Abstract
The Na+-Ca2+ exchanger 1 (NCX1) is reduced in stroke by the RE1-silencing transcription factor (REST), whereas it is increased in ischemic brain preconditioning (PC) by hypoxia-inducible factor 1 (HIF-1). Because ncx1 brain promoter (ncx1-Br) has five putative consensus sequences, named Sp1A–E, for the specificity protein (Sp) family of transcription factors (Sp1–4), we investigated the role of this family in regulating ncx1 transcription in rat cortical neurons. Here we found that Sp1 is a transcriptional activator, whereas Sp3 is a transcriptional repressor of ncx1, and that both bind ncx1-Br in a sequence-specific manner, modulating ncx1 transcription through the Sp1 sites C–E. Furthermore, by transient middle cerebral artery occlusion (tMCAO) in rats, the transcriptional repressors Sp3 and REST colocalized with the two histone-deacetylases (HDACs) HDAC1 and HDAC2 on the ncx1-Br, with a consequent hypoacetylation. Contrarily, in PC+tMCAO the transcriptional activators Sp1 and HIF-1 colocalized with histone acetyltransferase p300 on ncx1-Br with a consequent hyperacetylation. In addition, in neurons silenced with siRNA of NCX1 and subjected to oxygen and glucose deprivation (OGD) (3 h) plus reoxygenation (RX) (24 h), the neuroprotection of Class I HDAC inhibitor MS-275 was counteracted, whereas in neurons overexpressing NCX1 and subjected to ischemic preconditioning (PC+OGD/RX), the neurotoxic effect of p300 inhibitor C646 was prevented. Collectively, these results demonstrate that NCX1 expression is regulated by the Sp3/REST/HDAC1/HDAC2 complex in tMCAO and by the Sp1/HIF-1/p300 complex in PC+tMCAO and that epigenetic intervention, by modulating the acetylation of ncx1-Br, may be a strategy for the development of innovative therapeutic intervention in stroke.
Keywords: acetylation, brain ischemia, epigenetic, ischemic brain preconditioning, REST\NRSF, sodium/calcium exchanger
Introduction
Changes in the Na+-Ca2+ exchanger 1 (ncx1) gene expression, a ubiquitous plasma membrane protein regulating cellular calcium and sodium homeostasis in the brain, is important for the progression of stroke (Annunziato et al., 2004; Pignataro et al., 2004) and for the reduction of the cerebral infarct damage elicited by ischemic preconditioning (Valsecchi et al., 2011; Pignataro et al., 2012). Indeed, studies have shown that ncx1 ablation markedly increases infarct volume after stroke (Pignataro et al., 2004; Boscia et al., 2006) and partially reverts preconditioning-induced neuroprotection (Pignataro et al., 2012). We previously demonstrated that in an in vivo model of stroke, obtained by transient middle cerebral artery occlusion (tMCAO), NCX1 reduction is due to RE1-Silencing Transcription factor (REST) (Formisano et al., 2013), which regulates global gene expression after stroke (Noh et al., 2012; Schweizer et al., 2013; Kaneko et al., 2014). Instead, NCX1 increase in ischemic preconditioning plus ischemia (PC+tMCAO) is determined by hypoxia-inducible factor 1 (HIF-1) (Valsecchi et al., 2011). Interestingly, the ncx1 brain promoter (ncx1-Br) sequence contains several consensus binding sites for Specific protein 1 (Sp1). Sp proteins comprise four isoforms, Sp1-Sp4 (Suske, 1999), which recognize the same Sp1 sequence with similar affinities (Hwang et al., 2001). The only exception is Sp2, which binds to a GT-rich element instead (Philipsen and Suske, 1999). Sp1, Sp3, and Sp4 can act as either activators (Ammanamanchi et al., 2003; Ishimaru et al., 2007; Ravache et al., 2010) or repressors (Ammanamanchi and Brattain, 2001; Ramos et al., 2009; Law et al., 2011) of gene expression. Therefore, both the presence of several binding sites for Sp1 on the ncx1-Br region and that Sp1 is involved in NGF-induced regulation of NCX1 (Sirabella et al., 2012) prompted us to investigate the relationship between the members of the Sp family and its role in modulating NCX1 expression during tMCAO and PC+tMCAO. Furthermore, because REST reduces NCX1 expression during tMCAO (Formisano et al., 2013) and HIF-1 increases NCX1 expression during PC+tMCAO, we also investigated the possible interaction between REST and Sp3 and between HIF-1 and Sp1 after tMCAO and PC+tMCAO, respectively. In addition, because both REST and Sp3 modulate their target genes via the histone deacetylase (HDAC) family (Won et al., 2002; Rodenas-Ruano et al., 2012), we investigated whether the interaction between REST and Sp3 forms a protein complex which, in turn, recruits HDAC1 and HDAC2, thus repressing ncx1 expression after tMCAO. Moreover, considering that histone acetyl transferase (HAT) is involved in gene expression induced by HIF-1 and Sp1 (Billon et al., 1999; Ke and Costa, 2006), we investigated whether the interaction between HIF-1 and Sp1 forms a protein complex that recruits the HAT p300, and activates ncx1 expression after PC+tMCAO. Finally, we assessed the effect of the Class I HDAC inhibitor MS-275 (Lanzillotta et al., 2013; Formisano et al., 2015b) and of the HAT p300 inhibitor C646 (Min et al., 2010) on cell survival and its correlation with NCX1 expression in neurons subjected to either oxygen and glucose deprivation followed by reoxygenation (OGD/RX) or PC plus OGD/RX.
Materials and Methods
Materials
All restriction enzymes and DNA-modifying enzymes were purchased from New England Biolabs or Promega. Luciferase reporter kits and luciferase vectors were from Promega. Synthetic oligonucleotides were from Primm. siRNAs against rat REST and HIF-1 were performed as already published (Formisano et al., 2013), whereas siRNAs for Sp1 (siSp1) (SI02039044), Sp3 (siSp3) (SI05434387), Sp4 (siSp4) (SI02039114), p300 (sip300) (SI02989693), and negative control siCONTROL (siCTL) (1027280) were from QIAGEN. To knock down rat HDAC1 and HDAC2,MISSION siRNAs from Sigma were used. The sequences of siRNAs were as follows: for HDAC1 forward, 5′-CUUUGAAUACUUUGGACCA(dT) (dT)-3′ and reverse, 5′-UGGUCCAAAGUAUUCAAAG(dT) (dT)-3′; for HDAC2 forward, 5′-GAUAUCGGGAAUUAUUAUU(dT) (dT)-3′ and reverse, 5′-AAUAAUAAUUCCCGAUAUC(dT) (dT)-3′. Two sets of siRNAs were tested for Sp1, Sp3, Sp4, p300, HDAC1, and HDAC2. The set that was more effective was chosen for the experiments (data not shown). The construct used to silence NCX1 (siNCX1) and the mismatch sequence cloned in the same vector (MS siNCX1) were used as previously described (Formisano et al., 2008; Sirabella et al., 2009). The constructs pN3 and pN3-Sp1, pN3-Sp3, and pN3-Sp4, carrying Sp1, Sp3, and Sp4 cDNAs, were kindly provided by Prof. G. Suske (Marburg, Germany). The constructs pKCRH-NCX1 overexpressing canine NCX1 and the empty vector (pKCRH) were kindly provided by Prof. Iwamoto (Fukuoka, Japan) (Iwamoto et al., 2004). The HDAC inhibitor MS-275 (EPS002) and the HAT-inhibitor C646 (SML002) were obtained from Sigma. Both were dissolved in DMSO and diluted with cell culture medium. The final concentration of DMSO was <0.2%. All common reagents were of the highest quality and were purchased from Sigma.
Cell cultures
Human neuroblastoma SH-SY5Y cells were prepared as previously described (Guida et al., 2014). Primary cortical neurons were prepared from 17-day-old Wistar rat embryos (Charles River) and used after 7 d. Cytosine arabinoside (2.5 μm) was added on the second day to reduce glial contamination. The experiments on primary cortical neurons were performed according to the experimental protocols approved by the Ethics Committee of “Federico II” University of Naples. Briefly; dissection and dissociation were performed in Ca2+/Mg2+-free PBS containing glucose (30 mm). Tissues were incubated with papain for 10 min at 37°C and dissociated by trituration in Earl's Balanced Salt Solution containing DNase (0.16 U/ml), BSA (10 mg/ml), and ovomucoid (10 mg/ml). Neurons, plated in plastic Petri dishes (Falcon Becton-Dickinson) precoated with poly-d-lysine (20 μg/ml), were grown in MEM/F12 containing glucose, deactivated FBS (5%), horse serum (5%), glutamine (2 mm), penicillin (50 U/ml), and streptomycin (50 μg/ml) (Invitrogen). For LDH assay, cells were plated in 24 well plates at a density of 1 × 106 cells/well; for luciferase assay, they were plated in 12 well plates at a density of 2 × 106 cells/well; for qRT-PCR, they were plated in 60 mm well plates at a density of 5 × 106 cells/well; for Western blot and chromatin immunoprecipitation (ChIP) analyses, they were plated in 100 mm well plates at a density of 15 × 106 cells/well.
Transfection with expression plasmids or small interfering RNA (siRNA) and luciferase reporter assay, in cortical neurons
Cortical neurons were transfected with 50 nm of rat siCTL, siSp1, siSp3, or siSp4 (for catalog numbers, see Materials). To overexpress pN3, Sp1, Sp3, and Sp4, pKCRH and pKCRH-NCX1, neurons were transfected with the above mentioned constructs in the following amounts: 0.5 μg for 24 well plates, 1.3 μg for 12 well plates, 7 μg for 60 mm plates, and 15 μg for 100 mm plates. Each transfection was performed at 7 DIV in Opti-MEM together with Lipofectamine LTX (15338–100, Invitrogen), as suggested by the producer. After a 2 h transfection, the medium was replaced with a fresh one. To silence NCX1 with siNCX1 and MS-NCX1 (siCTL) neurons were transfected as previously reported (Sirabella et al., 2009). In particular, cells were transfected with 0.5 μg for 24 well plates (for LDH assay) and 15 μg for 100 mm plates (for Western blot analysis) of the above mentioned constructs. For luciferase assay experiments, cortical neurons were transfected in 12 well plates. Cells were cotransfected with 2 μg of total DNA vectors; the reporters (560 ng each) were as follows: (1) the pGL3-Basic (pGL3) construct; (2) the pGL3-ncx1 (−340/151) contained a 350 bp fragment, named (short ncx1 promoter) (Valsecchi et al., 2011); (3) the pGL3-ncx1/Sp1Bmut (GGCGGCGGGC); (4) the pGL3-ncx1/Sp1Cmut (CGGGCGGGG); (5) the pGL3-ncx1/Sp1Dmut (GGGAGGGG); (6) the pGL3-ncx1/Sp1Emut (GGCCCCGGC); and (7) the pGL3-ncx1/Sp1CDEmut. This last reporter contained the simultaneous mutations of the Sp1 binding sites C, D, and E. Each italicized base represents the mutated sequences in pGL3. Each ncx1/Sp1 site was mutated by GG >TT substitution. Mutagenesis of the Sp1 sites in the promoter was performed using the QuikChangeSite-Directed Mutagenesis Kit from Stratagene. To overexpress Sp1, Sp3, and Sp4, we used expression vectors and the empty vector pN3 (1.3 μg each). For RNA interference, a concentration of 50 nm of specific siCTL, siSp1, siSp3, or siSp4 was used. Each transfection mix also contained 140 ng of the pRL-TK control vector expressing the Renilla luciferase gene. After 2 h of incubation, the medium was replaced with a fresh one and analyzed after 24 h with the Dual-Luciferase Reporter Assay System kit (E1910) (Promega), as already reported (Formisano et al., 2013). SHSY-5Y cells (70% of confluence) were plated in 100 mm dishes and transiently cotransfected with 9 μg of constructs carrying cDNA for Sp1 and Sp3 or with pN3 and with 6 μg of (1) pGL3-ncx1, (2) pGL3-ncx1 Sp1/C mut, (3) pGL3-ncx1 Sp1/D mut, (4) pGL3-ncx1 Sp1/E mut, and (5) pGL3-ncx1 Sp1/CDE mut using lipofectamine in Opti-MEM, for 6 h. After this time, the medium was changed to normal DMEM. Cells were harvested 48 h after transfection and lysed for Western blot analysis. Transfection efficiency was ≅60% for SH-SY5Y cells and ≅40% for cortical neurons (data not shown).
Combined OGD and RX
OGD was performed in cortical neurons incubated in a medium previously saturated with 95% N2 and 5% CO2 for 20 min and containing the following: 116 mm NaCl, 5.4 mm KCl, 0.8 mm MgSO4, 26.2 mm NaHCO3, 1 mm NaH2PO4, 1.8 mm CaCl2, 0.01 mm glycine, and 0.001% w/v phenol red; afterward, cells were placed in a hypoxic chamber for 3 h (Billups Rothemberg) (37°C, 5% CO2, and 95% N2) (Sisalli et al., 2014). To terminate OGD, neurons were removed from the hypoxic chamber and placed in a normal medium for 24 h of RX. For PC, neurons were exposed to 30 min of OGD, as described above, and then placed in a normal medium. Twenty-four hours after PC, cultures were again subjected to OGD for 3 h followed by 24 h of RX. Twenty-four hours after transfection with pKCRH-NCX1, pKCRH, siNCX1, and MS siNCX1 cells were subjected to OGD/RX or PC+OGD/RX. MS-275 (1 μm) was added at the end of OGD for 2 h, whereas C646 (20 μm) was added to the medium 30 min before PC and throughout the PC phase.
qRT-PCR analysis
First-strand cDNA and qRT-PCR were performed as previously described (Formisano et al., 2007). Using one-tenth of the cDNAs as a template, qRT-PCR was performed in a 7500-fast real-time PCR system (Applied Biosystems) by Fast SYBR Green Master Mix (cod 4385610; Applied Biosystems). Samples were amplified simultaneously in triplicate in one assay as follows: heating 2 min at 50°C, denaturation 10 min at 95°C, amplification and quantification 35 cycles of 15 s at 95°C; 1 min at 60°C with a single fluorescence measurement. PCR data were collected using ABI Prism 7000 SDS software (Applied Biosystems). After PCR, products were electrophoretically separated on 1.5% agarose gels, and the bands were visualized with ethidium bromide and documented using a Gel Doc Imaging System (Bio-Rad). The data were normalized by hypoxanthine phosphoribosyl-transferase as an internal control. Differences in mRNA content between groups were calculated as normalized values by using 2−ΔΔct formula, and results were tested for significance using the relative expression software tool (REST) (Formisano et al., 2007). The oligonucleotide sequences for NCX1 (forward 1716–1736 and reverse 3311–3292) (GenBank accession number NM_019268.3), NCX3 (forward 3229–3250 and reverse 1916–1899) (GenBank accession number NM_078620.1), and hypoxanthine phosphoribosyl-transferase (forward 444–467 and reverse 528–507) (GenBank accession number NM_078620.1) were performed as previously described (Pignataro et al., 2011).
