Abstract
Mutations in amyloid β precursor protein (APP) gene alter APP processing, either causing familial Alzheimer's disease (AD) or protecting against dementia. Under normal conditions, β-site APP cleaving enzyme 1 (BACE1) cleaves APP at minor Asp1 site to generate C99 for amyloid β protein (Aβ) production, and predominantly at major Glu11 site to generate C89, resulting in truncated Aβ production. We discovered that A673V mutation, the only recessive AD-associated APP mutation, shifted the preferential β-cleavage site of BACE1 in APP from the Glu11 site to the Asp1 site both in male and female transgenic mice in vivo and in cell lines and primary neuronal culture derived from timed pregnant rats in vitro, resulting in a much higher C99 level and C99/C89 ratio. All other mutations at this site, including the protective Icelandic A673T mutation, reduced C99 generation, and decreased the C99/C89 ratio. Furthermore, A673V mutation caused stronger dimerization between mutant and wild-type APP, enhanced the lysosomal degradation of the mutant APP, and inhibited γ-secretase cleavage of the mutant C99 to generate Aβ, leading to recessively inherited AD. The results demonstrate that APP673 regulates APP processing and the BACE1 cleavage site selection is critical for amyloidogenesis in AD pathogenesis, and implicate a pharmaceutical potential for targeting the APP673 site for AD drug development.
SIGNIFICANCE STATEMENT β-site APP cleaving enzyme 1 (BACE1) is essential for amyloid β protein production. We discovered that A673V mutation shifted the BACE1 cleavage site from the Glu11 to the Asp1 site, resulting in much higher C99 level and C99/C89 ratio. All other mutations at this site of amyloid β precursor protein (APP) reduced C99 generation and decreased the C99/C89 ratio. Furthermore, A673V mutation resulted in stronger dimerization between mutant and wild-type APP, enhanced the lysosomal degradation of the mutant APP, and inhibited γ-secretase cleavage of the mutant C99 to generate amyloid β protein, leading to recessively inherited Alzheimer's disease (AD). The results demonstrate that APP673 regulates APP processing, and the BACE1 cleavage site selection is critical for amyloidogenesis in AD pathogenesis, and implicate a pharmaceutical potential for targeting the APP673 site for AD drug development.
Keywords: Alzheimer's disease, BACE1
Introduction
Deposition of amyloid β protein (Aβ) to form neuritic plaques is the unique neuropathological feature of Alzheimer's disease (AD). Aβ is derived from amyloid β precursor protein (APP). Under normal conditions, the majority of APP proteins are processed in the nonamyloidogenic pathway: cleaved first by α-secretase at Leu17 site of Aβ domain to generate the secreted sAPPα and the membrane-bound C83 (Esch et al., 1990). C83 is subsequently cleaved by γ-secretase to generate CTFγ and p3 fragment, precluding Aβ production (Song et al., 1999; Zhang et al., 2000; Qing et al., 2008). In the amyloidogenic pathway, APP is cleaved first by β-secretase at Asp1 site to generate the secreted sAPPβ and the membrane-bound fragment C99. C99 is further cleaved by γ-secretase to generate CTFγ and Aβ. β-cleavage is a rate-limiting step and the β-secretase cleavage product C99 is directly associated with the overall production of heterogeneous Aβ species (Li et al., 2006; Sun et al., 2012).
β-site APP cleaving enzyme 1 (BACE1) cleaves APP at two β-secretase sites. Under physiological conditions, BACE1 predominantly cleaves APP at the major Glu11 site within Aβ, yielding the nonamyloidogenic C89 and resulting in truncated Aβ production; whereas it cleaves APP at the minor Asp1 site, generating C99 and leading to Aβ generation (Deng et al., 2013). Enhancing BACE1 cleavage of APP or shifting its preferential cleavage site from Glu11 to Asp1 resulted in higher C99 and Aβ production and hence facilitated neuritic plaque formation under pathological conditions (Mullan et al., 1992; Sun et al., 2006a, b; Deng et al., 2013; Ly et al., 2013; Zhang et al., 2014).
Whereas most AD cases are sporadic with late onset at the age of 65 years and older, <1% are early-onset familial AD (FAD) cases. The FAD cases are caused by mutations in APP, presenilin 1 (PSEN1), or presenilin 2 (PSEN2) genes. More than 30 FAD-associated APP point mutations have been discovered on 17 residues, which are in close vicinity of secretase cleavage sites. These mutations have been shown to cause FAD by altering APP processing, including enhancing C99 production (Swedish mutation KM670/671NL) (Mullan et al., 1992; Felsenstein et al., 1994; Perez et al., 1996), inhibiting nonamyloidogenic α-cleavage of APP (Arctic mutation E693G) (Nilsberth et al., 2001; Stenh et al., 2002; Sahlin et al., 2007), increasing Aβ42 level or the ratio of Aβ42 to Aβ40 (London mutation V717I and Florida mutation I716V) (Goate et al., 1991; Eckman et al., 1997; De Jonghe et al., 2001), or accelerating Aβ40 fibril formation (Dutch mutation E693Q and Iowa mutation D694N) (Wisniewski et al., 1991; Van Nostrand et al., 2001). Our laboratory further demonstrated that APP Swedish mutation strongly shifted the BACE1 cleavage site from the Glu11 site to the Asp1, resulting in a higher C99/C89 ratio and promoting Aβ generation (Deng et al., 2013).
All the known pathogenic mutations in the APP gene are dominantly inherited, except an A673V mutation (APPrIta) identified in an Italian family (Di Fede et al., 2009). This alanine to valine missense mutation occurs at the 673rd amino acid position of APP770 (i.e., the second amino acid of Aβ) and acts in a recessively inherited manner in AD pathogenesis. APPrIta mutation increased Aβ production, and the resulting mutant AβA2V displayed altered oligomerization, aggregation, and toxicity (Di Fede et al., 2009; Benilova et al., 2014; Maloney et al., 2014). However, the mechanism underlying the recessive inheritance of this mutant remains elusive. The unique recessiveness of this APP mutation might provide insights into how altered APP processing and possible quantitative change of Aβ accumulation lead to Alzheimer's development. Intriguingly, a recently identified mutation at the same site changing alanine into threonine (APPIce) was shown to be protective against AD in the Icelandic population (Peacock et al., 1993; Jonsson et al., 2012), highlighting the close involvement of altered APP processing in AD pathogenesis. In this study, we demonstrated that the alanine at the 673rd amino acid position of APP770 was critical for regulating β-secretase cleavage site selection and the BACE1-mediated amyloidogenesis in AD pathogenesis.
Materials and Methods
Plasmid constructs.
Mutations of APP were introduced into the cDNA coding sequence of APP695 by PCR-based site-directed mutagenesis, and the mutated cDNAs were cloned into expression vector pcDNA4-mycHis (Invitrogen). Human APPWt, APPrIta, or APPSwe cDNAs were also cloned into an inducible mammalian expression vector pIND (SP1)/Hygro to generate pIND-APPWt,, pIND-APPrIta, and pIND-APPSwe. pIND (SP1)/Hygro vector contains modified ecdysone response elements and SP1 enhancers. Plasmid pVgRXR expresses the heterodimeric ecdysone receptor (VgEcR) and the retinoid X receptor (RXR) (Invitrogen). Endoplasmic reticulum (ER) retention signal was introduced into pcDNA4-APPWt-ER and pcDNA4-APPrIta-ER by mutating QMQN in the C-terminal tail of APP to KKQN. To generate plasmid pFlag-APP, the coding sequence of the FLAG tag was inserted between the signal peptide and the luminal/extracellular domain of APP695, and the resulting DNA was cloned into pcDNA4-myc-His. For pulse-chase assay, APPWt and APPrIta were cloned into pSNAP (New England Biolabs). cDNAs coding for C99, C89, and C83 with a methionine in the N termini were cloned into pcDNA4, confirmed by DNA sequencing and expressed in HEK293 cells as CTF markers. CTFs derived from APP processing can be resolved by the comigration with these markers.
Immunoblot analysis.
