Abstract
Transcriptional regulation is crucial for neuronal activity-dependent processes that govern neuronal circuit formation and synaptic plasticity. An intriguing question is how neuronal activity influences the spatiotemporal interactions between transcription factors and their target sites. Here, using a single-molecule imaging technique, we investigated the activity dependence of DNA binding and dissociation events of cAMP-response element binding protein (CREB), a principal factor in activity-dependent transcription, in mouse cortical neurons. To visualize CREB at the single-molecule level, fluorescent-tagged CREB in living dissociated cortical neurons was observed by highly inclined and laminated optical sheet microscopy. We found that a significant fraction of CREB spots resided in the restricted locations in the nucleus for several seconds (dissociation rate constant: 0.42 s−1). In contrast, two mutant CREBs, which cannot bind to the cAMP-response element, scarcely exhibited long-term residence. To test the possibility that CREB dynamics depends on neuronal activity, pharmacological treatments and an optogenetic method involving channelrhodopsin-2 were applied to cultured cortical neurons. Increased neuronal activity did not appear to influence the residence time of CREB spots, but markedly increased the number of restricted locations (hot spots) where CREB spots frequently resided with long residence times (>1 s). These results suggest that neuronal activity promotes CREB-dependent transcription by increasing the frequency of CREB binding to highly localized genome locations.
SIGNIFICANCE STATEMENT The transcription factor, cAMP response element-binding protein (CREB) is known to regulate gene expression in neuronal activity-dependent processes. However, its spatiotemporal interactions with the genome remain unknown. Single-molecule imaging in cortical neurons revealed that fluorescent-tagged CREB spots frequently reside at fixed nuclear locations in the time range of several seconds. Neuronal activity had little effect on the CREB residence time, but increased the rapid and frequent reappearance of long-residence CREB spots at the same nuclear locations. Thus, activity-dependent transcription is attributable to frequent binding of CREB to specific genome loci.
Keywords: cortex, CREB, gene expression, imaging, neuronal activity, single molecule
Introduction
Neuronal activity plays an important role in cell differentiation and circuit formation in the cerebral cortex. In these processes, various genes are transcriptionally regulated in an activity-dependent fashion (West et al., 2002; Flavell and Greenberg, 2008). The transcription factor cAMP response element-binding protein (CREB) is involved in these regulatory processes (Lonze and Ginty, 2002). Biochemical and structural analyses have demonstrated that a CREB homodimer binds to a cAMP-response element (CRE) in the promoter regions of its target genes (Roesler et al., 1988; Mayr and Montminy, 2001). Furthermore, it has also been established that neuronal activity induces cofactor binding to CREB to enhance transcription. For example, activity-induced phosphorylation of CREB promotes the binding of CREB-binding protein (CBP)/p300 to CREB (Mayr and Montminy, 2001; Lonze and Ginty, 2002). cAMP-regulated transcriptional co-activator 1 (CRTC1) also binds to CREB through activity-dependent translocation from the cytoplasm to the nucleus (Zhou et al., 2006; Kovács et al., 2007; Li et al., 2009). Subsequently, basal transcriptional machinery components, such as transcription factors IIA–IIH and RNA polymerase II, are assembled (Ferreri et al., 1994; Saluja et al., 1998).
Assembly and disassembly of these factors are governed by protein–DNA and protein–protein interactions and by control spatiotemporal transcription within the nucleus (Ptashne and Gann, 1997). Recently, to elucidate these dynamic events, several studies have succeeded in monitoring the single-molecule dynamics of transcription factors (Speil et al., 2011; Gebhardt et al., 2013; Chen et al., 2014; Groeneweg et al., 2014; Morisaki et al., 2014; Sugo et al., 2015). We have demonstrated the spatiotemporal dynamic behavior of single-molecule CREB both in noncellular conditions and in living Neuro2a cells; these experiments indicated that CREB binds specifically to CRE for several seconds (Sugo et al., 2015). In accord with this view, other mammalian transcription factors transiently reside on their target sequences in the nucleus with a similar time course of the order of seconds (Speil et al., 2011; Gebhardt et al., 2013; Chen et al., 2014; Groeneweg et al., 2014; Morisaki et al., 2014). However, the kinetics of CREB–CRE interaction in activity-dependent mechanisms remain unknown.
In the present study, we investigated how neuronal activity influences CREB dynamics by real-time imaging of fluorescent-tagged CREB at the single-molecule level in living cortical neurons. Our results show that neuronal activity does not detectably alter the residence time distribution of CREB spots in the nucleus, but more markedly increases the number of locations where CREB frequently resides with long residence times (>1 s). These findings suggest that neuronal activity promotes the frequency of CREB–CRE binding events in particular subnuclear positions.
Materials and Methods
Animals.
ICR (Institute for Cancer Research) mice of either sex were used to obtain cortical cultures (Japan SLC). Noon of the day on which the vaginal plug was detected in the morning was designated embryonic day (E) 0. All experiments were conducted under the guidelines for laboratory animals of the Graduate School of Frontier Biosciences, Osaka University.
Plasmids.
pFN21AB5414, containing HaloTag-human CREB1 cDNA, and pFN21AB9936, containing HaloTag-human CRTC1 cDNA, were purchased from Promega. The HaloTag-human CREB1 R301L and L318/325V mutants were generated from pFN21AB5414 (Sugo et al., 2015).
To generate the bicistronic tetracycline (Tet)-inducible expression vector pTet-On Advance/TRE-Tight, a XhoI-digested fragment containing the Ptight-inducible promoter with a multiple cloning site from pTRE-Tight (Clontech) was ligated to XhoI-digested pTet-On Advance (Clontech) containing the rtTA2S-M2 transactivator. To generate the Tet-inducible HaloTag-CREB expression vector pTet-On Advance/TRE-Tight HaloTag-CREB, HaloTag-human CREB cDNA was inserted into the multiple cloning site in pTet-on Advance/TRE-Tight. The neuron-specific βIII tubulin promoter-driven EGFP expression vector pTα1-EGFP was used for identifying neurons in primary dissociated cultures (Hatanaka and Murakami, 2002).
Primary cortical neuron culture and pharmacological treatment.
