Abstract
Here, we have asked about post-transcriptional mechanisms regulating murine developmental neurogenesis, focusing upon the RNA-binding proteins Smaug2 and Nanos1. We identify, in embryonic neural precursors of the murine cortex, a Smaug2 protein/nanos1 mRNA complex that is present in cytoplasmic granules with the translational repression proteins Dcp1 and 4E-T. We show that Smaug2 inhibits and Nanos1 promotes neurogenesis, with Smaug2 knockdown enhancing neurogenesis and depleting precursors, and Nanos1 knockdown inhibiting neurogenesis and maintaining precursors. Moreover, we show that Smaug2 likely regulates neurogenesis by silencing nanos1 mRNA. Specifically, Smaug2 knockdown inappropriately increases Nanos1 protein, and the Smaug2 knockdown-mediated neurogenesis is rescued by preventing this increase. Thus, Smaug2 and Nanos1 function as a bimodal translational repression switch to control neurogenesis, with Smaug2 acting in transcriptionally primed precursors to silence mRNAs important for neurogenesis, including nanos1 mRNA, and Nanos1 acting during the transition to neurons to repress the precursor state.
SIGNIFICANCE STATEMENT The mechanisms instructing neural stem cells to generate the appropriate progeny are still poorly understood. Here, we show that the RNA-binding proteins Smaug2 and Nanos1 are critical regulators of this balance and provide evidence supporting the idea that neural precursors are transcriptionally primed to generate neurons but translational regulation maintains these precursors in a stem cell state until the appropriate developmental time.
Keywords: nanos1, neurogenesis, radial precursor development, Smaug2, stem cells, translational repression
Introduction
During mammalian brain development, embryonic neural precursors must generate the correct numbers and types of neurons and glia without exhausting themselves prematurely (Miller and Gauthier, 2007). The importance of this balance is exemplified by the observation that, when neural cell genesis is perturbed genetically in humans, this can cause aberrant development and cognitive dysfunction (for example, see Gauthier et al., 2007; Wang et al., 2010). However, despite the fact that regulated cell genesis underlies the establishment of appropriate neural circuitry, the mechanisms controlling neural precursor differentiation are still not well understood.
Studies in model organisms, such as Drosophila melanogaster, have established post-transcriptional regulation as important for cellular differentiation. For example, during D. melanogaster oogenesis and early development, translational repression of unlocalized mRNAs together with active translation of these same transcripts after localization play crucial roles in pattern specification and germ plasm formation (Becalska and Gavis, 2009; Lasko, 2011; Lai and King, 2013). Indeed, in metazoan embryos, bulk degradation of maternal mRNAs may be essential for appropriate spatial and temporal regulation of gene expression (for review, see Tadros and Lipshitz, 2009; Walser and Lipshitz, 2011). Is translational regulation similarly important in mammalian stem cells? A few recent studies in neural stem cells suggest that it may be. In particular, the RNA binding protein Staufen2 asymmetrically segregates mRNAs associated with neural cell fate decisions (Kusek et al., 2012; Vessey et al., 2012). Moreover, we recently identified a repressive translational complex involving the eIF4E-binding protein 4E-T that regulates mammalian neurogenesis by sequestering and silencing mRNAs encoding proneurogenic transcription factors (Yang et al., 2014). However, while these studies indicate that translational repression is important, the molecular players involved in this regulation are largely uncharacterized. For example, while Staufen2 and 4E-T are both part of translational regulatory complexes in mammalian neural precursors, neither protein is thought to be an mRNA-specific repressor protein; Staufen2 is a double-stranded RNA binding protein involved in mRNA transport, localization, and stability (Miki et al., 2005; Tosar et al., 2012; Heraud-Farlow et al., 2013; Laver et al., 2013) and 4E-T does not itself bind to mRNAs (Kamenska et al., 2014). Thus, while translational repression is emerging as a key regulatory mechanism in neural precursors, many of the relevant molecular players have not been identified.
We have addressed this issue here, focusing on the Smaug and Nanos families of RNA-binding proteins. In Drosophila, Smaug is a key translational repressor of mRNAs, such as that encoding Nanos in the bulk cytoplasm of early embryos (Dahanukar and Wharton, 1996; Dahanukar et al., 1999; Smibert et al., 1996, 1999). Furthermore, Smaug functions during the maternal-to-zygotic transition in both the soma and the primordial germ cells of early embryos via translational repression and/or degradation of its target mRNAs (Tadros et al., 2007; Benoit et al., 2009; Siddiqui et al., 2012; Chen et al., 2014a). Smaug does this by binding target mRNAs via stem-loop structures known as Smaug Recognition Elements (SREs) (Smibert et al., 1996, 1999; Dahanukar et al., 1999; Aviv et al., 2003) and recruiting proteins involved in mRNA repression and/or degradation (Chen et al., 2014a). Despite this broad and essential role for Smaug during Drosophila development, virtually nothing is known about Smaug during vertebrate development. Here, we show that murine Smaug2 is a key regulator of mammalian neural development, where it acts to maintain embryonic neural precursors in a stem cell state by binding to and repressing translation of the mRNA encoding a second repressive RNA-binding protein, Nanos1. Smaug2 silences nanos1 mRNA by recruiting it into a repressive Processing (P)-body-like granule in association with 4E-T. Moreover, we show that Nanos1, a known translational repressor, functions to promote the differentiation of precursors into neurons, likely by silencing translation of proteins associated with maintaining the stem cell state.
Materials and Methods
Animals.
All animal use was approved by the Animal Care Committee of the Hospital for Sick Children in accordance with the Canadian Council of Animal Care policies. CD1 mouse embryos of either sex (Charles River Laboratories) were used for all experiments.
Primary cultures and cell lines.
Primary cortical precursor cultures were prepared as previously described (Gauthier et al., 2007). Briefly, dissociated E12.5 CD1 cortical cells were plated at 250,000 cells/ml on glass coverslips precoated with 2% laminin (BD Biosciences) and 1% poly-d-lysine (Sigma) in Neurobasal medium (Invitrogen) supplemented with 2% B27 (Invitrogen), 0.5 mm lglutamine (Invitrogen), and 40 ng/ml FGF2 (BD Biosciences). For transfections, 1 μg of DNA in 100 μl of Opti-MEM was mixed with Lipofectamine LTX according to the manufacturer's instructions and added upon plating. In cotransfection experiments, EGFP expression vector was mixed with shRNA or overexpression vectors at a 1:3 ratio; when three plasmids were used, the ratio was 1:1.5:1.5. Cells were immunostained 3 d later. Clonal analysis was performed as previously described (Gallagher et al., 2013). Briefly, cortical cultures were prepared as described above and then transfected with 1.5 μg of DNA at a 1:1:3 ratio of PCAG-PB-GFP, pCyL43-Pbase, and shRNA or control plasmids. HEK-293T cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were transfected with Lipofectamine 2000 according to the manufacturer's instructions and used 16–24 h later.
In utero electroporation.
In utero electroporations were performed as previously described (Gauthier et al., 2007; Yang et al., 2014). Briefly, a nuclear EGFP plasmid (pEF-EGFP) was coelectroporated with shRNA or overexpression vectors at a 1:3 ratio, or when two additional plasmids were coelectroporated, at a 1:2:2 ratio for a total final DNA concentration of 4.0 μg/μl. Before injection, plasmids were mixed with 0.5% trypan blue. Following injection into the lateral ventricles, the square electroporator CUY21 EDIT (TR Tech) was used to deliver five 50 ms pulses of 40–50 V with 950 ms intervals per embryo. For all experiments, we analyzed at least 3 embryos per condition, each from an independent experiment with a different mother.
Plasmids and shRNAs.
The Flag-tagged expression constructs for mouse and human Smaug2 and Smaug1 and mouse Nanos1, Nanos 2, and Nanos3 and human Nanos1 were obtained from OriGene. shRNA vectors were obtained from EZBiolab and had the following sequences: Smaug2 shRNA-1 5′-GAG GAG AAC ATC ACC AGT TAC T-3′, Smaug2 shRNA-2 5′-GGG CTG GAA TGA GTG TGA ACAT-3′, and Nanos1 shRNA-1 5′-GCACATACCATCAAGTATTGCT-3′. The 4E-T shRNA was previously described (Yang et al., 2014), and the sequence was 5′-CCG TTA TAC CAA AGA ACA A-3′. All clones were verified by sequencing.
Immunostaining and quantitative analysis.
Immunocytochemistry on cultured cells was performed as previously described (Vessey et al., 2012). Briefly, cells on glass coverslips were fixed for 15 min with 4% buffered PFA, permeabilized with 0.2% NP40 in HBSS, blocked with 5% bovine serum albumin in HBSS (Jackson ImmunoResearch Laboratories) for 1 h, and incubated with primary antibodies in HBSS for 2 h at room temperature. Samples were washed 3× with HBSS, and secondary antibodies (1:1000) diluted in HBSS were added for an additional hour. Coverslips were mounted on glass slides with the ProLong Gold Antifade Reagent with DAPI (Invitrogen). For immunostaining of cortical sections, embryonic brains were dissected in ice-cold HBSS, fixed in 4% buffered PFA at 4°C overnight, cryopreserved with 30% sucrose overnight, placed in OCT, and kept at −80°C for a few hours to overnight, and cryosectioned coronally with a thickness of 16 μm. Sections were blocked at room temperature with 5% BSA (Jackson ImmunoResearch Laboratories) and 0.3% Triton X in PBS, and incubated with primary antibodies in ½ blocking buffer overnight at 4°C, followed by incubation with appropriate secondary antibodies (1:1000) in PBS at room temperature for 1 h. Nuclei were stained with Hoechst 33258 (Sigma).
Quantification of immunostained cell cultures and brain sections was performed as previously described (Wang et al., 2010). Briefly, cells grown on glass coverslips were analyzed with a Zeiss Axioplan2 microscope. For cell identity analysis, at least 300 cells from different fields were counted per condition and results from at least three independent experiments were analyzed together. For in utero electroporation, three or four anatomically matched sections per brain from at least three embryos from different mothers were imaged with a 20× objective on an Olympus IX81 fluorescence microscope equipped with a Hamamatsu C9100-13 EM-CCD camera and Okogawa CSU X1 spinning disk confocal scan head. Images were processed by using the Volocity software (PerkinElmer). Pax6, Tbr2, and Hoechst staining was used to define the ventricular zone (VZ), subventricular zone (SVZ), and cortical plate (CP).
Proximity ligation assays (PLA).
Immunocytochemistry was performed as above up to primary antibody incubation, which was performed at 4°C overnight. Coverslips were then washed 3 times for 5 min with HBSS. Proximity ligation assays were performed as previously described (Gallagher et al., 2015) with a DuoLink in situ Red Starter Kit Mouse/Rabbit (Sigma, DUO92101) following the manufacturer's instructions. Briefly, the coverslips were incubated with the secondary antibodies provided in the kit for 1 h, followed by ligation reaction for 30 min and signal amplification reaction for 1 h and 40 min. All incubation steps were performed in a humidified chamber at 37°C. Washes were performed between each step as per the manufacturer's instructions. Following signal amplification and final washes, coverslips were mounted with the DAPI-containing mounting medium provided in the kit.
FISH.
