Abstract
Chronic stress-induced aberrant gene expression in the brain and subsequent dysfunctional neuronal plasticity have been implicated in the etiology and pathophysiology of mood disorders. In this study, we examined whether altered expression of small, regulatory, noncoding microRNAs (miRNAs) contributes to the depression-like behaviors and aberrant neuronal plasticity associated with chronic stress. Mice exposed to chronic ultra-mild stress (CUMS) exhibited increased depression-like behaviors and reduced hippocampal expression of the brain-enriched miRNA-124 (miR-124). Aberrant behaviors and dysregulated miR-124 expression were blocked by chronic treatment with an antidepressant drug. The depression-like behaviors are likely not conferred directly by miR-124 downregulation because neither viral-mediated hippocampal overexpression nor intrahippocampal infusion of an miR-124 inhibitor affected depression-like behaviors in nonstressed mice. However, viral-mediated miR-124 overexpression in hippocampal neurons conferred behavioral resilience to CUMS, whereas inhibition of miR-124 led to greater behavioral susceptibility to a milder stress paradigm. Moreover, we identified histone deacetylase 4 (HDAC4), HDAC5, and glycogen synthase kinase 3β (GSK3β) as targets for miR-124 and found that intrahippocampal infusion of a selective HDAC4/5 inhibitor or GSK3 inhibitor had antidepressant-like actions on behavior. We propose that miR-124-mediated posttranscriptional controls of HDAC4/5 and GSK3β expressions in the hippocampus have pivotal roles in susceptibility/resilience to chronic stress.
SIGNIFICANCE STATEMENT Depressive disorders are a major public health concern worldwide. Although a clear understanding of the etiology of depression is still lacking, chronic stress-elicited aberrant neuronal plasticity has been implicated in the pathophysiology of depression. We show that the hippocampal expression of microRNA-124 (miR-124), an endogenous small, noncoding RNA that represses gene expression posttranscriptionally, controls resilience/susceptibility to chronic stress-induced depression-like behaviors. These effects on depression-like behaviors may be mediated through regulation of the mRNA or protein expression levels of histone deacetylases HDAC4/5 and glycogen synthase kinase 3β, all highly conserved miR-124 targets. Moreover, miR-124 contributes to stress-induced dendritic hypotrophy and reduced spine density of dentate gyrus granule neurons. Modulation of hippocampal miR-124 pathways may have potential antidepressant effects.
Keywords: anxiety, depression, epigenetics, hippocampus, microRNA, stress
Introduction
Stressful life events are potent adverse environmental factors that can predispose individuals to psychiatric disorders such as clinical depression and anxiety disorders (Krishnan and Nestler, 2008; Duman and Aghajanian, 2012; Franklin et al., 2012; McEwen et al., 2015). Although the pathophysiology of depression remains to be addressed fully, stress-induced aberrant synaptic and structural neuroplasticity may be key underlying mechanisms (Krishnan and Nestler, 2008; Duman and Aghajanian, 2012; Franklin et al., 2012; Chattarji et al., 2015; McEwen et al., 2015). Neuroplasticity is regulated by a complex gene expression program (Flavell and Greenberg, 2008) and multiple lines of evidence implicate aberrant transcriptional regulation of neuroplasticity-associated genes in the pathophysiology of depression (Krishnan and Nestler, 2008; Feder et al., 2009).
In addition to the potential contribution of dysregulated transcription, recent evidence has emerged showing that altered posttranscriptional regulation of gene expression is pivotal to the development of depression. MicroRNAs (miRNAs) are a class of small, noncoding RNAs able to mediate cleavage or translational repression of target mRNAs by binding preferentially to 3′ untranslated regions (UTRs) (Kosik, 2006; Bartel, 2009). There is a growing body of evidence that miRNAs regulate neuronal and synaptic plasticity (Schratt et al., 2006; Rajasethupathy et al., 2009; O'Carroll and Schaefer, 2013). Both preclinical and clinical studies have demonstrated that miRNAs contribute to the pathophysiology of depression and to antidepressant drug actions (Im and Kenny, 2012; O'Connor et al., 2012; Dwivedi, 2014; Geaghan and Cairns, 2015; Issler and Chen, 2015; Kocerha et al., 2015). Indeed, environmental stressors and antidepressant drugs alter the expression levels of miRNAs in prefrontal cortex, amygdala, nucleus accumbens, locus ceruleus, and dorsal raphe nucleus, all regions intimately involved in mood regulation (Baudry et al., 2010; Uchida et al., 2010; Haramati et al., 2011; Dias et al., 2014; Issler et al., 2014).
The hippocampal formation is vulnerable to damage from a variety of psychological stressors (McEwen, 2001; Schoenfeld and Gould, 2012) and aberrant structural and functional changes in this brain structure have been implicated in the pathophysiology of depression (Nestler et al., 2002; McEwen, 2007; Schmidt and Duman, 2007). In addition, there is growing evidence indicating that environmental challenges alter miRNAs in the brain, including the hippocampus (O'Connor et al., 2012; Dwivedi, 2014; Issler and Chen, 2015). Nevertheless, the involvement of hippocampal miRNA-mediated gene dysregulation in chronic stress-induced aberrant neuronal plasticity and depression-related behaviors remains largely unexplored.
To assess the potential contributions of hippocampal miRNAs to depression-like behaviors and aberrant neuronal plasticity, we measured the expression levels of several brain-enriched miRNAs in the hippocampus of mice exposed to chronic stress, a widely studied animal model of depression. We found that miRNA-124 (miR-124) was markedly suppressed, so we examined the effects of hippocampal miR-124 gain- and loss-of-function on depression-like behaviors and dendritic structure and identified downstream miR-124 target genes contributing to these effects. Collectively, our results suggest that stress-induced downregulation of hippocampal miR-124 and concomitant dysregulation of target genes encoding histone deacetylase 4 (HDAC4), HDAC5, and glycogen synthase kinase 3β (GSK3β) contribute to dendritic remodeling, depression-like behaviors, and antidepressant-like action.
Materials and Methods
Animals.
Adult male BALB/c (BALB) mice (Charles River Laboratories) were maintained on a 12 h/12 h light/dark cycle with ad libitum access to mouse chow and water. Mice were 8 weeks old at the start of the experiments (i.e., at the time of stereotaxic surgery or stress exposure). All experimental procedures were performed according to the Guidelines for Animal Care and Use of Yamaguchi University Graduate School of Medicine.
Chronic ultra-mild stress (CUMS).
The CUMS procedure used in this study was a modified version of chronic mild stress (Willner et al., 1987). The CUMS was based solely on environmental and social stressors without food/water deprivation or any nociceptive event (Uchida et al., 2011a; Abe-Higuchi et al., 2016). In brief, mice were subjected to a variety of mild stressors such as cage tilt (30°), paired housing, confinement to small cages (11 × 8 × 8 cm), and a soiled cage (50 ml of water/L of sawdust bedding). A reversed light/dark cycle was used from Friday evening to Monday morning. This procedure was scheduled over a 1 week period and repeated 6 times, but the reversed light/dark cycle was omitted during the weekend of the last session. Mice were subjected to a 1 h period of morning stress, a 2 h period of afternoon stress, and an overnight stress. Nonstressed (NS) mice were handled daily for weighing.
Repeated restraint stress procedure.
Adult mice were weighed and individually subjected to mild restraint stress (1 h/d) in a 50 ml plastic centrifuge tube for 14 consecutive days (resilience session; see Figs. 2I, 3D) or to severe restraint stress (4 h/d) in a 30 ml plastic centrifuge tube for 10 consecutive days (susceptible session; see Figs. 5H, 6H). Naive control animals were left in their home cages and handled for 14 or 10 consecutive days during weighing.
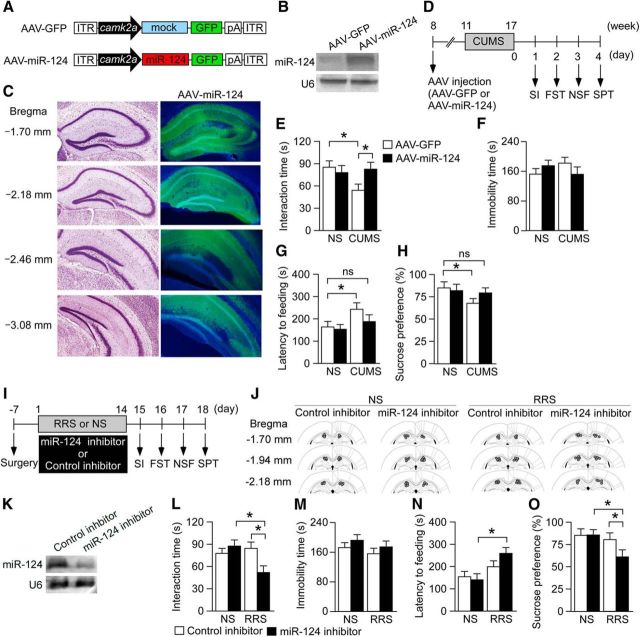
Figure 2.
Hippocampal miR-124 mediates behavioral responses to chronic stress. A, Schematics of AAV engineered to overexpress GFP together with either miR-124 or mock under the control of the Camk2a promoter. B, C, Successful transduction of miR-124 using AAV-mediated gene transfer. B, Northern blotting shows overexpression of miR-124 in the hippocampus. C, Left, Coronal brain sections with distance from bregma, adapted from Franklin and Paxinos (2008). Right, Corresponding images showing transgene expression (GFP, green) after vector microinjection (counterstained with 4′,6-diamidino-2-phenylindole, blue). D, Schematic of the experimental design used to demonstrate reversal of CUMS-induced depression-like behaviors by miR-124 overexpression in the hippocampus. Mice were injected with the AAV–miR-124 overexpression vector or AAV–GFP control vector in the bilateral hippocampus. After 3 weeks, they were subjected to CUMS for 6 weeks and then tested in the behavioral assays. E–H, Behavioral results. E, SI time in the SI. F, Immobility time in the FST. G, Latency to feeding in the NSF. H, Sucrose preference in the SPT. miR-124 overexpression blocked the CUMS-induced decrease in SI (E), increased latency to feeding (G), and reduction in sucrose preference (H). n = 10–13 mice per group. *p < 0.05. I, Schematic of the experimental design for testing the effects of miR-124 downregulation on susceptibility to stress-induced depression-like behaviors. Mice were bilaterally implanted with cannulae directed to the hippocampus. One week after surgery, mice were subjected to mild RRS (1 h/d for 14 d). The miR-124 inhibitor or negative control miRNA was infused every 3 d during the mild RRS session. After the mild RRS session, behavioral tests were performed. J, Location of the cannula tips. K, Northern blotting showing reduced mature miR-124 expression in mice injected with miR-124 inhibitor. L–O, Results of behavioral tests. L, SI time in the SI. M, Immobility time in the FST. N, Latency to feeding in the NSF. O, Sucrose preference in the SPT. This mild RRS protocol did not induce depression-like behaviors compared with the NS condition and the miR-124 inhibitor did not affect baseline responses (NS groups). However, the miR-124 inhibitor reduced interaction time (L), prolonged latency to feeding (N), and decreased sucrose preference (O) in mild RRS-exposed mice, indicative of enhanced susceptibility to depression-like behavior effects. n = 10–13 mice per group. *p < 0.05. All data are presented as mean ± SEM.