Pregnant mice were deeply anesthetized with pentobarbital (50 mg/kg, i.p.). Cortical lobes were dissected from E16 mouse embryos in ice-cold HBSS and minced with fine scissors. The minced tissues were incubated with 0.125% trypsin and 0.02% EDTA in PBS for 5 min and dissociated thoroughly by pipetting. After a brief centrifugation, the cells were resuspended in DMEM/F12 medium (Life Technologies) supplemented with B27 (Life Technologies) and 5% fetal bovine serum (Hyclone). Aliquots of the cell suspension containing 1–5 × 105 cells were plated with culture medium on a 35 mm glass-bottom culture dish (Greiner Bio-One), which had been coated with 0.1 mg/ml poly-L-ornithine (Sigma-Aldrich). The cultures were maintained at 37°C in an environment of 5% CO2 and humidified 95% air. To induce expression of HaloTag-CREB in neurons transfected with pTet-On Advance/TRE-Tight HaloTag-CREB, the cells were treated with doxycycline (0.05 ng/ml) for 17 h before observation. To label HaloTag-CREB, HaloTag-CREB cDNA-transfected cells were incubated with growth medium containing 10 nm tetramethylrhodamine (TMR)-conjugated HaloTag Ligand (Promega) for 15 min in a CO2 incubator, after which the medium was replaced.
To depolarize the cultured cells, 0.41 volumes of KCl depolarization solution (170 mm KCl, 1.3 mm MgCl2, 0.9 mm CaCl2, 10 mm HEPES, pH 7.4) was added to the culture medium. To block action potentials, tetrodotoxin (TTX; Sankyo) was added to the culture medium (final concentration, 100 nm).
Transfection.
Cultured cortical neurons were transfected by electroporation. Plasmid solution (0.5 μg/μl DNA in Ca2+-free, Mg2+-free, and phenol red-free HBSS) was added to the culture dish, and electric pulses were delivered with plate electrodes (LF513-5, BEX) connected to a square-pulse generator (CUY21EX, BEX). One 275 V pulse of 10 ms duration and 10 30 V pulses of 50 ms duration were applied at 50 ms intervals. After pulse application, the plasmid solution was replaced with culture medium as described above.
Photostimulation.
Photostimulation was performed as described previously (Boyden et al., 2005; Malyshevskaya et al., 2013). In brief, pCAGGS-hChR2(H134R)-EYFP was used to induce firing activity of cortical neurons. pCAGGS-EYFP was used as a control for the optogenetic experiment. A solid-state illuminator (475 nm wavelength; Spectra, Lumencor) was used for photostimulation through the objective lens. The duration and frequency of the light stimulus were controlled by a pulse generator (Master-8, AMPI).
Immunostaining.
Cortical cultures were fixed in 4% paraformaldehyde in PBS at room temperature for 10 min or in ice-cold methanol for 15 min. They were then permeabilized and blocked for 10 min in buffer G, composed of 5% normal goat serum (Vector Laboratories) and 0.1% Triton X-100 in PBS. The cells were then incubated overnight at 4°C with anti-lamin A/C mouse monoclonal antibody (39288, clone 3A6-4C11; Active Motif) at 1:1000, anti-CREB mouse monoclonal antibody (Cell Signaling Technology) at 1:200, anti-c-Fos rabbit monoclonal antibody (#2250; Cell Signaling Technology) at 1:100, or anti-RNAP II C-terminal domain phospho-Ser5 rabbit polyclonal antibody (Active Motif) at 1:10,000 in buffer G. Primary antibodies were detected by incubation with Cy5-conjugated anti-mouse IgG (Jackson ImmunoResearch) at 1:400 or Cy5-conjugated anti-rabbit IgG (Jackson ImmunoResearch) at 1:400 in buffer G at room temperature for 2 h. Cells were mounted with a medium containing 50% glycerol and 2.3% 1,4-diazabicyclo[2.2.2]octane (Sigma-Aldrich), and 0.1% 4′,6-diamidino-2-phenylimdole (DAPI, Sigma-Aldrich) in 50 mm Tris-HCl, pH 8.0.
Microscopy.
An inverted microscope (Ti-E, Nikon) with an oil-immersion objective (100×; numerical aperture, 1.49; Nikon) was used for all experiments. EGFP, TMR, and Cy5 were excited by highly inclined and laminated optical sheet (HILO) illumination with 488 (20 mW; Coherent), 561 (20 mW; Coherent), and 640 nm (40 mW; Coherent) lasers, respectively. All fluorescence live images were obtained at 10 frames per second using an EM-CCD (iXon897, Andor Technology) with Solis software (Andor Technology). To observe the dynamics of TMR-CREB in living cells, the cell culture dish was mounted on a stage top incubator (Tokai Hit) maintained at 37°C in an environment of humidified 5% CO2, 20% O2, and 75% N2. All images were analyzed by ImageJ software with a self-made plugin (Particle Track and Analysis). Fluorescence images of fixed cells were acquired and processed with NIS Element software linked to a deconvolution module (Nikon).
Statistical analysis.
Significant differences were determined with the Mann–Whitney U test, paired t test, or ANOVA with Tukey–Kramer post hoc test. Origin 9.1 software (OriginLab) and Excel 2010 (Microsoft) were used for statistical analysis and data plotting. The biexponential function fitting distributions of residence time is F(t) = A0 + A1 exp(−t/t1) + A2 exp(−t/t2) (Eq. 1), where t is time and A0 is constant. A1 and A2 are the fractions with dissociation rate constants 1/t1 and 1/t2 for the short-residence and long-residence component, respectively.
Results
Single-molecule CREB dynamics in living cortical neurons
To study the dynamics of CREB in the nuclei of living neurons at the single-molecule level, HaloTag protein fused to CREB (HaloTag–CREB) was used. A single molecule of HaloTag–CREB is specifically and covalently labeled with a TMR-conjugated HaloTag ligand in living cells (TMR-CREB; Sugo et al., 2015). HILO microscopy was used for single-molecule observation of TMR-CREB (Fig. 1A; Tokunaga et al., 2008; Sugo et al., 2015).
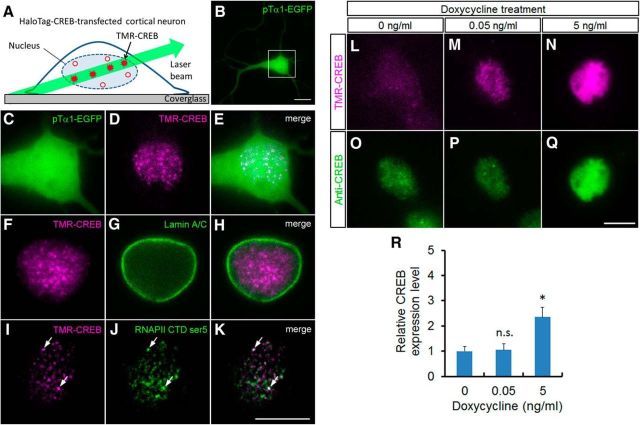
Figure 1.