The single molecule FISH was performed as previously described (Yang et al., 2014) using the RNAscope kit (Advanced Cell Diagnostics), according to the manufacturer's instructions. Briefly, freshly dissected embryonic brains were fixed overnight at 4°C in RNase-free 4% PFA, cryopreserved overnight in 30% RNase-free sucrose and placed in OCT at −80°C overnight. Brains were cryosectioned coronally at 14 μm. Sections were postfixed with 4% PFA and washed with ethanol, followed by tissue pretreatment, probe hybridization, and signal amplification. Alternatively, cortical precursor cultures from E12-E13 cortices were maintained for 3 d before fixation, ethanol pretreatment, probe hybridization, and signal amplification. In both cases, positive staining was identified as punctate dots. For simultaneous immunodetection of a protein of interest after FISH, sections or cultures were blocked and incubated with the relevant primary antibody overnight at 4°C, followed by 1 h incubation with the appropriate Alexa secondary antibodies at room temperature before DAPI staining. Z-stacks of confocal images were taken with optical slice thickness of 0.1 μm; 200–800 mRNA granules in 80–300 Z-stacked images from random regions of the VZ/SVZ were used for 4E-T or Smaug2 colocalization quantification. Bright and clear mRNA granules that overlapped with immunostained 4E-T or Smaug2 granules were counted. Cortical precursor cultures were imaged with a 40× objective. Z-stacks of confocal images were taken with optical slice thickness of 0.1 μm and for each culture condition 20–30 random fields were imaged. For the current study, the following probes were used: nanos1 (NM_174421.3) catalog #431391, nanos2 (NM_194064.2) catalog # 450691, nanos3 (NM_194059.2) catalog #450721, and smaug2/samd4b (NM_175021.3) catalog #450731.
Antibodies.
The primary antibodies used were mouse anti-4E-T (Abnova, H00056,478-B01P, 1:2000), mouse anti-GFP (Invitrogen, GF28R, 1:1000), rabbit anti-GFP (Abcam, AB290, 1:2000), chicken anti-GFP (Abcam, AB13970, 1:2000), mouse anti-Ki67 (BD Biosciences PharMingen, 556003, 1:500), mouse anti-Satb2 (Abcam, AB51502, 1:400), mouse anti-βIII-tubulin (Covance, MMS-435P-250, 1:2000), rabbit anti-βIII-tubulin (Covance, PRB-435P, 1:2000), rabbit anti-Pax6 (Covance, PRB-278P, 1:1000), mouse anti-Pax6 (Millipore MAB5552, 1:1000), rabbit anti-Sox2 (Cell Signaling Technology, 3728, 1:1000), goat anti-Sox2 (Santa Cruz Biotechnology, sc17320, 1:500), rabbit anti-Tbr2 (Abcam, AB23345, 1:500), mouse anti-Dcp1 (Abnova, H00055802-M06, 1:2000), rabbit anti-ERK1/2 (Santa Cruz Biotechnology, SC93, 1:10,000), mouse anti-Flag (Origene, TA50011-100, 1:1000), rabbit anti-Nanos1 (Abcam, AB83417, 1:1000), rabbit anti-Smaug2 (Sigma, HPA016800-100UL, 1:1000). The Alexa488-, Alexa555-, and Alexa647-conjugated secondary antibodies were obtained from Invitrogen. HRP-conjugated goat anti-mouse or anti-rabbit secondary antibodies were purchased from Boehringer Mannheim, and HRP-conjugated chicken anti-goat secondary antibody was obtained from Millipore.
Coimmunoprecipitations.
Freshly dissected E12-E13 murine cortices were lysed with Gentle Lysis Buffer containing 25 mm Tris-HCl, pH 7.4, 2 mm EDTA, 1 mm EGTA, 10 mm NaCl, 0.5% Triton X-100, and 10% glycerol supplemented with the Complete Protease Inhibitor Tablets (Roche Applied Science) and 1 mm PMSF. Total protein concentration was determined with the Protein Quantitation kit (Abcam). Lysates were first precleared by incubating with Protein A/G magnetic beads (Millipore) for 1–2 h at 4°C, followed by incubation for 3 h with rabbit anti-Smaug2 or normal rabbit IgG (Santa Cruz Biotechnology) at 4°C, and a 1 h incubation with protein A/G magnetic beads at 4°C. Immunoprecipitates were washed three times with Gentle Lysis Buffer, boiled in 2× sample buffer with 1 mm DTT for 3 min, and analyzed with SDS-PAGE as described previously (Yang et al., 2014). Alternatively, freshly dissected E12-E13 murine cortices were dissociated and plated as described earlier and maintained in culture for 3 d, after which immunoprecipitation was performed as described above. The same procedure was followed for 4E-T immunoprecipitation; normal mouse IgG was used as control (Santa Cruz Biotechnology).
RNA immunoprecipitation.
Freshly dissected E12-E13 cortices were homogenized in Brain Extraction Buffer (Vessey et al., 2012) containing 25 mm HEPES, pH 7.3, 150 mm KCl, 8% glycerol, and 0.1% NP-40 supplemented with Complete Protease Inhibitor Tablets (Roche Applied Science), 1 mm PMSF, and RiboLock RNase Inhibitor (2 μl/ml, Thermo Scientific). A total of 1 mg of protein extract was precleared with 40 μl of 50% protein-A/G Sepharose beads (Sigma) for 1 h at 4°C; 2 μg of Smaug2 antibody or 4E-T antibody or 2 μg of the relevant nonspecific IgG was added to the precleared lysates and incubated at 4°C with rocking for 2–3 h. Antibody-bound protein was isolated by adding 40 μl of 50% protein-A/G Sepharose beads for 1 h at 4°C. Following 3 washes with Brain Extraction Buffer, mRNA was isolated with Trizol reagent following the instructions of the manufacturer.
RT-PCR.
mRNA from E12-E13 cortices or from RNA immunoprecipitation was isolated using the Trizol reagent (Invitrogen). cDNA was generated using the First Strand cDNA Synthesis Kit (Thermo Scientific) following the manufacturer's protocol and used for PCR. The annealing temperature of all primers was ∼60°C and amplified products were between 100 and 300 nt. All reactions were subjected to 35 cycles and products resolved on 2% agarose gels. Products were verified by sequence analysis. The primers used for RT-PCR were as follows: smaug2, forward 5′-CGAGGAGAACATCACCAGTTACC-3′ and reverse 5′-CGGAGGAGTTTCAGCACTTGCT-3′ (OriGene); smaug1, forward 5′-TGCGCTCTTCTCGCAGATGACT-3′ and reverse 5′-CCCTTTCCAAAGACTTCAGGAGG-3′ (OriGene); gapdh, forward 5′-GGGTGTGAACCACGAGAAATA-3′ and reverse 5′-CTGTGGTCATGAGCCCTTC-3′ (PrimerBank, Spandidos et al., 2010); nanos1 primer1, forward 5′-GTGTGTGTTTTGCCGGAAC-3′ and reverse 5′-CTAGCGCAGCTTCTTGCTG-3′, nanos1 primer2, forward 5′-GGAGCTTCAGGTGTGTGTGTT-3′ and reverse 5′-CTAGCGCAGCTTCTTGCTG-3′; nanos2, forward 5′-ACCCTGGATGTCTGCCTACCAT-3′ and reverse 5′-CACATAGTGCCTCAGGATGGGA-3′ (Origene); and nanos3, forward 5′-TCTGCAGGCAAAAAGCTGACCC-3′ and reverse 5′-GGGCTTCCTGCCACTTTTGGAA-3′ (Origene). For qRT-PCR, 10 μl PCR mixture containing FastStart DNA Master SYBR Green I (Roche Molecular Biochemicals) was prepared according to the manufacturer's instructions and loaded on to a 96 multiwell plate. The CFX96 thermocycler (Bio-Rad) was used with a protocol involving an initial activation cycle (2 min, 95°C), 45 cycles of denaturation (10 s, 95°C), annealing (20 s, 60°C), and elongation (20 s, 72°C). A single fluorescence reading was acquired at the end of each elongation step. A melting curve analysis cycle was performed after the PCR amplification. The primers used in qRT-PCR were nanos1, forward 5′- CTACACCACACACATCCTCAAGG-3′ and reverse 5′- GCACTTTGGAGAGCGGGCAATA-3′ (OriGene); and nanos2, and nanos3 (OriGene, same as above). All primers for qRT-PCR were validated in accordance with MIQE guidelines (Bustin et al., 2009).
Statistics.
All data are expressed as the mean ± SEM. Statistic analysis was performed with a two-tailed Student's t test or, where relevant, ANOVA with Tukey's post hoc multiple comparisons test unless otherwise indicated.
Results
Smaug2 is expressed in embryonic cortical precursors during development
To ask whether Smaug is expressed in neural precursor cells, we focused upon embryonic murine cortical radial glial precursors during the neurogenic period from embryonic day 12 (E12) until birth. RT-PCR analysis showed that the mRNAs encoding the mammalian Smaug homologs Smaug1 and Smaug2 were expressed in the embryonic cortex over this time period (Fig. 1A). Western blots confirmed that Smaug2 protein was also readily detectable (Fig. 1B). However, Smaug1 protein was almost absent at these early stages and was instead expressed in the adult cortex (Fig. 1C).
Figure 1.
