Abstract
Objectives
We investigated the associations with HLA and microtubule-associated protein tau (MAPT) H1 haplotype in anti-IgLON5 disease, a recently identified disorder characterized by gait instability, brainstem dysfunction, and a prominent sleep disorder in association with IgLON5 antibodies and pathologic findings of a novel neuronal-specific tauopathy.
Methods
We compared the HLA alleles and MAPT H1/H1 genotype of 35 patients with anti-IgLON5 with healthy controls. The on-line server tool NetMHCIIpan 3.1 was used to predict the IgLON5 peptide binding to HLA Class II molecules.
Results
The HLA-DRB1*10:01-DQB1*05:01 haplotype was overrepresented in patients with anti-IgLON5 disease (OR = 54.5; 95% CI: 22.2–133.9, p < 0.0001). In addition, HLA-DQA was genotyped in 27 patients, and 25 (92.6%) of them had DQ molecules composed by DQA1*01 and DQB1*05 chains compared with 148/542 (27.3%) controls (OR = 43.9; 95% CI: 10.4–185.5, p < 0.0001). Patients DRB1*10:01 positive developed more frequently sleep or bulbar symptoms than those carrying other HLA alleles (70.0% vs 26.7%; p = 0.011). Prediction algorithms identified 2 IgLON5 peptides (1 located in the signal sequence) that showed strong binding to HLA-DRB1*10:01 and other HLA-DRB1, but not to HLA-DQA and HLA-DQB molecules. The MAPT H1/H1 homozygous genotype was present in 20/24 (83.3%) anti-IgLON5 Caucasian patients compared with 54/116 (46.5%) healthy controls (p = 0.0007).
Conclusions
The robust association of anti-IgLON5 disease with distinct HLA Class II molecules supports a primary autoimmune origin. The significant association of MAPT H1 haplotype also suggests that an underlying neurodegenerative process could be involved in anti-IgLON5 disease.
The anti-IgLON5 disease is a recently identified neurologic disorder, characterized by progressive gait instability, brainstem dysfunction, and a characteristic sleep disorder. The biological hallmark of this disease is the presence of antibodies against IgLON5, a neural cell adhesion molecule of unknown function.1 Neuropathologic studies demonstrated a novel neuronal tauopathy with predominant involvement of the hypothalamus and brainstem.2 Although these findings favor a primary tauopathy, the presence of highly specific antibodies against a neuronal surface protein and the tight association of the disease with HLAs DRB1*10:01and DQB1*05:01 (previously shown in 15 patients) alternatively suggested an immune-mediated pathogenesis.3
Familial forms of tauopathies are caused by mutations in the microtubule-associated protein tau (MAPT) gene.4 In addition, the frequency of the MAPT H1/H1 homozygous genotype has been described to be consistently increased in sporadic tauopathies including progressive supranuclear palsy (PSP) and corticobasal degeneration (CBD), but its frequency in IgLON5 disease is unknown.5,6
It is also unknown whether the HLA alleles associated with anti-IgLON5 disease can present IgLON5 peptides to generate a specific autoimmune response.7 We report here the identification of IgLON5 peptides with potential binding to HLA Class II molecules using in silico techniques. In addition, we performed DNA sequencing analyses of the MAPT gene and determined the association with the H1 haplotype and examined whether the most prevalent HLA allele (HLA-DRB1*10:01) in patients with anti-IgLON5 disease segregated with a specific clinical phenotype.
Methods
Patients
The study includes 35 patients with IgLON5 antibodies identified in the Neuroimmunology Laboratory of the Institute of Biomedical Research August Pi i Sunyer (IDIBAPS), Hospital Clinic (Barcelona, Spain). Genomic DNA of 28 patients was available for HLA and MAPT analysis. For the remaining 7 patients, HLA-DR genotyping was performed elsewhere and the information used for analysis of HLA-DR frequencies and association with specific neurologic symptoms. Methods of detection of IgLON5 antibodies and collection of clinical information have been previously reported.1,3 The frequency of HLA and MAPT haplotypes in patients was compared with that of healthy Caucasian Spanish controls (542 controls for the HLA study and 116 for the MAPT genotyping).
Standard protocol approvals, registrations, and patient consents
The study was approved by the Hospital Clinic Ethics Committee. All patients or proxies gave written informed consent for the storage and use of serum, CSF, and clinical information for research purposes.
HLA genotyping
HLA Class I and Class II genotyping was performed by techniques based on DNA-PCR and polymorphism identification by reverse hybridization with specific probes and fluorescence labeling of hybridized fragments (polymerase chain reaction [PCR]-sequence-specific oligonucleotide probes) (Immucor GTI Diagnostics Inc., Waukesha) in combination with genomic DNA sequencing by Sanger methodology (PCR-sequenced-based typing) (SuBiTo, Inno-Train Diagnostik GmbH and the software HiType, Kronberg, Germany), providing an unambiguous resolution of the 2 fields that define the HLA molecule polymorphism according to the WHO official current nomenclature (hla.alleles.org/nomenclature/nomenc_updates.html).