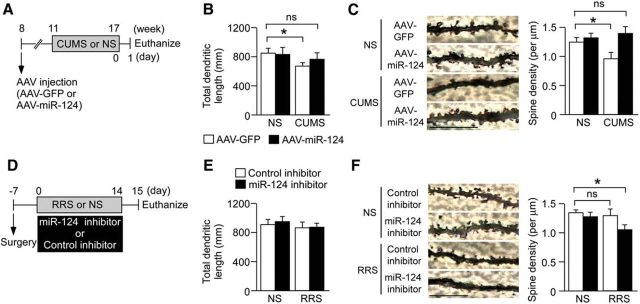
Figure 3.
miR-124 is associated with chronic stress-induced changes in dendritic morphology and spine density. A, Schematic of the experimental design used to assess the effects of miR-124 overexpression on stress-induced neuroplastic changes. Mice were injected with AAV–miR-124 or AAV–GFP into the bilateral hippocampus. Three weeks later, mice were subjected to a 6-week CUMS session. B, Bar graph showing the total dendrite length of DG granule neurons (n = 5–6 mice per group). CUMS reduced dendritic length in AAV–GFP-injected mice. Overexpression of miR-124 had no effect on dendritic length in NS mice but blocked the CUMS-induced decrease. *p < 0.05. C, Representative images of dendritic spines from DG granule neurons. Bar graph shows mean spine densities of DG granule neurons (n = 5–6 mice per group). CUMS reduced spine density compared with NS mice injected with the control vector, whereas miR-124 overexpression blocked this effect. *p < 0.05 versus NS. Scale bars, 10 μm. D, Schematic of the experimental design used to evaluate the effects of miR-124 inhibition on stress-induced neuroplastic changes. Mice were injected with miR-124 inhibitor or control inhibitor into the bilateral hippocampus every 3 d during 14 d of mild RRS (1 h/d). E, Bar graph showing the total dendrite length of DG granule neurons (n = 5–6 mice per group). This mild stress protocol had no effect on dendritic length. *p < 0.05. F, Representative images of dendritic spines from DG granule neurons. Bar graph shows spine densities of DG granule neurons after mild RRS with miR-124 inhibitor or control inhibitor cotreatment (n = 5–6 mice per group). The mild RRS regimen alone did not reduce spine density. Spine density was reduced in mild RRS-exposed mice injected with the miR-124 inhibitor, indicating enhanced susceptibility to stress-induced neuroplastic changes. *p < 0.05 versus NS. Scale bars, 10 μm. Data are presented as mean ± SEM.
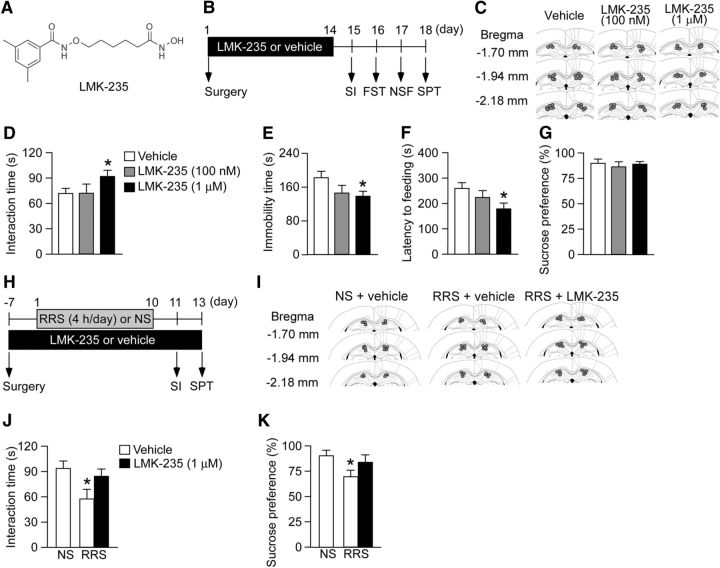
Figure 5.
Pharmacological inhibition of HDAC4/5 induces antidepressant-like behaviors. A, Structure of LMK-235, a selective HDAC4/5 inhibitor. B, Schematic of the experimental design to show that HDAC4/5 inhibition reduces depression-like behaviors. BALB/c mice were continuously infused with LMK-235 or vehicle into the bilateral hippocampus using osmotic minipumps. Two weeks after pump implantation, mice were tested using behavioral assays. C, Location of the cannula tips. D–G, Behavioral results. D, SI time in the SI. E, Immobility time in the FST. F, Latency to feeding in the NSF. G, Sucrose preference in the SPT. n = 10–12 mice per group. LMK-235 enhanced SI (D) and reduced both immobility time (E) and latency to feeding (F). *p < 0.05 versus vehicle control. H, Schematic of the experimental design to demonstrate that HDAC4/5 inhibition reduces depression-like behaviors in stressed mice. BALB/c mice were infused with LMK-235 or vehicle into the hippocampus using osmotic minipumps. One week after pump implantation surgery, mice were subjected to severe RRS (4 h/d) for 10 d. The mice were then tested using behavioral assays. I, Location of the cannula tips. J, K, Behavioral results. J, SI time in the SI. K, Sucrose preference in the SPT. n = 11–14 per group. *p < 0.05 versus NS mice receiving vehicle. All data are presented as mean ± SEM.
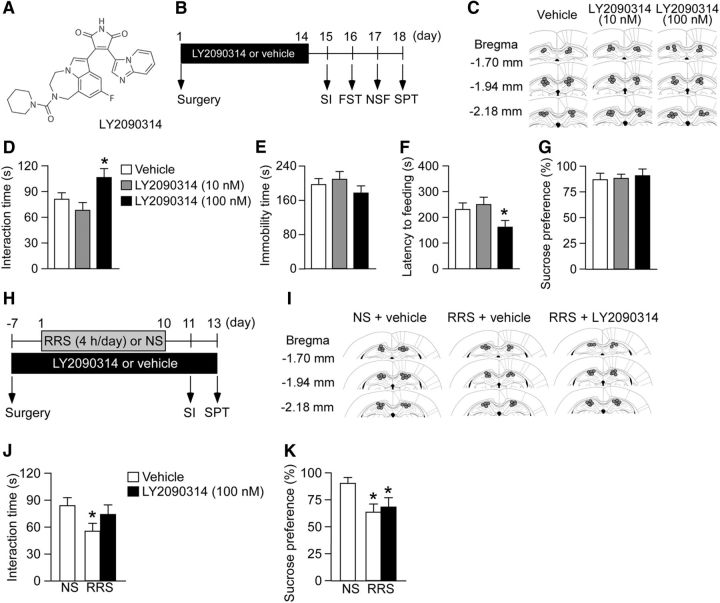
Figure 6.
Pharmacological inhibition of GSK3 induces antidepressant-like behaviors. A, Structure of LY2090314, a selective GSK3 inhibitor. B, Schematic of the experimental design to show that GSK3 inhibition suppresses depression-like behaviors. BALB/c mice were bilaterally infused with LY2090314 or vehicle into the hippocampus using osmotic minipumps. Two weeks after pump implantation surgery, mice were tested using behavioral assays. C, The location of the cannula tips. D–G, Behavioral results. D, SI time in the SI. E, Immobility time in the FST. F, Latency to feeding in the NSF. G, Sucrose preference in the SPT. n = 10–12 mice per group. Inhibition of GSK3 increased SI (D) and reduced latency to feeding (F). *p < 0.05 versus vehicle control. H, Schematic of the experimental design to show that GSK3 inhibition suppresses depression-like behaviors in stressed mice. BALB/c mice were infused with LY2090314 or vehicle into the bilateral hippocampus using osmotic minipumps. One week after pump implantation surgery, mice were subjected to severe RRS (4 h/d) for 10 d and then tested using behavioral assays. I, Location of the cannula tips. J, K, Behavioral results. J, SI time in the SI. K, Sucrose preference in the SPT. n = 12–15 per group. *p < 0.05 versus NS mice receiving vehicle. All data are presented as mean ± SEM.
Drug treatments.
Imipramine (IMI; Sigma-Aldrich) was dissolved in tap water at a concentration of 160 mg/L (Uchida et al., 2011a). LMK-235 was synthesized using a previously reported procedure (Marek et al., 2013) from benzyl 6-bromohexanoate and N-hydroxyphtalimide in six steps: (1) O-alkylation of N-hydroxyphthalimide with benzyl 6-bromohexanoate, (2) deprotection of the benzyl group, (3) amide formation between the carboxylic acid and O-benzylhydroxylamine, (4) methylhydrazine-mediated cleavage of the phthaloyl-protection group, (5) amide formation between the hydroxylamine and 3,5-dimethylbenzoic acid, and (6) deprotection of the benzyl group. LMK-235 is a selective inhibitor of HDAC4 and HDAC5 with IC50 values of 11.9 nm and 4.2 nm, respectively (Marek et al., 2013). LY2090314 is a potent GSK3 inhibitor with IC50 values of 1.5 nm for GSK3α and 0.9 nm for GSK3β. LMK-235 (100 nm or 1 μm), LY2090314 (10 or 100 nm), or vehicle were continuously infused bilaterally into the hippocampus using osmotic minipumps (Alzet, model 1002 or model 1004) as described previously (Abe-Higuchi et al., 2016). These pumps provide up to 2 weeks (model 1002) or 4 weeks (model 1004) of continuous infusion.
Behavioral procedures.
Behavioral tests were performed during the light phase (9:00 A.M. to 3:00 P.M.) as described previously (Uchida et al., 2011a; Uchida et al., 2011b). For the social interaction (SI) test, an adult mouse was placed in a measuring cage for 120 min. A male juvenile (4–5 weeks old) was then introduced into the cage and the amount of time spent in SI such as grooming, licking, sniffing, or crawling over or under the other mouse was recorded during a 3 min session. For the sucrose preference test (SPT), mice were subjected to water deprivation for 16 h and then two preweighed bottles, one containing tap water and the other containing 1.5% sucrose solution, were presented for 1.5 h. The positions of the water and sucrose bottles (left or right) were switched every 30 min. The bottles were weighed again and the weight difference during the last 60 min was used to calculate the volume intake from each bottle. The sum of the water and sucrose intake was defined as the total intake and the sucrose preference was expressed as the percentage of sucrose intake relative to the total intake. For the forced swim test (FST), a mouse was placed in a water tank (20 cm height × 14 cm diameter filled with 23–24°C water to a depth of 13 cm) for 5 min and the total duration of floating/immobility (i.e., the time during which the mouse performed only small movements necessary to keep its head above water) and the time to the first episode of immobility were measured. For the novelty-suppressed feeding test, mice were housed individually and food pellets were removed from their cages the day before testing. Twenty-four hours after food removal, the percentage loss of body weight was measured and the test mouse was transferred to a clean holding cage in the testing room. The testing apparatus consisted of a square open field (25 × 20 × 20 cm). A piece of chow was placed in the center of the testing apparatus. The mouse was placed in the testing apparatus and the time until the first feeding episode was recorded for up to 10 min. After termination of the test, the mouse was returned to its home cage with food pellets and the amount of food consumed was measured for 30 min.
Quantitative real-time PCR.