Cells were lysed in RIPA-DOC lysis buffer (1% Triton X-100, 1% sodium deoxycholate, 0.4% SDS, 0.15 m NaCl, 0.05 m Tris-HCl, pH 7.2) supplemented with complete mini protease inhibitor mixture tablet (Roche Diagnostics). The samples were resolved by SDS-PAGE on 8% Tris-glycine or 16% Tris-tricine gels, then transferred to Immobilon-FL PVDF membranes (Millipore). C20, a rabbit antibody against the last 20 amino acids of APP C terminus, was used to detect APP and its CTF products (Ly et al., 2013). β-actin was detected using monoclonal antibody AC-15 (Sigma). The membranes were incubated with IRDye 800CW-labeled goat anti-mouse or anti-rabbit antibodies in PBS with 0.1% Tween 20 at 22°C for 1 h, and visualized on the Odyssey system (LI-COR Biosciences). All quantifications were performed using the LI-COR Odyssey system or ImageJ. To perform coimmunoprecipitation, plasmids coding for FLAG-APP and C99Wt or C99A2V were cotransfected into HEK293 cells, and the detailed procedure was performed as described previously (Eggert et al., 2009).
Intracranial injection and immunoprecipitation.
All the protocols of animal procedure were approved by University of British Columbia Animal Care Center. Adeno-associated virus-9 viruses expressing human APPWt or human APPrIta at the titer of ∼2 × 1012 genome copies/ml were generated by Vector Core, University of Pennsylvania. The brains of 3 neonatal male and 3 female mice within 12 h were intracranially injected with 1 μl of the virus into the ventricle. Two weeks after injection, pups were killed. Half brains were homogenized in lysis buffer (10 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA) supplemented with complete protease inhibitors (Roche). After centrifugation, the supernatant was added to the C20 antibody immobilized on protein-G agarose bead, and incubated at 4°C for overnight. Proteins were eluted from the beads by boiling in SDS sample buffer.
Cell culture and transfection.
HEK293 cells (RRID:CVCL_0045) were cultured in DMEM containing 10% FBS, 1 mm sodium pyruvate, 2 mm l-glutamine, 50 U/ml penicillin G sodium, and 50 μg/ml streptomycin sulfate (Invitrogen). PC12 cells (RRID: CVCL_0481) were cultured in DMEM supplemented with 15% FBS. Stable cell lines were maintained in media containing zeocin (50 μg/ml). All cells were maintained at 37°C in an incubator containing 5% CO2. For transfection, cells were grown to 70% confluence in a 35 mm plate and transfected with 2 μg plasmid DNA with 4uL of Lipofectamine 2000 Reagent (Invitrogen). To establish APP-inducible cell lines, BACE1-stable cell line 293B2 cells were cotransfected with pVgRXR and pIND-APPWt,, pIND-APPrIta, or pIND-APPSwe. Activation of the exogenous APP gene transcription in the transfected cells was dependent on the binding of a heterodimer of VgEcR and RXR receptors in the presence of a ligand, such as the ecdysone analog ponasterone A (Invitrogen). The expression level of APP variants was controlled by the different dosages of ponasterone A treatment (Li et al., 2006).
Primary neurons and virus transduction.
E18 cortical neurons derived from pregnant rats were maintained in neurobasal medium supplemented with B27 in poly-D-lysine-coated plates. For each 35 mm dish, 4 μl of viruses expressing either human APPWt or human APPrIta at the titer of ∼2 × 1012 genome copies/ml was added to the culture on DIV 3. Half of the medium was replaced with fresh medium every 3 d. Chloroquine (CHL, from Sigma) (100 μg/ml) or solvent control (H2O) was added to the neurons on DIV 9, and cells were harvested for immunoblot assay after overnight CHL treatment.
Pharmacological treatments of cells.
L-685,458 (Sigma), a potent γ-secretase inhibitor, was dissolved in DMSO and applied to cell culture medium at 1 μm final concentration for 3 h. Cycloheximide (Sigma), an inhibitor for protein translation, was used at 100 μg/ml for the indicated times to inhibit APP synthesis. CHL (Sigma) is a lysosomal degradation inhibitor. MG132 (from Millipore) is a potent, reversible, and cell-permeable proteasome inhibitor. To determine the effect of APP mutations on APP degradation, HEK293 were treated with CHL at 100 μm or MG132 at 10 μm.
Aβ ELISA.
Conditioned medium from cell culture was collected, and protease inhibitors and 4-(2-aminoethyl)-benzenesulfonyl fluoride (Roche) were added to prevent Aβ degradation. The concentration of Aβ42 was detected using β-amyloid 1–42 colorimetric ELISA kit (Invitrogen) according to the manufacturer's instructions. For enrichment of Aβ immobilized on the ELISA plate, 300 μl of the total ∼1 ml conditioned medium was added to the plate. After incubation at room temperature for 3 h, the medium was replaced by another 300 μl of the same conditioned medium to repeat the 3 h reaction. After all the medium was applied to the plate, secondary antibody was added. Conditioned media from PC12 cells without any overexpression were used as control to avoid Aβ contamination.
Statistical analysis.
The numbers of replication for each experiment were at least three times as indicated in the figure legends. All results are expressed as mean ± SEM. Two-tailed Student's t test was used to analyze the difference between two groups; and multiple-comparison test was analyzed by ANOVA followed by post hoc Newman–Keuls test, unless otherwise specified in figure legends.
Results
APPrIta mutation shifted BACE1 cleavage site and increased Aβ production
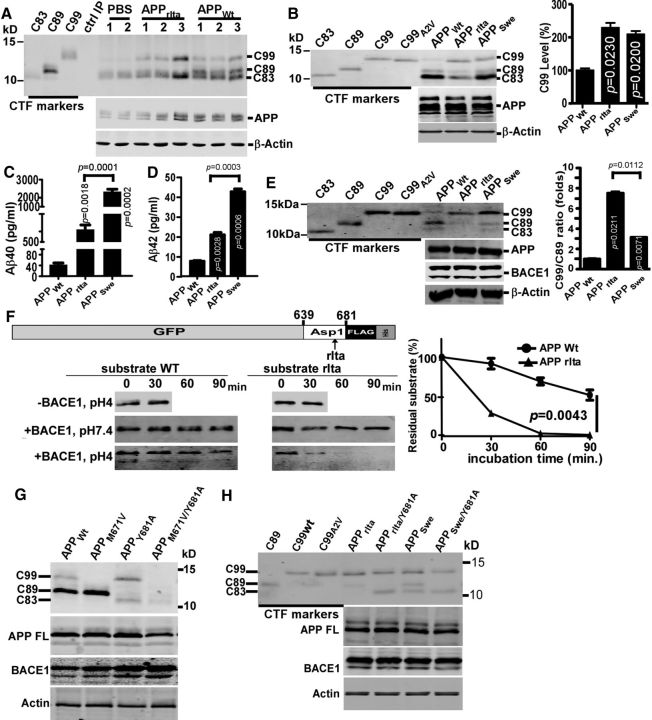
To investigate the effect of the Italian APP A673V mutation (hereafter referred as APPrIta) on APP processing, adeno-associated viruses carrying human APPWt and APPrIta were injected into neonatal mouse brains. After immunoprecipitation, APPrIta displayed clear C99 overexpression but not C89, and APPWt apparently produced more C89 than C99 (Fig. 1A), suggesting that A673V mutation shifted the major BACE1 cleaving site in APP from the Glu11 site to Asp1 site. A similar result was also reported previously (Kimura et al., 2016).
Figure 1.