MAPT genetic studies
Genotyping of genomic DNA was performed using the predesigned TaqMan assay for MAPT rs1800547 (A/G), which defines the H1/H2 haplotypes and run on a Step One Plus Real-time PCR System (Applied Biosystems, Foster City, CA) as previously described.8 Twenty-seven patients with DNA available and 116 healthy controls were genotyped. All but 3 patients were of European Caucasian origin. In addition, in 7 patients with anti-IgLON5, we performed a mutational screening of MAPT exons 1, 9, 10, 11, 12, and 13, where pathogenic mutations have been previously identified in several neurodegenerative diseases including frontotemporal dementia or PSP.4 We performed a touchdown PCR amplification on a PTC-100 PCR (MJ Research; Watertown, MA) using 20–50 ng of total genomic DNA and using primer pairs as previously described.9 We used the BigDye Terminator v3.1 Cycle Sequencing Kit according to the manufacturer's instructions (Applied Biosystems, Foster City, CA). Sequences were run on an ABI3100 automatic sequencer (Applied Biosystems, Foster City, CA) and analyzed using the DNA baser v4.36.0 software (dnabaser.com).
HLA-IgLON5 peptide binding prediction
The human IgLON5 sequence was retrieved from UNIPROT database entry A6NGN9 (uniprot.org) encompassing the signal peptide (amino acid positions 1–30) and the mature protein (amino acid positions 31–336). The on-line server tool NetMHCIIpan 3.1 was used for the prediction of IgLON5 peptide binding to HLA Class II molecules (cbs.dtu.dk/services/NetMHCIIpan/).10 The full protein sequence was submitted as 15-residue overlapping fragments with 1 gap residue at the N-terminal flanking position. Binding was considered strong if the predicted IC50 (half maximal inhibitory concentration) value was <50 nM and ranked <1% among 200.000 random peptides. Only strong binders were taken for analysis.10–12 Because the 9-residue binding core is contained in several 15-mer peptides, only the strongest predicted binder of any series of 15-mer peptides with a 9-residue or longer overlap was used for calculations.
Binding prediction was calculated with the 15 different HLA-DRB1 alleles present in patients (*10:01, *01:01, *01:02, *03:01, *04:01, *04:03, *07:01, *09:01, *11:01, *12:01, *13:01, *14:01, *14:04, *15:01, and *16:01). Because of the strong linkage disequilibrium between DRB1 and DQ alleles, the NetMHCIIpan 3.1 algorithm was also applied to the DQ alleles. The 4 possible combinations for each individual were considered, combining each DQA1 with DQB1 alleles present in 1 patient in cis and/or trans, considering both chromosomes.
Molecular modeling of HLA Class II-IgLON5 peptide interactions
To confirm the HLA-IgLON5 peptide binding predictions generated by the NetMHCIIpan 3.1 program, molecular modeling of complexes including HLA-DRB1*10:01 and *01:01 and the 2 IgLON5 peptides identified as strong binders was performed using a simulation protocol (appendix e-1, links.lww.com/NXI/A136).
Statistical analysis
OR with 95% CI was calculated to measure the association of HLA alleles and MAPT haplotypes with anti-IgLON5 disease. For MAPT haplotypes, we performed a genotypic association analysis using unconditional logistic regression models, as implemented in SNPstat software, to compare the frequencies of MAPT H1/H1—which is the risk genotype described in other tauopaties (CBD or PSP)—with the other 2 genotypes H1/H2 and H2/H2 in patients with anti-IgLON5 and controls (dominant model). The χ2 test and Fisher exact probability test were used to determine the significance of differences in the distribution of the clinical features between patients who were HLA-DRB1*10:01 positive or carried other DRB1* alleles. The nonparametric Mann-Whitney U test was used to compare medians. p Values < 0.05 were considered significant. The statistical analyses were performed using commercially available software (SPSS, Version 22.0).
Data availability
Data from patients reported within the article are available and will be shared anonymously by request from any qualified investigator.