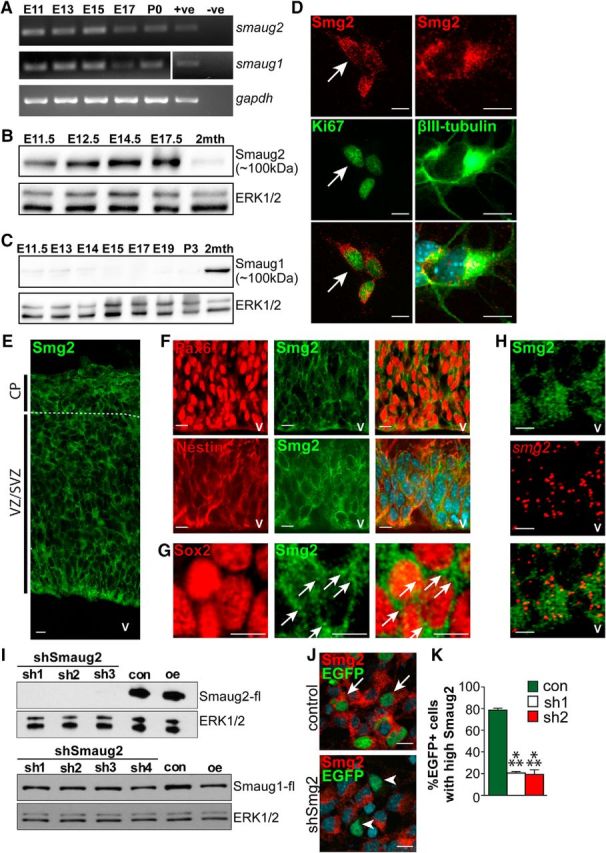
Smaug2, but not Smaug1, protein is expressed in apical precursors and newborn neurons during embryonic cortical neurogenesis. A, RT-PCR for smaug1 and smaug2 mRNAs in the developing murine cortex from embryonic days 11 to 17 (E11–E17) and at birth (P0). gapdh mRNA expression was monitored as a control. +ve, Sample with known expression of target mRNA and used as a positive control for the reaction; -ve, sample generated in the absence of reverse transcriptase. B, C, Western blots for Smaug2 (B) and Smaug1 (C) in E11.5 to 2-month-old (2mth) cortices. Blots were reprobed for ERK1/2 as a loading control. D, Images of cortical precursors isolated at E12.5, cultured for 3 d and immunostained for Smaug2 (red) and the proliferation marker Ki67 or the early neuronal marker βIII-tubulin (both green). Arrows indicate a double-labeled cell. Scale bar, 10 μm. E, Confocal image of a coronal E12.5 cortical section immunostained for Smaug2 (green). Boundaries of the VZ/SVZ and CP are denoted. v, Ventricle. Scale bar, 10 μm. F, G, Confocal images of the E12.5 cortical VZ/SVZ immunostained for Smaug2 (green) and the radial precursor marker Pax6 (F, red) or the neural precursor markers nestin (F, red) or Sox2 (G, red) at higher magnification. F, Bottom, Counterstained with Hoechst to highlight nuclei. Arrows highlight Smaug2-positive granules. Scale bars: F, 10 μm; G, 5 μm. H, Confocal images of the E12.5 cortical VZ immunostained for Smaug2 (green) and subjected to FISH for smaug2 mRNA (red). Bottom, Merge. v, Ventricle. Scale bar, 10 μm. I, Western blot analysis with anti-FLAG of HEK-293T cell lysates cotransfected with FLAG-tagged mouse Smaug2 or Smaug1 expression constructs and a control shRNA (con) or one of four different Smaug2 shRNAs (sh1, sh2, sh3, and sh4). Cells were transfected with the expression construct alone as a positive control (oe). Blots were reprobed with ERK1/2. J, Images of cultured precursors cotransfected with a nuclear EGFP construct and a control shRNA (top) or Smaug2 shRNA #2 (bottom, shSmg2), and immunostained 3 d later for Smaug2 (red) and EGFP (green). Arrows and arrowheads indicate EGFP-positive, Smaug2 positive cells and EGFP-positive, Smaug2-negative cells, respectively. Scale bar, 10 μm. K, Quantification of transfected precursors as shown in J transfected with Smaug2 shRNA1 or shRNA2 (sh1 or sh2) and analyzed for their relative levels of immunodetectable Smaug2. ***p < 0.001. n = 3 experiments. K, Statistics were performed with ANOVA and Tukey's post hoc multiple comparisons test. Error bars indicate SEM.
To determine whether Smaug2 was expressed in embryonic neural precursors and/or neurons, we examined E12.5 cortical cultures, which are comprised of proliferating cortical precursors that generate neurons in vitro. Smaug2 immunoreactivity was present in cycling precursors that were positive for the proliferation marker Ki67, and in newborn neurons expressing βIII-tubulin (Fig. 1D). In both cell types, Smaug2 was expressed in a punctate cytoplasmic fashion, consistent with localization in granule-like structures. Immunostaining of E12.5 embryonic cortical sections showed a similar pattern; Smaug2 was detectable in most cells in the precursor regions of the cortex, the VZ/SVZ, and in the CP, which contains newborn neurons (Fig. 1E). Double-labeling for the radial precursor marker Pax6 or the neural precursor markers Sox2 and nestin confirmed the presence of punctate Smaug2-positive granule-like structures in precursors in vivo (Fig. 1F,G). FISH confirmed that smaug2 mRNA was expressed in Smaug2-immunoreactive precursors in the VZ and SVZ (Fig. 1H).
Smaug2 regulates the genesis of cortical neurons
To understand the function of Smaug2 during cortical development, we performed acute knockdown experiments with shRNAs targeted to Smaug2. Initially, we tested the efficacy and specificity of these Smaug2 shRNAs by cotransfecting them into HEK-293T cells together with an expression vector for either mouse Smaug2 or mouse Smaug1. Western blot analysis 1 d later showed that the Smaug2 shRNAs efficiently knocked down Smaug2 expression but did not affect expression of Smaug1 (Fig. 1I). We showed that these shRNAs were equally effective in precursors by cotransfecting them into cultured E12.5 cortical precursors together with a nuclear EGFP plasmid. Immunostaining 3 d later showed that the Smaug2 shRNAs, but not a control shRNA, caused a robust decrease in the proportion of EGFP-positive cells expressing detectable Smaug2 (Fig. 1J,K).
We used these Smaug2 shRNAs to ask about a potential biological role for Smaug2, initially in cultured cortical precursors. E12.5 cortical cultures were cotransfected with one of the two Smaug2 shRNAs and a nuclear EGFP plasmid, and immunostained 3 d later for Pax6, Ki67, or βIII-tubulin (Fig. 2A). This analysis showed that the proportion of EGFP-positive and Pax6-positive proliferating precursors was significantly reduced by Smaug2 knockdown (Fig. 2B,C) while at the same time the proportion of EGFP-positive newborn neurons increased (Fig. 2D). These changes were not due to cell death because the proportion of cells with condensed, apoptotic nuclei was statistically similar between control and Smaug2 knockdown at days 1, 2 (p > 0.05 for all comparisons), and 3 (Fig. 2E) after transfection.
Figure 2.
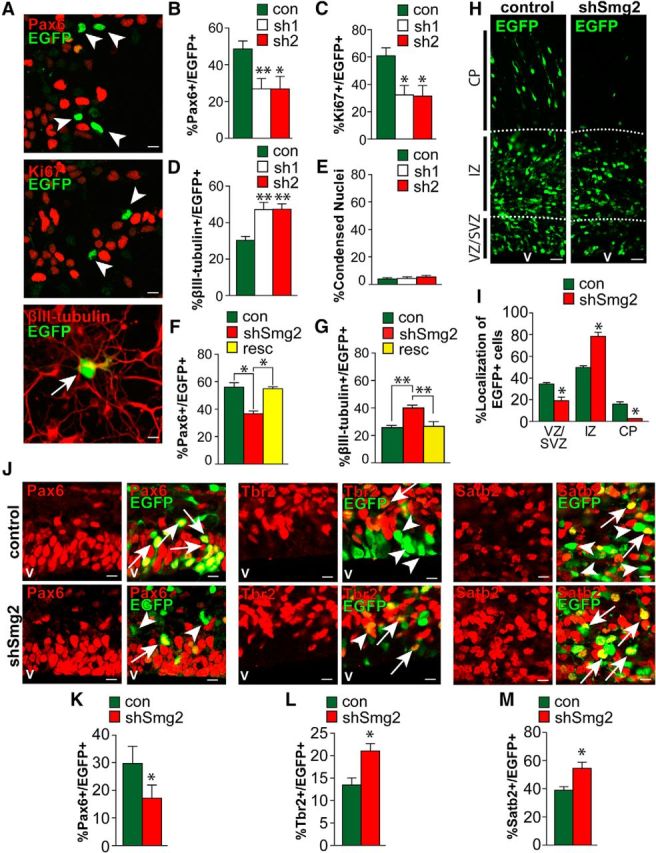
Smaug2 knockdown in culture and in vivo increases neurogenesis and depletes cycling precursors. A–E, Cultured cortical precursors were cotransfected with a nuclear EGFP construct and a control shRNA (con) or one of two Smaug2 shRNAs (sh1 and sh2), and analyzed by immunostaining 3 d later. A, Images of precursors transfected with Smaug2 shRNA #1 and immunostained for EGFP (green) and Pax6, Ki67, or βIII-tubulin (all red). Arrows and arrowheads indicate EGFP-positive, marker-positive cells and EGFP-positive, marker-negative cells, respectively. Scale bar, 10 μm. B–D, Quantification of cultures as in A for the percentage of EGFP-positive cells expressing Pax6 (B), Ki67 (C), or βIII-tubulin (D). *p < 0.05. **p < 0.01. n = 3 experiments. E, Quantification of condensed nuclei to assess cell death in cultures as in A. F, G, Cultured precursors were cotransfected with a nuclear EGFP construct and a control shRNA (con) or Smaug2 shRNA #2 (shSmg2) with or without an expression vector encoding an shRNA-resistant human Smaug2 (resc). Three days later, cultures were immunostained and quantified for the percentage of EGFP-positive cells expressing Pax6 (F) or βIII-tubulin (G). *p < 0.05. **p < 0.01. n = 3 experiments. H–M, E13/E14 murine cortices were coelectroporated with a nuclear EGFP construct together with control (con) or Smaug2 shRNA #2 (shSmg2), and coronal sections were analyzed 3 d later at E16/E17. H, Confocal images of electroporated cortices immunostained for EGFP (green). v, Ventricle. Scale bar, 30 μm. I, Quantification of sections as in H for the percentage of EGFP-positive cells located in the different cortical regions. *p < 0.05. n = 3 embryos each, at least 3 sections per embryo. J, Confocal images of the VZ/SVZ of electroporated sections immunostained for EGFP (green) and Pax6, the intermediate progenitor marker Tbr2, or the neuronal marker Satb2 (all red). Arrows indicate double-labeled cells. Arrowheads indicate EGFP-positive, marker-negative cells. Scale bar, 10 μm. K–M, Quantification of sections as in J for the percentage of EGFP-positive cells expressing Pax6 (K), Tbr2 (L), or Satb2 (M). *p < 0.05. n = 3 embryos each, at least 3 sections per embryo. B–G, Statistics were performed with ANOVA and Tukey's post hoc multiple comparisons test. I–M, Statistics were performed with Student's t test. Error bars indicate SEM.
These data indicate that, in culture, Smaug2 maintains the neural precursor state and prevents neuronal differentiation. To ensure the specificity of these shRNA-dependent phenotypes, we performed rescue experiments using a human Smaug2 construct that did not contain the sequences targeted by the shRNAs. We transfected cultured precursors with EGFP and Smaug2 shRNA with or without the human Smaug2 expression vector. Quantification of Pax6-positive precursors and βIII-tubulin-positive newborn neurons 3 d later showed that coincident expression of human Smaug2 rescued both the decrease in precursors and the increase in neurons caused by Smaug2 knockdown (Fig. 2F,G).
To ask whether Smaug2 was important for embryonic cortical development in vivo, we next used in utero electroporation to cotransfect E13/E14 cortical radial precursors that line the lateral ventricles with Smaug2 shRNA #2 and a nuclear EGFP expression plasmid. These transfected precursors generate neurons either directly or indirectly (the latter via intermediate progenitor transit amplifying cells), and the newborn neurons then migrate from the VZ/SVZ through the intermediate zone (IZ) to the CP. Three days after electroporation, coronal sections through the embryonic cortex were analyzed by immunostaining for EGFP (Fig. 2H). This analysis demonstrated that Smaug2 knockdown altered the location of transfected cells; fewer EGFP-positive cells were located in the VZ/SVZ and the CP, with coincidentally more in the IZ (Fig. 2I). We asked about the cellular basis of these alterations by immunostaining similar sections for EGFP and the radial precursor marker Pax6, the intermediate progenitor marker Tbr2, or the neuronal marker Satb2 (Fig. 2J). We chose Satb2 because our previous work showed that >90% of neurons born from cortical precursors over this timeframe express this marker (Tsui et al., 2013). Quantification demonstrated a significant decrease in the proportion of EGFP-positive, Pax6-positive precursors following Smaug2 knockdown (Fig. 2J,K) with a coincident increase in Tbr2-positive intermediate progenitors and Satb2-positive neurons (Fig. 2J,L,M). Thus, as seen in culture, Smaug2 is required in vivo to maintain Pax6-positive precursors and to prevent their differentiation into neurons.
