaerts M, Vasse M, Al-Mahmood S. CD9P-1 expression correlates with the metastatic status of lung cancer, and a truncated form of CD9P-1, GS-168AT2, inhibits in vivo tumour growth. Br J Cancer. 2011;104(3):496–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orlicky DJ, Lieber JG, Morin CL, Evans RM. Synthesis and accumulation of a receptor regulatory protein associated with lipid droplet accumulation in 3T3-L1 cells. J Lipid Res. 1998;39(6):1152–61. [PubMed] [Google Scholar]
- 13.Chambrion C, Le Naour F. The tetraspanins CD9 and CD81 regulate CD9P1-induced effects on cell migration. PloS one. 2010;5(6):e11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yanez-Mo M, Barreiro O, Gordon-Alonso M, Sala-Valdes M, Sanchez-Madrid F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 2009;19(9):434–46. [DOI] [PubMed] [Google Scholar]
- 15.Andre M, Chambrion C, Charrin S, Soave S, Chaker J, Boucheix C, Rubinstein E, Le Naour F. In situ chemical cross-linking on living cells reveals CD9P-1 cis-oligomer at cell surface. J Proteomics. 2009;73(1):93–102. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Wu C, Chen N, Gu H, Yen A, Cao L, Wang E, Wang L. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget. 2016;7(22):33440–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nature reviews Cancer. 2002;2(7):489–501. [DOI] [PubMed] [Google Scholar]
- 18.Chakravarti A, Zhai G, Suzuki Y, Sarkesh S, Black PM, Muzikansky A, Loeffler JS. The prognostic significance of phosphatidylinositol 3-kinase pathway activation in human gliomas. J Clin Oncol. 2004;22(10):1926–33. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura JL, Karlsson A, Arvold ND, Gottschalk AR, Pieper RO, Stokoe D, Haas-Kogan DA. PKB/Akt mediates radiosensitization by the signaling inhibitor LY294002 in human malignant gliomas. J Neurooncol. 2005;71(3):215–22. [DOI] [PubMed] [Google Scholar]
- 20.Saito Y, Tachibana I, Takeda Y, Yamane H, He P, Suzuki M, Minami S, Kijima T, Yoshida M, Kumagai T, Osaki T, Kawase I. Absence of CD9 enhances adhesion-dependent morphologic differentiation, survival, and matrix metalloproteinase-2 production in small cell lung cancer cells. Cancer Res. 2006;66(19):9557–65. [DOI] [PubMed] [Google Scholar]
- 21.Sugiura T, Berditchevski F. Function of alpha3beta1-tetraspanin protein complexes in tumor cell invasion. Evidence for the role of the complexes in production of matrix metalloproteinase 2 (MMP-2). J Cell Biol. 1999;146(6):1375–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64(19):7011–21. [DOI] [PubMed] [Google Scholar]
- 23.Spina R, Voss DM, Asnaghi L, Sloan A, Bar EE. Atracurium Besylate and other neuromuscular blocking agents promote astroglial differentiation and deplete glioblastoma stem cells. Oncotarget. 2016;7(1):459–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voss DM, Spina R, Carter DL, Lim KS, Jeffery CJ, Bar EE. Disruption of the monocarboxylate transporter-4-basigin interaction inhibits the hypoxic response, proliferation, and tumor progression. Sci Rep. 2017;7(1):4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Z, Huang Z and Lung ML. Subcellular Fractionation of Cultured Human Cell Lines. Bio-protocol 2013;3(9):e754. [Google Scholar]
- 26.Kass EM, Helgadottir HR, Chen CC, Barbera M, Wang R, Westermark UK, Ludwig T, Moynahan ME, Jasin M. Double-strand break repair by homologous recombination in primary mouse somatic cells requires BRCA1 but not the ATM kinase. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(14):5564–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleisig H, Wong J. Measuring cell cycle progression kinetics with metabolic labeling and flow cytometry. J Vis Exp. 2012(63):e4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan AM. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC medical genomics. 2009;2:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN, Cancer Genome Atlas Research N. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Behnan J, Finocchiaro G, Hanna G. The landscape of the mesenchymal signature in brain tumours. Brain : a journal of neurology. 2019;142(4):847–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Cancerous stem cells can arise from pediatric brain tumors. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(25):15178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39(3):193–206. [DOI] [PubMed] [Google Scholar]
- 34.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. [DOI] [PubMed] [Google Scholar]
- 35.Pastrana E, Silva-Vargas V, Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell. 2011;8(5):486–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Awan MJ, Dorth J, Mani A, Kim H, Zheng Y, Mislmani M, Welford S, Yuan J, Wessels BW, Lo SS, Letterio J, Machtay M, Sloan A, Sohn JW. Development and Validation of a Small Animal Immobilizer and Positioning System for the Study of Delivery of Intracranial and Extracranial Radiotherapy Using the Gamma Knife System. Technol Cancer Res Treat. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maecker HT, Todd SC, Levy S. The tetraspanin superfamily: molecular facilitators. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1997;11(6):428–42. [PubMed] [Google Scholar]