Results
HLA subtypes in anti-IgLON5 disease
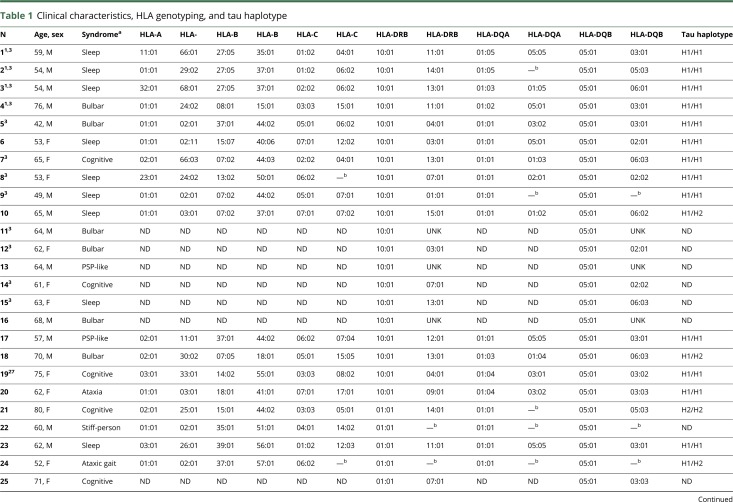
General information and the HLA and MAPT genotyping of the 35 patients with anti-IgLON5 disease are summarized in table 1. The median age at disease onset was 63 years (range: 42–81 years), and 51% were female. Thirty-two patients were of European Caucasian origin. The countries of origin were Spain (12 patients), Austria (8), Germany (7), Brazil (2), France (1), Belgium (1), Italy (1), Australia (1), India (1), and Philippines (1). Twenty of the 35 patients (57.1%) carried the HLA-DRB1*10:01-DQB1*05:01 haplotype compared with 2.4% of healthy controls (OR = 54.5; 95% CI: 22.2–133.9, p < 0.0001). Another 7 patients (20%) carried the DRB1*01:01 allele (2 were homozygous, and 1 heterozygous with DRB1*10:01) representing a similar frequency seen in controls (14.1%) (p = 0.34). The DRB1*03:01 allele was present in 8 patients (22.9%) (2 were also DRB1*10:01 positive) representing a similar frequency observed in controls (20.6%) (p = 0.75). The remaining 3 patients had other HLA-DRB1 (table 1). HLA-DQA was genotyped in 27 patients, and 25 (92.6%) of them had DQ molecules composed by DQA1*01 and DQB1*05 chains compared with 148/542 (27.3%) controls (OR = 43.9; 95% CI: 10.4–185.5, p < 0.0001). Although the high frequency of these alleles could be partially explained by its genetic linkage with the HLA-DRB1*10:01 allele (allelefrequencies.net), the DQA1*01/DQB1*05 molecule was present in 11/13 (84.6%) patients without DRB1*10:01.
Table 1.
Clinical characteristics, HLA genotyping, and tau haplotype
HLA Class I alleles were studied in 27 patients with anti-IgLON5 (table 1). Five patients (18.5%) were HLA-B*37:01 compared with 17 controls (3.1%) (OR = 7.0; 95% CI: 2.4–20.8; p = 0.0004). The allele HLA-A*01:01 was also overrepresented in patients with anti-IgLON5 compared with controls (47.4% vs 20.5%; OR = 2.7; 95% CI: 1.2–5.9; p = 0.01). These associations were expected considering the strong linkage disequilibrium of these 2 alleles with DRB1*10:01 that conform a common haplotype in Caucasians.13
Association of the HLA-DRB1*10:01 allele with neurologic symptoms
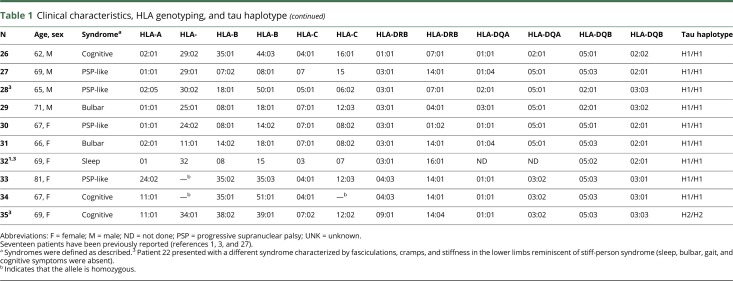
Next, we investigated whether the presence of the most prevalent HLA-DRB1*10:01 allele associated with distinct clinical features (table 2). At disease onset, 14 DRB1*10:01-positive patients (70%) presented with sleep or bulbar clinical phenotypes. Conversely, these 2 phenotypes tended to be less frequent (26.7%) in patients with other HLA alleles, who often presented with a PSP-like phenotype or cognitive impairment. Accordingly, a trend to a higher frequency of parasomnia, sleep breathing alterations, and bulbar dysfunction was observed in DRB1*10:01 carriers. Dysautonomia tended also to be more frequent in DRB1*10:01 carriers, whereas cognitive impairment occurred more frequently in non-DRB1*10:01 carriers. Improvement with immunotherapy occurred in 5 (26.3%) of 19 patients. DRB1*10:01-negative patients showed a trend to a better response to immunotherapy (3 of 5 cases improved). IgG subclass predominance was IgG4 in all DRB1*10:01-negative patients compared with 70% in DRB1*10:01-positive cases (p = 0.027).
Table 2.