Quantitative real-time PCR was performed as described previously (Uchida et al., 2010; Uchida et al., 2011a). Total RNA from dissected tissues was extracted using TRIzol Reagent (Life Technologies) and treated with DNase (DNA-free; Life Technologies). The quality of RNA was determined based on the A260/A280 ratio, which was 1.81–1.96 for all RNA preparations used for subsequent expression analyses. One microgram of total RNA was reverse transcribed to cDNA using the QuantiTect Reverse Transcription Kit (Qiagen). The kit primer mix contains oligo-dT and random primers to ensure cDNA synthesis from all regions of RNA transcripts, including primary/precursor (pri/pre) miRNAs (Uchida et al., 2008; Uchida et al., 2010). cDNA was stored at −80°C until use. Real-time PCR was performed using the Applied Biosystems StepOne Real-Time PCR System with SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer's protocol. PCR conditions were 15 min at 95°C, followed by 45 cycles of 15 s at 95°C and 30 s at 60°C. The following primers were used for quantitative PCR: pri/miR-124-1, forward (5′-TCTCTCTCCGTGTTCACAGC-3′) and reverse (5′-ACCGCGTGCCTTAATTGTAT-3′); pri/pre-miR-29a, forward (5′-CCCCTTAGAGGATGACTGATTTC-3′) and reverse (5′-AACCGATTTCAGATGGTGCT-3′); pri/pre-miR-132, forward (5′-ACCGTGGCTTTCGATTGTTA-3′) and reverse (5′-CGACCATGGCTGTAGACTGTT-3′); pri/pre-miR-9-1, forward (5′-CGGGGTTGGTTGTTATCTTT-3′) and reverse (5′-TGGGGTTATTTTTACTTTCGGTTA-3′); pri/pre-miR-9-3, forward (5′-GCCCGTTTCTCTCTTTGGTT-3′) and reverse (5′-TCTAGCTTTATGACGGCTCTGTGG-3′); pri/pre-miR-212, forward (5′-GGCACCTTGGCTCTAGACTG-3′) and reverse (5′-GCCGTGACTGGAGACTGTTA-3′); pri/pre-miR-330, forward (5′-GACCCTTTGGCGATCTCTG-3′) and reverse (5′-CTGTGCTTTGCTCGTTGGAT-3′); pri/pre-miR-153, forward (5′-TCATTTTTGTGACGTTGCAG-3′) and reverse (5′-TGACTATGCAACTGGGCTCAT-3′); pri/pre-miR-135b, forward (5′-TGGCCTATGGCTTTTCATTC-3′) and reverse (5′-GAGCTTGCCCCTCACTGTAG-3′); pri/pre-miR-134, forward (5′-TGTGACTGGTTGACCAGAGG-3′) and reverse (5′-GTGACTAGGTGGCCCACAG-3′); pri/pre-miR-181a-1, forward (5′-GCTGTCGGTGAGTTTGGAAT-3′) and reverse (5′-AATCAACGGTCGATGGTTTT-3′); pri/pre-miR-128b, forward (5′-GGCCGATGCACTGTAAGAGA-3′) and reverse (5′-TTCACTGTGAGACCTGCTACTC-3′); pri/pre-miR-188, forward (5′-GGAGGGTGAGCTCTCTGAAA-3′) and reverse (5′-CTGCAAACCCTGCATGTG-3′); U6 snRNA, forward (5′-CGCTTCGGCAGCACATATAC-3′) and reverse (5′-TTCACGAATTTGCGTGTCAT-3′); Hdac4, forward (5′-CAATCCCACAGTCTCCGTGT-3′) and reverse (5′-CAGCACCCCACTAAGGTTCA-3′); Hdac5, forward (5′-TGTCACCGCCAGATGTTTTG-3′) and reverse (5′-TGAGCAGAGCCGAGACACAG-3′); Gsk3b, forward (5′-CCTTATCCCTCCACATGCTC-3′) and reverse (5′-ATTGGTCTGTCCACGGTCTC-3′); Pde4b, forward (5′-GCACACAGCTTCTGTGGAAA-3′) and reverse (5′-ATCTTGAGCATCCGGTTGAA-3′); and Gapdh, forward (5′-AGGTCGGTGTGAACGGATTTG-3′) and reverse (5′-TGTAGACCATGTAGTTGAGGTCA-3′). Amplification curves were inspected visually to set a suitable baseline range and threshold level. The relative quantification method was used for the estimation of target mRNA or miRNA expression according to the manufacturer's protocol, in which the ratio of each target nucleotide amount to the reference nucleotide within the same sample was calculated. All measurements were performed in duplicate. Levels of Gapdh mRNA or U6 snRNA were used to normalize the relative expression levels of target mRNAs and pri/pre-miRNAs, respectively.
Northern blotting.
Northern blotting was performed as described previously (Uchida et al., 2010). Twenty micrograms of total RNA was separated on 15% denaturing polyacrylamide gels and transferred onto Hybond N+ membranes (GE Healthcare). After RNA fixation by UV crosslinking, hybridization with appropriate probes was performed using the Gene Images AlkPhos Direct Labeling and Detection System (GE Healthcare) according to the manufacturer's protocol. After hybridization, the membrane was washed to remove excess probe and incubated with CDP-star chemiluminescent detection reagent (GE Healthcare). Probe sequences for miR-124 and miR-29a are 5′-TGGCATTCACCGCGTGCCTTAA-3′ and 5′-ATAACCGATTTCAGATGGTGCTAG-3′, respectively. Levels of ribosomal RNA were visualized on gels with ethidium bromide staining. A U6 snRNA probe (5′-GAATTTGCGTGTCATCCTTGCGCAGGGGCCATGCTAA-3′) was applied to normalize relative miRNA expression levels. Densitometric analysis was performed using Image Studio Lite software (LI-COR Biosciences) after scanning with the GE Healthcare Imager 600.
Western blotting and immunohistochemistry.
Western blotting was performed as described previously (Uchida et al., 2014). Equal amounts of protein were separated on 7% or 10% Bis-Tris gels (Life Technologies) and transblotted onto polyvinylidene difluoride membranes (GE Healthcare). After blocking with 5% skim milk, membranes were incubated with the indicated primary antibody. Primary antibodies used were rabbit polyclonal antibodies against HDAC4 (1:1000; Millipore), HDAC5 (1:1000; Abcam), GSK3β (1:1000; Cell Signaling Technology), and PDE4B (1:1000; Cell Signaling Technology) and a mouse monoclonal antibody against β-actin (1:10000; Sigma-Aldrich). After incubation with appropriate horseradish peroxidase-conjugated secondary antibodies (1:4000;Cell Signaling Technology), the blots were developed using Pierce ECL Western Blotting Substrate Plus. Densitometric analysis was performed using Image Studio Lite software (LI-COR Biosciences) after scanning with the GE Healthcare Imager 600.
Immunohistochemistry.
Immunohistochemistry was performed as described previously (Uchida et al., 2011a; Martel et al., 2016). Mice were deeply anesthetized with sodium pentobarbital (50 mg/kg) and transcardially perfused with 4% paraformaldehyde in 0.1 m phosphate buffer, pH 7.4. Immunofluorescence staining was performed on free-floating sections (30 μm) using an antibody against green fluorescent protein (GFP, 1:500; Life Technologies). Secondary antibodies were conjugated with Alexa Fluor 488 (1:500; Life Technologies). Images were acquired using a Keyence BZ-9000 fluorescence microscope.
miRNA inhibitor infusion.
Mice were infused with a single-stranded, chemically modified RNA designed to inhibit endogenous miR-124 specifically, referred to herein as the miRNA inhibitor (anti-miR-124, AM10691; Life Technologies) or with a control (anti-miR negative control, AM17010; Life Technologies). The anti-miR negative control is an oligonucleotide with a random base sequence that has been demonstrated extensively by the manufacturer to cause no measureable effects on known miRNA functions. Mice were anesthetized with sodium pentobarbital and bilateral cannulae (Plastics One) aimed at the hippocampus were implanted stereotaxically (AP, −2.0 mm; ML, ±1.5 mm; DV, −1.5 mm from bregma; Franklin and Paxinos, 2008). Seven days after surgery, bilateral intrahippocampal (0.5 μl/side) injection of anti-miRNAs was performed on freely moving mice via injection cannulae (projecting 0.5 mm beyond the tip of the guide cannula) attached to 10 μl Hamilton syringes by polyethylene catheter tubing. The oligonucleotides were dissolved in in vivo jetSI (Polyplus Transfection) reagent at a final concentration of 10 μm as described previously (Uchida et al., 2010). The syringes were held in a constant-rate infusion pump and bilateral injections were conducted over a 5 min period. The cannulae were left in place for an additional 2 min before removal. Mice were injected with anti-miRNA/jetSI complexes every 3 d during the experiments. At the end of the experiments, all mice were killed and coronal brain sections prepared to check cannula placement.
Adeno-associated virus (AAV)-mediated gene transfer.
AAV-mediated gene transfer was performed as described previously (Uchida et al., 2011a; Uchida et al., 2011b). A DNA fragment encoding a mouse Camk2a promoter (1.3 kb) was amplified from C57BL/6J mouse genomic DNA and inserted into the pAAV-ΔCMV-MCS vector, which was constructed by removing the CMV promoter from pAAV-CMV-MCS (Agilent Technologies), yielding the pAAV-Camk2a-MCS vector. Then, EGFP cDNA amplified from pEGFP-C1 plasmid (Clontech) was inserted into the pAAV-Camk2a-MCS vector, yielding the pAAV-Camk2a-EGFP vector. To generate the miR-124 expression plasmids, a 519 bp DNA fragment containing the mouse pri-miR-124-1 sequences was PCR amplified from mouse genomic DNA using the forward primer 5′-CGCGTCGCCAGCTTTTTCTTTCT-3′ and the reverse primer 5′-CACAGAATGAGTTGTTTGCTT-3′. The fragments were inserted in front of an EGFP-coding region within the pAAV-Camk2a-EGFP vector, yielding the pAAV–Camk2a-miR-124-EGFP vector. Recombinant viruses (AAV serotype 8) were generated at Vector Laboratories. The genomic titer of each virus was determined using real-time PCR. The titers of AAV8-Camk2a-EGFP (AAV–GFP) and AAV8–Camk2a-miR-124-EGFP (AAV–miR-124) were measured as 6.0 × 1013 and 6.7 × 1013 viral genomes/ml, respectively. For viral vector injections, mice were anesthetized intraperitoneally with sodium pentobarbital (50 mg/kg) and placed in a stereotaxic frame. The skull was exposed and a small portion of the skull over the hippocampus was removed bilaterally with a dental drill. Subsequently, AAV vectors were dissolved in physiological saline (0.5 μl) and injected bilaterally into the hippocampus (AP, −2.0 mm; ML, ±1.5 mm; DV, −2.0 mm; Franklin and Paxinos, 2008) at 0.1 μl/min. The needle was slowly withdrawn after 3 min. Mice were allowed to recover for 3 weeks for maximum transgene induction. Successful transduction in the hippocampus was confirmed by immunohistochemistry with an antibody against GFP.
Golgi staining.
Golgi staining was performed using the Rapid Golgi Stain kit according to the manufacturer's protocol (FD NeuroTechnologies) as described previously (Uchida et al., 2011b). The brains were sliced using a Vibratome to a thickness of 100 μm. Bright-field microscopy images (Nikon, 20× or 60×) of granule neurons in the dentate gyrus (DG) were obtained (6–12 neurons per animal). Only fully Golgi-impregnated neurons displaying dendritic trees without obvious truncations and well separated from neighboring impregnated neurons were retained for analysis. The total dendrite length was quantified by tracing entire granule neurons. Quantification of spine density was limited to dendrites that were 100–150 μm from the soma. The spine density is expressed as the number of spines per μm of dendritic length.
Cell cultures.
Primary hippocampal neurons were prepared from embryonic day 17 (E17) mouse embryos. After removal of the brain, hippocampi were dissected and dissociated and the tissue pieces were incubated in 2.5% trypsin (Life Technologies) for 20 min at 37°C. Then, the cells were incubated with a trypsin inhibitor (Worthington) and subsequently resuspended in DMEM containing 10% fetal bovine serum. Viable cells, as indicated by trypan blue exclusion, were seeded on poly-d-lysine-coated 24-well dishes. Four hours after plating, the medium was replaced with Neurobasal medium (Life Technologies) containing 1% B27 supplement (Life Technologies) and 50 μg/ml streptomycin. The cultures were maintained at 37°C in a 5% CO2 humidified atmosphere. On day 2 in vitro (2 DIV), hippocampal neurons were treated with 2 μm 1-β-d-arabinofuranosylcytosine (Sigma-Aldrich) to reduce the number of proliferating non-neuronal cells.
Luciferase assay.
The mouse Hdac4 3′ UTR (617 bp fragment), Hdac5 3′ UTR (999 bp fragment), and Gsk3b 3′ UTR (843 bp fragment) containing putative target sites of miR-124 were PCR amplified from mouse brain cDNA using the following primers: Hdac4 3′ UTR forward (5′-CACATGGAACACAGGACAGC-3′) and reverse (5′-CCACAGGACACTGTCACACC-3′); Hdac5 3′ UTR forward (5′-CCTCTGGGCTTCATCATTGT-3′) and reverse (5′-AGCCTCTGGACACAAGGCTA-3′); and Gsk3b 3′ UTR forward (5′-GACTTGGTGTGCGTGCTAGA-3′) and reverse (5′-GACTGCCAACATGGTTTCCT-3′). PCR products were cloned into the pmirGLO vector (Promega) downstream of the firefly luciferase coding sequence. Mutant plasmids for the 3′ UTRs were made using a KOD Plus Mutagenesis kit (TOYOBO). Hippocampal neurons at 5 DIV were infected with AAV–miR-124 or control AAV–GFP. The neurons at 10 DIV were transfected with a luciferase reporter vector containing wild-type or mutated Hdac4, Hdac5, or Gsk3b 3′ UTR using the calcium phosphate method according to the manufacturer's protocol (Promega). Forty-eight hours after transfection, the Firefly and Renilla luciferase activities were measured using a Dual-GLO Luciferase Assay System (Promega). In all cases, the total amount of transfected plasmid DNA per well was matched with that of the empty or mutant plasmid. At least three independent experiments were performed on separate culture, each with triplicate wells for all treatments.
