INTRODUCTION
The epidermal growth factor receptor/human epidermal growth factor receptor (HER) family plays a critical role in the growth of numerous human cancers. Multiple therapeutics targeted to members of the HER family have been developed, including the epidermal growth factor receptor–targeting monoclonal antibody cetuximab, which is FDA approved for treatment of head and neck squamous cell carcinoma (HNSCC). HER3/ErbB3 is expressed in most solid tumors, including HNSCC, and has been shown to correlate with a poor prognosis.1,2 Phosphorylation of ErbB3 (pErbB3) affects numerous pathways, including activation of the PI3K/AKT pathway, driving cell survival and growth. For this reason, blocking ErbB3 activity could be an effective strategy to decrease tumor growth and resistance to other tyrosine kinase inhibitors.3-5
CDX-3379 is a human immunoglobulin G1 lambda monoclonal antibody that binds ErbB3 and inhibits its activity by multiple mechanisms. In vivo, CDX-3379 exhibits significant antitumor activity in HNSCC cell lines and xenograft models.6,7 Results from a phase IB trial of CDX-3379 in combination with other targeted therapeutics showed favorable antitumor and adverse effect profiles.8 To further explore the activity of CDX-3379 in HNSCC, a phase I window of opportunity study in surgically resectable HNSCC was executed (NCT02473731).9
CASE REPORT
A 45-year-old white male user of smokeless tobacco presented with a cT4N2CM0 oral cavity SCC of the floor of the mouth. After pathologic confirmation by biopsy, informed consent was obtained, the patient was enrolled in NCT02473731 and given two 1,000 mg intravenous doses of CDX-3379. A significant clinical response was noted within the first 48 hours after administration of the first dose, with concomitant reduction in pain from 8 of 10 to 2 of 10. On examination before surgical extirpation, the primary tumor was found to have a nearly complete response, demonstrating a 92% reduction in greatest dimension. Final pathology revealed a pT1N1M0 oral cavity SCC. The patient subsequently received adjuvant chemoradiotherapy as indicated by his pre–CDX-3379 staging. Eighteen-month follow-up revealed no evidence of disease. Here, we present genomic and in vivo molecular correlates of this exceptional response to CDX-3379.
Sequencing and Informatics
Formalin-fixed paraffin-embedded (FFPE) tissue from pre- and post-treatment tumor specimens was microdissected to ensure more than 95% tumor cells. Tumor DNA and RNA were extracted using QIAGEN FFPE DNA and RNeasy protocols. Germline DNA was extracted from peripheral blood using the QIAGEN DNA blood mini kit (Germantown, MD). Tumor and germline DNA underwent whole exome sequencing (WES) library construction with the Illumina TruSeq kit (San Diego, CA), with exonic regions captured using Agilent SureSelect v4 (Santa Clara, CA). Two × 100 paired-end sequencing was performed on an Illumina HiSequation 2500, with average coverage of 95×. RNA underwent library preparation with the Illumina TruSeq RNA Access kit followed by paired-end (2 × 76) sequencing on an Illumina NextSeq 500, with average coverage of 45 million reads per sample. Primary processing of sequence data was performed using Illumina CASAVA software v1.8. Somatic mutations and copy number variants (CNVs) were identified by VariantDx as previously described.10 All nonsynonymous coding variants, splice site variants, indels, and CNVs above or below 2.0 were considered potentially deleterious. RNA-Seq reads were assessed using FastQC v0.11.3. After adapter trimming with cutadapt v1.5, reads were mapped to the human reference genome GRCh37/38 using HISAT2 v2.0.4, and gene expression was quantitated using HTSeq v0.9. Differential gene expression was performed using edgeR v3.20.9.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was isolated from human HNSCC cell lines (Te6, KYSE510, Te9, FaDu, PECAPJ34, Cal33, SCC4, BHY, OSC19, SCC25) using the QIAGEN RNeasy kit. cDNA was synthetized using the Bio-Rad iScript Reverse Transcription Supermix (Hercules, CA). Real-time polymerase chain reaction (PCR) was performed using the Bio-Rad iQ SYBR Green Supermix and Applied Biosystems 7900HT Fast Real-Time PCR System. The following primers were used to detect Fzd3 mRNA: 5′-gcttccacagtgacacaagg-3′ and 5′-accatacactgccagccata-3′. To detect ErbB3 mRNA, the following primers were used: 5′-actctccatatcccttcctctc-3′ and 5′-gtttcctccctttcctctct-3′. PCR amplifications were run in triplicate for each cell line. Comparative quantification of gene expression was performed using the ΔΔCt method, and values were referenced to GAPDH mRNA expression.