Clinical features in patients HLA-DRB1*10:01 positive and negative
HLA-IgLON5 peptide binding prediction
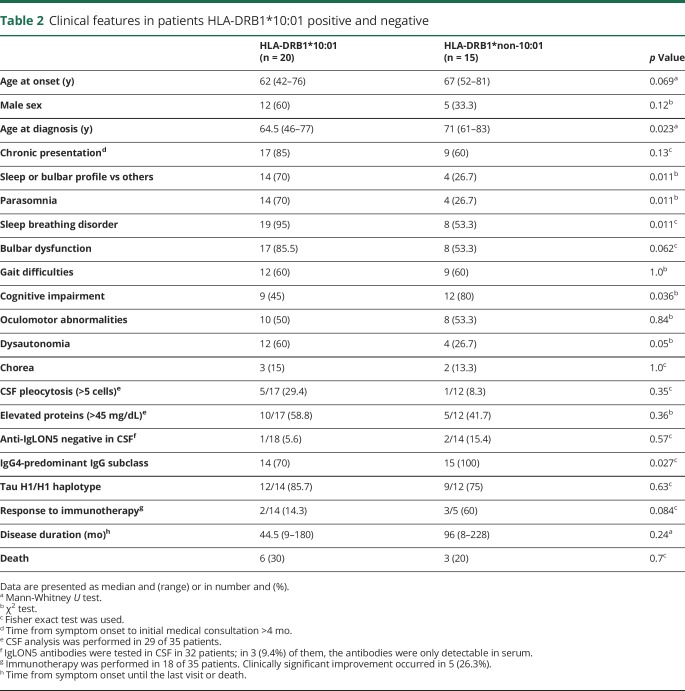
The binding ability of IgLON5 peptide sequences to 15 HLA-DRB1 alleles using the NetMHCIIpan 3.1 server showed that only 3 DRB1 molecules, the DRB1*01:01, DRB1*10:01, and DRB1*09:01, were strong binders for the same 2 IgLON5-derived peptides (table 3). The epitope with the strongest predicted affinity was LRLLAAAAL, belonging to the signal peptide of IgLON5. The second epitope, IVHVPARIV, is located in the Ig-like 2 domain of the protein. Seven different HLA-DRB1 molecules also had a predicted binding capacity for one of these peptides, mainly IVHVPARIV. Another peptide (WTSDPRVRL) showed binding capacity for only HLA-DRB1*03:01 present in 8 patients (table 3). No IgLON5 peptide sequences were defined as strong binders considering all possible, cis or trans, combinations of the DQA and DQB molecules. Similarly, no IgLON5 peptide sequences were binders to HLA Class I–associated alleles A*01:01 and B*37:01 using the NetMHC 3.4 server.14
Table 3.
IgLON5 peptides with strong binding affinity for HLA-DRB1 molecules in anti-IgLON5 disease
Given that the signal peptide sequence may vary among different family members and isoforms of a protein family, we determined the prediction of binding to HLA Class II molecules the signal peptide sequences of the other members of the IgLON family, IgLON1, 2, 3, and 4. In contrast to the IgLON5 peptide LRLLAAAAL, none of the other signal peptide sequences showed strong predicted binding affinity. Only sequences from isoforms 1, 2, 5, 7, and 9 of IgLON2 (Uniprot accession NP-057606.1, NP_001041674.1, NP_001338930.1, NP_001338932.1, and NP_001338934.1, respectively) resulted in weak predicted binding affinities to HLA-DRB1 alleles. The second epitope, IVHVPARIV, was not present in any of the other members of the IgLON family.
Because tau protein deposits have been described in the brain of patients with anti-IgLON5 antibodies who underwent autopsy, a prediction study was also made on the binding ability of tau protein sequences (Uniprot P10636) to the indicated HLA-DRB1 and DQA-DQB alleles. No sequences with high affinity for HLA molecules were identified.
Molecular modeling of HLA Class II-IgLON5 peptide interactions
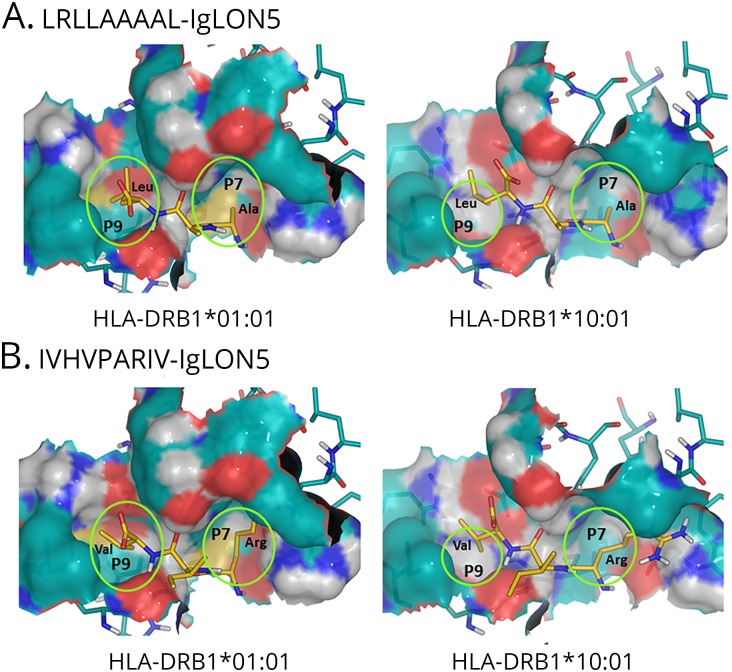
We next modeled the interaction of the strong binder IgLON5 peptide LRLLAAAAL with DRB1*01:01 and DRB1*10:01. The computed structure-based scores for the binding of this peptide to DRB1*01:01 and DRB1*10:01 were −97 and −91 kJ/mol, respectively. Although based on fundamentally different principles, this finding agrees with the NetMHCIIpan 3.1 binding affinity predictions, thus suggesting a slightly higher affinity of LRLLAAAAL for DRB1*01:01 than for DRB1*10:01. Taking the structure into account, the clearest differences between the pockets in the peptide-binding grooves of DRB1*01:01 and DRB1*10:01 are probably found in pocket P9, where there is absence of a well-defined pocket in DRB1*10:01, whereas a shallow pocket is visible in DRB1*01:01 (figure).