Western blotting
For Western blot analysis, cells (or tissues) were collected in ice-cold lysis buffer (Formisano et al., 2013) containing anti-protease mixture (P8340 Sigma). For HDAC1, HDAC2, and acetyl-histone H3 expression, proteins (50 μg) were separated on 12% SDS polyacrylamide gels, whereas for NCX1, NCX3, Sp1, Sp3, Sp4, REST, HIF-1 and p300 expression, proteins (100 μg) were separated on 8% SDS-polyacrylamide gels. Both were transferred onto Hybond ECL nitrocellulose membranes (GE Healthcare). Membranes were first blocked with 5% nonfat dry milk in 0.1% Tween 20 (Sigma) (2 mm Tris-HCl and 50 mm NaCl, pH 7.5) for 2 h at room temperature. Then, they were incubated overnight at 4°C in the blocking buffer with the 1:1000 monoclonal antibodies against HDAC1 (5356) and HDAC2 (5113) (Cell-Signaling, EuroClone), p300 (sc-48343; Santa Cruz Biotechnology), HIF-1 (MAB5382; Millipore), and 1:2000 β-actin (A 4700; Sigma), either with the 1:1000 polyclonal antibodies against REST (Iannotti et al., 2013) and acetyl-histone H3 (06–866) (Millipore), NCX1 (p 11–13; Swant), NCX3 (Boscia et al., 2012), Sp1 (sc-14027), Sp3 (sc-644), or Sp4 (sc-645) (Santa Cruz Biotechnology). Finally, after incubation with primary antibodies, membranes were first washed with 0.1% Tween 20 and then incubated with secondary antibodies for 1 h at room temperature. The immunoreactive bands were detected with the ECL reagent (GE Healthcare). The optical density of the bands, normalized to β-actin, was determined by Chemi-Doc Imaging System (Bio-Rad).
ChIP and re-ChiP assay
Brain tissue and cortical neurons were processed into chromatin according to previous protocols (Formisano et al., 2007) with some modifications. Cells and tissues were cross-linked with 1% formaldehyde, and then reaction was stopped by adding glycine to a final concentration of 0.125 M. Brain tissue and cells were washed three times in cold PBS containing proteinase inhibitors and then collected in a buffer containing 50 mm Tris, pH 8.1, 1% SDS, 10 mm EDTA, and anti-protease mixture. For cell and tissue samples, chromatin was fragmented by sonication into 200–500 bp fragments (6 rounds for cells and 15 rounds for brain tissue of 10 s pulses at 50% of maximum potency) by a Bandelin Sonopuls HD 2070 ultrasonic homogenizer (Bandelin). Equal amounts of chromatin lysates (50 μg for cells and 70 μg for tissues) were incubated overnight with 5 μg of antibody for Sp1 (sc-14027), Sp3 (sc-644), Sp4 (sc-645), p300 (sc-48343) (Santa Cruz Biotechnology), REST (Iannotti et al., 2013), acetyl-histone H4 (06–866), hypoxia inducible factor 1 α (MAB5382) (Millipore), HDAC1 (5356), HDAC2 (5113) (Cell-Signaling, EuroClone), and RNA POL II (R1530; Sigma). Normal rabbit or mouse IgG was used as negative control. After immunoprecipitation, the DNA-histone complex was collected with 40 μl of salmon sperm DNA/protein A or G-agarose beads (16–157, 16–201; Millipore). After rotating for 2 h at 4°C on a spinning wheel, the beads were washed once with each of the following buffers in the order shown: high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris HCl, pH 8.1, 500 mM NaCl); low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris HCl, pH 8.1, 150 mm NaCl); LiCl buffer (0.25 M LiCl, 1% NP40, 1% deoxycholate, 1 mm EDTA, 10 mm Tris HCl, pH 8.1). They were then washed twice at room temperature with TE buffer (10 mm Tris, pH 8.1, and 1 mm EDTA). The precipitated fragments were eluted with a buffer containing 1% SDS and 0.1 M NaHCO3. DNA was analyzed by qRT-PCR using Fast SYBR Green Master Mix (cod 4385610; Applied Biosystems). In particular, DNA sequences/samples were heated for 2 min at 50°C and denatured for 10 min at 95°C. Amplification and quantification, achieved after 35 cycles at 95°C for 30 s, were followed by single fluorescence measurements for 1 min at 60°C. The binding activity was graphically represented as the percentage of the total input of chromatin, and the results were analyzed using a previously described formula (Renthal et al., 2007). The following oligonucleotides were used for the amplification of immunoprecipitated DNA: ncx1-Br (I) forward 5′-CCGCTGGGGAAACCCCTGCC-3′ and reverse 5′-GCGCTGCAACTTTTCTTTTGAACG-3′, ncx1-Br (II) forward 5′-GGGTGCAGAAGAGAGCGCTGGC-3′ and reverse 5′- GCACAAAGCGCGGCGGCCCG-3′ and Neurotrophin-3 (NT3) (Ishimaru et al., 2007). For each amplification, melting curves and gel electrophoresis of the PCR product were used to verify their identities. Samples were amplified simultaneously in triplicate in one assay run. Re-ChIP experiments were performed to detect the simultaneous presence of Sp1 with HIF-1 and p300 or of Sp3 with REST, HDAC1, and HDAC2. Beads from the first ChIP analysis with anti-Sp1 and anti-Sp3 were incubated with an equal volume of 10 nm dithiothreitol at 37°C for 30 min and centrifuged at 12,000 rpm for 1 min to elute DNA-bound proteins. Elution was repeated twice. The final elute was diluted 1:10 in lysis buffer containing a protease inhibitor mixture and reimmunoprecipitated with anti-REST, anti-HIF-1, anti-HDAC1, anti-HDAC2, anti-p300 antibodies or IgG. ChIP and input DNA were analyzed with PCR: denaturation for 10 min at 95°C, 35 cycles of 30 s at 95°C; for 1 min at 60°C; for 1 min at 72°C. PCR products were then electrophoresed on a 1.5% agarose gel and stained with ethidium bromide. For ChIP and Re-ChIP experiments, we used ncx1-Br (II) primers because the amplified sequence of ncx1-Br contains the binding sites for REST (−18/1) (Formisano et al., 2013), HIF-1 (−331/−327 and −164/−160) (Valsecchi et al., 2011), Sp1, and Sp3.
Transient-transfection ChIP assay
Cell culture and ChIP from SHSY-5Y cells was performed as described above. At 48 h after transfection, the cells were harvested by cross-linking with 1% formaldehyde, and ChIP was performed with antibodies for Sp1, Sp3, and IgG as negative control (data not shown). The input and the precipitated DNA was amplified using ncx1-Br forward (II) 5′-GGGTGCAGAAGAGAGCGCTGGC-3′ and a LucNrev primer 5′- CCTTATGCAGTTGCTCTCC −3.
Electrophoretic mobility shift assay (EMSA)
EMSA experiments were performed as previously described (Valsecchi et al., 2011). In particular, 5 μg of nuclear extracts was incubated with double-stranded DNA oligonucleotides (PRIMM) labeled with cy5 at the 5′ termini of both strands. Sense strand sequences were as follows: Cy5-Sp1 consensus sequence: 5′-ATTCGATCGGGGCGGGGCGAGC-3′ (Ko et al., 1998; Ryu et al., 2003); ncx1-Sp1B: 5′-AGGAGGCGGCGGGCTGGGCT-3′ and ncx1-Sp1Bmut: 5′-AGGAGGCTTCGGGCTGGGCT-3′; ncx1-Sp1C:5′-GGGCGAGCGGGCGGGGTGCG′-3′ and ncx1-Sp1Cmut: 5′-GGGCGAGCGTTCGGGGTGCG-3′; ncx1-Sp1D:5′-GGGAGGGAGGGGAGGGAGCG′-3′ and ncx1-Sp1Dmut: 5′-GGGAGGGATTGGAGGGAGCG-3′; ncx1-Sp1E:5′-GCCGCCCGGCCCCGGCCCTC-3′ and ncx1-Sp1Emut: 5′-GCCGCCCGGCCCCTTCCCTC-3′. Each italicized base represents the mutated bases. For supershift experiments, 3 μg of antibody against Sp1, Sp3, or Sp4 (Santa Cruz Biotechnology) was incubated on ice for 30 min before the addition of the probe. The reaction was run on a 5% nondenaturing acrylamide gel at 200 V at 4°C in Tris borate-EDTA (30 mm Tris, 30 mm boric acid, 0.6 mm EDTA). The gels were scanned at 600 V on a Typhoon 9400 imager (GE Healthcare), using a green laser (633 nm) for excitation and a 670BP30 emission filter.
Quantification of neuronal injury
Neuronal injury was assessed by measurement of LDH efflux into the medium after OGD/RX or PC+OGD/RX. In neuronal cultures, LDH activity is correlated with the number of damaged cells in the medium (Koh and Choi, 1987). Cytosolic levels of LDH in the extracellular medium were measured with the LDH Cytotoxicity Kit (1000882 Cayman, DBA). Briefly, after induction of OGD/RX or PC+OGD/RX, the medium was removed and sampled for LDH content by measuring absorbance at 490 nm using a spectrophotometer BioPhotometer (Eppendorf). Cell lysate, prepared 1% Triton X-100, was used as a positive control and its value was considered 100%.
In vivo studies
Experimental groups.
Male Sprague Dawley rats (Charles River) weighing 250–300 g were housed under diurnal lighting conditions (12 h darkness/light). Experiments were performed according to the international guidelines for care and use of experimental animals of the European Community Council directive (86/609/EEC). All experiments were approved by the Institutional Animal Care and Use Committee of “Federico II” University of Naples, IT.
Transient focal ischemia and ischemic preconditioning.
Transient focal ischemia was induced by suture occlusion of middle cerebral artery (MCA) in male rats with 1.5% sevoflurane, 70% N2O, and 28.5% O2 (Valsecchi et al., 2011) Achievement of ischemia was confirmed by monitoring regional cerebral blood flow through laser Doppler (PF5001; Perimed). Animals not showing cerebral blood flow reduction of at least 70%, as well as those dying after ischemia induction, were excluded from the study. Rats were divided into four experimental groups: (1) sham-operated rats (CTL); (2) preconditioned rats (PC); (3) ischemic rats, subjected to tMCAO; and (4) preconditioned ischemic rats (PC+tMCAO). The sham-operated animals underwent the same experimental conditions, except that the filament was not introduced; in the ischemic group, the MCA was occluded for 100 min; in the preconditioned ischemic group, rats were subjected to 30 min of tMCAO 72 h before 100 min of tMCAO. The rat siRNAs used in ischemic and preconditioned ischemic rats were intracerebroventricularly administered (1 μl) at concentrations of 20 μm for REST (Formisano et al., 2013), 3 μm for HIF-1 (Valsecchi et al., 2011), and 10 μm for Sp1, Sp3, HDAC1, HDAC2, and p300 (the catalog numbers are reported in Materials). For tMCAO and PC+tMCAO experiments, intracerebroventricular injections were performed three times (i.e., 18 and 6 h before and 6 h after ischemia induction) and 24, 18, and 6 h before ischemic preconditioning induction. All animals were killed 24 h after 100 min tMCAO. Rectal temperature was maintained at 37 ± 0.5°C with a thermostatically controlled heating pad, and a catheter was inserted into the femoral artery to measure arterial blood gases before and after ischemia (Rapid Laboratory 860, Chiron Diagnostic). All surgical procedures were performed under an operating stereomicroscope.
Statistical analysis
The data were evaluated as mean ± SEM. Statistically significant differences among means were determined by ANOVA followed by Student-Newman-Keuls test. The threshold for statistical significance data was set at p < 0.05.