Single-molecule imaging of CREB in cortical neurons. A, Experimental design of single-molecule imaging to visualize TMR-CREB in the nuclei of cortical neurons. A limited number of TMR-CREB molecules in the nucleus are observed with low background using HILO microscopy. B, A cortical neuron transfected with pTα1-EGFP and HaloTag–CREB was observed at 7 DIV. C–K, Subcellular localization of TMR-CREB in fixed HaloTag–CREB-transfected cortical neurons using HILO fluorescence microscopy. C, Magnified image of the box in B. The cortical cells were immunostained with anti-lamin A/C antibody (G, H), anti-Ser5-RNAPII C-terminal domain (J, K), and CREB signals were visualized by TMR-conjugated HaloTag ligand (D, E, F, H, I, K). I–K, Series of images (0.2 μm × 50 sections) were subjected to a deconvolution algorithm. Arrows indicate TMR-CREB colocalized with Ser5-phosphorylated RNAP II C-terminal domain. Scale bar, 10 μm. L–Q, The endogenous and exogenous CREB expression level in the HaloTag–CREB-expressing neurons. Fluorescence images of TMR-CREB (L–N) and immunostaining with anti-CREB antibody (O–Q) in fixed HaloTag–CREB-transfected cortical neurons when adding different concentration of doxycycline (0, 0.05, 5 ng/ml). R, The quantitative analysis shows the relative expression level of CREB in transfected neurons treated with different doxycycline concentrations. Bars represent the mean ± SD. The number of neurons analyzed is 21 (0 ng/ml), 15 (0.05 ng/ml), and 15 (5 ng/ml) cells, respectively. The asterisk indicates a significant difference in untreated neurons (0 ng/ml; ANOVA with Tukey–Kramer post hoc tests, *p < 0.001).
A neuron-specific, βIII-tubulin promoter Tα1-driven EGFP expression vector was cotransfected in cortical neurons with a Tet-inducible promoter-driven HaloTag–CREB expression vector. Low-level expression of HaloTag–CREB was induced by adding a low concentration of doxycycline (0.05 ng/ml) and labeled with cell membrane-permeable TMR-conjugated HaloTag ligand (TMR-CREB). At 7 days in vitro (DIV), TMR-CREB signals were observed in EGFP-expressing cortical neurons (Fig. 1B–E). First, immunocytochemical analysis was performed to examine the subcellular localization of TMR-CREB in the transfected neurons. The signals were predominantly localized within the nucleus, delineated by lamin A/C, a nuclear envelope component (Gerace et al., 1978; Fig. 1F–H). Moreover, some TMR-CREB spots colocalized with the signals of Ser5-phosphorylated RNAP II C-terminal domain, suggesting that these TMR-CREB molecules are present at actively transcribing loci (Fig. 1I–K). These results in primary cortical neurons were consistent with our previous findings in Neuro2a cells, a neuroblastoma cell line (Sugo et al., 2015).
To evaluate the expression level of HaloTag–CREB against endogenous CREB, we performed immunostaining with CREB antibody, which can recognize both endogenous CREB and HaloTag–CREB (Fig. 1L–Q). The immunostaining analysis showed that the CREB expression level was approximately the same between the untreated neurons and those treated with the low concentration of doxycycline (0.05 ng/ml), although the expression level significantly increased with a higher concentration of doxycycline (5 ng/ml; Fig. 1O–R). Thus, HaloTag–CREB expression induced by the low concentration of doxycycline, which is the concentration used in the present study, was very low compared with the endogenous level.
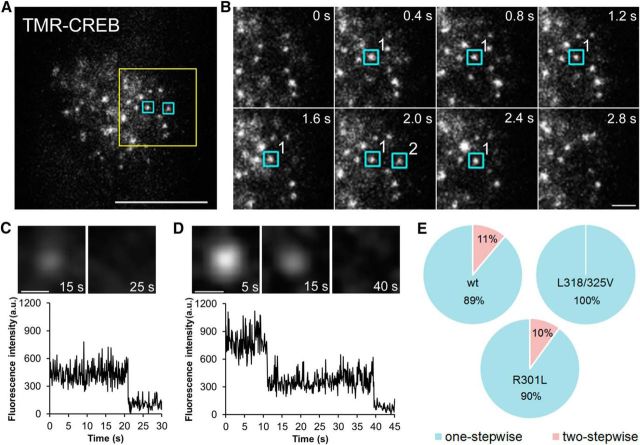
Next the movements of TMR-CREB spots were investigated in living cortical neurons. To do this, real-time imaging was performed at 10 frames per second, which enables us to monitor individual spots interacting with DNA (Sugo et al., 2015). Numerous TMR-CREB spots were visible in the nuclei of living cortical neurons (Movie 1; Fig. 2A). We studied whether the observed TMR-CREB spots represent single molecules by examining the manner in which light spots diminished when the excitation light was applied continuously after fixation. As shown in Figure 2C,D, one-stepwise or two-stepwise reduction in their fluorescence intensities was found in the TMR-CREB spots. The majority (89%) of CREB spots showed a one-stepwise reduction, whereas the remaining fraction (11%) exhibited a two-stepwise reduction (Fig. 2E). The single-molecule aspect of TMR-CREB spots was further confirmed by using two mutant CREBs, which cannot bind to CRE because they either lack the ability to dimerize (L318/325V; Dwarki et al., 1990) or possess a disrupted DNA-binding domain despite of the dimerization ability (R301L; Walton et al., 1992). The two-stepwise disappearance was never observed in the mutant CREB (L318/325V; Fig. 2E). On the other hand, in another mutant CREB (R301L), 10% of TMR-CREB spots disappeared with two steps, which is similar to that of wild-type CREB (Fig. 2E). This result indicates that TMR-CREB spots are visualized in cortical neurons at the single-molecule level.
Figure 2.
Binding dynamics of CREB in the nuclei of living cortical neurons. A, HILO image showing individual TMR-CREB spots in the nucleus at 5 DIV. Scale bar, 10 μm. B, Time-lapse images of the yellow box in A. The blue boxes indicate TMR-CREB spots with long (>1 s, spot 1) and short (<1 s, spot 2) residence times at restricted locations. Scale bar, 2 μm. C, D, TMR-CREB spots exhibit one-stepwise (C) or two-stepwise (D) reductions of the fluorescence intensities. Scale bar, 0.5 μm. E, The pie graph shows the ratio of one-stepwise and two-stepwise reduction of the fluorescence intensities in wild-type and mutant CREB (L318/325V, R301L).
Movie 1.
Single-molecule dynamics of CREB in living cortical neurons at 7 DIV. HILO images show individual TMR-CREB spots in a living neuron nucleus (left). TMR-CREB spots were tracked with the ImageJ software plugin Particle Track and Analysis (right). Movie speed is real time.