Statistical analyses.
Multiple group means were compared using an appropriate ANOVA (one-way or two-way). Significant effects were then evaluated with Tukey's post hoc tests or Bonferroni correction. Unpaired t tests were used for two-group comparisons. In all cases, the tests were two-tailed and considered significant when p < 0.05. All data are presented as mean ± SEM.
Results
Chronic stress reduces the expression of hippocampal miR-124
We reported previously that BALB mice show vulnerability to a CUMS regimen consisting solely of environmental and social stressors without food/water deprivation or nociceptive stimuli (Uchida et al., 2011a). We subjected BALB mice to a 6-week CUMS regimen or a control period with or without continuous oral administration of the tricyclic antidepressant IMI during the last 3 weeks (Fig. 1A) and assessed depression-like behaviors. In the SI test, which uses decreased SI time with a novel cage mate as an index of depression- and anxiety-like phenotypes (File and Seth, 2003; Berton et al., 2006; Uchida et al., 2011b), mice exposed to CUMS showed reduced SI time compared with NS mice, and this reduction was reversed by IMI (Fig. 1B; two-way ANOVA, stress × drug, F(1,59) = 6.99, p < 0.05; post hoc, NS-water vs CUMS-water, p < 0.05; CUMS-water vs CUMS-IMI, p < 0.05). In the FST, which uses decreased immobility time as an index of antidepressant efficacy (Porsolt et al., 1977; David et al., 2009; Uchida et al., 2011a), CUMS exposure tended to increase immobility time, although not significantly, whereas IMI treatment decreased immobility time in both NS and CUMS-stressed mice (Fig. 1C; two-way ANOVA, drug, F(1,59) = 20.61, p < 0.001; post hoc, NS-water vs NS-IMI, p < 0.05; CUMS-water vs CUMS-IMI, p < 0.05). The novelty-suppressed feeding (NSF) test uses the latency period to begin eating food placed in the center of an open field as an index of anxiety and antidepressant-like response (Santarelli et al., 2003; David et al., 2009; Uchida et al., 2011b). Mice exposed to CUMS showed increased latency to feeding versus NS mice, which was reversed by IMI treatment (Fig. 1D; two-way ANOVA, stress, F(1,59) = 10.75, p < 0.01; drug, F(1,59) = 22.96, p < 0.01; post hoc, NS-water vs NS-IMI, p < 0.05; NS-water vs CUMS-water, p < 0.05; CUMS-water vs CUMS-IMI, p < 0.05). We also administered the SPT to investigate anhedonia (diminished interest or pleasure) and found decreased sucrose preference in mice exposed to CUMS, which again was blocked by IMI (Fig. 1E; two-way ANOVA, stress, F(1,59) = 16.13, p < 0.001; drug, F(1,59) = 4.67, p < 0.05; post hoc, NS-water vs CUMS-water, p < 0.05). These behavioral data suggest that stressed BALB mice are robust animal models of depression.
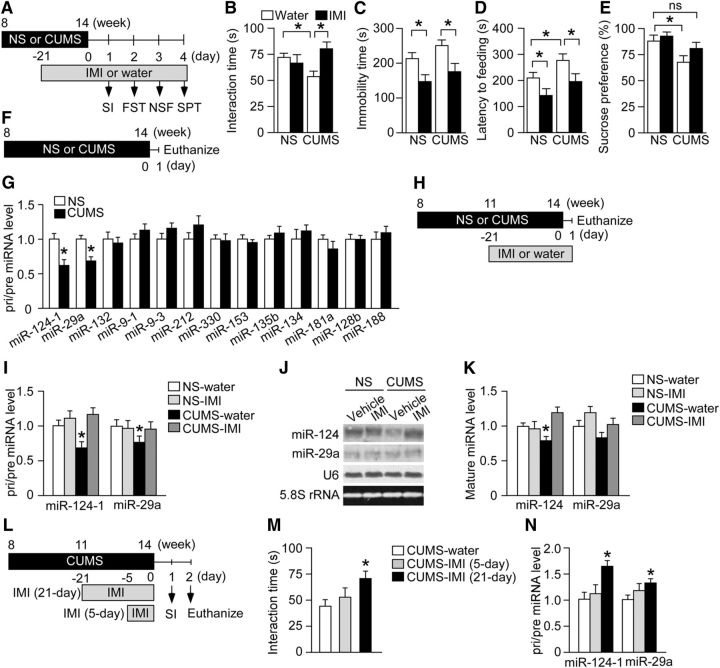
Figure 1.
Chronic stress induces depression-like behaviors and miR-124 downregulation in the hippocampus. A, Schematic of the experimental design for assessing the effects of CUMS and antidepressant treatment on depression-like behaviors. Mice were subjected to CUMS for 6 weeks. The antidepressant IMI was administered orally during the last 3 weeks of the CUMS session. Stressed and NS mice receiving aqueous IMI or water were tested in behavioral assays. B–E, Results of behavioral tests. B, SI time in the SI. C, Immobility time in the FST. D, Latency to feeding in the NSF. E, Sucrose preference in the SPT. CUMS induced multiple depression-like behaviors compared with NS mice received water: reduced SI (B), enhanced latency to feeding (D), and reduced sucrose preference (E). n = 14–18 mice per group. *p < 0.05. ns, Nonsignificant. F, Schematic of the experimental design for analysis of stress-induced changes in hippocampal expression of brain-enriched pri/pre-miRNAs. Mice were subjected to CUMS for 6 weeks. Stressed and NS mice were then killed for hippocampal miRNA expression analysis. G, Levels of pri/pre-miRNAs in the hippocampus of mice subjected to CUMS or the NS condition (n = 6 mice per group). Pri/pre-miR-124-1 and -miR-29a expression levels were reduced by CUMS. *p < 0.05. H, Schematic of the experimental design for testing the effects of IMI on hippocampal miR-124 and miR-29a expression levels. Mice were subjected to a 6-week CUMS protocol with administration of water or IMI during the last 3 weeks and then killed for expression analysis. I, Levels of pri/pre-miR-124-1 and pri/pre-miR-29a in the hippocampus of mice subjected to CUMS or the NS condition and receiving either water or IMI for the last 3 weeks (n = 6 per group). CUMS-induced downregulation of both pri/pre miRNAs was blocked by IMI. *p < 0.05 versus NS-water. J, K, Levels of mature miR-124 and miR-29a in the hippocampus of mice subjected to CUMS or the NS condition and receiving either water or IMI for the last 3 weeks (n = 6 mice per group). Mature miR-124 expression, but not mature miR-29a expression, was downregulated by CUMS. This effect was blocked by IMI. *p < 0.05 versus NS-water. L, Schematic of the experimental design to test the effects of subchronic IMI on depression-like behavior and hippocampal miRNA expression. Mice were subjected to a 6-week CUMS session. IMI was administered during the last 21 d (chronic administration) or 5 d (subchronic administration) of the CUMS session. M, SI time of mice subjected to a 6-week CUMS session with or without IMI treatment (n = 12–14 mice per group). Only chronic IMI administration (21 d) significantly increased the SI time. *p < 0.05 versus CUMS-water. N, Levels of pri/pre-miR-124-1 and pri/pre-miR-29a in the hippocampus of stressed mice receiving subchronic or chronic IMI (n = 6 mice per group). Only chronic administration (21 d) significantly increased these miRNAs. *p < 0.05 versus CUMS-water. All data are presented as mean ± SEM.
We next addressed the role of miRNAs in behavioral responses to chronic stress and antidepressants. We investigated whether chronic stress alters the expression of 13 miRNAs that are known to be enriched in the brain and to be associated with neuronal function and synaptic plasticity and/or suggested to be involved in psychiatric disorders (Smirnova et al., 2005; O'Carroll and Schaefer, 2013; Geaghan and Cairns, 2015; Issler and Chen, 2015; Kocerha et al., 2015). To this end, BALB mice were subjected to the 6-week CUMS regimen or a control period (Fig. 1F) and then killed for real-time PCR analysis to measure the levels of pre/pri-miRNAs in the hippocampus. Expression levels of pri/pre-miR-124-1 and miR-29a were significantly reduced in stressed mice compared with NS control mice (Fig. 1G; pri/pre-miR-124-1, unpaired t test, t = 3.63, p < 0.05; pri/pre-miR-29a, unpaired t test, t = 3.60, p < 0.05) and these effects were blocked by IMI (Fig. 1H,I; pri/pre-miR-124-1: two-way ANOVA, drug, F(1,20) = 16.31, p < 0.01; stress × drug, F(1,20) = 16.31, p < 0.01; post hoc, NS-water vs CUMS-water, p < 0.05, NS-water vs CUMS-IMI, p > 0.05; pri/pre-miR-29a: two-way ANOVA, stress × drug, F(1,20) = 5.12, p < 0.05; post hoc, NS-water vs CUMS-water, p < 0.05). Northern blotting revealed that CUMS exposure reduced mature miR-124, but not miR-29a, expression in the hippocampus and this miR-124 downregulation was blocked by IMI (Figs. 1J,K; miR-124: two-way ANOVA, drug, F(1,20) = 13.85, p < 0.05; stress × drug, F(1,20) = 6.4, p < 0.05; post hoc, NS-water vs CUMS-water, p < 0.05, CUMS-water vs CUMS-IMI, p < 0.05). We also examined the effect of subchronic IMI treatment (5 d) on hippocampal miR-124 expression under the stressed condition (Fig. 1L). Subchronic IMI treatment did not affect the SI time [Fig. 1M; one-way ANOVA, F(2,35) = 6.35, p < 0.05; post hoc, CUMS-water vs CUMS-IMI (5-day), p > 0.05; CUMS-water vs CUMS-IMI (21-day), p < 0.05] and did not alter the expression levels of pri/pre-miR-124-1 and pri/pre-miR-29a compared with stressed mice receiving tap water (Fig. 1N; pri/pre-miR-124-1: one-way ANOVA, F(2,15) = 5.00, p < 0.05; post hoc, CUMS-water vs CUMS-IMI (5-day), p > 0.05; CUMS-water vs CUMS-IMI (21-day), p < 0.05; pri/pre-miR-29a: one-way ANOVA, F(2,15) = 5.09, p < 0.05; post hoc, CUMS-water vs CUMS-IMI (5-day), p > 0.05; CUMS-water vs CUMS-IMI (21-day), p < 0.05). Together, these results suggest an association between stress-induced downregulation of hippocampal miR-124 expression and depression-like behaviors.
Effects of miR-124 overexpression on depression-like behavior
To explore the role of hippocampal miR-124 in the depressive phenotype, we established a mouse model that specifically overexpresses miR-124 in hippocampal neurons. BALB mice were injected with either an AAV expressing both pre-miR-124 and GFP under control of the Camk2a promoter, limiting expression to excitatory neurons, or a control vector expressing GFP into the bilateral hippocampus (Fig. 2A–C). Three weeks after the surgery, mice were subjected to the 6-week CUMS regimen or an equivalent control period and depression-like behaviors were assessed (Fig. 2D). Although NS mice overexpressing miR-124 showed no behavioral changes (Figs. 2E–H), stressed mice overexpressing miR-124 showed significant resilience to the development of depression-like behaviors compared with control CUMS-stressed mice. In the SI test, miR-124 overexpressing mice exposed to CUMS spent more time in SI than control CUMS-stressed mice infected with the negative control vector (Fig. 2E; two-way ANOVA, stress, F(1,42) = 5.06, p < 0.05; stress × virus, F(1,42) = 6.24, p < 0.05; post hoc, NS-GFP vs CUMS-GFP, p < 0.05, CUMS-GFP vs CUMS-miR-124, p < 0.05). Similar results were observed in the NSF and SPT because control mice exposed to CUMS exhibited increased latency to feeding (Fig. 2G) and decreased sucrose preference (Fig. 2H) compared with NS control mice, whereas CUMS-exposed mice overexpressing miR-124 showed no significant differences in these tests compared with NS control mice (Fig. 2G,H; NSF: two-way ANOVA, stress, F(1,42) = 7.78, p < 0.05; post hoc, NS-GFP vs CUMS-GFP, p < 0.05; SPT: two-way ANOVA, stress, F(1,42) = 9.46, p < 0.05; post hoc, NS-GFP vs CUMS-GFP, p < 0.05). In the FST, there was no effect of miR-124 overexpression in CUMS-exposed mice (Fig. 2F). Overall, overexpression of miR-124 in the hippocampus enhanced resilience to CUMS-induced depression-like behaviors.