Results
Sp1 and Sp3 bind to a specific region of ncx1-Br promoter sequence
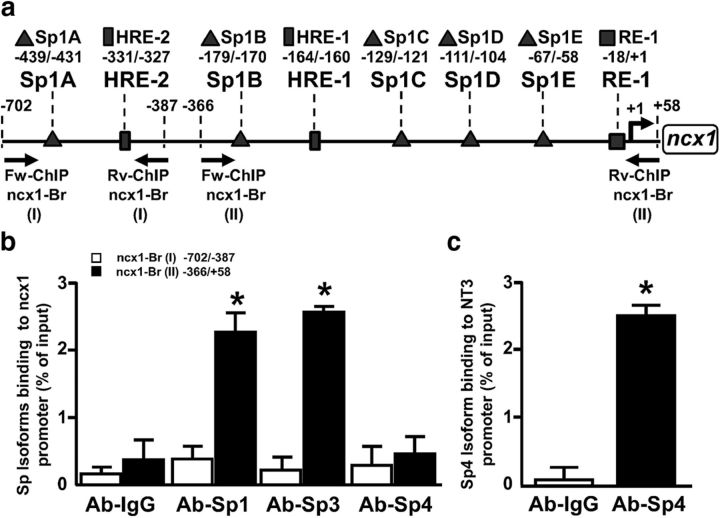
Because Sp1 and Sp3 both recognize the GC-rich sequences known as GC boxes (Isomura et al., 2005), we searched for Sp1 sequences at the level of the ncx1-Br promoter (GenBank accession no. U95138) and subjected the ncx1-Br sequence to a computational prediction of transcriptional factors using the database TF SEARCH, version 1.3 (Cassimere et al., 2009). As shown in Fig. 1a, five binding sites, named Sp1A–E, were identified within the ncx1-Br sequence at −439/−431, −179/−170, −129/−121, −111/−104, and −67/−58 from the transcriptional start site. All these sequences were characterized by the TF search threshold score, a parameter of sequence identity, >85.0. To prove a direct association of Sp transcription factors with the ncx1-Br promoter in the nuclear environment, we performed ChIP assays on extracts from cortical neurons that basally express Sp1, Sp3 (Ryu et al., 2003), and Sp4 (Ishimaru et al., 2007). To this aim, we divided ncx1-Br into two different fragments, termed ncx1-Br (I) (−702/−387) and ncx1-Br (II) (−366/58), and experiments were performed using specific primers recognizing these two parts of the ncx1-Br promoter. As shown in Fig. 1b, when chromatin was precipitated with Sp1, Sp3, and Sp4 antibodies, ncx1-Br (II), but not ncx1-Br (I), was amplified with Sp1 and Sp3 antibodies, compared with control IgG. Importantly, no signal was detected with the Sp4 antibody in the ncx1-Br (I) and ncx1-Br (II) fragments (Fig. 1b),whereas it was able to bind its known target gene neurotrophin-3 (NT3) (Ishimaru et al., 2007) in cortical neurons (Fig. 1c). This result suggests that Sp4 does not bind to the ncx1-Br promoter and that only the ncx1-Br (II) sequence is important for Sp1 and Sp3 binding in cortical neurons.
Figure 1.
Transcription factors Sp1 and Sp3, but not Sp4, bind to ncx1 brain promoter sequence from −366 to 58. a, Map of the rat ncx1 gene indicating location of Sp1, HIF-1 (HRE), REST (RE-1) sequences contained in distal [ncx1-Br (I)] and proximal [ncx1-Br (II)] regions of ncx1 brain promoter and of PCR primers used to detect the presence of specific DNA sequences in ChIP complexes. Triangles represent the 5 Sp1 identified putative motifs in ncx1-Br, named Sp1A–E. Rectangles and square represent the 2 HRE sites and RE-1 site, respectively. b, ChIP analysis of the ncx1-Br (I) and ncx1-Br B (II) regions performed with anti-Sp1, anti-Sp3, and anti-Sp4. Anti-IgG was used as negative control. The binding activity of Sp1, Sp3, and Sp4 is graphically represented as the percentage of total input of chromatin DNA. c, ChIP analysis of the NT3 promoter region performed with anti-Sp4. Anti-IgG was used as negative control. The binding activity of Sp4 is graphically represented as the percentage of total input of chromatin DNA. *p < 0.05 versus IgG. Each column represents the mean ± SEM (n = 3).
Sp1 and Sp3 have an opposite effect on ncx1 gene promoter activity, mRNA, and protein expression
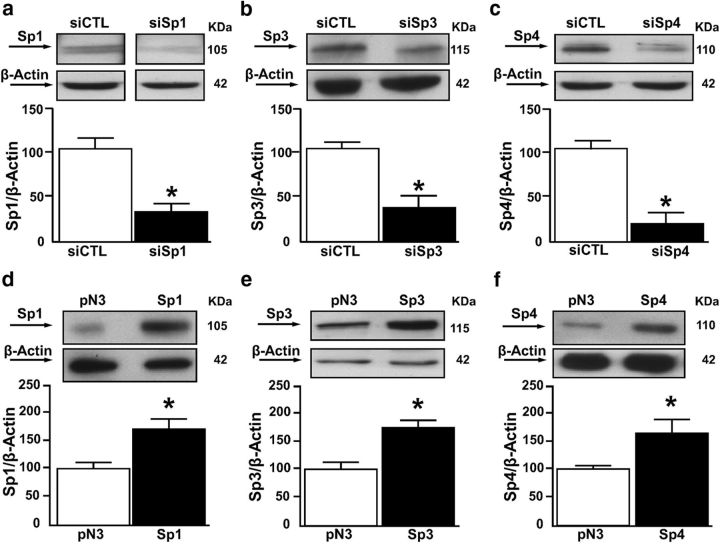
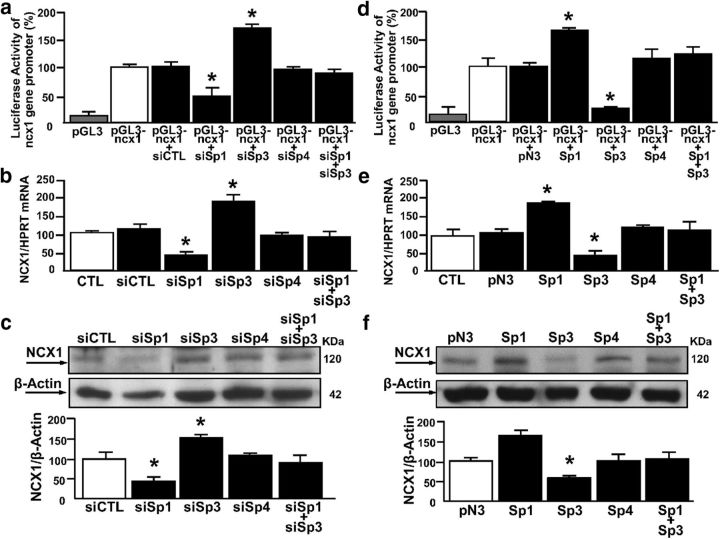
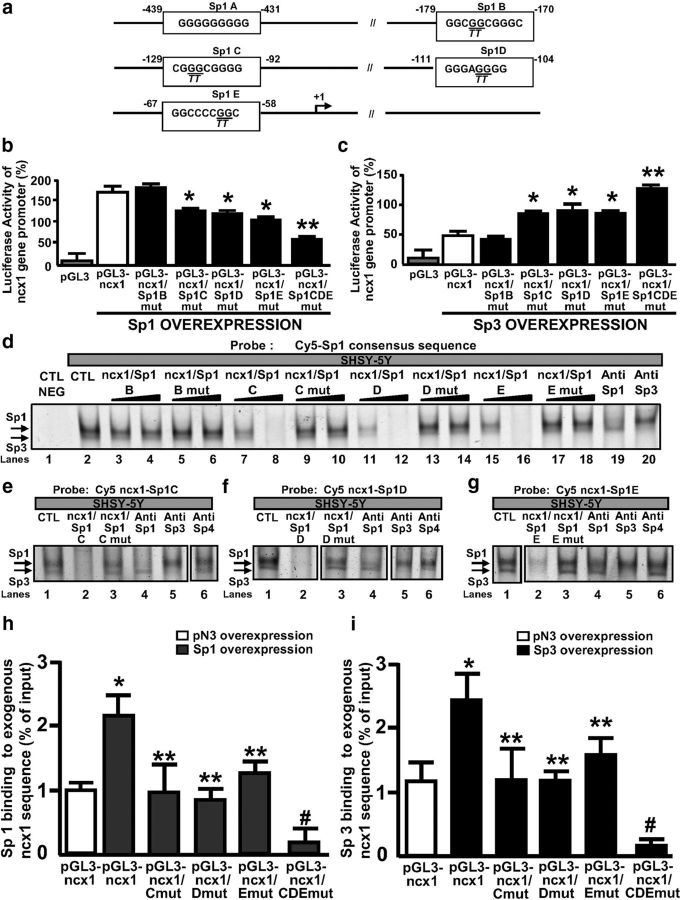
To evaluate the effect of Sp transcription factors on the ncx1 gene promoter activity (evaluated by luciferase assay), as well as mRNA and protein expression levels, neurons were silenced by specific siRNAs, namely, siSp1, siSp3, and siSp4. These were able to decrease Sp1, Sp3, and Sp4 protein expression by 68%, 63%, and 70%, respectively (Fig. 2a–c). In particular, for luciferase experiments, we used a pGL3 construct containing the 5′-flanking region upstream of the brain ncx1 transcriptional start site (+1), named short ncx1 promoter (pGL3-ncx1), already used in a previous study (Valsecchi et al., 2011). This construct contained the native Sp1 consensus sequences from B to E, which are present in the sequence of ncx1-Br (II) fragment. As shown in Figure 3a–c, Sp1 silencing reduced ncx1 gene promoter activity, mRNA levels (24 h after the treatment), and protein expression (48 h after treatment) by 45%, 48%, and 37%, respectively. By contrast, Sp3 silencing increased ncx1 gene promoter activity, mRNA levels (24 h after treatment), and protein expression (48 h after the treatment) by 68%, 65%, and 53%, respectively. The concomitant silencing of Sp1 and Sp3 or the knockdown of Sp4 did not affect the ncx1 gene promoter, mRNA levels, or protein expression, compared with cells transfected with siCTL. To further confirm the role of Sp transcription factors in modulating NCX1 expression, cortical neurons were transfected with constructs overexpressing Sp1, Sp3, and Sp4 (Fig. 2d–f). Neurons overexpressing Sp1 showed an increase in ncx1 gene promoter activity, mRNA levels (24 h after the treatment), and protein expression (48 h after the treatment) by 64%, 77%, and 61%, respectively. By contrast, Sp3 overexpression decreased ncx1 gene promoter activity, mRNA levels (24 h after the treatment), and protein expression (48 h after the treatment) by 76%, 58%, and 42%, respectively. The concomitant overexpression of Sp1 and Sp3 or Sp4 transfection alone did not affect the ncx1 gene promoter, mRNA levels, or protein expression, compared with control (Fig. 3d–f). To identify the functional Sp1 sequences within the ncx1 promoter, the Sp1 binding sites B-E were mutated in the pGL3-ncx1 construct by site-directed mutagenesis. Thus, four different luciferase reporter constructs (pGL3-ncx1/Sp1 B mut, pGL3-ncx1/Sp1 C mut, pGL3-ncx1/Sp1 D mut, and pGL3-ncx1/Sp1 E mut; Fig. 4a) were generated and cotransfected into cortical neurons with an expression plasmid of Sp1 or Sp3 (Fig. 4b,c). Interestingly, the cells transfected with the three constructs pGL3-ncx1/Sp1C–E mut significantly reduced the increase of luciferase activity induced by Sp1 overexpression by 30% (Fig. 4b). By contrast, transfection of the three constructs in cells overexpressing Sp3 significantly reverted by 35% Sp3-decreased ncx1 luciferase activity (Fig. 4c). Under both experimental conditions, mutation of Sp1 sequence B (pGL3-ncx1/Sp1 B mut) had no effect on ncx1 transcriptional activity (Fig. 4b,c). Interestingly, in neurons overexpressing Sp1 or Sp3, transfection of pGL3-ncx1/Sp1 CDE construct containing mutations of all three Sp1 sites on ncx1-Br sequence, further reduced ncx1 promoter-luciferase activity by 65% or increased it by 85%, respectively, compared with pGL3-ncx1 alone (Fig. 4b,c). Because the ncx1-Br (II) fragment contains four (Sp1B–E) of the five putative Sp1 binding elements identified, EMSA experiments were performed to determine which of these Sp1 sites were involved in the binding with Sp1 and Sp3 proteins. These experiments were conducted in SH-SY5Y cell lines that have been already used to study the binding of Sp transcription factors on specific gene promoter sequence (Formisano et al., 2015a). Therefore, nuclear extracts from SH-SY5Y were incubated with a 5′-Cy5 fluorescent double-stranded oligonucleotide (Cy5-Sp1 consensus sequence), which specifically binds Sp1 and Sp3 as previously reported (Ko et al., 1998; Ryu et al., 2003). As shown in Figure 4d, lane 2 protein binding to the Cy5-Sp1 consensus sequence oligonucleotide produced two major closely migrating complexes (as indicated by the arrowheads). Interestingly, only the unlabeled oligonucleotides of ncx1/Sp1C–E sites competed for binding of both protein complexes (Fig. 4d, lanes 7, 8, 11, 12, 15, and 16); however, this competition did not occur in the presence of excess mutated probes (Fig. 4d, lanes 9, 10, 13, 14, 17, and 18). Furthermore, as shown in Figure 4d (lanes 3–6), the excess of ncx1/Sp1 B probes wild-type partially competed with Cy5-Sp1 consensus sequence, whereas the mutated were not able to modify the intensity of the doublet bands protein/DNA complexes. Moreover, the addition of antibodies against Sp1 and Sp3 was able to decrease the appearance of the upper and lower band of the formed complexes, respectively (Fig. 4d, lanes 19 and 20). Furthermore, EMSA experiments performed with fluorescent probes containing ncx1/Sp1C or ncx1/Sp1D or ncx1/Sp1E sequences showed the formation of specific DNA-protein complexes (Fig. 4e–g, lanes 1), that completely diminished in the presence of an excess of unlabeled probes (Fig. 4e–g, lanes 2), but not in the presence of an excess of mutated probes (Fig. 4e–g, lanes 3). This suggests that mutations of ncx1-Sp1C–E sites at these positions alter binding of proteins to the ncx1-Br promoter sequence. Interestingly, incubation of the nuclear extract with an anti-Sp1 and anti-Sp3 decreased the intensity of the upper and lower band for ncx1-Sp1 C (Fig. 4e, lanes 4 and 5), ncx1-Sp1D (Fig. 4f, lanes 4 and 5), and ncx1-Sp1E sites (Fig. 4g, lanes 4 and 5), indicating that the binding of Sp1 and Sp3, detected by the upper and lower band, was inhibited by the antibodies. Notably, experiments with antibody against Sp4 were not able to modify both bands of the complex formed between nuclear extracts and the probes (Fig. 4c–e, lane 7). To further confirm that Sp1 and Sp3 bind the ncx1-Sp1C–E sequences as suggested by EMSA, SHSY-5Y cells were cotransfected with plasmid containing the wild-type ncx1-Br promoter sequence (pGL3-ncx1) or the luciferase reporter constructs: pGL3-ncx1/Sp1 C mut, pGL3-ncx1/Sp1 D mut, pGL3-ncx1/Sp1 E mut, and pGL3-ncx1/Sp1 CDE mut, together with the Sp1- and Sp3-expressing plasmids to overexpress Sp1 and Sp3, respectively. Sp1 and Sp3 constructs were able to increase Sp1 and Sp3 protein expression by almost 50%, respectively (data not shown). Then, transfected cells were subjected to ChIP assay with Sp1 and Sp3 antibodies. As shown in Figure 4h, i, exogenous wild-type chromatin was immunoprecipitated by the Sp1 and Sp3 and amplified using as forward primer an oligonucleotide recognized the ncx1-Br sequence [ncx1-Br forward (II)] and as reverse primer an oligonucleotide recognized specifically the luciferase gene of pGL3 construct (LucNrev). In particular, overexpression of Sp1 or Sp3 increased their binding on ncx1 exogenous promoter sequence, compared with cells transfected with the empty vector pN3. By contrast, PCR product containing the mutated ncx1-Sp1C–E sequences was reduced, whereas transfection of a construct containing ncx1-Sp1C–E sites jointly mutated completely abolished Sp1 and Sp3 binding on ncx1 exogenous sequence, compared with cells overexpressing Sp1 or Sp3 and transfected with pGL3-ncx1 construct. Thus, Sp1 and Sp3 are efficiently recruited to the ncx1-br promoter sequence by binding ncx1-Sp1C–E sites. Together, these results demonstrate that Sp1 and Sp3 interact with the ncx1-Br promoter within three specific sequences, thereby increasing or decreasing its expression, respectively.