Many TMR-CREB spots disappeared immediately, while a fraction of them resided in the same locations with a time course of a few seconds (Movie 1; Fig. 2A,B). The spots with a longer residence time (>1 s) may represent specific binding of TMR-CREB to CRE sites (Sugo et al., 2015). To test the possibility, we compared the residence time distributions of wild-type CREB with those of the two mutant CREBs (L318/325V or R301L). Although the majority of TMR-CREB spots disappeared by 1 s in both wild-type CREB (94%) and mutant CREBs (>97%; Fig. 3A), those with the longer residence times (>1 s) were found more frequently in wild-type CREB (Fig. 3B).
Figure 3.
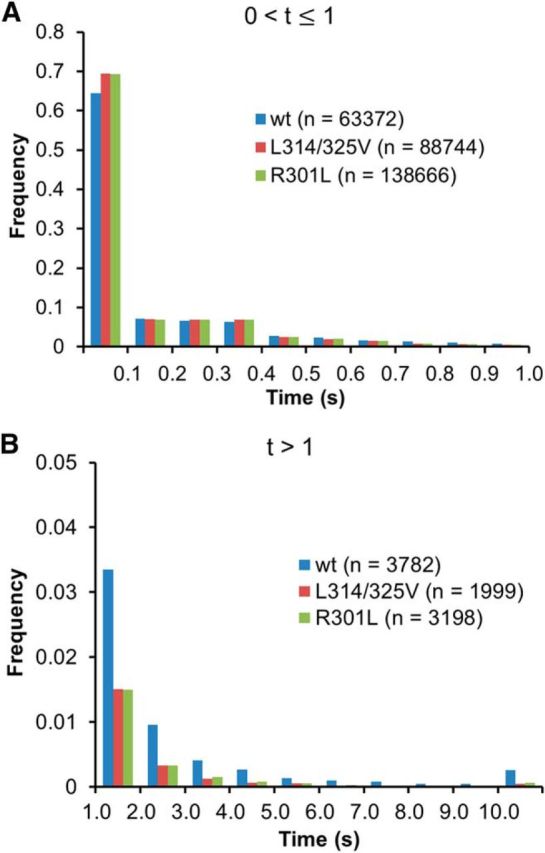
The behavior of wild-type CREB compared with two mutant CREBs. A, B, Distribution histograms show the short (≤1 s; A) and long (>1 s; B) residence times of TMR-CREB, TMR-mutant CREB (R301L), and TMR-mutant CREB (L318/325V) in living neurons. The number of spots analyzed in living cells is presented in parentheses.
To evaluate this difference quantitatively, the residence time distribution was fitted with a biexponential function (short-residence and long-residence components) for wild-type and mutant CREBs, and the ratio of the long-residence component to the short-residence component (A2/A1; see Materials and Methods, Eq. 1) and the dissociation rate constants (1/t1 and 1/t2) were calculated for each case. From the fitting data, the dissociation rate constants for wild-type CREB in cortical neurons at 5 DIV (3.18 ± 0.34 s−1 and 0.42 ± 0.13 s−1 for the short-residence and long-residence components, respectively) were comparable with those in Neuro2a cells (Sugo et al., 2015, their Table S1). The ratio of the long-residence to short-residence component was approximately 1:8 (0.120 ± 0.039 for wild type). In contrast, the ratio of the long-residence to short-residence component was significantly lower in both mutant CREBs (0.043 ± 0.037 for L318/325V, 0.050 ± 0.058 for R301L, p < 0.05 for Mann–Whitney U test with a Bonferroni correction; Table 1), although the dissociation rate constant for the long-residence component was not significantly different. Collectively, these results suggest that wild-type CREB but not mutant CREBs resided at fixed positions for several seconds.
Table 1.
Ratios of long-residence to short-residence component fractions of wild-type CREB and mutant CREB spots with different binding modes (short and long residence time) in cortical neurons
| CREB | Number of cells | Dissociation rate constant (s−1; mean ± SD) |
Ratio of long-residence to short-residence component (mean ± SD) | |
|---|---|---|---|---|
| Short residence | Long residence | |||
| Wild type | 9 | 3.18 ± 0.34 | 0.42 ± 0.13 | 0.120 ± 0.039 |
| L318/325V | 7 | 3.81 ± 0.23 | 0.99 ± 1.37 | 0.043 ± 0.037* |
| R301L | 9 | 3.71 ± 0.15 | 0.77 ± 1.10 | 0.050 ± 0.058* |
The data were evaluated by fitting the distribution histogram of wild-type CREB or mutant CREBs. An asterisk indicates that the ratio of long-residence to short-residence component of mutant CREB (L318/325V or R301L) was significantly different from that of wild-type CREB (*p < 0.05, Mann–Whitney U test with a Bonferroni correction).
The residence time of CREB is almost unchanged when neuronal activity increases
A fundamental question is how neuronal activity influences the CREB–CRE interaction and thereby enhances transcription. We predicted that increased neuronal activity would affect the residence time of CREB. To test this, we examined the distribution of CREB residence times before and after pharmacological treatments that alter neuronal activity.
We first tested whether a depolarizing KCl treatment (50 mm) is sufficient to induce CREB-dependent transcription with nuclear translocation of CRTC1 (Zhou et al., 2006; Kovács et al., 2007; Li et al., 2009). In cortical neurons transfected with HaloTag–CRTC1, HaloTag–CRTC1 labeled with TMR (TMR-CRTC1) was predominantly localized in the cytoplasm at 7 DIV, when spontaneous firing activity is low in the present culture (Uesaka et al., 2005; Sugo et al., 2010; Fig. 4A). After KCl treatment, TMR-CRTC1 translocated from the cytoplasm to the nucleus (Fig. 4A, left). Simultaneously, expression of c-fos, a CREB-dependent immediate early gene, was also increased (Fig. 4A, middle). Thus, this KCl treatment promotes CREB-mediated downstream activity-dependent gene expression. Under these conditions, we next examined the CREB residence time in HaloTag–CREB-transfected cortical cultures.
Figure 4.