Smaug2 is sufficient to maintain cortical precursors
The preceding data indicate that Smaug2 is necessary to promote Pax6-positive cortical precursor maintenance. To ask whether it is also sufficient, we overexpressed Smaug2. Initially, we performed this experiment in culture by cotransfecting E12.5 cortical precursors with a nuclear EGFP plasmid together with a murine Smaug2 expression construct. Immunostaining 3 d later demonstrated that Smaug2 overexpression increased the proportion of EGFP-positive, Ki67-positive proliferating precursors, and decreased the proportion of EGFP-positive, βIII-tubulin-positive newborn neurons (Fig. 3A–C). These data indicate that in culture Smaug2 is sufficient to promote precursor maintenance. To confirm this conclusion, we performed clonal analysis using the piggybac (PB) transposon system, which indelibly marks precursors and their progeny (Gallagher et al., 2013; Tsui et al., 2013). E12.5 cortical precursors were cotransfected at low efficiency with Smaug2 or control expression vectors, together with plasmids encoding PB transposase and a PB EGFP reporter. Three days later, cultures were immunostained for EGFP. Smaug2 overexpression significantly increased the number of multicellular clones (Fig. 3D).
Figure 3.

Smaug2 overexpression in vitro and in vivo is sufficient to enhance cortical precursor self-renewal. A–C, Cultured E12.5 precursors were cotransfected with a nuclear EGFP construct and control (con) or murine Smaug2 (Smg2-OE) expression vectors, and analyzed by immunostaining 3 d later. A, Images of transfected precursors immunostained for EGFP (green) and Ki67 or βIII-tubulin (both red). Arrows and arrowheads indicate EGFP-positive, marker-positive cells and EGFP-positive, marker-negative cells, respectively. Scale bar, 10 μm. B, C, Quantification of cultures as in A for the percentage of EGFP-positive cells expressing Ki67 (B) or βIII-tubulin (C). **p < 0.01. n = 3 experiments. D, Cortical precursor cultures were cotransfected with the piggybac EGFP labeling system plus control (con) or murine Smaug2 (Smg-OE) expression vectors. Cultures were immunostained for EGFP 3 d later and clone size was scored. *p < 0.05. n = 3 experiments. E–J, E13/E14 murine cortices were coelectroporated with a nuclear EGFP construct and control (con) or mouse Smaug2 (Smg2-OE) expression vectors and coronal cortical sections were analyzed 3 d later at E16/E17. E, Confocal images of electroporated sections immunostained for EGFP (green). v, Ventricle. Scale bar, 30 μm. F, Quantification of sections similar to those in E for the percentage of EGFP-positive cells located in the different cortical regions. *p < 0.05. ns, Nonsignificant. n = 3 embryos each, at least 3 sections per embryo. G, Confocal images of the VZ/SVZ (two top rows) or CP (bottom row) of electroporated sections immunostained for EGFP (green) and Pax6, Tbr2, or Satb2 (all red). Arrows indicate double-labeled cells. Arrowheads indicate EGFP-positive, marker-negative cells. v, Ventricle. Scale bar, 10 μm. H–J, Quantification of sections as in G for the percentage of EGFP-positive cells that were positive for Pax6 (H), Tbr2 (I), or Satb2 (J). *p < 0.05. **p < 0.01. ***p < 0.001. n = 3 embryos each, at least 3 sections per embryo. Statistics were performed with Student's t test. Error bars indicate SEM.
To ask whether Smaug2 was also sufficient to promote precursor maintenance in vivo, we electroporated E13/E14 cortices with an EGFP plasmid and the Smaug2 expression construct, and analyzed coronal cortical sections 3 d later. Overexpression of Smaug2 caused a significant increase in the proportion of EGFP-positive cells in the VZ/SVZ (Fig. 3E,F). Immunostaining demonstrated that this increase in electroporated cells in the VZ/SVZ was due to an increase in EGFP-positive, Pax6-positive precursors (Fig. 3G,H). The increase in precursors occurred at the expense of differentiation because there was a coincident decrease in EGFP-positive, Tbr2-positive intermediate progenitors and Satb2-positive neurons (Fig. 3G,I,J). Thus, Smaug2 is sufficient to maintain Pax6-positive precursors and to repress their differentiation into neurons.
The mRNA encoding Nanos1, but not Nanos2 or Nanos3, is a target of Smaug2 in the embryonic cortex
In Drosophila, Smaug mediates its embryonic function, in part, by binding and repressing the translation of nanos mRNA, which itself encodes a repressive RNA-binding protein. In mammals, there are three Nanos homologs, Nanos1, Nanos2, and Nanos3 (Jaruzelska et al., 2003; Tsuda et al., 2003; Lolicato et al., 2008), and RT-PCR analysis showed that mRNAs encoding all three of them were expressed in the embryonic cortex from E11 to birth (Fig. 4A). We asked whether any of these nanos mRNAs might be Smaug2 binding targets by searching for potential SREs using the consensus loop sequence of CNGGN(0–3) flanked by nucleotides that have the potential to form a loop on a 4 nt nonspecific stem (Aviv et al., 2003, 2006). These analyses showed that nanos1 mRNA contained five potential SREs (Fig. 4B) with the following sequences: #1, cguccgggggcg starting at base 71; #2, gggcccgggccc starting at base 472; #3, acggccgggugcugu starting at base 911; #4, gcuccaggucugagu starting at base 3029; and #5, ggcacagguugugcu starting at base 3755. In contrast, neither nanos2 nor nanos3 mRNAs contained SREs. A similar result was obtained when we applied a computational approach that includes a thermodynamic-based calculation of the likelihood that an SRE will fold properly, to generate an SRE score for a transcript (Chen et al., 2014a). nanos1 mRNA had an SRE score of 88, similar to the scores for Drosophila Smaug's two best-characterized target mRNAs hsp83 and nanos, which have SRE scores of 70 and 80, respectively. In contrast, nanos2 and nanos3 had SRE scores of 1.9 and 22, respectively.
Figure 4.
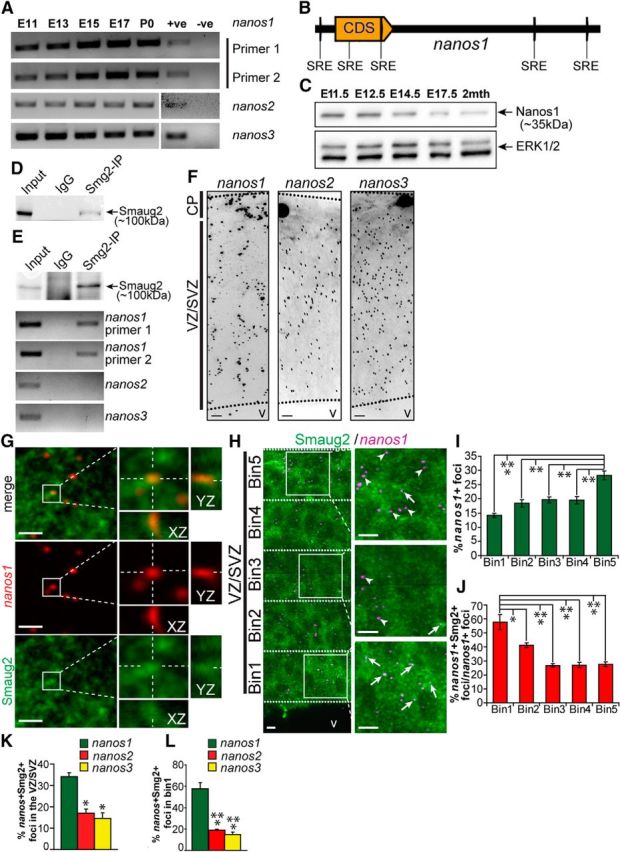
nanos1 mRNA is a Smaug2 target in embryonic cortical precursors. A, RT-PCR for nanos1, nanos2, and nanos3 mRNAs in murine cortices from E11 to birth (P0). nanos1 mRNA expression was detected using two different primer sets. PCR products were sequenced to confirm specificity. +ve, Sample with known expression of target mRNA and used as a positive control for the reaction; -ve, sample generated in the absence of reverse transcriptase. B, Schematic of SREs in the nanos1 mRNA transcript. Yellow arrow labeled CDS represents the protein-coding region. C, Western blot analysis for Nanos1 in E11.5 to 2-month-old cortices. The blot was reprobed for ERK1/2 as a loading control. D, Western blot of HEK-293T cells transfected with a Flag-tagged mouse Smaug2 construct and immunoprecipitated with anti-Smaug2 or with control nonspecific rabbit IgG, probed with antibodies for Smaug2. As a control, 10% of the input homogenate was loaded. E, Western blot (top) of E12.5 cortical lysates immunoprecipitated with the same Smaug2 antibody as in D or with control, nonspecific rabbit IgG and probed with anti-Smaug2. As a positive control, 10% of the input homogenate was loaded. Similar immunoprecipitates were generated in parallel, mRNA was extracted, and the samples were analyzed for nanos1, nanos2, and nanos3 mRNAs using RT-PCR (second to bottom panels). F, Confocal images of FISH for nanos1 (left), nanos2 (center), and nanos3 (right) mRNAs (black granules) in coronal sections of the E12.5 cortex. v, Ventricle. Scale bar, 10 μm. G, Higher-magnification confocal images of the VZ/SVZ of an E13.5 cortical section showing FISH for nanos1 mRNA (red) and immunostaining for Smaug2 (green). Top, Merge. Boxed regions are shown at higher magnification in the right panels, which also show colocalization of Smaug2 and nanos1 mRNA on the z-axis (XZ and YZ), as indicated by the hatched white lines. Scale bar, 10 μm. H, Confocal images of the E12.5 cortex showing FISH for nanos1 mRNA (magenta) and immunostaining for Smaug2 (green). The VZ/SVZ is divided into five bins of identical width, as denoted by the hatched white lines, and boxed regions within some of these bins are shown at higher magnification in the right panels. Arrows indicate foci with colocalized nanos1 mRNA and Smaug2. Arrowheads indicate foci with only nanos1 mRNA. v, Ventricle. Scale bar, 10 μm. I, J, Quantification of sections similar to that shown in H for the distribution of total nanos1 mRNA-positive foci (I) and the relative proportion of nanos1 mRNA-positive foci that colocalize with Smaug2 in each bin (J). *p < 0.05. **p < 0.01. ***p < 0.001. n = 3. K, L, Quantification of sections similar to those shown in H for the proportion of nanos1, nanos2, or nanos3 mRNA foci that colocalize with Smaug2 across the entire E12.5 VZ/SVZ (K) or only in Bin1 (L), the apical-most region of the VZ. *p < 0.05. ***p < 0.001. n = 3. Statistics were performed with ANOVA and Tukey's post hoc multiple comparisons test. Error bars indicate SEM.