Effects of miR-124 inhibition on depression-like behaviors
To further determine the importance of hippocampal miR-124 in mediating depression-like behaviors, we injected an anti-miR-124 oligonucleotide (miR-124 inhibitor) into the bilateral hippocampus of BALB mice subjected to a mild repeated restraint stress (RRS) protocol (1 h/d for 14 d) or a control period (NS condition) (Fig. 2I,J). Northern blotting showed successful downregulation of mature miR-124 expression in the hippocampus of mice injected with miR-124 inhibitor (Fig. 2K). BALB mice injected with miR-124 inhibitor showed no behavioral changes in the NS condition (Fig. 2L–O). In addition, BALB mice injected with a negative control nucleotide showed no behavioral changes in response to mild RRS (Fig. 2L–O). Therefore, the use of the mild RRS paradigm enabled us to investigate the effect of miR-124 downregulation on stress resilience. We found that mice injected with miR-124 inhibitor showed decreased SI (Fig. 2L), increased latency to feeding in the NSF test (Fig. 2N), and decreased sucrose preference (Fig. 2O) after mild RRS, but no change in the FST compared with control RRS-stressed mice (Fig. 2M) (SI: two-way ANOVA, stress × drug, F(1,42) = 7.07, p < 0.05; post hoc, NS-miR-124 inhibitor vs RRS-miR-124 inhibitor, p < 0.05, RRS-control inhibitor vs RRS-miR-124 inhibitor, p < 0.05; NSF: two-way ANOVA, stress, F(1,42) = 8.50, p < 0.05; post hoc, NS-miR-124 inhibitor vs RRS-miR-124 inhibitor, p < 0.05; SPT: two-way ANOVA, stress, F(1,42) = 6.06, p < 0.05; post hoc, NS-miR-124 inhibitor vs RRS-miR-124 inhibitor, p < 0.05, RRS-control inhibitor vs RRS-miR-124 inhibitor, p < 0.05). Therefore, decreasing hippocampal miR-124 appears to enhance susceptibility to depression-like behaviors after a normally benign mild chronic stress regimen.
Involvement of miR-124 in hippocampal structural plasticity
Dynamic alterations in synaptic and dendritic structure and function are associated with depressive disorders (Krishnan and Nestler, 2008; Duman and Aghajanian, 2012; Licznerski and Duman, 2013; McEwen et al., 2016). Previous studies reported that chronic stress episodes induced dendritic remodeling in rodent hippocampus that was reversed by antidepressants (Watanabe et al., 1992; Pittenger and Duman, 2008; Gray et al., 2013; Chattarji et al., 2015; McEwen et al., 2016). Indeed, we reported previously that CUMS reduced the dendritic spine density of DG granule neurons in BALB mice and that this effect was blocked by chronic antidepressant treatment (Abe-Higuchi et al., 2016). We thus investigated here whether CUMS-induced changes in dendritic structure are blocked by miR-124 overexpression. To this end, we injected AAV–miR-124 or control AVV–GFP into the bilateral hippocampus (Fig. 3A). miR-124 overexpression had no effect on DG granule neuron dendritic length or spine density in NS mice (Fig. 3B,C). However, miR-124 overexpression blocked the decrease in total dendritic length observed in CUMS-stressed mice injected with control vector (AAV–GFP) (Fig. 3B; two-way ANOVA, stress, F(1,18) = 13.48, p < 0.05; stress × virus, F(1,18) = 10.29, p < 0.05; post hoc, NS-GFP vs CUMS-GFP, p < 0.05). Moreover, the DG granule neuron spine density in stressed mice injected with AAV–GFP was significantly lower than that in NS mice given AAV–GFP, whereas AAV–miR-124 injection reversed this stress-induced effect (Fig. 3C; two-way ANOVA, virus, F(1,18) = 4.82, p < 0.05; post hoc, NS-GFP vs CUMS-GFP, p < 0.05). We then investigated whether blocking miR-124 alters the dendritic morphology of DG granule neurons in the presence and absence of mild RRS. BALB mice were injected with miR-124 inhibitor or control nucleotide into the bilateral hippocampus every 3 d during the 14 d mild RRS or control period (Fig. 3D). Mild RRS had no effect on dendritic length (Fig. 3E). Similarly, treatment with miR-124 inhibitor did not affect total dendritic length or spine density under the NS condition (Fig. 3F). However, mice subjected to mild RRS together with intrahippocampal injection of miR-124 inhibitor showed significantly lower spine density, but normal dendritic length, compared with NS mice injected with control nucleotide (Fig. 3E,F; spine density: two-way ANOVA, stress, F(1,18) = 7.53, p < 0.05; drug, F(1,18) = 9.43, p < 0.05; post hoc, NS-control inhibitor vs RRS-miR-124 inhibitor, p < 0.05). Consistent with behavior effects (Fig. 2I–O), miR-124 downregulation enhanced susceptibility to stress-induced changes in neuroplasticity. These results suggest that miR-124 contributes to the dendritic remodeling induced by chronic stress.
Identifying miR-124 target genes
Because the effects of miRNAs are mediated by posttranslational suppression of gene expression, we screened predicted miR-124 target genes for effects on stress-induced behavior and neuroplasticity. Computational algorithms with Targetscan (Friedman et al., 2009), PiTar (Krek et al., 2005), and/or RNA22 (Miranda et al., 2006) indicate that Hdac4, Hdac5, Gsk3b, and Pde4b, all implicated in depression and antidepressant action (Kaidanovich-Beilin et al., 2004; O'Donnell and Zhang, 2004; Tsankova et al., 2006; Renthal et al., 2007; Sailaja et al., 2012; Liu et al., 2013; Sarkar et al., 2014), are potential targets of miR-124. Moreover, target sites of these genes are conserved among humans, rats, and mice (Fig. 4A–D). We hypothesized that alteration of miR-124 expression by chronic stress or chronic IMI treatment could mediate these behavioral and neuroplastic effects by controlling mRNA and/or protein levels of HDAC4, HDAC5, GSK3β, and/or PDE4B. Indeed, this hypothesis is supported in part by findings that chronic IMI treatment significantly decreased the levels of HDAC4 and HDAC5 proteins (but not of corresponding mRNAs) in the hippocampus of BALB mice exposed to CUMS (Fig. 4E,F; HDAC4 protein: two-way ANOVA, drug, F(1,20) = 6.30, p < 0.05; stress × drug, F(1,20) = 5.00, p < 0.05; post hoc, NS-water vs CUMS-IMI, p < 0.05, CUMS-water vs CUMS-IMI, p < 0.05; HDAC5 protein: two-way ANOVA, stress, F(1,20) = 5.33, p < 0.05; stress × drug, F(1,20) = 5.06, p < 0.05; post hoc, NS-water vs CUMS-IMI, p < 0.05). CUMS did not affect either mRNA or protein expression levels of HDAC4 and HDAC5, but did increase mRNA and protein expression levels of GSK3β, changes reversed by IMI (Fig. 4E,F; GSK3β mRNA: two-way ANOVA, stress × drug, F(1,20) = 6.56, p < 0.05; post hoc, NS-water vs CUMS-water, p < 0.05; GSK3β protein: two-way ANOVA, drug, F(1,20) = 4.04, p < 0.05; stress × drug, F(1,20) = 7.85, p < 0.05; post hoc, NS-water vs CUMS-water, p < 0.05, CUMS-water vs CUMS-IMI, p < 0.05). We did not observe any changes in the mRNA or protein level of PDE4B in mice subjected to CUMS regardless of IMI treatment (Fig. 4E,F). These results suggest that the miR-124–HDAC4/5 pathway may contribute to the behavioral response to IMI, whereas the miR-124–GSK3β pathway could be involved in both depression-like behaviors and antidepressant drug effects.
Figure 4.
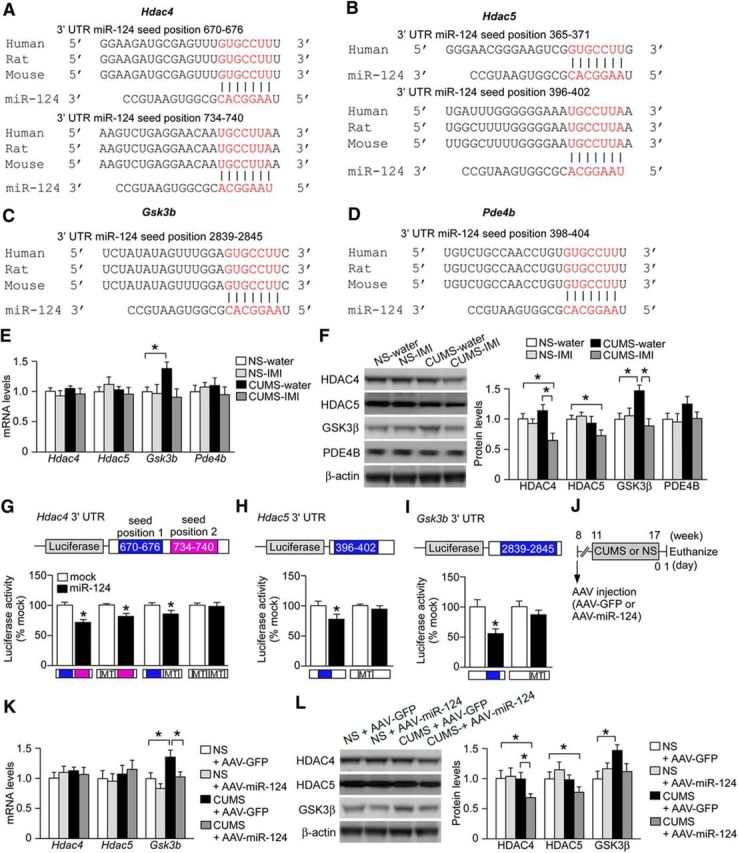
miR-124 represses HDAC4/5 and GSK3β expression. A–D, Alignment of potential binding sites for miR-124 in the 3′ UTRs of Hdac4 (A), Hdac5 (B), Gsk3b (C), and Pde4b (D) mRNAs. All of these potential target sequences of miR-124 are conserved among human, mouse, and rat. E, Levels of mRNAs for Hdac4, Hdac5, Gsk3b, and Pde4b in the hippocampus of mice subjected to a 6-week CUMS session or the NS condition receiving either water or IMI (n = 6 mice per group). CUMS-exposed mice receiving water showed elevated Gsk3b mRNA levels. This overexpression was reversed by IMI. *p < 0.05. F, Protein levels of HDAC4, HDAC5, GSK3β, and PDE4B in the hippocampus of mice subjected to 6-week CUMS or the NS condition receiving water or continuous IMI (n = 6 mice per group). CUMS-exposed mice receiving water showed elevated GSK3β protein level, which was reversed by IMI. IMI also reduced HDAC4 and HDAC5 expressions in CUMS-exposed mice compared with water-treated CUMS-exposed mice. *p < 0.05. G–I, Luciferase reporter assays showing that miR-124 targets Hdac4 3′ UTR, Hdac5 3′ UTR, and Gsk3b 3′ UTR. Mutations (MT) in both the miR-124 seed matches (seed positions 1 and 2) in Hdac4 3′ UTR, seed match in Hdac5 3′ UTR, and seed match in Gsk3b 3′ UTR blocked the inhibitory effects of miR-124. *p < 0.05 versus mock control. J. Schematic of the experimental design to demonstrate that miR-124 overexpression also alters the expression levels of HDAC4, HDAC5, and GSK3β in CUMS-exposed mice. Mice were injected with AAV–miR-124 or AAV–GFP into the bilateral hippocampus. After 3 weeks, they were subjected to CUMS for 6 weeks and then killed for expression analyses. K, Levels Hdac4, Hdac5, and Gsk3b mRNAs in the hippocampus of stressed mice injected with AAV–miR-124. CUMS upregulated Gsk3b mRNA in AAV–GFP-injected mice, an effect reversed by AAV–miR-124-induced miR-124 overexpression. n = 6 mice per group. *p < 0.05. L, Levels of HDAC4, HDAC5, and GSK3β proteins in the hippocampus of stressed mice injected with AAV–miR-124 (n = 6 mice per group). CUMS upregulated GSK3β expression in AAV–GFP-injected mice, an effect blocked by AAV–miR-124-induced miR-124 overexpression. miR-124 overexpression also suppressed HDAC4/5 expression in CUMS-exposed mice. *p < 0.05. All data are presented as mean ± SEM.