ErbB3 Phosphorylation
ErbB3 phosphorylation was measured by VeraTag assay (San Francisco, CA) on FFPE tissue.6
Colony-Formation Assays
Five thousand cells per well were seeded in six-well plates and allowed to attach overnight. CDX-3379 or IgG control was added to a concentration of 100 nM. After 7 to 10 days, cells were fixed and stained with crystal violet. The area covered by stained cells was measured using ImageJ. Statistical analysis was performed using GraphPad Prism 7.
pErbB3 was assessed by VeraTag assay to examine changes in activated ErbB3 pretreatment to post-treatment, revealing a 69% decrease from 0.45 to less than 0.14 (lower limit of quantitation of the assay), demonstrating successful blockage of ErbB3 activation by CDX-3379. Because ErbB3 expression and changes in pErbB3 levels pretreatment to post-treatment have been shown not to correlate with clinical response to CDX-3379 (unpublished data), WES and RNA-Seq were performed on both pretreatment and post-treatment tumor samples to investigate how additional genomic alterations could relate to treatment response.
WES revealed 103 potentially deleterious somatic alterations (101 single nucleotide variants and two CNVs) in the pretreatment tumor. Ninety-six single nucleotide variants and no CNVs were present in the post-treatment tumor (Data Supplement). No additional alterations were identified in the post-treatment biopsy that were not present pretreatment, suggesting tight clonal architecture. We hypothesized that loss of somatic alterations from pretreatment to post-treatment, in combination with a significant reduction in clinical tumor volume, could represent selective killing of clones more sensitive to CDX-3379 and thus give insight to the mechanism of sensitivity. Interrogating the list of alterations that were present in the tumor pretreatment but not post-treatment (on the basis of known mechanisms of action of each gene) identified amplification of FZD3 (frizzled class receptor 3), a receptor in the Wnt signaling pathway, as