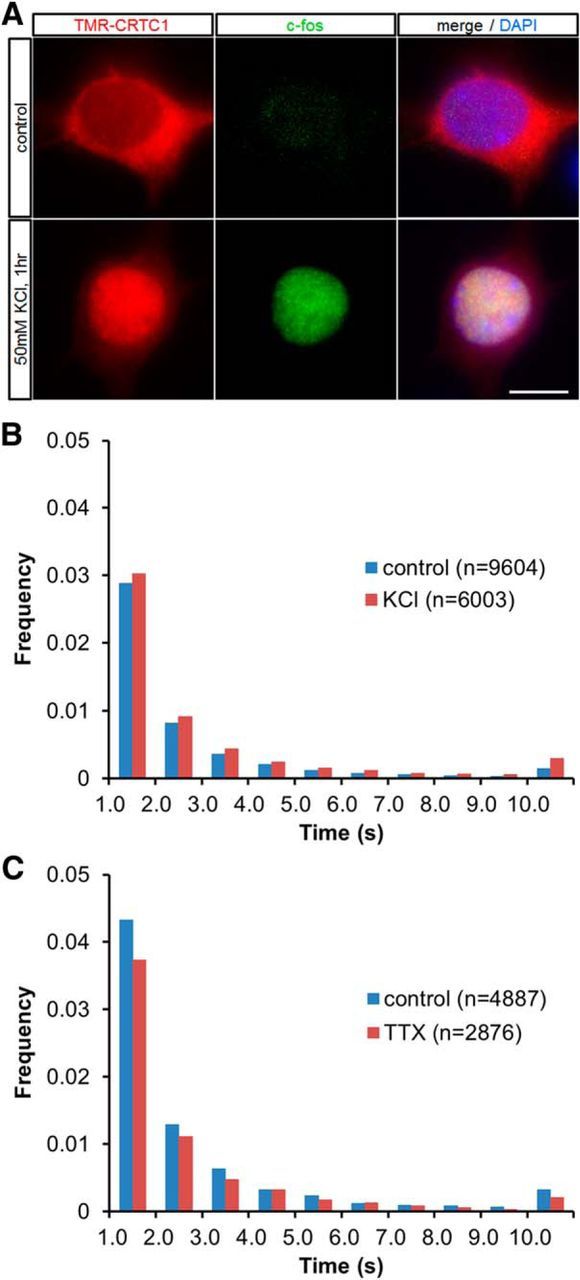
Dynamics of CREB in response to changing neuronal activity by pharmacological treatment. A, Subcellular localization of TMR-CRTC1 in fixed HaloTag–CRTC1-transfected neurons with or without KCl treatment (50 mm) for 1 h at 7 DIV. Immunocytochemical staining was also performed with c-fos antibody and DAPI. Scale bar, 10 μm. B, Distribution histograms show the long (>1 s) residence times of TMR-CREB spots in untreated (control) cortical neurons at 7 DIV or in those treated with 50 mm KCl for 1 h. The number of spots analyzed is presented in parentheses. C, Distribution histograms show the long (>1 s) residence times of TMR-CREB spots in untreated (control) cortical neurons at 14 DIV and in those treated with TTX for 24 h. The number of spots analyzed is presented in parentheses.
One hour after the KCl treatment, the distribution of CREB residence times was slightly shifted to increasing the population of spots with long residence times (Fig. 4B). Quantitative analysis showed that the dissociation rate constant for the long-residence component decreased significantly (p < 0.01 for Mann–Whitney U test) in cortical neurons, but the ratio of the long-residence to short-residence component was not significantly different (Table 2). To study the effect of reducing neuronal activity, the cortical cultures at 14 and 15 DIV, when spontaneous firing occurs frequently, were treated with the sodium channel blocker TTX (Uesaka et al., 2005; Sugo et al., 2010). We found that 24 h after TTX treatment, the residence time distribution of TMR-CREB was not different from that without the treatment. In fact, neither the ratio of the long-residence to short-residence component nor the dissociation rate constant was significantly different (Fig. 4C; Table 2).
Table 2.
TMR-CREB dissociation rate constants in neurons in response to KCl depolarization and TTX
| DIV | Number of cells | Treatment | Dissociation rate constant (s−1; mean ± SD) |
Ratio of long-residence to short-residence component (mean ± SD) | |
|---|---|---|---|---|---|
| Short residence | Long residence | ||||
| 7 | 22 | — | 3.21 ± 0.43 | 0.46 ± 0.07 | 0.124 ± 0.049 |
| 7 | 18 | KCl | 3.02 ± 0.44 | 0.36 ± 0.10** | 0.140 ± 0.059 |
| 14 | 6 | — | 2.74 ± 0.43 | 0.42 ± 0.11 | 0.152 ± 0.043 |
| 15 | 9 | TTX | 3.07 ± 0.43 | 0.46 ± 0.08 | 0.136 ± 0.037 |
The data were evaluated by fitting the distribution histogram of TMR-CREB with or without pharmacological treatments. The asterisks show that the dissociation rate constant of the long component after KCl treatment was significantly different from that in untreated neurons (**p < 0.01, Mann–Whitney U test).
To reveal a possible causal relationship between CREB dynamics and neuronal activity, we further studied the residence time of CREB by stimulating cortical neurons using an optogenetic method with the light-gated channel channelrhodopsin-2 (ChR2; Boyden et al., 2005; Malyshevskaya et al., 2013). In HaloTag–CRTC1 and ChR2-EYFP cotransfected neurons, repetitive photostimulation (0.1 Hz, 1 s duration, 60 pulses) induced the rapid translocation of TMR-CRTC1 from the cytoplasm to the nucleus, but did not in EYFP-transfected control neurons (Fig. 5A,B). Thus, optogenetic stimulation effectively increased neuronal activity and induced TMR-CRTC1 translocation. Under these conditions, we examined the behavior of CREB after a 5 min period of repetitive photostimulation (0.1 Hz, 1 s duration, 30 pulses; Fig. 5D). Similar to the result for KCl treatment (Fig. 4B; Table 2), the dissociation rate constant of the long-residence component was slightly different from that before the photostimulation (p < 0.05 for paired t test), but the ratio of the long-residence to short-residence component was unchanged (Fig. 5E; Table 3). Different stimulation (2 Hz, 50 ms duration, 600 pulses; Fig. 5C) also did not influence the residence time distribution (Fig. 5F). The distribution of mutant-CREB (R301L) was also unaffected by photostimulation (Fig. 5G). These results indicate that neuronal activity does not detectably influence the residence time distribution of CREB.
Figure 5.