To validate the functional relevance of these putative miRNA–mRNA interactions, primary mouse hippocampal neurons infected with either AAV–miR-124 or control AAV–GFP were transfected with luciferase reporters containing mouse Hdac4, Hdac5, or Gsk3b 3′ UTRs. Overexpression of miR-124 suppressed the expression of the luciferase reporter gene with the 3′ UTR of Hdac4, whereas reporter expression was not suppressed when we mutated two miR-124 seed match sequences [seed positions 1 and 2; Fig. 4G; unpaired t test, WT 3′ UTR, t = 2.83, p < 0.05; mutant 3′ UTR (seed position 1), t = 3.60, p < 0.05; mutant 3′ UTR (seed position 2), t = 4.19, p < 0.05; double mutant 3′ UTR, t = 0.48, p > 0.05]. Similarly, the expression levels of luciferase reporter genes with the 3′ UTRs of Hdac5 and Gsk3b were repressed by miR-124 overexpression, but these effects were not observed when we transfected reporter constructs containing Hdac5 or Gsk3b 3′ UTR mutated at the seed positions for miR-124 binding (Fig. 4H,I; Hdac5 3′ UTR: unpaired t test, WT 3′ UTR, t = 3.55, p < 0.05; mutant 3′ UTR, t = 0.98, p > 0.05; Gsk3b 3′ UTR: unpaired t test, WT 3′ UTR, t = 7.17, p < 0.05; mutant 3′ UTR, t = 0.46, p > 0.05).
Subsequently, we investigated whether the expression levels of HDAC4, HDAC5, and GSK3β are regulated by miR-124 in vivo. BALB mice were injected with either AAV–miR-124 or AAV–GFP into the bilateral hippocampus before a 6-week CUMS session or control period (Fig. 4J). We found no significant effects of miR-124 overexpression on HDAC4 or HDAC5 mRNA expression levels, whereas increased mRNA expression of GSK3β by CUMS was repressed by miR-124 overexpression (Fig. 4K; two-way ANOVA, stress, F(1,20) = 9.96, p < 0.05; post hoc, NS-GFP vs CUMS-GFP, p < 0.05; CUMS-GFP vs CUMS-miR-124, p < 0.05). At the protein level, miR-124 overexpression did not affect the expression of HDAC4 and HDAC5 in NS mice, whereas protein levels were significantly reduced by miR-124 overexpression in CUMS-stressed mice compared with both NS and stressed mice injected with AAV–GFP (Fig. 4L; HDAC4: two-way ANOVA, stress, F(1,20) = 7.00, p < 0.05; stress × virus, F(1,20) = 7.31, p < 0.05; post hoc, NS-GFP vs CUMS-miR-124, p < 0.05; CUMS-GFP vs CUMS-miR-124, p < 0.05; HDAC5: two-way ANOVA, stress, F(1,20) = 8.07, p < 0.05; stress × virus, F(1,20) = 7.80, p < 0.05; post hoc, NS-GFP vs CUMS-miR-124, p < 0.05). In addition, CUMS-induced upregulation of GSK3β protein was blocked by miR-124 overexpression (Fig. 4L; two-way ANOVA, stress, F(1,20) = 5.66, p < 0.05; stress × virus, F(1,20) = 5.39, p < 0.05; post hoc, NS-GFP vs CUMS-GFP, p < 0.05). Overall, these results suggest that miR-124 regulates changes in HDAC4, HDAC5, and GSK3β expression levels induced by CUMS, but not basal expression levels.
Pharmacological inhibition of HDAC4/5 and GSK3β induce antidepressant-like behavioral responses
Finally, to test the importance of miR-124 targets in mediating antidepressant-like behavior, we synthesized LMK-235 (N-((6-(hydroxyamino)- 6-oxohexyl)oxy)-3,5-dimethylbenzamide), a potent and selective HDAC4/5 inhibitor (Marek et al., 2013), infused this drug into the bilateral hippocampus of BALB mice using osmotic pumps, and measured behaviors in the SI, FST, NSF, and SPT (Fig. 5A–C). NS mice infused with LMK-235 (1 μm) showed increased SI time (Fig. 5D; one-way ANOVA, F(2,30) = 3.73, p < 0.05; post hoc, vehicle vs LMK-235 (1 μm), p < 0.05), decreased immobility time in the FST (Fig. 5E; one-way ANOVA, F(2,30) = 4.01, p < 0.05; post hoc, vehicle vs LMK-235 (1 μm), p < 0.05), and shorter latency to feeding in the NSF compared with vehicle-infused NS mice (Fig. 5F; one-way ANOVA, F(2,30) = 3.90, p < 0.05; post hoc, vehicle vs LMK-235 (1 μm), p < 0.05), whereas this drug was not effective in the SPT (Fig. 5G; one-way ANOVA, F(2,30) = 0.20, p > 0.05). We also examined the antidepressant action of LMK-235 in mice exposed to a severe RRS regimen (4 h/d for 10 d), which can also be used as an animal model of depression (Abe-Higuchi et al., 2016). BALB mice implanted with osmotic minipumps containing LMK-235 or vehicle were subjected to severe RRS episodes (Fig. 5H,I). As expected, severe RRS exposure in mice infused with vehicle reduced SI and sucrose preference, and these effects of severe RRS were blocked in mice infused with LMK-235 (Fig. 5J,K; SI: one-way ANOVA, F(2,34) = 6.42, p < 0.05; post hoc, NS-vehicle vs RRS-vehicle, p < 0.05; NS-vehicle vs RRS-LMK-235, p > 0.05; SPT: one-way ANOVA, F(2,34) = 4.99, p < 0.05; post hoc, NS-vehicle vs RRS-vehicle, p < 0.05; NS-vehicle vs RRS-LMK-235, p > 0.05). These results suggest an antidepressant-like action of HDAC4/5 inhibition in the hippocampus.
We subsequently investigated the antidepressant effect of LY2090314, a potent and selective GSK3 inhibitor (Atkinson et al., 2015). We infused LY2090314 into the bilateral hippocampus of mice using osmotic pumps and measured these same behaviors (Fig. 6A–C). NS mice infused with LY2090314 (100 nm) showed increased SI time (Fig. 6D; one-way ANOVA, F(2,30) = 7.00, p < 0.05; post hoc, vehicle vs LY2090314 (100 nm), p < 0.05) and shorter latency to feeding in the NSF (Fig. 6F; one-way ANOVA, F(2,30) = 3.55, p < 0.05; post hoc, vehicle vs LY2090314 100 nm, p < 0.05) compared with vehicle-infused stressed mice, whereas there were no effects on the FST (Fig. 6E; one-way ANOVA, F(2,30) = 0.62, p > 0.05) or SPT (Fig. 6G; one-way ANOVA, F(2,30) = 0.97, p > 0.05). We also investigated whether LY2090314 has an antidepressant action in depressive stressed mice. BALB mice implanted with osmotic minipumps containing LY2090314 or vehicle were subjected to severe RRS episodes (Fig. 6H,I). LY2090314 infusion prevented the severe RRS-induced reduction in SI, but had no significant effect on reduced sucrose preference (Fig. 6J,K; SI: one-way ANOVA, F(2,38) = 3.92, p < 0.05; post hoc, NS-vehicle vs RRS-vehicle, p < 0.05; NS-vehicle vs RRS-LY2090314, p > 0.05; SPT: one-way ANOVA, F(2,38) = 5.89, p < 0.05; post hoc, NS-vehicle vs RRS-vehicle, p < 0.05; NS-vehicle vs RRS-LY2090314, p < 0.05). These results suggest that the inhibition of GSK3 can block some depression-like behaviors induced by severe RRS.
Discussion
There is accumulating evidence indicating a significant contribution of miRNAs to depression, anxiety, and antidepressant action. A recent preclinical study showed that overexpression of miR-34c in the central amygdala led to anxiolytic-like effects through the downregulation of corticotropin-releasing factor receptor type 1, a key mediator of stress responses (Haramati et al., 2011). Treatment with fluoxetine, a selective serotonin reuptake inhibitor, decreased miR-16 expression in the locus ceruleus and miR-16 inhibition in the locus ceruleus blocked chronic stress-induced depression-related behavior through modulation of serotonin transporter expression (Baudry et al., 2010). Moreover, treatment with antidepressants increased miR-135a levels in the dorsal raphe nucleus and overexpression of miR-135a led to resilience against chronic social defeat stress by targeting raphe mRNAs encoding the serotonin transporter and serotonin receptor 1A (Issler et al., 2014). More importantly, depressed individuals and suicide victims had reduced miR-135a levels in the blood and postmortem dorsal raphe nucleus, respectively, and blood miR-135a levels in depressed patients increased after antidepressant treatment (Issler et al., 2014). Therefore, posttranscriptional regulation by miRNA networks in various brain regions is likely involved in depression and antidepressant drug actions. Although the hippocampus is strongly associated with depression and antidepressant actions, to our knowledge, this is the first study demonstrating a critical role for hippocampal miRNAs in chronic stress-induced depression-like behaviors and aberrant neuroplasticity.
Here, we demonstrate that hippocampal miR-124 mediates behavioral responses to chronic stress in mice. Increasing hippocampal miR-124 blocked depression-like behaviors caused by chronic stress, resulting in a state of stress resilience. Conversely, inhibition of hippocampal miR-124 enhanced behavioral susceptibility to a mild RRS protocol. These behavioral effects were paralleled by changes in the dendritic structure of DG granule neurons. Chronic stress-induced spine loss was prevented by miR-124 overexpression and miR-124 inhibition reduced spine density under mild stress. Therefore, chronic-stress-induced aberrant dendritic plasticity may be caused by downregulation of miR-124. Moreover, we found that miR-124 targets Hdac4, Hdac5, and Gsk3b mRNAs and that pharmacological inhibition of HDAC4/5 and GSK3 had antidepressant-like behavioral effects. Collectively, our findings suggest that diminished hippocampal miR-124 mediates chronic stress-induced depression-like behaviors and aberrant structural plasticity, at least in part by ensuing upregulation of HDAC4/5 and/or GSK3β.