a possible candidate conferring sensitivity to CDX-3379 (Table 1). RNA-Seq demonstrated a 2.5-fold–higher expression of FZD3 pretreatment versus post-treatment (P < 4.5e-13), consistent with the 3.0 amplification identified on WES in the pretreatment tumor (Fig 1A). FZD3 alterations were investigated in HNSCCs from The Cancer Genome Atlas (TCGA). Six percent (28 of 496) of HNSCCs in TCGA possess an FZD3 alteration that could result in an increase in downstream Wnt signaling, consistent with the case reported here (one amplification and 27 mRNA upregulations). Patients with HNSCCs from TCGA that possess an FZD3 alteration have poorer overall survival, suggesting that alterations in this gene may be important in the biology of a subset of HNSCCs (Fig 1B).
TABLE 1.
Somatic Alterations in Pretreatment but Not Post-Treatment Tumor
FIG 1.
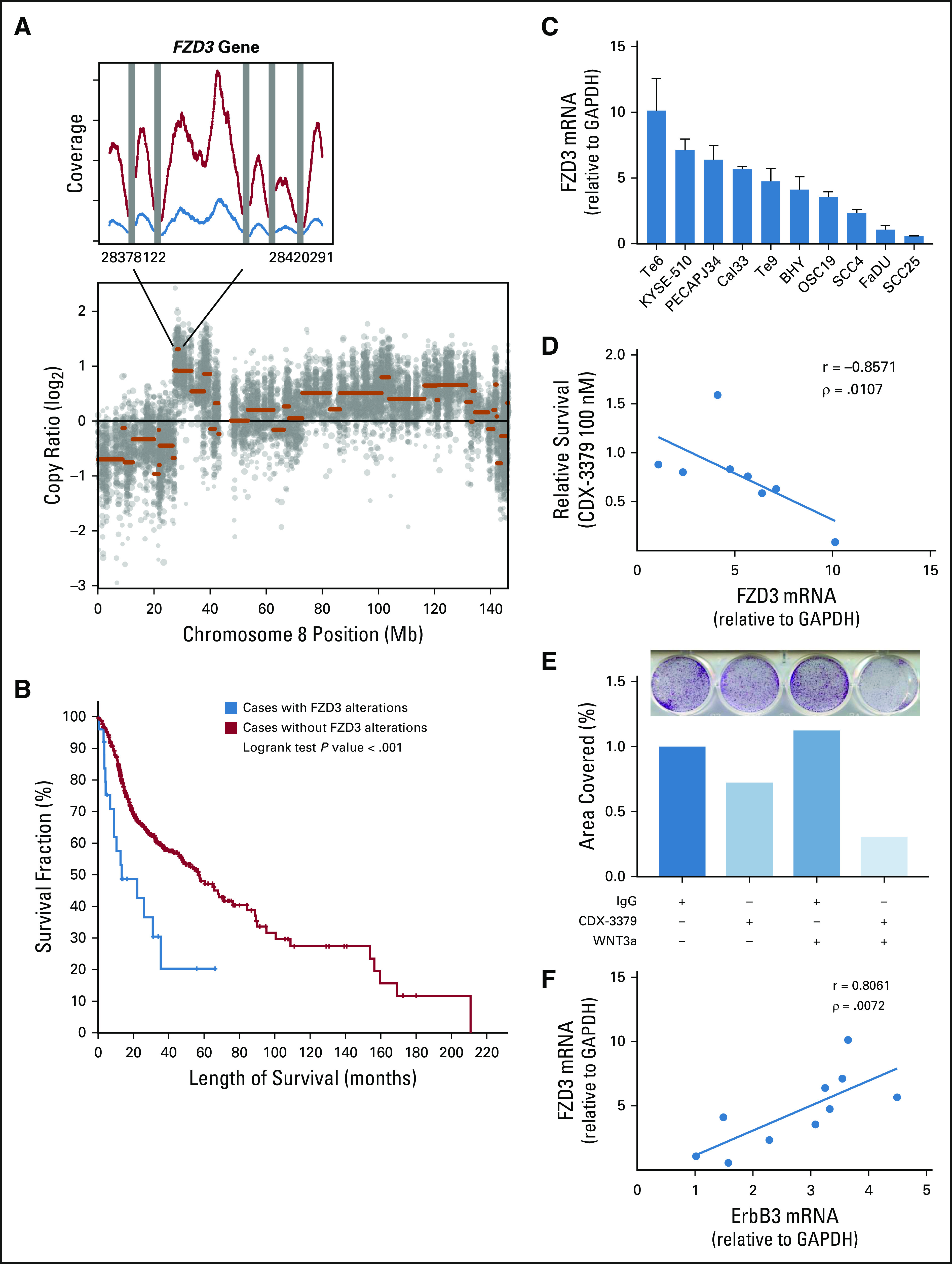
FZD3 in head and neck squamous cell carcinoma (HNSCC). (A) Chromosome 8 copy number variant plot and identified FZD3 gene amplification in pretreatment tumor.(B) Kaplan-Meier curve demonstrating worse survival in patients with HNSCC in The Cancer Genome Atlas whose tumors possess FZD3 amplification, deletion, or mRNA upregulation (red) compared with patients whose tumors do not have FZD3 alterations (blue; log rank P < .001). (C) FZD3 mRNA expression is variable among 10 HNSCC cell lines. (D) Negative correlation between FZD3 mRNA expression levels in HNSCC cell lines and survival in colony formation in the presence of CDX-3379 relative to IgG (Spearman correlation coefficient −0.86, P = .01). (E) CDX-3379 growth inhibition is potentiated in Te6 cells by the addition of Wnt3a. Images of a representative colony formation (top) with quantification of stained area represented in bar graph (middle) and culture additives (bottom) P < .001 for all comparisons by one-way analysis of variance. (F) Positive correlation between FZD3 and ErbB3 mRNA expression levels in HNSCC cell lines (Spearman correlation coefficient 0.81, P < .01).