Effects of APP A673V (rIta) mutation on APP processing and Aβ generation. A, APPWt and APPrIta adeno-associated viruses were transduced into the brains of 3 neonatal mice, and CTFs were analyzed after immunoprecipitation. PBS was injected into 2 mice as control. In the “control IP,” only lysis buffer, but not brain lysate, was added to the C20 antibody immobilized on protein-G beads. cDNAs coding for C99, C89, and C83 with a methionine in the N termini were cloned into pcDNA4, confirmed by DNA sequencing, and expressed in HEK293 cells as CTF markers. B, APP695WT, APP695rIta, and APP695Swe plasmids were transfected into PC12 cells. APP and its CTFs were detected by C20 antibody. C99 levels in APP variants were normalized to that in APPWT. C, D, Aβ40 and Aβ42 in the conditioned media of PC12 cells expressing APPWT, APPrIta and APPSwe were quantified by ELISA and expressed as pg/ml. E, 293B2 were transfected with APP695Wt, APP695rIta, or APP695Swe plasmids, and CTF markers and cell lysates were blotted for APP, BACE1, and CTFs. C99/C89 ratios in APP mutants were normalized to that of APPWt. F, In vitro BACE1 cleavage kinetics. APP fragments spanning from 639 to 681 (β-site between M671–D672) with or without the rIta mutation at A673 were inserted between GFP and FLAG-His tags and expressed in bacteria. The recombinant proteins were purified with His tag beads. The 0.35 μg of recombinant proteins in 35 μl buffered solutions were incubated at 37°C with or without 2.1 μl recombinant BACE1 (R&D Systems, 500 μg/ml) at different pH values for the indicated times. The Western blot bands by anti-FLAG antibody were quantified and plotted. G, APPWt and APPWt carrying M671V (to inhibit Asp-1-cleavage) and/or Y681A (to inhibit Glu-11-cleavage) mutations were expressed in 293B2 cell, and the whole-cell lysates were blotted for CTFs, full-length APP, and BACE1. H, APPrIta with or without the Y681A mutation and APPSwe with or without the Y681 mutation were expressed in 293B2 cells, and whole-cell lysates were blotted for CTFs, APP, and BACE1. Data are mean ± SEM; n = 3 technical replicates. Statistical analysis: one-way ANOVA with post hoc Newman–Keuls tests.
To confirm the in vivo results in vitro and to further investigate the mechanism, we transfected APPWt, APPrIta, or APPSwe into PC12 cells, a rat pheochromocytoma cell line expressing readily detectable endogenous BACE1. APP and its CTFs, including the α-secretase product C83 and BACE1 products C89 and C99, were examined by Western blot with C20 antibody. APPrIta and APPSwe significantly increased C99 levels to 228.8 ± 6.9% and 209.1 ± 9.8%, respectively, compared with APPWt (p < 0.001) (Fig. 1B). Additionally, APPrIta also exhibited much weaker C83, suggesting the reduction of α-cleavage of APP (Fig. 1B). To examine the effect of A673V mutation on Aβ production, APP variants were overexpressed in PC12 cells, and conditioned media were analyzed using Aβ ELISA assay. APPrIta and APPSwe increased the Aβ40 levels to 12.8 ± 0.2-fold and 56.4 ± 0.5-fold, respectively (p < 0.05) (Fig. 1C), and enhanced Aβ42 levels to 2.96 ± 0.09-fold and 6.73 ± 0.10-fold, respectively (p < 0.05) (Fig. 1D). The data suggested that the A673V mutation enhanced APP processing at the Asp1 β-site cleavage and promoted subsequent Aβ generation compared with APPWt.
Under physiological conditions, the majority of APP undergoes α-secretase pathway, with C83 (CTFα) being the predominant form of APP CTFs. Increasing BACE1 expression causes the majority of APP undergoing β-secretase pathway, resulting in CTFβ (C89 and C99) being predominant forms of APP CTFs (Deng et al., 2013). C89 and C99 are generated by BACE1 cleavage of APP at two β-secretase sites, Glu11(also known β′-site) and Asp1 (also known as β-site), respectively, and the Glu11 is the major β-secretase site to processing wild-type APP under normal conditions (Deng et al., 2013). To specifically assess BACE1 cleavage of APPrIta, APP plasmids were transfected into a BACE1-stable cell line 293B2. Consistent with our previous report, the major CTFs in APPWt-expressing cells was C89 derived by BACE1 cleavage at Glu11 site (Fig. 1E). The Swedish mutation APPSwe (APP K670N/M671L) altered BACE1-cleavage site selection, resulting in the Asp1 site as the preferential β-secretase cleavage site and C99 as the major product (Fig. 1E). Similar to the Swedish mutation, the A673V mutation in APPrIta also shifted β-secretase cleavage site from the normally predominant Glu11 site to Asp1 site, leading to C99 as the predominant β-secretase product (Fig. 1E). More strikingly, the A673V mutation was more potent than the Swedish mutation on shifting preferential β-secretase site from the Glu11 to Asp1, resulting in even higher C99/C89 ratio, 7.51 ± 0.17-fold for APP695rIta and 3.15 ± 0.06-fold for APP695Swe (p < 0.05) (Fig. 1E). These data unequivocally demonstrate that the A673Vmutation shifts the preferential β-secretase site from the Glu11 to Asp1, and the effect on the β-secretase site selection is even stronger than the Swedish mutation.
To confirm the enhanced C99 production from APPrIta by BACE1 was due to the higher cleavage efficiency at this site, but not the weak C89 and C83 production which may spare more full-length APPrIta for BACE1 to cleave at the Asp1 site, in vitro kinetics assay was performed by recombinant BACE1 and an artificial substrate spanning from residue 639–681 (with Asp1 site between 671 and 672) (Fig. 1F). As expected, the recombinant BACE1 did not cleave either the wild-type or the rIta substrate at neutral pH. At pH 4, which was an optimal pH for recombinant BACE1 activity, the rIta substrate showed much faster reduction than the wild-type substrate, suggesting that the rIta mutant was a better substrate of BACE1. Therefore, this mutation in full-length APP may directly enhance BACE1 cleavage at the Asp1 site. However, it should be noted that we did not detect the cleaved products from these artificial substrates by using anti-GFP antibody. This could be because the cleaved fragments were not stable in vitro at lower pH, or the fragments were further degraded by the recombinant BACE1 when they were mixed with BACE1 in the solution. The latter circumstance, if true, is unlikely to happen in cells because intracellular BACE1 is anchored at the membrane, and the N-terminal fragments of APP are free in the lumen of organelles and quickly secreted out of the cells, which is the reason why secreted APPβ is used as an indicator of β-cleavage.
As BACE1 cleaves full-length APP at Glu11 site within C99 region, it may also cleave C99 at the same site, which could alter the C99/C89 ratio. To examine whether such effects contribute to C99/C89 ratio in our cell-based system, several APP mutants were generated to abolish C99 (M671V) (Vetrivel et al., 2011) and/or C89 (Y681A) from APPWT, APPSwe, and APPrIta. In APPWT (Fig. 1G), M671V did not reduce C89, and Y681A only slightly increased C99, which may be derived from full-length APP spared from Glu11 cleavage. Another APP mutation (E682V) inhibiting Glu11 cleavage displayed a similar effect on CTF pattern (Vetrivel et al., 2011). For APPrIta and APPSwe (Fig. 1H), the Y681A mutation did not cause significant increase of C99, presumably because APPrIta and APPSwe (producing wild-type C99) do not produce much of C89 by BACE1, and there was not much of full-length APP being spared from Glu11 cleavage. Together, sequential cleavage of APP and C99 by BACE1 might happen, but it does not essentially alter the C99/C89 ratio. Therefore, the C99/C89 ratio indicates the portions of an APP variant being cleaved at Asp1 site and at Glu11 site. The cleavage preference may reflect the energy barrier for BACE1 cleavage. The lower barrier, the faster cleavage, and the more of CTF production; and inhibiting one of the two sites (Asp1 and Glu11) does not essentially alter the cleavage kinetics at the other one.