In contrast to our results, one study reported that social defeat stress increased miR-124 levels, which in turn downregulated the expression of another target, the mRNA-encoding brain derived neurotrophic factor (BDNF), in the rat hippocampus (Bahi et al., 2014). Furthermore, virus-mediated miR-124 overexpression exacerbated, whereas inhibition decreased, stress-induced depression-like behaviors (Bahi et al., 2014), which is in direct contradiction to our findings. However, another study found that overexpression of miR-124 rescued the reduced SI observed in a mouse model of frontotemporal dementia (Gascon et al., 2014), supporting our notion that enhancing hippocampal miR-124 pathways can sustain normal behaviors or reverse aberrant behaviors (such as mood and emotion). Differences in model species, genetic background, stress paradigm, and their interactions may account for the discrepancies between our results and those of Bahi et al. (2014). We used BALB mice subjected to CUMS, whereas they used rats exposed to chronic social defeat stress. In mice, C57BL/6 strain subjected to chronic social defeat stress exhibit reduced hippocampal Bdnf mRNA (Tsankova et al., 2006), whereas BALB strain mice subjected to CUMS do not (Uchida et al., 2011a). Therefore, individual stress protocols and genetic background may have unique effects on gene expression profiles. Future studies are necessary to clarify how genetic–environment interactions influence miRNA expression and the functions of these noncoding RNAs in depression. Another possible explanation for this discrepancy is the contribution of miR-124 in specific neural cell types to depression- and anxiety-like behaviors. We used AAV vectors with the Camk2a promoter to overexpress miR-124 specifically in excitatory neurons, whereas Bahi et al. (2014) used lentiviral vector in which expression was driven by the ubiquitous cytomegalovirus promoter. Therefore, miR-124 overexpression in different cell types may produce disparate behavioral outcomes. Indeed, a recent report demonstrated promoter-specific effects of viral-mediated gene transfer on synaptic transmission and plasticity in the hippocampus (López et al., 2016). Nonetheless, our results are generally consistent with human studies underscoring the importance of miRNA dysregulation in the pathogenesis of depression (O'Connor et al., 2012; Dwivedi, 2014; Geaghan and Cairns, 2015; Issler and Chen, 2015; Kocerha et al., 2015).
Multiple lines of evidence indicate that altered dendritic structure and neurogenesis result, at least in part, from a common posttranscriptional process mediated by miRNAs (Schratt et al., 2006; O'Carroll and Schaefer, 2013; Dwivedi, 2014). miRNA-124 is abundantly expressed in neurons, where it regulates adult neurogenesis and promotes dendritic maturation (Kuwabara et al., 2004; Smirnova et al., 2005; Cao et al., 2007; Makeyev et al., 2007; Cheng et al., 2009). Therefore, our finding that miR-124 overexpression prevented chronic stress-induced dendritic atrophy and spine loss in the DG suggests that downregulation of hippocampal miR-124 pathways is a causal mechanism underlying aberrant dendritic remodeling induced by chronic stress. Both altered dendritic structure and neurogenesis may be involved in depression (Krishnan and Nestler, 2008; Franklin et al., 2012; Schoenfeld and Gould, 2012; Chattarji et al., 2015; Gold, 2015; McEwen et al., 2015). Dendritic spines are the major site for dynamic structural plasticity of excitatory transmission (Holtmaat and Svoboda, 2009). Several preclinical studies reported that chronic stress exposure induced spine loss in hippocampal neurons and impaired synaptic transmission concomitant with the emergence of depressive behaviors (Kassem et al., 2013; Qiao et al., 2014). Further, downregulation of genes related to synaptic function and the decreased number of spine synapses were observed in postmortem brain from depressed patients (Kang et al., 2012). Therefore, there is increasing correlative evidence of spine dysfunction in depression. However, there is no causative evidence for synaptic spines in controlling the cellular physiology of disease states and subsequent affective behaviors. Studies using novel investigative tools such as synaptic optogenetics (Hayashi-Takagi et al., 2015) may uncover the mechanistic relationships between synaptic dysfunction and depression-like behaviors.
We identified three miR-124 target mRNAs in vitro and in vivo. Chronic stress increased both mRNA and protein levels of GSK3β and these increases were blocked by antidepressant treatment. In contrast, chronic stress did not affect mRNA and protein levels of HDAC4 and HDAC5, but antidepressant treatment decreased protein levels under the stressed condition. Considering that miRNAs mediate cleavage or translational repression of target mRNAs (Kosik, 2006; Bartel, 2009), our data suggest that miR-124 cleaves GSK3β mRNA in response to chronic stress, whereas translational inhibition of HDAC4/5 mRNAs by miR-124 is suppressed by antidepressant treatment under the stressed condition. Therefore, the target gene specificity and activity of miR-124 may be context dependent. Intriguingly, both GSK3β and HDAC4/5 have been associated with depression and antidepressant drug actions. Enhancement of GSK3 activity contributed to the reduction in synaptic spine density in response to stress (Collingridge et al., 2010; Duman and Aghajanian, 2012) and postmortem studies have shown increased GSK3β activity in patients with major depressive disorder (Li and Jope, 2010). A recent report found that depression-related behavior in rats induced by postnatal administration of fluoxetine was accompanied by HDAC4 recruitment to specific depression-linked genes (such as mTOR) and that these depressive behaviors were prevented by postnatal cotreatment with the HDAC inhibitor sodium butyrate (Sarkar et al., 2014). In addition, viral-mediated suppression of hippocampal HDAC4 completely abolished the behavioral effect of stress (Sailaja et al., 2012), whereas viral-mediated overexpression of HDAC5 suppressed the behavioral response to an antidepressant drug (Tsankova et al., 2006). A clinical study also found altered HDAC4/5 expression in mood disorder patients (Hobara et al., 2010). Moreover, in our present study, local infusion of an HDAC4/5 inhibitor or a GSK3 inhibitor induced antidepressant-like behavioral effects. Therefore, both preclinical and clinical studies strongly support our notion that GSK3β and HDAC4/5 contribute to stress-induced aberrant structural plasticity, depression-like behaviors, and antidepressant drug action.
Another important implication of this study is that miR-124 pathways may contribute to the epigenetic gene regulation required for antidepressant drug action. Both chronic stress and antidepressants induce epigenetic chromatin remodeling (Tsankova et al., 2006; Krishnan and Nestler, 2008; Uchida et al., 2011a). The contribution of epigenetic mechanisms to synaptic plasticity is also well documented (Guan et al., 2009). miRNA-124 has been shown to regulate gene expression through DNA methylation and histone deacetylation (Lujambio et al., 2007; Oikawa et al., 2015) and our data further suggest that downstream pathways involving miR-124 targets (such as HDAC4/5) also control gene expression through epigenetic mechanisms. Feedback and feedforward pathways between miR-124 and epigenetic gene regulation mechanisms may serve to sustain the molecular and behavioral responses to chronic stress that are associated with depression.
In conclusion, we propose that miR-124 is an important regulator of structural plasticity and behavioral responses to chronic stress. Detailed descriptions of the miR-124-associated gene networks involved in stress-induced epigenetic gene regulation, dendritic structural remodeling, and behavioral changes may reveal new aspects of depression pathophysiology. Moreover, the involvement of miRNAs in depression is strongly supported by this and previous studies showing that chronic stress and clinical antidepressants alter miRNA expression. Therefore, direct modulation of miRNA pathways may be an effective therapeutic strategy for depression.
Footnotes
This work was supported by grants-in-aid from the Japan Society for the Promotion of Science (F.H., S.U., Y.W.), Core Research for Evolutional Science and Technology and Japan Science and Technology Agency (S.U. and T. Suzuki), and the Integrated Research on Neuropsychiatric Disorders performed under the Strategic Research Program for Brain Sciences from the Ministry of Education, Culture, Sports, Science and Technology of Japan and Japan Agency for Medical Research and Development (H.Y.) and by research grants from the New Frontier Research Project in Yamaguchi University (S.U.), the SENSHIN Medical Research Foundation (Y.W.), the Uehara Memorial Foundation (S.U.), and the Takeda Science Foundation (S.U.). We thank Drs. Yoichi Mizukami, Koichi Udo, and Tomoaki Murata for technical advice.
The authors declare no competing financial interests.
References
- Abe-Higuchi N, Uchida S, Yamagata H, Higuchi F, Hobara T, Hara K, Kobayashi A, Watanabe Y. Hippocampal Sirtuin 1 signaling mediates depression-like behavior. Biol Psychiatry. 2016;pii:S0006–3223(16) doi: 10.1016/j.biopsych.2016.01.009. 00080-9. [DOI] [PubMed] [Google Scholar]
- Atkinson JM, Rank KB, Zeng Y, Capen A, Yadav V, Manro JR, Engler TA, Chedid M. Activating the Wnt/beta-catenin pathway for the treatment of melanoma–application of LY2090314, a novel selective inhibitor of glycogen synthase kinase-3. PLoS One. 2015;10:e0125028. doi: 10.1371/journal.pone.0125028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahi A, Chandrasekar V, Dreyer JL. Selective lentiviral-mediated suppression of microRNA124a in the hippocampus evokes antidepressants-like effects in rats. Psychoneuroendocrinology. 2014;46:78–87. doi: 10.1016/j.psyneuen.2014.04.009. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry A, Mouillet-Richard S, Schneider B, Launay JM, Kellermann O. miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science. 2010;329:1537–1541. doi: 10.1126/science.1193692. [DOI] [PubMed] [Google Scholar]
- Berton O, McClung CA, Dileone RJ, Krishnan V, Renthal W, Russo SJ, Graham D, Tsankova NM, Bolanos CA, Rios M, Monteggia LM, Self DW, Nestler EJ. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. 2006;311:864–868. doi: 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattarji S, Tomar A, Suvrathan A, Ghosh S, Rahman MM. Neighborhood matters: divergent patterns of stress-induced plasticity across the brain. Nat Neurosci. 2015;18:1364–1375. doi: 10.1038/nn.4115. [DOI] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingridge GL, Peineau S, Howland JG, Wang YT. Long-term depression in the CNS. Nat Rev Neurosci. 2010;11:459–473. doi: 10.1038/nrn2867. [DOI] [PubMed] [Google Scholar]
- David DJ, Samuels BA, Rainer Q, Wang JW, Marsteller D, Mendez I, Drew M, Craig DA, Guiard BP, Guilloux JP, Artymyshyn RP, Gardier AM, Gerald C, Antonijevic IA, Leonardo ED, Hen R. Neurogenesis-dependent and -independent effects of fluoxetine in an animal model of anxiety/depression. Neuron. 2009;62:479–493. doi: 10.1016/j.neuron.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias C, Feng J, Sun H, et al. beta-catenin mediates stress resilience through Dicer1/microRNA regulation. Nature. 2014;516:51–55. doi: 10.1038/nature13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman RS, Aghajanian GK. Synaptic dysfunction in depression: potential therapeutic targets. Science. 2012;338:68–72. doi: 10.1126/science.1222939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y. Emerging role of microRNAs in major depressive disorder: diagnosis and therapeutic implications. Dialogues Clin Neurosci. 2014;16:43–61. doi: 10.31887/DCNS.2014.16.1/ydwivedi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder A, Nestler EJ, Charney DS. Psychobiology and molecular genetics of resilience. Nat Rev Neurosci. 2009;10:446–457. doi: 10.1038/nrn2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- File SE, Seth P. A review of 25 years of the social interaction test. Eur J Pharmacol. 2003;463:35–53. doi: 10.1016/S0014-2999(03)01273-1. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K, Paxinos G. The mouse brain in stereotaxic coordinates. Ed 3. New York: Academic; 2008. [Google Scholar]