To explore the possibility that FZD3 plays a role in sensitivity to ErbB3 inhibition, we examined FZD3 mRNA expression by quantitative real-time PCR in 10 HNSCC cell lines, finding high variability among cell lines (Fig 1C). To test the hypothesis that increased FZD3 expression could render cells more sensitive to CDX-3379, akin to the pretreatment tumor, we performed colony-formation assays with CDX-3379. We found that cell lines with the highest expression of FZD3 were the most sensitive to CDX-3379 (Spearman correlation coefficient −0.86, P = .01; Fig 1D). We further stimulated FZD3 receptors by the addition of the FZD3 ligand Wnt3a to the high FZD3–expressing cell line Te6, demonstrating decreased colony formation in the presence of CDX-3379 plus Wnt3a, compared with control and CDX-3379 alone (P < .001 for all comparisons by one-way analysis of variance; Fig 1E). Because FZD3 mRNA expression seemed to correlate with sensitivity to ErbB3 inhibition (suggesting a convergence or crosstalk), ErbB3 mRNA levels were quantitated in the above cell lines and compared with FZD3 mRNA expression, demonstrating a tight correlation (Spearman correlation coefficient 0.81, P < .01; Fig 1F).
DISCUSSION
The FZD proteins are a family of seven transmembrane-spanning G protein–coupled receptors that bind Wnt ligands. Coupling of Wnt and FZD results in activation of the canonical and noncanonical Wnt signaling pathways, which regulate embryonal development, cell proliferation, motility, and polarity. The role of Wnt signaling pathways in cancer is well established.11 FZD upregulation has been identified in numerous cancers, correlates with poor outcomes, and is an active area of investigation for targeted therapeutics.11,12 FZD3, in particular, is believed to play a role in both the canonical and noncanonical Wnt pathways and has been found to be upregulated in lung, esophageal, and colon cancers.13-15
Here, we report FZD3 gene amplification and overexpression in pretreatment but not post-treatment tumor from an exceptional responder to CDX-3379. This suggests that FZD3 alterations, or alterations in the Wnt pathway, may play a role in mediating sensitivity to ErbB3 inhibition in HNSCC. Review of FZD3 in HNSCCs from TCGA supports the potential biologic importance of alterations in this gene. We further demonstrate that FZD3 expression levels correlate with sensitivity to CDX-3379 and that activation of FZD3 by its ligand Wnt3a further potentiates the effects of CDX-3379. The mechanism by which FZD3 confers sensitivity to ErbB3 inhibition is unclear; however, the Wnt and ErbB pathways have been reported to closely interact and collude in tumorigenesis.16,17 It remains to be seen whether this is by convergence on a common downstream molecule, such as β-catenin,18 or via effects on different target genes. In breast cancer, Wnt overexpression activates signaling via ErbB.19,20 Interestingly, we identified a tight correlation between ErbB3 and FZD3 mRNA expression levels in HNSCC cell lines, suggesting a similar relationship and highlighting the interconnected nature of these singling pathways. Additional investigation of alterations in FZD in HNSCC, and the relationship between FZD and ErbB signaling pathways, is warranted.
ACKNOWLEDGMENT
We thank the UPMC Hillman Cancer Center shared resource facility (Cancer Genomics Facility), which is supported in part by award P30CA047904 and Personal Genome Diagnostics.
Footnotes
Supported by Department of Veterans Affairs/I01 Merit Award/I01BX003456 (U.D.) and American Head and Neck Society Pilot Grant (D.L.F.).
AUTHOR CONTRIBUTIONS
Conception and design: Daniel L. Faden, Umamaheswar Duvvuri
Administrative support: Umamaheswar Duvvuri
Financial support: Umamaheswar Duvvuri
Provision of study material or patients: Daniel L. Faden, Diego Alvarado, Umamaheswar Duvvuri
Collection and assembly of data: Daniel L. Faden, Roberto Gomez-Casal, Umamaheswar Duvvuri
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or po.ascopubs.org/site/ifc.
Diego Alvarado
Employment: Celldex
Stock and Other Ownership Interests: Celldex
Patents, Royalties, Other Intellectual Property: Patents pending on antibodies targeting receptor tyrosine kinases
Umamaheswar Duvvuri
Consulting or Advisory Role: Medtronic
Research Funding: Kolltan Pharmaceuticals (Inst), Medrobotics (Inst)
No other potential conflicts of interest were reported.