CREB behavior in response to stimulation using ChR2 optogenetics. A, Subcellular localization of TMR-CRTC1 was observed by time-lapse imaging after the onset of photostimulation (0.1 Hz, 1 s duration) in an EYFP-expressing or ChR2-EYFP-expressing cortical neuron at 5 DIV. Representative images at 0, 200, 400, and 600 s are shown. Scale bar, 10 μm. B, Fluorescence intensities of TMR-CRTC1 in the nucleus of an EYFP-transfected or ChR2-EYFP-transfected cortical neuron were analyzed in the series of images represented in A. C, Subcellular localization of TMR-CRTC1 was observed before and after photostimulation (2 Hz, 50 ms duration) for 10 min in a ChR2-EYFP-expressing cortical neuron at 5 DIV. Scale bar, 10 μm. D, A TMR-CREB-expressing and ChR2-EYFP-expressing cortical neurons. Scale bar, 10 μm. E, F, Distribution histograms show the long (>1 s) residence times of TMR-CREB spots in cotransfected neurons at 5 DIV, before (pre) and after (post) photostimulation (E: 0.1 Hz, 1 s duration; F: 2 Hz, 50 ms duration) for 5 min. G, Distribution histograms show the long (>1 s) residence times of TMR-mutant CREB (R301L) spots in cotransfected neurons at 5 DIV, before (pre) and after (post) photostimulation (0.1 Hz, 1 s duration) for 5 min. The number of spots analyzed is presented in parentheses.
Table 3.
TMR-CREB dissociation rate constants in ChR2-transfected neurons in response to photostimulation
| Photostimulation | Number of cells | Dissociation rate constant (s−1; mean ± SD) |
Ratio of long-residence to short-residence component (mean ± SD) | |
|---|---|---|---|---|
| Short residence | Long residence | |||
| − | 15 | 3.07 ± 0.22 | 0.54 ± 0.13 | 0.139 ± 0.049 |
| + | 15 | 3.09 ± 0.20 | 0.46 ± 0.08* | 0.171 ± 0.057 |
The data were evaluated by fitting the distribution histogram of TMR-CREB in cortical neurons at 5 DIV before and after photostimulation. The asterisk shows that the dissociation rate constant of the long component was significantly different before and after photostimulation (*p < 0.05, paired t test).
Neuronal activity increases the frequency of CREB–CRE binding events in subnuclear areas
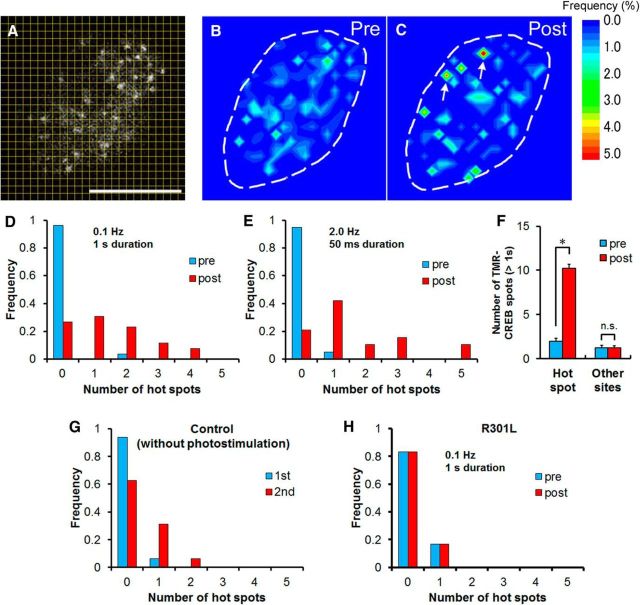
In our previous study using Neuro2a cells, we found that TMR-CREB spots with long residence times (>1 s) repeatedly appeared at highly restricted positions in the nucleus (Sugo et al., 2015). We finally investigated the possibility that neuronal activity regulates the spatial distribution of TMR-CREB spots in HaloTag–CREB and ChR2-EYFP cotransfected neurons. The number of TMR-CREB spots with long residence times was counted in each subnuclear area (0.8 × 0.8 μm; Fig. 6A). A subnuclear area in which >5% of the total TMR-CREB spots accumulated was defined as a “hot spot.” As exemplified in Figure 6B, no hot spots were detected in a cortical neuron tested at 5 DIV. After photostimulation (0.1 Hz, 1 s duration, 30 pulses), two different hot spots emerged (Fig. 6C). Hot spots were found in only one neuron (1 of 26) before photostimulation, whereas hot spots emerged in the majority of the neurons tested (19 of 26) after stimulation. Quantitative analysis revealed that photostimulation significantly increased the number of hot spots in an optical section per cell (0.1 ± 0.1 before stimulation; 1.4 ± 0.2 after stimulation, n = 26 cells, p < 0.001 for Mann–Whitney U test; Fig. 6D). Another stimulation (2 Hz, 50 ms duration, 600 pulses) also increased the number of hot spots (0.1 ± 0.1 before stimulation; 1.6 ± 0.4 after stimulation, n = 19 cells, p < 0.001 for Mann–Whitney U test; Fig. 6E). In these hot spots, TMR-CREB spots with long residence times (>1 s) appeared more frequently after photostimulation (before: 2.0 ± 0.3; after: 10.2 ± 0.4 during 60 s of observation time, Mann–Whitney U test, p < 0.001), but not in other sites (before: 1.3 ± 0.3; after: 1.2 ± 0.2 during 60 s of observation time, Mann–Whitney U test, p = 0.894; Fig. 6F). In contrast, there was no significant difference in the number of hot spots between two successive measurements without photostimulation (0.1 ± 0.1 for the first observation; 0.4 ± 0.2 for the second observation, n = 16 cells, p = 0.686 for Mann–Whitney U test; Fig. 6G). To further investigate whether CREB–CRE binding is required for the formation of hot spots, the TMR-mutant CREB (R301L) was examined. The result of photostimulation in the TMR-mutant CREB (R301L) was a relative absence of hot spots (Fig. 6H) compared with wild type (Fig. 6D).
Figure 6.