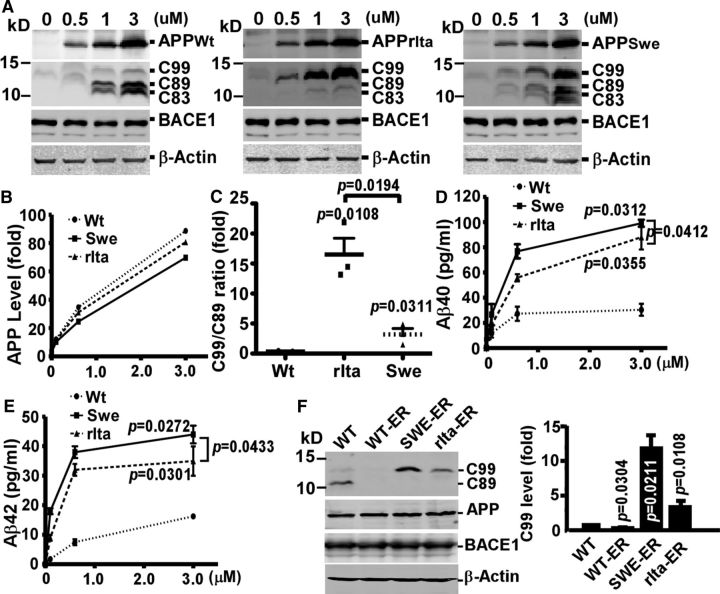
To determine whether APPrIta expression levels affect β-cleavage site selection and Aβ generation, an inducible APP expression system (Li et al., 2006) was introduced to 293B2 cells to control APP expression at different levels. While the expression levels of APP variants are similar (p > 0.05), APPrIta consistently displayed the highest C99/C89 ratio (16.7 ± 2.8-fold) compared with 3.2 ± 0.2-fold of APPSwe relative to APPWT (Fig. 2A–C). These data indicated that the effect of A673V mutation on β-cleavage shift from Glu11 to Asp1 was intrinsic and independent of the expression level of APP. Meanwhile, although APP levels followed a linear pattern upon the induction of expression, Aβ40 and Aβ42 levels reached plateau for all the APP variants (Fig. 2D,E). Consistent with the result in PC12 cells, Aβ40 and Aβ42 levels in APPrIta were significantly lower than APPSwe, but higher than APPWt, whether Aβ levels reached plateau or not (p < 0.05). The data suggested the intrinsic nature of A673V mutation on Aβ generation.
Figure 2.
The A673V mutation altered APP processing independently of expression level and enabled BACE1 to cleave nascent APP. To determine whether APPrIta expression levels affect β-cleavage site selection and Aβ generation, an inducible APP expression system (Li et al., 2006) was introduced to 293B2 cells to control APP expression at different levels. A, pIND-APPWt, pIND-APPrIta, and pIND-APPSwe were transfected into 293B2 cells. After ponasterone A treatment at different concentrations, cell lysates were blotted for full-length APP, CTFs, and BACE1. Full-length APPs (B), CTFs (C), Aβ40 (D), and Aβ42 (E) were quantified. F, APPWt, APPSwe, and APPrIta bearing the KKQN ER retention signal were expressed in 293B2 cells. C99s were quantified. Data are mean ± SEM; n = 3 technical replicates. Statistical analysis: one-way ANOVA with post hoc Newman–Keuls tests.
To investigate the mechanism underlying the altered β-cleavage site selection by the A673V mutation, we generated APP-ER retention plasmids by mutating APP C-terminal sequence QMQN to KKQN in APPWt, APPrIta, and APPSwe plasmids. The KKXX-COOH motif (where X refers to any amino acids) retains nascent APP in the ER in the default structure, excluding the influence from possible intracellular trafficking and further modification of these APP variants upon exit from the ER. When expressed in 293B2 cells, both APPSwe-ER and APPrIta-ER generated a remarkable amount of C99, with 11.8 ± 2.7-fold for APPSwe-ER and 4.0 ± 0.9-fold for APPrIta-ER compared with APPWt, whereas APPWt-ER produced a trace amount, if any, of C99. C89 and C83 were hardly detected. (Fig. 2F). These results suggest that the nascent APPSwe and APPrIta are structurally more susceptible to β-cleavage at Asp1 site. Although it remains unclear at this stage whether later trafficking and modifications alter this susceptibility, the higher C99 generation from the nascent mutants in the ER may partly contribute to the higher total C99 and Aβ production of APPrIta and APPSwe.
Wild-type APP suppressed amyloidogenic processing of APPrIta
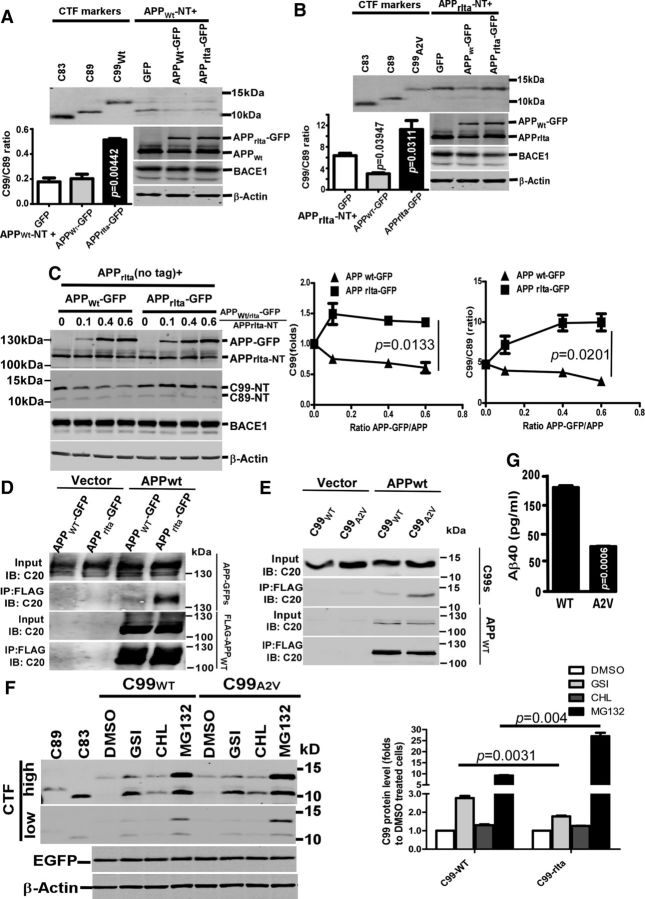
To examine the β-cleavage under the APPWt/rIta heterozygous state, we coexpressed nontagged APPWt or APPrIta (APPWt-NT and APPrIta-NT) with APPWt-GFP or APPrIta-GFP in 293B2 cells. This way, CTF-GFP from APP-GFP variants were easily separated with the CTF-NT from APP-NT variants, and the effects of one of the two variants on another could be unambiguously demonstrated. Surprisingly, APPrIta-GFP significantly increased C99/C89 ratios of APPWt-NT and APPrIta-NT. By contrast, APPWt-GFP overexpression only reduced C99/C89 ratio of APPrIta-NT but not that of APPWt-NT (Fig. 3A,B). APPWt-GFP also proportionally reduced C99 and C89 from APPWt-NT, which could be due to competition for BACE1 cleavage. A dosage course further demonstrated that APPWt-GFP gradually downregulated C99 and decreased C99 to C89 ratio of APPrIta-NT. In contrast, APPrIta-GFP increased both C99 and C99/C89 ratio of APPrIta-NT Figure 3C).
Figure 3.
Enhanced dimerization suppressed C99 production from APPrIta and γ-cleavage of C99rIta. Nontagged APPWt (APPWt-NT) (A) or APPrIta (APPrIta-NT) (B) were coexpressed with GFP, or APPWt-GFP, or APPrIta-GFP in 293B2 cells, and the cell lysates were blotted for CTFs-NT, APPs, and BACE1. The ratios of C99-NT/C89-NT were plotted. C, APPrIta-NT (nontagged APPrIta) plasmid was cotransfected into 293B2 cells with increasing amount of either APPWt-GFP or APPrIta-GFP plasmid. C20 antibody was used to detect APPs and CTFs derived from APrIta-NT, and 9E10 for BACE1. Protein bands for full-length APPWt/rIta-GFP and full-length APPrIta-NT were quantified, and the ratios of APPWt/rIta-GFP/APPrIta-NT were indicated. The ratios of C99rIta in APP-GFP expressing cells to C99rIta in cells without APP-GFP expression were plotted. The ratios of C99rIta/C89 over the APPWt/rIta-GFP dosage courses were plotted as well. D, Flag-APPWt plasmid and empty vector were transfected with APPWt-GFP or APPrIta-GFP. The immunoprecipitated Flag-APPWt by the FLAG-beads and the coprecipitated APP-GFPs were both detected with C20 antibody. E, Flag-APPWt and empty vector were transfected with C99Wt or C99A2V. The immunoprecipitated FLAG-APPWt by the FLAG-beads and the coprecipitated C99s were detected with C20 antibody. F, pcDNA4–C99WT or -C99A2V with a signal peptide were cotransfected with pEGFP plasmid (as an internal control for transfection) into HEK293 and then treated with indicated catabolism inhibitors overnight. DMSO, control solvent; GSI, γ-secretase inhibitor L-685,458; MG132, proteasome inhibitor. GFP was used for transfection efficiency control, and β-actin was used as for protein loading control. C99 level was quantified and compared between C99WT and C99A2V. G, Conditioned medium was collected to determine the amount of Aβ40 in HEK293 with C99WT (WT) and C99A2V (A673V) overexpression. Data are mean ± SEM; n = 3 technical replicates. Statistical analysis: one-way ANOVA with post hoc Newman–Keuls tests.