Figure. Modeling of the interaction between Class II molecules and IgLON5 peptides.
Detail of the pockets P7 and P9 in the modeled complexes between peptides LRLLAAAAL (A) and IVHVPARIV (B) of IgLON5 and molecules HLA-DRB1*01:01 and HLA-DRB1*10:01. Residues 7–9 (from right to left) of the peptides are represented as sticks, with the carbons in golden yellow. Protein residues forming the pockets are represented as sticks, with carbon atoms in teal blue. Covering the protein atoms, their Connolly surface is represented to facilitate the visualization of the pockets. Noncarbon atoms are colored as follows: N-blue, H-light gray, O-red, and S-yellow. It can be observed that P9 (cavity at the left of each panel) is shallower in HLA-DRB1*10:01 than in HLA-DRB1*01:01. As a result, the aliphatic residues at position 9 of both peptides bind more deeply in HLA-DRB1*01:01. The models also show that the Arg (R) residue at position 7 in IVHVPARIV is too bulky for the small P7 and has to orient its side chain toward previous pockets, particularly in HLA-DRB1*10:01.
The computed scores for the binding of the other strong binder peptide IVHVPARIV to DRB1*01:01 and DRB1*10:01 were −75 and −57 kJ/mol, respectively. In this case, the difference of 18 kJ/mol is significantly larger than that predicted by NetMHCIIpan 3.1. Nevertheless, using any of these 2 methods, a higher affinity for DRB1*01:01 is predicted. Comparing the values obtained for the 2 peptides, we also found that both methods predicted a higher affinity of LRLLAAAAL than IVHVPARIV for either of the 2 HLA molecules. Considering the binding scores of IVHVPARIV relative to LRLLAAAAL, and given that the other anchoring residues are significantly conserved, we conclude that Arg (R) in pocket P7 is likely the main responsible for the lower binding affinity of IVHVPARIV relative to LRLLAAAAL for the 2 HLA molecules.
Genetic analysis of the MAPT gene in anti-IgLON5 disease
The MAPT H1/H1 homozygous genotype was significantly overrepresented with respect to H1/H2 and H2/H2 in 27 anti–IgLON5-positive patients (81.5% were H1/H1) compared with 116 healthy controls (46.5% H1/H1), OR 5.05 (95% CI: 1.79–14.25), p = 0.0007. Similar results were found when the sample was limited to the 24 anti–IgLON5-positive patients of European Caucasian origin (83% H1/H1) (p = 0.0007). Finally, we did not find mutations in the commonly mutated MAPT exons 1, 9, 10, 11, 12, and 13 in a subset of 7 patients with anti-IgLON5 disease.
Discussion
Anti-IgLON5 disease was the first among multiple CNS disorders associated with antibodies against synaptic receptors or neuronal surface proteins in which an association with HLA Class II alleles was identified.1 Here, we confirm the association with HLA-DRB1*10:01-DQB1*05:01 alleles in a larger series of patients and provide several novel findings that help to understand the interplay between the autoimmune and neurodegenerative processes of the disease. First, the HLA-DRB1*10:01 was strongly associated with anti-IgLON5 disease and was found more prevalent in patients who presented with parasomnias, sleep breathing disorder, or bulbar symptoms. Second, 2 IgLON5 peptides showed high affinity for the HLA-DRB1 molecules identified in patients with the disease as predicted by an algorithm using artificial neural networks. These findings were confirmed by molecular modeling using the 2 IgLON5 peptides and DRB1*01:01 and DRB1*10:01, the 2 alleles that showed the strongest affinity for these peptides. Last, the MAPT H1/H1 genotype was overrepresented in patients with anti-IgLON5 disease.