Figure 2.
Cortical neurons transfected with siRNAs or constructs for Sp1, Sp3, and Sp4. a–c, Representative WB and quantification of Sp1, Sp3, and Sp4 protein expression in neurons after treatment with siCTL, siSp1, siSp3, and siSp4. d–f, Representative WB and quantification of Sp1, Sp3, and Sp4 protein expression in neurons after transfection with empty vector (pN3), and after Sp1, Sp3, and Sp4 overexpression. Each column represents the mean ± SEM (n = 3). *p < 0.05 versus respective siCTL or pN3.
Figure 3.
Sp1 is a transcriptional activator, whereas Sp3 is a transcriptional repressor of ncx1 in cortical neurons. a–c, Luciferase assay of ncx1-Br, qRT-PCR, and representative WB with quantification of NCX1 under control conditions (CTL) and after treatment with siCTL, siSp1, siSp3, siSp4, and in combination with siSp1 and siSp3 (n = 4). Each column represents the mean ± SEM. *p < 0.05 versus pGL3 ncx1, siCTL. d–f, Luciferase assay of ncx1-Br, qRT-PCR, and representative WB with quantification of NCX1 under control conditions (CTL) and after transfection with pN3 or overexpression of Sp1, Sp3, Sp4, and after combination of Sp1 and Sp3 (n = 3). Each column represents the mean ± SEM. *p < 0.05 versus pGL3 ncx1, pN3.
Figure 4.
Effect of Sp1 and Sp3 on the transcriptional activity of ncx1-Br after site-directed mutagenesis of the putative Sp1 sites and analysis of their binding on ncx1-Br sequence. a, The core consensus sequences of the five putative Sp1A–E sites are boxed, and the locations relative to the reported transcription start site are separately numbered. Mutated nucleotides of the particular sequences are underlined, and the replaced nucleotides are shown in italics below the wild-type sequences. The bent arrows indicate the reported transcription start site. b, c, Cortical neurons transiently overexpressing Sp1 or Sp3 were cotransfected with pGL3-ncx1 or pGL3-ncx1/Sp1Bmut, pGL3-ncx1/Sp1Cmut, pGL3-ncx1/Sp1Dmut, pGL3-ncx1/Sp1Emut, and pGL3-ncx1/Sp1CDEmut constructs. Twenty-four hours after transfection, neurons were lysed in 1× passive lysis buffer. Lysates were analyzed for luciferase activity. Luciferase activity was expressed as firefly-to-renilla ratio. Each column represents the mean ± SEM (n = 4). *p < 0.05 vs pGL3-ncx1. **p < 0.05 versus all. d, EMSA was performed incubating Cy5′-Sp1 consensus sequence probe without or with nuclear extracts from SH-SY5Y cells (CTL NEG and CTL, respectively, lanes 1 and 2). Competition experiments were performed with increasing molar excess (20- and 100-fold) of the unlabeled wild-type ncx1/Sp1B–E (ncx1/Sp1B, lanes 3 and 4; ncx1/Sp1C, lanes 7 and 8; ncx1/Sp1D, lanes 11 and 12; ncx1/Sp1E, lanes 15 and 16) and the mutant ncx1/Sp1B–E probes (ncx1/Sp1B, lanes 5 and 6; ncx1/Sp1C, lanes 9 and 10; ncx1/Sp1D, lanes 13 and 14; ncx1/Sp1E, lanes 17 and 18). Antibodies specific for Sp1 and Sp3 were added to the EMSA reaction as indicated (lanes 19 and 20). e–g, Nuclear extracts from SH-SY5Y cells were incubated with the Cy5′-tagged ncx1-Sp1C–E (CTL, lanes 1). Competition experiments were performed with increasing molar excess (100-fold) of the unlabeled wild-type ncx1/Sp1C–E (ncx1/Sp1C–E, lanes 2) and the mutant ncx1/Sp1C–E (ncx1/Sp1C–E, lanes 3). Nuclear extracts were incubated in the presence of anti-Sp1, anti-Sp3, and anti-Sp4 (lanes 4–6). h, i, Chromatin was prepared from SHSY-5Y cells transiently transfected with Sp1, Sp3, and pN3 (empty vector) expressing plasmids and with the pGL3-ncx1 or pGL3-ncx1/Sp1Cmut, pGL3-ncx1/Sp1Dmut, pGL3-ncx1/Sp1Emut, and pGL3-ncx1/Sp1CDEmut constructs. Chromatin from transfected cells was immunoprecipitated with Sp1 and Sp3 antibodies. Exogenous DNA containing either the ncx1-Br fragment (−340/151, pGL3-ncx1) or the mutated Sp1C–E sequence alone or in combination (pGL3-ncx1/Sp1Cmut, pGL3-ncx1/Sp1Dmut, pGL3-ncx1/Sp1Emut, and pGL3-ncx1/Sp1CDEmut constructs) was amplified using ncx1-Br forward II promoter-specific forward primer and a luciferase gene-specific reverse primer (LucNrev primer). The binding activity of Sp1 is graphically represented as the percentage of total input of chromatin DNA. *p < 0.05 versus cells cotransfected with pN3 and pGL3-ncx1. **p < 0.05 versus cells cotransfected with constructs overexpressing Sp1 or Sp3 and pGL3-ncx1. #p < 0.05 versus all. Each column represents the mean ± SEM (n = 3).
REST/Sp3 and HIF-1/Sp1 complexes colocalize on ncx1 brain promoter after tMCAO and PC+tMCAO, respectively
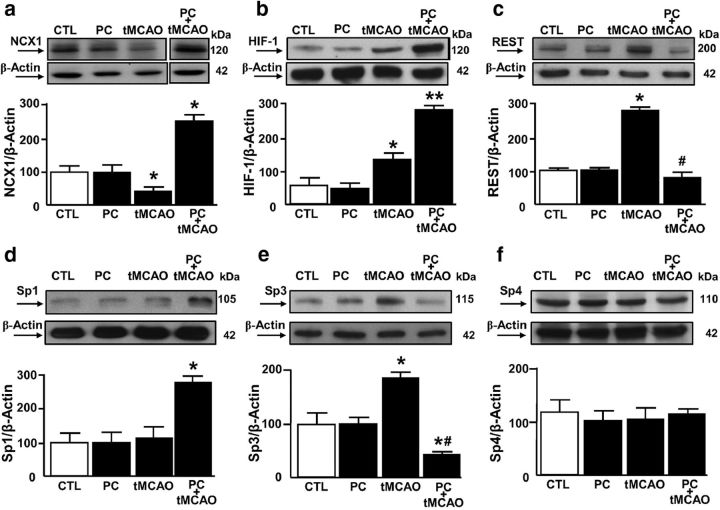
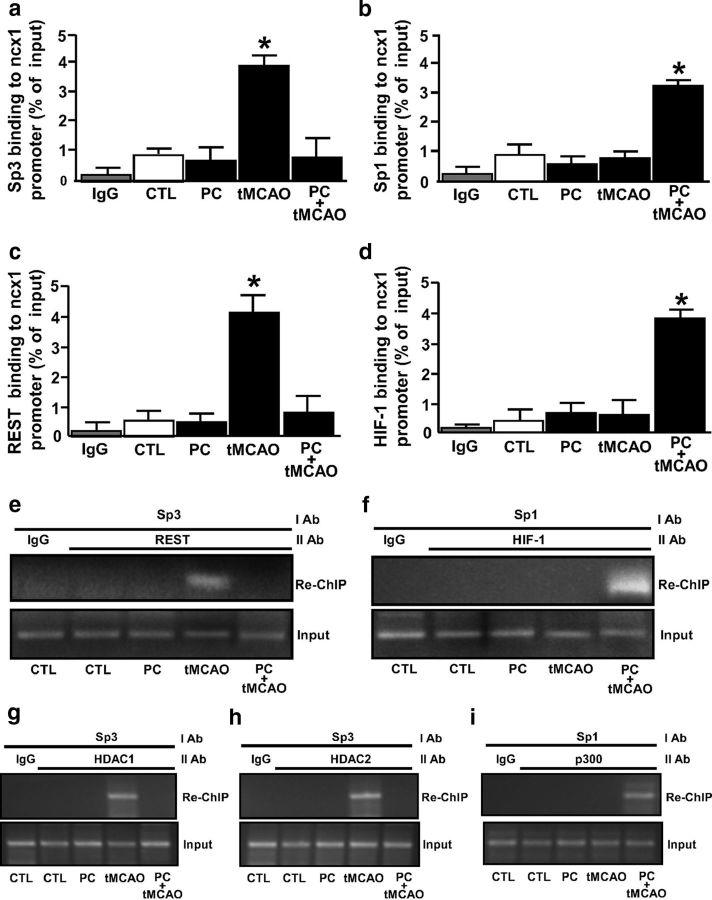
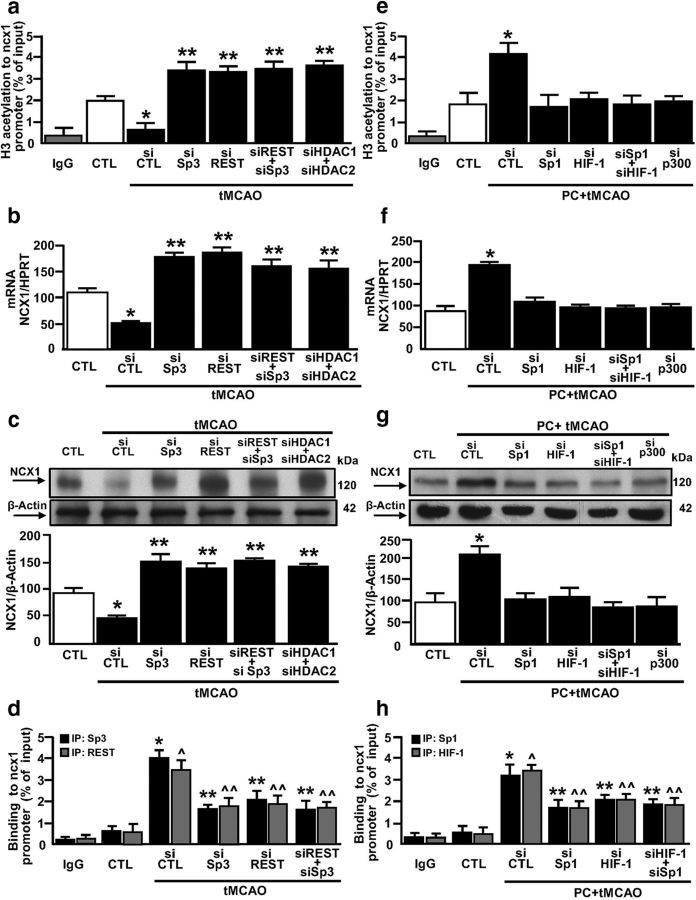
The three Sp1 sequences we identified on the ncx1-Br promoter are located near the binding sites of both the ncx1 transcriptional repressor REST and the ncx1 transcriptional activator HIF-1. Whereas the former is responsible for NCX1 reduction in cerebral ischemia (tMCAO) (Formisano et al., 2013), the latter is responsible for NCX1 increase after ischemic preconditioning plus tMCAO (PC+tMCAO) (Valsecchi et al., 2011). The temporoparietal brain cortex has been chosen as a region of interest because after tMCAO ischemic damage it involves both striatum and cortex, which represent the brain areas irrigated by MCA. In particular, the temporoparietal cortex is a unique region because after tMCAO it includes the ischemic core, the area irreversibly damaged, and the ischemic penumbra, the area that can be still rescued from cell death (Barone et al., 1998; Pignataro et al., 2012) and intact tissue. In addition, the same cortical area is spared by the ischemic damage when a preconditioning stimulus is applied. Therefore, the processes triggered by preconditioning and able to modify the extent of ischemic brain lesion are usually evaluated in this area. Interestingly, Western blot analysis, performed in the ipsilesional–temporoparietal cortex of rats at 24 h after PC alone, tMCAO, or PC+tMCAO revealed that PC alone did not significantly modify the expression of NCX1, REST, HIF-1, Sp1, or Sp3 compared with CTL (Fig. 5a–e). On the contrary, tMCAO reduced NCX1 by 44% while simultaneously increasing HIF-1 by 75%, REST by 180%, and Sp3 by 100%. However, it did not affect Sp1 (Fig. 5d). Finally, PC+tMCAO remarkably increased NCX1 by 140%, HIF-1 by 450%, and Sp1 by 180% compared with CTL, whereas it completely blocked tMCAO-induced REST and Sp3 increase, compared with tMCAO values (Fig. 5a–e). In all the above experimental conditions, Sp4 protein expression was not affected (Fig. 5f). To evaluate the possible interaction between REST and Sp3 and between HIF-1 and Sp1, their binding activity to the ncx1-Br sequence was measured in rats subjected to tMCAO and PC+tMCAO. Interestingly, in rats subjected to cerebral ischemia, REST and Sp3 binding, but not HIF-1 and Sp1, increased, compared with CTL (Fig. 6a–d). On the contrary, whereas HIF-1 and Sp1 binding to the ncx1-Br sequence increased after PC+tMCAO, REST and Sp3 did not (Fig. 6a–d). We further conducted re-ChIP assay to test whether the transcriptional repressors REST and Sp3 or the transcriptional activators HIF-1 and Sp1 all colocalized on the ncx1 Br sequence during tMCAO and PC+tMCAO, respectively. Re-ChIP assay revealed that Sp3, REST, HDAC1, and HDAC2 (Fig. 6e,g,h, lanes 4), and Sp1, HIF-1, and p300 (Fig. 6f,i, lanes 5) were bound together on the ncx1 promoter after tMCAO and after PC+tMCAO, respectively. These results suggest that Sp3 REST, HDAC1, and HDAC2, and Sp1, HIF-1, and p300 form two functional complexes on the ncx1-Br sequence during tMCAO and PC+tMCAO, respectively.