We therefore focused on nanos1 mRNA as a potential Smaug2 binding target. Western blot analysis showed that Nanos1 protein, like its mRNA, was expressed in the embryonic cortex from E11.5 through to adulthood (Fig. 4C). We asked whether nanos1 mRNA copurified with Smaug2 in the embryonic cortex. Initially, we confirmed that the Smaug2 antibody was able to immunoprecipitate FLAG-tagged overexpressed Smaug2 from transfected HEK-293T cells (Fig. 4D). We then used this antibody to immunoprecipitate Smaug2 from the embryonic cortex at E12.5 (Fig. 4E). RT-PCR analysis showed that nanos1 mRNA was present in the Smaug2 immunoprecipitates, but not in control immunoprecipitates (Fig. 4E). In addition, as predicted, neither nanos2 nor nanos3 mRNAs were present in the Smaug2 immunoprecipitates (Fig. 4E).
These data indicate that nanos1 mRNA, but not nanos2 or nanos3 mRNAs, is associated with Smaug2 in the embryonic cortex. To determine whether this occurs in precursors, we performed histological analyses, comparing nanos1, nanos2, and nanos3 mRNAs. FISH demonstrated that all three mRNAs were expressed throughout the E12.5 cortex (Fig. 4F). To ask whether nanos1 mRNA and Smaug2 colocalized during this period of development, we combined FISH with immunocytochemistry for Smaug2. This analysis showed that nanos1 mRNA partially colocalized with Smaug2-positive foci (Fig. 4G). This localization was not exclusive; Smaug2 was present in many granules that did not contain nanos1 mRNA, and the converse was also true, with nanos1 mRNA present in granules that did not contain Smaug2. We confirmed this colocalization with Z-stack analysis (Fig. 4G).
Next, we quantified the nanos1 mRNA/Smaug2 foci in the VZ/SVZ of E12.5 cortical sections (Fig. 4H–J); 34 ± 2% of nanos1 mRNA foci colocalized with Smaug2. These complexes were not, however, evenly distributed across the VZ/SVZ. Of all of the nanos1 mRNA-positive foci, the highest proportion was found in the basal-most region of the SVZ, at the border of the newly formed cortical plate (Fig. 4H,I; Bin 5). In contrast, the highest percentage of nanos1 mRNA-positive foci that were colocalized with Smaug2 was in the apical-most part of the VZ (Fig. 4H,J; Bin 1). In this region, which is predominantly composed of radial precursors, almost 60% of nanos1 mRNA foci were colocalized with Smaug2 (Fig. 4J). Thus, nanos1 mRNA is physically associated with and colocalizes with Smaug2, particularly in apical precursor cells.
As a control for the specificity of this analysis, we performed similar studies for nanos2 and nanos3 mRNAs, quantifying the proportion of mRNA-positive foci that colocalized with Smaug2 in E12.5 cortical sections. While ∼35% of total nanos1 mRNA foci were colocalized with Smaug2, this was decreased to ∼15% for nanos2 and nanos3 mRNAs (Fig. 4K). Moreover, when only the apical-most radial precursor region of the VZ was considered (Bin 1), ∼60% of nanos1 mRNA foci versus 15%–20% of nanos2 and nanos3 mRNA foci were colocalized with Smaug2 (Fig. 4L).
Nanos1 promotes the genesis of neurons from cortical precursors
These findings define a nanos1 mRNA/Smaug2 complex in embryonic cortical precursors. We therefore next asked about a potential biological role for Nanos1 in cortical development. To do this, we generated a Nanos1 shRNA and verified that it knocked down Nanos1 but not Nanos2 or Nanos3 when cotransfected with the relevant expression constructs into HEK-293T cells (Fig. 5A). We then used this shRNA to assess a functional role for Nanos1 by electroporating it into the E13/E14 embryonic cortex. Analysis of coronal cortical sections 3 d after electroporation demonstrated that Nanos1 knockdown resulted in an accumulation of transfected EGFP-positive cells in the VZ/SVZ, and fewer cells in the CP (Fig. 5B,C). Consistent with this increase in EGFP-positive cells in the precursor zones, immunostaining demonstrated an increase in transfected Pax6-positive and Ki67-positive cortical precursors following Nanos1 knockdown (Fig. 5D–F). At the same time, EGFP-positive, Tbr2-positive intermediate progenitors and Satb2-positive neurons were decreased by Nanos1 knockdown (Fig. 5D,G,H). To verify the specificity of these phenotypes, we performed rescue experiments with a human Nanos1 expression vector that was resistant to the shRNA. Analysis 3 d later showed that coincident expression of Nanos1 rescued the perturbations in location of EGFP-positive cells observed following Nanos1 knockdown (Fig. 5I), as well as the increase in Pax6-positive precursors and decrease in Satb2-positive neurons (Fig. 5J,K), thereby confirming the specificity of the knockdown phenotypes.
Figure 5.

Nanos1 is necessary and sufficient to promote neurogenesis in vivo. A, Western blots of HEK-293T cell lysates cotransfected with murine Nanos1 or Flag-tagged murine Nanos2 or Nanos3 expression constructs and a control shRNA (Con) or a Nanos1 shRNA (shNos1) and probed with anti-Nanos1 or anti-Flag, as indicated. The blots were reprobed with ERK1/2 as a loading control. B–H, E13/E14 murine cortices were coelectroporated with a nuclear EGFP construct, and either a control (con) or Nanos1 shRNA (shNos1) and coronal sections were analyzed 3 d later at E16/E17. B, Images of electroporated sections immunostained for EGFP (green). v, Ventricle. Scale bar, 10 μm. C, Quantification of sections similar to those in B for the percentage of EGFP-positive cells located in the different cortical regions. **p < 0.01. n = 3 embryos each, at least 3 sections per embryo. D, Confocal micrographs of the VZ/SVZ (three top rows) or CP (bottom row) of electroporated sections immunostained for EGFP (green) and Pax6, Ki67, Tbr2, or Satb2 (all red). Arrows indicate double-labeled cells. v, Ventricle. Scale bar, 10 μm. E–H, Quantification of sections similar to those in D for the percentage of EGFP-positive cells that expressed Pax6 (E), Ki67 (F), Tbr2 (G), or Satb2 (H). **p < 0.01. ***p < 0.001. n = 3 embryos each, at least 3 sections per embryo. I–K, E13/E14 cortices were coelectroporated with a nuclear EGFP construct and a control (con) or Nanos1 shRNA (shNos1) ± an shRNA-resistant human Nanos1 expression vector (resc) and coronal sections were analyzed 3 d later at E16/E17. I, Images of electroporated sections immunostained for EGFP (green). v, Ventricle. Scale bar, 10 μm. J, K, Sections similar to those in I were immunostained for EGFP and Pax6 or Satb2 and the proportion of EGFP-positive cells that were also positive for the marker was quantified. **p < 0.01. ***p < 0.001. n = 3 embryos each, at least 3 sections per embryo. L–R, E13/E14 cortices were coelectroporated with a nuclear EGFP construct and either a control (con) or murine Nanos1 (Nos1-OE) expression vector, and coronal sections were analyzed 3 d later at E16/E17. L, Images of electroporated sections immunostained for EGFP (green). v, Ventricle. Scale bar, 10 μm. M, Quantification of sections as in L for the percentage of EGFP-positive cells located in the different cortical regions. *p < 0.05. ns, Nonsignificant. n = 3 embryos each, at least 3 sections per embryo. N, Confocal images of the VZ/SVZ (top three rows) or CP (bottom row) of electroporated sections similar to those in L immunostained for EGFP (green) and Pax6, Ki67, Tbr2, or Satb2 (all red). Arrows indicate double-labeled cells. Arrowheads indicate EGFP-positive, marker-negative cells. v, Ventricle. Scale bar, 10 μm. O–R, Quantification of sections as in N for the percentage of EGFP-positive cells that were also positive for Pax6 (O), Ki67 (P), Tbr2 (Q), or Satb2 (R). *p < 0.05. **p < 0.01. ***p < 0.001. n = 3 embryos each, at least 3 sections per embryo. J, K, Statistics were performed with ANOVA and Tukey's post hoc multiple comparisons test. All other panels, Statistics were performed with Student's t test. Error bars indicate SEM.
These data indicate that Nanos1 normally functions to promote the genesis of neurons from precursors. We therefore asked whether ectopic expression of Nanos1 was sufficient to increase neurogenesis. We coelectroporated plasmids encoding murine Nanos1 and nuclear-localized EGFP into the E13/E14 cortex and analyzed coronal sections 3 d later. Immunostaining showed that EGFP-positive cells were mislocalized following Nanos1 overexpression, with a significant decrease in transfected cells in the VZ/SVZ (Fig. 5L,M). This loss of transfected cells in the precursor zones was due to a decrease in Ki67-positive, Pax6-positive precursors and a coincident increase in Tbr2-positive intermediate progenitors and Satb2-positive transfected neurons (Fig. 5N–R). Thus, Nanos1 depletion and overexpression result in reciprocal phenotypes: loss of Nanos1 results in increased apical precursors and fewer neurons, whereas overexpression of Nanos1 results in fewer apical precursors and more neurons.
Smaug2 and nanos1 mRNA are present in RNP granules containing the repressors Dcp1 and 4E-T
These data are consistent with the hypothesis that Smaug2 and Nanos1 mediate opposing functions during the precursor to neuron transition, and that Smaug2 maintains cortical precursors in a stem cell state, perhaps by associating with nanos1 mRNA. In this regard, we (Yang et al., 2014) recently identified a repressive P-body-like RNA granule in embryonic cortical precursors involving the eIF4E-binding protein 4E-T, which is distantly related to Drosophila Cup, an eIF4E-binding protein that interacts with Smaug and contributes to Smaug-mediated translational repression (Nelson et al., 2004). We therefore asked whether the Smaug2/nanos1 mRNA complex might be associated with the 4E-T repression complex. Initially, we asked whether Smaug2 was associated with 4E-T in the embryonic cortex. We immunoprecipitated endogenous Smaug2 from cultured cortical precursors and found that 4E-T coimmunoprecipitated with Smaug2 (Fig. 6A). This association was confirmed by immunoprecipitating 4E-T and showing coimmunoprecipitation of Smaug2 (Fig. 6B). We further assessed association between these two proteins using two additional approaches. First, we immunostained cultured cortical precursors. As we previously reported (Yang et al., 2014), much of the 4E-T immunoreactivity was present in fairly large, punctate cytoplasmic granules (Fig. 6C). Many of these particles were also positive for Smaug2, although much of the Smaug2 was also present in smaller cytoplasmic puncta (Fig. 6C). Second, we used the PLA, which is based upon antibodies binding to protein targets that are within 30–40 nm of each other (Weibrecht et al., 2010). This analysis showed that Smaug2 and 4E-T interacted in multiple bright cytoplasmic puncta in cortical precursors (Fig. 6D). No or very few puncta were observed when we performed similar experiments with antibodies for 4E-T or Smaug2 and proteins, such as Tbr2, Neuropeptide Y, and Pax6 (data not shown).
Figure 6.