The robust association between anti-IgLON5 disease and distinct HLA alleles supports a primary autoimmune event as cause of the disease. One of the hypotheses to explain the link between antibody-mediated diseases and specific HLA-DR or DQ alleles is that these molecules present peptides of the antibody-targeted antigen to autoimmune-prone CD4+ T cells, which in turn induce B-cell proliferation and differentiation into antibody-secreting cells.15 In anti-IgLON5 disease, there is an overrepresentation of DRB1*10:01; this is an infrequent allele that in all population studies represents less than 5% of individuals (allelefrequencies.net). The DRB1*10:01 allele has been linked to an increased risk of several rheumatological autoimmune diseases.16 Indeed, in HLA-DRB1*10:01–positive patients with rheumatoid arthritis and antibodies against citrullinated type II collagen, CD4+ T cells are activated when a peptide from citrullinated type II collagen protein is presented to T cells by DRB1*10:01 but not by other DRB1 molecules showing the fine specificity of the peptide-binding motifs of the HLA molecules.17
The present study shows that the DRB1 alleles identified in patients with anti-IgLON5 disease, DRB1*01:01 and DRB1*10:01, are predicted to bind with high affinity 2 peptides of IgLON5. Molecular modeling of the interaction of the 2 IgLON5 peptides (LRLLAAAAL and IVHVPARIV) with the strongest binding with DRB1*01:01 and DRB1*10:01 confirmed the high affinity of this binding. LRLLAAAAL corresponds to a sequence of the signal peptide of IgLON5. Unlike other members of the IgLON family, only the IgLON5 signal peptide region showed a high binding affinity for DRB1 molecules. Signal peptides contain numerous hydrophobic residues that help to anchor the nascent polypeptide to the membrane.18 Once the protein is anchored in the membrane, the signal sequence is cleaved and degraded in the cytosol, and the resulting peptides are known to be strong binders of the T-cell activating MHC Class I molecules. Indeed, signal peptides have been proposed as adjuvants in vaccination strategies.19 Thus, although the epitope recognized by the helper T cell (e.g., signal recognition peptide) must be linked to that recognized by the B cell (surface peptide of the same antigen, recognized by antibodies), the 2 cells do not need to recognize identical epitopes, a phenomenon called linked recognition.20 The in silico techniques to assess potential binding of specific IgLON5 peptides to HLA molecules used in the present study represent an exploratory computational tool that will require its confirmation in future immunologic experiments assessing the proliferative T-cell response to the selected IgLON5 peptides.
Despite the DQA1*01/DQB1*05 haplotype was strongly associated with anti-IgLON5 disease, no IgLON5 peptide could be defined as a strong binder for these HLA alleles. This association can only be partially explained by linkage disequilibrium with the DRB1*10:01 allele, as 84.6% of the patients non-DRB1*10:01 carriers were positive for DQA1*01/DQB1*05. No IgLON5 peptide could be defined either to be a binder to HLA Class I–associated alleles (HLA-A*01:01, B*37:01). In this case, the association is probably due to linkage disequilibrium of these Class I alleles with the DRB1*10:01 allele. Peptide binding to HLA Class I molecules implies antigenic presentation to CD8+ cytotoxic T cells,21 but there are no data suggesting a role of CD8+ T cells in anti-IgLON5 disease.
Although this series is relatively small, we noted a trend to an association between DRB1*10:01 and sleep and bulbar dysfunction as predominant symptoms at disease onset.1 Indeed, although 78% (14/18) of patients with these clinical presentations had DRB1*10:01, this haplotype was only observed in 36% (5/14) of patients who initially presented with PSP-like symptoms or cognitive deterioration. With the increasing number of reports on patients with IgLON5 antibodies, it is becoming evident that this disease is heterogeneous and that the sleep disorder that was characterized in the initial series is not always present.22 Clinical heterogeneity occurs in many diseases, and the cause is probably multifactorial; however, the genetic background plays an important role in some disorders. For example, distinct HLA-DRB1 alleles appear to influence the clinical phenotype of inclusion body myositis or the spinal cord lesion load in MS.23,24
DRB1*10:01 is in strong linkage disequilibrium with DQB1*05 alleles that associate with some IgG4-related diseases such as MuSK antibody–mediated myasthenia.25 IgLON5 antibodies are predominantly of the IgG4 subclass, but we found that in up to 30% of DRB1*10:01-positive patients, the percentage of IgG1 was higher than that of IgG4. Whether the IgG subclass predominance in anti-IgLON5 disease associates with different HLA haplotypes should also be assessed in future studies.