- Franklin TB, Saab BJ, Mansuy IM. Neural mechanisms of stress resilience and vulnerability. Neuron. 2012;75:747–761. doi: 10.1016/j.neuron.2012.08.016. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascon E, Lynch K, Ruan H, Almeida S, Verheyden JM, Seeley WW, Dickson DW, Petrucelli L, Sun D, Jiao J, Zhou H, Jakovcevski M, Akbarian S, Yao WD, Gao FB. Alterations in microRNA-124 and AMPA receptors contribute to social behavioral deficits in frontotemporal dementia. Nat Med. 2014;20:1444–1451. doi: 10.1038/nm.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geaghan M, Cairns MJ. MicroRNA and posttranscriptional dysregulation in psychiatry. Biol Psychiatry. 2015;78:231–239. doi: 10.1016/j.biopsych.2014.12.009. [DOI] [PubMed] [Google Scholar]
- Gold PW. The organization of the stress system and its dysregulation in depressive illness. Mol Psychiatry. 2015;20:32–47. doi: 10.1038/mp.2014.163. [DOI] [PubMed] [Google Scholar]
- Gray JD, Milner TA, McEwen BS. Dynamic plasticity: the role of glucocorticoids, brain-derived neurotrophic factor and other trophic factors. Neuroscience. 2013;239:214–227. doi: 10.1016/j.neuroscience.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramati S, Navon I, Issler O, Ezra-Nevo G, Gil S, Zwang R, Hornstein E, Chen A. MicroRNA as repressors of stress-induced anxiety: the case of amygdalar miR-34. J Neurosci. 2011;31:14191–14203. doi: 10.1523/JNEUROSCI.1673-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Takagi A, Yagishita S, Nakamura M, Shirai F, Wu YI, Loshbaugh AL, Kuhlman B, Hahn KM, Kasai H. Labelling and optical erasure of synaptic memory traces in the motor cortex. Nature. 2015;525:333–338. doi: 10.1038/nature15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobara T, Uchida S, Otsuki K, Matsubara T, Funato H, Matsuo K, Suetsugi M, Watanabe Y. Altered gene expression of histone deacetylases in mood disorder patients. J Psychiatr Res. 2010;44:263–270. doi: 10.1016/j.jpsychires.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci. 2009;10:647–658. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325–334. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issler O, Chen A. Determining the role of microRNAs in psychiatric disorders. Nat Rev Neurosci. 2015;16:201–212. doi: 10.1038/nrn3879. [DOI] [PubMed] [Google Scholar]
- Issler O, Haramati S, Paul ED, Maeno H, Navon I, Zwang R, Gil S, Mayberg HS, Dunlop BW, Menke A, Awatramani R, Binder EB, Deneris ES, Lowry CA, Chen A. MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron. 2014;83:344–360. doi: 10.1016/j.neuron.2014.05.042. [DOI] [PubMed] [Google Scholar]
- Kaidanovich-Beilin O, Milman A, Weizman A, Pick CG, Eldar-Finkelman H. Rapid antidepressive-like activity of specific glycogen synthase kinase-3 inhibitor and its effect on beta-catenin in mouse hippocampus. Biol Psychiatry. 2004;55:781–784. doi: 10.1016/j.biopsych.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Voleti B, Hajszan T, Rajkowska G, Stockmeier CA, Licznerski P, Lepack A, Majik MS, Jeong LS, Banasr M, Son H, Duman RS. Decreased expression of synapse-related genes and loss of synapses in major depressive disorder. Nat Med. 2012;18:1413–1417. doi: 10.1038/nm.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassem MS, Lagopoulos J, Stait-Gardner T, Price WS, Chohan TW, Arnold JC, Hatton SN, Bennett MR. Stress-induced grey matter loss determined by MRI is primarily due to loss of dendrites and their synapses. Mol Neurobiol. 2013;47:645–661. doi: 10.1007/s12035-012-8365-7. [DOI] [PubMed] [Google Scholar]
- Kocerha J, Dwivedi Y, Brennand KJ. Noncoding RNAs and neurobehavioral mechanisms in psychiatric disease. Mol Psychiatry. 2015;20:677–684. doi: 10.1038/mp.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Krishnan V, Nestler EJ. The molecular neurobiology of depression. Nature. 2008;455:894–902. doi: 10.1038/nature07455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116:779–793. doi: 10.1016/S0092-8674(04)00248-X. [DOI] [PubMed] [Google Scholar]
- Li X, Jope RS. Is glycogen synthase kinase-3 a central modulator in mood regulation? Neuropsychopharmacology. 2010;35:2143–2154. doi: 10.1038/npp.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licznerski P, Duman RS. Remodeling of axo-spinous synapses in the pathophysiology and treatment of depression. Neuroscience. 2013;251:33–50. doi: 10.1016/j.neuroscience.2012.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu RJ, Fuchikami M, Dwyer JM, Lepack AE, Duman RS, Aghajanian GK. GSK-3 inhibition potentiates the synaptogenic and antidepressant-like effects of subthreshold doses of ketamine. Neuropsychopharmacology. 2013;38:2268–2277. doi: 10.1038/npp.2013.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López AJ, Kramár E, Matheos DP, White AO, Kwapis J, Vogel-Ciernia A, Sakata K, Espinoza M, Wood MA. Promoter-specific effects of DREADD modulation on hippocampal synaptic plasticity and memory formation. J Neurosci. 2016;36:3588–3599. doi: 10.1523/JNEUROSCI.3682-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Git A, Gitt A, Spiteri I, Das PP, Caldas C, Miska E, Esteller M. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek L, Hamacher A, Hansen FK, Kuna K, Gohlke H, Kassack MU, Kurz T. Histone deacetylase (HDAC) inhibitors with a novel connecting unit linker region reveal a selectivity profile for HDAC4 and HDAC5 with improved activity against chemoresistant cancer cells. J Med Chem. 2013;56:427–436. doi: 10.1021/jm301254q. [DOI] [PubMed] [Google Scholar]
- Martel G, Uchida S, Hevi C, Chévere-Torres I, Fuentes I, Park YJ, Hafeez H, Yamagata H, Watanabe Y, Shumyatsky GP. Genetic demonstration of a role for stathmin in adult hippocampal neurogenesis, spinogenesis, and NMDA receptor-dependent memory. J Neurosci. 2016;36:1185–1202. doi: 10.1523/JNEUROSCI.4541-14.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS. Plasticity of the hippocampus: adaptation to chronic stress and allostatic load. Ann N Y Acad Sci. 2001;933:265–277. doi: 10.1111/j.1749-6632.2001.tb05830.x. [DOI] [PubMed] [Google Scholar]
- McEwen BS. Physiology and neurobiology of stress and adaptation: central role of the brain. Physiol Rev. 2007;87:873–904. doi: 10.1152/physrev.00041.2006. [DOI] [PubMed] [Google Scholar]
- McEwen BS, Nasca C, Gray JD. Stress effects on neuronal structure: hippocampus, amygdala, and prefrontal cortex. Neuropsychopharmacology. 2016;41:3–23. doi: 10.1038/npp.2015.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS, Bowles NP, Gray JD, Hill MN, Hunter RG, Karatsoreos IN, Nasca C. Mechanisms of stress in the brain. Nat Neurosci. 2015;18:1353–1363. doi: 10.1038/nn.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Barrot M, DiLeone RJ, Eisch AJ, Gold SJ, Monteggia LM. Neurobiology of depression. Neuron. 2002;34:13–25. doi: 10.1016/S0896-6273(02)00653-0. [DOI] [PubMed] [Google Scholar]
- O'Carroll D, Schaefer A. General principals of miRNA biogenesis and regulation in the brain. Neuropsychopharmacology. 2013;38:39–54. doi: 10.1038/npp.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor RM, Dinan TG, Cryan JF. Little things on which happiness depends: microRNAs as novel therapeutic targets for the treatment of anxiety and depression. Mol Psychiatry. 2012;17:359–376. doi: 10.1038/mp.2011.162. [DOI] [PubMed] [Google Scholar]
- O'Donnell JM, Zhang HT. Antidepressant effects of inhibitors of cAMP phosphodiesterase (PDE4) Trends Pharmacol Sci. 2004;25:158–163. doi: 10.1016/j.tips.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Oikawa H, Goh WW, Lim VK, Wong L, Sng JC. Valproic acid mediates miR-124 to down-regulate a novel protein target, GNAI1. Neurochem Int. 2015;91:62–71. doi: 10.1016/j.neuint.2015.10.010. [DOI] [PubMed] [Google Scholar]
- Pittenger C, Duman RS. Stress, depression, and neuroplasticity: a convergence of mechanisms. Neuropsychopharmacology. 2008;33:88–109. doi: 10.1038/sj.npp.1301574. [DOI] [PubMed] [Google Scholar]
- Porsolt RD, Bertin A, Jalfre M. Behavioral despair in mice: a primary screening test for antidepressants. Arch Int Pharmacodyn Ther. 1977;229:327–336. [PubMed] [Google Scholar]
- Qiao H, An SC, Ren W, Ma XM. Progressive alterations of hippocampal CA3-CA1 synapses in an animal model of depression. Behav Brain Res. 2014;275:191–200. doi: 10.1016/j.bbr.2014.08.040. [DOI] [PubMed] [Google Scholar]
- Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil SV, Russo JJ, Sander C, Tuschl T, Kandel E. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63:803–817. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, 3rd, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, Kerstetter KA, Neve RL, Haggarty SJ, McKinsey TA, Bassel-Duby R, Olson EN, Nestler EJ. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Sailaja BS, Cohen-Carmon D, Zimmerman G, Soreq H, Meshorer E. Stress-induced epigenetic transcriptional memory of acetylcholinesterase by HDAC4. Proc Natl Acad Sci U S A. 2012;109:E3687–E3695. doi: 10.1073/pnas.1209990110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarelli L, Saxe M, Gross C, Surget A, Battaglia F, Dulawa S, Weisstaub N, Lee J, Duman R, Arancio O, Belzung C, Hen R. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science. 2003;301:805–809. doi: 10.1126/science.1083328. [DOI] [PubMed] [Google Scholar]
- Sarkar A, Chachra P, Kennedy P, Pena CJ, Desouza LA, Nestler EJ, Vaidya VA. Hippocampal HDAC4 contributes to postnatal fluoxetine-evoked depression-like behavior. Neuropsychopharmacology. 2014;39:2221–2232. doi: 10.1038/npp.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HD, Duman RS. The role of neurotrophic factors in adult hippocampal neurogenesis, antidepressant treatments and animal models of depressive-like behavior. Behav Pharmacol. 2007;18:391–418. doi: 10.1097/FBP.0b013e3282ee2aa8. [DOI] [PubMed] [Google Scholar]
- Schoenfeld TJ, Gould E. Stress, stress hormones, and adult neurogenesis. Exp Neurol. 2012;233:12–21. doi: 10.1016/j.expneurol.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Smirnova L, Gräfe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- Uchida S, Nishida A, Hara K, Kamemoto T, Suetsugi M, Fujimoto M, Watanuki T, Wakabayashi Y, Otsuki K, McEwen BS, Watanabe Y. Characterization of the vulnerability to repeated stress in Fischer 344 rats: possible involvement of microRNA-mediated down-regulation of the glucocorticoid receptor. Eur J Neurosci. 2008;27:2250–2261. doi: 10.1111/j.1460-9568.2008.06218.x. [DOI] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Funato H, Hobara T, Otsuki K, Yamagata H, McEwen BS, Watanabe Y. Early life stress enhances behavioral vulnerability to stress through the activation of REST4-mediated gene transcription in the medial prefrontal cortex of rodents. J Neurosci. 2010;30:15007–15018. doi: 10.1523/JNEUROSCI.1436-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Otsuki K, Yamagata H, Hobara T, Suzuki T, Miyata N, Watanabe Y. Epigenetic status of Gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron. 2011a;69:359–372. doi: 10.1016/j.neuron.2010.12.023. [DOI] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Fujimoto M, Otsuki K, Yamagata H, Hobara T, Abe N, Higuchi F, Shibata T, Hasegawa S, Kida S, Nakai A, Watanabe Y. Impaired hippocampal spinogenesis and neurogenesis and altered affective behavior in mice lacking heat shock factor 1. Proc Natl Acad Sci U S A. 2011b;108:1681–1686. doi: 10.1073/pnas.1016424108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Martel G, Pavlowsky A, Takizawa S, Hevi C, Watanabe Y, Kandel ER, Alarcon JM, Shumyatsky GP. Learning-induced and stathmin-dependent changes in microtubule stability are critical for memory and disrupted in ageing. Nat Commun. 2014;5:4389. doi: 10.1038/ncomms5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Gould E, McEwen BS. Stress induces atrophy of apical dendrites of hippocampal CA3 pyramidal neurons. Brain Res. 1992;588:341–345. doi: 10.1016/0006-8993(92)91597-8. [DOI] [PubMed] [Google Scholar]
- Willner P, Towell A, Sampson D, Sophokleous S, Muscat R. Reduction of sucrose preference by chronic unpredictable mild stress, and its restoration by a tricyclic antidepressant. Psychopharmacology (Berl) 1987;93:358–364. doi: 10.1007/BF00187257. [DOI] [PubMed] [Google Scholar]