Smaug2 and nanos1 mRNA are associated with 4E-T in a P-Body-like granule in Pax6-positive apical precursors. A, Western blot analysis for Smaug2 (Smg2) and 4E-T in lysates of E12.5 cortical precursors cultured for 3 d and immunoprecipitated with anti-Smaug2 or with control, nonspecific rabbit IgG. As a positive control, 10% of the input homogenate was loaded. B, Western blot analysis for Smaug2 and 4E-T in lysates of E12.5 cortical precursors cultured for 3 d and immunoprecipitated with anti-4E-T or with control, nonspecific mouse IgG. As a positive control, 10% of the input homogenate was loaded. C, Confocal images of E12.5 cortical precursors cultured for 3 d and immunostained for Smaug2 (green) and 4E-T (magenta). Cultures were also counterstained with Hoechst (blue). Top, Boxed areas are shown at higher magnification in the bottom panels. Arrows indicate granules that are positive for both Smaug2 and 4E-T. Scale bar, 5 μm. D, Confocal images of E12.5 3 d cortical precursor cultures after the PLA with Smaug2 and 4E-T antibodies. Cultures were also counterstained with Hoechst (blue). Left, Boxed areas are shown at higher magnification to the right. Scale bar, 10 μm. E, Confocal images of E12.5 cortical precursors cultured for 3 d and immunostained for Smaug2 (red) and Dcp1 (green). Cultures were also counterstained with Hoechst (blue). Top, Boxed areas are shown at higher magnification in the bottom panels. Arrows indicate granules that are double labeled for Smaug2 and Dcp1. Scale bar, 10 μm. F, Confocal images of cortical precursor cultures after PLA with Smaug2 and Dcp1 antibodies. Cultures were also counterstained with Hoechst (blue). Left, Boxed areas are shown at higher magnification on the right. Scale bar, 10 μm. G, RT-PCR analysis for nanos1 mRNA in 4E-T immunoprecipitates (4E-T IP) from the E12.5 cortex. As a control, similar lysates were immunoprecipitated with a control, nonspecific mouse IgG (IgG). H, qRT-PCR analysis for nanos1 mRNA enrichment in multiple independent 4E-T immunoprecipitates from the E12.5 cortex, in comparison with control IgG immunoprecipitates. I, Confocal images of E12.5 cortical precursors cultured for 3 d and analyzed by FISH for nanos1 mRNA (red or magenta) and immunostaining for 4E-T or Smaug2 (both green). Cultures were also counterstained with Hoechst (blue). Arrows and arrowheads indicate nanos1 mRNA-positive foci that are or are not positive for the relevant protein, respectively. Scale bar, 5 μm. J, Quantification of cultures as in I for the percentage of total nanos1 mRNA-positive foci that also colocalized with Smaug2 (Smg2) or 4E-T alone, or with both together. *p < 0.05. **p < 0.01. ***p < 0.01. n = 3. K, Confocal images of the E12.5 cortical VZ immunostained with 4E-T (green) and subjected to FISH (magenta) with a nanos1 mRNA probe shown at low magnification (left) and high magnification (right). Cell nuclei were counterstained with Hoechst (blue). Bottom, Merge. Left, Boxed regions are shown at high magnification to the right. Arrows indicate foci positive for both nanos1 mRNA and 4E-T. Arrowheads indicate nanos1 mRNA foci that are negative for 4E-T. v, Ventricle. Scale bar, 10 μm. L, Quantification of sections similar to that shown in K for the relative proportion of nanos1 mRNA-positive foci that colocalized with 4E-T in each bin of the VZ/SVZ, as defined in Figure 4H. **p < 0.01. n = 3. M, N, Quantification of sections similar to those shown in K for the proportion of nanos1, nanos2, or nanos3 mRNA foci that colocalized with Smaug2 across the entire E12.5 VZ/SVZ (M) or only in Bin1 (N), the apical-most region of the VZ. *p < 0.05. n = 3. Statistics were performed with ANOVA and Tukey's post hoc multiple comparisons test. Error bars indicate SEM.
We previously showed that these large 4E-T-positive granules in cortical precursors contained other P-body proteins (Yang et al., 2014). Double-label immunostaining showed that Smaug2 was indeed colocalized with another P-body protein, Dcp1, in large foci (Fig. 6E). Moreover, PLA with antibodies for Dcp1 and Smaug2 showed multiple bright cytoplasmic foci (Fig. 6F), consistent with this colocalization.
These data are consistent with the idea that Smaug2 is associated, in part, with 4E-T-containing, P-body-like repressive granules in cortical precursors. We therefore asked whether nanos1 mRNA was also present in these granules. Immunoprecipitation of 4E-T from the E12.5 cortex followed by qRT-PCR demonstrated that nanos1 mRNA was enriched in 4E-T immunoprecipitates relative to controls (Fig. 6G,H). We then asked whether nanos1 mRNA, Smaug2, and 4E-T were all colocalized, performing FISH and immunostaining on cultured cortical precursors. This analysis showed that almost 70% of nanos1 mRNA was colocalized with 4E-T in large, P-body-like granules (Fig. 6I,J). Approximately 45% of these also contained Smaug2 protein, and almost all of the Smaug2/nanos1 mRNA complexes were also associated with 4E-T (Fig. 6I,J). In contrast, only ∼20% of nanos1 mRNA was not associated with either protein.
We also asked whether nanos1 mRNA was colocalized with 4E-T in vivo, performing FISH and immunostaining on E12.5 cortical sections (Fig. 6K). For comparison, we performed similar studies with nanos2 and nanos3 mRNAs. Approximately 40% of nanos1 mRNA foci colocalized with 4E-T in the VZ/SVZ (Fig. 6L,M). These complexes were not, however, evenly distributed, and the highest percentage, ∼55%, were in the apical-most part of the VZ (Fig. 6L,N; Bin 1). In contrast, many fewer nanos2 and nanos3 mRNA foci were colocalized with 4E-T in the entire VZ/SVZ (Fig. 6M) or in the apical-most region of the VZ (Bin 1; Fig. 6N).
Enhanced neurogenesis following Smaug2 knockdown is caused by derepression of nanos1 mRNA translation
Together, these data suggest that Smaug2 binds and represses nanos1 mRNA in cortical apical precursors and that disruption of this complex might promote neurogenesis by causing aberrant Nanos1 translation. To test this hypothesis, we in utero electroporated E13/E14 cortices with Smaug2 shRNA and nuclear EGFP, and immunostained cortices 3 d later for Nanos1 protein. In control cortices, Nanos1 immunoreactivity was most robust in the CP in newborn neurons (Fig. 7A), although some scattered positive cells with cytoplasmic staining were also observed in the VZ/SVZ. Quantification showed that only ∼10% of EGFP-positive control cells expressed detectable Nanos1 3 d after electroporation (Fig. 7B,C). In contrast, knockdown of Smaug2 increased the proportion of EGFP-positive, Nanos1-positive cells to almost 30% (Fig. 7B,C), consistent with derepression of nanos1 mRNA.
Figure 7.
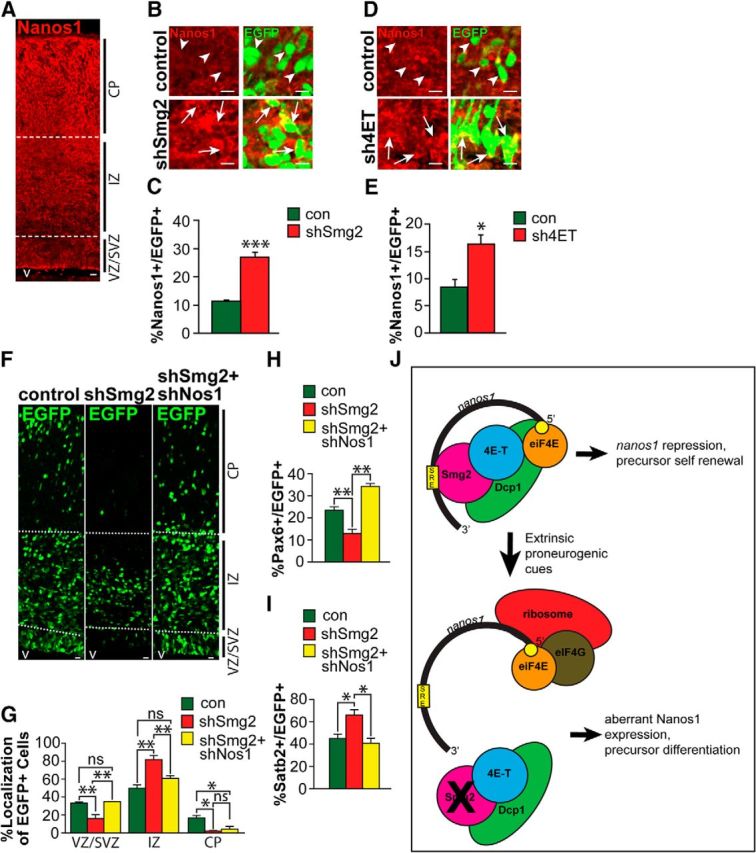
Knockdown of Smaug2 or 4E-T causes aberrant Nanos1 expression, and this is responsible for the Smaug2 knockdown-mediated increase in neurogenesis. A, Nanos1 immunoreactivity in a coronal section of the E16/E17 cortex. v, Ventricle. Scale bar, 10 μm. B, Confocal images of cells at the border of the SVZ and the IZ of E16/E17 murine cortices that were coelectroporated 3 d earlier with a nuclear EGFP construct and control (con) or Smaug2 (shSmg2) shRNAs. Sections were immunostained for EGFP (green) and Nanos1 (red). Arrows and arrowheads indicate EGFP-positive cells that do or do not express Nanos1, respectively. Scale bar, 10 μm. C, Quantification of the proportion of EGFP-positive cells expressing detectable Nanos1 in sections similar to those in B. ***p < 0.001. n = 3 embryos each, at least 3 sections per embryo. D, Confocal images of cells at the border of the SVZ and the IZ of E15/E16 murine cortices that were coelectroporated 2 d earlier with a nuclear EGFP construct and control (con) or 4E-T (sh4ET) shRNAs. Sections were immunostained for EGFP (green) and Nanos1 (red). Arrows and arrowheads indicate EGFP-positive cells that do or do not express Nanos1, respectively. Scale bar, 10 μm. E, Quantification of the proportion of EGFP-positive cells expressing detectable Nanos1 in sections similar to those in D. *p < 0.05. n = 3 embryos each, at least 3 sections per embryo. F–I, E13/E14 cortices were coelectroporated with a nuclear EGFP construct and control (con) or Smaug2 shRNA (shSmg2) ± Nanos1 shRNA (shNos1), and coronal cortical sections were analyzed 3 d later at E16/E17. F, Images of electroporated sections immunostained for EGFP (green). v, Ventricle. Scale bar, 10 μm. G, Quantification of sections similar to those in F for the percentage of EGFP-positive cells located in the different cortical regions. *p < 0.05. **p < 0.01. ns, Nonsignificant. n = 3 embryos each, at least 3 sections per embryo. H, I, Quantification of EGFP-positive, marker-positive cells in sections as in F immunostained for EGFP and either Pax6 (H) or Satb2 (I). *p < 0.05. **p < 0.01. n = 3 embryos each, at least 3 sections per embryo. J, Schematic showing the proposed repressive complex involving Smg2, 4E-T, Dcp1, and nanos1 mRNA (top). When the complex is disrupted, either by environmental signals or by knockdown of complex components such as Smaug2, this causes aberrant translation of Nanos1, thereby promoting neurogenesis (bottom). G–I, Statistics were performed with ANOVA and Tukey's post hoc multiple comparisons test. Other panels, Statistics were performed with Student's t test. Error bars indicate SEM.