An unexpected observation in anti-IgLON5 disease was that the brain of the first 3 autopsied patients showed a novel tauopathy with neuronal accumulation of hyperphosphorylated tau preferentially involving the hypothalamus and the tegmental nuclei of the brainstem.2 A subsequent case report suggested that the tauopathy is not a constant finding and that it could be a late event in the evolution of the disease.26 The absence of neuronal tau deposits in early stages of the disease perhaps could explain that some patients improve with immunotherapy and that brain biopsies showed inflammatory infiltrates without tau pathology.27,28
Independently of whether autoimmunity or neurodegeneration is the first pathogenic event, our current findings suggest that patients with anti-IgLON5 disease may have a genetic association with the MAPT H1/H1 genotype. Patients with this genotype are susceptible to several neurodegenerative diseases, particularly tauopathies (PSP and CBD) and synucleinopathies such as Parkinson disease and multiple system atrophy.29 There is robust evidence that inflammation plays a critical role in some neurodegenerative diseases, and it has been postulated that it is required for the progression of the degenerative process.30 Thus, it is conceivable that in anti-IgLON5 disease, the initial event is an inflammatory autoimmune response that results in a stress situation in patients susceptible to neurodegenerative processes, eventually leading to the indicated tauopathy. However, our finding related to MAPT association must be taken with caution due to the relative low number of patients and the potential genetic heterogeneity among different populations. Future validation studies of the MAPT H1/H1 genotype association with anti-IgLON5 disease are warranted.
In summary, the tight association of anti-IgLON5 disease with the DRB1*10:01-DQB1*05:01 haplotype and its high affinity for IgLON5 peptides, particularly the peptide in the signal sequence of IgLON5, strongly support that the disease is autoimmune. Neuronal tau deposits might occur due to a genetic predisposition of the patients, supported by the association with the MAPT H1/H1 genotype. We have previously shown that IgLON5 antibodies cause an irreversible antibody-mediated internalization of surface IgLON5 in cultured hippocampal neurons.31 Therefore, a task for the future is to determine how antibody disruption of IgLON5 ultimately leads to neuronal accumulation of hyperphosphorylated tau.
Acknowledgment
The authors thank all physicians who have contributed by providing clinical information of their patients and all patients for their generous contribution to research. They acknowledge support provided by the technician Manel Fernandez.
Glossary
- CBD
corticobasal degeneration
- MAPT
microtubule-associated protein tau
- PSP
progressive supranuclear palsy
Appendix. Authors

Study funding
This study was supported in part by Fondo de Investigaciones Sanitarias (FIS), FEDER, Spain (FIS 15/00377, FG; FIS 14/00203, J. Dalmau, FIS 18/00067, L. Sabater and C. Gaig), NIH RO1NS077851 (J. Dalmau), Fundació Cellex (J. Dalmau), Centros de Investigación Biomédica en Red de enfermedades neurodegenerativas (CIBERNED) (C. Gaig, M. Ezquerra, R. Fernández-Santiago, A. Iranzo, and J. Santamaria) Jóvenes Investigadores (JIN) grant of the Spanish Ministry of Economy and Competitiveness (MINECO) (grant # SAF2015-73508-JIN) (Ruben Fernández-Santiago), and Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER) (L. Sabater and J. Dalmau).
Disclosure
J. Dalmau receives royalties from Athena Diagnostics for the use of Ma2 as autoantibody tests and from Euroimmun for the use of NMDAR and GABAB receptor as autoantibody tests and licensing fees from Euroimmun for the use of DPPX, GABAA receptor, and IgLON5 as diagnostic tests. F. Graus receives royalties from Euroimmun for the use of IgLON5 as a diagnostic test. The rest of the authors declare that they have no competing interests. Go to Neurology.org/NN for full disclosures.
References
- 1.Sabater L, Gaig C, Gelpi E, et al. A novel non-rapid-eye movement and rapid-eye-movement parasomnia with sleep breathing disorder associated with antibodies to IgLON5: a case series, characterisation of the antigen, and post-mortem study. Lancet Neurol 2014;13:575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gelpi E, Höftberger R, Graus F, et al. Neuropathological criteria of anti-IgLON5-related tauopathy. Acta Neuropathol 2016;132:531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaig C, Graus F, Compta Y, et al. Clinical manifestations of the anti-IgLON5 disease. Neurology 2017;88:1736–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghetti B, Oblak AL, Boeve BF, Johnson KA, Dickerson BC, Goedert M. Invited review: frontotemporal dementia caused by microtubule-associated protein tau gene (MAPT) mutations: a chameleon for neuropathology and neuroimaging. Neuropathol Appl Neurobiol 2015;41:24–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker M, Litvan I, Houlden H, et al. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum Mol Genet 1999;8:711–715. [DOI] [PubMed] [Google Scholar]
- 6.Pastor P, Ezquerra M, Tolosa E, et al. Further extension of the H1 haplotype associated with progressive supranuclear palsy. Mov Disord 2002;17:550–556. [DOI] [PubMed] [Google Scholar]
