Abstract
Establishing topographical maps of the external world is an important but still poorly understood feature of the vertebrate sensory system. To study the selective innervation of hindbrain regions by sensory afferents in the zebrafish embryo, we mapped the fine-grained topographical representation of sensory projections at the central level by specific photoconversion of sensory neurons. Sensory ganglia located anteriorly project more medially than do ganglia located posteriorly, and this relates to the order of sensory ganglion differentiation. By single-plane illumination microscopy (SPIM) in vivo imaging, we show that (1) the sequence of arrival of cranial ganglion inputs predicts the topography of central projections, and (2) delaminated neuroblasts differentiate in close contact with the neural tube, and they never loose contact with the neural ectoderm. Afferent entrance points are established by plasma membrane interactions between primary differentiated peripheral sensory neurons and neural tube border cells with the cooperation of neural crest cells. These first contacts remain during ensuing morphological growth to establish pioneer axons. Neural crest cells and repulsive slit1/robo2 signals then guide axons from later-differentiating neurons toward the neural tube. Thus, this study proposes a new model by which the topographical representation of cranial sensory ganglia is established by entrance order, with the entry points determined by cell contact between the sensory ganglion cell bodies and the hindbrain.
Keywords: axon navigation, inner ear, neural crest cells, neuron differentiation, sensory systems, somatotopy
Introduction
The ability of the brain to build an internal representation of the external world based on sensory information, which relies on the establishment of topographic projections, is essential for the accurate transmission of environmental stimuli to processing centers in the brain (for review, see Luo and Flanagan, 2007). For instance, in the inner ear, vestibular or acoustic signals acquired by hair cells are transmitted to bipolar afferent neurons that project axons to the corresponding nuclei in the hindbrain. This first mechanosensory relay contains a topographic neural map, in which the afferent central projections are stratified along the dorsoventral (DV) and mediolateral (ML) axes reflecting the spatial distribution of the sensory patches (Sapède and Pujades, 2010).
Specialized sensory organs in the vertebrate head, the cranial sensory placodes, originate from thickenings in the embryonic ectoderm. Placodes give rise to two key cell types that underlie the function of sensory systems: cells that receive the stimuli, such as the hair cells in the inner ear or the lateral line, and sensory afferent neurons, which conduct the extracted information to the brainstem (for review, see Patthey et al., 2014). Several studies tried to unveil how peripheral ganglia “send” afferent projections to “reach” their entry points in the hindbrain. The mechanisms proposed include (1) cranial neural crest cells (cNCCs) form corridors, which provide a passive mechanism for sensory axons to migrate toward the CNS (Freter et al., 2013), and (2) guidance molecules, such as in robo/slit signaling, which direct axons toward the hindbrain. Chemorepulsion plays several roles in organizing sensory systems: maintaining the spatial restriction of distinct sensory neuron assemblies (Wang et al., 2013), controlling axonal arborization (Campbell et al., 2007), forming placode-derived ganglia (Shiau et al., 2008; Shiau and Bronner-Fraser, 2009), and regulating specific afferent projection patterns (Pan et al., 2012). However, these views consider differentiated sensory neurons as a population of cells arising far from the neural tube extending their axons through the mesenchyme toward the hindbrain.
To investigate the mechanism by which external stimuli are relayed to the brain, we mapped the topographical organization of the sensory axonal projections of the trigeminal (TGg), statoacoustic (SAg), and lateral line ganglia (LLg) within the hindbrain. We show that anterior sensory ganglia project more medially than those located more posteriorly, and this recapitulates the order of ganglion differentiation. We unveil the importance of the site of differentiation in the positioning of the sensory entry points into the hindbrain: placode-derived neurons differentiate in close apposition with the neural ectoderm, and their afferent entry points are established by membrane interactions between these first differentiated sensory neurons and cells at the border of the neural tube, before afferent sensory axon formation. This first contact of pioneer neurons with the CNS needs the input from NCCs to establish the afferent entry sites. Only then, NCCs and slit1/robo2 guidance cues organize the architecture of the sensory system, by maintaining ganglion coalescence and by guiding the later-differentiated sensory neurons, thereby controlling axonal branching and the fasciculation of the nerve bundle.
Materials and Methods
Zebrafish strains and maintenance
Zebrafish embryos of either sex were obtained by mating adult fish using standard methods. All fish strains were maintained individually as inbred lines. All procedures used have been approved by the Barcelona Biomedical Research Park Animal Care and Use Ethics Committee and implemented according to national rules and European regulations. The Tg[neuroD:GFP] line expresses GFP in neuronal progenitors (Obholzer et al., 2008) and differentiating cells (see Fig. 7), and the Tg[Isl3:GFP] line (also called Isl2b) expresses GFP in the afferent sensory neurons of cranial ganglia (Pittman et al., 2008). The Tg[hspGFF53A] line was generated by random integration of an enhancer-trap construct and expresses Gal4FF in afferent neurons of the trigeminal ganglia, inner ear, and the lateral line, with background expression in axial muscle (Asakawa and Kawakami, 2008). It was crossed with the Tg[UAS:KAEDE] line for photoconversion (PhC) experiments (Pujol-Martí et al., 2010). The use of Tg[hspGFF53A]xTg[UAS:KAEDE] crosses results in delayed expression of KAEDE in sensory neurons, due to the time needed for Gal4 to bind to UAS and activate KAEDE transcription, causing mosaic expression of KAEDEGreen at 48 h postfertilization (hpf). Mü4127 is an enhancer trap line, in which mCherry is under the control of krx20 regulatory elements, and therefore labeling r3 and r5 (Distel et al., 2009). cxcr4b mutants (odyJI049) are null-functional mutants (Knaut et al., 2003), and sdf1 mutants (also called medusa) carry a non-sense mutation (Valentin et al., 2007). Tg[cldnb:lynGFP] was described previously by Haas and Gilmour (2006).
Figure 7.
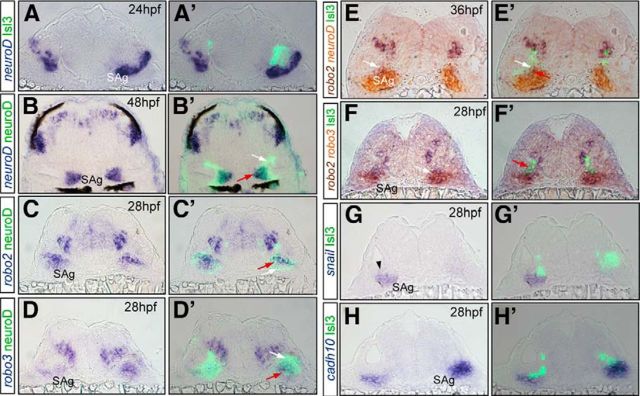
robo2 and robo3 label different SAg neuronal populations according to their differentiation state. A, A′, Tg[Isl3:GFP] embryos analyzed for neuroD. B-D′, Tg[neuroD:GFP] embryos hybridized with neuroD (B, B′), robo2 (C, C′), and robo3 (D, D′); note that all neuroD-expressing neuroblasts display neuroD:GFP (B′, red arrow), but neuroD is not expressed in the early differentiated neuronal population (still GFP positive due to its high stability; B′, white arrow). robo2 is expressed only in a subpopulation of neuroD:GFP cells (C′, red arrows). robo3 is expressed in a subpopulation of neuro:GFP cells (D′, red arrow), but not in the earliest differentiated ones (D′, white arrow). E, E′, Tg[Isl3:GFP] embryos hybridized with robo2/neuroD. Within the robo2-positive population, some cells express Isl3:GFP (white arrows) and some neuroD (red arrow). F, F′, robo2/robo3. Note that GFP-positive cells expressing robo2 do not express robo3 (F′, red arrow), and cells expressing robo2/robo3 do not display GFP (F, white arrow). G, G′, snail in delaminating neuroblasts (black arrowhead). H, H′, cadh10 in a subpopulation of nondifferentiated neuroblasts. All images are transverse sections of embryos at the level of the otic vesicle.
Photoconversion experiments
PhC of KAEDE protein was performed on a Leica SP5 inverted confocal microscope scanning one or few focal planes in a region of interest (ROI) centered on TGg, SAg, or LLg, with 8 to 16 frame averages per image, under 405 nm laser excitation. Before PhC, ganglia were visualized for KAEDEGreen under 488 nm laser excitation. Proper PhC was monitored by the appearance of a strong KAEDERed signal, under excitation with a 543 nm laser. As an internal control, we checked disappearance of KAEDEGreen upon appearance of KAEDERed (Fig. 1A).
Figure 1.