Dynamics of CREB hot spots in the nuclei of living cortical neurons. A–C, The number of TMR-CREB spots for the entire observation time (60 s) was counted in each subarea. For the nucleus depicted here, eight and 11 of 157 spots (>5%) accumulated in the two different subnuclear areas defined as hot spots (red), indicated by the arrows. A, The area covering the whole nucleus in a cortical neuron was divided into 625 (25 × 25) equal subareas (0.8 × 0.8 μm). Scale bar, 10 μm. Color contour maps represent the spatial distribution of the frequency of appearance of TMR-CREB spots having long residence times (>1 s) in the living neuron shown in A before (pre, B) and after (post, C)photostimulation. The dashed line marks the perimeter of the nucleus. D, The distribution histogram shows the number of hot spots before (pre) and after (post) photostimulation (0.1 Hz, 1 s duration; n = 26 cells). The average number of hot spots was significantly different before (0.1 ± 0.1) and after stimulation (1.4 ± 0.2; Mann–Whitney U test, p < 0.001). E, The distribution histogram shows the number of hot spots of TMR-CREB before (pre) and after (post) photostimulation (2 Hz, 50 ms duration; n = 19 cells) for 5 min. The average number of hot spots was significantly different before (0.1 ± 0.1) and after stimulation (1.6 ± 0.4; Mann–Whitney U test, p < 0.001). F, The histogram shows the number of TMR-CREB spots with long-residence times (>1 s) observed in hot spots (n = 37) or other sites (n = 32) in 26 cells. Bars represent the mean ± SEM. The asterisk indicates a significant difference between before (pre) and after (post) photostimulation (Mann–Whitney U test, *p < 0.001). G, The distribution histogram shows the number of hot spots in two successive control measurements without photostimulation (n = 16 cells). The average number of hot spots in the first (1st; 0.1 ± 0.1) and the second (2nd; 0.4 ± 0.2) measurement was not significantly different (Mann–Whitney U test, p = 0.686). H, The distribution histogram shows the number of hot spots of mutant CREB (R301L) before (pre) and after (post) photostimulation (0.1 Hz, 1 s duration; n = 6 cells) for 5 min. The average number of hot spots was not significantly different before (0.2 ± 0.2) and after stimulation (0.2 ± 0.2; Mann–Whitney U test, p = 1.000).
Taken together, these results indicate that neuronal activity promotes the formation of hot spots, where TMR-CREB appears to more frequently bind to highly restricted genome locations than to other sites.
Discussion
present study demonstrates, for the first time, activity-dependent behavior of CREB in living cortical neurons at the single-molecule level. The results revealed that a small but significant fraction of TMR-CREB molecules resided in fixed locations over a time range of several seconds. Furthermore, we found that neuronal activity induced the frequent reappearance of TMR-CREB spots with long residence times (>1 s) at the same nuclear locations, whereas the residence time distribution was not significantly affected. Therefore, we propose that neuronal activity contributes to CREB-dependent gene expression by increasing the frequency of CREB binding to specific gene loci.
Our previous study using noncellular assays demonstrated that CREB spots reside on a CRE sequence-coated substratum with much longer residence times than on substrates coated with unrelated sequences, suggesting that spots with long residence times (>1 s) reflect specific binding of CREB to CRE while those with short residence times represent nonspecific binding to DNA (Sugo et al., 2015). CREB spot appearance with short and long residence times was further demonstrated in the nuclei of Neuro2a cells (Sugo et al., 2015). In the present study, we discovered similar CREB dynamics in cortical neurons (Table 1). Thus, these temporal properties may be common in many cell types (Lanctôt et al., 2007; Hager et al., 2009).
Our previous study also suggested that hot spots represent actively transcribing gene loci (Sugo et al., 2015). Some TMR-CREB spots were also found to colocalize with active RNA polymerase loci in cortical neurons (Fig. 1I–K). Supporting this view, the present results showed an increase in the hot spots after photostimulation in the wild-type HaloTag–CREB-transfected neurons, but did not in the mutant HaloTag–CREB-transfected neurons (Fig. 6), suggesting that neuronal activity enhances the frequency of CREB binding to specific CRE sites and the subsequent downstream gene expression. Fluorescence recovery after photobleaching analyses have suggested that nuclear receptors also bind frequently to artificial tandem gene arrays that include their binding sites in response to signals, such as those from hormones (McNally et al., 2000; Rayasam et al., 2005; Sharp et al., 2006). On the other hand, heat-shock transcription factors have been shown to promote transcription in Drosophila by extending their residence time on target sites up to the order of minutes in response to heat shock (Yao et al., 2006). Thus, different transcription factors may control gene expression by distinct modes of interaction with their target sites (Hager et al., 2009), although further study is also need to quantify transcriptional activity in the hot spots of living neurons.
Which genes are activated by the formation of hot spots in response to neuronal activity? CREB regulates the expression of numerous genes involved in neuronal development and plasticity (Impey et al., 2004; Zhang et al., 2005). Among them, the transcription of immediate early genes (IEGs), such as c-fos, is rapidly induced, within a few minutes, in response to neuronal activity (Greenberg et al., 1986). Considering that the CREB hot spots increased only 5 min after photostimulation (Fig. 6), the hot spots observed in the present study may correspond to loci of CREB-regulated IEGs.
The molecular mechanism of how neuronal activity increases the number of hot spots remains unclear. Chromatin remodeling is a strong candidate to explain CREB behavior, because specific chromatin states have been shown to inhibit the binding of CREB to the CRE (Iguchi-Ariga and Schaffner, 1989; Chen et al., 2003; Martinowich et al., 2003; Zhang et al., 2005). Indeed, neuronal activity affects chromatin regulation to modulate transcription (Chawla et al., 2003; Korzus et al., 2004; Sugo et al., 2010; Cohen et al., 2011; Saha et al., 2011). In particular, CBP possesses histone acetyltransferase activity and can alter the chromatin structure (Bannister and Kouzarides, 1996; Ogryzko et al., 1996). Activity-dependent formation of CREB/CBP complexes may increase the probability of CREB binding to the CRE by relaxing local chromatin structure around CRE sites.
Footnotes
This work was supported by Ministry of Education, Culture, Sports, Science, and Technology (MEXT) KAKENHI (Grants in Aid for Scientific Research) grants on Innovative Areas “Adaptive Circuit Shift” (Grant No. 15H01436 to N.Y.), “Cross-talk between Moving Cells and Microenvironment as a Basis of Emerging Order in Multicellular System” (Grant No. 23111516 to N.S.), and Grant No. 20200009 (to N.S.). This work was also supported by the Japan Society for the Promotion of Science KAKENHI Grant Nos. 23700447, 25640035, and 15K14350 to N.S. and 20300110 and 23300118 to N.Y., and by the MEXT Global Centers of Excellence Program. We thank Aya Ohkuni for technical assistance and discussions. We also thank Dr. Ian Smith and Gabriel Hand for critical reading of the manuscript.
The authors declare no competing financial interests.