The enhanced coimmunoprecipitation between APPWt and APPrIta (Fig. 3D) suggested that APPWt was more likely to form oligomers with APPrIta than APPWt itself, and the APPWt/APPrIta hetero-oligomerization may account for the altered BACE1 cleavage preference in APPrIta (Kaden et al., 2008). Moreover, C99rIta also displayed stronger coimmunoprecipitation with APPWt than C99Wt (Fig. 3E), implicating that A2V mutation in C99 promoted the oligomerization through the Aβ region, but not allosterically through the E1 or E2 dimerization domains of APP.
Given that the APP dimers and C99 dimers inhibit γ-cleavage and Aβ production (Eggert et al., 2009) and the production of Aβ was lower in APPrIta than APPSwe, despite similar C99 levels between the two mutants (Fig. 3B--E), we examined whether the γ-secretase cleavage of C99 was affected by this mutation. C99Wt (also produced from APPSwe) and C99A2V were expressed in HEK293 cells, and the cells were treated with each of γ-secretase inhibitor L-685,458, lysosome inhibitor CHL, and proteasome inhibitor MG132 for 3 h (Fig. 3F). To equalize C99Wt and C99rIta levels without inhibitor treatment, we transfected ∼10 times more of C99rIta expression vector, which suggests that the catabolism of C99rIta was much stronger than that of C99Wt. Regarding γ-secretase inhibition, C99Wt level was increased by 183.6 ± 11.3%, but C99rIta was only increased by 53 ± 4.3%. By contrast, MG132 hugely increased C99rIta by ∼25-fold and increased C99Wt by a much milder ∼8-fold. Hence, compared with C99Wt, C99rIta was more efficiently degraded by proteasome, and only a relatively small portion was cleaved by γ-secretase (Fig. 2D). Consistently, when the same amount of basal C99 proteins was expressed in HEK293 cells, Aβ40 secretion from C99rIta-expressing cells was only 36.0 ± 3.5% of that from C99Wt-expressing cells (p < 0.05; Fig. 3G). Both changes of C99 level upon γ-secretase inhibition and Aβ production revealed a weaker γ-cleavage of C99rIta compared with C99Wt. The weaker γ-cleavage of C99A2V, which is at least partially due to potent proteasome degradation, is presumably the most important mechanism underlying the disproportional Aβ and C99 productions from APPrIta, compared with APPSwe.
APPrIta mutation directed mature APP to its lysosome-dependent degradation
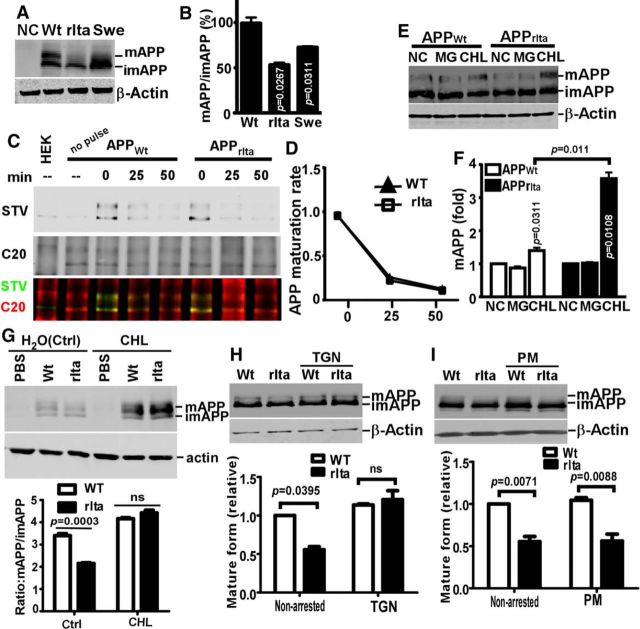
The maturation of APP is required for APP processing and Aβ generation (Perez et al., 1999; Steinhilb et al., 2001). We found that APPrIta proteins displayed significantly less mature form (53 ± 5%) than APPWt (p < 0.05) (Fig. 4A,B). To examine whether the A673V mutation affected APP maturation, pulse chase assay was performed for overexpressed APPWt and APPrIta in HEK293 cells (Fig. 4C,D). The reduction of APPrIta did not show apparent difference from that of APPWt, suggesting that the A673V mutation did not affect the maturation or the ER exportation of the nascent immature APP.
Figure 4.
Lysosome-dependent degradation of mature APPA673V. A, Plasmid pcDNA4-APPWt, pcDNA4-APPrIta, or pcDNA4-APPSwe were transfected into HEK293 cells. The nascent APP (imAPP) and modified APP (mAPP) in cell lysate were resolved on 8% glycine SDS-PAGE gel and detected by C20 antibody. B, The ratios of mAPP/imAPP in A were quantified and normalized to that in APPWt. C, D, APPWt-SNAP and APPrIta-SNAP were overexpressed in HEK293 cells and pulse-labeled. After indicated chasing time, cells lysate was blotted for pulse-labeled APP with streptavidin (green) and total APP with C20 (red). The maturation rate (as indicated by the reduction of pulse-labeled immature APP) of APPWt and APPrIta were plotted after normalization with total APP. E, pcDNA4-APPWt and pcDNA4-APPrIta were introduced into HEK293 cells and then treated with10 μm MG132 or 100 μm CHL for 45 min. F, Mature APP protein levels were quantified. Data are mean ± SEM; n = 3 technical repeats. Statistical analysis: ANOVA with post hoc Newman–Keuls test. G, Viruses expressing human APPWt or APPrIta were tranduced into primary rat neurons on DIV 3, and neuronal cells with overnight treatment of CHL orsolvent H2O (Ctrl) were harvested for blotting of full-length APP on DIV 10. Ratios of mature APP to immature APP were plotted. H, The SDYQRL trans-Golgi targeting motif (Wong and Hong, 1993) was fused to APPWt or APPrIta, and the fusion proteins were expressed in HEK293 cells, full-length APPs were blotted with C20 antibody, and the levels of mature APP variants after normalization with immature APP variants were plotted. I, The NPTY internalization signal (Perez et al., 1999) was deleted from APPWt and APPrIta to retain them on the plasma membrane (PM). The resulting APP variants with a FLAG tag fused to the N termini after signal peptide removal were overexpressed in HEK293 cells. Full-length APPs were blotted with anti-FLAG antibody as the NPTY motif was within the epitope of C20 antibody. Levels of mature APP variants after normalization with immature APP variants were plotted. G–I, Data are mean ± SEM; n = 2 technical repeats. Statistical analysis: two-way ANOVA with Newman–Keuls tests.
Next, we examined whether the weak mature form of APPrIta was due to faster degradation. HEK293 cells were transfected with APPWt or APPrIta and then treated with proteasome inhibitor MG132 or lysosome inhibitor CHL (Fig. 4E). The mature forms of both APPWt and APPrIta were accumulated upon acute lysosomal inhibition by CHL but not the proteasomal inhibition by MG132 (Fig. 4E,F). Moreover, the accumulation of mature APPrIta was stronger than that of mature APPWt (1.31 ± 0.09-fold for APPWt vs 3.38 ± 0.28-fold for APPrIta) (Fig. 3F), indicating that mature APPrIta underwent faster lysosome-dependent degradation. The weaker mature APPrIta and the CHL-induced rescue of mature APPrIta were also observed in primary rat neurons transduced with viruses expressing APPWt or APPrIta (Fig. 4G). While APPrIta arrested in trans-Golgi network (APPrIta-TGN) displayed similar amounts of mature form with APPWt-TGN, the mature form of APPrIta arrested at the plasma membrane (APPrIta-PM) appeared to be much weaker compared with APPWt-PM (Fig. 4H,I). It is very likely that a significant amount of mature APPrIta is directly transported to the lysosome from the trans-Golgi network for degradation instead of being targeted to the plasma membrane.