Figure 5.
Effect of tMCAO and PC+tMCAO on NCX1, HIF-1, REST, Sp1, Sp3, and Sp4 in the rat ipsilateral–temporoparietal cortex. a–f, Representative WB with quantification of NCX1, HIF-1, REST, Sp1, Sp3, and Sp4 protein expression in: (1) control group (CTL), (2) PC, (3) tMCAO, and (4) PC+tMCAO. Each column represents the mean ± SEM (n = 5 animals for each column). *p < 0.05 versus CTL. **p < 0.05 versus all. #p < 0.05 versus tMCAO.
Figure 6.
tMCAO and PC+tMCAO rat models promote REST/Sp3/HDAC1/HDAC2 and HIF-1/Sp1/p300 colocalization on ncx1-Br in the ipsilateral–temporoparietal cortex, respectively. a–d, ChIP analysis of ncx1-Br in the ipsilateral–temporoparietal cortex in (1) CTL, (2) PC, (3) tMCAO, and (4) PC+tMCAO. The binding activity of HIF-1, REST, Sp1, and Sp3 is graphically represented as the percentage of total input of chromatin DNA. Anti-IgG was used as negative control. Each column represents the mean ± SEM (n = 6 animals for each column). *p < 0.05 versus CTL. e–i, Re-ChIP analysis of ncx1-Br in the ipsilateral–temporoparietal cortex in (1) CTL, (2) PC, (3) tMCAO, and (4) PC+tMCAO. Primary ChIP products for anti-Sp3 (I Ab) were subjected to re-ChIP with anti-REST, anti-HDAC1, and anti-HDAC2 (II Ab) (e, g, h) but for anti-Sp1 (I Ab) were subjected to re-ChIP with anti-p300 (II Ab) (f, i). Anti-IgG was used as negative control. The input DNA lane represents 5% of the precleared chromatin used in each ChIP reaction. The figure is representative of two independent experiments.
Epigenetic remodeling of the ncx1-Br promoter by HDAC1 and HDAC2 after tMCAO and by p300 after PC+tMCAO
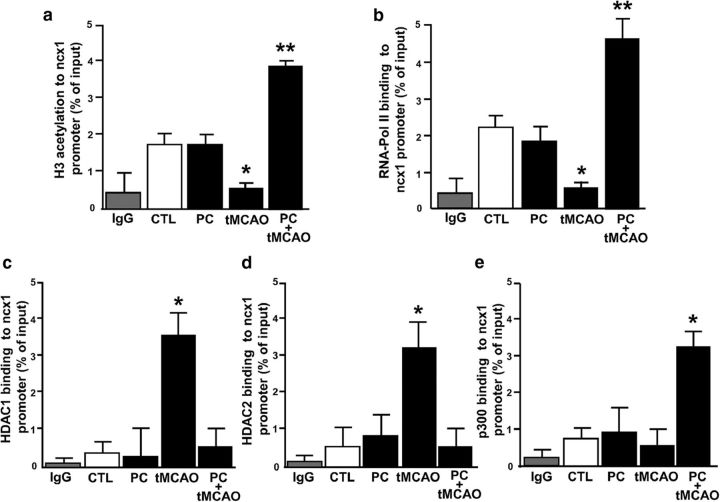
The transcription factors REST, HIF-1, Sp1, and Sp3 regulate their target genes by epigenetic modifications, such as acetylation (Won et al., 2002; Formisano et al., 2007; Azahri et al., 2012; Noh et al., 2012; Pawlus and Hu, 2013). Thus, by performing ChIP assay, we evaluated the following: (1) the modification of the acetylated histone protein H3; (2) the binding of HDAC1, HDAC2, and histone acetyltransferase p300; and (3) the binding of RNA polymerase II (RNA-Pol II) on ncx1-Br promoter sequence in the ipsilesional–temporoparietal cortex of rats subjected to PC, tMCAO, and PC+tMCAO and compared it with CTL. PC alone did not significantly modify the amount of the ncx1 promoter associated with acetylated H3, RNA-Pol II, HDAC1, HDAC2, and p300 (Fig. 7a–e).
Figure 7.
Effect of tMCAO and PC+tMCAO rat models on H3 acetylation, RNA-Pol II, HDAC1, HDAC2, and p300 binding to ncx1-Br. a–e, ChIP analysis of ncx1-Br in the ipsilateral–temporoparietal cortex in (1) CTL, (2) PC, (3) tMCAO, and (4) PC+tMCAO. The binding activity of H3 acetyl, RNA-Pol II, HDAC1, HDAC2, and p300 is graphically represented as the percentage of total input of chromatin DNA. IgG was used as negative control. Each column represents the mean ± SEM (n = 6 animals for each column). *p < 0.05 versus CTL. **p < 0.05 versus all.
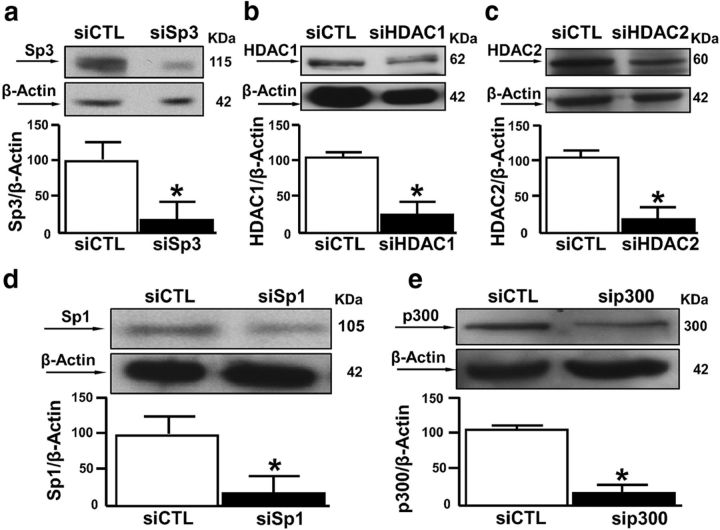
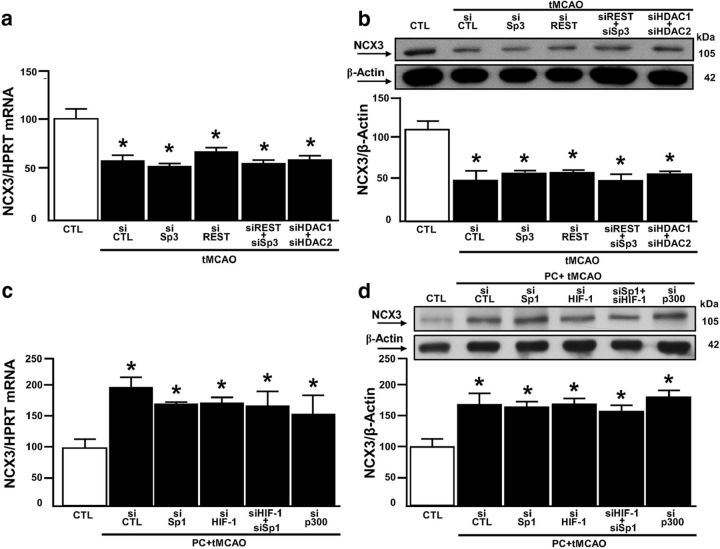
On the other hand, our results showed that tMCAO reduced the amount of the ncx1 promoter associated with acetylated H3 and RNA-Pol II (Fig. 7a,b). Concomitantly, the same experimental condition increased the binding activity of HDAC1 and HDAC2 on the ncx1 gene promoter (Fig. 7c,d), compared with CTL. After tMCAO, p300 binding on the ncx1-Br promoter was absent (Fig. 7e). Interestingly, HDAC1 and HDAC2 did not bind to the ncx1-Br promoter in animals subjected to PC+tMCAO (Fig. 7c,d). Contrarily, our results showed that PC+tMCAO increased not only the amount of ncx1-Br associated with acetylated H3, but also the binding activity of RNA-Pol II and p300 to the ncx1-Br promoter, compared with CTL (Fig. 7a,b,e). To better understand the role of REST, Sp3, HDAC1, and HDAC2 in tMCAO-induced NCX1 reduction, we evaluated the acetylation status of the ncx1-Br promoter, the ncx1 gene level, and protein expression, after intracerebroventricular injection of siSp3, siREST, alone or in combination, and siHDAC1 and siHDAC2 in combination. In particular, siSp3, siHDAC1, and siHDAC2 were able to reduce protein expression by 80%, 75%, and 81%, respectively, whereas expression for siREST was already published (Formisano et al., 2013) (Fig. 8a–c). As shown in Figure 9a–c, tMCAO-induced ncx1-Br deacetylation. In addition, the ncx1 gene and protein reduction were reverted when all the proteins were silenced, compared with CTL. Furthermore, tMCAO-induced Sp3 and REST binding to ncx1-Br sequence was reverted when Sp3 and REST were silenced alone or in combination, compared with siCTL (Fig. 9d). Interestingly, in the same experimental conditions, tMCAO-induced ncx3 gene and protein decrease was not reverted by siSp3, siREST, alone or in combination, and by double silencing for HDAC1 and HDAC2, compared with the CTL (Fig. 10a,b). Furthermore, we studied the role of HIF-1, Sp1, and p300 in the PC+tMCAO-induced NCX1 increase after intracerebroventricular injection of siSp1, siHIF-1, alone or in combination, and sip300. In particular, siSp1 and sip300 were able to reduce protein expression by 80% and 85% (Fig. 8d,e), respectively, whereas the decrease of protein expression obtained by siHIF-1 was already published (Valsecchi et al., 2011). Our results showed that hyperacetylation of histone H3 on the ncx1-Br promoter sequence, induced by PC+tMCAO, was reverted when we administered siSp1 (Fig. 9e), siHIF-1, alone or in combination, and sip300 (Fig. 9e), compared with the CTL (Fig. 9e). Likewise, the ncx1 gene and protein increase, induced by PC+tMCAO, was also reverted (Fig. 9f,g). In addition, PC+tMCAO-induced Sp1 and HIF-1 binding to ncx1-Br sequence was reverted when Sp1 and HIF-1 were silenced alone or in combination, compared with siCTL (Fig. 9h). Importantly, in the same experimental conditions, PC+tMCAO-induced ncx3 gene and protein increase was not reverted by silencing the above mentioned transcriptional factors, compared with the CTL (Fig. 10c,d).
Figure 8.
Sp1, Sp3, HDAC1, HDAC2, and p300 protein expression in rat brain after siRNA treatment. a–e, Representative Western blots of Sp3, HDAC1, HDAC2, Sp1, and P300 protein levels in the temporoparietal cortex of rats intracerebroventricularly treated with siSp3, siHDAC1, siHDAC2, siSp1, and siP300. Each column represents the mean ± SEM (n = 3). *p < 0.05 versus respective siCTL.
Figure 9.