References
- Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Chawla S, Vanhoutte P, Arnold FJ, Huang CL, Bading H. Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J Neurochem. 2003;85:151–159. doi: 10.1046/j.1471-4159.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, Tjian R, Liu Z. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Cohen S, Gabel HW, Hemberg M, Hutchinson AN, Sadacca LA, Ebert DH, Harmin DA, Greenberg RS, Verdine VK, Zhou Z, Wetsel WC, West AE, Greenberg ME. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron. 2011;72:72–85. doi: 10.1016/j.neuron.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwarki VJ, Montminy M, Verma IM. Both the basic region and the ‘leucine zipper’ domain of the cyclic AMP response element binding (CREB) protein are essential for transcriptional activation. EMBO J. 1990;9:225–232. doi: 10.1002/j.1460-2075.1990.tb08099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreri K, Gill G, Montminy M. The cAMP-regulated transcription factor CREB interacts with a component of the TFIID complex. Proc Natl Acad Sci U S A. 1994;91:1210–1213. doi: 10.1073/pnas.91.4.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt JC, Suter DM, Roy R, Zhao ZW, Chapman AR, Basu S, Maniatis T, Xie XS. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat Methods. 2013;10:421–426. doi: 10.1038/nmeth.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace L, Blum A, Blobel G. Immunocytochemical localization of the major polypeptides of the nuclear pore complex-lamina fraction. Interphase and mitotic distribution. J Cell Biol. 1978;79:546–566. doi: 10.1083/jcb.79.2.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ME, Ziff EB, Greene LA. Stimulation of neuronal acetylcholine receptors induces rapid gene transcription. Science. 1986;234:80–83. doi: 10.1126/science.3749894. [DOI] [PubMed] [Google Scholar]
- Groeneweg FL, van Royen ME, Fenz S, Keizer VI, Geverts B, Prins J, de Kloet ER, Houtsmuller AB, Schmidt TS, Schaaf MJ. Quantitation of glucocorticoid receptor DNA-binding dynamics by single-molecule microscopy and FRAP. PLoS One. 2014;9:e90532. doi: 10.1371/journal.pone.0090532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka Y, Murakami F. In vitro analysis of the origin, migratory behavior, and maturation of cortical pyramidal cells. J Comp Neurol. 2002;454:1–14. doi: 10.1002/cne.10421. [DOI] [PubMed] [Google Scholar]
- Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács KA, Steullet P, Steinmann M, Do KQ, Magistretti PJ, Halfon O, Cardinaux JR. TORC1 is a calcium- and cAMP-sensitive coincidence detector involved in hippocampal long-term synaptic plasticity. Proc Natl Acad Sci U S A. 2007;104:4700–4705. doi: 10.1073/pnas.0607524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Li S, Zhang C, Takemori H, Zhou Y, Xiong ZQ. TORC1 regulates activity-dependent CREB-target gene transcription and dendritic growth of developing cortical neurons. J Neurosci. 2009;29:2334–2343. doi: 10.1523/jneurosci.2296-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Malyshevskaya O, Shiraishi Y, Kimura F, Yamamoto N. Role of electrical activity in horizontal axon growth in the developing cortex: a time-lapse study using optogenetic stimulation. PLoS One. 2013;8:e82954. doi: 10.1371/journal.pone.0082954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- McNally JG, Müller WG, Walker D, Wolford R, Hager GL. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- Morisaki T, Müller WG, Golob N, Mazza D, McNally JG. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat Commun. 2014;5:4456. doi: 10.1038/ncomms5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/S0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Rayasam GV, Elbi C, Walker DA, Wolford R, Fletcher TM, Edwards DP, Hager GL. Ligand-specific dynamics of the progesterone receptor in living cells and during chromatin remodeling in vitro. Mol Cell Biol. 2005;25:2406–2418. doi: 10.1128/MCB.25.6.2406-2418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler WJ, Vandenbark GR, Hanson RW. Cyclic AMP and the induction of eukaryotic gene transcription. J Biol Chem. 1988;263:9063–9066. [PubMed] [Google Scholar]
- Saha RN, Wissink EM, Bailey ER, Zhao M, Fargo DC, Hwang JY, Daigle KR, Fenn JD, Adelman K, Dudek SM. Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat Neurosci. 2011;14:848–856. doi: 10.1038/nn.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saluja D, Vassallo MF, Tanese N. Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators. Mol Cell Biol. 1998;18:5734–5743. doi: 10.1128/MCB.18.10.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp ZD, Mancini MG, Hinojos CA, Dai F, Berno V, Szafran AT, Smith KP, Lele TP, Lele TT, Ingber DE, Mancini MA. Estrogen-receptor-alpha exchange and chromatin dynamics are ligand- and domain-dependent. J Cell Sci. 2006;119:4101–4116. doi: 10.1242/jcs.03161. [DOI] [PubMed] [Google Scholar]
- Speil J, Baumgart E, Siebrasse JP, Veith R, Vinkemeier U, Kubitscheck U. Activated STAT1 transcription factors conduct distinct saltatory movements in the cell nucleus. Biophys J. 2011;101:2592–2600. doi: 10.1016/j.bpj.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugo N, Oshiro H, Takemura M, Kobayashi T, Kohno Y, Uesaka N, Song WJ, Yamamoto N. Nucleocytoplasmic translocation of HDAC9 regulates gene expression and dendritic growth in developing cortical neurons. Eur J Neurosci. 2010;31:1521–1532. doi: 10.1111/j.1460-9568.2010.07218.x. [DOI] [PubMed] [Google Scholar]
- Sugo N, Morimatsu M, Arai Y, Kousoku Y, Ohkuni A, Nomura T, Yanagida T, Yamamoto N. Single-molecule imaging reveals dynamics of CREB transcription factor bound to its target sequence. Sci Rep. 2015;5:10662. doi: 10.1038/srep10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- Uesaka N, Hirai S, Maruyama T, Ruthazer ES, Yamamoto N. Activity dependence of cortical axon branch formation: a morphological and electrophysiological study using organotypic slice cultures. J Neurosci. 2005;25:1–9. doi: 10.1523/JNEUROSCI.3855-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KM, Rehfuss RP, Chrivia JC, Lochner JE, Goodman RH. A dominant repressor of cyclic adenosine 3′,5′-monophosphate (cAMP)-regulated enhancer-binding protein activity inhibits the cAMP-mediated induction of the somatostatin promoter in vivo. Mol Endocrinol. 1992;6:647–655. doi: 10.1210/me.6.4.647. [DOI] [PubMed] [Google Scholar]
- West AE, Griffith EC, Greenberg ME. Regulation of transcription factors by neuronal activity. Nat Rev Neurosci. 2002;3:921–931. doi: 10.1038/nrn987. [DOI] [PubMed] [Google Scholar]
- Yao J, Munson KM, Webb WW, Lis JT. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 2006;442:1050–1053. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
- Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, Kadam S, Ecker JR, Emerson B, Hogenesch JB, Unterman T, Young RA, Montminy M. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U S A. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wu H, Li S, Chen Q, Cheng XW, Zheng J, Takemori H, Xiong ZQ. Requirement of TORC1 for late-phase long-term potentiation in the hippocampus. PLoS One. 2006;1:e16. doi: 10.1371/journal.pone.0000016. [DOI] [PMC free article] [PubMed] [Google Scholar]