- 39.Chu G Double strand break repair. The Journal of biological chemistry. 1997;272(39):24097–100. [DOI] [PubMed] [Google Scholar]
- 40.Kumar A, Fernandez-Capetillo O, Carrera AC. Nuclear phosphoinositide 3-kinase beta controls double-strand break DNA repair. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(16):7491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pridham KJ, Varghese RT, Sheng Z. The Role of Class IA Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunits in Glioblastoma. Front Oncol. 2017;7:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orlicky DJ. Negative regulatory activity of a prostaglandin F2 alpha receptor associated protein (FPRP). Prostaglandins Leukot Essent Fatty Acids. 1996;54(4):247–59. [DOI] [PubMed] [Google Scholar]
- 43.Sun SQ, Gu X, Gao XS, Li Y, Yu H, Xiong W, Yu H, Wang W, Li Y, Teng Y, Zhou D. Overexpression of AKR1C3 significantly enhances human prostate cancer cells resistance to radiation. Oncotarget. 2016;7(30):48050–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S, Yang Q, Fung KM, Lin HK. AKR1C2 and AKR1C3 mediated prostaglandin D2 metabolism augments the PI3K/Akt proliferative signaling pathway in human prostate cancer cells. Mol Cell Endocrinol. 2008;289(1–2):60–6. [DOI] [PubMed] [Google Scholar]
- 45.Pridham KJ, Le L, Guo S, Varghese RT, Algino S, Liang Y, Fajardin R, Rodgers CM, Simonds GR, Kelly DF, Sheng Z. PIK3CB/p110beta is a selective survival factor for glioblastoma. Neuro Oncol. 2018;20(4):494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pu P, Kang C, Zhang Z, Liu X, Jiang H. Downregulation of PIK3CB by siRNA suppresses malignant glioma cell growth in vitro and in vivo. Technol Cancer Res Treat. 2006;5(3):271–80. [DOI] [PubMed] [Google Scholar]
- 47.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Regulation of the p85/p110 phosphatidylinositol 3’-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol. 1998;18(3):1379–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar A, Redondo-Munoz J, Perez-Garcia V, Cortes I, Chagoyen M, Carrera AC. Nuclear but not cytosolic phosphoinositide 3-kinase beta has an essential function in cell survival. Mol Cell Biol. 2011;31(10):2122–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hemler ME. Tetraspanin proteins promote multiple cancer stages. Nature Reviews Cancer. 2013;14(1):49–60. [DOI] [PubMed] [Google Scholar]
- 50.Shi Y, Zhou W, Cheng L, Chen C, Huang Z, Fang X, Wu Q, He Z, Xu S, Lathia JD, Ping Y, Rich JN, Bian XW, Bao S. Tetraspanin CD9 stabilizes gp130 by preventing its ubiquitin-dependent lysosomal degradation to promote STAT3 activation in glioma stem cells. Cell Death Differ. 2017;24(1):167–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stipp CS, Orlicky D, Hemler ME. FPRP, a major, highly stoichiometric, highly specific CD81- and CD9-associated protein. The Journal of biological chemistry. 2001;276(7):4853–62. [DOI] [PubMed] [Google Scholar]
- 52.Zhao HF, Wang J, Shao W, Wu CP, Chen ZP, To ST, Li WP. Recent advances in the use of PI3K inhibitors for glioblastoma multiforme: current preclinical and clinical development. Mol Cancer. 2017;16(1):100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Netland IA, Forde HE, Sleire L, Leiss L, Rahman MA, Skeie BS, Miletic H, Enger PO, Goplen D. Treatment with the PI3K inhibitor buparlisib (NVP-BKM120) suppresses the growth of established patient-derived GBM xenografts and prolongs survival in nude rats. J Neurooncol. 2016;129(1):57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wen PY, Yung WKA, Mellinghoff IK, Ramkissoon S, Alexander BM, Rinne ML, Colman H, Omuro AMP, DeAngelis LM, Gilbert MR, Groot JFD, Cloughesy TF, Chi AS, Lee EQ, Nayak L, Batchelor T, Chang SM, Prados M, Reardon DA, Ligon KL. Phase II trial of the phosphatidyinositol-3 kinase (PI3K) inhibitor buparlisib (BKM120) in recurrent glioblastoma. Journal of Clinical Oncology. 2014;32(15_suppl):2019-. [Google Scholar]
- 55.Foster P, Yamaguchi K, Hsu PP, Qian F, Du X, Wu J, Won KA, Yu P, Jaeger CT, Zhang W, Marlowe CK, Keast P, Abulafia W, Chen J, Young J, Plonowski A, Yakes FM, Chu F, Engell K, Bentzien F, Lam ST, Dale S, Yturralde O, Matthews DJ, Lamb P, Laird AD. The Selective PI3K Inhibitor XL147 (SAR245408) Inhibits Tumor Growth and Survival and Potentiates the Activity of Chemotherapeutic Agents in Preclinical Tumor Models. Mol Cancer Ther. 2015;14(4):931–40. [DOI] [PubMed] [Google Scholar]
- 56.Cloughesy TF, Mischel PS, Omuro AMP, Prados M, Wen PY, Wu B, Rockich K, Xu Y, Lager JJ, Mellinghoff IK. Tumor pharmacokinetics (PK) and pharmacodynamics (PD) of SAR245409 (XL765) and SAR245408 (XL147) administered as single agents to patients with recurrent glioblastoma (GBM): An Ivy Foundation early-phase clinical trials consortium study. Journal of Clinical Oncology. 2013;31(15_suppl):2012-. [Google Scholar]
- 57.Koul D, Shen R, Kim YW, Kondo Y, Lu Y, Bankson J, Ronen SM, Kirkpatrick DL, Powis G, Yung WK. Cellular and in vivo activity of a novel PI3K inhibitor, PX-866, against human glioblastoma. Neuro Oncol. 2010;12(6):559–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.