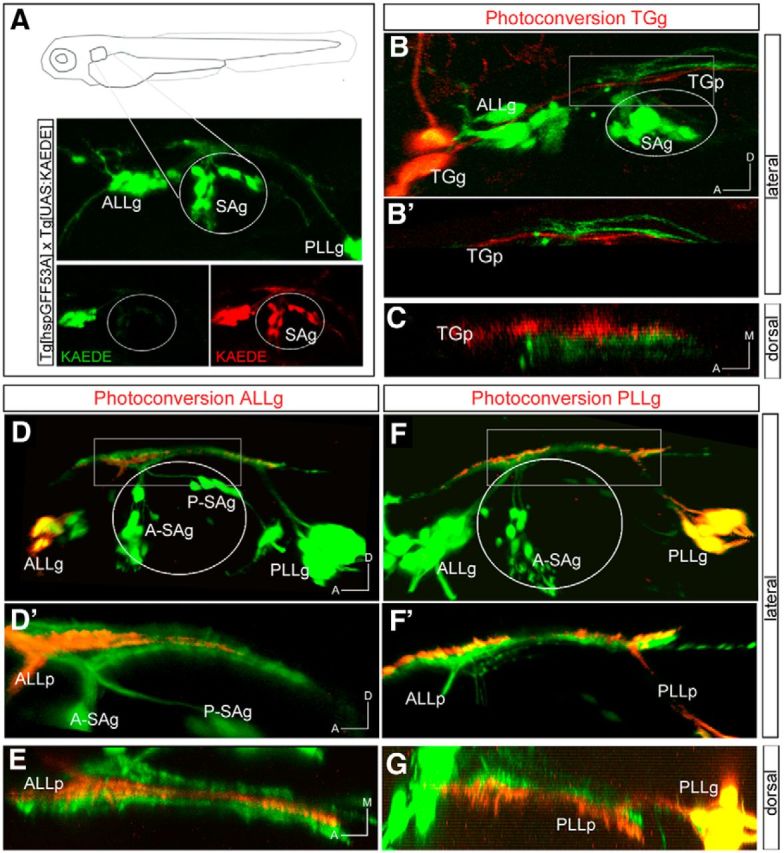
PhC of specific sensory neuronal pools in Tg[hspGFF53A]xTg[UAS:KAEDE] embryos. A, Depiction of the experiment: 48 hpf embryos expressed KAEDEGreen in sensory neurons; KAEDEGreen was photoconverted, and expression of KAEDERed in the sensory projection was assessed a few hours later. B–C, PhC of neurons from the TGg. B′, Magnification of the boxed region in B. C, Dorsal view of B′. D–E, PhC of neurons from the ALLg. D′, Magnification of the boxed region in D. E, Dorsal view of D′. F–G, PhC of neurons from the PLLg. F′, Magnification of the boxed region in F. G, Dorsal view of F′. Anterior is to the left. Axes are indicated in the figure. The contour of the otic vesicle is indicated with white circles.
Ablation experiments
Tg[neuroD:GFP] and Tg[Isl3:GFP] embryos at 18–20 hpf were used for ablation of the first differentiated neurons from the anterior LLg (ALLg)/SAg. For this purpose, the small ROI for each ganglion underwent high-intensity irradiation (910 nm) using the laser from a multiphoton microscope (tunable Mai Tai broadband laser, 710–990 nm) connected to an upright Leica TCS SP5 confocal microscope. Successful ablations were monitored for absence of the first differentiated neurons and axonal degradation during the following hour. Afterwards, embryos were incubated at 28°C and imaged 24 h later on a Leica SP5 inverted confocal microscope to monitor the reinnervation established by the late-differentiating neurons.
Leflunomide treatment
Embryos at 50% epiboly were dechorionated and grown until the desired stage in 6.5 μm leflunomide (L5025; Sigma) in EB buffer solution (10 mM TRIS/HCl, pH 8.5). The EB buffer containing the drug was renewed every 12 h. Leflunomide is a pharmacological agent that inhibits the transcriptional elongation of genes required for neural crest development (White et al., 2011).
In situ hybridization and immunolabeling
Whole-mount in situ hybridization (ISH) was performed as described previously (Hauptmann and Gerster, 1994). For chromogenic ISH, fluorescein- and digoxigenin (DIG)-labeled probes were detected with INT-BCIP and NBT/BCIP substrates (Roche), respectively. For fluorescent in situ hybridization, embryos were first equilibrated in 0.1 m Tris-HCl, pH 8.2, and then the DIG-labeled probe was detected with Fast Red (Roche) dissolved in 0.1 m Tris-HCl, pH 8.2. Probes were as follows: cadh6 and cadh10 (Liu et al., 2006), crestin (Berndt and Halloran, 2006), neuroD (also called neuroD1; Itoh and Chitnis, 2001), slit1a and slit1b (Hutson et al., 2003), snail2 (Thisse et al., 1995), and robo2 and robo3 (Lee et al., 2001).
For GFP immunolabeling, staged embryos were fixed in 4% paraformaldehyde (PFA) at room temperature for 20 min, washed in 0.1% Tween 20/PBS, and incubated overnight at 4°C with anti-GFP primary antibody (1:400; Clontech) in blocking solution followed by secondary antibodies conjugated with Alexa Fluor 488.
Cryostat sectioning
Embryos were fixed in 4% PFA, cryoprotected in 15% sucrose, and embedded in 7.5% gelatin/15% sucrose. Blocks were frozen in 2-methylbutane (Sigma) to improve tissue preservation, and then 20 μm sections were cut on a LeicaCM1510-1 cryostat.
Antisense morpholino injections
For morpholino knockdowns, embryos were injected with translation-blocking morpholino oligomers (MOs) obtained from GeneTools. MO injections were as follows: 5 ng/μl MO-Robo2, 5′-AAG GAC CCA TCC TGT CAT AGT CCA C-3′ (Zhang et al., 2012); 5 ng/μl MO-Slit1a, 5′-GAC AAC ATC CTC CTC TCG CAG GCA T-3′ (Barresi et al., 2005); 5 ng/μl MO-Slit1b 5′-GCT CGG TGT CCG GCA TCT CCA AAA G-3′ (Kastenhuber et al., 2009); 7.5 ng/μl MO-p53 as control, 5′-GCG CCA TTG CTT TGC AAG AAT TG-3′ (Langheinrich et al., 2002). MO-p53 was used to avoid off-target events (Robu et al., 2007). No significant differences were observed among MO-p53-injected embryos and those injected with a scrambled morpholino (data not shown). Thus, MO-p53 was used as control and included in all MO injections. In the case of double MO-Slit1a and MO-Slit1b injection, a 2 ng/μl concentration was used for each MO. To trace the injected embryos, they were coinjected at the one-cell stage with 80 ng/μl H2B–mCherry mRNA (Olivier et al., 2010) or lyn-TdTomato mRNA (Ingham, 2009). They were left to develop at 28°C until desired stages.
Phenotype analysis
To allow statistical treatment of phenotype occurrence, a score of 0 or 1 was given to the absence or presence, respectively, of each phenotype for each injected embryo. The expressivity of the observed defects in terms of combination of phenotypes is depicted in Figure 8I. Defasciculation was further analyzed by comparing nerve bundle width between control embryos and morphants (data not shown).
Figure 8.
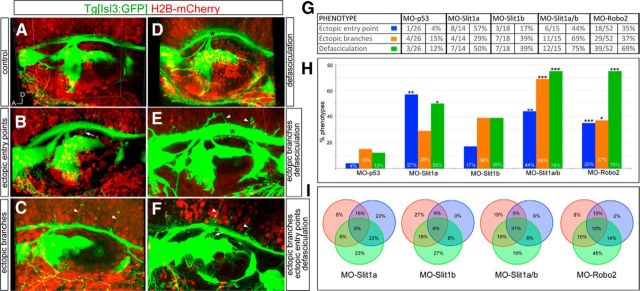
Robo2/Slit1 signaling regulates axonal branching and nerve bundle fasciculation. Tg[Isl3:GFP] embryos were coinjected with MO-CTRL (MO-p53), mRNA for H2B-mCherry or lyn-TdTomato, and MO-Slit1a, MO-Slit1b, MO-Robo2, or double MO-Slit1a/b. A–F, Examples of phenotypes observed at 48 hpf. Note the variety of effects ranging from ectopic entry points (white arrows), ectopic branches (white arrow heads), defasciculation (black asterisks), and combinations of primary phenotypes (E, F). G, Statistics of MO injections. H, Analyses of the percentages of different phenotypes with different MO combinations. *p < 0.1; **p < 0.01; ***p < 0.001. I, Analysis of the percentage of morphant embryos displaying different combinations of phenotypes. Orange circles correspond to embryos displaying ectopic branches, blue circles to embryos with ectopic entry points, and green circles to embryos with defasciculation. Note that many embryos display a combination of phenotypes.
Imaging and image processing
Confocal imaging.
Embryos were anesthetized in tricaine and mounted laterally on glass-bottomed Petri dishes (Mattek) in 1% low melting point-agarose, or fixed and mounted in 100% glycerol. Confocal imaging was performed on a Leica TCS SP5 II CW-STED inverted confocal microscope system (without stimulated emission depletion) using hybrid detectors and a 20× objective. In the case of x–y–z confocal cross-sections, z-stacks were acquired with a 1.5 μm z distance.