We performed similar experiments knocking down 4E-T in the E13/E14 cortex with an shRNA that we previously characterized (Yang et al., 2014), and then analyzing Nanos1 protein-positive cells 2 d later. This analysis showed that the proportion of EGFP-positive cells expressing detectable Nanos1 doubled following 4E-T knockdown (Fig. 7D,E). These findings are consistent with the idea that a Smaug2/4E-T complex represses nanos1 mRNA translation in apical precursors.
Finally, we asked whether the enhanced neurogenesis observed following Smaug2 knockdown might be due to this aberrant derepression of Nanos1 translation. To do this, we electroporated E13/E14 cortices with nuclear EGFP and Smaug2 shRNA, either with or without Nanos1 shRNA. Analysis 3 d later demonstrated that knockdown of Nanos1 substantially rescued the aberrant distribution of EGFP-positive cells that occurred following Smaug2 knockdown (Fig. 7F,G) while also rescuing the decrease in Pax6-positive precursors and the increase in Satb2-positive neurons (Fig. 7H,I). These results are consistent with a rescue of the Smaug2 knockdown phenotype by knocking down Nanos1, although we cannot rule out the possibility that nanos1 knockdown is genetically dominant with regard to cellular phenotype. Together, these findings support a model where Smaug2-mediated translational repression regulates expression of Nanos1 (Fig. 7J) and, in doing so, ensures appropriate generation of neurons during cortical development.
Discussion
The mechanisms regulating genesis of appropriate numbers and types of neurons are a key issue in mammalian development. While transcriptional mechanisms determining neurogenesis have been well studied (Miller and Gauthier, 2007; Imayoshi and Kageyama, 2014), little is known about post-transcriptional regulatory mechanisms. Here, we identify a key role for translational repression, showing that two RNA-binding proteins, Smaug2 and Nanos1, regulate the balance of self-renewal versus differentiation of embryonic neural precursors and, in doing so, determine the timing and numbers of neurons that are generated. We show that these two repressive proteins do this by functioning in opposition, with Smaug2 maintaining the precursor state and inhibiting differentiation, and Nanos1 promoting differentiation and depleting precursors. Moreover, we show that these antagonistic activities are coordinated, with Smaug2 associating with and silencing nanos1 mRNA, potentially by localizing it to P-body-like granules in association with the translational repressor 4E-T.
Based upon these findings, we propose a translational repression “switch” model for the precursor to neuron transition. In this model, developing neural precursors are transcriptionally primed to generate neurons, but Smaug2 and 4E-T-dependent mRNA repression/silencing maintains them in a stem cell state. In this model, extrinsic proneurogenic cues would disrupt these repressive complexes, thereby releasing the relevant mRNAs and allowing for rapid, precise, and coordinated translation of proteins that promote neurogenesis. This environmentally driven dissociation of Smaug2 repressive complexes would also derepress nanos1 mRNA, and the newly translated Nanos1 would then act to repress mRNAs associated with and necessary for the stem cell state. Thus, the RNA-binding proteins Smaug2 and Nanos1 act as a “switch”: when Smaug2 is active or “turned on,” this keeps Nanos1 “turned off,” thereby maintaining translation of precursor proteins and silencing neuronal translation. However, when Smaug2 is turned off, this turns Nanos1 on, allowing it to silence translation of precursor proteins. Coincident with this Nanos1-dependent silencing of the precursor state, other derepressed proteins, such as the neurogenic bHLHs (Yang et al., 2014), would promote establishment of a neuronal phenotype.
One attractive feature of this model is that it would ensure that cells did not become “confused” by adopting a neuronal phenotype while at the same time maintaining features of cycling precursors. Thus, adoption of one phenotype would coincide with repression of the other. A second key feature of this model is that it involves transcriptional “priming” of precursors, which would allow for phenotypic preprogramming of different neuronal phenotypes because these mRNAs could be sequestered by Smaug2/4E-T complexes until the appropriate developmental time point. Support for this translational repression switch model comes from our data showing that (1) Smaug2 was necessary and sufficient to maintain neural precursors in a stem cell state; (2) Nanos1 was necessary and sufficient to promote neurogenesis and deplete precursors; (3) nanos1 mRNA was highly colocalized with Smaug2 and 4E-T; (4) knockdown of Smaug2 or 4E-T caused aberrant Nanos1 expression, presumably because of complex disruption; and (5) coincident Nanos1 knockdown largely rescued the increase in neurogenesis and loss of Pax6-positive precursors caused by Smaug2 knockdown. Further support for this model comes from our recent work showing that 4E-T, which does not itself bind mRNAs, is necessary to maintain radial precursors in a stem cell state, and that when it is knocked down, this derepresses translation of the proneurogenic bHLHs Neurogenin1, Neurogenin2, and NeuroD1 (Yang et al., 2014).
How does Smaug2 silence nanos1 mRNA and its other potential target mRNAs? In Drosophila, Smaug silences its bound mRNAs in several ways. First, it associates with the eIF4E-binding protein Cup, which prevents eIF4E from interacting with its binding partner eIF4G and, in so doing, inhibits recruitment of the 40S subunit of the ribosome to the bound mRNA (Nelson et al., 2004). Second, Smaug can recruit Argonaute 1 to a target mRNA in an miRNA-independent fashion to repress translation (Pinder and Smibert, 2013a,b). Finally, Smaug can recruit the CCR4-NOT deadenylase complex, leading to mRNA deadenylation and destabilization (Semotok et al., 2005, 2008; Zaessinger et al., 2006). Our data do not distinguish these different potential mechanisms with regard to developing radial precursors. We have, however, shown that almost all of the nanos1 mRNA that is associated with Smaug2 in Pax6-positive precursors is also colocalized with 4E-T, which is distantly related to the Drosophila Cup. These large 4E-T-containing granules also contain Dcp1, eIF4E, Rck, and Lsm1 (shown in Yang et al., 2014) and thus resemble the P-bodies that are seen in other cells (Dostie et al., 2000; Ferraiuolo et al., 2005). Thus, P-body-like, 4E-T-containing granules are a major site for the Smaug2/nanos1 mRNA complex and likely for many other repressive complexes in radial precursors. This localization does not, however, allow us to distinguish Smaug2-mediated mRNA repression versus degradation because both processes are thought to occur in P-bodies (Balagopal and Parker, 2009). Interestingly, this localization is distinct from that seen in Drosophila embryos, where Smaug is found in foci that appear to be distinct from P-bodies (Zaessinger et al., 2006). Moreover, one of the few studies on Smaug in mammals shows that in adult hippocampal neurons Smaug1 is localized in foci that lack Dcp1 (Baez and Boccaccio, 2005; Baez et al., 2011). Thus, it appears that Smaug proteins can associate with different granules in different cell types.
While we have focused here on a function for Smaug2 in apical precursors, our data have shown that it is also expressed in newborn cortical neurons. Moreover, Smaug2 knockdown caused enhanced genesis of neurons, but these neurons did not reach the cortical plate. While there are a number of potential explanations for this neuronal mislocalization, these data might indicate that Smaug2 plays an important role in migration of newborn neurons. In this regard, while a function for Smaug2 in the adult brain has not been described, previous work has shown that the related family member Smaug1 forms mRNA-silencing foci that reversibly release their associated mRNAs upon specific synaptic activation by NMDA in dendrites of hippocampal neurons (Baez and Boccaccio, 2005; Baez et al., 2011). In addition, mutation of Smaug1 causes a lean phenotype in mice, likely due to deregulation of the mTOR pathway (Chen et al., 2014b) consistent with the recent finding that Drosophila Smaug regulates a large set of metabolic transcripts (Chen et al., 2014a). Thus, mammalian Smaug 1 and 2 are likely to play diverse roles in multiple cell types, in part dependent upon the constellation of target mRNAs that are expressed in each case.
One key remaining issue involves the molecular mechanism(s) responsible for the proneurogenic effects of Nanos1. In Drosophila, Nanos associates with two other RNA-binding proteins, Pumilio and Brain tumor (Loedige et al., 2014). This complex binds to and represses target mRNAs, a function that is essential for germ cell development (Hayashi et al., 2004; Lai and King, 2013). This role as a translational repressor in germ cells is conserved in vertebrates, including mice, where there are three family members, Nanos1, Nanos2, and Nanos3 (Jaruzelska et al., 2003; Tsuda et al., 2003; Lolicato et al., 2008). We therefore propose that Nanos1 regulates neurogenesis by associating with and repressing mRNAs associated with the neural precursor state. What then are the relevant Nanos1 target mRNAs in newborn neurons? In Drosophila and Xenopus, Nanos associates with Pumilio to repress cyclin B mRNA and to thereby lock cells out of the cell cycle (Nakahata et al., 2001; Kadyrova et al., 2007). This repression might be equally essential for newborn neurons, which, unlike their precursor parents, are postmitotic. Another potential target is Sox2. In murine germ cells, Nanos2 has been shown to associate with sox2 mRNA (Saba et al., 2014), and Sox2 is a key stem cell gene in cortical radial precursors (Julian et al., 2013). Thus, we propose that Nanos1-mediated translational repression of mRNAs encoding proteins, such as Cyclin B and Sox2, represses their previous precursor identity in newborn neurons, thereby allowing them to fully establish a neuronal phenotype.
In conclusion, our findings identify an essential role for translational repression in the neural precursor to neuron transition. The bimodal translational regulation we identify here may provide a widespread mechanism for ensuring that newborn progeny completely adopt their differentiated phenotype during development.
Footnotes
This work was supported by Canadian Institutes of Health Research Grant MOP-13958 to F.D.M. and D.R.K., who are Canada Research Chairs. F.D.M. is a Howard Hughes Medical Institute Senior International Research Scholar. G.A. was supported by studentships from Ontario Graduate Scholarships University of Toronto, and Hospital for Sick Children. G.Y. was supported by a Brain Canada fellowship.
The authors declare no competing financial interests.