REFERENCES
- 1.Amin DN, Campbell MR, Moasser MM. The role of HER3, the unpretentious member of the HER family, in cancer biology and cancer therapeutics. Semin Cell Dev Biol. 2010;21:944–950. doi: 10.1016/j.semcdb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takikita M, Xie R, Chung JY, et al. Membranous expression of Her3 is associated with a decreased survival in head and neck squamous cell carcinoma. J Transl Med. 2011;9:126. doi: 10.1186/1479-5876-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee-Hoeflich ST, Crocker L, Yao E, et al. A central role for HER3 in HER2-amplified breast cancer: Implications for targeted therapy. Cancer Res. 2008;68:5878–5887. doi: 10.1158/0008-5472.CAN-08-0380. [DOI] [PubMed] [Google Scholar]
- 4.Sergina NV, Rausch M, Wang D, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445:437–441. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brand TM, Hartmann S, Bhola NE, et al. Cross-talk signaling between HER3 and HPV16 E6 and E7 mediates resistance to PI3K inhibitors in head and neck cancer. Cancer Res. 2018;78:2383–2395. doi: 10.1158/0008-5472.CAN-17-1672. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 6.Alvarado D, Ligon GF, Lillquist JS, et al. ErbB activation signatures as potential biomarkers for anti-ErbB3 treatment in HNSCC. PLoS One. 2017;12:e0181356. doi: 10.1371/journal.pone.0181356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao Z, Carrasco RA, Schifferli K, et al. A potent HER3 monoclonal antibody that blocks both ligand-dependent and -independent activities: Differential impacts of PTEN status on tumor response. Mol Cancer Ther. 2016;15:689–701. doi: 10.1158/1535-7163.MCT-15-0555. [DOI] [PubMed] [Google Scholar]
- 8.Falchook GS, Bauer TM, LoRusso P, et al. Safety, pharmacokinetics (PK), pharmacodynamics (Pd), and antitumor activity in a phase 1b study evaluating anti-ErbB3 antibody KTN3379 in adults with advanced tumors alone and with targeted therapies. J Clin Oncol. 2016;34 [Google Scholar]
- 9.Duvvuri U, George J, Kim S, et al. Molecular and clinical activity of CDX-3379, an anti-ErbB3 monoclonal antibody, in head and neck squamous cell carcinoma: A preoperative “window of opportunity” study. AACR Annual Meeting Proceedings. 2018;78 [Google Scholar]
- 10.Anagnostou V, Smith KN, Forde PM, et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 2017;7:264–276. doi: 10.1158/2159-8290.CD-16-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueno K, Hirata H, Hinoda Y, et al. Frizzled homolog proteins, microRNAs and Wnt signaling in cancer. Int J Cancer. 2013;132:1731–1740. doi: 10.1002/ijc.27746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong SC, He CW, Chan CM, et al. Clinical significance of frizzled homolog 3 protein in colorectal cancer patients. PLoS One. 2013;8:e79481. doi: 10.1371/journal.pone.0079481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka S, Akiyoshi T, Mori M, et al. A novel frizzled gene identified in human esophageal carcinoma mediates APC/beta-catenin signals. Proc Natl Acad Sci USA. 1998;95:10164–10169. doi: 10.1073/pnas.95.17.10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee EH, Chari R, Lam A, et al. Disruption of the non-canonical WNT pathway in lung squamous cell carcinoma. Clin Med Oncol. 2008;2008:169–179. [PMC free article] [PubMed] [Google Scholar]
- 16.Paul I, Bhattacharya S, Chatterjee A, et al. Current understanding on EGFR and Wnt/β-catenin signaling in glioma and their possible crosstalk. Genes Cancer. 2013;4:427–446. doi: 10.1177/1947601913503341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu T, Li C. Convergence between Wnt-β-catenin and EGFR signaling in cancer. Mol Cancer. 2010;9:236. doi: 10.1186/1476-4598-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schroeder JA, Adriance MC, McConnell EJ, et al. ErbB-beta-catenin complexes are associated with human infiltrating ductal breast and murine mammary tumor virus (MMTV)-Wnt-1 and MMTV-c-Neu transgenic carcinomas. J Biol Chem. 2002;277:22692–22698. doi: 10.1074/jbc.M201975200. [DOI] [PubMed] [Google Scholar]
- 19.Faivre EJ, Lange CA. Progesterone receptors upregulate Wnt-1 to induce epidermal growth factor receptor transactivation and c-Src-dependent sustained activation of Erk1/2 mitogen-activated protein kinase in breast cancer cells. Mol Cell Biol. 2007;27:466–480. doi: 10.1128/MCB.01539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musgrove EA. Wnt signalling via the epidermal growth factor receptor: A role in breast cancer? Breast Cancer Res. 2004;6:65–68. doi: 10.1186/bcr737. [DOI] [PMC free article] [PubMed] [Google Scholar]