Single-Plane Illumination Microscopy (SPIM) imaging.
Embryos were injected with lyn-TdTomato mRNA (Ingham, 2009) at the one-cell stage to label plasma membranes and grown at 28°C. Embryos at the desired stage were anesthetized and mounted in 0.75% agarose in glass capillaries (size 2; volume, 20 μl; Brand). Imaging was performed on a Zeiss Lightsheet Z.1 microscope using a 20× objective. Image processing was done using the Zeiss ZEN software and involved dual illumination side fusion and deconvolution (regularized inverse method). Movies and stills were generated from the 4D datasets using FIJI. Embryos were at 26°C during imaging, and developmental stages were corrected accordingly (hours postfertilization × 0.7).
Fluorescence microscope imaging.
Cryostat sections were imaged on a Leica DM6000B fluorescence microscope with a DFC300KX camera under the control of the Leica Application Suite, Advanced Fluorescence 1.8, using 20× and 40× objectives. ISH and fluorescent image processing were done with FIJI.
Photoconversion experiments, imaging of drug-treated embryos, morphant phenotype analyses, and bundle width measurements were done using Imaris software (Bitplane).
Results
Mapping the neurosensory network
Spatial and temporal differences in axonal projections can provide clues as to the connectivity patterns of the overall neural circuits. Thus, to understand the early differences among sensory cranial ganglion projections, we first explored their topographical organization in the hindbrain using double transgenic Tg[hspGFF53A]Tg[UAS:KAEDE] embryos, which express photoconvertible KAEDEGreen early in the developing sensory ganglia. Distinct sensory ganglion neurons were photoconverted at 48 hpf, and the expression of KAEDERed in the sensory projection toward the CNS was assessed (Fig. 1A). We found that when the photoconversion was performed in TGg neurons, photoconverted axonal projections were located very ventral within the neural tube (Fig. 1B,B′). In fact, the KAEDERed bundle was the most ventrally located compared with all KAEDEGreen projections (Fig. 1B′). When the analysis was performed along the ML axis, TGg projections were positioned completely medially compared with the rest of sensory projections (Fig. 1C). Second, we photoconverted the ALLg/posterior LLg (PLLg) and observed that KAEDERed ALLg projections were in an intermediate position along the DV axis: they are more dorsal to the non-photoconverted axons of the SAg, but more ventral to another non-photoconverted sensory bundle (Fig. 1D,D′). Similar results were obtained along the ML axis: the ALLg projection (ALLp) is allocated in a middle position (Fig. 1E). On the other hand, the PLLg projection (PLLp) is the most dorsal and lateral of the sensory projections (Fig. 1F–G). These observations support previous studies describing that PLLg neurons projected more dorsally than ALLg neurons, before sensory organ innervation (Gompel et al., 2001), and are located more dorsal and lateral to the TGg projection.
We next investigated the positions of the two different neuronal populations of the SAg: the anterior SAg (A-SAg) and posterior SAg (P-SAg). Photoconverted A-SAg neurons projected quite ventrally and medially (Fig. 2A,A',B). However, the bundle is not the most ventrally positioned, considering the allocation of the TGg projection (TGp; compare Fig. 1B,B′). When we followed the KAEDERed P-SAg projection (P-SAp), we observed it to be more dorsally and laterally located than the A-SAg projection (A-SAp; Fig. 2C–D). These results suggest that the TGg projection is the most ventral and medially located, whereas the PLLg projection the most dorsally and laterally positioned. However, to decipher the relative position of the projections of the distinct SAg neuronal populations and the ALLg, we photoconverted two different ganglia at once (Fig. 2E–F′). We observed that the ALLg neurons project more dorsally than the A-SAg neurons (Fig. 2E,E′); on the other hand, the P-SAg is allocated ventral to the PLLg and separated by projections from the ALLg (Fig. 2F,F′). A summary of the fine mapping of the neurosensory network is depicted in Figure 2G, which reveals a highly ordered connectivity map with the dorsal/lateral to ventral/medial organization as follows: PLLp, ALLp, P-SAp, A-SAp, TGp.
Figure 2.
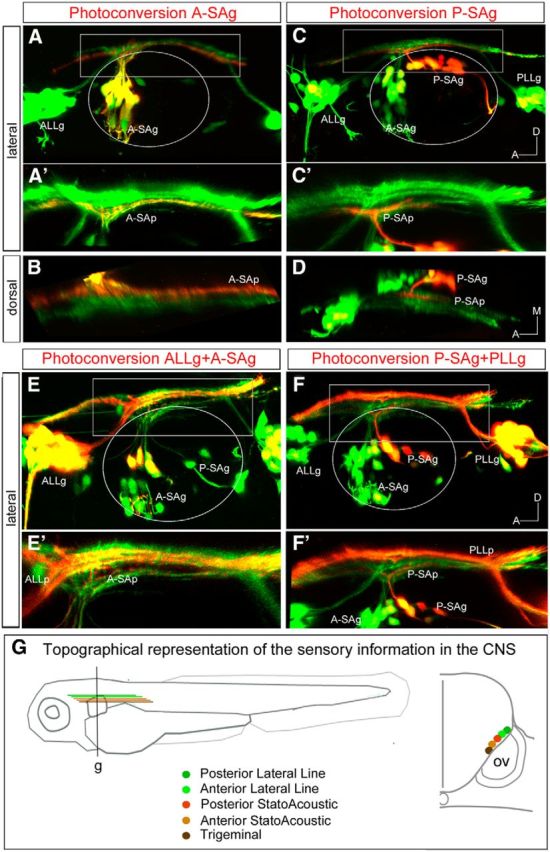
PhC of specific sensory neuronal pools in Tg[hspGFF53A]xTg[UAS:KAEDE] embryos. A–D, PhC of KAEDEGreen of neurons from the A-SAg (A–B) and P-SAg (C–D). A′, Magnification of the boxed region in A. B, Dorsal view of projections in A′. Note that red projection runs very ventral and medially, although it is not the most ventrally positioned, considering the allocation of the TGp. C′, Magnification of the boxed region in C. D, Dorsal view of projections in C′. Note that KAEDERed P-SAg projections are more dorsal and lateral than the A-SAp. E–F′, Double PhC of neurons from the ALLg and A-SAg (E, E′) and the P-SAg and PLLg (F, F′). E′, F′, Magnification of the boxed regions in E and F, respectively. G, Scheme depicting the neurosensory network with the highly ordered connectivity map, with DL/VM organization as follows: PLLg, ALLg, P-SAg, A-SAg, TGg. The right drawing represents a transverse section at the level of g. Anterior is always to the left. Axes are indicated in the figure. The contour of the otic vesicle is indicated with white circles.
Membrane contacts between pioneer sensory neurons and neural tube border cells prefigure the entry point at the central level
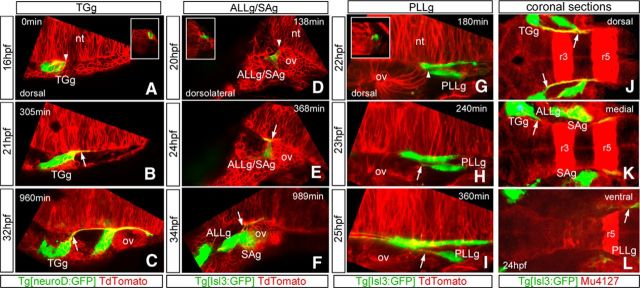
With the organization of the cranial sensory ganglia projections in the hindbrain established, we next asked how their entrance points are chosen and how this topography is built up. Previous works studying how early axonal contacts from cranial sensory systems were established observed that inputs from the separate cranial systems arrive sequentially in the order TGg–SAg–LLg (Kimmel et al., 1990). Our thought was that this sequence of arrival could determine the ML organization of the sensory bundles within the hindbrain. With this in mind, we first investigated how pioneer sensory axons navigate to the entrance point by in vivo imaging studies using the SPIM system (for review, see Weber and Huisken, 2011). Embryos expressing GFP either in differentiated (Tg[Isl3:GFP]) or differentiating (Tg[neuroD:GFP]) sensory neurons were injected with lyn-TdTomato mRNA to label the plasma membranes and live-imaged for several hours. As expected, the order of neuronal differentiation is sequential, as is the appearance of the projections toward the hindbrain: first the TGg differentiates, then the ALLg/SAg, and finally the PLLg (Movies 1, 2), and this relates to the topography of central projections. But most interestingly, the first neurons of the TGg, SAg, and PLLg to differentiate do so in close contact with the hindbrain cells and at the same anteroposterior (AP) level where the entrance point will be (Fig. 3A,D,G, white arrowheads; Movies 3–5). Specifically, the plasma membranes of the sensory neurons establish close interactions with the plasma membranes of the neural tube border cells at the level of the future nerve entry point (Fig. 3A,D,G, insets), and this happens before formation of afferent sensory axonal processes. Pioneer neurons maintain these established contacts with the neural tube, even when they are pushed away due to morphogenetic growth (Fig. 3B,E,H), so that although the sensory neuron is pushed toward the periphery, it leaves a trailing axon behind. Later-differentiating neurons reach the same entrance point (Fig. 3C,F,I, white arrows). For better comprehension, see Movies 3–5, which show TGg, ALLg/SAg, and PLLg neuronal differentiation, respectively. We mapped the positions of the contact/entrance points along the AP axis using transgenic embryos with rhombomeric landmarks, and allocated the entry points to r2 for the TGg, r4 for ALLg/SAg, and r6 for the PLLg (Fig. 3J–L), as suggested by the in vivo imaging experiments.