NCX1 is epigenetically modulated by Sp3/REST/HDAC1/2 complex in tMCAO and by Sp1/HIF-1/p300 in PC+tMCAO. a, ChIP analysis with anti-H3 acetyl of ncx1-Br in CTL and in tMCAO after intracerebroventricular injection of siCTL, siSp3, siREST, siREST+siSp3, and siHDAC1+siHDAC2. IgG was used as negative control. Each column represents the mean ± SEM (n = 5 animals for each column). *p < 0.05 versus CTL. **p < 0.05 versus all. b, c, qRT-PCR and representative WB with quantification of NCX1 in CTL and in tMCAO after intracerebroventricular injection of siCTL, siSp3, siREST, siREST+siSp3, and siHDAC1+siHDAC2. Each column represents the mean ± SEM (n = 6 animals for each column). *p < 0.05 versus CTL. **p < 0.05 versus all. d, ChIP analysis with anti-Sp3 (black bars) and anti-REST (gray bars) of ncx1-Br in CTL and in tMCAO after intracerebroventricular injection of siCTL, siSp3, siREST, siREST+siSp3, and siHDAC1+siHDAC2. IgG was used as negative control. Each column represents the mean ± SEM (n = 6 animals for each column). *,^p < 0.05 versus CTL. *,^^p < 0.05 versus all. e, ChIP analysis with an anti-H3 acetyl of ncx1-Br in CTL and in PC+tMCAO after intracerebroventricular injection of siCTL, siSp1, siHIF-1, siHIF-1+siSp1, and sip300. IgG was used as negative control. Each column represents the mean ± SEM (n = 6 animals for each column). *p < 0.05 versus CTL. f, g, qRT-PCR and representative WB with quantification of NCX1 in CTL and PC+tMCAO after intracerebroventricular injection of siCTL, siSp1, siHIF-1, siHIF-1+siSp1, and sip300. Each column represents the mean ± SEM (n = 6 animals for each column). *p < 0.05 versus CTL. h, ChIP analysis with anti-Sp1 (black bars) and anti-HIF-1 (gray bars) of ncx1-Br in CTL and in PC+tMCAO after intracerebroventricular injection of siCTL, siSp1, siHIF-1, and siHIF-1+siSp1. IgG was used as negative control. Each column represents the mean ± SEM (n = 5 animals for each column). *,^p < 0.05 versus CTL. *,^^p < 0.05 versus all.
Figure 10.
NCX3 is not modulated by the Sp3/REST/HDAC1/2 complex in tMCAO and by Sp1/HIF-1/p300 in PC+tMCAO. a, b, qRT-PCR and representative WB with quantification of NCX3 in CTL and in tMCAO after intracerebroventricular injection of siCTL, siSp3, siREST, siREST+siSp3, and siHDAC1+siHDAC2. Each column represents the mean ± SEM (n = 5 animals for each column). *p < 0.05 versus CTL. **p < 0.05 versus all. c, d, qRT-PCR and representative WB with quantification of NCX3 in CTL and PC+tMCAO after intracerebroventricular injection of siCTL, siSp1, siHIF-1, siSp1+siHIF-1, and sip300. Each column represents the mean ± SEM (n = 5 animals for each column). *p < 0.05 versus CTL.
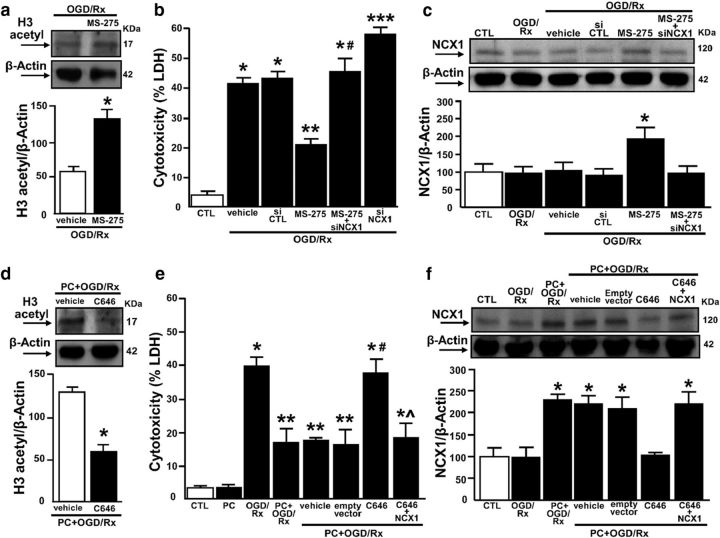
Class I HDAC inhibitor MS-275 reduces neuronal cell death by increasing NCX1 expression during OGD/RX
Considering that HDAC1 and HDAC2 belong to Class I histone deacetylases (Chuang et al., 2009), we evaluated the effect of the Class I HDAC inhibitor MS-275 (1 μm) in cortical neurons exposed to OGD/RX (Lanzillotta et al., 2013; Formisano et al., 2015b). First, we validated the specificity of MS-275 to inhibit Class I HDAC by measuring the levels of the acetylated forms of histone H3 (indicating the activity of Class I) (Simonini et al., 2006; Galmozzi et al., 2013). Results showed that MS-275 increased histone H3 acetylation (Fig. 11a). In addition, MS-275 exerted a remarkable neuroprotective effect by preventing cell death during OGD/RX, an effect that was though counteracted by siNCX1. Interestingly, siNCX1 alone exacerbated the neuronal death induced by OGD/RX (Fig. 11b).
Figure 11.
Effect of Class I HDAC inhibitor MS-275 and of HAT p300 inhibitor C646 on cell survival and NCX1 protein expression in cortical neurons exposed to OGD/RX or to PC+OGD/RX, respectively. a, Representative WB with quantification of H3 acetyl in neurons subjected to OGD/RX alone or with MS-275. Each column represents the mean ± SEM (n = 3). *p < 0.05 versus OGD/Rx. b, LDH assay in (1) control condition (CTL), (2) OGD/RX+vehicle, (3) OGD/RX+siCTL, (4) OGD/RX+MS-275 (1 μm), (5) OGD/RX+MS-275+siNCX1, and (6) OGD/RX+siNCX1. Each column represents the mean ± SEM (n = 5). *p < 0.05 versus CTL. **p < 0.05 versus OGD/Rx+vehicle and OGD/Rx+siCTL. #p < 0.05 versus OGD/Rx+MS-275. ***p < 0.05 versus all. c, NCX1 protein expression with quantification in (1) control condition (CTL), (2) OGD/RX, (3) OGD/RX+vehicle, (4) OGD/RX+siCTL, (5) OGD/RX+MS-275 (1 μm), and (6) OGD/RX+MS-275+siNCX1. Each column represents the mean ± SEM (n = 3). *p < 0.05 versus all. d, Representative WB with quantification of H3 acetyl in neurons subjected to PC+OGD/RX alone or with C646. Each column represents the mean ± SEM (n = 3). *p < 0.05 versus OGD/Rx. e, f, LDH assay and NCX1 protein expression with quantification in (1) CTL, (2) PC, (3) OGD/RX, (4) PC+OGD/RX, (5) PC+OGD/RX+vehicle, (6) PC+OGD/RX+empty vector, (7) PC+OGD/RX+C646, and (8) PC+OGD/RX+C646+NCX1. Each column represents the mean ± SEM (n = 3 for LDH assay and for WB). *p < 0.05 versus CTL. **p < 0.05 versus all. #p < 0.05 versus PC+OGD/RX alone or with vehicle and empty vector. ^p < 0.05 versus PC+OGD/RX+C646.
To identify the role of NCX1 in MS-275-induced neuroprotection, Western blot analysis was performed for NCX1 under the same experimental conditions. Furthermore, exposure of cortical neurons to OGD/RX+MS-275 induced an increase in NCX1 protein expression compared with the OGD/RX+vehicle (Fig. 11c). As expected, the MS-275-induced NCX1 increase was counteracted by siNCX1 (Fig. 11c).
Hystone acetyl transferase p300 inhibitor C646 reverts PC-induced neuroprotection by counteracting NCX1 increase
We investigated whether PC could exert a neuroprotective effect in cortical neurons exposed to OGD/RX by increasing p300 and NCX1 expression. The specificity of C646 to inhibit hystone acetyl transferase p300 was validated by measuring the levels of the acetylated form of histone H3 (Federman et al., 2013). Results showed that C646 decreased histone H3 acetylation (Fig. 11d). In particular, in neurons exposed to PC+OGD/RX, the p300 inhibitor C646 (20 μm) (Min et al., 2010), increased cell death compared with vehicle. Interestingly, NCX1 overexpression was able to counteract C646-induced neuronal death, compared with vector (Fig. 11e). In addition, C646 prevented the increase in NCX1 in cortical neurons exposed to PC+OGD/RX, an effect that was counteracted by NCX1 overexpression (Fig. 11f).
Discussion
This study has shown, for the first time, that NCX1 is epigenetically downregulated in brain ischemia by the REST/Sp3 complex and upregulated in brain ischemic preconditioning by the HIF-1/Sp1 complex. Particularly, our results suggest that these proteins form two functional complexes on the ncx1-Br sequence in brain ischemia and in brain ischemic preconditioning. We showed that REST/Sp3-induced NCX1 downregulation after tMCAO 24 h and HIF-1/Sp1-induced NCX1 upregulation after PC+tMCAO 24 h are associated with these transcriptional factors expression. Particularly, 24 h after tMCAO and 24 h after PC+tMCAO was the first time points in which Sp3 (data not shown) and REST (Formisano et al., 2013) or HIF-1 (Valsecchi et al., 2011) and Sp1 (data not shown), respectively, were increased. Indeed, intracerebroventricular injections of siREST and siSp3, alone or in combination, in tMCAO, and of siHIF-1 and siSp1, alone or in combination, in PC+tMCAO completely reverted the decreases and the increases, respectively, in NCX1 mRNA and protein levels. Previous studies have proposed that a crosstalk between Sp1, Sp3, and REST regulates the expression of some genes. For instance, hypoxia-induced expression of the regulation in development and DNA damage response (Redd1) gene is mediated by the coactivation of Sp1 and HIF-1 (Jin et al., 2007). Interestingly, in our study, we report, for the first time, that a crosstalk between Sp1, Sp3, and REST specifically regulates ncx1 expression. In particular, whereas Sp1 activates the expression of REST target genes (Plaisance et al., 2005), Sp3 interacts with REST by binding to the GC box, thereby repressing the expression of mu opioid receptor (Kim et al., 2006). Moreover, within the rat ncx1-Br, we discovered five Sp1 motifs (Sp1A–E) located at −439, −179, −129, −111, and −67 from the transcriptional start site, and indentified them as possible molecular determinants involved in Sp1 isoform-induced ncx1 regulation. Furthermore, we found that, among the Sp family of transcription factors, Sp1 and Sp3, but not Sp4, bind to ncx1-Br in a very selective manner, as revealed by ChIP analysis. These results are in accordance with other studies showing that not all the genes containing GC boxes in their promoter are regulated by all Sp1 transcription factor isoforms (Lerner et al., 2002) and that some genes are target of Sp1 and Sp3, but not of Sp4 (Nguyen-Huynh and Schaffer, 1998; Khalil et al., 2008). Importantly, even though Sp4 in mammals is predominantly expressed in tissues of neuronal origin (Lerner et al., 2005), regulating the developmental patterning of dendrites (Ramos et al., 2007), and has been linked to schizophrenia (Pinacho et al., 2011), our experiments show that Sp4 does not bind the ncx1-Br promoter sequence in cortical neurons. Conversely, our results on the regulation of NCX1 by Sp1 and Sp3 are in line with data showing that Sp1 and Sp3 regulate the transcription of target genes by binding to identical cognate DNA elements with similar affinities (Hagen et al., 1992). Indeed, transfection of specific siRNAs for Sp1 and Sp3 in cortical neurons produced a decrease or an increase, respectively, in the ncx1 promoter activity, mRNA levels, and protein expression. These results demonstrate that Sp1 is an activator, whereas Sp3 is a repressor of ncx1 transcription, a mechanism also reported for other Sp1 target genes (Stains et al., 2003). Notably, in the present paper, we identified three Sp1 binding sites (C–E) on ncx1-Br, which showed the highest promoter activity, and found that mutations at the level of the Sp1C–E sites in neurons overexpressing Sp1 and Sp3 could significantly reduce or increase, respectively, the activity of ncx1 brain promoter, whereas Sp1B had no effect. As regard the Sp1-B site, because the binding to this putative Sp1 site of nuclear factors from SHSY-5Y cells is partially competed by the Cy5-Sp1 consensus sequence, we cannot exclude the possibility that other additional transcription factors recognizing GC elements different from Sp1 family (Carrasco-Serrano et al., 1998), such as the Early growth response protein 1 (Mora-López et al., 2008), may participate to bind such complexes. Accordingly, luciferase experiments show that Sp1 and Sp3 factors are not involved in the regulation of ncx1-Br promoter activity through the Sp1-B site. Importantly, as shown by ChIP, this effect was associated with an increase of Sp1 or of Sp3 binding on Sp1C–E sites, which was completely abolished when the three sequences were jointly mutated. Collectively, these results show that three of the five putative Sp1 elements present on the ncx1-Br sequence modulate the promoter activity of the ncx1 gene. Indeed, studies report that global transcriptional repression upon ischemia is similarly reflected by a decrease in histone acetylation and that HDAC inhibition reduces ischemia-induced alterations in HDAC expression, thereby determining neuroprotection (Abel and Zukin, 2008; Noh et al., 2012). Consistently, data emerging from our study seem to suggest that both REST during ischemia and HIF-1 during preconditioning interfere with NCX1 gene expression through the participation of HDAC1, HDAC2, and p300. In particular, our ChIP analyses reveal that in brain ischemia REST and Sp3, together with the corepressors HDAC1 and HDAC2, are recruited to ncx1-Br, thus decreasing H3 acetylation, RNA-Pol II binding, and determining a reduction in ncx1 mRNA and protein levels. Interestingly, PC+tMCAO induces an increase in the abundance of ncx1-Br associated with acetylated H3, along with increases in RNA-Pol II, HIF-1, and p300 binding, thus determining an increase in NCX1 mRNA levels and protein expression. Furthermore Re-ChIP experiments using Sp3 as first antibody and REST, HDAC1, or HDAC2 as second antibody, or Sp1 as first antibody and HIF-1 or p300 as second antibody, further confirm that REST/Sp3/HDAC1/HDAC2 and HIF-1/Sp1/p300 are present on ncx1-Br sequence during tMCAO or PC+tMCAO, respectively. All these data suggest that the REST/Sp3/HDAC1/HDAC2 complex decreases NCX1 expression during brain ischemia, whereas the HIF-1/Sp1/p300 complex increases its expression during preconditioning. Notably, this regulation regards exclusively NCX1 and not NCX3, another NCX isoform involved in brain ischemia and ischemic brain preconditioning (Pignataro et al., 2012). Indeed, changes in NCX3 mRNA and protein expression were not associated with either the REST/Sp3/HDAC1/HDAC2 complex in tMCAO or the PC+tMCAO complex in ischemic brain preconditioning. Furthermore, the rat Sp1C–E, repressor element 1 (RE-1) and hypoxia-responsive element-1 (HRE-1) sequences have 60%, 75%, 70%, 85%, and 85% of similarity with human sequences, respectively. Fascinatingly, the positions of all the above mentioned sequences on ncx1 human and rat promoters are conserved. This high percentage of homology between rat and human ncx1 brain promoter sequences indicates the relevance of our results obtained in animal models of stroke to human brain ischemia. On the other hand, as REST (Calderone et al., 2003; Formisano et al., 2007; Ooi and Wood, 2007; Yeo et al., 2009), Sp1 and Sp3 (Bigger et al., 1997; Lee et al., 2006), and HIF-1 (Matrone et al., 2004) regulate also other genes, on the basis of our results, it cannot rule out that other proteins in coordination with NCX1 can participate in the physiopathology of stroke. About the mechanism(s) that trigger(s) the increase of REST and Sp3 by tMCAO and of HIF-1 and Sp1 in PC+tMCAO, it has been reported that the increasing of REST expression in stroke has been associated with a decrease of casein kinase 1 (CK1) activation, which reduces REST phosphorylation blocking its proteasomal degradation (Kaneko et al., 2014), whereas the transcriptional repressor Sp3 is associated with CK2-phosphorylated histone deacetylase 2 (Sun et al., 2002), it could be suggested a role of CK1 to increase Sp3 and REST protein expression and their consequent binding on ncx1-Br promoter sequence. On the other hand, it has been found in PC+tMCAO that p-AKT inhibition prevented the upregulation of NCX1 (Pignataro et al., 2012) and that Sp1 and HIF-1 are both targets of AKT (Jiang et al., 2001; Tan and Khachigian, 2009); therefore, it could be hypothesized that activation of this protein kinase during PC+tMCAO determines an increase of Sp1 and HIF-1 binding on ncx1-Br promoter sequence. Furthermore, we investigated the role of HDAC1, HDAC2, and p300 in NCX1-induced neuroprotection in neurons subjected to either OGD/RX or PC+OGD/RX. We observed that neuroprotection induced by the HDAC1 and HDAC2 inhibitor MS-275 was completely abolished during OGD/RX by silencing NCX1, a finding that further supports the tight relationship between HDAC1, HDAC2, and NCX1 in the pathophysiological processes leading to stroke. These results are in accordance with Noh et al. (2012), showing that HDAC1 and HDAC2 are recruited to the AMPA receptor subunit GluA2 promoter sequence and that injection of the pan-HDAC inhibitor thrichostatin A after an ischemic episode ameliorates neuronal injury. In addition, p300 inhibitor C646-induced cell death was completely reverted in PC+OGD/RX by NCX1 overexpression, thus suggesting a close relationship between the histone acetyltransferase p300 and NCX1 in ischemic preconditioning. In summary, we have shown that, under brain ischemia and brain ischemic preconditioning, NCX1 expression is epigenetically modulated, respectively, by two functional protein complexes: REST/Sp3/HDAC1/HDAC2 and HIF-1/Sp1/p300. In particular, whereas the former downregulates NCX1 expression during brain ischemia, the latter upregulates it during preconditioning. Notably, the development of drugs that epigenetically regulate NCX1 by preventing its downregulation in stroke might be a new pharmacological avenue to ameliorate neuronal damage during brain ischemia.