- 7.Rock KL, Reits E, Neefjes J. Present yourself by MHC class I and MHC class II molecules. Trends Immunol 2016;37:724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ezquerra M, Pastor P, Gaig C, et al. Different MAPT haplotypes are associated with Parkinson's disease and progressive supranuclear palsy. Neurobiol Aging 2011;32:547.e11–16. [DOI] [PubMed] [Google Scholar]
- 9.Rizzu P, Van Swieten JC, Joosse M, et al. High prevalence of mutations in the microtubule-associated protein tau in a population study of frontotemporal dementia in the Netherlands. Am J Hum Genet 1999;64:414–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreatta M, Karosiene E, Rasmussen M, Stryhn A, Buus S, Nielsen M. Accurate pan-specific prediction of peptide-MHC class II binding affinity with improved binding core identification. Immunogenetics 2015;67:641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karosiene E, Rasmussen M, Blicher T, Lund O, Buus S, Nielsen M. NetMHCIIpan-3.0, a common pan-specific MHC class II prediction method including all three human MHC class II isotypes, HLA-DR, HLA-DP and HLA-DQ. Immunogenetics 2013;65:711–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen KK, Andreatta M, Marcatili P, et al. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 2018;154:394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang X, Kushekhar K, Nolte I, et al. HLA associations in classical Hodgkin lymphoma: EBV status matters. PLoS One 2012;7:e39986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res 2008;36:509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sollid LM, Pos W, Wucherpfennig KW. Molecular mechanisms for contribution of MHC molecules to autoimmune diseases. Curr Opin Immunol 2014;31:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevens AM, Kanaan SB, Torok KS, et al. Brief report: HLA-DRB1, DQA1, and DQB1 in Juvenile-onset systemic sclerosis. Arthritis Rheumatol 2016;68:2772–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chemin K, Pollastro S, James E, et al. A novel HLA-DRB1*10:01-restricted T cell epitope from citrullinated type II collagen relevant to rheumatoid arthritis. Arthritis Rheumatol 2016;68:1124–1135. [DOI] [PubMed] [Google Scholar]
- 18.Hegde RS, Bernstein HD. The surprising complexity of signal sequences. Trends Biochem Sci 2006;31:563–571. [DOI] [PubMed] [Google Scholar]
- 19.Kovjazin R, Volovitz I, Daon Y, et al. Signal peptides and trans-membrane regions are broadly immunogenic and have high CD8+ T cell epitope densities: implications for vaccine development. Mol Immunol 2011;48:1009–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snow EC, Noelle RJ, Uhr JW, Vitetta ES. Activation of antigen-enriched B cells. II. Role of linked recognition in B cell proliferation to thymus-dependent antigens. J Immunol 1983;130:614–618. [PubMed] [Google Scholar]
- 21.Picascia S, Sidney J, Camarca A, et al. Gliadin-specific CD8+ T cell responses restricted by HLA class I A*0101 and B*0801 molecules in Celiac disease patients. J Immunol 2017;198:1838–1845. [DOI] [PubMed] [Google Scholar]
- 22.Gaig C, Iranzo A, Santamaria J, Graus F. The sleep disorder in anti-lgLON5 disease. Curr Neurol Neurosci Rep 2018;18:41. . [DOI] [PubMed] [Google Scholar]
- 23.Mastaglia FL, Needham M, Scott A, et al. Sporadic inclusion body myositis: HLA-DRB1 allele interactions influence disease risk and clinical phenotype. Neuromuscul Disord 2009;19:763–765. [DOI] [PubMed] [Google Scholar]
- 24.DeLuca GC, Alterman R, Martin JL, et al. Casting light on multiple sclerosis heterogeneity: the role of HLA-DRB1 on spinal cord pathology. Brain 2013;136:1025–1034. [DOI] [PubMed] [Google Scholar]
- 25.Bartoccioni E, Scuderi F, Augugliaro A, et al. HLA class II allele analysis in MuSK-positive myasthenia gravis suggests a role for DQ5. Neurology 2009;72:195–197. [DOI] [PubMed] [Google Scholar]
- 26.Cagnin A, Mariotto S, Fiorini M, et al. Microglial and neuronal TDP-43 pathology in anti-IgLON5-related tauopathy. J Alzheimers Dis 2017;59:13–20. [DOI] [PubMed] [Google Scholar]
- 27.Montagna M, Amir R, De Volder I, Lammens M, Huyskens J, Willekens B. IgLON5-Associated encephalitis with atypical brain magnetic resonance imaging and cerebrospinal fluid changes. Front Neurol 2018;9:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honorat JA, Komorowski L, Josephs KA, et al. IgLON5 antibody: neurological accompaniments and outcomes in 20 patients. Neurol Neuroimmunol Neuroinflamm 2017;4:e385 doi:10.1212/NXI.0000000000000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caffrey TM, Wade-Martins R. Functional MAPT haplotypes: bridging the gap between genotype and neuropathology. Neurobiol Dis 2007;27:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fakhoury M. Immune-mediated processes in neurodegeneration: where do we stand? J Neurol 2016;263:1683–1701. [DOI] [PubMed] [Google Scholar]
- 31.Sabater L, Planagumà J, Dalmau J, Graus F. Cellular investigations with human antibodies associated with the anti-IgLON5 syndrome. J Neuroinflammation 2016;13:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data from patients reported within the article are available and will be shared anonymously by request from any qualified investigator.