Figure 3.
Sensory neuron differentiation and establishment of hindbrain afferents entrance points. A–I, Still images of the SPIM time-lapse analysis of Tg[neuroD:GFP] (A–C) or Tg[Isl3:GFP] (D–I) embryos injected with lyn-TdTomato mRNA are shown. The first differentiated sensory neurons of the TGg (A), SAg (D) and PLLg (G) are in close contact with neural tube border cells through plasma membranes, at the level of the future nerve entry point (white arrowheads). Insets in A, D, and G are z-resliced images with dorsal to the top to show as transverse views the contact point of the respective sensory neurons with the border cells of the hindbrain. Primary sensory neurons maintain contacts with the neural tube, even when they are pushed away by morphogenetic growth (B, E, H). Note that they leave trailing axons (C, F, I, white arrows). Images are single confocal planes except for PLLg images, which are MIPs of several confocal planes. J–L, Serial coronal sections of Tg[Isl3:GFP]xMu4127 embryo from dorsal to ventral. Note that the TGg entry point is located in r2, the SAg one in r4, and the PLLg one in r6. nt, Neural tube; r, rhombomere; ov, otic vesicle. Anterior is always to the left except for insets.
Movie 1.
Sequential order of sensory neuron differentiation. SPIM time-lapse analysis of Tg[Isl3:GFP] embryos injected with lyn-TdTomato mRNA to visualize differentiated neurons. Note that the order of neuronal differentiation is sequential, TGg/SAg/LLg; sensory neurons from the ALLg differentiate a bit later (21.0 hpf) and more anteriorly (white arrowhead) than those from the SAg primordium (19.3 hpf). Then, ALLg cells migrate close to the SAg where they rest for a while, before complete segregation of the ganglia is accomplished. First PLLg differentiated cells (yellow arrowhead) are visible at 21.2 hpf when ALLp/SAp are already entering the hindbrain (white arrow). PLLp enters the hindbrain at ∼22.8 hpf (yellow arrow). Embryos were imaged from 19 hpf onward at 5 min intervals. Dorsal view with anterior to the left.
Movie 2.
Dynamics of differentiation of TGg, ALLg, and SAg neurons. SPIM time-lapse analysis of Tg[neuroD:GFP] embryos injected with lyn-TdTomato mRNA to see differentiating neurons. Note that ALLg/SAg/PLLg neurons are already differentiating at 16 hpf (white and yellow arrowheads for ALLg and PLLg, respectively). At 18.7 hpf, the projection of the ALLg/SAg (white arrow) reaches the hindbrain; meanwhile the PLLp does so a bit later (20.1 hpf; yellow arrow). The PLLp enters at 22.6 hpf (see it better in the right side of the embryo). The PLLp from the right embryonic side runs anteriorly at 26.4 hpf (yellow arrow), and the PLLp from the left side does so at 29.2 hpf (yellow arrow). Embryos were imaged from 16 hpf onward at 5 min intervals.
Movie 3.
Dynamics of differentiation of TGg neurons. SPIM time-lapse analysis of Tg[neuroD:GFP] embryos injected with lyn-TdTomato mRNA. The arrowhead indicates the TGg, and the arrow the appearance of the TGp. Embryos were imaged from 16 hpf onward at 5 min intervals. A dorsal view with the anterior to the left is shown.
Movie 4.
Dynamics of differentiation of ALLg/SAg neurons. SPIM time-lapse analysis of Tg[Isl3:GFP] embryos injected with lyn-TdTomato mRNA. The arrowhead indicates the first ALLg/SAg cells, and the arrow the appearance of the SAp. Embryos were imaged from 18 hpf onward at 23 min intervals. A dorsolateral view with the anterior to the left is shown.
Movie 5.
Dynamics of differentiation of PLLg neurons. SPIM time-lapse analysis of Tg[Isl3:GFP] embryos injected with lyn-TdTomato mRNA. The arrowhead indicates the first PLLg cells, and the arrow the appearance of the PLLp. Embryos were imaged from 18 hpf onward at 5 min intervals. A dorsal view with the anterior to the left is shown.
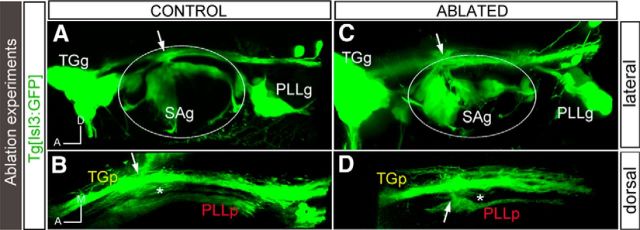
To investigate the importance of the original plasma membrane contact for the establishment of the entry points, we ablated the pioneer axon of the first-differentiating neurons of the SAg in Tg[neuroD:GFP] and Tg[Isl3:GFP] embryos. Ablation of the first sensory axons did not result in alterations in the entry point (Fig. 4, white arrows; results not shown). However, several defects in nerve bundle elongation were observed: otic axons enter the central system (Fig. 4C,D, white arrows), but once there, they do not elongate the nerve bundle projection, leaving an empty space between the TGp and the PLLp (n = 5/9; Fig. 4, comapre B, D, asterisks). These results indicate that although first differentiating sensory neurons (pioneer neurons) directly contact the neural tube border cells at the site of the future entry point, these contacts by themselves are not sufficient to define the central entry point, and/or that another player is involved in the maintenance of the established entry points. However, these data suggest that once the pioneer contact has been abolished, the elongation of the central nerve bundles is impeded.
Figure 4.
Ablation of the first differentiated neurons results in defects in the sensory projections at the central level. Tg[Isl3:GFP] embryos were used for the ablation of the first differentiated neurons from the ALLg/SAg using the laser of a multiphoton microscope. A, C, Lateral views of control and ablated embryos, respectively, showing no ectopic entry points after pioneer axon ablation. B, D, Dorsal views of A and C, respectively, showing the sensory projections at the central levels. Note the defects in SAg nerve bundle elongation upon ablation (compare B, D, asterisks). White arrows indicate the entry point. White asterisks indicate the location of the SAp. The contour of the otic vesicle is indicated with white circles. Anterior is always to the left.