Footnotes
This work was supported by Ministero dell'Istruzione, dell'Università e della Ricerca Grant PON PON01_01602 and PON03PE_00146_1 all to L.A. We thank Dr Paola Merolla for editorial revision and all the members of the L.A. laboratory for their constructive comments on the data and the manuscript; Prof. Antonella Scorziello and Dr Maria Josè Sisalli for teaching us the rat neuronal culture and for many stimulating discussions; and Prof. Nicola Zambrano for helping us to perform and analyze EMSA experiments.
The authors declare no competing financial interests.
References
- Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8:57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammanamanchi S, Brattain MG. Sp3 is a transcriptional repressor of transforming growth factor-beta receptors. J Biol Chem. 2001;276:3348–3352. doi: 10.1074/jbc.M002462200. [DOI] [PubMed] [Google Scholar]
- Ammanamanchi S, Freeman JW, Brattain MG. Acetylated sp3 is a transcriptional activator. J Biol Chem. 2003;278:35775–35780. doi: 10.1074/jbc.M305961200. [DOI] [PubMed] [Google Scholar]
- Annunziato L, Pignataro G, Di Renzo GF. Pharmacology of brain Na+/Ca2+ exchanger: from molecular biology to therapeutic perspectives. Pharmacol Rev. 2004;56:633–654. doi: 10.1124/pr.56.4.5. [DOI] [PubMed] [Google Scholar]
- Azahri NS, Di Bartolo BA, Khachigian LM, Kavurma MM. Sp1, acetylated histone-3 and p300 regulate TRAIL transcription: mechanisms of PDGF-BB-mediated VSMC proliferation and migration. J Cell Biochem. 2012;113:2597–2606. doi: 10.1002/jcb.24135. [DOI] [PubMed] [Google Scholar]
- Barone FC, White RF, Spera PA, Ellison J, Currie RW, Wang X, Feuerstein GZ. Ischemic preconditioning and brain tolerance: temporal histological and functional outcomes, protein synthesis requirement, and interleukin-1 receptor antagonist and early gene expression. Stroke. 1998;29:1937–1950. doi: 10.1161/01.STR.29.9.1937. discussion 1950–1951. [DOI] [PubMed] [Google Scholar]
- Bigger CB, Melnikova IN, Gardner PD. Sp1 and Sp3 regulate expression of the neuronal nicotinic acetylcholine receptor beta4 subunit gene. J Biol Chem. 1997;272:25976–25982. doi: 10.1074/jbc.272.41.25976. [DOI] [PubMed] [Google Scholar]
- Billon N, Carlisi D, Datto MB, van Grunsven LA, Watt A, Wang XF, Rudkin BB. Cooperation of Sp1 and p300 in the induction of the CDK inhibitor p21WAF1/CIP1 during NGF-mediated neuronal differentiation. Oncogene. 1999;18:2872–2882. doi: 10.1038/sj.onc.1202712. [DOI] [PubMed] [Google Scholar]
- Boscia F, Gala R, Pignataro G, de Bartolomeis A, Cicale M, Ambesi-Impiombato A, Di Renzo G, Annunziato L. Permanent focal brain ischemia induces isoform-dependent changes in the pattern of Na+/Ca2+ exchanger gene expression in the ischemic core, periinfarct area, and intact brain regions. J Cereb Blood Flow Metab. 2006;26:502–517. doi: 10.1038/sj.jcbfm.9600207. [DOI] [PubMed] [Google Scholar]
- Boscia F, D'Avanzo C, Pannaccione A, Secondo A, Casamassa A, Formisano L, Guida N, Sokolow S, Herchuelz A, Annunziato L. Silencing or knocking out the Na(+)/Ca(2+) exchanger-3 (NCX3) impairs oligodendrocyte differentiation. Cell Death Differ. 2012;19:562–572. doi: 10.1038/cdd.2011.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone A, Jover T, Noh KM, Tanaka H, Yokota H, Lin Y, Grooms SY, Regis R, Bennett MV, Zukin RS. Ischemic insults derepress the gene silencer REST in neurons destined to die. J Neurosci. 2003;23:2112–2121. doi: 10.1523/JNEUROSCI.23-06-02112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco-Serrano C, Campos-Caro A, Viniegra S, Ballesta JJ, Criado M. GC- and E-box motifs as regulatory elements in the proximal promoter region of the neuronal nicotinic receptor alpha7 subunit gene. J Biol Chem. 1998;273:20021–20028. doi: 10.1074/jbc.273.32.20021. [DOI] [PubMed] [Google Scholar]
- Cassimere EK, Pyndiah S, Sakamuro D. The c-MYC-interacting proapoptotic tumor suppressor BIN1 is a transcriptional target for E2F1 in response to DNA damage. Cell Death Differ. 2009;16:1641–1653. doi: 10.1038/cdd.2009.98. [DOI] [PubMed] [Google Scholar]
- Chuang DM, Leng Y, Marinova Z, Kim HJ, Chiu CT. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 2009;32:591–601. doi: 10.1016/j.tins.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federman N, de la Fuente V, Zalcman G, Corbi N, Onori A, Passananti C, Romano A. Nuclear factor κB-dependent histone acetylation is specifically involved in persistent forms of memory. J Neurosci. 2013;33:7603–7614. doi: 10.1523/JNEUROSCI.4181-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formisano L, Noh KM, Miyawaki T, Mashiko T, Bennett MV, Zukin RS. Ischemic insults promote epigenetic reprogramming of mu opioid receptor expression in hippocampal neurons. Proc Natl Acad Sci U S A. 2007;104:4170–4175. doi: 10.1073/pnas.0611704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formisano L, Saggese M, Secondo A, Sirabella R, Vito P, Valsecchi V, Molinaro P, Di Renzo G, Annunziato L. The two isoforms of the Na+/Ca2+ exchanger, NCX1 and NCX3, constitute novel additional targets for the prosurvival action of Akt/protein kinase B pathway. Mol Pharmacol. 2008;73:727–737. doi: 10.1124/mol.107.042549. [DOI] [PubMed] [Google Scholar]
- Formisano L, Guida N, Valsecchi V, Pignataro G, Vinciguerra A, Pannaccione A, Secondo A, Boscia F, Molinaro P, Sisalli MJ, Sirabella R, Casamassa A, Canzoniero LM, Di Renzo G, Annunziato L. NCX1 is a new rest target gene: role in cerebral ischemia. Neurobiol Dis. 2013;50:76–85. doi: 10.1016/j.nbd.2012.10.010. [DOI] [PubMed] [Google Scholar]
- Formisano L, Guida N, Laudati G, Boscia F, Esposito A, Secondo A, Di Renzo G, Canzoniero LM. Extracellular signal-related kinase 2/specificity protein 1/specificity protein 3/repressor element-1 silencing transcription factor pathway is involved in Aroclor 1254-induced toxicity in SH-SY5Y neuronal cells. J Neurosci Res. 2015a;93:167–177. doi: 10.1002/jnr.23464. [DOI] [PubMed] [Google Scholar]
- Formisano L, Guida N, Laudati G, Mascolo L, Di Renzo G, Canzoniero LM. MS-275 inhibits aroclor 1254-induced SH-SY5Y neuronal cell toxicity by preventing the formation of the HDAC3/REST complex on the synapsin-1 promoter. J Pharmacol Exp Ther. 2015b;352:236–243. doi: 10.1124/jpet.114.219345. [DOI] [PubMed] [Google Scholar]
- Galmozzi A, Mitro N, Ferrari A, Gers E, Gilardi F, Godio C, Cermenati G, Gualerzi A, Donetti E, Rotili D, Valente S, Guerrini U, Caruso D, Mai A, Saez E, De Fabiani E, Crestani M. Inhibition of class I histone deacetylases unveils a mitochondrial signature and enhances oxidative metabolism in skeletal muscle and adipose tissue. Diabetes. 2013;62:732–742. doi: 10.2337/db12-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guida N, Laudati G, Galgani M, Santopaolo M, Montuori P, Triassi M, Di Renzo G, Canzoniero LM, Formisano L. Histone deacetylase 4 promotes ubiquitin-dependent proteasomal degradation of Sp3 in SH-SY5Y cells treated with di(2-ethylhexyl)phthalate (DEHP), determining neuronal death. Toxicol Appl Pharmacol. 2014;280:190–198. doi: 10.1016/j.taap.2014.07.014. [DOI] [PubMed] [Google Scholar]
- Hagen G, Müller S, Beato M, Suske G. Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res. 1992;20:5519–5525. doi: 10.1093/nar/20.21.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CK, D'Souza UM, Eisch AJ, Yajima S, Lammers CH, Yang Y, Lee SH, Kim YM, Nestler EJ, Mouradian MM. Dopamine receptor regulating factor, DRRF: a zinc finger transcription factor. Proc Natl Acad Sci U S A. 2001;98:7558–7563. doi: 10.1073/pnas.121635798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannotti FA, Barrese V, Formisano L, Miceli F, Taglialatela M. Specification of skeletal muscle differentiation by repressor element-1 silencing transcription factor (REST)-regulated Kv7.4 potassium channels. Mol Biol Cell. 2013;24:274–284. doi: 10.1091/mbc.E11-12-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru N, Tabuchi A, Hara D, Hayashi H, Sugimoto T, Yasuhara M, Shiota J, Tsuda M. Regulation of neurotrophin-3 gene transcription by Sp3 and Sp4 in neurons. J Neurochem. 2007;100:520–531. doi: 10.1111/j.1471-4159.2006.04216.x. [DOI] [PubMed] [Google Scholar]
- Isomura H, Stinski MF, Kudoh A, Daikoku T, Shirata N, Tsurumi T. Two Sp1/Sp3 binding sites in the major immediate-early proximal enhancer of human cytomegalovirus have a significant role in viral replication. J Virol. 2005;79:9597–9607. doi: 10.1128/JVI.79.15.9597-9607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto T, Inoue Y, Ito K, Sakaue T, Kita S, Katsuragi T. The exchanger inhibitory peptide region-dependent inhibition of Na+/Ca2+ exchange by SN-6 [2-[4-(4-nitrobenzyloxy)benzyl]thiazolidine-4-carboxylic acid ethyl ester], a novel benzyloxyphenyl derivative. Mol Pharmacol. 2004;66:45–55. doi: 10.1124/mol.66.1.45. [DOI] [PubMed] [Google Scholar]
- Jiang BH, Jiang G, Zheng JZ, Lu Z, Hunter T, Vogt PK. Phosphatidylinositol 3-kinase signaling controls levels of hypoxia-inducible factor 1. Cell Growth Differ. 2001;12:363–369. [PubMed] [Google Scholar]
- Jin HO, An S, Lee HC, Woo SH, Seo SK, Choe TB, Yoo DH, Lee SB, Um HD, Lee SJ, Park MJ, Kim JI, Hong SI, Rhee CH, Park IC. Hypoxic condition- and high cell density-induced expression of Redd1 is regulated by activation of hypoxia-inducible factor-1alpha and Sp1 through the phosphatidylinositol 3-kinase/Akt signaling pathway. Cell Signal. 2007;19:1393–1403. doi: 10.1016/j.cellsig.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Kaneko N, Hwang JY, Gertner M, Pontarelli F, Zukin RS. Casein kinase 1 suppresses activation of REST in insulted hippocampal neurons and halts ischemia-induced neuronal death. J Neurosci. 2014;34:6030–6039. doi: 10.1523/JNEUROSCI.4045-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Q, Costa M. Hypoxia-inducible factor-1 (HIF-1) Mol Pharmacol. 2006;70:1469–1480. doi: 10.1124/mol.106.027029. [DOI] [PubMed] [Google Scholar]
- Khalil MI, Hay J, Ruyechan WT. Cellular transcription factors Sp1 and Sp3 suppress varicella-zoster virus origin-dependent DNA replication. J Virol. 2008;82:11723–11733. doi: 10.1128/JVI.01322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CS, Choi HS, Hwang CK, Song KY, Lee BK, Law PY, Wei LN, Loh HH. Evidence of the neuron-restrictive silencer factor (NRSF) interaction with Sp3 and its synergic repression to the mu opioid receptor (MOR) gene. Nucleic Acids Res. 2006;34:6392–6403. doi: 10.1093/nar/gkl724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JL, Liu HC, Minnerath SR, Loh HH. Transcriptional regulation of mouse mu-opioid receptor gene. J Biol Chem. 1998;273:27678–27685. doi: 10.1074/jbc.273.42.27678. [DOI] [PubMed] [Google Scholar]