References
- Aviv T, Lin Z, Lau S, Rendl LM, Sicheri F, Smibert CA. The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat Struct Biol. 2003;10:614–621. doi: 10.1038/nsb956. [DOI] [PubMed] [Google Scholar]
- Aviv T, Lin Z, Ben-Ari G, Smibert CA, Sicheri F. Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p. Nat Struct Mol Biol. 2006;13:168–176. doi: 10.1038/nsmb1053. [DOI] [PubMed] [Google Scholar]
- Baez MV, Boccaccio GL. Mammalian Smaug is a translational repressor that forms cytoplasmic foci similar to stress granules. J Biol Chem. 2005;280:43131–43140. doi: 10.1074/jbc.M508374200. [DOI] [PubMed] [Google Scholar]
- Baez MV, Luchelli L, Maschi D, Habif M, Pascual M, Thomas MG, Boccaccio GL. Smaug1 mRNA-silencing foci respond to NMDA and modulate synapse formation. J Cell Biol. 2011;195:1141–1157. doi: 10.1083/jcb.201108159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becalska AN, Gavis ER. Lighting up mRNA localization in Drosophila oogenesis. Development. 2009;136:2493–2503. doi: 10.1242/dev.032391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit B, He CH, Zhang F, Votruba SM, Tadros W, Westwood JT, Smibert CA, Lipshitz HD, Theurkauf WE. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development. 2009;136:923–932. doi: 10.1242/dev.031815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Chen L, Dumelie JG, Li X, Cheng MHK, Yang Z, Laver JD, Siddiqui NU, Westwood JT, Morris Q, Lipshitz HD, Smibert CA. Global regulation of mRNA translation and stability in the early Drosophila embryo by the Smaug RNA-binding protein. Genome Biol. 2014a;15:1–21. doi: 10.1186/gb-2014-15-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Holland W, Shelton JM, Ali A, Zhan X, Won S, Tomisato W, Liu C, Li X, Moresco EM, Beutler B. Mutation of mouse Samd4 causes leanness, myopathy, uncoupled mitochondrial respiration, and dysregulated mTORC1 signaling. Proc Natl Acad Sci U S A. 2014b;111:7367–7372. doi: 10.1073/pnas.1406511111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahanukar A, Wharton RP. The Nanos gradient in Drosophila embryos is generated by translational regulation. Genes Dev. 1996;10:2610–2620. doi: 10.1101/gad.10.20.2610. [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Walker JA, Wharton RP. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol Cell. 1999;4:209–218. doi: 10.1016/S1097-2765(00)80368-8. [DOI] [PubMed] [Google Scholar]
- Dostie J, Ferraiuolo M, Pause A, Adam SA, Sonenberg N. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J. 2000;19:3142–3156. doi: 10.1093/emboj/19.12.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraiuolo MA, Basak S, Dostie J, Murray EL, Schoenberg DR, Sonenberg N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher D, Norman AA, Woodard CL, Yang G, Gauthier-Fisher A, Fujitani M, Vessey JP, Cancino GI, Sachewsky N, Woltjen K, Fatt MP, Morshead CM, Kaplan DR, Miller FD. Transient maternal IL-6 mediates long-lasting changes in neural stem cell pools by deregulating an endogenous self-renewal pathway. Cell Stem Cell. 2013;13:564–576. doi: 10.1016/j.stem.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Gallagher D, Voronova A, Zander MA, Cancino GI, Bramall A, Krause MP, Abad C, Tekin M, Neilsen PM, Callen DF, Scherer SW, Keller GM, Kaplan DR, Walz K, Miller FD. Ankrd11 is a chromatin regulator involved in autism that is essential for neural development. Dev Cell. 2015;32:31–42. doi: 10.1016/j.devcel.2014.11.031. [DOI] [PubMed] [Google Scholar]
- Gauthier AS, Furstoss O, Araki T, Chan R, Neel BG, Kaplan DR, Miller FD. Control of CNS cell-fate decisions by SHP-2 and its dysregulation in Noonan syndrome. Neuron. 2007;54:245–262. doi: 10.1016/j.neuron.2007.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Hayashi M, Kobayashi S. Nanos suppresses somatic cell fate in Drosophila germ line. Proc Natl Acad Sci U S A. 2004;101:10338–10342. doi: 10.1073/pnas.0401647101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heraud-Farlow JE, Sharangdhar T, Li X, Pfeifer P, Tauber S, Orozco D, Hörmann A, Thomas S, Bakosova A, Farlow AR, Edbauer D, Lipshitz HD, Morris QD, Bilban M, Doyle M, Kiebler MA. Staufen2 regulates neuronal target RNAs. Cell Rep. 2013;5:1115–1118. doi: 10.1016/j.celrep.2013.11.039. [DOI] [PubMed] [Google Scholar]
- Imayoshi I, Kageyama R. bHLH factors in self-renewal, multipotency, and fate choice of neural progenitor cells. Neuron. 2014;82:9–23. doi: 10.1016/j.neuron.2014.03.018. [DOI] [PubMed] [Google Scholar]
- Jaruzelska J, Kotecki M, Kusz K, Spik A, Firpo M, Reijo Pera RA. Conservation of a Pumilio-Nanos complex from Drosophila germ plasm to human germ cells. Dev Genes Evol. 2003;213:120–126. doi: 10.1007/s00427-003-0303-2. [DOI] [PubMed] [Google Scholar]
- Julian LM, Vandenbosch R, Pakenham CA, Andrusiak MG, Nguyen AP, McClellan KA, Svoboda DS, Lagace DC, Park DS, Leone G, Blais A, Slack RS. Opposing regulation of Sox2 by cell-cycle effectors E2f3a and E2f3b in neural stem cells. Cell Stem Cell. 2013;12:440–452. doi: 10.1016/j.stem.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Kadyrova LY, Habara Y, Lee TH, Wharton RP. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development. 2007;134:1519–1527. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- Kamenska A, Lu WT, Kubacka D, Broomhead H, Minshall N, Bushell M, Standart N. Human 4E-T represses translation of bound mRNAs and enhances microRNA-mediated silencing. Nucl Acids Res. 2014;42:3298–3313. doi: 10.1093/nar/gkt1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusek G, Campbell M, Doyle F, Tenenbaum SA, Kiebler M, Temple S. Asymmetric segregation of the double-stranded RNA binding protein Staufen2 during mammalian neural stem cell divisions promotes lineage progression. Cell Stem Cell. 2012;11:505–516. doi: 10.1016/j.stem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, King ML. Repressive translational control in germ cells. Mol Reprod Dev. 2013;80:665–676. doi: 10.1002/mrd.22161. [DOI] [PubMed] [Google Scholar]
- Lasko P. Posttranscriptional regulation in Drosophila oocytes and early embryos. Wiley Interdiscip Rev RNA. 2011;2:408–416. doi: 10.1002/wrna.70. [DOI] [PubMed] [Google Scholar]
- Laver JD, Li X, Ancevicius K, Westwood JT, Smibert CA, Morris QD, Lipshitz HD. Genome-wide analysis of Staufen-associated mRNAs identifies secondary structures that confer target specificity. Nucleic Acids Res. 2013;41:9438–9460. doi: 10.1093/nar/gkt702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loedige I, Stotz M, Qamar S, Kramer K, Hennig J, Schubert T, Löffler P, Längst G, Merkl R, Urlaub H, Meister G. The NHL domain of BRAT is an RNA-binding domain that directly contacts the hunchback mRNA for regulation. Genes Dev. 2014;28:749–764. doi: 10.1101/gad.236513.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolicato F, Marino R, Paronetto MP, Pellegrini M, Dolci S, Geremia R, Grimaldi P. Potential role of Nanos3 in maintaining the undifferentiated spermatogonia population. Dev Biol. 2008;313:725–738. doi: 10.1016/j.ydbio.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Miki T, Takano K, Yoneda Y. The role of mammalian Staufen on mRNA traffic: a view from its nucleocytoplasmic shuttling function. Cell Struct Funct. 2005;30:51–56. doi: 10.1247/csf.30.51. [DOI] [PubMed] [Google Scholar]
- Miller FD, Gauthier AS. Timing is everything: making neurons versus glial in the developing cortex. Neuron. 2007;54:357–369. doi: 10.1016/j.neuron.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Nakahata S, Katsu Y, Mita K, Inoue K, Nagahama Y, Yamashita M. Biochemical identification of Xenopus pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J Biol Chem. 2001;276:20945–209453. doi: 10.1074/jbc.M010528200. [DOI] [PubMed] [Google Scholar]
- Nelson MR, Leidal AM, Smibert CA. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J. 2004;23:150–159. doi: 10.1038/sj.emboj.7600026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinder BD, Smibert CA. Smaug: an unexpected journey into the mechanisms of post-transcriptional regulation. Fly (Austin) 2013a;3:142–145. doi: 10.4161/fly.24336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinder BD, Smibert CA. microRNA-independent recruitment of Argonaute 1 to nanos mRNA through the Smaug RNA-binding protein. EMBO Rep. 2013b;14:80–86. doi: 10.1038/embor.2012.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba R, Kato Y, Saga Y. NANOS2 promotes male germ cell development independent of meiosis suppression. Dev Biol. 2014;385:32–40. doi: 10.1016/j.ydbio.2013.10.018. [DOI] [PubMed] [Google Scholar]
- Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol. 2005;15:284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- Semotok JL, Luo H, Cooperstock RL, Karaiskakis A, Vari HK, Smibert CA, Lipshitz HD. Drosophila maternal Hsp83 mRNA destabilization is directed by multiple SMAUG recognition elements in the open reading frame. Mol Cell Biol. 2008;28:6757–6772. doi: 10.1128/MCB.00037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui NU, Li X, Luo H, Karaiskakis A, Hou H, Kislinger T, Westwood JT, Morris Q, Lipshitz HD. Genome-wide analysis of the maternal-to-zygotic transition in Drosophila primordial germ cells. Genome Biol. 2012;13:R11. doi: 10.1186/gb-2012-13-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert CA, Wilson JE, Kerr K, Macdonald PM. Smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev. 1996;10:2600–2609. doi: 10.1101/gad.10.20.2600. [DOI] [PubMed] [Google Scholar]
- Smibert CA, Lie YS, Shillinglaw W, Henzel WJ, Macdonald PM. Smaug, a novel and conserved protein, contributes to repression of nanos mRNA translation in vitro. RNA. 1999;5:1535–1547. doi: 10.1017/S1355838299991392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38:792–799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell. 2007;12:143–155. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Tosar LJ, Thomas MG, Baez MV, Ibanez I, Chernomoretz A, Boccaccio GL. Staufen: from embryo polarity to cellular stress and neurodegeneration. Front Biosci (Schol Ed) 2012;4:432–452. doi: 10.2741/277. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- Tsui D, Vessey JP, Tomita H, Kaplan DR, Miller FD. FoxP2 regulates neurogenesis during embryonic cortical development. J Neurosci. 2013;33:244–258. doi: 10.1523/JNEUROSCI.1665-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey JP, Amadei G, Burns SE, Kiebler MA, Kaplan DR, Miller FD. An asymmetrically localized Staufen2-dependent RNA complex regulates maintenance of mammalian neural stem cells. Cell Stem Cell. 2012;11:517–528. doi: 10.1016/j.stem.2012.06.010. [DOI] [PubMed] [Google Scholar]
- Walser CB, Lipshitz HD. Transcript clearance during the maternal-to-zygotic transition. Curr Opin Genet Dev. 2011;4:431–434. doi: 10.1016/j.gde.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Wang J, Weaver IC, Gauthier-Fisher A, Wang H, He L, Yeomans J, Wondisford F, Kaplan DR, Miller FD. CBP histone acetyltransferase activity regulates embryonic neural differentiation in the normal and Rubinstein-Taybi syndrome brain. Dev Cell. 2010;18:114–125. doi: 10.1016/j.devcel.2009.10.023. [DOI] [PubMed] [Google Scholar]
- Weibrecht I, Leuchowius KJ, Clausson CM, Conze T, Jarvius M, Howell WM, Kamali-Moghaddam M, Söderberg O. Proximity ligation assays: a recent addition to the proteomics toolbox. Exp Rev Proteom. 2010;7:401–409. doi: 10.1586/epr.10.10. [DOI] [PubMed] [Google Scholar]
- Yang G, Smibert CA, Kaplan DR, Miller FD. An eIF4E1/4E-T complex determines the genesis of neurons from precursors by translationally repressing a proneurogenic transcription program. Neuron. 2014;84:723–739. doi: 10.1016/j.neuron.2014.10.022. [DOI] [PubMed] [Google Scholar]
- Zaessinger S, Busseau I, Simonelig M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133:4573–4583. doi: 10.1242/dev.02649. [DOI] [PubMed] [Google Scholar]