Pioneer axons and neural crest cells cooperate in the establishment of entry points
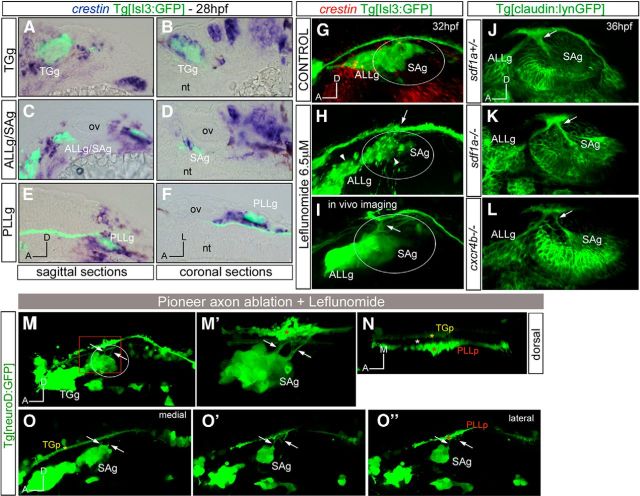
cNCCs are specified in the dorsal part of the neural tube and begin their migration at ∼14–15 hpf, segregating into three distinct streams lateral to r2, r4, and r6. Since cNCCs are present at the time when entry points are chosen and they have previously been shown to interact with sensory ganglia supporting coalescence (Freter et al., 2013; Sandell et al., 2014), we next asked whether cNCCs were involved in instructing the sensory neurons to find the position of the central entry point. We first assessed the spatial relationship between cNCCs and placode-derived neurons by in situ hybridization experiments with crestin, which labels early NCC derivatives, in embryos expressing GFP in the sensory ganglia (Fig. 5A–F). NCCs were observed primarily on the exterior surface of the aggregated ganglia, with crestin-positive cells usually adjacent to (or surrounding the) GFP neurons in all sensory ganglia (Fig. 5A–F). To better understand the relationship between NCCs and the centrally projecting axons, crestin-stained embryos were imaged by confocal microscopy. No overlap between crestin and GFP was observed in single z-planes (data not shown), and only some overlap was obtained when maximal intensity projections (MIPs) were analyzed (Fig. 5G), indicating that cNCCs envelop sensory ganglia to maintain coalescence, as reported previously (Freter et al., 2013; Sandell et al., 2014), and suggesting that cNCCs may help instruct sensory axons to reach the hindbrain. To determine their possible role in defining/instructing sensory neurons, we blocked the formation of NCC precursors using leflunomide, an inhibitor of NCC development (White et al., 2011), or inhibited the migration of NCCs by genetic mutations in the sdf1a/cxcr4b pathway (Olesnicky Killian et al., 2009; Theveneau et al., 2013). Embryos treated with leflunomide did not display any crestin expression (Fig. 5H), and they exhibited defects in the coalescence of the three ganglia (TGg, SAg, LLg) as expected, although no effect on the entry point position (either at 28 or 32 hpf) or in the differentiation of the ganglia was observed (Fig. 5H; n = 0/9) compared with control embryos (Fig. 5G). To better illustrate this, we also imaged live embryos treated with leflunomide, and no entry point defects were observed (Fig. 5I). Furthermore, mutant embryos for either the sdf1a ligand or cxcr4b receptor did not show ectopic entry points (Fig. 5J–L; n = 15 for sdf1a−/−; n = 30 for cxcr4b−/−), although they did display defects in collective migration of the lateral line primordium cells (Knaut et al., 2003; Valentin et al., 2007) and in the migration of NCCs (Olesnicky Killian et al., 2009; Theveneau et al., 2013; data not shown). These results strongly support the hypothesis that cNCCs envelop the sensory ganglia to maintain coalescence and may support and guide their late-differentiating axons, as suggested previously (Freter et al., 2013; Sandell et al., 2014), but do not instruct the first sensory neurons to find the central entrance points. We next asked whether NCCs, in combination with pioneer axon contacts, are necessary for establishing the afferent entry point. In fact, when the pioneer neurons from the ALLg/SAg were ablated in the absence of NCCs, we did observe ectopic entry points into the hindbrain (Fig. 5M,M′; n = 3/6). This phenotype was accompanied by nerve bundle elongation defects (Fig. 5N; n = 6/6), the same phenotype was observed when the pioneer axon was ablated. In addition, ALLg/SAg axons wrongly innervate the PLLp at central levels (Fig. 5O,O″; n = 5/6). Thus, these results suggest that cooperation of NCCs with the neuronal pioneer contacts is important for the establishment and maintenance of the entry point.
Figure 5.
Cooperation of pioneer axonal contacts and NCCs in the establishment of the entry points. A–F, Tg[Isl3:GFP] embryos were assayed for crestin in situ hybridization (blue). A, B, TGg. C, D, ALLg/SAg. E, F, PLLg. G–I, Tg[Isl3:GP] embryos were treated with DMSO (G) or leflunomide (H, I) and hybridized with crestin probe (G, H, red) or observed in vivo (I). Note that crestin expression is abolished in leflunomide-treated embryos, whereas sensory ganglia present defects in coalescence (H, white arrowheads). No effects in the entry points are observed, even in in vivo embryos (H, I, white arrows). J–L, Inhibition of NCC migration does not affect the entry point of SAg sensory axons at the central level: sdf1a−/−Tg[cldnb:lynGFP] (K) and cxcr4b−/−Tg[cldnb:lynGFP] (L) embryos were analyzed for ectopic entry points and compared with control embryos sdf1a+/−Tg[cldnb:lynGFP] (J). M–N, Tg[neuroD:GFP] embryos were treated with leflunomide and the first differentiated neurons from the ALLg/SAg were ablated using multiphoton microscopy. M, Lateral view showing ectopic entry points (white arrows). M′, Magnification of boxed region in M showing the ectopic entry points (arrows) in contact with the PLLp (red asterisk). N, Dorsal view of M′ showing the TGp (yellow asterisk), PLLp (red asterisk), and a lack of ALLp/SAp nerve bundle elongation (empty space marked with white asterisk). O–O″, Different single confocal planes from medial (O) to lateral (O″) showing that ectopic entry points now contact with the PLLp (red asterisk). Anterior is always to the left. Axes are indicated in the figure. The contour of the otic vesicle is indicated in white circles. ov, otic vesicle.
robo2 and slit1a/b genes are expressed in SAg sensory neurons and in their afferent target field, respectively
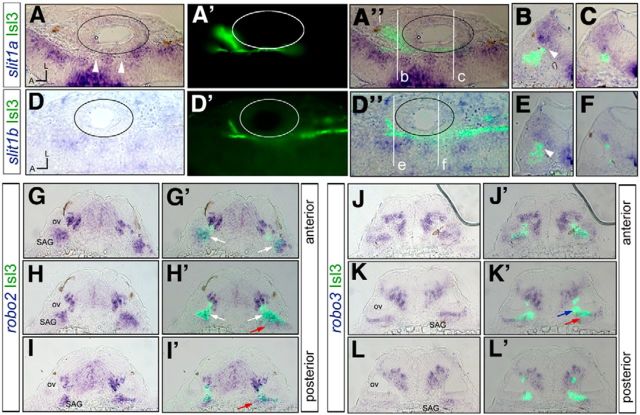
To understand how later-differentiating sensory axons reach the proper entry site and whether slit/robo signaling guides them, we studied the expression of slit and robo molecules in the hindbrain and in the cranial sensory placodes, focusing on the otic vesicle. The expression profile analysis of slits (the secreted ligands for robos) showed that in the hindbrain, all four zebrafish slits were expressed (Fig. 6A–F; Pan et al., 2012); however, only slit1a and slit1b were expressed adjacent to the sensory afferent bundle (Fig. 6A–F). Both slit1s are expressed in similar regions, with slit1b being expressed in a more restricted domain than slit1a (Fig. 6A,D). Although the expression of slits is quite diffuse in the hindbrain, there are regions devoid of slit expression (Fig. 6A, white arrowheads). Neither slit1a nor slit1b overlap with sensory axonal projections (Fig. 6B,C,E,F). These results reveal that slit1a and slit1b are enriched in the vicinity of the sensory neuron afferents in the hindbrain, suggestive of a role in entry point selection and/or axon guidance.
Figure 6.
robo2 and slit1 are expressed in SAg neurons and afferent target fields, respectively. A–L′, Tg[Isl3:GFP] embryos were analyzed for slit1a (A–C), slit1b (D–F), robo2 (G–I′), and robo3 (J–L′) expression at 28 hpf. Note the expression of slit1a/b along the hindbrain, with zones devoid of slit1 corresponding to the places where central projection enters (A, B, E, white arrowheads). robo2 is expressed in the differentiated Isl3 sensory neurons (G′, H′, white arrows), and robo3 in the SAg neuroblasts not yet differentiated (K′, red arrows), but never in differentiated sensory neurons (blue arrow). A–A″, D–D″, Coronal sections corresponding to half-sided embryos. B, C, E, F, Transverse sections corresponding to b, c, e, and f, respectively. G–I, J–L, Serial transverse sections along the AP axis.
Analysis of the expression of robo receptors revealed that only robo2 and robo3 are expressed in the sensory neurons (Fig. 6G–L′; data not shown). robo2 was dynamically expressed in otic sensory neurons and became restricted to the Isl3:GFP-positive population (Fig. 6G,G′, white arrows), although cells within the SAg not yet differentiated display robo2 as well (Fig. 6H′,I′, red arrows). On the other hand, robo3 has a complementary expression profile to robo2 within the SAg: it is mainly present in the nondifferentiated SAg neuroblasts (Fig. 6J–L′, red arrows) and in very few or no Isl3-positive differentiated neurons (Fig. 6K,K′, blue arrow). To better characterize the expression of robo2/3 in the SAg neuronal populations, we did a more thorough analysis using different neuronal markers (Fig. 7). We took advantage of neuroD, which labels otic placode-delaminated neuroblasts and is downregulated in differentiated neurons (Fig. 7A,A′), and the Tg[neuroD:GFP] fish line, where neuroD-positive cells display neuroD:GFP, although not all GFP-positive cells express neuroD due to a difference in the stability of the neuroD mRNA and GFP protein (Fig. 7B′, white arrow). The fact that neuroD expression and GFP do not fully overlap in differentiated neuroblasts provides a useful marker for the latest differentiated neurons (neuroD negative, neuroD:GFP positive). robo2 is expressed in a subpopulation of neuroD:GFP cells (Fig. 7C,C′, red arrow), supporting our previous observation that the robo2 receptor is present in differentiated and nondifferentiated SAg populations. Triple staining for robo2/neuroD and Isl3:GFP shows three neuronal populations: one expressing Isl3:GPF/robo2 (Fig. 7E,E′, white arrow), another expressing robo2/neuroD (Fig. 7E′, red arrow), and a third one displaying solely neuroD (Fig. 7E,E′). robo3 is expressed in most of the neuroD:GFP population, with few GFP-cells lacking robo3 expression (Fig. 7D,D′, red and white arrows, respectively). A previous study illustrated the various stages of SAg development, which involves a sequential process of specification, delamination, proliferative expansion, and differentiation of precursor cells to form the mature SAg (Vemaraju et al., 2012). When we combined the analysis of the robo2/3 and neuroD/Isl3:GFP expression territories with markers such as snail and cadh6/cadh10, which label subpopulations of otic neurons, we could ascribe robo2 and robo3 expression to different SAg neurons according to their differentiation state (Fig. 7): (1) neuroblasts that just delaminated from the otic epithelium (snail, neuroD), (2) transit-amplifying neurons (snail, cadh6, cadh10, neuroD, robo3), (3) neurons ready to undergo differentiation (neuroD, robo2, robo3), and (4) differentiated neurons (Isl3:GFP, neuroD:GFP, robo2). In summary, robo2/slit1 genes are expressed in SAg neurons and afferent target fields consistent with a role in guiding the later-differentiated sensory neurons to the proper target site.