- Koh JY, Choi DW. Quantitative determination of glutamate mediated cortical neuronal injury in cell culture by lactate dehydrogenase efflux assay. J Neurosci Methods. 1987;20:83–90. doi: 10.1016/0165-0270(87)90041-0. [DOI] [PubMed] [Google Scholar]
- Lanzillotta A, Pignataro G, Branca C, Cuomo O, Sarnico I, Benarese M, Annunziato L, Spano P, Pizzi M. Targeted acetylation of NF-kappaB/RelA and histones by epigenetic drugs reduces post-ischemic brain injury in mice with an extended therapeutic window. Neurobiol Dis. 2013;49:177–189. doi: 10.1016/j.nbd.2012.08.018. [DOI] [PubMed] [Google Scholar]
- Law AY, Yeung BH, Ching LY, Wong CK. Sp1 is a transcription repressor to stanniocalcin-1 expression in TSA-treated human colon cancer cells, HT29. J Cell Biochem. 2011;112:2089–2096. doi: 10.1002/jcb.23127. [DOI] [PubMed] [Google Scholar]
- Lee J, Kosaras B, Aleyasin H, Han JA, Park DS, Ratan RR, Kowall NW, Ferrante RJ, Lee SW, Ryu H. Role of cyclooxygenase-2 induction by transcription factor Sp1 and Sp3 in neuronal oxidative and DNA damage response. FASEB J. 2006;20:2375–2377. doi: 10.1096/fj.06-5957fje. [DOI] [PubMed] [Google Scholar]
- Lerner LE, Gribanova YE, Whitaker L, Knox BE, Farber DB. The rod cGMP-phosphodiesterase beta-subunit promoter is a specific target for Sp4 and is not activated by other Sp proteins or CRX. J Biol Chem. 2002;277:25877–25883. doi: 10.1074/jbc.M201407200. [DOI] [PubMed] [Google Scholar]
- Lerner LE, Peng GH, Gribanova YE, Chen S, Farber DB. Sp4 is expressed in retinal neurons, activates transcription of photoreceptor-specific genes, and synergizes with Crx. J Biol Chem. 2005;280:20642–20650. doi: 10.1074/jbc.M500957200. [DOI] [PubMed] [Google Scholar]
- Matrone C, Pignataro G, Molinaro P, Irace C, Scorziello A, Di Renzo GF, Annunziato L. HIF-1alpha reveals a binding activity to the promoter of iNOS gene after permanent middle cerebral artery occlusion. J Neurochem. 2004;90:368–378. doi: 10.1111/j.1471-4159.2004.02483.x. [DOI] [PubMed] [Google Scholar]
- Min SW, Cho SH, Zhou Y, Schroeder S, Haroutunian V, Seeley WW, Huang EJ, Shen Y, Masliah E, Mukherjee C, Meyers D, Cole PA, Ott M, Gan L. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron. 2010;67:953–966. doi: 10.1016/j.neuron.2010.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-López F, Pedreño-Horrillo N, Delgado-Pérez L, Brieva JA, Campos-Caro A. Transcription of PRDM1, the master regulator for plasma cell differentiation, depends on an SP1/SP3/EGR-1 GC-box. Eur J Immunol. 2008;38:2316–2324. doi: 10.1002/eji.200737861. [DOI] [PubMed] [Google Scholar]
- Nguyen-Huynh AT, Schaffer PA. Cellular transcription factors enhance herpes simplex virus type 1 oriS-dependent DNA replication. J Virol. 1998;72:3635–3645. doi: 10.1128/jvi.72.5.3635-3645.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh KM, Hwang JY, Follenzi A, Athanasiadou R, Miyawaki T, Greally JM, Bennett MV, Zukin RS. Repressor element-1 silencing transcription factor (REST)-dependent epigenetic remodeling is critical to ischemia-induced neuronal death. Proc Natl Acad Sci U S A. 2012;109:E962–E971. doi: 10.1073/pnas.1121568109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi L, Wood IC. Chromatin crosstalk in development and disease: lessons from REST. Nat Rev Genet. 2007;8:544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- Pawlus MR, Hu CJ. Enhanceosomes as integrators of hypoxia inducible factor (HIF) and other transcription factors in the hypoxic transcriptional response. Cell Signal. 2013;25:1895–1903. doi: 10.1016/j.cellsig.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignataro G, Gala R, Cuomo O, Tortiglione A, Giaccio L, Castaldo P, Sirabella R, Matrone C, Canitano A, Amoroso S, Di Renzo G, Annunziato L. Two sodium/calcium exchanger gene products, NCX1 and NCX3, play a major role in the development of permanent focal cerebral ischemia. Stroke. 2004;35:2566–2570. doi: 10.1161/01.STR.0000143730.29964.93. [DOI] [PubMed] [Google Scholar]
- Pignataro G, Esposito E, Cuomo O, Sirabella R, Boscia F, Guida N, Di Renzo G, Annunziato L. The NCX3 isoform of the Na+/Ca2+ exchanger contributes to neuroprotection elicited by ischemic postconditioning. J Cereb Blood Flow Metab. 2011;31:362–370. doi: 10.1038/jcbfm.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignataro G, Boscia F, Esposito E, Sirabella R, Cuomo O, Vinciguerra A, Di Renzo G, Annunziato L. NCX1 and NCX3: two new effectors of delayed preconditioning in brain ischemia. Neurobiol Dis. 2012;45:616–623. doi: 10.1016/j.nbd.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Pinacho R, Villalmanzo N, Lalonde J, Haro JM, Meana JJ, Gill G, Ramos B. The transcription factor SP4 is reduced in postmortem cerebellum of bipolar disorder subjects: control by depolarization and lithium. Bipolar Disord. 2011;13:474–485. doi: 10.1111/j.1399-5618.2011.00941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisance V, Niederhauser G, Azzouz F, Lenain V, Haefliger JA, Waeber G, Abderrahmani A. The repressor element silencing transcription factor (REST)-mediated transcriptional repression requires the inhibition of Sp1. J Biol Chem. 2005;280:401–407. doi: 10.1074/jbc.M411825200. [DOI] [PubMed] [Google Scholar]
- Ramos B, Gaudillière B, Bonni A, Gill G. Transcription factor Sp4 regulates dendritic patterning during cerebellar maturation. Proc Natl Acad Sci U S A. 2007;104:9882–9887. doi: 10.1073/pnas.0701946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos B, Valín A, Sun X, Gill G. Sp4-dependent repression of neurotrophin-3 limits dendritic branching. Mol Cell Neurosci. 2009;42:152–159. doi: 10.1016/j.mcn.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravache M, Weber C, Mérienne K, Trottier Y. Transcriptional activation of REST by Sp1 in Huntington's disease models. PLoS One. 2010;5:e14311. doi: 10.1371/journal.pone.0014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, 3rd, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, Kerstetter KA, Neve RL, Haggarty SJ, McKinsey TA, Bassel-Duby R, Olson EN, Nestler EJ. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Rodenas-Ruano A, Chávez AE, Cossio MJ, Castillo PE, Zukin RS. REST-dependent epigenetic remodeling promotes the developmental switch in synaptic NMDA receptors. Nat Neurosci. 2012;15:1382–1390. doi: 10.1038/nn.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Lee J, Zaman K, Kubilis J, Ferrante RJ, Ross BD, Neve R, Ratan RR. Sp1 and Sp3 are oxidative stress-inducible, antideath transcription factors in cortical neurons. J Neurosci. 2003;23:3597–3606. doi: 10.1523/JNEUROSCI.23-09-03597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer S, Meisel A, Märschenz S. Epigenetic mechanisms in cerebral ischemia. J Cereb Blood Flow Metab. 2013;33:1335–1346. doi: 10.1038/jcbfm.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonini MV, Camargo LM, Dong E, Maloku E, Veldic M, Costa E, Guidotti A. The benzamide MS-275 is a potent, long-lasting brain region-selective inhibitor of histone deacetylases. Proc Natl Acad Sci U S A. 2006;103:1587–1592. doi: 10.1073/pnas.0510341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirabella R, Secondo A, Pannaccione A, Scorziello A, Valsecchi V, Adornetto A, Bilo L, Di Renzo G, Annunziato L. Anoxia-induced NF-kappaB-dependent upregulation of NCX1 contributes to Ca2+ refilling into endoplasmic reticulum in cortical neurons. Stroke. 2009;40:922–929. doi: 10.1161/STROKEAHA.108.531962. [DOI] [PubMed] [Google Scholar]
- Sirabella R, Secondo A, Pannaccione A, Molinaro P, Formisano L, Guida N, Di Renzo G, Annunziato L, Cataldi M. ERK1/2, p38, and JNK regulate the expression and the activity of the three isoforms of the Na(+) /Ca(2+) exchanger, NCX1, NCX2, and NCX3, in neuronal PC12 cells. J Neurochem. 2012;122:911–922. doi: 10.1111/j.1471-4159.2012.07838.x. [DOI] [PubMed] [Google Scholar]
- Sisalli MJ, Secondo A, Esposito A, Valsecchi V, Savoia C, Di Renzo GF, Annunziato L, Scorziello A. Endoplasmic reticulum refilling and mitochondrial calcium extrusion promoted in neurons by NCX1 and NCX3 in ischemic preconditioning are determinant for neuroprotection. Cell Death Differ. 2014;21:1142–1149. doi: 10.1038/cdd.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stains JP, Lecanda F, Screen J, Towler DA, Civitelli R. Gap junctional communication modulates gene transcription by altering the recruitment of Sp1 and Sp3 to connexin-response elements in osteoblast promoters. J Biol Chem. 2003;278:24377–24387. doi: 10.1074/jbc.M212554200. [DOI] [PubMed] [Google Scholar]
- Sun JM, Chen HY, Moniwa M, Litchfield DW, Seto E, Davie JR. The transcriptional repressor Sp3 is associated with CK2-phosphorylated histone deacetylase 2. J Biol Chem. 2002;277:35783–35786. doi: 10.1074/jbc.C200378200. [DOI] [PubMed] [Google Scholar]
- Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/S0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- Tan NY, Khachigian LM. Sp1 phosphorylation and its regulation of gene transcription. Mol Cell Biol. 2009;29:2483–2488. doi: 10.1128/MCB.01828-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsecchi V, Pignataro G, Del Prete A, Sirabella R, Matrone C, Boscia F, Scorziello A, Sisalli MJ, Esposito E, Zambrano N, Di Renzo G, Annunziato L. NCX1 is a novel target gene for hypoxia-inducible factor-1 in ischemic brain preconditioning. Stroke. 2011;42:754–763. doi: 10.1161/STROKEAHA.110.597583. [DOI] [PubMed] [Google Scholar]
- Won J, Yim J, Kim TK. Sp1 and Sp3 recruit histone deacetylase to repress transcription of human telomerase reverse transcriptase (hTERT) promoter in normal human somatic cells. J Biol Chem. 2002;277:38230–38238. doi: 10.1074/jbc.M206064200. [DOI] [PubMed] [Google Scholar]
- Yeo M, Berglund K, Augustine G, Liedtke W. Novel repression of Kcc2 transcription by REST-RE-1 controls developmental switch in neuronal chloride. J Neurosci. 2009;29:14652–14662. doi: 10.1523/JNEUROSCI.2934-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]