Residue 673 of APP was critical for BACE1 cleavage of APP
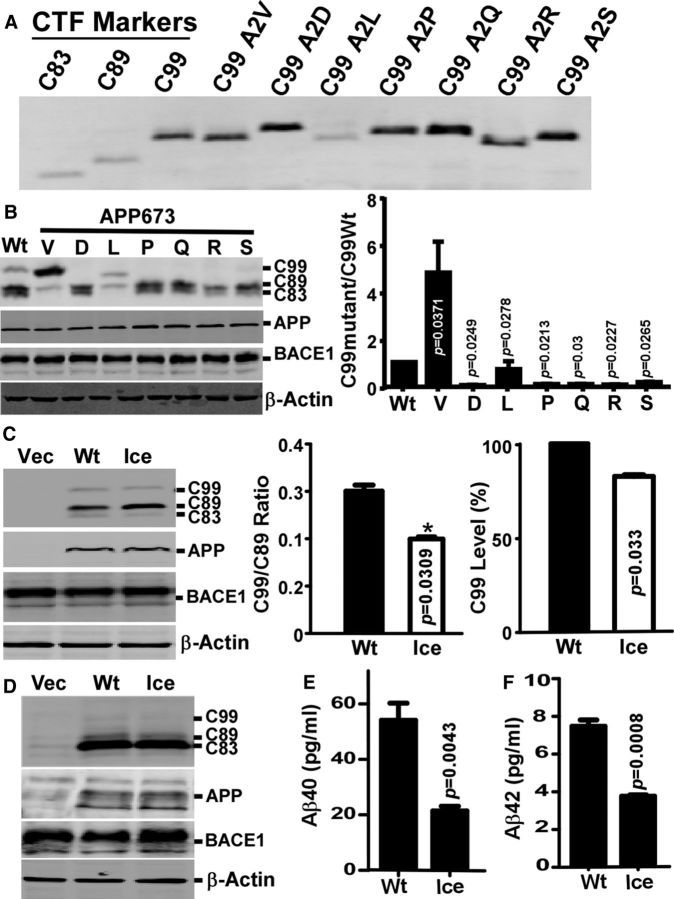
Our data had shown that the recessive Italian mutation A673V altered the preferential BACE1 cleavage site, resulting in higher C99/C89 ratio and promoting C99 production. Meanwhile, a newly identified missense mutation of alanine to threonine on the same residue (A673T) in the Icelandic population, in contrast, was protective against AD development (Jonsson et al., 2012), suggesting that the amino acid at position 673 of APP might be critical for BACE1 cleavage site selection for APP processing. To examine this issue, we generated additional APP expression plasmids carrying different missense mutations at 673rd amino acid position of APP770, replacing the alanine at 673 site with negatively charged aspartic acid (D), positively charged arginine (R), hydrophobic leucine (L), hydrophobic and structurally rigid proline (P), hydrophilic glutamine (Q), or serine (S). It was worth mentioning that hydrophilic glutamine (Q) and serine (S) were selected for their structural similarity to hydrophobic alanine. Some of the amino acid substitutions altered migration rate of C99 markers by SDS-PAGE, implying an SDS-resistant structural change (Fig. 5A). Substitution of the alanine at 673 with D, P, Q, R, or S almost abolished C99 generation (Fig. 5B). In APPA673L, BACE1 produced weak but clearly detectable C99A2L (A2 in C99 was A673 in APP770) (Fig. 5B) that comigrated with C99A2L marker (data not shown). Meanwhile, C89 (without the mutation site) from the APPA673L mutant was hugely decreased compared with wild-type and other artificial mutants, resulting in high C99A2L/C89 ratio (Fig. 5B), which was similar to A673V. L is the only residue structurally similar to V among the six substituting amino acids. Presumably, short alkane side chains at the position 673 of APP suppress BACE1 cleavage of APP at the Glu11 site. Therefore, with the exception of the pathogenic A673V mutation that enhances C99 production, all other artificial mutations inhibited, instead of enhanced, BACE1 cleavage at Asp1 site and suppressed C99 production.
Figure 5.
Effects of amino acid substitution of A673 on β-cleavage. A, C99 with the indicated mutations at position 2 (position 673 in full-length APP) displayed altered gel mobility by SDS-PAGE. B, A673 in APP were substituted with V, D, L, P-R, or S, and mutants were expressed in 293B2 cells. C99 levels were quantified. C, APPWt and APPIce were expressed in 293B2 cells, and CTFs were quantified. D, APPWt and APPIce were expressed in PC12 cells, and C99s and C89s were quantified. Secreted Aβ40 (E) and Aβ42 (F) were measured by ELISA. Data are mean ± SEM; n = 3 technical repeats. Statistical analysis: one-way ANOVA with post hoc Newman–Keuls tests.
Next, we examined the β-cleavage of the Icelandic mutant A673T in 293B2 cells (Fig. 5C) and PC12 cells (Fig. 5D). Interestingly, this mutation only mildly reduced C99 and C99/C89 ratio in both cell lines and suppressed Aβ40 and Aβ42 secretion from PC12 cells by ∼60% and ∼40%, respectively (Fig. 5D–F). Thus, APP amino acid at position 673 is critical for BACE1 cleavage and a single amino acid change at residue 673 can change BACE1 cleavage site preference to Asp1 or Glu11, resulting in either increasing or decreasing C99 and Aβ generation.
Discussion
The A673V mutation in APP gene is the only known recessive mutation causing AD. Both parental linkages of the proband were heterozygous carriers and the heterozygous family members did not show any AD symptoms by the age of 88. By contrast, the homozygous proband developed dementia symptoms by the age of 36 and died at 46, and his homozygous sister also showed mild cognitive impairment (Giaccone et al., 2010). All other FAD-associated mutations in APP are dominant, and the majority result in increasing Aβ level. However, how the mutations affect APP processing leading to AD pathogenesis remains elusive. A recently identified APP mutation (A673T) was shown to be inversely correlated to AD incidence, which implicated a protective role of this mutation. Thus, different mutations on the same site could have opposite effects on AD occurrence due to its differential effect on APP processing. Under physiological conditions, the wild-type APP protein is preferentially cleaved by BACE1 at the Glu11β-secretase site to generate C89 and truncated Aβ. In this report, we clearly showed that the recessive Italian A673V mutation altered the β-secretase cleavage site selection, resulting in Asp1 site as the preferential site for the mutant APPrIta protein. By facilitating BACE1 cleavage at Asp1 site, this A673V mutation produced more Aβ substrate C99. The effect on BACE1 cleavage site shifting by A673V mutation is even stronger than that by the Swedish mutation, resulting in much higher C99/C89 ratio. On the other hand, BACE1 cleavage at the Asp1 site was compromised by the protective Icelandic A673T mutation. Therefore, the difference in β-cleavage is in perfect agreement with the opposite effects of these two mutants on AD pathogenesis. It should be noted that the ratios of C99/C89 presented do not necessarily reflect the real relative amount of C99 to C89 generated by BACE1 because the catabolism of CTFs was not blocked. CTFs could be degraded or further processed by proteasome, lysosome, γ-secretase, α-secretases, etc. If these catabolism pathways have a preference to one of C99 and C89 over the other, the ratio of C99/C89 can no longer indicate the BACE1 cleavage preference. Nonetheless, our conclusion that the A673V mutation enhances C99 and suppresses C89 productions is based on the comparison with the C99 and C89 from APPWt, and is therefore not affected by the preferential catabolism of different CTFs.