Slit1/Robo2 signaling regulates the number of sensory branches and coalescence of the sensory bundle
To assess the effects of downregulation of Slit1/Robo2 signaling, we used morpholinos against the ligands (slit1a/slit1b; Barresi et al., 2005; Kastenhuber et al., 2009) and the receptor robo2 (Zhang et al., 2012). Tg[Isl3:GFP] embryos were injected, and the effects were analyzed at 24 and 48 hpf. We focused on three main phenotypes: the number/position of SAg nerve entry points at central levels, branching of the sensory nerves toward the hindbrain, and the coalescence of the sensory nerves bundles along the AP path in the hindbrain (Fig. 8). Interestingly, no effects were observed at 24 hpf (data not shown), suggesting that Slit1/Robo2 pathway plays a role only in late-differentiated neurons. At 48 hpf, morphants displayed ectopic entry points (Fig. 8B), ectopic branches (C), and defects in fasciculation (D); in many cases, a mix of phenotypes was obtained (E–F,I). Negligible effects were observed when control morpholino was injected (Fig. 8A,G,H, MO-CTRL; see Materials and Methods). Upon downregulation of Slit1a, over 57% of embryos displayed ectopic entry points, and half of the embryos showed defasciculation of the sensory nerve bundles compared with control embryos (Fig. 8G). Only 4/14 embryos displayed ectopic branches toward the hindbrain, and this result was not statistically significant when compared with control embryos (n = 4/26). No effects were observed in any of the analyzed phenotypes upon downregulation of Slit1b by itself (Fig. 8G). However, when the function of both ligands was inhibited (MO-Slit1a/b), over 44% of embryos had ectopic entry points, 69% displayed ectopic branches, and 75% had problems in nerve bundle fasciculation (Fig. 8G). Overall, these results suggest that Slit1 repulsion signals are involved in keeping the coalescence of the sensory bundle. To verify that Slit1a function was executed through Robo2 receptor, we knocked-down Robo2 in the sensory neurons and analyzed the embryos for similar phenotypes as seen with the Slit1a/b morphants. We observed a consistent increase in the number of embryos presenting ectopic entry points (n = 18/52), 35% of the embryos had ectopic branches, and 75% of embryos displayed ML defasciculation of the sensory nerves bundle along the anteroposterior path through the hindbrain (Fig. 8G–I). No problems in the primary entry point were observed in any of the analyzed embryos (Fig. 8A–F). These results support our hypothesis that the Slit1/Robo2 pathway does not play a role in the early differentiating neurons; however, it is important in guiding the late-differentiating neurons to the proper site. In addition, Slit1/Robo2 signaling plays a crucial role in keeping the fasciculation of the bundle (Fig. 8G–I), most probably due to the repulsion cues sent by Slit1s to sensory axons to avoid their expansion through the ML axis. In line with this, the width of the sensory bundle increases upon loss of function of Slit1/Robo2 signaling by the use of Slit1a/b and Robo2 morpholinos (data not shown). Interestingly, the observed phenotype of ectopic branches with MO-Slit1a/b might involve another receptor, since the effects of MO-Robo2 are quite mild (MO-Slit1a/b, 69%; MO-Robo2, 37%).
In summary, we show that although Slit1/Robo2 axon repulsion signaling is apparently not involved in establishing the pioneer axons, it does play an important role for later-differentiating sensory neurons to (1) reach the proper entry point established by the pioneer axons, (2) maintain fasciculation of nerve bundles to avoid bundle expansion, and (3) restrain sensory central afferents at the border of the neural tube to avoid branching into the hindbrain in incorrect places.
Discussion
The fine mapping of the neurosensory network displays a highly ordered connectivity map at the central level, and the order of cranial sensory ganglion differentiation is important for this organization. This topographic arrangement is laid out very early, according to studies suggesting that somatotopy in the PLLg is achieved in the absence of sensory input (Gompel et al., 2001). The order of afferent differentiation establishes the sequence of central projections within the PLLg in axolotl and in zebrafish (Fritzsch et al., 2005; Pujol-Martí et al., 2012). This is consistent with our observations: TGg neurons differentiate earlier than SAg and LLg neurons, and therefore their projections are allocated more ventrally and medially. However, we cannot discard that genes expressed or active in a DV gradient in the hindbrain could act as molecular landmarks of somatotopy, determining the molecular identity of the projecting sensory neurons as they do in other systems (Fariñas et al., 2001; Schuster et al., 2010; Wang et al., 2013). Indeed, Eph receptor tyrosine kinases and their ligands regulate axon guidance and contribute to the establishment of topographic projections in several areas of the nervous system. Eph proteins are extensively expressed in structures of the inner ear as well as in neurons in the peripheral and central components of the auditory system (Cramer and Gabriele, 2014). They are involved in the formation of auditory system connections between the hindbrain and the diencephalon, and in the innervation of hair cells of the sensory patches by SAG neurons; however, they do not instruct how sensory neurons find their target region in the hindbrain (for review, see Cramer, 2005). The sensory bundle is located within the pax7a/pax6/pax3-positive domain, suggesting the transcription factor code may play a role in placing the bundle along the DV axis. However, this can play only an early role because pax expression around the bundle is downregulated from 36 hpf onward (data not shown).
In vivo imaging confirmed that neuronal differentiation is a key aspect in the topographical organization. The differentiation order correlates with the ML topography of sensory bundles, whereas the site of differentiation correlates with the location of the entry point of the sensory neurons into the CNS (in collaboration with NCCs). First traced neurons differentiate in close apposition to the neural tube cells, establish a very robust membrane contact with the neural tube cells and change their shape. This first contact remains even when challenged by morphogenetic growth, resulting in a trailing axon that is likely used by late-differentiating neurons to reach the same entrance point. This mechanism operates in the TGg, SAg, and PLLg, suggesting that it is a common strategy for posterior and dorsal cranial placodes; whether this mechanism operates in other placodes is an open question. In our opinion, this raises the question of whether the specification of central pathways of distinct types of sensory information depends on the order of differentiation of peripheral sensory neurons. For the ALLg, the scenario is a bit more complex, as our high-resolution imaging data show: first differentiated ALLg neuroblasts arise several cell diameters away from the rudiment of the SAg, and later they migrate close to the SAg, where they remain for a while before complete segregation of the ganglia (Movies 1, 2). Interestingly, previous work describing placode neurogenesis suggested that the LLg neurons specify before SAg neurons according to neurog1 and neuroD1 expression (Andermann et al., 2002). The fact that in our movies we observed differentiation of the SAg before the LLg is probably due to the different technologies we used; neurog1 clearly labels cells undergoing neuronal specification, whereas neuroD:GFP is expressed in differentiating neurons that have already started to downregulate neuroD (Fig. 7), therefore unveiling different cell states. What positions the neuroblasts in this specific Cartesian grid along the AP axis is not known. Embryos with gross defects in the AP patterning of the hindbrain, such as the vhnf1 mutants (Hernandez et al., 2004), do not display any defects in the entry points of the sensory axons (D. Sapède and C. Pujades, unpublished results). Thus, sensory cells may sense positional information from the surrounding tissues or may have intrinsic cues responsible for this.