The cleavages of BACE1 at the Asp1 and Glu11 sites in a certain APP variant are very likely independent of each other. This is evident concerning the fact that inhibition of C99 did not affect C89 and vice versa. Hence, the ratio of C99 to C89 is not necessarily a consequence of the “competition” of the two sites for BACE1 cleavage. It may rather reflect the relative energy barriers for BACE1 to cleave APP at the two sites that are preset for each APP variant. The higher energy barrier results in lower cleavage efficiency, and lower C99 or C89 generation.
While the A673V mutation markedly increased C99 production and significantly suppressed C89, it was pathogenic only in homozygotes. By contrast, the APP mutation E682K having a similar effect as A673V but with milder C99 increase is autosomal-dominant (Zhou et al., 2011). The decreased pathogenic effect of A673V mutation might be due to the impaired processing of C99rIta into Aβ compared with C99Wt; the higher tendency of oligomerization suppressing β-cleavage of full-length APPrIta and γ-cleavage of C99rIta (Eggert et al., 2009), the faster degradation of mature APPrIta reducing the amount of APP as BACE1 substrate or the hugely reduced C89 diminishing Aβ 11-x. Aβ 11-x was also suggested to be toxic Aβ species (Pike et al., 1995; Cai et al., 2001; Barritt and Viles, 2015). In the heterozygotes, the overall effect of all the above mechanisms caused by A673V mutation may suppress Aβ production; however, both alleles at this site need to be mutated to increase Aβ production to trigger AD pathogenesis.
Our data demonstrate that the amino acids adjacent to the Asp1 site are critical for directing BACE1's preferential cleavage site selection. A single amino acid change at residue 673 can affect β-cleavage of APP and γ-cleavage of C99. Valine substitution (recessive Italian mutation) enhances C99 production, whereas threonine (Icelandic mutation), aspartic acid, proline, glutamine, arginine, or serine substitution inhibits C99 generation. These data indicate that the position of APP 673 appears to serve as a “switch” governing the BACE1-mediated amyloidogenesis. The differential effects of the pathogenic Italian A673V mutation and the protective Icelandic A673T mutation on β-cleavage site selection further highlight the key roles of β-cleavage in AD pathogenesis. Given the importance of position 673 in controlling BACE1-mediated amyloidogenesis, targeting this site to direct BACE1 cleavage away from Aβ-generating Asp1 site without affecting other functions of BACE1 could be promising strategy for AD drug development.
Footnotes
This work was supported by Canadian Institutes of Health Research Operating Grant MOP-97825. W.S. is the holder of the Tier 1 Canada Research Chair in Alzheimer's Disease. S.Z. was the recipient of the China Scholarship Council Ph.D. Student award. Z.W. was the recipient of Alzheimer's Society of Canada Postdoctoral Fellowship. M.Z. was supported by University of British Columbia 4YF Scholarship.
The authors declare no competing financial interests.
References
- Barritt JD, Viles JH (2015) Truncated amyloid-beta(11–40/42) from Alzheimer disease binds Cu2+ with a femtomolar affinity and influences fiber assembly. J Biol Chem 290:27791–27802. 10.1074/jbc.M115.684084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benilova I, Gallardo R, Ungureanu AA, Castillo Cano V, Snellinx A, Ramakers M, Bartic C, Rousseau F, Schymkowitz J, De Strooper B (2014) The Alzheimer disease protective mutation A2T modulates kinetic and thermodynamic properties of amyloid-beta (Abeta) aggregation. J Biol Chem 289:30977–30989. 10.1074/jbc.M114.599027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Wang Y, McCarthy D, Wen H, Borchelt DR, Price DL, Wong PC (2001) BACE1 is the major beta-secretase for generation of Abeta peptides by neurons. Nat Neurosci 4:233–234. 10.1038/85064 [DOI] [PubMed] [Google Scholar]
- De Jonghe C, Esselens C, Kumar-Singh S, Craessaerts K, Serneels S, Checler F, Annaert W, Van Broeckhoven C, De Strooper B (2001) Pathogenic APP mutations near the gamma-secretase cleavage site differentially affect Abeta secretion and APP C-terminal fragment stability. Hum Mol Genet 10:1665–1671. 10.1093/hmg/10.16.1665 [DOI] [PubMed] [Google Scholar]
- Deng Y, Wang Z, Wang R, Zhang X, Zhang S, Wu Y, Staufenbiel M, Cai F, Song W (2013) Amyloid-beta protein (Abeta) Glu11 is the major beta-secretase site of beta-site amyloid-beta precursor protein-cleaving enzyme 1(BACE1), and shifting the cleavage site to Abeta Asp1 contributes to Alzheimer pathogenesis. Eur J Neurosci 37:1962–1969. 10.1111/ejn.12235 [DOI] [PubMed] [Google Scholar]
- Di Fede G, Catania M, Morbin M, Rossi G, Suardi S, Mazzoleni G, Merlin M, Giovagnoli AR, Prioni S, Erbetta A, Falcone C, Gobbi M, Colombo L, Bastone A, Beeg M, Manzoni C, Francescucci B, Spagnoli A, Cantù L, Del Favero E, et al. (2009) A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science 323:1473–1477. 10.1126/science.1168979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckman CB, Mehta ND, Crook R, Perez-tur J, Prihar G, Pfeiffer E, Graff-Radford N, Hinder P, Yager D, Zenk B, Refolo LM, Prada CM, Younkin SG, Hutton M, Hardy J (1997) A new pathogenic mutation in the APP gene (I716V) increases the relative proportion of A beta 42(43). Hum Mol Genet 6:2087–2089. 10.1093/hmg/6.12.2087 [DOI] [PubMed] [Google Scholar]
- Eggert S, Midthune B, Cottrell B, Koo EH (2009) Induced dimerization of the amyloid precursor protein leads to decreased amyloid-beta protein production. J Biol Chem 284:28943–28952. 10.1074/jbc.M109.038646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch FS, Keim PS, Beattie EC, Blacher RW, Culwell AR, Oltersdorf T, McClure D, Ward PJ (1990) Cleavage of amyloid beta peptide during constitutive processing of its precursor. Science 248:1122–1124. 10.1126/science.2111583 [DOI] [PubMed] [Google Scholar]
- Felsenstein KM, Hunihan LW, Roberts SB (1994) Altered cleavage and secretion of a recombinant beta-APP bearing the Swedish familial Alzheimer's disease mutation. Nat Genet 6:251–255. 10.1038/ng0394-251 [DOI] [PubMed] [Google Scholar]
- Giaccone G, Morbin M, Moda F, Botta M, Mazzoleni G, Uggetti A, Catania M, Moro ML, Redaelli V, Spagnoli A, Rossi RS, Salmona M, Di Fede G, Tagliavini F (2010) Neuropathology of the recessive A673V APP mutation: Alzheimer disease with distinctive features. Acta Neuropathol 120:803–812. 10.1007/s00401-010-0747-1 [DOI] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L (1991) Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature 349:704–706. 10.1038/349704a0 [DOI] [PubMed] [Google Scholar]
- Jonsson T, Atwal JK, Steinberg S, Snaedal J, Jonsson PV, Bjornsson S, Stefansson H, Sulem P, Gudbjartsson D, Maloney J, Hoyte K, Gustafson A, Liu Y, Lu Y, Bhangale T, Graham RR, Huttenlocher J, Bjornsdottir G, Andreassen OA, Jönsson EG, et al. (2012) A mutation in APP protects against Alzheimer's disease and age-related cognitive decline. Nature 488:96–99. 10.1038/nature11283 [DOI] [PubMed] [Google Scholar]
- Kaden D, Munter LM, Joshi M, Treiber C, Weise C, Bethge T, Voigt P, Schaefer M, Beyermann M, Reif B, Multhaup G (2008) Homophilic interactions of the amyloid precursor protein (APP) ectodomain are regulated by the loop region and affect beta-secretase cleavage of APP. J Biol Chem 283:7271–7279. 10.1074/jbc.M708046200 [DOI] [PubMed] [Google Scholar]
- Kimura A, Hata S, Suzuki T (2016) Alternative selection of beta-site APP-cleaving enzyme 1 (BACE1) cleavage sites in amyloid beta-protein precursor (APP) harboring protective and pathogenic mutations within the Abeta sequence. J Biol Chem 291:24041–24053. 10.1074/jbc.M116.744722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhou W, Tong Y, He G, Song W (2006) Control of APP processing and Abeta generation level by BACE1 enzymatic activity and transcription. FASEB J 20:285–292. 10.1096/fj.05-4986com [DOI] [PubMed] [Google Scholar]
- Ly PT, Wu Y, Zou H, Wang R, Zhou W, Kinoshita A, Zhang M, Yang Y, Cai F, Woodgett J, Song W (2013) Inhibition of GSK3beta-mediated BACE1 expression reduces Alzheimer-associated phenotypes. J Clin Invest 123:224–235. 10.1172/JCI64516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney JA, Bainbridge T, Gustafson A, Zhang S, Kyauk R, Steiner P, van der Brug M, Liu Y, Ernst JA, Watts RJ, Atwal JK (2014) Molecular mechanisms of Alzheimer disease protection by the A673T allele of amyloid precursor protein. J Biol Chem 289:30990–31000. 10.1074/jbc.M114.589069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullan M, Crawford F, Axelman K, Houlden H, Lilius L, Winblad B, Lannfelt L (1992) A pathogenic mutation for probable Alzheimer's disease in the APP gene at the N-terminus of beta-amyloid. Nat Genet 1:345–347. 10.1038/ng0892-345 [DOI] [PubMed] [Google Scholar]
- Nilsberth C, Westlind-Danielsson A, Eckman CB, Condron MM, Axelman K, Forsell C, Stenh C, Luthman J, Teplow DB, Younkin SG, Näslund J, Lannfelt L (2001) The ‘Arctic’ APP mutation (E693G) causes Alzheimer's disease by enhanced Abeta protofibril formation. Nat Neurosci 4:887–893. 10.1038/nn0901-887 [DOI] [PubMed] [Google Scholar]
- Peacock ML, Warren JT Jr, Roses AD, Fink JK (1993) Novel polymorphism in the A4 region of the amyloid precursor protein gene in a patient without Alzheimer's disease. Neurology 43:1254–1256. 10.1212/WNL.43.6.1254 [DOI] [PubMed] [Google Scholar]
- Perez RG, Squazzo SL, Koo EH (1996) Enhanced release of amyloid beta-protein from codon 670/671 “Swedish” mutant beta-amyloid precursor protein occurs in both secretory and endocytic pathways. J Biol Chem 271:9100–9107. 10.1074/jbc.271.15.9100 [DOI] [PubMed] [Google Scholar]
- Perez RG, Soriano S, Hayes JD, Ostaszewski B, Xia W, Selkoe DJ, Chen X, Stokin GB, Koo EH (1999) Mutagenesis identifies new signals for beta-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Abeta42. J Biol Chem 274:18851–18856. 10.1074/jbc.274.27.18851 [DOI] [PubMed] [Google Scholar]
- Pike CJ, Overman MJ, Cotman CW (1995) Amino-terminal deletions enhance aggregation of beta-amyloid peptides in vitro. J Biol Chem 270:23895–23898. 10.1074/jbc.270.41.23895 [DOI] [PubMed] [Google Scholar]
- Qing H, He G, Ly PT, Fox CJ, Staufenbiel M, Cai F, Zhang Z, Wei S, Sun X, Chen CH, Zhou W, Wang K, Song W (2008) Valproic acid inhibits Abeta production, neuritic plaque formation, and behavioral deficits in Alzheimer's disease mouse models. J Exp Med 205:2781–2789. 10.1084/jem.20081588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlin C, Lord A, Magnusson K, Englund H, Almeida CG, Greengard P, Nyberg F, Gouras GK, Lannfelt L, Nilsson LN (2007) The Arctic Alzheimer mutation favors intracellular amyloid-beta production by making amyloid precursor protein less available to alpha-secretase. J Neurochem 101:854–862. 10.1111/j.1471-4159.2006.04443.x [DOI] [PubMed] [Google Scholar]
- Song W, Nadeau P, Yuan M, Yang X, Shen J, Yankner BA (1999) Proteolytic release and nuclear translocation of Notch-1 are induced by presenilin-1 and impaired by pathogenic presenilin-1 mutations. Proc Natl Acad Sci U S A 96:6959–6963. 10.1073/pnas.96.12.6959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhilb ML, Turner RS, Gaut JR (2001) The protease inhibitor, MG132, blocks maturation of the amyloid precursor protein Swedish mutant preventing cleavage by beta-secretase. J Biol Chem 276:4476–4484. 10.1074/jbc.M008793200 [DOI] [PubMed] [Google Scholar]
- Stenh C, Nilsberth C, Hammarbäck J, Engvall B, Näslund J, Lannfelt L (2002) The Arctic mutation interferes with processing of the amyloid precursor protein. Neuroreport 13:1857–1860. 10.1097/00001756-200210280-00005 [DOI] [PubMed] [Google Scholar]
- Sun X, Tong Y, Qing H, Chen CH, Song W (2006a) Increased BACE1 maturation contributes to the pathogenesis of Alzheimer's disease in Down syndrome. FASEB J 20:1361–1368. 10.1096/fj.05-5628com [DOI] [PubMed] [Google Scholar]
- Sun X, He G, Qing H, Zhou W, Dobie F, Cai F, Staufenbiel M, Huang LE, Song W (2006b) Hypoxia facilitates Alzheimer's disease pathogenesis by up-regulating BACE1 gene expression. Proc Natl Acad Sci U S A 103:18727–18732. 10.1073/pnas.0606298103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Bromley-Brits K, Song W (2012) Regulation of beta-site APP-cleaving enzyme 1 gene expression and its role in Alzheimer's disease. J Neurochem 120 [Suppl 1]:62–70. [DOI] [PubMed] [Google Scholar]
- Van Nostrand WE, Melchor JP, Cho HS, Greenberg SM, Rebeck GW (2001) Pathogenic effects of D23N Iowa mutant amyloid beta -protein. J Biol Chem 276:32860–32866. 10.1074/jbc.M104135200 [DOI] [PubMed] [Google Scholar]
- Vetrivel KS, Barman A, Chen Y, Nguyen PD, Wagner SL, Prabhakar R, Thinakaran G (2011) Loss of cleavage at beta′-site contributes to apparent increase in beta-amyloid peptide (Abeta) secretion by beta-secretase (BACE1)-glycosylphosphatidylinositol (GPI) processing of amyloid precursor protein. J Biol Chem 286:26166–26177. 10.1074/jbc.M111.260471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski T, Ghiso J, Frangione B (1991) Peptides homologous to the amyloid protein of Alzheimer's disease containing a glutamine for glutamic acid substitution have accelerated amyloid fibril formation. Biochem Biophys Res Commun 180:1528. 10.1016/S0006-291X(05)81370-1 [DOI] [PubMed] [Google Scholar]
- Wong SH, Hong W (1993) The SXYQRL sequence in the cytoplasmic domain of TGN38 plays a major role in trans-Golgi network localization. J Biol Chem 268:22853–22862. [PubMed] [Google Scholar]
- Zhang M, Cai F, Zhang S, Song W (2014) Overexpression of ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) delays Alzheimer's progression in vivo. Sci Rep 4:7298. 10.1038/srep07298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Nadeau P, Song W, Donoviel D, Yuan M, Bernstein A, Yankner BA (2000) Presenilins are required for gamma-secretase cleavage of beta-APP and transmembrane cleavage of Notch-1. Nat Cell Biol 2:463–465. 10.1038/35017108 [DOI] [PubMed] [Google Scholar]
- Zhou L, Brouwers N, Benilova I, Vandersteen A, Mercken M, Van Laere K, Van Damme P, Demedts D, Van Leuven F, Sleegers K, Broersen K, Van Broeckhoven C, Vandenberghe R, De Strooper B (2011) Amyloid precursor protein mutation E682K at the alternative beta-secretase cleavage beta′-site increases Abeta generation. EMBO Mol Med 3:291–302. 10.1002/emmm.201100138 [DOI] [PMC free article] [PubMed] [Google Scholar]