Previous experiments in amniotes demonstrated that cNCC streams transform into corridors that are subsequently associated with migrating sensory neuroblasts, bridging the domain between the placodal epithelium and the CNS for neuroblasts to extend their axons (Freter et al., 2013). Despite the intimate relationship between NCCs and sensory neurons (NCCs envelop the sensory ganglia), in our system NCCs are necessary to maintain coalescence of the ganglia. But by themselves are not sufficient to define the entry point, since inhibition of NCC formation by pharmacological reagents or mutants for the sdf1a/cxcr4b pathway that display defects in NCC migration do not result in defects in the number or position of the pioneer axons. Still, this mechanism could be used by epibranchial neurons residing far away from the neural tube, the fact that neurons of dorsolateral ganglia differentiate in close contact with the neural tube borders, with no need to migrate toward the CNS, may make NCC corridors less needed. As the ganglia are pushed away from the neural tube, first differentiated sensory neurons leave behind the axons that later-differentiating axons can use as a scaffold to migrate to the hindbrain. When first axonal contact is ablated and NCCs inhibited, ectopic entry points are generated, strongly supporting an intimate collaboration between these two mechanisms to establish pioneer axonal contacts. The question remains as to what information the cNCCs provide to the sensory neurons as they differentiate. They may be involved in direct cell-to-cell contact with the first axon, they may secrete some instructive signals that do not involve Slit molecules, or they may provide corridors that simply restrain the pioneer axons as the ganglia move; however, further investigation will be needed to unveil these putative signals.
Loss-of-function experiments show that Robo2 and Slit1a/b have late effects in the control of entrance points into the hindbrain, in restraining sensory central afferents at the border of the neural tube, and as inhibitors of defasciculation of the sensory bundles. No effects were observed at 24 hpf, in accordance with the observation that first entrance points are established upon sensory neuron differentiation, with no need for other cues except the right positioning of the neuroblasts and the cooperation of NCCs. Thus, Slit1 repulsion signals are probably involved in avoiding expansion of the sensory bundle; their expression around the sensory projection running along the AP axis of the hindbrain may repulse stray axons and keep them in a compacted fascicle restraining the range of central neurons that the sensory bundle would contact. Previous work has shown nicely that the vertebrate hindbrain contains stripes of neurons with shared neurotransmitter phenotypes that extend throughout the hindbrain of young zebrafish, reflecting a structural and functional patterning (Kinkhabwala et al., 2011). Accordingly, the sensory bundle would contact a single neuronal stripe displaying the appropriate transcription factors, and therefore with a given molecular identity.
Although Slit1a/b controls the arborization of sensory branches, our results suggest this effect may also be mediated through a Robo2-independent pathway. As Robo2 is the only roundabout family member expressed in differentiated SAg neurons, this indicates that Slit1 may act through an additional, non-Robo receptor. Previous work in the retinotectal system also suggested a role for Slits inhibiting arborization and synaptogenesis in the CNS via a Robo2-independent mechanism (Campbell et al., 2007). Slit2/Robo2 promote the axonal elongation and branching of the TGg sensory neurons (Yeo et al., 2004), and Slit3/Robo2 signaling has been proposed to prevent erroneous innervation of these neurons (Pan et al., 2012). However, our results suggest that in the inner ear, the synergy between Slit1a and Slit1b is the main regulator of sensory branch arborization.
Slit/Robo signaling has pleiotropic functions. In chicks and mice, perturbation of slit1 or robo2 disrupted proper ganglion formation (Shiau et al., 2008; Shiau and Bronner-Fraser, 2009). Interestingly, slit1 is expressed in NCCs in these species, which is not the case in zebrafish (data not shown). Thus, inhibition of NCC migration in zebrafish phenocopies the effect of downregulation of slit1/robo2 in amniotes, namely, a lack of ganglia coalescence. Although Slit/Robo signaling plays a role in the establishment of the DV topology of the longitudinal tract of the forebrain (Devine and Key, 2008), it seems that it is not the case in the sensory bundle of the hindbrain. These results support a model in which Robo2-dependent Slit1a/b activity maintains the normal spread of fascicles and controls the arborization of sensory branches.
Overall, our data reveal that establishing the proper topographical organization of the cranial sensory afferents is a multistep process. First, the entry point in the hindbrain is established by close apposition between sensory neurons and neural tube cell membranes, in a sequential order that correlates with sensory bundle topography. Second, sensory neurons are pushed away, leaving the pioneer axonal contact as a trailing cue for late-differentiating neurons. Third, instructions from cNCCs and Slit1/Robo2 signaling help maintain this topographical organization when challenged by morphogenetic growth. This third step is important for helping later-differentiated axons navigate, and for building a more complex system upon the initial scaffold.
Footnotes
This work was supported by predoctoral Formació d'Investigadors fellowships from Agencia de Gestio d'Ajuts Universitaris i de Recerca (Generalitat de Catalunya; A.Z., S.D.), a predoctoral fellowship from Obra Social La Caixa (A.V.), and Spanish Ministry of Economy and Competitiveness Grant BFU2012-31994 (C.P.). We thank Thomas Pujol for his valuable help in SPIM imaging, and Zeiss for use of the Lightsheet Z.1 microscope. We thank M. Linares and M. Verges for excellent technical assistance, members of Pujades lab for insightful discussions, and those who kindly provided us with transgenic fish lines and reagents, especially M. Brand, C. B. Chien, R. Koster, V. Lecaudey, H. Lopez-Schier, A. Nechiporuk, and A. Pan. We thank S. Schneider-Maunoury for critical reading of this manuscript.
References
- Andermann P, Ungos J, Raible DW. Neurogenin1 defines zebrafish cranial sensory ganglia precursors. Dev Biol. 2002;251:45–58. doi: 10.1006/dbio.2002.0820. [DOI] [PubMed] [Google Scholar]
- Asakawa K, Kawakami K. Targeted gene expression by the Gal4-UAS system in zebrafish. Dev Growth Differ. 2008;50:391–399. doi: 10.1111/j.1440-169X.2008.01044.x. [DOI] [PubMed] [Google Scholar]
- Barresi MJ, Hutson LD, Chien CB, Karlstrom RO. Hedgehog regulated Slit expression determines commissure and glial cell position in the zebrafish forebrain. Development. 2005;132:3643–3656. doi: 10.1242/dev.01929. [DOI] [PubMed] [Google Scholar]
- Berndt JD, Halloran MC. Semaphorin 3d promotes cell proliferation and neural crest cell development downstream of TCF in the zebrafish hindbrain. Development. 2006;133:3983–3992. doi: 10.1242/dev.02583. [DOI] [PubMed] [Google Scholar]
- Campbell DS, Stringham SA, Timm A, Xiao T, Law MY, Baier H, Nonet ML, Chien CB. Slit1a inhibits retinal ganglion cell arborization and synaptogenesis via Robo2-dependent and -independent pathways. Neuron. 2007;55:231–245. doi: 10.1016/j.neuron.2007.06.034. [DOI] [PubMed] [Google Scholar]
- Cramer KS. Eph proteins and the assembly of auditory circuits. Hear Res. 2005;206:42–51. doi: 10.1016/j.heares.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Cramer KS, Gabriele ML. Axon guidance in the auditory system: Multiple functions of Eph receptors. NEUROSCIENCE. 2014:1–11. doi: 10.1016/j.neuroscience.2014.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine CA, Key B. Robo-Slit interactions regulate longitudinal axon pathfinding in the embryonic vertebrate brain. Dev Biol. 2008;313:371–383. doi: 10.1016/j.ydbio.2007.10.040. [DOI] [PubMed] [Google Scholar]
- Distel M, Wullimann MF, Köster RW. Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc Natl Acad Sci U S A. 2009;106:13365–13370. doi: 10.1073/pnas.0903060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fariñas I, Jones KR, Tessarollo L, Vigers AJ, Huang E, Kirstein M, de Caprona DC, Coppola V, Backus C, Reichardt LF, Fritzsch B. Spatial shaping of cochlear innervation by temporally regulated neurotrophin expression. J Neurosci. 2001;21:6170–6180. doi: 10.1523/JNEUROSCI.21-16-06170.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freter S, Fleenor SJ, Freter R, Liu KJ, Begbie J. Cranial neural crest cells form corridors prefiguring sensory neuroblast migration. Development. 2013;140:3595–3600. doi: 10.1242/dev.091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Gregory D, Rosa-Molinar E. The development of the hindbrain afferent projections in the axolotl: Evidence for timing as a specific mechanism of afferent fiber sorting. Zoology. 2005;108:297–306. doi: 10.1016/j.zool.2005.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N, Dambly-Chaudière C, Ghysen A. Neuronal differences prefigure somatotopy in the zebrafish lateral line. Development. 2001;128:387–393. doi: 10.1242/dev.128.3.387. [DOI] [PubMed] [Google Scholar]
- Haas P, Gilmour D. Chemokine signaling mediates self-organizing tissue migration in the zebrafish lateral line. Dev Cell. 2006;10:673–680. doi: 10.1016/j.devcel.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Hauptmann G, Gerster T. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 1994;10:266. doi: 10.1016/0168-9525(90)90008-T. [DOI] [PubMed] [Google Scholar]
- Hernandez RE, Rikhof HA, Bachmann R, Moens CB. vhnf1 integrates global RA patterning and local FGF signals to direct posterior hindbrain development in zebrafish. Development. 2004;131:4511–4520. doi: 10.1242/dev.01297. [DOI] [PubMed] [Google Scholar]
- Hutson LD, Jurynec MJ, Yeo SY, Okamoto H, Chien CB. Two divergent slit1 genes in zebrafish. Dev Dyn. 2003;228:358–369. doi: 10.1002/dvdy.10386. [DOI] [PubMed] [Google Scholar]
- Ingham PW. The power of the zebrafish for disease analysis. Hum Mol Genet. 2009;18:R107–R112. doi: 10.1093/hmg/ddp091. [DOI] [PubMed] [Google Scholar]
- Itoh M, Chitnis AB. Expression of proneural and neurogenic genes in the zebrafish lateral line primordium correlates with selection of hair cell fate in neuromasts. Mech Dev. 2001;102:263–266. doi: 10.1016/S0925-4773(01)00308-2. [DOI] [PubMed] [Google Scholar]
- Kastenhuber E, Kern U, Bonkowsky JL, Chien CB, Driever W, Schweitzer J. Netrin-DCC, Robo-Slit, and heparan sulfate proteoglycans coordinate lateral positioning of longitudinal dopaminergic diencephalospinal axons. J Neurosci. 2009;29:8914–8926. doi: 10.1523/JNEUROSCI.0568-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Hatta K, Metcalfe WK. Early axonal contacts during development of an identified dendrite in the brain of the zebrafish. Neuron. 1990;4:535–545. doi: 10.1016/0896-6273(90)90111-R. [DOI] [PubMed] [Google Scholar]
- Kinkhabwala A, Riley M, Koyama M, Monen J, Satou C, Kimura Y, Higashijima SI, Fetcho J. A structural and functional ground plan for neurons in the hindbrain of zebrafish. Proc Natl Acad Sci U S A. 2011;108:1164–1169. doi: 10.1073/pnas.1012185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaut H, Werz C, Geisler R, Nüsslein-Volhard C, Tübingen 2000 Screen Consortium A zebrafish homologue of the chemokine receptor Cxcr4 is a germ-cell guidance receptor. Nature. 2003;421:279–282. doi: 10.1038/nature01338. [DOI] [PubMed] [Google Scholar]
- Langheinrich U, Hennen E, Stott G, Vacun G. Zebrafish as a model organism for the identification and characterization of drugs and genes affecting p53 signaling. Curr Biol. 2002;12:2023–2028. doi: 10.1016/S0960-9822(02)01319-2. [DOI] [PubMed] [Google Scholar]
- Lee JS, Ray R, Chien CB. Cloning and expression of three zebrafish roundabout homologs suggest roles in axon guidance and cell migration. Dev Dyn. 2001;221:216–230. doi: 10.1002/dvdy.1136. [DOI] [PubMed] [Google Scholar]
- Liu Q, Duff RJ, Liu B, Wilson AL, Babb-Clendenon SG, Francl J, Marrs JA. Expression of cadherin10, a type II classic cadherin gene, in the nervous system of the embryonic zebrafish. Gene Expr Patterns. 2006;6:703–710. doi: 10.1016/j.modgep.2005.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Flanagan JG. Development of continuous and discrete neural maps. Neuron. 2007;56:284–300. doi: 10.1016/j.neuron.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Obholzer N, Wolfson S, Trapani JG, Mo W, Nechiporuk A, Busch-Nentwich E, Seiler C, Sidi S, Söllner C, Duncan RN, Boehland A, Nicolson T. Vesicular glutamate transporter 3 is required for synaptic transmission in zebrafish hair cells. J Neurosci. 2008;28:2110–2118. doi: 10.1523/JNEUROSCI.5230-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesnicky Killian EC, Birkholz DA, Artinger KB. A role for chemokine signaling in neural crest cell migration and craniofacial development. Dev Biol. 2009;333:161–172. doi: 10.1016/j.ydbio.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier N, Luengo-Oroz MA, Duloquin L, Faure E, Savy T, Veilleux I, Solinas X, Débarre D, Bourgine P, Santos A, Peyriéras N, Beaurepaire E. Cell lineage reconstruction of early zebrafish embryos using label-free nonlinear microscopy. Science. 2010;329:967–971. doi: 10.1126/science.1189428. [DOI] [PubMed] [Google Scholar]
- Pan YA, Choy M, Prober DA, Schier AF. Robo2 determines subtype-specific axonal projections of trigeminal sensory neurons. Development. 2012;139:591–600. doi: 10.1242/dev.076588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patthey C, Schlosser G, Shimeld SM. The evolutionary history of vertebrate cranial placodes—I: Cell type evolution. Dev Biol. 2014;389:82–97. doi: 10.1016/j.ydbio.2014.01.017. [DOI] [PubMed] [Google Scholar]
- Pittman AJ, Law MY, Chien CB. Pathfinding in a large vertebrate axon tract: isotypic interactions guide retinotectal axons at multiple choice points. Development. 2008;135:2865–2871. doi: 10.1242/dev.025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol-Martí J, Baudoin JP, Faucherre A, Kawakami K, López-Schier H. Progressive neurogenesis defines lateralis somatotopy. Dev Dyn. 2010;239:1919–1930. doi: 10.1002/dvdy.22320. [DOI] [PubMed] [Google Scholar]
- Pujol-Martí J, Zecca A, Baudoin JP, Faucherre A, Asakawa K, Kawakami K, López-Schier H. Neuronal birth order identifies a dimorphic sensorineural map. J Neurosci. 2012;32:2976–2987. doi: 10.1523/JNEUROSCI.5157-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Butler Tjaden NE, Barlow AJ, Trainor PA. Cochleovestibular nerve development is integrated with migratory neural crest cells. Dev Biol. 2014;385:200–210. doi: 10.1016/j.ydbio.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapède D, Pujades C. Hedgehog signaling governs the development of otic sensory epithelium and its associated innervation in zebrafish. J Neurosci. 2010;30:3612–3623. doi: 10.1523/JNEUROSCI.5109-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster K, Dambly-Chaudière C, Ghysen A. Glial cell line-derived neurotrophic factor defines the path of developing and regenerating axons in the lateral line system of zebrafish. Proc Natl Acad Sci U S A. 2010;107:19531–19536. doi: 10.1073/pnas.1002171107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau CE, Bronner-Fraser M. N-cadherin acts in concert with Slit1-Robo2 signaling in regulating aggregation of placode-derived cranial sensory neurons. Development. 2009;136:4155–4164. doi: 10.1242/dev.034355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau CE, Lwigale PY, Das RM, Wilson SA, Bronner-Fraser M. Robo2-Slit1 dependent cell-cell interactions mediate assembly of the trigeminal ganglion. Nat Neurosci. 2008;11:269–276. doi: 10.1038/nn2051. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Steventon B, Scarpa E, Garcia S, Trepat X, Streit A, Mayor R. Chase-and-run between adjacent cell populations promotes directional collective migration. Nat Cell Biol. 2013;15:763–772. doi: 10.1038/ncb2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Postlethwait JH. Expression of snail2, a second member of the zebrafish snail family, in cephalic mesendoderm and presumptive neural crest of wild-type and spadetail mutant embryos. Dev Biol. 1995;172:86–99. doi: 10.1006/dbio.1995.0007. [DOI] [PubMed] [Google Scholar]
- Valentin G, Haas P, Gilmour D. The chemokine SDF1a coordinates tissue migration through the spatially restricted activation of Cxcr7 and Cxcr4b. Curr Biol. 2007;17:1026–1031. doi: 10.1016/j.cub.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Vemaraju S, Kantarci H, Padanad MS, Riley BB. A spatial and temporal gradient of Fgf differentially regulates distinct stages of neural development in the zebrafish inner ear. PLoS Genet. 2012;8:e1003068. doi: 10.1371/journal.pgen.1003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SZ, Ibrahim LA, Kim YJ, Gibson DA, Leung HC, Yuan W, Zhang KK, Tao HW, Ma L, Zhang LI. Slit/Robo signaling mediates spatial positioning of spiral ganglion neurons during development of cochlear innervation. J Neurosci. 2013;33:12242–12254. doi: 10.1523/JNEUROSCI.5736-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Huisken J. Light sheet microscopy for real-time developmental biology. Curr Opin Genet Dev. 2011;21:566–572. doi: 10.1016/j.gde.2011.09.009. [DOI] [PubMed] [Google Scholar]
- White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, Chen F, Long HK, Kramer M, Datta S, Neuberg D, Granter S, Young RA, Morrison S, Wheeler GN, Zon LI. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471:518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo SY, Miyashita T, Fricke C, Little MH, Yamada T, Kuwada JY, Huh TL, Chien CB, Okamoto H. Involvement of Islet-2 in the Slit signaling for axonal branching and defasciculation of the sensory neurons in embryonic zebrafish. Mech Dev. 2004;121:315–324. doi: 10.1016/j.mod.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Zhang C, Gao J, Zhang H, Sun L, Peng G. Robo2–slit and Dcc–netrin1 coordinate neuron axonal pathfinding within the embryonic axon tracts. J Neurosci. 2012;32:12589–12602. doi: 10.1523/JNEUROSCI.6518-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]